Abstract
Objective:
This work was undertaken in order to identify Parkinson’s disease (PD) risk variants in a Latino cohort, to describe the overlap in the genetic architecture of PD in Latinos compared to European-ancestry subjects, and to increase the diversity in PD genome-wide association (GWAS) data.
Methods:
We genotyped and imputed 1497 PD cases and controls recruited from nine clinical sites across South America. We performed a GWAS using logistic mixed models; variants with a p-value < 1×10−5 were tested in a replication cohort of 1,234 self-reported Latino PD cases and 439,522 Latino controls from 23andMe, Inc. We also performed an admixture mapping analysis where local ancestry blocks were tested for association with PD status.
Results:
One locus, SNCA, achieved genome-wide significance (p-value < 5×10−8); rs356182 achieved genome-wide significance in both the discovery and the replication cohorts (discovery, G allele: 1.58 OR, 95% CI 1.35–1.86, p-value 2.48×10−8; 23andMe, G allele: 1.26 OR, 95% CI 1.16–1.37, p-value 4.55×10−8). In our admixture mapping analysis, a locus on chromosome 14, containing the gene STXBP6, achieved significance in a joint test of ancestries and in the Native American single-ancestry test (p-value < 5×10−5). A second locus on chromosome 6, containing the gene RPS6KA2, achieved significance in the African single-ancestry test (p-value < 5×10−5).
Interpretation:
This study demonstrated the importance of the SNCA locus for the etiology of PD in Latinos. By leveraging the demographic history of our cohort via admixture mapping, we identified two potential PD risk loci that merit further study.
Introduction
Parkinson disease (PD) is truly a global disease, impacting all ethnic groups and imposing an increasing social and economic burden worldwide.1,2 Despite this, GWAS efforts to date have been limited to individuals of European and East Asian ancestry.1,3–5 This under-representation is not limited to PD; nearly 80% of all study participants represented in the GWAS Catalog are of European descent.6 As of 2018, only 1.3% of study participants in the GWAS Catalog are Hispanics/Latinos, 0.03% are Native American, and 2.4% are African.6 This risks missing population-specific variation, and creating biased polygenic risk scores due to differences in linkage disequilibrium structure.6–8
PD incidence rates are rising in nearly every global region2, highlighting the need for greater diversity in PD consortiums. In the United States, studies of Medicare beneficiaries and of a private insurance company’s members found the age-adjusted PD incidence rate to be highest in Hispanics/Latinos among the surveyed ancestries.9,10 Furthermore, few genetic studies have been done in Latino and the existing studies have exclusively utilized candidate gene approaches.11,12 The Latin American Research Consortium on the Genetics of PD (LARGE-PD) formed in 2009 to fill this gap.13 LARGE-PD is an ongoing effort of 35 institutions in 12 countries across the Americas and the Caribbean. Here we performed the first GWAS of Latino PD patients from South America composed of 1,497 subjects from LARGE-PD and 8.7 million variants obtained using a genotyping array and an imputation reference panel optimized for diverse subjects.14
Materials and Methods
Sample Recruitment and Genotyping
1,504 LARGE-PD samples from Uruguay, Peru, Chile, Brazil, and Colombia were recruited from 2007 to 2015 and genotyped using the Multi-Ethnic Genotyping Array (MEGA) from Illumina14 at the Genomics Core at the University of Washington. The MEGA array was designed to accurately genotype diverse samples and provides suitable coverage for imputation. After quality control (see below), the discovery cohort consisted of 807 PD cases and 690 controls (see Table 1). PD patients were evaluated by a local movement disorder specialist using the UK PD Society Brain Bank clinical diagnostic criteria (UKPDSBB).15 Individuals who did not exhibit neurological symptoms were selected as controls. All participants provided written informed consent according to their respective locale’s national requirements.
Table 1:
Cohort description
| SITE | N | N CASES |
MEAN (SD) AGE | SEX RATIO | MEAN (SD) AAO | MEAN (SD) AFR | MEAN (SD) EUR | MEAN (SD) NAT_AM | MEAN (SD) OTHER |
|---|---|---|---|---|---|---|---|---|---|
| Brazil: Porto Alegre | 13 | 3 | 64.46 (9.12) | 0.15 | 64.33 (10.02) | 0.080 (0.15) | 0.821 (0.21) | 0.095 (0.12) | 0.004 (0.01) |
| Brazil: Ribeirao Preto | 195 | 126 | 57.15 (15.12) | 0.49 | 51.13 (14.13) | 0.092 (0.12) | 0.825 (0.18) | 0.063 (0.06) | 0.020 (0.11) |
| Brazil: Sao Paulo | 19 | 19 | 59.37 (9,04) | 0.74 | 50.21 (9.93) | 0.143 (0.19) | 0.757 (0.22) | 0.096 (0.07) | 0.004 (0.008) |
| Chile | 13 | 13 | 63.67 (9.59) | 0.75 | 56.83 (10.06) | 0.016 (0.008) | 0.614 (0.16) | 0.340 (0.17) | 0.030 (0.07) |
| Colombia: Bogota | 23 | 23 | 49.09 (10.18) | 0.39 | 40.70 (10.22) | 0.025 (0.03) | 0.596 (0.14) | 0.374 (0.14) | 0.005 (0.006) |
| Colombia: Medellin | 328 | 134 | 60.34 (12.44) | 0.44 | 51.83 (15.70) | 0.095 (0.09) | 0.645 (0.15) | 0.260 (0.10) | 0.001 (0.002) |
| Peru: Lima | 670 | 437 | 60.15 (13.57) | 0.46 | 56.77 (13.68) | 0.021 (0.05) | 0.188 (0.15) | 0.783 (0.18) | 0.009 (0.05) |
| Peru: Puno | 45 | 0 | 43.00 (19.80) | 0.24 | NA | 1x10−5 (0) | 0.0067 (0.02) | 0.993 (0.02) | 2.00E–05 (0) |
| Uruguay | 191 | 52 | 61.13 (12.62) | 0.35 | 50.77 (14.51) | 0.051 (0.06) | 0.825 (0.13) | 0.120 (0.11) | 0.004 (0.01) |
| TOTAL | 1497 | 807 | 59.30 (13.90) | 0.44 | 54.09 (14.35) | 0.052 (0.08) | 0.472 (0.33) | 0.469 (0.35) | 0.008 (0.06) |
| TOTAL: CASES | 807 | NA | 61.70 (12.81) | 0.53 | 54.09 (14.35) | 0.05 (0.08) | 0.44 (0.32) | 0.50 (0.34) | 0.01 (0.07) |
| TOTAL: CONTROLS | 690 | NA | 56.48 (14.59) | 0.33 | NA | 0.06 (0.08) | 0.50 (0.33) | 0.43 (0.35) | 0.004 (0.02) |
Description of LARGE-PD cohort. SITE, recruitment site. N, sample size from each site. N CASES, number of cases from each site. MEAN (SD) AGE, mean and standard deviation of age at data collection. SEX RATIO, the proportion of the cohort that is male. MEAN (SD) AGE ONSET, the mean and standard deviation of age at disease diagnosis. MEAN (SD) AFR, the mean and standard deviation of African ancestry proportions. MEAN (SD) EUR, the mean and standard deviation of European ancestry proportions. MEAN (SD) NAT_AMR, the mean and standard deviation of Native American ancestry proportions. MEAN (SD) OTHER, the mean and standard deviation of the combination of the inferred East Asian and South Asian components.
Quality Control
We converted the raw genotype data to PLINK format and carried out quality control (QC) steps using PLINK 1.9.16 We removed unaligned, duplicated, non-autosomal, monomorphic variants prior to filtering. We also filtered for HWE using a p-value threshold of less than 1×10−6 in controls and 1×10−10 in cases17 and a genotype missingness filter of 5%. No samples failed due to missing greater than 5% of genotyped sites and the ascertained sex of all samples matched the sex inferred from the X chromosome. Overall, 1,497 samples and 1,240,909 bi-allelic variants passed QC with an overall genotyping rate of 0.999. Out of these samples, 1481 (798 cases) have complete age and sex records. Though initially removed from the data set to ensure the highest quality variants were used for imputation, the 79 variants with a p-value less than 1×10−10 in cases for the HWE exact test were later included in our GWAS.
Imputation
We imputed the LARGE-PD dataset using the TOPMed Imputation Server (version r1) which utilizes MINIMAC4 and a reference panel of 125,568 haplotypes from diverse samples.18 This imputation panel has been shown to improve imputation for Hispanics/Latinos.18,19 Variants unable to be lifted over to GRC38 or rectified via strand flips were removed by the Imputation Server pipeline. We retained imputed variants if they had a minimum imputation R2 greater than 0.3.
Characterization of LARGE-PD Population Structure
To improve inference of LARGE-PD population structure, we merged LARGE-PD genotyped variants with sequenced variants from the 1000 Genomes Project20; the intersection consisted of 606,977 variants. We then filtered the merged dataset for a minimum minor allele frequency (MAF) of 1% and linkage disequilibrium (LD) pruning using PLINK’s indep-pairwise with a window of 50 variants, a step of 5 variants and a maximum R2 of 0.2 as its parameters. For the admixture analysis, we resolved pairs of relatives by randomly removing one relative from each pair using KING’s unrelated algorithm21 and a threshold of second-degree relatedness. We ran ADMIXTURE22 with K equal to 5, repeating the analysis 20 times using the random seed option and retaining the repetition with the highest log-likelihood.
We performed principal component analysis (PCA) on all LARGE-PD subjects using the PC-AiR23 and PC-Relate24 methods that are implemented in the GENESIS package (https://www.bioconductor.org/packages/release/bioc/html/GENESIS.html). We first estimated a kinship matrix using KING-robust21, which PC-AiR then utilizes to partition the samples into a mutually unrelated and ancestry-representative set of subjects, as well as a related set. PC-AiR performs standard PCA on the set of unrelated individuals and then projects the components of variation for the related set. We then estimated a kinship matrix using PC-Relate with adjustment of the ancestry-representative PCs derived by PC-AiR. Finally, we performed a second round of analyses of PC-AiR and PC-Relate using the kinship matrix obtained in the previous step in order to generate the final PCs.
Estimation of Additive Heritability (h2)
We estimated heritability using GCTA25 and imputed LARGE-PD variants and a method developed by Yang et al. to correct for the bias due to LD.26 Imputed variants with a MAF of at least 1% are stratified into four groups based on their LD score, followed by the estimation of genetic relatedness matrices (GRMs) corresponding to each of the strata. We restricted our heritability analysis to the unrelated subset of LARGE-PD up to the second degree, as determined via KING21 in the same manner described in the admixture analysis. We then estimated narrow-sense heritability using AI-REML in GCTA and the four stratified GRMs, assuming a prevalence of 0.5% and including age, sex, the first five PCs, and recruitment site as fixed effects.
Genome-Wide Association Study
We conducted a GWAS utilizing all samples from the imputed LARGE-PD cohort and logistic mixed models implemented in the GENESIS R package.27 We included age, sex, the first five PCs, and the GRM estimated using GCTA in our null model. We tested imputed dosages against the null via a score test.
Fine Mapping
We identified the variants previously associated with PD in the GWAS Catalog.28 For novel loci, we used FUMA to gather annotations and perform an eQTL mapping analysis with GTEx data.29,30 We determined the LD structure of the chromosome 4 peak using PLINK 1.9. We also utilized this LD information to create custom LocusZoom-style plots. We determined the 95% credible set using PAINTOR 3.031 with neuronal and brain annotations.
Conditional Analysis
We performed a conditional analysis where we adjusted for rs356182, the lead SNCA variant in European-ancestry PD analyses along with age, sex, and the first 5 PCs using logistic mixed models implemented with the GMMAT package32 in R. We evaluated p-values using two different p-value thresholds: the number of GWAS-significant variants and the number of independent tests in the SNCA region.33 We then performed a stepwise conditional analysis, adjusting for rs356182 and additional significant SNPs until no SNPs remained statistically significant.
23andMe Replication of LARGE-PD GWAS Primary Results
We provided 23andMe with a list of variants to test as per their replication pipeline guidelines. We selected 180 variants for replication with a minimum p-value of 1×10−5 provided they met one of the following criteria: the top variant at a genomic locus (+/− 500 KB) or in the 95% credible set at the NRROS and SNCA loci. 23andMe tested the set of identified variants via their replication pipeline and an independent cohort of 1,234 Hispanic/Latino subjects with self-reported PD status and 439,522 controls. All self-reported PD cases and controls from 23andMe provided informed consent and answered surveys online according to 23andMe’s protocol, which was reviewed and approved by Ethical & Independent Review Services, a private institutional review board (http://www.eandireview.com). Samples were genotyped on one of five genotyping platforms; for inclusion, samples needed a minimal call rate of 98.5%. Genotyped samples were then phased using either Finch or Eagle234 and imputed using Minimac3 and a reference panel of 1000 Genomes Phase III4 and UK10K data.35 For this replication study, samples were classified as Latino using a genotype-based pipeline36 consisting of a support vector machine and a hidden Markov model, followed by a logistic classifier to differentiate Latinos from African Americans. Unrelated individuals were included in the analysis, as determined via identity-by-descent (IBD). Variants were tested for association with PD status using logistic regression, adjusting for age, sex, the first five PCs, and genotyping platform. Reported p-values were from a likelihood ratio test.
Replication of Previously Identified PD Risk Variants
We attempted to test 90 independent PD risk variants, previously identified by Nalls et al. 20191, in LARGE-PD for association with PD. We successfully imputed 84 of the 90 variants. Five of the six variants that we were unable to impute were absent from the TOPMed imputation reference panel due to failing TOPMed’s QC protocol; the remaining variant was absent from the dataset. For this variant look-up, we applied the approximation of the Wald test to the score test results from our primary GWAS in order to obtain beta coefficients. In order to ensure fair comparisons, we removed strand ambiguous (CG/AT) sites with a MAF greater than 30%. We also removed rare variants with a minor allele count (MAC) of less than or equal to 10 in LARGE-PD. Beta coefficient correlations were performed using Pearson’s method. In addition to the variants from Nalls et al. 2019, we also performed a variant look-up of additional PD GWAS results from European and East Asian-ancestry studies.3–5
Quality Control- Admixture Mapping
For the admixture mapping, we employed a slightly modified quality control pipeline. We converted the Illumina files to binary PLINK37 format. We excluded SNPs with missing genotype > 0.10, HWE p-value <0.0001, and monomorphic SNPs, with a final genotyping rate of 0.998. We did not need to exclude any of the subjects for low genotyping (maximum missing genotype data of 0.10). The final admixture mapping analysis included all 1,497 subjects with both genotype and phenotype data, and 1,294,079 SNPs that passed quality control filtering.
Admixture Mapping Analysis
We selected 63 unrelated individuals from CEU (Utah residents with Northern and Western European ancestry from the CEPH collection) and YRI (Yoruba in Ibadan, Nigeria) samples from the HapMap project phase III38 (International HapMap Consortium, 2003), and Native American (Pima, Maya and Colombian) samples from the HGDP project (https://www.hagsc.org/hgdp/) to be used as references for European, African, and Native American ancestral populations. We then merged the HapMap and HGDP reference datasets with our 1,497 LARGE-PD samples using PLINK, keeping 164,651 autosomal SNPs in common to all datasets with an overall genotyping rate of 0.999. We performed a joint phasing of LARGE-PD and reference samples using Shapeit239 and an additional reference panel of phased haplotypes from 1000 Genomes project, phase III.
We performed the local ancestry estimation using RFMix40, version 1.5.4, considering the trihybrid ancestry (European, African, and Native American) of the samples. We prepared the input files for RFMix using auxiliary Python scripts of the Ancestry Pipeline developed by Martin et al. 2017.8
We performed admixture mapping through a joint test implemented in the GENESIS R package (https://github.com/UW-GAC/GENESIS), in which all European, African, and Native American ancestries are tested jointly in an admixture mapping logistic mixed model. We fit a null model including sex, age, and the first five PCs as fixed effects and the GRM as random effects. Then, we tested the joint ancestry term at each locus against the null using a multivariate score test. Secondary admixture mapping analyses were performed for each European, African, and Native American ancestry separately in order to identify which ancestral population was driving the significant signal.
Based on previous studies, a p-value of 5×10−5 controls the type I error at level of 0.05.41 Recent admixture, such as observed in Latinos, creates long-range LD and thus the significance threshold does not need to be as stringent as that used for the association mapping. We also estimated the significance level empirically by fitting an autoregressive model to the admixture mapping p-values, summing the results across each chromosome.42 From the empirical autoregression model, we obtained a significance level of 7.7×10-5. We elected to utilize the more conservative p-value threshold of 5×10−5.
We fine mapped the suggestive admixture peaks by overlaying our GWAS results (as described above) with admixture mapping peaks. Significance levels were determined via Bonferroni’s correction for the number of imputed SNPs with minimum MAF of 1% in each peak.
Results
Cohort Description and Ancestry Analysis
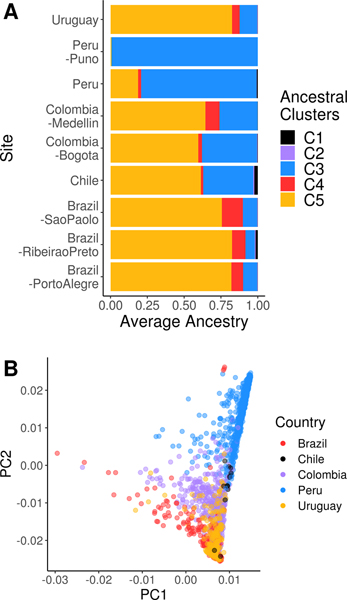
Genotyped LARGE-PD samples came from PD cases and healthy controls across nine sites in five countries: Uruguay, Brazil, Colombia, Peru, and Chile (see Table 1). PD cases were 53% male and had a mean age of 61.7 years (±12.8 years) and a mean age at onset of 54.1 years (± 14.4 years); controls were 33% male and had a mean age of 56.5 years (± 14.6 years). Though the sex ratio of PD cases is lower in LARGE-PD than that of U.S.-based Hispanic/Latinos,10 it appears to be concordant with that of other studies in the region.43,44 Hispanic/Latino populations tend to have a three-way admixture pattern with contributions from African, European, and Native American ancestry. Restricting LARGE-PD to unrelated subjects, the mean proportion of Native American ancestry was 0.47, European ancestry was 0.47, African ancestry was 0.0517, and other ancestries were 0.0076 (see Figure 1; Table 1).
Figure 1: LARGE-PD ancestry.
A: Mean ancestry proportions by LARGE-PD site. We estimated ancestry proportions using ADMIXTURE and a K of 5 in a joint dataset that included LARGE-PD and 1000 Genomes Project samples. Using 1000 Genomes super-population codes to infer the ancestry underlying each cluster, C1 represents East Asian, C2 represents South Asian, C3 represents Native American, C4 represents African, and C5 represents European ancestry B: PCA plot of LARGE-PD subjects. We conducted a principal components analysis using PC-AiR in the merged 1000 Genomes-LARGE-PD dataset. Note the preponderance of individuals with high Amerindian and European ancestries. Principal components were calculated using the PC-AiR algorithm from the GENESIS package in R.
Additive Heritability of PD
Using GCTA and all imputed SNPs with a minor allele frequency (MAF) of at least 1%, we estimated the additive heritability (h2) of PD in LARGE-PD to be 0.38 (SE 0.068) with an assumed prevalence of 0.5% (see Methods).
Genome-wide Association Study
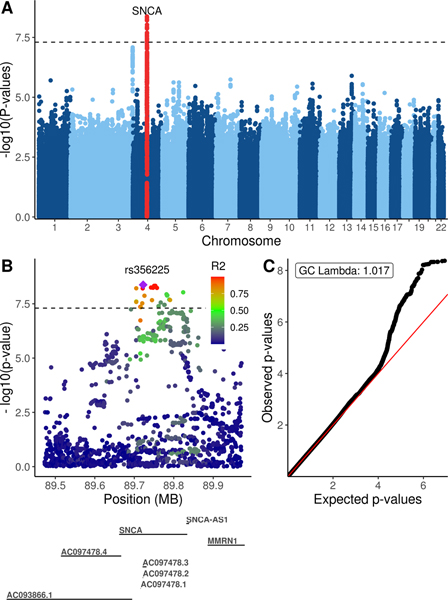
One locus achieved genome-wide significance: the SNCA locus on chromosome 4 (see Figure 2A) with rs356225 achieving the lowest p-value (0.62 OR, 95% CI 0.53–0.73, p-value 4.22×10−9). The SNCA locus is well-characterized in PD literature and a number of SNPs have been put forth as contributing to PD risk.1,33 In LARGE-PD, 28 SNCA SNPs achieved genome-wide significance (see Table 2, supplementary table 1). By utilizing LD information, we observed three LD blocks. An overall pattern of higher LD was observed in the Peruvian subset than in the entire LARGE-PD cohort. The lead SNP, rs356225, is in strong LD with known PD risk SNPs, including an R2 of 0.63 with rs356182, the lead variant in large-scale European meta-analyses (see Figure 2B). Overall, we observed minimal inflation (GC lambda 1.017) and did not correct for this inflation factor (see Figure 2C).
Figure 2: LARGE-PD GWAS results.
A. Manhattan plot of log-transformed p-values by chromosome. P-values were obtained via a logistic mixed model adjusting for age, sex, and the first five principal components using the GENESIS package in R. The significant peak is located within SNCA on chromosome 4. The suggestive peak one near chromosome 3 is near NRROS. B. QQ-plot of GWAS p-values. GC lambda was 1.017. C. Locuszoom-style plot of the chromosome 4 locus using LARGE-PD linkage disequilibrium data.
Table 2:
LARGE-PD GWAS results
| SNP | GENE | AF | IMP R2 | BETA | SE | PVAL | CHR | POS | Effect Allele | AA |
|---|---|---|---|---|---|---|---|---|---|---|
| rs356182 | SNCA | 0.557 | 0.948 | −0.460 | 0.083 | 2.48x10−8 | 4 | 89704960 | A | . |
| rs356225 | SNCA | 0.470 | 0.985 | −0.471 | 0.080 | 4.22x10−9 | 4 | 89722606 | G | . |
| rs6830166 | SNCA | 0.326 | 0.988 | 0.504 | 0.088 | 9.35x10−9 | 4 | 89823842 | T | . |
| rs2870004 | SNCA | 0.846 | 0.965 | 0.077 | 0.114 | 0.499 | 4 | 89550094 | A | . |
| rs763443 | MMRN1 | 0.580 | 0.990 | 0.266 | 0.084 | 0.001 | 4 | 89898810 | C | . |
| rs78820950 | NRROS (nearest) | 0.104 | 0.976 | 0.746 | 0.139 | 8.25x10−8 | 3 | 196630255 | T | . |
| rs1800547 | MAPT | 0.127 | 1.000 | −0.432 | 0.127 | 0.001 | 17 | 45974480 | G | . |
| rs34311866 | TMEM175 | 0.102 | 0.970 | 0.045 | 0.139 | 0.748 | 4 | 958159 | C | p.M383T |
| rs34637584 | LRRK2 | 0.002 | 0.978 | . | . | . | 12 | 40340400 | A | p.G2019S |
| rs421016 | GBA | . | . | . | . | . | 1 | 155235252 | C/G | p.L444P |
| rs76763715 | GBA | 0.003 | 0.817 | . | . | . | 1 | 155235843 | C/G | p.N370S |
| rs2230288 | GBA | 0.005 | 0.944 | . | . | . | 1 | 155236376 | T | p.E326K |
| rs773409311 | GBA | . | . | . | . | . | 1 | 155238186 | C | p.K198E |
| rs149171124 | GBA | 0.000 | 0.988 | . | . | . | 1 | 155235790 | A/T | p.E388K |
| rs439898 | GBA | . | . | . | . | . | 1 | 155238630 | A | p.R120W |
| rs398123530 | GBA | . | . | . | . | . | 1 | 155238597 | A | p.R131C |
| rs35749011 | GBA (nearest) | 0.005 | 0.998 | . | . | . | 1 | 155162560 | A | . |
SNP, rs ID. GENE, gene or nearest gene. AF, effect allele frequency in LARGE-PD. IMP R2, imputation R2 of SNP. BETA, beta effect size. SE, standard error of the beta. PVAL, p-value. CHR, chromosome. POS, physical position (build hg38). Effect Allele, effect allele (or A1) in the association test. AA, amino acid change, if applicable. Note: variants without data in AF-PVAL columns were not imputed or genotyped. Variants that have AF and IMP R2 data and lack BETA-PVAL data were too rare to test in LARGE-PD.
A second locus in chromosome 3 approached genome-wide significance with rs78820950 achieving the lowest p-value (2.11 OR, 95% CI 1.61–2.77, 8.25 ×10−8). This locus is located in an intergenic region between FBXO45 and NRROS and has not been previously reported in the PD literature. Out of the 46 SNPs with a GWAS p-value < 1×10−6, 44 were mapped to NRROS using eQTL (mean p-value 8.5×10−6) using FUMA and GTEx expression data.29,30 The SNP rs78820950 has a MAF of 10.3% in LARGE-PD. However, this variant was more than three times as frequent in Peru than other LARGE-PD sites (16.8% vs. 4.5%).
Conditional Analysis
Using a logistic mixed model, we performed a conditional analysis adjusting for rs356182 to test if this known PD risk variant was driving the observed signal. When correcting for the number of GWAS-significant variants, 8 SNPs remain significant, though attenuated, after adjusting for rs356182, with rs6830166 having the smallest adjusted p-value (0.012). None of the SNPs remain statistically significant when adjusting for both rs356182 and rs6830166, despite LD patterns showing evidence of three blocks. However, if we utilized a more stringent threshold, such as the regional correction implemented by Pihlstrøm et al. (n=220) in their conditional analysis of SNCA33, we found minimal evidence of independence from rs356182.
23andMe Replication
23andMe tested 171 variants (p-value < 1×10−5 in the LARGE-PD GWAS) using their replication pipeline (see Methods) in a cohort of 1,234 self-reported PD cases and 439,522 controls, all identified as Latinos in a genotype-based manner (see Methods). Only the chromosome 4 locus replicated in the 23andMe cohort with 20 SNCA variants that replicated (p-value < 0.00029) and one, rs356182, that also achieved genome-wide significance (see Table 2, supplementary table 2).
Replication of Known PD Loci
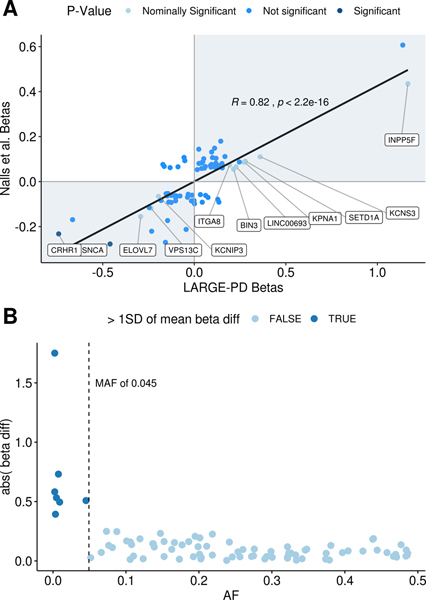
The largest PD-GWAS meta-analysis to date identified 90 independent GWAS-significant PD risk variants in subjects of European ancestry.1 To determine whether these SNPs conferred risk in the LARGE-PD cohort, we looked up 84 of the 90 SNPs in our primary GWAS (see supplementary table 3). Seventy-six of these variants passed our frequency and CG/AT filters (see Methods). Sixty-three of the 76 variants (82.9%) had concordant direction of effect with a Pearson’s correlation of 0.82 (p < 2×10−16; see Figure 3A). Ten variants were nominally significant (p < 0.05 and > 5.95×10−4), and two were significant after correction for 84 tests (SNCA-rs356182 and CRHR1-rs117615688, p < 5.95×10−4). All variants with a difference in beta coefficient greater than one standard deviation from the mean had a MAF less than 4.52% (Figure 3B). If we remove these variants, the concordance rate improves to 86.3%.
Figure 3: Replication of GWAS significant results from Nalls et al. 2019.
A:Beta-beta plot of study beta coefficients. On the scatterplot of beta coefficients, the x-axis corresponds to betas obtained from LARGE-PD and the y-axis corresponds to beta coefficients from Nalls et al. 20191 for 76 of the 90 GWAS significant variants. In LARGE-PD, we successfully imputed 84 of the 90 variants; this figure excludes three variants with a MAC less than 10 and five strand ambiguous (CG/AT) sites that did not pass our filters (see Methods). The color scheme represents p-values obtained from LARGE-PD. Significant (p-value < 5.9×10−4) and nominally significant (p-value < 0.05) variants are labeled by their respective nearest genes. B: Difference in study beta coefficients by MAF. Six variants demonstrated a difference in beta coefficients from LARGE-PD and Nalls et al.1 greater than one standard deviation of the mean. All of the variants have a MAF lower than 4.52%; three have a MAC lower than 10. This was partially due to the larger effect sizes of rare variants, but also could be attributed to inaccurate beta estimates due to insufficient sample size.
In addition to the replication of Nalls et al. 2019, we also looked up variants of particular interest regarding the genetic etiology of PD (see Table 2, Supplementary Table 3). Foo et al. 2020 performed the largest PD GWAS of East Asian ancestry to date and described two novel loci.5 Both variants were consistent in their direction of effect, though neither were nominally significant (p > 0.05). We also looked up the three independent PD risk variants in SNCA that were identified by Pihlstrøm et al.33 One, rs356182, was already included in our replication study. The other two, rs2870004 and rs763443, were not genome-wide significant (p=0.5 and p=0.0015) but were consistent in effect size direction. Neither were in LD with rs356182 in LARGE-PD (R2 0.08 and 0.01, respectively). The MAPT locus did appear to play a role in the etiology of PD in LARGE-PD, with rs1800547 nominally significant (p < 0.05) and rs117615688 (p= 2.29×10−4) replicating from the Nalls et al. 2019 study. Other rare coding variants such as rs2230288 (p.E326K) in GBA and rs34637584 (p.G2019S) in LRRK2 are too rare in LARGE-PD to reliably estimate effect sizes via GWAS and were not directly genotyped.
Admixture Mapping
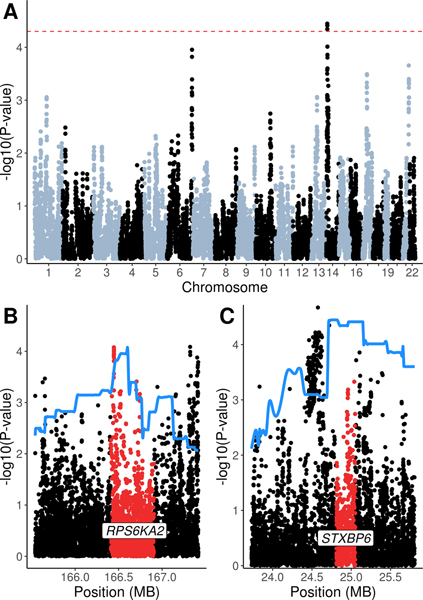
Admixture mapping can be employed if a phenotype shows evidence of differential risk by ancestral background or if we observe allele frequency differences across ancestral populations. For PD, we do see global patterns of PD incidence and prevalence suggestive of differential PD risk.2,9 We found that African ancestry was significantly associated with lower PD risk (OR 0.85, 95% CI 0.75–0.97, p-value 0.017). Given this result, we performed admixture mapping to test local ancestry blocks for associations with PD risk by employing a joint test of ancestries followed by a single-ancestry analysis to determine the ancestry driving each signal (see Methods). In the joint test, a locus on chromosome 14 was significantly associated with PD status (p-value < 5×10−5; see Figure 4A). In the single-ancestry tests, the chromosome 6 locus was significant in the African-ancestry model and the chromosome 14 locus was significant in the Native American model (see Table 3).
Figure 4: LARGE-PD Admixture mapping results.
A: Admixture mapping result of a joint test as implemented by the GENESIS package in R. The significance level of 5×10–5 is indicated by the dashed line. B and C: The admixture mapping results are fit using spline interpolation (solid line) and overlaid on the GWAS results in that region. The gene co-localized with the admixture mapping peak is labeled and highlighted.
Table 3:
Admixture mapping results
| CHR | PEAK | P (ADJ) | AFR PVAL (ADJ) | NAM PVAL (ADJ) | SNP | GENE | P_SNP (ADJ) | MAF |
|---|---|---|---|---|---|---|---|---|
| 6 | 166465311 – 166607482 | 1.11 x10−4 (0.111) | 2.02x10−5 (0.02) | 0.11 (1.00) | rs75880521 | RPS6KA2 | 6.1 x10−4 (0.352) | AFR:0.22; EUR: 0.0; AMR: 0.014 |
| 14 | 24713480 – 25147976 | 3.57x10−5 (0.036) | 0.012 (1.00) | 1.9 x10−5 (0.02) | rs79647551 | STXBP6 | 4.5 x10−5 (0.070) | AFR:0.007; EUR:0.11; AMR:0.31 |
| 17 | 33970400 – 34509873 | 3.22 x10−4 (0.322) | 0.346 (1.00) | 9.2 x10−5 (0.09) | rs4795926 | NA | 1.7 x10−2 (1.00) | AFR:0.09; EUR:0.22; AMR:0.412 |
| 21 | 44767470 – 46068473 | 2.21x10−4 (0.221) | 0.073 (1.00) | 6.9 x10−5 (0.07) | rs183517 | ITGB2 | 1.66 x10−4 (0.111) | AFR:0.67; EUR: 0.55; AMR: 0.37 |
LARGE-PD Admixture mapping (AM) results. CHR, the chromosomal local of the AM peak. PEAK, the physical position in build hg38. P (ADJ), the p-values from the joint test. AFR_PVAL (ADJ), the p-values for the African single-ancestry test; NAM_PVAL (ADJ), the p-values for the Native American single-ancestry test. All adjusted p-values (ADJ) are corrected for a significance threshold of 5x10−5 (see methods). SNP, the variant with the lowest p-value within the AM peak. GENE, the gene co-localized with the peak. POS, the variant position in hg38 coordinates. P_SNP (ADJ), the p-value from the LARGE-PD GWAS for the top variant, with the ADJ the p-value adjusted for the number of imputed variants in the peak. MAF, the super-population frequencies of the variant from the 1000 Genomes Project.
To fine map the admixture mapping signal, we performed a look-up of the GWAS summary statistics of variants co-localized within each peak (see Table 3). The chromosome 6 admixture mapping peak contains RPS6KA2 (Figure 4B); an intronic variant, rs75880521, achieved the lowest p-value (6.05×10−4). This variant had a MAF of 22% in Africans in 1000 Genomes but is virtually absent in populations without African ancestry. The chromosome 14 locus encompasses STXBP6 (Figure 4C) and rs79647551 achieved the lowest p-value (4.5 ×10−5). This variant is intergenic with a frequency of 31% in Admixed Latin American populations in 1000 Genomes but was considerably less frequent in other populations. Neither SNP remained statistically significant when performing a Bonferroni correction.
Discussion
To our knowledge, we have conducted the first GWAS of PD in Latinos, giving the most comprehensive examination of PD genetics in this population to date. In LARGE-PD, we estimated the additive heritability of PD to be 0.38 (SE 0.068). The heritability estimate is higher than that of European cohorts1,45, though this has a number of potential explanations. Nalls et al. utilized LD score regression for their heritability estimate, which is known to be conservative compared to GCTA1, the software we utilized. Keller et al. used GCTA to estimate the heritability of PD in European-ancestry cohorts; the 95% confidence interval of their overall estimate overlapped with the heritability estimated in LARGE-PD, while several cohorts had point estimates that were even higher than that estimated in LARGE-PD.45 This suggests that the estimated heritability in LARGE-PD was reasonable. In addition, the choice of relatedness threshold could have impacted the heritability estimates. We removed relatives up to the 2nd degree, as did Keller et al., while Nalls et al. used a more conservative 3rd degree threshold.1,45 Finally, it is possible that PD heritability is somewhat higher in Latinos than that of European cohorts. This is concordant with a study where more familial aggregation of PD was observed in Hispanics/Latinos that in other population classifications.46
The SNCA locus in chromosome 4 achieved genome-wide significance in both LARGE-PD and the 23andMe replication cohort (Figure 2A, supplementary table 2). In large-scale PD GWAS and meta-analyses, the strongest associations were consistently within the SNCA locus, though such studies have been limited to populations of European and East Asian ancestry.1,5 In LARGE-PD, 28 variants in SNCA achieved genome-wide significance, with 20 replicating in 23andMe. This includes rs356182, the lead variant at the SNCA locus in the European-ancestry studies. The differences in the GWAS summary statistics at this locus between cohorts is likely attributable to variation in sample size, allele frequencies, and LD. Worth noting is the higher LD between SNCA variants in the Peruvian subjects who made up over half of LARGE-PD subjects (see supplementary table 1). Fourteen of the 28 significant variants were tightly correlated with rs356182 (R2 > 0.8) and all tested variants were at least moderately correlated with rs356182 in Peruvian subjects (R2 > 0.41). This suggests that the signal we observed in the SNCA locus was primarily driven by rs356182. In addition, rs356182 was the only genome-wide significant variant in both LARGE-PD (G allele: 1.58 OR, 95% CI 1.35–1.86, p-value 2.48×10−8) and the 23andMe replication cohort (G allele: 1.26 OR, 95% CI 1.16–1.37, p-value 4.55×10−8).
A second locus on chromosome 3 approached genome-wide significance in our GWAS (Figure 2A); this signal was driven by Peruvians of primarily Amerindian ancestry where cases had an allele frequency of 18.8% compared to 9.7% in controls for the lead SNP. Variants at this locus were mapped to NRROS via an eQTL analysis (see Methods). NRROS, is biologically plausible as a potential PD risk gene. NRROS knockout mice display neurological abnormalities including motor deficits47 and a neurodegenerative phenotype has recently been identified in patients who are homozygous for loss-of-function NRROS variants.48 In addition, NRROS appears to be critical for microglial development.47 This locus did not replicate in the 23andMe cohort and the lead SNP was not significant in European-ancestry cohorts49 (p > 0.05), so this may be a false positive. However, we must still consider the possibility of population-specific variation that was not captured by either the array or the subsequent imputation. In addition, the mean ancestral proportions differ between 23andMe (19% Native American) and LARGE-PD (46.9% Native American; see Table 1). Consequently, an additional replication in a cohort with greater Native American ancestry coupled with a sequencing of the region might be necessary.
In our replication of the independent GWAS-significant variants identified by Nalls et al.1, we found that 82% of the tested variants were concordant in their effect size direction in LARGE-PD. Two of the variants, rs356182 (SNCA) and rs117615688 (nearest gene CRHR1 in the MAPT locus), replicated, with rs356182 achieving genome-wide significance. Though we were able to identify that MAPT and SNCA loci contribute to etiology of PD in LARGE-PD, negative results do not necessarily mean other loci, such as for the TMEM175 locus, do not contribute to PD risk in Latinos due to our sample size limitations. Nevertheless, we found evidence of a substantial overlap in the genetic architecture of PD between Latinos and Europeans even with sample size limitations.
In our exploration of the relationship between ancestry and PD risk, we found evidence that African ancestry was protective against PD risk and there was a statistically significant locus on chromosome 6 in the African-ancestry admixture mapping model (see Table 3). Fine mapping the chromosome 6 locus found rs58837225, an intronic variant in RPS6KA2 that was common in individuals of African ancestry but rare in other populations. A variant in RPS6KA2 was recently shown to be in an three-way epistatic relationship with variants in SNCA and RPTOR in an age at PD onset study.50 However, this admixture peak did not achieve significance in the joint test nor was the fine-mapped variant regionally significant (see Methods). The mean proportion of African ancestry in LARGE-PD was under 0.06, which suggests that we were underpowered to detect African-specific variation. A second locus on chromosome 14 achieved significance in the joint test and in the Native American-ancestry model (Figure 3A, Table 3). This locus contains the gene STXBP6 which is highly expressed in the brain.51 While our admixture mapping results provide information for hypothesis generation, replication of our results is necessary, ideally in a cohort enriched in African ancestry for the chromosome 6 result and a cohort enriched in Native American ancestry for our chromosome 14 result.
A limitation of this study was its sample size. In the case of our heritability estimate, further refinement is necessary with a larger cohort. In our GWAS, we currently lack the power to identify rare variants and variants of small effect size. At the SNCA locus, we also lack the power to definitively distinguish independent signals. A regional stepwise conditional analysis in a large diverse dataset is necessary to determine the number of independent PD risk variants in SNCA. Finally, replicating the admixture mapping proved challenging due to the lack of available replication pipelines and genotyped Latino PD cohorts of sufficient sample size.
LARGE-PD is a significant step towards increasing the diversity in PD GWAS data. This project is an ongoing effort; in addition to a parallel study investigating familial forms of PD, 6000 more individuals will be genotyped over the course of two years in order to refine the results presented here and increase our overall power. Increasingly, GWAS summary statistics have been used in the form of polygenic risk scores to construct disease risk models and clinical applications have been proposed.52 The GWAS summary statistics from LARGE-PD could improve such models in Latino populations. Our findings in SNCA and the results of our look-up of known PD risk variants are notable, especially considering our sample size limitations. Through LARGE-PD, we provide critical insights into the genetic architecture of PD in Latino populations.
Supplementary Material
Summary for Social Media if Published.
Twitter handles: @nachogenePD, @OcOutlier, @doug_loesch, @LARGE_PD
What is the current knowledge on the topic? Large-scale GWAS meta-analyses have identified 90 Parkinson disease (PD) risk variants, but an overwhelming European-ancestry bias persists in GWAS data.
What question did this study address? This study addressed two major questions: are there population-specific variants conferring risk to PD and to what degree is European-ancestry PD GWAS data concordant in Latinos.
What does this study add to your knowledge? This study characterizes for the first time the overlap in genetic architecture between Latino and European-ancestry cohorts, affirms the importance of SNCA in the etiology of PD across ancestries, and identified several potential novel risk loci.
How might this potentially impact on the practice of neurology? Representation in research is critical for the practice of neurology to ensure both the generalizability and equitable clinical translation of research.
Acknowledgements
We thank all of the individuals who participated in LARGE-PD. We also want to thank all the support staff at the different Latin American sites for their efforts and support building this incredible resource. Finally, we would like to thank both the 23andMe research team (see supplementary table 4 for research team member names) and the 23andMe customers who consented to participate in research.
This work was supported by a Stanley Fahn Junior Faculty Award (PI: IFM) and an International Research Grants Program award from the Parkinson’s Foundation (PI: IFM), by a research grant from the American Parkinson’s Disease Association (PI: IFM), and with resources and the use of facilities at the Veterans Affairs Puget Sound Health Care System. This project was partially supported by “The Committee for Development and Research” (Comite para el desarrollo y la investigación-CODI)-Universidad de Antioquia grant #2020-31455 to CV-P and MJ-D-R. TDO was supported by National Human Genome Research Institute of the National Institutes of Health under Award Number R35HG010692. DPL was supported by the National Heart, Lung, And Blood Institute of the National Institutes of Health under Award Number T32HL007698.
Footnotes
Potential Conflict of Interest:
The authors do not declare any potential conflicts of interest.
Data and Code Availability
The full summary statistics from LARGE-PD are available upon request. In addition, LARGE-PD GWAS summary statistics can be found in the PD GWAS Browser: https://pdgenetics.shinyapps.io/GWASBrowser/. Scripts used for the GWAS and related analyses can be found in the GitHub repository https://github.com/dloesch/largePD_GWAS. The summary statistics from the 23andMe replication study are already provided in full in the supplementary data.
References
- 1.Nalls MA, Blauwendraat C, Vallerga CL, et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 2019;18(12):1091–1102. doi: 10.1016/S1474-4422(19)30320-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dorsey ER, Elbaz A, Nichols E, et al. Global, regional, and national burden of Parkinson’s disease, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 2018;17(11):939–953. doi: 10.1016/S1474-4422(18)30295-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nalls MA, Pankratz N, Lill CM, et al. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat Genet. 2014;46(9):989–993. doi: 10.1038/ng.3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang D, Nalls MA, Hallgrímsdóttir IB, et al. A meta-analysis of genome-wide association studies identifies 17 new Parkinson’s disease risk loci. Nat Genet. 2017;49(10):1511–1516. doi: 10.1038/ng.3955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foo JN, Chew EGY, Chung SJ, et al. Identification of Risk Loci for Parkinson Disease in Asians and Comparison of Risk Between Asians and Europeans: A Genome-Wide Association Study. JAMA Neurol. Published online April 20, 2020. doi: 10.1001/jamaneurol.2020.0428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sirugo G, Williams SM, Tishkoff SA. The Missing Diversity in Human Genetic Studies. Cell. 2019;177(1):26–31. doi: 10.1016/j.cell.2019.02.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duncan L, Shen H, Gelaye B, et al. Analysis of polygenic risk score usage and performance in diverse human populations. Nat Commun. 2019;10. doi: 10.1038/s41467-019-11112-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin AR, Gignoux CR, Walters RK, et al. Human Demographic History Impacts Genetic Risk Prediction across Diverse Populations. Am J Hum Genet. 2017;100(4):635–649. doi: 10.1016/j.ajhg.2017.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wright Willis A, Evanoff BA, Lian M, Criswell SR, Racette BA. Geographic and Ethnic Variation in Parkinson Disease: A Population-Based Study of US Medicare Beneficiaries. Neuroepidemiology. 2010;34(3):143–151. doi: 10.1159/000275491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Den Eeden SK, Tanner CM, Bernstein AL, et al. Incidence of Parkinson’s Disease: Variation by Age, Gender, and Race/Ethnicity. Am J Epidemiol. 2003;157(11):1015–1022. doi: 10.1093/aje/kwg068 [DOI] [PubMed] [Google Scholar]
- 11.Cornejo-Olivas M, Torres L, Velit-Salazar MR, et al. Variable frequency of LRRK2 variants in the Latin American research consortium on the genetics of Parkinson’s disease (LARGE-PD), a case of ancestry. NPJ Park Dis. 2017;3. doi: 10.1038/s41531-017-0020-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Velez-Pardo C, Lorenzo-Betancor O, Jimenez-Del-Rio M, et al. The distribution and risk effect of GBA variants in a large cohort of PD patients from Colombia and Peru. Parkinsonism Relat Disord. 2019;63:204–208. doi: 10.1016/j.parkreldis.2019.01.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zabetian CP, Mata IF, Latin American Research Consortium on the Genetics of PD (LARGE-PD). LARGE-PD: Examining the genetics of Parkinson’s disease in Latin America. Mov Disord Off J Mov Disord Soc. 2017;32(9):1330–1331. doi: 10.1002/mds.27081 [DOI] [PubMed] [Google Scholar]
- 14.Johnston HR, Hu Y-J, Gao J, et al. Identifying tagging SNPs for African specific genetic variation from the African Diaspora Genome. Sci Rep. 2017;7(1):1–9. doi: 10.1038/srep46398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibb WR, Lees AJ. The relevance of the Lewy body to the pathogenesis of idiopathic Parkinson’s disease. J Neurol Neurosurg Psychiatry. 1988;51(6):745–752. doi: 10.1136/jnnp.51.6.745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. 2015;4. doi: 10.1186/s13742-015-0047-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marees AT, Kluiver H de, Stringer S, et al. A tutorial on conducting genome-wide association studies: Quality control and statistical analysis. Int J Methods Psychiatr Res. 2018;27(2):e1608. doi: 10.1002/mpr.1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taliun D, Harris DN, Kessler MD, et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature. 2021;590(7845):290–299. doi: 10.1038/s41586-021-03205-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kowalski MH, Qian H, Hou Z, et al. Use of >100,000 NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium whole genome sequences improves imputation quality and detection of rare variant associations in admixed African and Hispanic/Latino populations. PLOS Genet. 2019;15(12):e1008500. doi: 10.1371/journal.pgen.1008500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Consortium T 1000 GP. A global reference for human genetic variation. Nature. 2015;526(7571):68. doi: 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen W-M. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26(22):2867–2873. doi: 10.1093/bioinformatics/btq559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19(9):1655–1664. doi: 10.1101/gr.094052.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conomos MP, Miller M, Thornton T. Robust Inference of Population Structure for Ancestry Prediction and Correction of Stratification in the Presence of Relatedness. Genet Epidemiol. 2015;39(4):276–293. doi: 10.1002/gepi.21896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Conomos MP, Reiner AP, Weir BS, Thornton TA. Model-free Estimation of Recent Genetic Relatedness. Am J Hum Genet. 2016;98(1):127–148. doi: 10.1016/j.ajhg.2015.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang J, Benyamin B, McEvoy BP, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42(7):565–569. doi: 10.1038/ng.608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang J, Bakshi A, Zhu Z, et al. Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index. Nat Genet. 2015;47(10):1114–1120. doi: 10.1038/ng.3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gogarten SM, Sofer T, Chen H, et al. Genetic association testing using the GENESIS R/Bioconductor package. Bioinformatics. 2019;35(24):5346–5348. doi: 10.1093/bioinformatics/btz567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacArthur J, Bowler E, Cerezo M, et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 2017;45(Database issue):D896–D901. doi: 10.1093/nar/gkw1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8(1):1826. doi: 10.1038/s41467-017-01261-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.GTEx Consortium, Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group, Statistical Methods groups—Analysis Working Group, et al. Genetic effects on gene expression across human tissues. Nature. 2017;550(7675):204–213. doi: 10.1038/nature24277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kichaev G, Yang W-Y, Lindstrom S, et al. Integrating Functional Data to Prioritize Causal Variants in Statistical Fine-Mapping Studies. PLOS Genet. 2014;10(10):e1004722. doi: 10.1371/journal.pgen.1004722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen H, Wang C, Conomos MP, et al. Control for Population Structure and Relatedness for Binary Traits in Genetic Association Studies via Logistic Mixed Models. Am J Hum Genet. 2016;98(4):653–666. doi: 10.1016/j.ajhg.2016.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pihlstrøm L, Blauwendraat C, Cappelletti C, et al. A comprehensive analysis of SNCA-related genetic risk in sporadic parkinson disease. Ann Neurol. 2018;84(1):117–129. doi: 10.1002/ana.25274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loh P-R, Palamara PF, Price AL. Fast and accurate long-range phasing in a UK Biobank cohort. Nat Genet. 2016;48(7):811–816. doi: 10.1038/ng.3571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.The UK10K project identifies rare variants in health and disease. Nature. 2015;526(7571):82–90. doi: 10.1038/nature14962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Durand EY, Do CB, Mountain JL, Macpherson JM. Ancestry Composition: A Novel, Efficient Pipeline for Ancestry Deconvolution. bioRxiv. Published online October 18, 2014:010512. doi: 10.1101/010512 [DOI] [Google Scholar]
- 37.Purcell S, Neale B, Todd-Brown K, et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am J Hum Genet. 2007;81(3):559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.International HapMap Consortium. The International HapMap Project. Nature. 2003;426(6968):789–796. doi: 10.1038/nature02168 [DOI] [PubMed] [Google Scholar]
- 39.Delaneau O, Marchini J, Zagury J-F. A linear complexity phasing method for thousands of genomes. Nat Methods. 2011;9(2):179–181. doi: 10.1038/nmeth.1785 [DOI] [PubMed] [Google Scholar]
- 40.Maples BK, Gravel S, Kenny EE, Bustamante CD. RFMix: A Discriminative Modeling Approach for Rapid and Robust Local-Ancestry Inference. Am J Hum Genet. 2013;93(2):278–288. doi: 10.1016/j.ajhg.2013.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sofer T, Baier LJ, Browning SR, et al. Admixture mapping in the Hispanic Community Health Study/Study of Latinos reveals regions of genetic associations with blood pressure traits. PLOS ONE. 2017;12(11):e0188400. doi: 10.1371/journal.pone.0188400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gignoux CR, Torgerson DG, Pino-Yanes M, et al. An admixture mapping meta-analysis implicates genetic variation at 18q21 with asthma susceptibility in Latinos. J Allergy Clin Immunol. 2019;143(3):957–969. doi: 10.1016/j.jaci.2016.08.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Orozco JL, Valderrama-Chaparro JA, Pinilla-Monsalve GD, et al. Parkinson’s disease prevalence, age distribution and staging in Colombia. Neurol Int. 2020;12(1). doi: 10.4081/ni.2020.8401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bauso DJ, Tartari JP, Stefani CV, Rojas JI, Giunta DH, Cristiano E. Incidence and prevalence of Parkinson’s disease in Buenos Aires City, Argentina. Eur J Neurol. 2012;19(8):1108–1113. doi: 10.1111/j.1468-1331.2012.03683.x [DOI] [PubMed] [Google Scholar]
- 45.Keller MF, Saad M, Bras J, et al. Using genome-wide complex trait analysis to quantify ‘missing heritability’ in Parkinson’s disease. Hum Mol Genet. 2012;21(22):4996–5009. doi: 10.1093/hmg/dds335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.My S, V M, Sk VDE, et al. Familial Aggregation of Parkinson’s Disease in a Multiethnic Community-Based Case-Control Study. Movement disorders : official journal of the Movement Disorder Society. doi: 10.1002/mds.23361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wong K, Noubade R, Manzanillo P, et al. Mice deficient in NRROS show abnormal microglial development and neurological disorders. Nat Immunol. 2017;18(6):633–641. doi: 10.1038/ni.3743 [DOI] [PubMed] [Google Scholar]
- 48.Dong X, Tan NB, Howell KB, et al. Bi-allelic LoF NRROS Variants Impairing Active TGF-β1 Delivery Cause a Severe Infantile-Onset Neurodegenerative Condition with Intracranial Calcification. Am J Hum Genet. 2020;106(4):559–569. doi: 10.1016/j.ajhg.2020.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lill CM, Roehr JT, McQueen MB, et al. Comprehensive research synopsis and systematic meta-analyses in Parkinson’s disease genetics: The PDGene database. PLoS Genet. 2012;8(3):e1002548. doi: 10.1371/journal.pgen.1002548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fernández-Santiago R, Martín-Flores N, Antonelli F, et al. SNCA and mTOR Pathway Single Nucleotide Polymorphisms Interact to Modulate the Age at Onset of Parkinson’s Disease. Mov Disord Off J Mov Disord Soc. 2019;34(9):1333–1344. doi: 10.1002/mds.27770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fagerberg L, Hallström BM, Oksvold P, et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics MCP. 2014;13(2):397–406. doi: 10.1074/mcp.M113.035600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Khera AV, Chaffin M, Aragam KG, et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet. 2018;50(9):1219–1224. doi: 10.1038/s41588-018-0183-z [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.