Abstract
Mediators of the initiation, development, and recurrence of periodontitis include the oral microbiome embedded in subgingival plaque and the host immune response to a dysbiosis within this dynamic and complex microbial community. Although mediators have been studied extensively, researchers in the field have been unable to fully ascribe certain clinical presentations of periodontitis to their nature. Emergence of high‐throughput sequencing technologies has resulted in better characterization of the microbial oral dysbiosis that extends beyond the extensively studied putative bacterial periodontopathogens to a shift in the oral virome composition during disease conditions. Although the biological dark matter inserted by retroviruses was once believed to be nonfunctional, research has revealed that it encodes historical viral‐eukaryotic interactions and influences host development. The objective of this review is to evaluate the proposed association of herpesviruses to the etiology and pathogenesis of periodontal disease and survey the highly abundant prokaryotic viruses to delineate their potential roles in biofilm dynamics, as well as their interactions with putative bacterial periodontopathogens and eukaryotic cells. The findings suggest that potential novel periodontal therapies targeting or utilizing the oral virome can alleviate certain clinical presentations of periodontitis. Perhaps it is time to embrace the viral dark matter within the periodontal environment to fully comprehend the pathogenesis and systemic implications of periodontitis.
Keywords: biofilms, cytomegalovirus, Epstein‐barr virus, oral microbiome, oral virome, periodontopathogens, phage therapy
1. INTRODUCTION
1.1. Oral ecology
The oral cavity is a diverse and dynamic environment. The three domains of life (Archaea, Bacteria, and Eukarya) along with viruses constitute the human oral microbiome. The ratio of prokaryotic organisms to human cells is reported to range from 1:1 to 10:1 while viral‐like particles surpass this prokaryotic ratio and lie closer to 100:1.1 The formation of our commensal microbiome begins shortly after birth and develops continuously throughout our lifetimes.2 Within this ecological unit, there is compositional variation of the human microbiota between body sites as a result of distinct selective pressures. For example, the taxonomic and genomic composition of a microbial community on the coronal portion of a tooth is more similar among individuals than those of the same individuals’ tongues.3
Coexistence with our diverse microbiome equips us with crucial biological functions and traits to protect us from invasion by transient or pathogenic microorganisms. Despite how stable the oral landscape may be, rupture of this symbiosis contributes to oral diseases, such as dental caries, periodontitis, and oral mucosal diseases, and is implicated in a number of systemic diseases.4, 5, 6, 7 Traditionally, investigations describing these oral diseases have focused on a bacterial or fungal etiology despite the advancement of viral metagenomics and cultivation methods, which have demonstrated viruses to be drivers of oral blisters, ulcers, and tumors.8 More recently, extensive reviews have detailed the intimate topographic relationship between common herpesviruses and known periodontopathogens and their potential role in the development of periodontal diseases.9
1.2. Periodontal inflammation
Within clinical dental practices, treatment is generally centered on dental and periodontal diseases. Periodontitis continues to be a leading chronic polymicrobial inflammatory condition that affects over 30% of the adult population. Periodontal health can be disrupted when there are shifts in the complex interactions between the microbiota in a biofilm state and the host immune defenses. This dysbiosis can arise from modifications of biological, social, and environmental factors, such as diabetes, alimentary habits, or smoking. Although bacteria are considered the primary etiologic factors in the initiation of periodontitis, host factors influence the progression of periodontitis. Thus, although plaque is necessary for the development of periodontal disease, on its own it is insufficient to drive all the destructive processes that are seen.
Viruses as potential drivers of periodontal disease have been implicated in studies using murine models of disease and human oral subgingival plaque, where virome composition and diversity differ with disease progression.10, 11 Additionally, characterizing the oral virome cannot be limited to eukaryotic viruses without taking into account the most abundant entity, namely, prokaryotic viruses. Prokaryotic viruses, more commonly known as bacteriophages, are the natural predators of bacteria. After utilizing the bacterial machinery for replication, bacteriophages become lytic or lysogenic and consequently may influence bacterial composition and genetic diversity.12 Bacteriophages have a more defined role in the management of biofilms, which in periodontitis has revolved around the more established periodontopathogens, such as those bacterial members of the red complex (Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia) and the “adhesive” bacterium Fusobacterium nucleatum. Successful bacteriophage‐based antibiofilm strategies in animal models have been employed in other systemic conditions as the growth of antibiotic resistance limits the efficacy of standard of care treatment.13 While several bacteriophages have been isolated within the oral cavity and subgingival plaque, the compositional shift that has been described in inflammatory conditions has yet to be linked to changes within the periodontopathogens they infect.
Understanding the effects that certain risk factors have on the development of periodontitis enables better prevention and treatment of periodontitis.14, 15 Consequently, identifying potential etiologic viruses and unraveling their roles in health and periodontitis becomes crucial for improving the outcomes of periodontal therapy. The purpose of this review is to describe how eukaryotic and prokaryotic viruses interact with periodontopathogens and the host immune response, and to examine their potential role in periodontal therapy.
2. EUKARYOTIC VIRUSES
Any ecosystem absent of viruses would look foreign or cease to exist. Similarly, human evolution has been influenced greatly by these obligate parasites. Approximately 10% of identifiable functional elements and more than 50% of the “dark matter” in the human genome sequence have been linked to retroviruses’ ability for viral genome integration.16 The long‐standing presence of the human virome thus becomes essential to our development. Despite these benefits, the mention of viruses connotes morbidity and mortality, as eukaryotic viruses have the potential to promote cellular degradation, dysregulation, and carcinogenesis.
Compulsory to the life cycle of viruses is the production of messenger RNA that can be translated by host ribosomes. Variable and specialized viral replication strategies, which are derived from environmental pressures that eukaryotic viruses face, drive human disease states. Because of the oral cavity's exposure to the external environment (eg, diet, mechanical abrasion) and variable physiologic states (eg, pH, salivary composition), a diverse and highly individualized oral microbiome has developed.17 Decoding the human oral virome will characterize the environmental stresses that modulate host‐viral interactions18 and the composition of bacterial and eukaryotic members in the oral cavity. Members of the human herpesvirus and human papillomavirus families cause the most common primary viral infections of the oral cavity. Below, we describe a number of these putative eukaryotic viruses that have an established role in systemic diseases and oral cancers, but are now being associated with periodontitis.
2.1. Herpesvirus family
Herpesviridae are large DNA viruses that are highly host specific but have the potential to cross host species barriers.19 More than 100 herpesviruses have been identified; however, only eight of them routinely infect primarily humans and these are divided into three subfamilies, alpha‐, beta‐, and gamma‐herpesviruses because of their replicative cycle and range of host infectivity. All herpesviruses can establish latent infection within specific tissues, which are characteristic for each virus. Latency and active replication have been demonstrated to occur within various microenvironments of the oral cavity and the different cell types that comprise the periodontium. Herpesviruses are sensitive to a number of environmental stresses that lead to reactivation of viral replication, cellular lysis, and clinical symptoms. Along with the associations between marginal periodontitis and two herpesviruses, Epstein‐Barr virus and cytomegalovirus, the involvement of these herpesviruses is implicated in the inflammatory process of periapical bone destruction.20, 21 Additional studies associate lytic proteins with activation of cellular signaling pathways, such as Notch signaling and increased pro‐inflammatory cytokine expression, which promote receptor activator of nuclear factor kappa‐B ligand transcription and consequently osteoclastogenesis, leading to bone resorption.22, 23 Similar mechanisms may appear in periodontitis, thus this review will briefly illustrate the proposed contributions of Epstein‐Barr virus and cytomegalovirus in the development of periodontitis.
2.1.1. Epstein‐Barr virus
Epstein‐Barr virus is classified as part of the gamma‐herpesviruses subfamily because of its highly restrictive host range. Persistent infection and latency of Epstein‐Barr virus occurs in epithelial cells of the oropharynx and in beta‐lymphocytes. It is estimated that more than 90% of adult humans present with a latent infection of Epstein‐Barr virus and are subject to Epstein‐Barr virus reactivation causing classic mononucleosis and several life‐threatening diseases.24, 25 The mechanisms that promote and regulate the reactivation of Epstein‐Barr virus are still being uncovered and are critical for potential prophylactic treatment.
Salivary transmission of Epstein‐Barr virus indicates a site of productive infection in the oral cavity, either directly from epithelial cells or from beta‐lymphocyte infection.26 Cycling between these cell types is mediated through a glycoprotein complex (glycoprotein H/glycoprotein L/glycoprotein 42) and allows Epstein‐Barr virus to travel from the oral cavity to the peripheral blood in asymptomatic carriers.27 Several studies have demonstrated elevated Epstein‐Barr virus DNA present in salivary samples in patients with lymphoproliferative disorders and immuno‐suppressive conditions.28, 29
Similar studies have indicated that the levels of Epstein‐Barr virus in saliva may reflect the status of periodontal inflammation, because a significantly elevated difference of Epstein‐Barr virus levels in patients with periodontitis was recorded compared with healthy patients.30, 31, 32 In a number of studies, the gingival crevicular fluid of patients with periodontitis reflects a similar association of Epstein‐Barr virus with periodontal inflammation.33, 34 However, these findings have been countered by contrasting results that indicate a lack of association between Epstein‐Barr virus and periodontitis.35, 36, 37
The role of Epstein‐Barr virus in the pathogenesis of periodontitis is not devalued by the lack of association reported from these studies. Instead, they highlight novel associations between Epstein‐Barr virus and periodontitis that warrant further research. For example, one study demonstrated that visfatin is associated with P. gingivalis in patients with chronic periodontitis, but did not find a significant increase in Epstein‐Barr virus in the same patients.37 However, the investigators did note that, regardless of periodontal inflammation status, an increase in Epstein‐Barr virus levels denoted an increase of gingival crevicular fluid visfatin levels. Visfatin participates in immunity and inflammation by modulating the production of inflammatory mediators, which is suggested to be a link to a variety of metabolic conditions and the pathogenesis of periodontitis.38 Production of visfatin occurs in periodontal ligament cells and fibroblasts and is stimulated by the presence of periodontal pathogens such as P. gingivalis.39 The interactions between P. gingivalis and Epstein‐Barr virus in periodontal pockets have been emphasized in other studies as potentially accelerating the destructive nature of periodontitis.40, 41, 42 Thus, exploring the function of visfatin and its interactions may lead to a more definitive role for Epstein‐Barr virus in periodontitis.
Latency of Epstein‐Barr virus has been reported in the gingival epithelium and subgingival plaque in healthy individuals.43 In addition, these tissues follow the trend of elevated levels of Epstein‐Barr virus in periodontal inflammation that has been described in salivary and gingival crevicular fluid samples of patients with periodontitis.43, 44, 45 Subgingival plaque is required for the initiation of periodontal inflammation. As periodontal inflammation is sustained, the periodontal pockets of patients increase in depth as a result of periodontal connective tissue being degraded from the release of collagenases and dysregulation of the host immune response. In one study, Japanese periodontal patients with deepened pockets (≥5 mm) had subgingival plaque composed of higher detection rates of Epstein‐Barr virus (66%) than shallow pockets (48%) and healthy pockets (45%) in patients with chronic periodontitis.44 Within these same patients with chronic periodontitis, coinfection of Epstein‐Barr virus with P. gingivalis was detected in 44% of deep pockets, while shallow and healthy pockets had coinfection rates of 14% and 13%, respectively.44 Another study highlighted the ability of Epstein‐Barr virus to modulate the host immune response, as, compared with shallow or healthy sites, deep pockets with increased levels of Epstein‐Barr virus DNA also demonstrated elevated levels of monocyte chemoattractant protein‐1, a known chemokine released in Epstein‐Barr virus‐infected tumors, within periodontal epithelial cells.43 Interestingly, 38% of deep pockets without Epstein‐Barr virus DNA also demonstrated elevated levels of monocyte chemoattractant protein‐1, which could be explained by the ability of P. gingivalis to regulate expression of this chemokine.46 However, in deep pockets with a coinfection of Epstein‐Barr virus and P. gingivalis, monocyte chemoattractant protein‐1 levels were consistently higher than epithelial cells infected with just one pathogen. All of the studies described demonstrate that the presence of Epstein‐Barr virus may exacerbate periodontal inflammatory conditions by promoting pro‐inflammatory response in infected cells, either directly or indirectly.
In addition to the synergistic effects seen in coinfected periodontal pocket sites, periodontopathogens may interact with Epstein‐Barr virus to cause reactivation of these viruses within the periodontium. The life cycle stage which the Epstein‐Barr virus is at during detection is determined by measuring the expression of latency transcripts (Epstein‐Barr nuclear antigen 1, Epstein‐Barr nuclear antigen 2, latent membrane protein 1, and latent membrane protein 2), transactivator BamHI Z fragment leftward open reading frame 1, and lytic transcripts. A potent lytic inducer of Epstein‐Barr virus via the activation of BamHI Z fragment leftward open reading frame 1 is the short chain fatty acid, butyric acid, which is seen in high concentrations within the gingival crevicular fluid of periodontally inflamed pockets.47 The high concentration of butyric acid may be attributed to its fermentation from periodontopathogens such as P. gingivalis and F. nucleatum.48 The close association between Epstein‐Barr virus and these butyric acid‐producing periodontopathogens suggests that tissues of the periodontium and the microbiome associated with it provide Epstein‐Barr virus with an environment that facilitates its replication and latency. Regulation of the life cycle of Epstein‐Barr virus within the periodontium thus becomes critical, not only for the progression of periodontitis, but for other Epstein‐Barr virus‐related malignancies.
2.1.2. Cytomegalovirus
Cytomegalovirus is classified as part of the beta‐herpesviruses subfamily because of its long replicative cycle and restricted host range. Individuals with cytomegalovirus can potentially transmit the virus through bodily secretions, such as breast milk, saliva, blood, and urine. The seroprevalence rate of cytomegalovirus is approximately 83% in immunocompetent adults and 31% in children aged 0‐7 years.49, 50 Vertical or transplacental transmission of cytomegalovirus may occur and results in congenital cytomegalovirus infection in approximately 35% of primary maternal cytomegalovirus infections.51 Depending on the stage of pregnancy during which cytomegalovirus is contracted, congenital cytomegalovirus may result in a range of conditions in the neonate, such as hearing loss, mental retardation, and microcephaly.52 Significant morbidity or mortality can occur in immunocompromised individuals from an active cytomegalovirus infection but is self‐limiting in immunocompetent adults.
Cytomegalovirus residence and viral production occurs within epithelial cells, fibroblasts, endothelial cells, monocytes, and T lymphocytes.53 Comparable with Epstein‐Barr virus, salivary levels of cytomegalovirus are used as a diagnostic marker for systemic conditions aggravated by cytomegalovirus54 and it has been detected in the saliva, gingival crevicular fluid, and subgingival plaque of individuals with periodontal inflammation.55, 56, 57 A recent meta‐analysis of 26 studies with periodontal patients calculated statistically significantly increased odds of periodontitis with the detection of subgingival cytomegalovirus (odds ratio 5.31; 95% confidence interval 3.15‐8.97).58 Lack of cytomegalovirus prevalence and association to disease states have also been indicated,59 while one study found elevated levels of cytomegalovirus in healthy controls compared with periodontally diseased sites.60
It has been suggested that herpesviruses, along with certain gram‐negative periodontopathogens, are associated with more severe forms of periodontal disease, such as aggressive periodontitis.61, 62, 63 A study of 34 Sudanese adolescents examined the subgingival plaque of 17 adolescents with localized aggressive periodontitis and 17 adolescents with no clinical attachment loss.64 Four putative periodontopathogens (Aggregatibacter actinomycetemcomitans, P. gingivalis, T. forsythia, and T. denticola) and two herpesviruses (Epstein‐Barr virus and cytomegalovirus) were detected in the subgingival plaque samples via loop‐mediated isothermal amplification. Detection of these microorganisms was seen in both aggressive periodontitis and healthy samples although there was a significant association of A. actinomycetecomitans, P. gingivalis, and cytomegalovirus with aggressive periodontitis.64 However, A. actinomycetemcomitans demonstrated the highest association with aggressive periodontitis (odds ratio 38.0; 95% confidence interval 3.9‐373.1) and the strongest dual infection association belonged to A. actinomycetemcomitans with cytomegalovirus (odds ratio 39.1; 95% confidence interval 2.0‐754.6).
Another similar case‐control study, which used subgingival plaque samples to determine the presence of periodontopathic bacteria and herpesviruses in 100 Jamaican adolescents, found that the strongest association with aggressive periodontitis was in a dual infection with P. gingivalis and cytomegalovirus (odds ratio 51.4; 95% confidence interval 5.4‐486.5).65 The same dual infection also had the strongest association in attachment loss (odds ratio 3.9; 95% confidence interval 1.3‐12.0).65 Thus, dual infection with cytomegalovirus may allude to additive or synergistic effects of herpesviruses in aggressive periodontitis‐diseased sites.
The course of aggressive periodontitis is multifactorial as it is dependent on genetic factors, microbial composition, and the host response. Bacterial pathogens such as A. actinomycetemcomitans and P. gingivalis share the ability of cytomegalovirus to invade and infect epithelial cells and may benefit from cytomegalovirus modulation of the immune defenses by upregulating chemotaxis and inhibiting apoptosis of neutrophils.61, 66 Cytokine production can be altered in periodontal tissues as innate cytokine production of interleukin‐1‐beta and tumor necrosis factor‐alpha were elevated in cytomegalovirus‐infected gingival tissues, and interleukin‐8, monocyte chemoattractant protein 1, macrophage inhibitory protein 1‐alpha, and macrophage inhibitory protein‐1‐beta followed an elevating expression pattern.67 Upregulating bacterial virulence has been described in cytomegalovirus‐infected renal transplant patients as cytomegalovirus infections promote expression of ASA, a plasmid‐encoded surface protein that increases enterococcal adherence to renal epithelial cells.68 Furthermore, a theory has been proposed that early infant cytomegalovirus infection in tissues surrounding tooth germ may alter tooth morphology, which may increase the susceptibility to development of periodontitis after complete development.69
3. ANTIVIRALS
The innate antiviral immune response of periodontal tissues also alludes to the presence and involvement of certain herpesviruses in both healthy and periodontitis patients. Interferons are a family of cytokines that possess immunomodulatory and antiviral properties that are critical for dampening immunopathic mechanisms.70 Interferons are rapidly produced after pattern‐recognition receptor and toll‐like receptor stimulation in natural killer cells, dendritic cells, and monocytes.71 Type III interferons share the same antiviral effects as other interferons but are produced more abundantly at mucosal sites by epithelial and myeloid cells in response to viral infections.72
Gingival tissue also displays expression of interferon‐gamma as one study measured interferon‐gamma mRNA transcripts from healthy, chronic periodontitis, and aggressive periodontitis patients, and found significantly elevated interferon‐gamma mRNA expression in chronic periodontitis and aggressive periodontitis gingival samples.73 The findings of this study, however, cannot be used to implicate the presence of herpesviruses within the periodontium as we have discussed. Additionally, the study lacked any attempt to detect herpesviruses. It is critical to address that type III interferons can be induced by toll‐like receptors expressed on mucosal plasma membranes and detect bacterial products, such as lipopolysaccharides.74 For example, impairment of negative regulation of type I interferon expression occurred in gingival epithelium in a murine model that was repetitively inoculated with P. gingivalis.75 This dysregulation and elevated levels of interferons induced alveolar bone loss as type I interferon overstimulated receptor activator of nuclear factor kappa‐B ligand expression from cluster of differentiation 4 and T cells.75
However, another study measured a significant difference in interferon‐gamma levels and detected several herpesviruses in the gingival crevicular fluid of patients with chronic periodontitis. Of 30 patients with chronic periodontitis, 50% had detectable levels of one of four herpesviruses (ie, Epstein‐Barr virus, cytomegalovirus, herpes simplex virus‐1, and herpes simplex virus‐2), and these 15 patients demonstrated significantly lower interferon‐gamma levels than those patients with chronic periodontitis who were herpesvirus‐negative.76 The investigators in this study suggested that interferon‐gamma levels were inversely correlated with increased detection of herpesvirus, but they did not quantify the level of herpesvirus in each herpesvirus‐positive patient with chronic periodontitis to evaluate this association. Despite the lack of association from this study, the antiviral effects of type III interferons have been well documented for herpesviruses.77, 78 Not surprisingly, however, herpesviruses such as Epstein‐Barr virus have developed mechanisms to evade the effects of the host immune response, including interferons. Early lytic protein‐2 and latent membrane protein 1 can suppress toll‐like receptor and Janus kinase‐signal transducer and activator of transcription signaling that promotes interferon production and binding.79, 80 The potential immunoregulation of interferon expression and function via herpesviruses needs to be further clarified before a definitive association between active herpesvirus infection and interferon levels can be made.
Considering the increased evidence of a synergistic relationship between herpesviruses and bacterial periodontopathogens, additional forms of periodontal therapy have been proposed. Type I interferons are considered a “standard of care” in patients with hepatitis C or hepatitis B infections but this immunotherapy does not come without any systemic effects, as interferon induces hyperactivity in the host response.81 Therefore, full characterization of interferon therapy and immunomodulation of herpesviruses should be documented prior to the use of specific autoimmune drugs.
Adjunctive antimicrobial regimens administered to enhance mechanical calculus removal in classical treatment of chronic periodontitis have been well established. Minocycline, in particular, has been administered as a systemic oral antibiotic or locally via minocycline microspheres, and has been proven to be more effective than other tetracyclines.82 Interestingly, the clinical potential of minocycline is not limited to only antibiotic activity, as it has also been described as having anti‐apoptotic, immunomodulatory, and antiviral effects. The antiviral effects of minocycline are demonstrated through in vitro inhibition of HIV reactivation and lytic transformation through its ability to reduce the activation of monocytes and their permissiveness to viral infection.83
Although not described in this review, the immuno‐suppressive state of HIV‐positive individuals may predispose them to increased herpesvirus viral loads and dampened immune host response, which putative periodontopathogens utilize to increase the risk of initiation of uncontrolled periodontal inflammation.84 A study conducted in South Africa on HIV‐infected children and adolescents with either gingival recession or localized aggressive periodontitis observed that children who started antiretroviral therapy earlier in life and were still in treatment were less likely to display gingival recession or localized aggressive periodontitis. Instead, clinical periodontal conditions were associated with antiretroviral therapy duration (odds ratio 0.9, 95% confidence interval 0.83‐0.97) and immuno‐suppression and/or virologic failure (odds ratio 1.77, 95% confidence interval 1.06‐2.96).85 Management of systemic viral infections thus become critical in the clinical therapy of immuno‐suppressed patients presenting with periodontal inflammation. Antiviral agents are effective in treating systemic conditions in active herpesvirus infections. An in vitro study of cytomegalovirus‐infected gingival tissues illustrated that viral replication can be halted in the oral mucosa with the treatment of a nucleoside analog, ganciclovir, as used with other in vivo anti‐cytomegalovirus therapies.86 Other antivirals have been utilized in periodontal patients, as one case report documents a definitive clearance of excessive Epstein‐Barr virus levels and improved clinical presentation following valacyclovir (500 mg twice daily for 10 days) within a patient with chronic periodontitis who had not responded to conventional nonsurgical therapy.87 The improvement seen from this case report and the lowered prevalence of periodontitis in immuno‐suppressed adolescents receiving antiretroviral therapy may indicate the potential use of antivirals in a specific subset of periodontal patients.
4. ORAL PHAGEOME
4.1. Bacteriophage classifications
Prokaryotic viruses that infect bacteria are known as bacteriophages and they are the most abundant biological entities known to date.88 A general rule regarding bacteriophages is that they are present in all environments in coexistence with their highly specific bacterial hosts. Interestingly, bacteriophages constitute a larger known portion of the human virome than their eukaryotic counterparts, despite limited taxonomic sequences in current genomic databases.89 Similar to eukaryotic viruses, taxonomic classification of bacteriophages is based on several properties, including molecular composition of the viral genome (ssDNA/dsDNA, ssRNA/dsRNA), the structure of the viral capsid, the presence of the viral envelope, host range, and shared genomic sequencing.90 Bacteriophages are classified into 19 families and are characterized in regard to their morphology and size; however, although approximately 96% of bacteriophages are tailed, filamentous and pleomorphic morphologies have also been described.91
Bacteriophages are distinct not only because of their abundance but also because of their highly developed life cycle of predation and lysogenic conversion, which regulates bacterial populations and biodiversity. Initial interaction between bacteriophages and their host occurs randomly by Brownian motion, dispersion, diffusion, or flow.92 Adsorption involves the recognition and attachment to a highly unique region of the bacterial cell wall, capsule, surface receptor, or appendages, and dictates the host range of bacteriophages, which typically involves only a single host species or strain.93 Pressure‐driven ejection of viral genetic material and auxiliary proteins into the host cytoplasm occurs through a conformational change in the multi‐protein tubular apparatus that attaches to and penetrates the host cell membrane.94 Following successful genomic infiltration, virulent bacteriophage replication occurs using the host cell’s machinery if host intracellular conditions are unfavorable, in a process called the lytic cycle. Alternatively, a temperate bacteriophage may enter a dormant state if conditions within the host cell are favorable, in a process known as the lysogenic cycle.
Our understanding of the physiologic significance of the human phageome is still incomplete because most viral genomes cannot be taxonomically classified or linked to a specific bacterial host. Prior to the emergence of the field of viral metagenomics, a very limited toolkit for the direct observation and counting of bacteriophages using transmission electron microscopy techniques or plaque assays existed. High‐throughput sequencing technologies have permitted a comprehensive characterization of bacterial communities and are now illuminating the diversity and membership of the human virome in health and disease.95, 96 The human gut microbiota has been one of the most surveyed human sites. Sequencing demonstrates that the gut phageome composition and dynamics vary with age97 and signal inflammatory conditions through altered bacteriophage community richness, specifically affecting enteric bacteriophages belonging to the Caudovirales order.98
4.2. Prevalence of bacteriophages in the oral cavity
Similar findings associated with the gut phageome are reflected in viral metagenomic studies of saliva, dental plaque, or oral swabs from both healthy individuals and those patients in a periodontally diseased state. The abundance of bacteriophages approximated in the gut phageome (109 virus‐like particles per gram of human feces)99 are comparable with salivary (108 virus‐like particles per milliliter of fluid)100 and dental plaque samples (1010 per milligram of plaque).101 Despite the abundance of identified bacteriophages within the oral cavity, viral contigs from salivary and dental plaque samples from healthy individuals reveal that only a small percentage of these bacteriophages are constant between individuals.100, 102, 103 Differences by sex in the oral virome have been previously reported,104 however, in a large study of 72 healthy Spanish young adults, the investigators did not see a significant difference by sex.103 Although the oral phageome has not been characterized in children, a study with salivary samples taken at different time points spanning 60 to 90 days in a healthy adult population demonstrated conservation of viral contig reads, suggesting that the human virome in adults is stable within individuals.100
Biogeographic sites of the oral cavity play an important role in the viral contigs that are recognized. The lower recognizable viral contigs found in dental plaque compared with salivary samples can be attributed in part to the limited viral sequence homologues in databases used to identify viral genomes.102, 105 This limitation has led various researchers to lower the homologous sequence thresholds used, to analyze viral‐like particles without purification, or to report only bacteriophage families rather than species. However, across studies, a high abundance of oral bacteriophages belonging to the Caudovirales order (Figure 1) has been documented, along with a shift in the composition of the well‐described Caudovirales families (Siphoviridae, Myoviridae, and Podoviridae) in periodontal disease states.11, 102, 103
FIGURE 1.
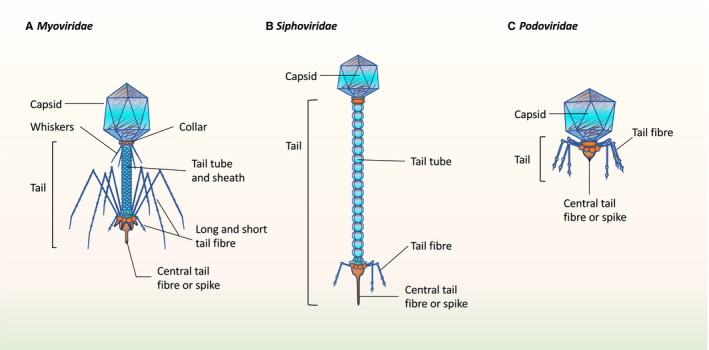
The majority of identified bacteriophages within the oral cavity pertain to the Caudovirales order, which contains three different families of tailed bacteriophages, all with an icosahedral capsid along with a linear dsDNA genome that is injected into bacterial hosts after penetration of cell membrane with tail. A, Myoviridae have a long contractile tail; B, Siphoviriadae have a long, noncontractile tail; and C, Podoviridae have a short tail
Variations in bacteriophage diversity and richness in gut inflammatory conditions consistently demonstrate homogenization of species composition in individuals in diseased states.106, 107 Periodontal inflammation mirrors the decreased species richness and diversity in plaque samples in diseased tissues compared with healthy tissue.11 Specifically, the temperate bacteriophage, Siphoviridae, was the most abundant family in healthy and diseased individuals for all sample types, while subgingival plaque from diseased pockets demonstrated the greatest shift in abundance for the virulent phage, Myoviridae.102 The dominance of temperate phages, such as Siphoviridae, is commonly found in health and is believed to be involved in host bacterial fitness and attenuation of bacterial virulence.108 Meanwhile, the surge of Myoviridae abundance in these diseased pockets may imply that a lytic cycle shifts the collective virulence of the complex biofilm that these bacteriophages are embedded in. However, these associations may currently be limited because of a lack of sequence data and need further validation by determining specific bacteriophage‐host interactions.
4.3. Bacteriophages of oral pathogens
As bacteriophages play a role in bacterial population control and DNA transfer, bacterial hosts have developed an adaptive defense mechanism within their genome, known as clustered regularly interspaced short palindromic repeats, along with associated proteins systems to protect against bacteriophages. Investigators have turned to the heterogeneous diversity of spacers within clustered regularly interspaced short palindromic repeats to trace the history of bacteriophage attacks and to match those bacteriophages or conjugative plasmids containing this sequence array that naturally infect the bacteria.109 Sequencing of clustered regularly interspaced short palindromic repeats and associated proteins systems within the dental plaque of four periodontally healthy individuals allowed for identification of various bacteriophages at a species level, including a high abundance of Streptococcus prophages (UCN34 and IS7493), Actinomyces bacteriophage AV‐1, Streptococcus bacteriophage DP‐1, Enterobacteria bacteriophage P7, and Enterobacteria bacteriophage lambda.101 Parallel to other studies, identification of bacteriophages using clustered regularly interspaced short palindromic repeats and associated proteins systems demonstrated that bacteriophage ecology is characteristic for each individual, despite approximately 50% of contigs remaining without homologous sequences.101
Clustered regularly interspaced short palindromic repeats and associated proteins systems act as a catalog of bacteriophage‐host interactions and demonstrate how these bacterial hosts become resistant to certain bacteriophages. In the same study described above, a unique finding in one individual's phageome included a transposon encoding tetracycline resistance within the Enterobacteria phage P7.101 P7 belongs to the Myoviridae family and is frequently described in coexistence with bacteriophage P1 carrying ampicillin or colistin resistance genes and extended spectrum beta‐lactamase genes.110 In a recent review, bacteriophages identified against their respective oral bacterial inhabitants were surveyed and exhibited a high volume of bacteriophages infecting the early colonizers for dental plaque formation, such as Actinomyces and Streptococcus species.111
Fewer bacteriophages infecting putative periodontopathogens have been described but those isolated suggest their involvement in pathogenesis. The arsenal of virulence factors for bacteria implicated in more aggressive forms of periodontitis, such as A. actinomycetemcomitans, may be attributed to temperate Myoviridae that, in vitro, transfers antibiotic resistance genes, induces serotype conversion, and stimulates production of leukotoxins.112, 113 Another identified temperate Myoviridae bacteriophage infecting T. denticola, designated as φtd1, was found as integrated prophage DNA and upregulated expression of prophage genes during biofilm growth.114 Curiously, unlike the extensively documented herpesvirus‐P. gingivalis relationship,41 no viral contigs or clustered regularly interspaced short palindromic repeats spacers have been isolated to suggest a bacteriophage that infects P. gingivalis.115
4.4. Bacteriophage‐host interactions
As optimization of metagenomic databases progresses steadily with increased viral sequence reads and contigs captured, more homologous sequences become available to identify resident oral bacteriophages with their bacterial hosts. Currently, genomic similarity between different bacteriophages paired with limited sequence data may result in an underestimation or misidentification of bacteriophages, as studies have described a fraction of the contigs that are unable to be classified.116 This observation should be taken into consideration with regard to both metagenomic and species‐specific bacteriophage studies, as the reported bacteriophages may reveal a wider host range of virulent factors than described earlier.117 For example, early identification of Aggregatibacter bacteriophages demonstrated a bacteriophage designated as φAa118 in leukotoxic strains of A. actinomycetemcomitans, but in subsequent studies the same strain was not isolated, and instead a novel bacteriophage was isolated, one which could infect six out of eight strains of A. actinomycetemcomitans, including the leukotoxic strain.119 These distinctions on bacteriophage host ranges have significance when describing the potential driving force bacteriophages have on either the progression or attenuation in clinical presentation of periodontitis.
As the second century of bacteriophage research begins, it is not sufficient to characterize bacteriophages in a pairwise manner with their bacterial hosts. Full characterization of the microbial shift seen in periodontal disease states will require the shaping of a network with the interactions between bacteriophages and all identified host cells.120 As with novel Aggregatibacter bacteriophage‐infecting multiple strains of A. actinomycetemcomitans, other known bacteriophages have the ability to cross‐infect between hosts of distinct species.117, 121 The existence of these cross‐infective bacteriophages can help to describe the highly individualized shift of the microbial community and aid in delineating the impact that bacteriophages have on the diversity of bacterial communities.
The first oral bacteriophage‐host interaction network was computed through analysis of the clustered regularly interspaced short palindromic repeats in 40 dental plaque and salivary samples from periodontally healthy and diseased individuals.11 Most of the findings reported reinforced the proposition that the majority of bacteriophages display a narrow host range or a one‐to‐one infection model. Hybrid contigs were used to identify cross‐infective bacteriophages that represented a small portion of bacteriophages but mainly infected Streptococcus and Actinomyces species.11 A key finding from this network suggests a regulatory role for a small subset of cross‐infective bacteriophages that infects both periodontopathogens (Campylobacter, Fusobacterium, and Prevotella) and commensal bacteria.11 Additional bacteriophage plaque assays paired with metagenomic analysis are necessary to reveal, at species level, those cross‐infective bacteriophages that potentially modulate the bacterial dysbiosis prevalent in the development of periodontitis.10 Future directions for these interaction networks should incorporate cross‐resistance, which is also encoded in clustered regularly interspaced short palindromic repeats and is identified from mutations targeting regulatory genes in order to evaluate novel ecologic adaptations and dynamics between bacteriophages and their host cells.122
4.5. Bacteriophage‐eukaryote interactions
Basic components of bacteriophage morphology and genomic organization include the nucleic acid genome that is encased in a capsid protein. Although bacteriophages are unable to infect eukaryotic cells, these basic components are foreign bodies sufficient enough to trigger an immune response, either directly or indirectly.123, 124 Bacteriophages can be recognized by the innate immune system through toll‐like receptors that recognize pathogen‐associated molecular patterns. Toll‐like receptor 9 specifically recognizes viral DNA after phagocytosis of bacteriophages and subsequently initiates the release of type I interferons.125 The phagocytosis of bacteriophages can inadvertently occur during lysogenic conversion or adsorption as either bacterial or viral molecular patterns are recognized. Complex interactions between bacteriophages, bacterial hosts, and phagocytes were described when the recognition of bacteriophage T4 by toll‐like receptors on neutrophils demonstrated mild inhibition of reactive oxygen species production, while significant neutrophil inhibition was recorded when T4 was phagocytosed with its bacterial host Escherichia coli.126 This in vitro study may suggest that bacteriophages can dampen the immune response during clearance of pathogenic infections.
Mucosal surfaces are sites that bacteriophages take residence in as they can bind to mucin glycoproteins to facilitate adsorption via immunoglobulin‐like domains commonly found on the surfaces of tailed bacteriophages.127 A subset of core bacteriophages found on these mucosal surfaces can confer metabolic and immune benefits to their human host, similar to other commensal microorganisms.128 Simply, these core lytic bacteriophages can act as an antimicrobial layer that reduces bacterial attachment while temperate bacteriophages express genes that increase bacterial fitness to shield them from lytic bacteriophages or pathogenic bacterial attacks. The antimicrobial effects that bacteriophages provide are enhanced further during colonization of pathogenic bacteria as gut and respiratory epithelium increase secretion of mucins, providing more attachment sites for core bacteriophages.129, 130 During in vitro studies, gut eukaryotic cells assisted the bacteriophage φCDHS1 in lysing the pathogenic bacterium Clostridium difficile by approximating the predator and prey through specific attachment surfaces that were not expressed in other cell lines.131 The distinct and elevated attachment of bacteriophage φCDHS1 in human colon cancer line HT‐29 cells compared with other cell lines may also indicate the influence that biogeographic sites have upon bacteriophage infectivity and homeostasis.
5. BACTERIOPHAGE‐MEDIATED BIOFILM DYNAMICS
Although the daily formation of dental plaque is unexceptional, it is a complex oral biofilm that mediates periodontal inflammation. Multicellular behavior exhibited in biofilms have evolved within the oral microbiome to overcome the environmental pressures faced. Biofilms are ubiquitous in the human body (Figure 2) and notoriously difficult to treat in chronic diseased states such as periodontal disease, endocarditis, chronic obstructive lung disorders, and inflammatory bowel disease.132, 133, 134, 135, 136 Certain oral bacteria collaborate in the colonization of hard tissue surfaces with adhesive appendages or supplement the polysaccharide matrix that suspends both commensal and opportunistic microbial community members.137 It is these initial and primary colonizers, mainly Streptococcus and Actinomyces species, which allow for periodontopathogens to aggregate and initiate microbial and immune dysbiosis.138 However, as noted earlier, the presence of these periodontopathogens alone do not describe all the clinical presentations of periodontitis. The role of bacteriophages within biofilm formation and reorganization may clarify some of the complex interactions occurring in biofilms that initiate periodontal tissue destruction.
FIGURE 2.
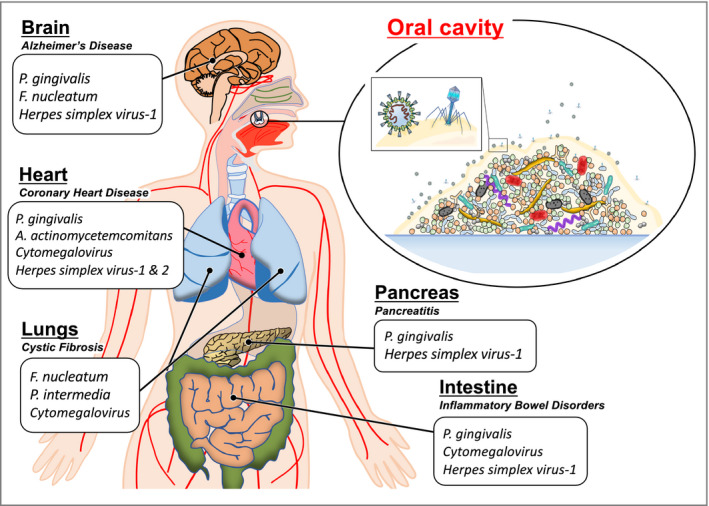
Bacterial and viral periodontopathogens are collaborative residents that form robust biofilms associated with other chronic systemic diseases
5.1. Biofilm formation
It is necessary for the development of a multispecies biofilm to share metabolic and virulence genes to enhance habitat range, improve metabolism, and develop resistance to antimicrobial agents and host immune responses. The exchange of bacterial genes can occur through natural transformation of DNA that is suspended in the biofilm matrix, conjugation between two compatible bacteria, or transduction of viral DNA.139 Bacteriophages support the dynamic exchange of genes as they are natural DNA reservoirs that can deliver nonviral DNA derived from bacterial chromosomes, transposons, and plasmids that encode antibiotic resistance genes.140 The degree to which the process of bacteriophage‐mediated horizontal gene transfer can impact virulence varies; however, one well‐known virulence factor, cholera toxin, arose from the coevolution of bacteriophage CTXφ along with its host bacteria.141 Vibrio cholerae also carries bacteriophage‐encoded virulence factors encoding toxin coregulated pilus that is critical for adherence to the small intestine.142
As bacteriophages attempt to infect bacterial hosts they represent environmental stressors to their prey, which may induce defense mechanisms that promote biofilm formation. Continuous encounters with lytic bacteriophage φ2 induced a change in phenotype in Pseudomonas fluorescens, resulting in overproduction of alginate and loss of motility caused by the mucoid conversion promoting adherence to surfaces.143 Direct contact between filamentous bacteriophage MDAφ and Neisseria meningitis promoted similar adherence to epithelial cells in the human nasopharynx prior to penetration of the blood‐brain barrier.144 The viral particles of bacteriophage MDAφ were actively expressed by N. meningitis instead of type IV pilus to form bundles of viral filaments on the apical surface of the host cell, which were directly attached to epithelial cells.144 Other filamentous bacteriophages serve as structures that strengthen the biofilm network by increasing the viscosity of the matrix and tolerance towards breakdown.145 Both lytic and temperate bacteriophages can strengthen and stabilize the biofilm matrix indirectly through the release of extracellular DNA after lysis of host cells.146 Actinomyces odontolyticus is a predominant Actinomyces species in developing subgingival plaque across all ages and has been isolated as multiple non‐oral lesions.147 The novel isolation of a Siphoviridae linear plasmid‐like prophage designed as xhp1 infects A. odontolyticus subspecies actinosynbacter strain XH001 and promotes biofilm formation through spontaneous induction and the release of host extracellular DNA compared with bacteriophage‐cured XH001.148
Biofilm communities are essential to the lifestyle of oral bacteria because of the relentless environmental and chemical stress the oral cavity presents. We can postulate that certain bacteriophages facilitate biofilm formation for similar evolutionary and protective reasons. A strengthened biofilm matrix protects bacteriophages from the hostile environment and biochemical attacks, while enriching the biofilm matrix with viral and bacterial DNA can further diversify bacteriophages present in these biofilms (Figure 3A).
FIGURE 3.
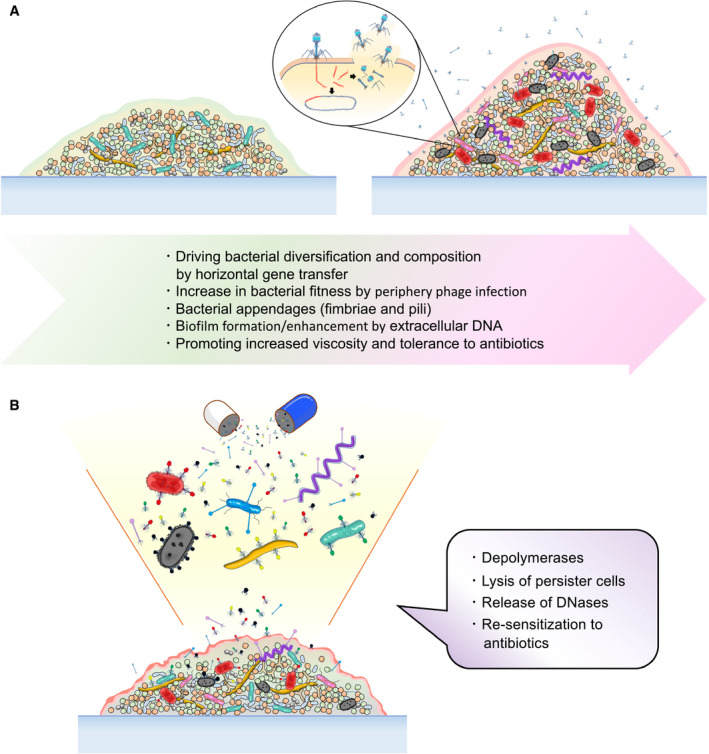
A, Bacteriophages can act as active and supportive members of a biofilm community promoting the development of resilience to environmental stressors, such as antibiotic therapies; and B, bacteriophages can disrupt biofilm dynamics through a variety of mechanisms evolved and may be utilized as an alternative to or adjunctive to conventional antimicrobial therapy
5.2. Bacteriophages as therapeutics
The taxonomic and genomic diversity harbored in biofilms creates a resilient microbial community that potentially limits the penetration of pathogenic microbes and antibiotics. Biofilms display resilience to cationic antimicrobials such as chlorhexidine because of the production of scaffolding extracellular polymeric substances that lowers diffusion of antimicrobials into the deeper layers of the biofilm.149 In addition, approximately 1% of the biofilm is composed of persister cells found in the deep layers of biofilms.150 Minimized metabolic activity equips persister cell tolerance towards antimicrobials and the capability of reinstating the biofilm after such challenges.151 The rise of antibiotic resistance can be attributed to these characteristics of biofilms and is enhanced by the horizontal gene transfer that bacteriophages facilitate. Even with the decline of tetracycline usage, investigators in Spain found early biofilm colonizers of the Streptococci species as carriers of tetracycline resistance genes within subgingival plaque in both healthy (73.5%) and periodontitis (42%) subjects, regardless of prior tetracycline exposure.152 As previously mentioned, bacteriophage P7 identified in the subgingival plaque of healthy individuals also expressed a transposon encoding tetracycline resistance,101 which suggests that bacteriophages can contribute to the transfer of these resistance genes within subgingival plaque and potentially between individuals.
Despite bacteriophage assistance in the development of antibiotic‐resistant biofilms, they also influence the destabilization of said biofilms. In fact, the use of bacteriophages as therapeutics (or phage therapy) was first suggested upon their discovery in 1915, but interest in phage therapy waned in less than 15 years with the discovery of penicillin. With the rise of bacterial resistance to both broad and narrow‐spectrum antibiotics, mechanisms that impair proper biofilm formation and development employed by bacteriophages are attractive alternatives to conventional antimicrobial therapy (Figure 3B).
5.2.1. Targeting biofilm scaffold
In the process of lysis from bacteriophage infection, contents of the intracellular cytoplasm of host cells including DNases are emptied into the biofilm matrix, which contains DNA filaments that reinforce the scaffold and where potential transfer of virulent genes between bacterial species occurs. Expression of DNases by certain bacteria enhances pathogenicity as it enables bacteria to evade immobilization and clearance from neutrophil extracellular traps that contain neutrophil DNA filaments.153 Albeit to a variable degree, periodontopathogens within the red and orange complex display DNase activity in both planktonic and biofilm conditions,154 probably to suppress the effects of neutrophil extracellular traps that are within infected periodontal tissue.155 Likewise, bacteriophage fitness can be enhanced by encoding DNase within its genome. Streptococcus pyogenes bacteriophage spd1 leases genes to express DNase and upregulates just prior to bacteriophage induction to digest bacterial chromosomal DNA released during bacteriophage lysis, allowing spd1 to disseminate freely.156 Furthermore, the potential of DNase to disrupt biofilms has been demonstrated to be more effective against developing and multispecies biofilms rather than matured or mono‐species biofilms, suggesting that the effects of DNase are time‐dependent.157
Depolymerases are enzymes that are capable of degrading capsular polysaccharides, lipopolysaccharides, and peptidoglycan, which are all components of bacterial capsules and are important components of the biofilm matrix. Most depolymerases are encoded in bacteriophage structural proteins such as tail fibers or baseplates and are classified as either hydrolases or lyases.158 Multi‐antibiotic resistant strains of Acinetobacter species were found to be susceptible to the tail fiber of bacteriophage Petty that is equipped with depolymerase activity that results in translucent halos on plaque assays, an apparent decrease in viscosity in the biofilm matrix, and production of reducing ends all of which are indicative of bacteriophage‐associated capsular depolymerase activity.159 In the same study, the depolymerase of bacteriophage Petty denoted as Dpo1 was applied to biofilms of different Acinetobacter calcoaceticus‐Acinetobacter baumannii complex stains and demonstrated the removal of approximately 20% of bacterial hosts.159 However, depolymerase Dpo7 derived from bacteriophage vB_SepiS‐philPLA7 was capable of reducing the biofilm biomass of Staphylococcus species by 53%‐85% in multiple strains.160
Putative periodontopathogens, such as P. gingivalis, evade host immune cells and mediate co‐aggregation with F. nucleatum when an encapsulated phenotype emerges.161 However, glycosyltransferase mutation, resulting in the loss of the polysaccharide capsule of P. gingivalis, perhaps emulating degradation through depolymerases, displayed increased auto‐aggregation and mono‐species biofilm formation.162 Although P. gingivalis currently lacks an identifiable bacteriophage,115 further studies on the effects of depolymerases on multispecies oral biofilms are required to fully understand bacteriophage dynamics within plaque‐induced periodontitis.
5.2.2. Targeting antimicrobial‐resistant microbial members
Targeting specific bacterial host cells within either developing or matured biofilms is another component of phage therapy that is being explored. Resistance to antimicrobials emerges within biofilms as a collaborative effort between microbial members. Specifically, persister cells are critical to the fastidious nature of biofilms as they are inherently inert to antibiotics, and during environmental stress can reinstate biofilms. However, bacteriophages, such as Sb‐1, can directly lyse methicillin‐resistant perister forms of Staphylococcus aureus strains at high titers (107 plaque‐forming units/milliliter) or upon activation of phage progeny formation, as these persister cells reactivate their metabolism within a mono‐species biofilm.163 Although total eradication of S. aureus biofilms was not achieved during Sb‐1 treatment, this study reinforced previous findings that phage therapy may be used as adjunctive therapy to conventional antibiotics because bacteriophages, such as Sb‐1 target, degrade the barriers, which lowers the efficacy of antibiotics.163, 164, 165 Identifying bacteriophages similar to Sb‐1 may become critical if effective phage therapy are developed for oral biofilms as antibiotic‐tolerant bacteria, and persister cells become more established in dental plaque.101, 152, 166, 167
5.2.3. Synthetic bacteriophages
An appreciation of the promising potency of phage therapy has led investigators to apply synthetic biologic concepts to enhance the mechanisms that bacteriophages already employ. Prevailing phage therapy formulations stress the strict use of obligatory lytic bacteriophages as their offensive strategies are more predictable than lysogeny.168 However, specific characteristics of temperate bacteriophages are attractive for biofilm removal and can be tailored by synthetic biology. Insertion of a clustered regularly interspaced short palindromic repeats gene array in bacteriophage lambda targeting bacterial plasmids with beta‐lactamase genes allowed bacteriophage lambda to resensitize E. coli to carbapenems.169 In addition, prevention of further carbapenem‐resistance surges was ensured after prophage induction and formation of progeny of the genetically modified bacteriophage lambda‐infected naive E. coli hosts.169 By extinguishing antibiotic‐resistant bacterial strains, the use of synthetic bacteriophages can enhance conventional antibiotic therapy, as seen with natural bacteriophages.163, 170
Currently, the majority of bacteriophages characterized from the oral cavity are temperate,102, 108 thus genetically modifying the life cycle of these temperate bacteriophages may expand the arsenal against hardy bacteria. Enterococcus faecalis is associated with biofilm formation in failed endodontic treatment at prevalence levels approaching 90%.171 Deletion of the repressor‐sensitive lytic cycle promoter of the natural lysogenic φEf11 E. faecalis bacteriophage and inserting an inducible nisin‐promoter of the lytic cycle produces a genetically modified bacteriophage capable of markedly lysing and reducing the biomass of vancomycin‐resistant E. faecalis strains in a mono‐species biofilm.172 The investigators suggested that exchanges in the introduced inducible promoter can alter the efficacy of phage therapy of genetically modified φEf11 thus increasing the already vast capabilities of bacteriophages.
5.2.4. Cocktail formulation
An abundance of oral bacteriophages allows for numerous modalities of phage therapy as a shift in the current management of periodontitis may be advocated to reduce the systemic effect of conventional antibiotic treatment.173, 174 Thus far, we have described distinct forms in which phage therapy can be delivered including single‐bacteriophage, bacteriophage‐derived enzymes, bacteriophage in combination with antibiotics, and synthetic bacteriophages. Each of these delivery systems has the potential to impact the current treatments prescribed in managing periodontitis. However, to combat the pathogenic shift in the polymicrobial consortium that dysregulates the host immune response within subgingival plaque, an assortment of the phage therapy modalities described is needed to effectively reduce pathogenic virulence and biomass.
One additional form of phage therapy that is significant for other polymicrobial inflammatory disorders is the formulation of a bacteriophage cocktail that targets multiple pathogenic bacteria in a synergistic manner.175, 176, 177, 178 Cocktail phage therapy takes advantage of the narrow host range bacteriophages exhibit to maintain a susceptible bacterial community within their surrounding environment. Selecting the correct combination of bacteriophages may be a challenge given that full characterization of both biofilm microbial members and their respective predator bacteriophages is required for effective therapy.179, 180 However, once full characterization occurs the bacteriophages utilized can spare commensal microbes, which may aid in the reestablishment of homeostasis after cocktail phage therapy.181 Moreover, cocktail formulation can reduce the emergence of bacteriophage‐resistant pathogenic bacteria and reinstatement of biofilms causing recurring inflammation.182
Ideal treatment for the polymicrobial model of periodontal pathogenesis will combine most, if not all, the modalities of phage therapy described. Currently, only a small fraction of identified bacteriophages within the oral cavity have been identified down to the species level.100, 101, 111 This limits the formulation of a bacteriophage cocktail that may prove effective against subgingival plaque. However, the oral bacteriophages against A. actinomycetemcomitans, E. faecalis, and Streptococcus mutans have been isolated at species level and have demonstrated effective disturbance of both planktonic and biofilm forms of their host bacteria.111 Full characterization of oral bacteriophages and their enzymes will not only affect phage therapy but clarify the extent of bacteriophage influence in biofilm formation and dysregulation in periodontitis.
6. CONCLUSIONS
This review alone cannot answer the question of the proposed association between periodontitis and the human oral virome, specifically those in the herpesvirus family. However, multiple studies have illustrated that both eukaryotic and prokaryotic viruses within subgingival plaque and periodontal tissues affect the more established mediators of periodontal inflammation, putative periodontopathogens, and the host immune response. In regard to the association of herpesviruses and periodontitis, the degree to which these viruses directly aggravate periodontal tissue damage still remains unanswered. Further studies demonstrating significant, reproducible, and density‐dependent effects of herpesviruses on putative periodontopathogens or host immune response are required to make this association into a causal relationship. Despite how potentially weak or underestimated the herpesvirus‐periodontitis association may be, the benefits described with prolonged administration of antivirals in immunocompromised or immunodeficient individuals demonstrates that specific populations are possibly more susceptible to viral periodontopathogens. Thus, it may be short‐sighted to neglect the initial implications of viral pathogen involvement in periodontitis.
In addition, the microbial ecology of the oral cavity is enriched and shaped by the omnipresence of bacteriophages. Characterizing novel bacteriophages continues to be a challenge as metagenomic analysis of bacteriophages is still developing. However, despite current limitations, novel bacteriophages with a higher prevalence in inflamed periodontal tissue than healthy tissue continue to be uncovered and provide a foundation for implicating bacteriophages as drivers of periodontitis.183 Understanding the variety of mechanisms that bacteriophages have evolved to infect their bacterial hosts suggests that bacteriophages potentially play a role in the microbial dysbiosis and immune dysregulation that occurs in periodontitis. Alternatively, full characterization of these mechanisms has been utilized or synthetically enhanced to combat fastidious bacterial and biofilm infections. Because of bacteriophage influence in the dynamics of biofilm formation and dispersal, phage therapy is promising for the management of the primary etiology of periodontal inflammation, that is, dental plaque.
There is still a paucity of information that can describe a causal relationship between the human oral virome and periodontitis. However, the biological dark matter inserted by retroviruses once believed to be nonfunctional encodes historical viral‐eukaryotic interactions and influences host development.184 Perhaps it is time to embrace the viral dark matter within the periodontal environment to fully comprehend the pathogenesis and systemic implications of periodontitis.
Martínez A, Kuraji R, Kapila YL. The human oral virome: Shedding light on the dark matter. Periodontol 2000. 2021;87:282–298. 10.1111/prd.12396
Funding information
This work was supported by funding from the AAP Sunstar Innovation Grant, NIH R01 DE025225 grant, and Larry Berkelhammer funds to YLK.
REFERENCES
- 1.Mokili JL, Rohwer F, Dutilh BE. Metagenomics and future perspectives in virus discovery. Curr Opin Virol. 2012;2:63‐77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Socransky SS, Manganiello SD. The oral microbiota of man from birth to senility. J Periodontol. 1971;42:485‐496. [DOI] [PubMed] [Google Scholar]
- 3.Kuczynski J, Costello EK, Nemergut DR, et al. Direct sequencing of the human microbiome readily reveals community differences. Genome Biol. 2010;11:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yost S, Duran‐Pinedo AE, Teles R, Krishnan K, Frias‐Lopez J. Erratum to: functional signatures of oral dysbiosis during periodontitis progression revealed by microbial metatranscriptome analysis. Genome Med. 2015;7:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mohammed H, Varoni EM, Cochis A, et al. Oral dysbiosis in pancreatic cancer and liver cirrhosis: a review of the literature. Biomedicines. 2018;6(4):115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pietiainen M, Liljestrand JM, Kopra E, Pussinen PJ. Mediators between oral dysbiosis and cardiovascular diseases. Eur J Oral Sci. 2018;126(Suppl 1):26‐36. [DOI] [PubMed] [Google Scholar]
- 7.He J, Li Y, Cao Y, Xue J, Zhou X. The oral microbiome diversity and its relation to human diseases. Folia Microbiol (Praha). 2015;60:69‐80. [DOI] [PubMed] [Google Scholar]
- 8.Slots J. Oral viral infections of adults. Periodontol 2000. 2009;49:60‐86. [DOI] [PubMed] [Google Scholar]
- 9.Chen C, Feng P, Slots J. Herpesvirus‐bacteria synergistic interaction in periodontitis. Periodontol 2000. 2020;82:42‐64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao L, Kang M, Zhang MJ, et al. Polymicrobial periodontal disease triggers a wide radius of effect and unique virome. NPJ Biofilms Microbiomes. 2020;6:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J, Gao Y, Zhao F. Phage‐bacteria interaction network in human oral microbiome. Environ Microbiol. 2016;18:2143‐2158. [DOI] [PubMed] [Google Scholar]
- 12.Wagner PL, Waldor MK. Bacteriophage control of bacterial virulence. Infect Immun. 2002;70:3985‐3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watanabe R, Matsumoto T, Sano G, et al. Efficacy of bacteriophage therapy against gut‐derived sepsis caused by Pseudomonas aeruginosa in mice. Antimicrob Agents Chemother. 2007;51:446‐452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leite FRM, Nascimento GG, Baake S, Pedersen LD, Scheutz F, Lopez R. Impact of smoking cessation on periodontitis: a systematic review and meta‐analysis of prospective longitudinal observational and interventional studies. Nicotine Tob Res. 2019;21:1600‐1608. [DOI] [PubMed] [Google Scholar]
- 15.Vieira AR, Albandar JM. Role of genetic factors in the pathogenesis of aggressive periodontitis. Periodontol 2000. 2014;65:92‐106. [DOI] [PubMed] [Google Scholar]
- 16.de Koning AP, Gu W, Castoe TA, Batzer MA, Pollock DD. Repetitive elements may comprise over two‐thirds of the human genome. PLoS Genet. 2011;7:e1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoare A, Marsh PD, Diaz PI. Ecological therapeutic opportunities for oral diseases. Microbiol Spectr. 2017;5:18015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herpesviruses GB. latency and reactivation – viral strategies and host response. J Oral Microbiol. 2013;5(1):22766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ehlers B, Dural G, Yasmum N, et al. Novel mammalian herpesviruses and lineages within the Gammaherpesvirinae: cospeciation and interspecies transfer. J Virol. 2008;82:3509‐3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sabeti M, Slots J. Herpesviral‐bacterial coinfection in periapical pathosis. J Endod. 2004;30:69‐72. [DOI] [PubMed] [Google Scholar]
- 21.Sabeti M, Simon JH, Nowzari H, Slots J. Cytomegalovirus and Epstein‐Barr virus active infection in periapical lesions of teeth with intact crowns. J Endod. 2003;29:321‐323. [DOI] [PubMed] [Google Scholar]
- 22.Jakovljevic A, Nikolic N, Carkic J, et al. Notch ‐ a possible mediator between Epstein‐Barr virus infection and bone resorption in apical periodontitis. Acta Odontol Scand. 2020;78:126‐131. [DOI] [PubMed] [Google Scholar]
- 23.Sabeti M, Kermani V, Sabeti S, Simon JH. Significance of human cytomegalovirus and Epstein‐Barr virus in inducing cytokine expression in periapical lesions. J Endod. 2012;38:47‐50. [DOI] [PubMed] [Google Scholar]
- 24.Jha HC, Banerjee S, Robertson ES. The role of gammaherpesviruses in cancer pathogenesis. Pathogens. 2016;5(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okano M. Epstein‐Barr virus in patients with immunodeficiency disorders. Biomed Pharmacother. 2001;55:353‐361. [DOI] [PubMed] [Google Scholar]
- 26.Ikuta K, Satoh Y, Hoshikawa Y, Sairenji T. Detection of Epstein‐Barr virus in salivas and throat washings in healthy adults and children. Microbes Infect. 2000;2:115‐120. [DOI] [PubMed] [Google Scholar]
- 27.Borza CM, Hutt‐Fletcher LM. Alternate replication in B cells and epithelial cells switches tropism of Epstein‐Barr virus. Nat Med. 2002;8:594‐599. [DOI] [PubMed] [Google Scholar]
- 28.Saito I, Nishimura S, Kudo I, Fox RI, Moro I. Detection of Epstein‐Barr virus and human herpes virus type 6 in saliva from patients with lymphoproliferative diseases by the polymerase chain reaction. Arch Oral Biol. 1991;36:779‐784. [DOI] [PubMed] [Google Scholar]
- 29.Braz‐Silva PH, Ortega KL, Rezende NP, Nunes FD, Magalhaes MH. Detection of Epstein‐Barr virus (EBV) in the oral mucosa of renal transplant patients. Diagn Cytopathol. 2006;34:24‐28. [DOI] [PubMed] [Google Scholar]
- 30.Idesawa M, Sugano N, Ikeda K, et al. Detection of Epstein‐Barr virus in saliva by real‐time PCR. Oral Microbiol Immunol. 2004;19:230‐232. [DOI] [PubMed] [Google Scholar]
- 31.Saygun I, Kubar A, Ozdemir A, Slots J. Periodontitis lesions are a source of salivary cytomegalovirus and Epstein‐Barr virus. J Periodontal Res. 2005;40:187‐191. [DOI] [PubMed] [Google Scholar]
- 32.Khosropanah H, Karandish M, Ziaeyan M, Jamalidoust M. Quantification of Epstein‐Barr virus and human cytomegalovirus in chronic periodontal patients. Jundishapur J Microbiol. 2015;8:e18691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klemenc P, Skaleric U, Artnik B, Nograsek P, Marin J. Prevalence of some herpesviruses in gingival crevicular fluid. J Clin Virol. 2005;34:147‐152. [DOI] [PubMed] [Google Scholar]
- 34.Shah R, Mehta DS. Prevalence of herpesviruses in gingivitis and chronic periodontitis: relationship to clinical parameters and effect of treatment. J Indian Soc Periodontol. 2016;20:279‐285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stein JM, Said Yekta S, Kleines M, et al. Failure to detect an association between aggressive periodontitis and the prevalence of herpesviruses. J Clin Periodontol. 2013;40:1‐7. [DOI] [PubMed] [Google Scholar]
- 36.Pallos D, Ruivo GF, Ferrari‐Junior SH, et al. Periodontal disease and detection of human herpesviruses in saliva and gingival crevicular fluid of chronic kidney disease patients. J Periodontol. 2020;91:1139‐1147. [DOI] [PubMed] [Google Scholar]
- 37.Ozcan E, Saygun NI, Serdar MA, Kubar A, Bengi VUPorphyromonas gingivalis and Epstein‐Barr virus are associated with increased levels of visfatin in gingival crevicular fluid. J Periodontol. 2016;87:443‐451. [DOI] [PubMed] [Google Scholar]
- 38.Bayani M, Pourali M, Keivan M. Possible interaction between visfatin, periodontal infection, and other systemic diseases: a brief review of literature. Eur J Dent. 2017;11:407‐410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Damanaki A, Nokhbehsaim M, Eick S, et al. Regulation of NAMPT in human gingival fibroblasts and biopsies. Mediators Inflamm. 2014;2014:912821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saygun I, Kubar A, Sahin S, Sener K, Slots J. Quantitative analysis of association between herpesviruses and bacterial pathogens in periodontitis. J Periodontal Res. 2008;43:352‐359. [DOI] [PubMed] [Google Scholar]
- 41.Slots J, Kamma JJ, Sugar C. The herpesvirus‐Porphyromonas gingivalis‐periodontitis axis. J Periodontal Res. 2003;38:318‐323. [DOI] [PubMed] [Google Scholar]
- 42.Kato A, Imai K, Sato H, Ogata Y. Prevalence of Epstein‐Barr virus DNA and Porphyromonas gingivalis in Japanese peri‐implantitis patients. BMC Oral Health. 2017;17:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vincent‐Bugnas S, Vitale S, Mouline CC, et al. EBV infection is common in gingival epithelial cells of the periodontium and worsens during chronic periodontitis. PLoS One. 2013;8:e80336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kato A, Imai K, Ochiai K, Ogata Y. Higher prevalence of Epstein‐Barr virus DNA in deeper periodontal pockets of chronic periodontitis in Japanese patients. PLoS One. 2013;8:e71990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dawson DR 3rd, Wang C, Danaher RJ, et al. Salivary levels of Epstein‐Barr virus DNA correlate with subgingival levels, not severity of periodontitis. Oral Dis. 2009;15:554‐559. [DOI] [PubMed] [Google Scholar]
- 46.Dommisch H, Chung WO, Rohani MG, et al. Protease‐activated receptor 2 mediates human beta‐defensin 2 and CC chemokine ligand 20 mRNA expression in response to proteases secreted by Porphyromonas gingivalis . Infect Immun. 2007;75:4326‐4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Niederman R, Buyle‐Bodin Y, Lu BY, Robinson P, Naleway C. Short‐chain carboxylic acid concentration in human gingival crevicular fluid. J Dent Res. 1997;76:575‐579. [DOI] [PubMed] [Google Scholar]
- 48.Lu R, Meng H, Gao X, Xu L, Feng X. Effect of non‐surgical periodontal treatment on short chain fatty acid levels in gingival crevicular fluid of patients with generalized aggressive periodontitis. J Periodontal Res. 2014;49:574‐583. [DOI] [PubMed] [Google Scholar]
- 49.Zuhair M, Smit GSA, Wallis G, et al. Estimation of the worldwide seroprevalence of cytomegalovirus: a systematic review and meta‐analysis. Rev Med Virol. 2019;29:e2034. [DOI] [PubMed] [Google Scholar]
- 50.Zheng QY, Huynh KT, van Zuylen WJ, Craig ME, Rawlinson WD. Cytomegalovirus infection in day care centres: a systematic review and meta‐analysis of prevalence of infection in children. Rev Med Virol. 2019;29:e2011. [DOI] [PubMed] [Google Scholar]
- 51.Chandler SH, Holmes KK, Wentworth BB, et al. The epidemiology of cytomegaloviral infection in women attending a sexually transmitted disease clinic. J Infect Dis. 1985;152:597‐605. [DOI] [PubMed] [Google Scholar]
- 52.Naing ZW, Scott GM, Shand A, et al. Congenital cytomegalovirus infection in pregnancy: a review of prevalence, clinical features, diagnosis and prevention. Aust N Z J Obstet Gynaecol. 2016;56:9‐18. [DOI] [PubMed] [Google Scholar]
- 53.Sing GK, Ruscetti FW. The role of human cytomegalovirus in haematological diseases. Baillieres Clin Haematol. 1995;8:149‐163. [DOI] [PubMed] [Google Scholar]
- 54.Waters S, Lee S, Lloyd M, Irish A, Price P. The detection of CMV in saliva can mark a systemic infection with CMV in renal transplant recipients. Int J Mol Sci. 2019;20:5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bilder L, Elimelech R, Szwarcwort‐Cohen M, Kra‐Oz Z, Machtei EE. The prevalence of human herpes viruses in the saliva of chronic periodontitis patients compared to oral health providers and healthy controls. Arch Virol. 2013;158:1221‐1226. [DOI] [PubMed] [Google Scholar]
- 56.Contreras A, Nowzari H, Slots J. Herpesviruses in periodontal pocket and gingival tissue specimens. Oral Microbiol Immunol. 2000;15:15‐18. [DOI] [PubMed] [Google Scholar]
- 57.Kubar A, Saygun I, Ozdemir A, Yapar M, Slots J. Real‐time polymerase chain reaction quantification of human cytomegalovirus and Epstein‐Barr virus in periodontal pockets and the adjacent gingiva of periodontitis lesions. J Periodontal Res. 2005;40:97‐104. [DOI] [PubMed] [Google Scholar]
- 58.Botero JE, Rodriguez‐Medina C, Jaramillo‐Echeverry A, Contreras A. Association between human cytomegalovirus and periodontitis: a systematic review and meta‐analysis. J Periodontal Res. 2020;55:551‐558. [DOI] [PubMed] [Google Scholar]
- 59.Nibali L, Atkinson C, Griffiths P, et al. Low prevalence of subgingival viruses in periodontitis patients. J Clin Periodontol. 2009;36:928‐932. [DOI] [PubMed] [Google Scholar]
- 60.Santangelo R, D'Ercole S, Graffeo R, et al. Bacterial and viral DNA in periodontal disease: a study using multiplex PCR. New Microbiol. 2004;27:133‐137. [PubMed] [Google Scholar]
- 61.Grundy JE, Lawson KM, MacCormac LP, Fletcher JM, Yong KL. Cytomegalovirus‐infected endothelial cells recruit neutrophils by the secretion of C‐X‐C chemokines and transmit virus by direct neutrophil‐endothelial cell contact and during neutrophil transendothelial migration. J Infect Dis. 1998;177:1465‐1474. [DOI] [PubMed] [Google Scholar]
- 62.Saygun I, Kubar A, Ozdemir A, Yapar M, Slots J. Herpesviral‐bacterial interrelationships in aggressive periodontitis. J Periodontal Res. 2004;39:207‐212. [DOI] [PubMed] [Google Scholar]
- 63.Sharma S, Tapashetti RP, Patil SR, Kalra SM, Bhat GK, Guvva S. Revelation of viral ‐ bacterial interrelationship in aggressive periodontitis via polymerase chain reaction: a microbiological study. J Int Oral Health. 2015;7:101‐107. [PMC free article] [PubMed] [Google Scholar]
- 64.Elamin A, Ali RW, Bakken V. Putative periodontopathic bacteria and herpes viruses interactions in the subgingival plaque of patients with aggressive periodontitis and healthy controls. Clin Exp Dent Res. 2017;3:183‐190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Michalowicz BS, Ronderos M, Camara‐Silva R, Contreras A, Slots J. Human herpesviruses and Porphyromonas gingivalis are associated with juvenile periodontitis. J Periodontol. 2000;71:981‐988. [DOI] [PubMed] [Google Scholar]
- 66.Pocock JM, Storisteanu DML, Reeves MB, et al. Human cytomegalovirus delays neutrophil apoptosis and stimulates the release of a prosurvival secretome. Front Immunol. 2017;8:1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Botero JE, Contreras A, Parra B. Profiling of inflammatory cytokines produced by gingival fibroblasts after human cytomegalovirus infection. Oral Microbiol Immunol. 2008;23:291‐298. [DOI] [PubMed] [Google Scholar]
- 68.Jarzembowski T, Daca A, Witkowski J, Rutkowski B, Gołębiewska J, Dębska‐Ślizień A. Does CMV infection impact the virulence of Enterococcus faecalis? Virulence. 2013;4:641‐645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Slots J. Update on human cytomegalovirus in destructive periodontal disease. Oral Microbiol Immunol. 2004;19:217‐223. [DOI] [PubMed] [Google Scholar]
- 70.Samuel CE. Antiviral actions of interferons. Clin Microbiol Rev. 2001;14:778‐809, table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee AJ, Ashkar AA. The dual nature of type I and type II interferons. Front Immunol. 2018;9:2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Baldridge MT, Lee S, Brown JJ, et al. Expression of Ifnlr1 on intestinal epithelial cells is critical to the antiviral effects of interferon lambda against norovirus and reovirus. J Virol. 2017;91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bilichodmath S, Nair SK, Bilichodmath R, Mangalekar SB. mRNA expression of IFN‐lambdas in the gingival tissue of patients with chronic or aggressive periodontitis: a polymerase chain reaction study. J Periodontol. 2018;89:867‐874. [DOI] [PubMed] [Google Scholar]
- 74.Odendall C, Voak AA, Kagan JC. Type III ifns are commonly induced by bacteria‐sensing TLRs and reinforce epithelial barriers during infection. J Immunol. 2017;199:3270‐3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mizraji G, Nassar M, Segev H, et al. Porphyromonas gingivalis promotes unrestrained type I interferon production by dysregulating TAM signaling via MYD88 degradation. Cell Rep. 2017;18:419‐431. [DOI] [PubMed] [Google Scholar]
- 76.Muzammil JD, Faizuddin M, Noor Ahamadi HM. Association of interferon lambda‐1 with herpes simplex viruses‐1 and ‐2, Epstein‐Barr virus, and human cytomegalovirus in chronic periodontitis. J Investig Clin Dent. 2017;8:e12200. [DOI] [PubMed] [Google Scholar]
- 77.Egli A, Levin A, Santer DM, et al. Immunomodulatory function of Interleukin 28B during primary infection with cytomegalovirus. J Infect Dis. 2014;210:717‐727. [DOI] [PubMed] [Google Scholar]
- 78.Zhou L, Li JL, Zhou Y, et al. Induction of interferon‐lambda contributes to TLR3 and RIG‐I activation‐mediated inhibition of herpes simplex virus type 2 replication in human cervical epithelial cells. Mol Hum Reprod. 2015;21:917‐929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wu L, Fossum E, Joo CH, et al. Epstein‐Barr virus LF2: an antagonist to type I interferon. J Virol. 2009;83:1140‐1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jangra S, Yuen KS, Botelho MG, Jin DY. Epstein‐Barr virus and innate immunity: friends or foes? Microorganisms. 2019;7:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang B, Kang W, Zuo J, Kang W, Sun Y. The significance of type‐I interferons in the pathogenesis and therapy of human immunodeficiency virus 1 infection. Front Immunol. 2017;8:1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Williams RC, Paquette DW, Offenbacher S, et al. Treatment of periodontitis by local administration of minocycline microspheres: a controlled trial. J Periodontol. 2001;72:1535‐1544. [DOI] [PubMed] [Google Scholar]
- 83.Zink MC, Uhrlaub J, DeWitt J, et al. Neuroprotective and anti‐human immunodeficiency virus activity of minocycline. JAMA. 2005;293:2003‐2011. [DOI] [PubMed] [Google Scholar]
- 84.Noguera‐Julian M, Guillen Y, Peterson J, et al. Oral microbiome in HIV‐associated periodontitis. Medicine. 2017;96:e5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Blignaut E, Rossouw TM, Becker PJ, Mavuso DS, Feucht UD. Gingival recession and localized aggressive periodontitis among HIV‐infected children and adolescents receiving antiretroviral therapy. Pediatr Infect Dis J. 2019;38:e112‐e115. [DOI] [PubMed] [Google Scholar]
- 86.Hai R, Chu A, Li H, Umamoto S, Rider P, Liu F. Infection of human cytomegalovirus in cultured human gingival tissue. Virol J. 2006;3:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sunde PT, Olsen I, Enersen M, Grinde B. Patient with severe periodontitis and subgingival Epstein‐Barr virus treated with antiviral therapy. J Clin Virol. 2008;42:176‐178. [DOI] [PubMed] [Google Scholar]
- 88.Mann NH. The third age of phage. PLoS Biol. 2005;3:e182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Aggarwala V, Liang G, Bushman FD. Viral communities of the human gut: metagenomic analysis of composition and dynamics. Mob DNA. 2017;8:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Simmonds P, Adams MJ, Benko M, et al. Consensus statement: Virus taxonomy in the age of metagenomics. Nat Rev Microbiol. 2017;15:161‐168. [DOI] [PubMed] [Google Scholar]
- 91.Ackermann HW. 5500 Phages examined in the electron microscope. Arch Virol. 2007;152:227‐243. [DOI] [PubMed] [Google Scholar]
- 92.Colwell RR. Release of genetically engineered micro‐organisms into the environment. MIRCEN J Appl Microbiol Biotechnol. 1986;2:41‐49. [Google Scholar]
- 93.Marks T, Sharp R. Bacteriophages and biotechnology: a review. J Chem Technol Biotechnol. 2000;75:6‐17. [Google Scholar]
- 94.Taylor NM, Prokhorov NS, Guerrero‐Ferreira RC, et al. Structure of the T4 baseplate and its function in triggering sheath contraction. Nature. 2016;533:346‐352. [DOI] [PubMed] [Google Scholar]
- 95.d'Humieres C, Touchon M, Dion S, et al. A simple, reproducible and cost‐effective procedure to analyse gut phageome: from phage isolation to bioinformatic approach. Sci Rep. 2019;9:11331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hayes S, Mahony J, Nauta A, van Sinderen D. Metagenomic approaches to assess bacteriophages in various environmental niches. Viruses. 2017;9:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Minot S, Bryson A, Chehoud C, Wu GD, Lewis JD, Bushman FD. Rapid evolution of the human gut virome. Proc Natl Acad Sci USA. 2013;110:12450‐12455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Norman JM, Handley SA, Baldridge MT, et al. Disease‐specific alterations in the enteric virome in inflammatory bowel disease. Cell. 2015;160:447‐460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hoyles L, McCartney AL, Neve H, et al. Characterization of virus‐like particles associated with the human faecal and caecal microbiota. Res Microbiol. 2014;165:803‐812. [DOI] [PubMed] [Google Scholar]
- 100.Pride DT, Salzman J, Haynes M, et al. Evidence of a robust resident bacteriophage population revealed through analysis of the human salivary virome. ISME J. 2012;6:915‐926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Naidu M, Robles‐Sikisaka R, Abeles SR, Boehm TK, Pride DT. Characterization of bacteriophage communities and CRISPR profiles from dental plaque. BMC Microbiol. 2014;14:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ly M, Abeles SR, Boehm TK, et al. Altered oral viral ecology in association with periodontal disease. MBio. 2014;5:e01133–01114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Perez‐Brocal V, Moya A. The analysis of the oral DNA virome reveals which viruses are widespread and rare among healthy young adults in Valencia (Spain). PLoS One. 2018;13:e0191867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Abeles SR, Robles‐Sikisaka R, Ly M, et al. Human oral viruses are personal, persistent and gender‐consistent. ISME J. 2014;8:1753‐1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Vazquez‐Castellanos JF, Garcia‐Lopez R, Perez‐Brocal V, Pignatelli M, Moya A. Comparison of different assembly and annotation tools on analysis of simulated viral metagenomic communities in the gut. BMC Genom. 2014;15:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Perez‐Brocal V, Garcia‐Lopez R, Vazquez‐Castellanos JF, et al. Study of the viral and microbial communities associated with Crohn's disease: a metagenomic approach. Clin Transl Gastroenterol. 2013;4:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhao G, Vatanen T, Droit L, et al. Intestinal virome changes precede autoimmunity in type I diabetes‐susceptible children. Proc Natl Acad Sci USA. 2017;114:E6166‐E6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chen Y, Yang L, Yang D, et al. Specific integration of temperate phage decreases the pathogenicity of host bacteria. Front Cell Infect Microbiol. 2020;10:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mojica FJ, Diez‐Villasenor C, Garcia‐Martinez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174‐182. [DOI] [PubMed] [Google Scholar]
- 110.Venturini C, Zingali T, Wyrsch ER, et al. Diversity of P1 phage‐like elements in multidrug resistant Escherichia coli . Sci Rep. 2019;9:18861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Szafranski SP, Winkel A, Stiesch M. The use of bacteriophages to biocontrol oral biofilms. J Biotechnol. 2017;250:29‐44. [DOI] [PubMed] [Google Scholar]
- 112.Stevens RH, Moura Martins Lobo dos Santos CD, Zuanazzi D, et al. Prophage induction in lysogenic Aggregatibacter actinomycetemcomitans cells co‐cultured with human gingival fibroblasts, and its effect on leukotoxin release. Microb Pathog. 2013;54:54‐59. [DOI] [PubMed] [Google Scholar]
- 113.Szafranski SP, Kilian M, Yang I, et al. Diversity patterns of bacteriophages infecting Aggregatibacter and Haemophilus species across clades and niches. ISME J. 2019;13:2500‐2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mitchell HL, Dashper SG, Catmull DV, et al. Treponema denticola biofilm‐induced expression of a bacteriophage, toxin‐antitoxin systems and transposases. Microbiology (Reading). 2010;156:774‐788. [DOI] [PubMed] [Google Scholar]
- 115.Watanabe T, Shibasaki M, Maruyama F, Sekizaki T, Nakagawa I. Investigation of potential targets of Porphyromonas CRISPRs among the genomes of Porphyromonas species. PLoS One. 2017;12:e0183752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hatfull GF. Bacteriophage genomics. Curr Opin Microbiol. 2008;11:447‐453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Flores CO, Valverde S, Weitz JS. Multi‐scale structure and geographic drivers of cross‐infection within marine bacteria and phages. ISME J. 2013;7:520‐532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Stevens RH, Hammond BF, Lai CH. Characterization of an inducible bacteriophage from a leukotoxic strain of Actinobacillus actinomycetemcomitans . Infect Immun. 1982;35:343‐349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Loftus A, Delisle AL. Inducible bacteriophages of Actinobacillus actinomycetemcomitans . Curr Microbiol. 1995;30:317‐321. [DOI] [PubMed] [Google Scholar]
- 120.Weitz JS, Poisot T, Meyer JR, et al. Phage‐bacteria infection networks. Trends Microbiol. 2013;21:82‐91. [DOI] [PubMed] [Google Scholar]
- 121.Sullivan MB, Waterbury JB, Chisholm SW. Cyanophages infecting the oceanic cyanobacterium Prochlorococcus. Nature. 2003;424:1047‐1051. [DOI] [PubMed] [Google Scholar]
- 122.Wright RCT, Friman VP, Smith MCM, Brockhurst MA. Cross‐resistance is modular in bacteria‐phage interactions. PLoS Biol. 2018;16:e2006057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Majewska J, Kazmierczak Z, Lahutta K, et al. Induction of phage‐specific antibodies by two therapeutic staphylococcal bacteriophages administered per os. Front Immunol. 2019;10:2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Carroll‐Portillo A, Lin HC. Bacteriophage and the innate immune system: access and signaling. Microorganisms. 2019;7:625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Noppert SJ, Fitzgerald KA, Hertzog PJ. The role of type I interferons in TLR responses. Immunol Cell Biol. 2007;85:446‐457. [DOI] [PubMed] [Google Scholar]
- 126.Shan J, Ramachandran A, Thanki AM, Vukusic FBI, Barylski J, Clokie MRJ. Bacteriophages are more virulent to bacteria with human cells than they are in bacterial culture; insights from HT‐29 cells. Sci Rep. 2018;8:5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Fraser JS, Yu Z, Maxwell KL, Davidson AR. Ig‐like domains on bacteriophages: a tale of promiscuity and deceit. J Mol Biol. 2006;359:496‐507. [DOI] [PubMed] [Google Scholar]
- 128.Devine DA, Marsh PD, Meade J. Modulation of host responses by oral commensal bacteria. J Oral Microbiol. 2015;7:26941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Barr JJ, Auro R, Furlan M, et al. Bacteriophage adhering to mucus provide a non‐host‐derived immunity. Proc Natl Acad Sci USA. 2013;110:10771‐10776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Roach DR, Leung CY, Henry M, et al. Synergy between the host immune system and bacteriophage is essential for successful phage therapy against an acute respiratory pathogen. Cell Host Microbe. 2017;22:38‐47.e4. [DOI] [PubMed] [Google Scholar]
- 131.Przerwa A, Zimecki M, Switala‐Jelen K, et al. Effects of bacteriophages on free radical production and phagocytic functions. Med Microbiol Immunol. 2006;195:143‐150. [DOI] [PubMed] [Google Scholar]
- 132.Nallapareddy SR, Singh KV, Sillanpaa J, et al. Endocarditis and biofilm‐associated pili of Enterococcus faecalis . J Clin Invest. 2006;116:2799‐2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hoiby N. Understanding bacterial biofilms in patients with cystic fibrosis: current and innovative approaches to potential therapies. J Cyst Fibros. 2002;1:249‐254. [DOI] [PubMed] [Google Scholar]
- 134.Srivastava A, Gupta J, Kumar S, Kumar A. Gut biofilm forming bacteria in inflammatory bowel disease. Microb Pathog. 2017;112:5‐14. [DOI] [PubMed] [Google Scholar]
- 135.Del Castillo E, Meier R, Chung M, et al. The microbiomes of pancreatic and duodenum tissue overlap and are highly subject specific but differ between pancreatic cancer and noncancer subjects. Cancer Epidemiol Biomarkers Prev. 2019;28:370‐383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Allen HB. Alzheimer's disease: assessing the role of spirochetes, biofilms, the immune system, and amyloid‐beta with regard to potential treatment and prevention. J Alzheimers Dis. 2016;53:1271‐1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Diaz PI, Chalmers NI, Rickard AH, et al. Molecular characterization of subject‐specific oral microflora during initial colonization of enamel. Appl Environ Microbiol. 2006;72:2837‐2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Cortelli JR, Aquino DR, Cortelli SC, et al. Etiological analysis of initial colonization of periodontal pathogens in oral cavity. J Clin Microbiol. 2008;46:1322‐1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Molin S, Tolker‐Nielsen T. Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of the biofilm structure. Curr Opin Biotechnol. 2003;14:255‐261. [DOI] [PubMed] [Google Scholar]
- 140.Calero‐Caceres W, Ye M, Balcazar JL. Bacteriophages as environmental reservoirs of antibiotic resistance. Trends Microbiol. 2019;27:570‐577. [DOI] [PubMed] [Google Scholar]
- 141.Boyd EF, Moyer KE, Shi L, Waldor MK. Infectious CTXPhi and the vibrio pathogenicity island prophage in Vibrio mimicus: evidence for recent horizontal transfer between V. mimicus and V. cholerae. Infect Immun. 2000;68:1507‐1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Karaolis DK, Somara S, Maneval DR Jr, Johnson JA, Kaper JB. A bacteriophage encoding a pathogenicity island, a type‐IV pilus and a phage receptor in cholera bacteria. Nature. 1999;399:375‐379. [DOI] [PubMed] [Google Scholar]
- 143.Scanlan PD, Buckling A. Co‐evolution with lytic phage selects for the mucoid phenotype of Pseudomonas fluorescens SBW25. ISME J. 2012;6:1148‐1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bille E, Meyer J, Jamet A, et al. A virulence‐associated filamentous bacteriophage of Neisseria meningitidis increases host‐cell colonisation. PLoS Pathog. 2017;13:e1006495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Secor PR, Sweere JM, Michaels LA, et al. Filamentous bacteriophage promote biofilm assembly and function. Cell Host Microbe. 2015;18:549‐559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Whitchurch CB, Tolker‐Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295:1487. [DOI] [PubMed] [Google Scholar]
- 147.Kononen E, Wade WG. Actinomyces and related organisms in human infections. Clin Microbiol Rev. 2015;28:419‐442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Shen M, Yang Y, Shen W, et al. A linear plasmid‐like prophage of actinomyces odontolyticus promotes biofilm assembly. Appl Environ Microbiol. 2018;84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Shen Y, Zhao J, de la Fuente‐Nunez C, et al. Experimental and theoretical investigation of multispecies oral biofilm resistance to chlorhexidine treatment. Sci Rep. 2016;6:27537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lewis K. Persister cells. Annu Rev Microbiol. 2010;64:357‐372. [DOI] [PubMed] [Google Scholar]
- 151.Kaldalu N, Hauryliuk V, Tenson T. Persisters‐as elusive as ever. Appl Microbiol Biotechnol. 2016;100:6545‐6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Arredondo A, Blanc V, Mor C, Nart J, León R. Tetracycline and multidrug resistance in the oral microbiota: differences between healthy subjects and patients with periodontitis in Spain. J Oral Microbiol. 2021;13:1847431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Buchanan JT, Simpson AJ, Aziz RK, et al. DNase expression allows the pathogen group A Streptococcus to escape killing in neutrophil extracellular traps. Curr Biol. 2006;16:396‐400. [DOI] [PubMed] [Google Scholar]
- 154.Palmer LJ, Chapple IL, Wright HJ, Roberts A, Cooper PR. Extracellular deoxyribonuclease production by periodontal bacteria. J Periodontal Res. 2012;47:439‐445. [DOI] [PubMed] [Google Scholar]
- 155.Vitkov L, Klappacher M, Hannig M, Krautgartner WD. Extracellular neutrophil traps in periodontitis. J Periodontal Res. 2009;44:664‐672. [DOI] [PubMed] [Google Scholar]
- 156.Broudy TB, Pancholi V, Fischetti VA. The in vitro interaction of Streptococcus pyogenes with human pharyngeal cells induces a phage‐encoded extracellular DNase. Infect Immun. 2002;70:2805‐2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Sharma K, Pagedar SA. Antibiofilm effect of dnase against single and mixed species biofilm. Foods. 2018;7(3):42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Pires DP, Oliveira H, Melo LD, Sillankorva S, Azeredo J. Bacteriophage‐encoded depolymerases: their diversity and biotechnological applications. Appl Microbiol Biotechnol. 2016;100:2141‐2151. [DOI] [PubMed] [Google Scholar]
- 159.Hernandez‐Morales AC, Lessor LL, Wood TL, et al. Genomic and biochemical characterization of acinetobacter podophage petty reveals a novel lysis mechanism and tail‐associated depolymerase activity. J Virol. 2018;92:e01064‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Gutierrez D, Briers Y, Rodriguez‐Rubio L, et al. Role of the pre‐neck appendage protein (Dpo7) from Phage vB_SepiS‐phiIPLA7 as an anti‐biofilm agent in staphylococcal species. Front Microbiol. 2015;6:1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Rosen G, Sela MN. Coaggregation of Porphyromonas gingivalis and Fusobacterium nucleatum PK 1594 is mediated by capsular polysaccharide and lipopolysaccharide. FEMS Microbiol Lett. 2006;256:304‐310. [DOI] [PubMed] [Google Scholar]
- 162.Davey ME, Duncan MJ. Enhanced biofilm formation and loss of capsule synthesis: deletion of a putative glycosyltransferase in Porphyromonas gingivalis . J Bacteriol. 2006;188:5510‐5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Tkhilaishvili T, Lombardi L, Klatt AB, Trampuz A, Di Luca M. Bacteriophage Sb‐1 enhances antibiotic activity against biofilm, degrades exopolysaccharide matrix and targets persisters of Staphylococcus aureus . Int J Antimicrob Agents. 2018;52:842‐853. [DOI] [PubMed] [Google Scholar]
- 164.Kumaran D, Taha M, Yi Q, et al. Does Treatment order matter? investigating the ability of bacteriophage to augment antibiotic activity against Staphylococcus aureus biofilms. Front Microbiol. 2018;9:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ryan EM, Alkawareek MY, Donnelly RF, Gilmore BF. Synergistic phage‐antibiotic combinations for the control of Escherichia coli biofilms in vitro. FEMS Immunol Med Microbiol. 2012;65:395‐398. [DOI] [PubMed] [Google Scholar]
- 166.Saleem HG, Seers CA, Sabri AN, Reynolds EC. Dental plaque bacteria with reduced susceptibility to chlorhexidine are multidrug resistant. BMC Microbiol. 2016;16:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Suppiger S, Astasov‐Frauenhoffer M, Schweizer I, Waltimo T, Kulik EM. Tolerance and persister formation in oral streptococci. Antibiotics (Basel). 2020;9:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Maciejewska B, Olszak T, Drulis‐Kawa Z. Applications of bacteriophages versus phage enzymes to combat and cure bacterial infections: an ambitious and also a realistic application? Appl Microbiol Biotechnol. 2018;102:2563‐2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Yosef I, Manor M, Kiro R, Qimron U. Temperate and lytic bacteriophages programmed to sensitize and kill antibiotic‐resistant bacteria. Proc Natl Acad Sci USA. 2015;112:7267‐7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Lu TK, Collins JJ. Engineered bacteriophage targeting gene networks as adjuvants for antibiotic therapy. Proc Natl Acad Sci USA. 2009;106:4629‐4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Rocas IN, Siqueira JF Jr, Santos KR. Association of Enterococcus faecalis with different forms of periradicular diseases. J Endod. 2004;30:315‐320. [DOI] [PubMed] [Google Scholar]
- 172.Tinoco JM, Buttaro B, Zhang H, Liss N, Sassone L, Stevens R. Effect of a genetically engineered bacteriophage on Enterococcus faecalis biofilms. Arch Oral Biol. 2016;71:80‐86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Jepsen K, Jepsen S. Antibiotics/antimicrobials: systemic and local administration in the therapy of mild to moderately advanced periodontitis. Periodontol 2000. 2016;71:82‐112. [DOI] [PubMed] [Google Scholar]
- 174.Merchant AT, Virani SS. Evaluating periodontal treatment to prevent cardiovascular disease: challenges and possible solutions. Curr Atheroscler Rep. 2017;19:4. [DOI] [PubMed] [Google Scholar]
- 175.Kifelew LG, Warner MS, Morales S, et al. Efficacy of Lytic phage cocktails on Staphylococcus aureus and Pseudomonas aeruginosa in mixed‐species planktonic cultures and biofilms. Viruses. 2020;12:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kifelew LG, Warner MS, Morales S, et al. Efficacy of phage cocktail AB‐SA01 therapy in diabetic mouse wound infections caused by multidrug‐resistant Staphylococcus aureus . BMC Microbiol. 2020;20:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Nale JY, Spencer J, Hargreaves KR, et al. Bacteriophage combinations significantly reduce clostridium difficile growth in vitro and proliferation in vivo. Antimicrob Agents Chemother. 2016;60:968‐981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Mendes JJ, Leandro C, Corte‐Real S, et al. Wound healing potential of topical bacteriophage therapy on diabetic cutaneous wounds. Wound Repair Regen. 2013;21:595‐603. [DOI] [PubMed] [Google Scholar]
- 179.Gibson SB, Green SI, Liu CG, et al. Constructing and characterizing bacteriophage libraries for phage therapy of human infections. Front Microbiol. 2019;10:2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Fernandez L, Gutierrez D, Garcia P, Rodriguez A. The perfect bacteriophage for therapeutic applications‐a quick guide. Antibiotics (Basel). 2019;8:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Cieplak T, Soffer N, Sulakvelidze A, Nielsen DS. A bacteriophage cocktail targeting Escherichia coli reduces E coli in simulated gut conditions, while preserving a non‐targeted representative commensal normal microbiota. Gut Microbes. 2018;9:391‐399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Yuan Y, Wang L, Li X, Tan D, Cong C, Xu Y. Efficacy of a phage cocktail in controlling phage resistance development in multidrug resistant Acinetobacter baumannii . Virus Res. 2019;272:197734. [DOI] [PubMed] [Google Scholar]
- 183.Zhang Y, Shan TL, Li F, et al. A novel phage from periodontal pockets associated with chronic periodontitis. Virus Genes. 2019;55:381‐393. [DOI] [PubMed] [Google Scholar]
- 184.Martin L, Chang HY. Uncovering the role of genomic "dark matter" in human disease. J Clin Invest. 2012;122:1589‐1595. [DOI] [PMC free article] [PubMed] [Google Scholar]


