Abstract
Innate lymphoid cells (ILCs), a critical component of the immune system, have recently been nominated as emerging players associated with tumor progression and inhibition. ILCs are classified into five groups: natural killer (NK) cells, ILC1s, ILC2s, ILC3s, and lymphoid tissue inducer (LTis) cells. NK cells and ILC1s are mainly involved in antitumor activities due to their cytotoxic and cytokine production capabilities, respectively. The current understanding of the heterogeneous behavior of ILC2s and ILC3s in tumors is limited and incomplete. Mostly, their dual roles are modulated by their resident tissues, released cytokines, cancer types, and plasticity. Based on overlap RORγt and cytokine expression, the LTi cells were previously considered part of the ILC3s ontogeny, which are essential for the formation of the secondary lymphoid organs during embryogenesis. Indeed, these facts highlight the urgency in understanding the respective mechanisms that shape the phenotypes and responses of ILCs, either on the repressive or proliferative side in the tumor microenvironment (TME). This review aims to provide an updated view of ILCs biology with respect to tumorigenesis, including a description of ILC plasticity, their interaction with other immune cells and communication with components of the TME. Taken together, targeting ILCs for cancer immunotherapy could be a promising approach against tumors that needs to be further study.
Keywords: Cancer immunotherapy, Innate lymphoid cells (ILCs), Plasticity Tumorigenesis, Tumor microenvironment (TME)
Innate lymphoid cells (ILCs) have been documenting as emerging players associated with tumor progression and inhibition. Therefore, understanding the underlying mechanisms of ILCs in shaping the immune response in tumor could be in favor of developing approaches against tumors.
Introduction
Innate lymphoid cells (ILCs) are considered a parallel lineage of innate immune cells to conventional T and B cells [1]. Unlike adaptive T and B lymphocytes, ILCs do not express somatically rearranged antigen receptors and, hence, do not possess any degree of antigen specificity [2, 3]. However, ILCs secrete a similar suite of inflammatory mediators as T lymphocytes [1, 2, 4, 5]. Compare to conventional T lymphocytes, ILCs develop in the fetal and perinatal stages, mainly reside in nonlymphoid tissues, have slow turnover rate and fast cytokine dependent activation in response to different stimuli [2]. ILCs are decorated with a range of cell surface receptor that help ILCs to sense, coordinate, and respond to respective stimuli generated by the cells and tissues in which they reside [6]. In both humans and mice, ILCs are present mainly in primary and secondary lymphoid organs, but also at barrier surfaces of the body including the skin, lungs, and gut [1, 4, 5]. ILCs are grouped into five categories depending on their developmental routes, which are defined by lineage‐specific transcription factors (TFs), activating modulators, and the cytokines they produce [1, 5]. ILCs are classified into NK cells, ILC1s, ILC2s, ILC3s, and lymphoid tissue inducer (LTi) cells [1, 7, 8]. ILC1s respond to tumors and viruses, ILC2s to extracellular parasites and allergens, and ILC3s to the extracellular microbial community such as fungi and bacteria [1].
NK cells, the founding member of the ILC family, express the TFs eomesodermin (Eomes) and Tbx21 (T‐bet) for their development. NK cells have the highest cytotoxic activity among ILCs and mirroring CD8+ T cells. NK cells produce granzymes, IFN‐γ, and perforin in response to external alarmins [9, 10, 11]. To define their developmental route, ILC1s also express T‐bet, produce IFN‐γ, but have less cytotoxic activity compared to NK cells. In both humans and mice, ILC1s express CD49a and TRAIL [1, 12]. In normal physiological conditions, ILC1s are rare in blood and primary lymphoid tissues; in contrast, they reside in other organs such as the liver, gut, and salivary glands [7]. However, conventional (Lin−CD127+CD117−CRTH2−NKp46−) ILC1s and CD103+ILC1‐like cells (ieILC1‐like) are significantly increased in the PBMCs and tumor microenvironment (TME) of some hematological malignancies and solid tumor, respectively [13, 14, 15, 16]. The development of ILC2s depends on the expression of GATA‐3, and in response to external stimuli, they produce cytokines: IL‐4, IL‐5, IL‐13, and IL‐19 along with amphiregulin (an EGF). ILC2s express CTHR2 and CD161 in both humans and mice [17]. Hence, ILC2s play a role in the type 2 inflammatory response against pathologies associated with allergy and asthma [18, 19]. Generally, ILC2s are localized in mucosal tissues where they are activated by different modulators such as IL‐25 and IL‐33 [20]. The expression of RORγt and AHR is a trademark of ILC3s. After maturation, they produce different cytokines, including IL‐17 and IL‐22, along with lymphotoxin and granulocyte‐macrophage colony stimulating factor (GM‐CSF). ILC3s are further subdivided into NCR+ILC3s and NCR−ILC3s depending upon the expression of natural cytotoxicity receptors (NCRs) such as NKp44 and NKp46 [1, 5, 6, 21, 22]. ILC3s present in the intestine, interact with commensal flora enabling them to monitor infections at mucosal barrier sites and this interaction is regulated by IL‐22 [23, 24]. LTis have a functional resemblance with NCR−ILC3s and produce considerable amounts of IL‐17 and IL‐22, although their developmental path is different from ILC3s and strictly depends on the expression of RORγt. LTis are vital for the formation of LNs, which is regulated by the release of lymphotoxins. In contrast to NCRs, LTis express CCR6 and c‐Kit [1, 25, 26].
ILCs have been comprehensively studied during the last decade, and now it is well established that they are crucial regulatory components of the immune system in different organs and stress conditions, specifically in tumorigenesis and therapy [27, 28]. Different groups have investigated the roles of ILCs in tumors and TME. NK cells are considered specifically fit for tumor cell removal due to their high cytotoxicity, as revealed during multiple preclinical trials [29, 30]. Recently, our group also reviewed the potential use of NK cell checkpoints as a target for cancer immunotherapeutics. Together with a previously described concept of exhausted T cells [31], we discussed the phenomenon of NK cell exhaustion. The upregulation of inhibitory receptors on NK cells surface leads to exhausted NK cells with poor cytotoxicity and compromised immune surveillance. We detailed the latest progress in NK cell checkpoint inhibitors and their clinical usage. In addition, we proposed that NK cell immunotherapies might give a second chance to ameliorate the limitations and nonresponsiveness of T‐cell immunotherapies [32]. In continuation of this review, and with a prime interest in immunotherapeutics responsible for the modulation of ILCs reactions in the TME, it is essential to have an indepth understanding of ILCs dynamicity in tumors and the TME.
Here, in this review, we will present the current understanding of the repressive and proliferative roles of ILCs in tumor biology. In addition, we will elaborate how ILCs link themselves with their environment and their communication with other immune cells, tumor cells, and the diverse constituents of the TME. Finally, we will discuss how these interactions lead to their pro‐ or antitumor behaviors and potential checkpoints, which can be targeted to develop new immunotherapies.
Role of ILCs during tumor progression
T cells kill tumor cells through synthesizing various biological molecules such as FAS ligand, perforin, and granzymes [33]. However, in the last decades, accumulating evidences have documented that the multifaceted immunosuppressive processes ongoing in the TME lead to T‐cell dysfunction, which is implicated in the multiple tumor development [34]. Like T cells, the antitumor effects of NK cells are well‐established and extensively studied and reviewed in several publications [35, 36, 37, 38, 39]. In short, the mechanism of NK cell‐mediated antitumor effects is related to their cytolytic activity and cytokine production. Similar to NK cells, ILC1s are considered to be antitumor fighters due to their considerable cytotoxic activity and ability to produce IFN‐γ, although this cytotoxic capacity is lesser than NK cells [40]. However, emerging findings indicate protumor roles of ILC1s [12, 41, 42]. Of note, besides conventional (Lin−CD127+CD117−CRTH2− NKp46−) ILC1s that are expanded in PBMC of hematological malignancies, a unique cluster of CD103+ILC1‐like cells (ieILC1‐like) have been identified in the TME of a mouse model of solid tumor [13–16, 43, 44]. Unlike CD127+ILC1s that can be converted to ILC3s under stimulation of polarization cytokines, CD103+ ILC1s seems to be refractory to such conversion [45]. In addition, the role of ILC2s and ILC3s in cancer immunity is poorly understood [46, 47, 48]. In this part, we will discuss the anti‐ and protumor roles of these helper ILCs during tumor progression (Fig. 1).
Figure 1.
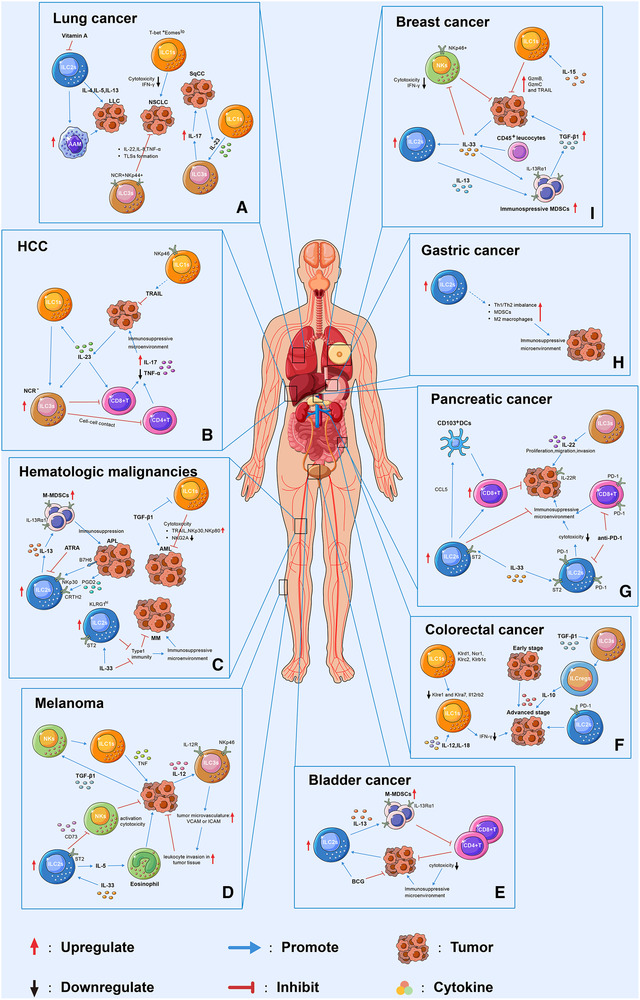
Schematic representation of the dynamic behaviour of ILCs in different tumors and their interactions with the TME. Loss of function of ILC1s was associated with the tumor progression. For instance, the decreased cytotoxicity of ILC1s were reported in NSCLC (A), and in AML(C), melanoma (D), and breast cancer (I) in the presence of IL‐15 or TGF‐β. In the presence of IL‐23, ILC1s transdifferentiated into ILC3s which exhibited tumor progression function through producing IL‐17 in SqCC (A) and inhibiting CD4+ or CD8+ T cells in HCC (B). ILC2s were found to facilitate tumor progression through interacting with MDSCs by secreting IL‐13 in APL (C), bladder (E) and breast cancers (I), and with eosinophil through IL‐5 in the presence of IL‐33 in melanoma (D). In addition, ILC2s were shown to inhibited the cytotoxicity of NKs cell through CD73 in melanoma (D). In pancreatic cancer (G), ILC2s exhibit antitumor effect through attracting DCs by secreting CCL5, which lead to increase cytotoxic CD8+T cells in tumor microenvironment. Moreover, IL‐33 was capable of regulating ILC2s function in MM (C), pancreatic cancer (G), and breast cancer (I) through binding to its receptor ST2 on ILC2s. ILC2s might be associated with immunosuppressive environment in gastric cancer. For instance, increased frequency of ILC2s was correlated with Th1/Th2 imbalance, increase MDSCs and M2 macrophages in gastric cancer tumor tissue (H). The role of ILC3s is cancer type dependent. In NSCLC (A), ILC3s inhibit tumor progression through producing IL‐22, IL‐8, TNF‐α, and tertiary lymphoid structures (TLSs) formation at tumor site. In melanoma (D), IL‐2R+ILC3s recognize IL‐12 secreted by tumor cells and upregulate adhesion molecules in tumor vasculature, such as ICAM and VCAM, which lead to accumulated leukocyte in tumor tissue. In pancreatic cancer (G), ILC3s was associated with tumor progression, migration, and invasion. Single‐cell RNA sequencing revealed that ILC1s express activating receptors, such as Klrd1, Ncr1, Klrc2, and Klrb1c, at an early stage of CRC (F), and inhibitory receptors at a late stage such as Klre1 and Klra7. PD‐1high ILC2s was found in advanced stage of CRC and might be associated with tumor progression. Moreover, upon TGF‐β stimulation, ILC3s transdifferentiated into ILCregs in CRC, and deletion of IL‐10 in ILCregs suppressed tumor development.
The antitumor roles of ILCs
In hematological malignancies
In PBMCs of chronic lymphocyte leukemia (CLL), the number of ILCs were increased at diagnosis compared to age‐matched healthy controls and progressively increased in patients with higher lymphocyte counts and shorter time to first treatment [13]. Notably, upon stimulatory signals, the production of TNF‐α in Lin−CRTH2–CD117−ILC1s was decreased in the presence of CLL tumor cells in vitro, reflecting an immunosuppressive effect of CLL cells and a distorted ILC1s function [13]. Similarly, an increased number of Lin–CD127+T‐bet+ILC1s and a concomitant remarkable reduction of NCR+ILC3 cells was reported in acute myeloid leukemia (AML) compared to healthy controls while the levels of ILC2 and NCR–ILC3 cells were comparable [14]. In this study, ILCs from AML patients were functionally impaired in the production of IFN‐γ and TNF‐α or type 2 cytokines compared to healthy controls [14]. A group of CD56+CD16−ILC1‐like cells possessing strong cytotoxicity were identified in AML [15]. The cytotoxicity of these CD56+CD16−ILC1‐like cells was impaired in AML patients at diagnosis due to decreased expression of TRAIL, NKp30, NKp80 and persistence of NKG2A, but was restored upon remission and might be modulated by TGF‐β1 (Fig. 1C) [15].
In lung cancers
ILC2s are developmentally and functionally dependent on IL‐33. In a mice xenograft model of lung cancer using TC1 and A9 cell lines, IL‐33 could elevate the frequency of tumor‐infiltrating ILC2s through inducing chemoattractants such as CCL5, CXCL10, and CXCL12. To investigate the role of ILC2s in the growth and metastasis of lung cancer, a mice model lacking ILC2s was constructed and transplanted with TC1 and A9 cell lines. The results showed that ILC2s deficiency led to a significantly increased tumor growth rate, a higher frequency of circulating tumor cells (CTCs), and metastasis to distal organs [49]. In non‐small cell lung cancer (NSCLC) patients, a tumor‐suppressing role of ILC3s has been documented [50]. ILC3s can recognize lung tumor cells and tumor‐associated fibroblasts via the NKp44 receptor and release IL‐22, TNF‐α, IL‐8, and IL‐2 once activated. In addition, ILC3s isolated from NSCLC tumor tissue showed LT‐inducing properties and might participate in the formation or maintenance of tertiary lymphoid structures (TLSs) or lymphoid aggregates at the tumor site (Fig. 1A) [50].
In breast and melanoma‐associated cancers
In the murine mammary tumor model polyoma middle T oncoprotein (PyMT), tumor growth elicits a group of innate lymphocytes expressing granzyme B, which is distinct from conventional ILC1s and cytotoxic NK (cNK) cells and further identified as TCR−NK1.1+CD49ahi cells type 1‐like ILCs (ILC1ls). In vitro and in vivo experiments showed that this new type of ILC1s induces cytotoxicity in tumor cells and inhibits tumor growth, respectively. Moreover, in PyMT mice, IL‐15 transgenic overexpression led to TCR−NK1.1+CD49ahiILC1l expansion and decreased tumor growth by expressing GzmB and GzmC as well as TRAIL, suggesting IL‐15 participated in regulating the generation and innate cytotoxicity of ILC1s (Fig. 1I) [16]. In melanoma lung metastasis, IL‐33 was shown to stimulate ILC2s to produce IL‐5 which led to eosinophil accumulation in the lungs and further inhibit the lung metastasis of B16F10 (Fig. 1D) [51]. Recently, a subset of NKp46+ ILC3 cells was identified and considered as a tumor suppressor in a melanoma animal model [52]. When stimulated by IL‐12, injected into or secreted by the tumor mass, NKp46+ ILC3 cells exhibited a tumor‐suppressive function independent of NK cells through upregulating adhesion molecules in tumor vasculature (such as ICAM and VCAM), which facilitate accumulation of leukocyte in tumor tissue (Fig. 1D) [52].
In gastrointestinal linked tumors
The liver is rich in Group 1 ILCs including intrahepatic NK cells or tissue‐resident NK cells and ILC1s [53, 54]. In hepatocellular carcinoma (HCC), the absolute number and immunosurveillance function of Group 1 ILCs were suppressed, which led to tumor progression [42]. Turchinovich et al. showed that prototypic hepatic CD3ε−NK1.1+ CD62L−ILC1s expressing NKp46 might exert tumoricidal activity when producing TRAIL (Fig. 1B) [55]. Previously, TRAIL has been shown to possess proapoptotic function and contribute to the suppression of tumor growth and metastasis in vitro and in vivo [56, 57]. In primary human pancreatic ductal adenocarcinoma (PDAC), ILC2s infiltration was observed in pancreatic cancer (PC) tissues, and this higher ILC2s frequency correlated with longer survival [58]. Notably, ILC2s expansion was accompanied by enhanced intratumoral CD8+ T‐cell cytokine production capacity, CD103+ expression in DCs, and programmed cell death protein 1 (PD‐1) upregulation. The study also revealed that recombinant IL‐33 (rIL33) treatment promotes ILC2s to produce CCL5 which recruit CD103+ DCs into tumours. In a PDAC animal model, combining rIL‐33 with αPD‐1 dramatically expanded ILC2s in tumors and enhanced tumor control, demonstrating an antitumor effect of ILC2s (Fig. 1G) [58].
The protumor roles of ILCs
In hematological malignancies
The phenotypic and functional alterations of ILC2s are associated with the progression of hematological malignancies [43, 59–61]. In acute promyelocytic leukemia (APL), CRTH2+NKp30+ILC2s were activated and released IL‐13, consequently driving the expansion and the immune suppressive function of IL‐13Rα1+ monocytic myeloid‐derived suppressor cells (M‐MDSCs) (Fig. 1C) [60]. In animal models of multiple myeloma (MM), ILC2s expressing KLRG1hi were identified in the liver and spleen of Il2rg−/− Rag2−/− mice reconstituted with BM ILC2s or IL‐33‐treated WT mice [43]. In the presence of IL‐33 combined with IL‐12 and IL‐18, KLRG1hi ILC2s were shown to promote MM progression and inhibit type 1 immunity in Rag‐deficient mice, indicating a subset of ILC2s are deficient in tumor immunosurveillance (Fig. 1C) [43]. In anaplastic large cell lymphoma (ALCL), gene expression analysis revealed that primary ALCL showed a gene expression pattern characteristic for ILC3s, implying that a minor subfraction of ALCL might originate from ILC3 [61].
In lung‐associated cancers
In peripheral blood samples of NSCLC, Eomes downregulation in NKp46+NK1.1+ group 1 ILCs was associated with increased NSCLC advancement [40]. This Eomeslo group 1 ILCs subset shares phenotypic similarities with ILC1s. Moreover, these Eomeslo ILC1s possessed reduced cytotoxicity and IFN‐γ production compared with Eomeshi cells, suggesting that the low Eomes levels in ILC1s might be associated with decreased cancer immunosurveillance (Fig. 1A) [40]. ILC1s were converted into ILC3s in the pulmonary squamous cell carcinomas (SqCC) TME [62]. The increased frequency of ILC3s and decreased percentage of ILC1s was associated with shortened patient survival, indicating that ILC3s might promote SqCC tumor progression (Fig. 1A) [62]. A potential protumor role of ILC2s was reported in an animal model with transplantation of Lewis lung cancer (LLC) cells [63]. In this study, an increase in ILC2s frequency was observed and associated with type II cytokines, such as IL‐4, IL‐5, and IL‐13, in tumor‐bearing mice fed on vitamin A‐deficient diet and led to a lower survival rate, larger tumor size, and alternatively activated macrophages (AAMs) in the lung tissues (Fig. 1A) [63].
In breast and melanoma‐related tumorigenesis
In human breast cancer, the number of ILC2s was reported to be higher in malignant, compared to benign tissue, indicating that ILC2s might be associated with tumor progression [64]. Jovanovic et al. reported that ILC2s trigger tumor progression and metastasis through producing IL‐13 in the presence of IL‐33 expressed by CD45+ leucocytes and tumor cells in the mammary carcinoma, sustaining an immunosuppressive milieu with increased TGF‐β1 producing MDSCs and reduced the number of IFN‐γ expressing NKp46+ cells in tumor‐bearing mice (Fig. 1I) [65]. Increased frequency of ILC3s was observed in the human breast cancer TME and had a positive correlation with an increased likelihood of LN metastasis [66]. In their mouse model, CCL21 was responsible for recruiting ILC3s into tumors, which induced the production of CXCL13 by the TME stromal cells. CXCL13 also promoted the ILC3‐stromal cells interaction, and production of the cancer cell motile factor RANKL. Moreover, depletion of ILC3s resulted in decreased LN metastasis [66].
Several studies indicated an immunosuppressive or impaired immunosurveillance role of ILC1s and ILC2s in melanoma [67, 68, 69, 70]. Gao et al. reported that the TGF‐β signaling pathway was capable of converting CD49a−CD49b+Eomes+NK cells into intermediate ILC1s cells with CD49a+CD49b+Eomes+intILC1s and CD49a+CD49b−Eomesint ILC1s phenotypes [67]. Their results showed that intILC1s and ILC1s were unable to control local tumor growth and metastasis, partially due to TNF produced by ILC1s (Fig. 1D) [67]. Similarly, deficiency of SMAD4, a signal transducer of the canonical TGF‐β signaling pathway, promoted NK cell transformation to an ILC1‐like gene signature and expressed an inhibitory receptor, which is associated with uncontrolled melanoma cell metastasis [68]. Regarding the role of ILC2s in melanoma leading to lung metastasis, IL‐33 dependent lung‐resident activation of ILC2s was associated with increased lung metastases and mortality [70]. Functionally, ILC2s activation was accompanied by local suppression of IFN‐γ production and cytotoxic function of lung NK cells [70]. In another study, IL‐33 injection significantly promoted the expansion of ILC2s in Rag1−/− mice bearing melanoma cells [69]. The increased frequency of ILC2s expressing immunosuppressive ectoenzyme CD73 which can inhibit the activation and cytotoxicity of NK cells, ultimately accelerated the time to tumor occurrence and the time until tumors reached their end stage (Fig. 1D) [69].
In gastrointestinal and urological cancers
In PBMCs of patients with gastric cancer, the increased frequency of ILC2s has been reported to be associated with Th2 cell‐mediated immunity, upregulation of MDSCs and M2 macrophages (Fig. 1H) [71]. Of note, ILC2s related genes, mRNAs, or cytokine molecules, such as RORα, GATA3, T1/ST2, IL‐17RB, CRTH2, IL‐33, and IL‐5, were also increased in the peripheral blood of patients with gastric cancer. These results indicated that a polarized ILC2s phenotype exists in the gastric cancer microenvironment and might play an immunosuppressive role in gastric cancer development [71]. In cholangiocarcinoma (CCA), Li et al. demonstrated that IL‐33 induced ILC2s to produce IL‐13, which promoted cholangiocyte hyperplasia [72]. Notably, activation of the IL‐33/ILC2s/IL‐13 arm was shown to induce CCA with liver metastases upon constructive activation of AKT and YAP in bile ducts [72].
Helicobacter hepaticus (H. hepaticus) enriched in the intestine from mice models is associated with the development of HBV‐infected HCC [73]. Helicobacter hepaticus activated ILCs expressing IL‐17 and IFN‐γ, exacerbates tumorigenesis in HBV‐associated HCC [73]. A subset of NCR−ILC3s has been identified and may contribute to HCC development in the presence of IL‐23 [74]. Functionally, IL‐23 induced the expansion of NCR−ILC3s and promoted the differentiation of NCR−ILC3s from ILC1s. Notably, NCR−ILC3 initiated IL‐17 production upon IL‐23 stimulation and directly inhibited TNF‐α production in CD4+T cells and the immunity of CD8+T cells by promoting lymphocyte apoptosis and limiting their proliferation (Fig. 1B) [74]. In patients with PC, IL‐22 and ILC3s were remarkably increased in the PBMCs and cancer tissues, and the receptor of IL‐22 was upregulated in PC cells [75]. Statistically, the increased frequency of ILC3s was positively correlated with tumor metastasis and vascular invasion [75]. In vitro experiments showed that IL‐22 secreted by ILC3s promoted the proliferation, invasion, and migration of PC cells (Fig. 1G) [75].
In the last decade, the role of ILCs in inflammation‐related tumorigenesis of gut cancer has been enthusiastically investigated [76, 77, 78, 79, 80, 81]. A group of IL‐17+IL‐22+ colonic innate lymphoid cells (cILCs) in bacteria‐induced colon cancer was described, which were phenotypically distinct from LTi and NK‐22 cells [78]. Depletion of these IL‐17+IL‐22+cILCs in mice with dysplastic inflammation prevented the development of invasive colon cancer, indicating a tumor‐promoting function of some specific ILCs [78]. In addition, it has been proposed that the IL‐23/IL‐23R+ILC3s/IL‐17 cascade is associated with gut tumorigenesis under a long‐term inflammation process in the gut. Moreover, ILC3s/IL‐22 shuttle has been confirmed to be associated with promoting colitis‐associated cancers [78, 81].
Recently, Wang et al. performed single‐cell RNA sequencing to analyze the role of ILC subgroups in the tumor immunity of colorectal cancer (CRC) (Fig. 1F) [41]. Their results showed that ILC1s express activating receptors, such as Klrd1, Ncr1, Klrc2, and Klrb1c, at an early stage of CRC, and inhibitory receptors, such as Klre1 and Klra7, at a late stage of CRC. Moreover, tumor‐infiltrating ILC1s in advanced CRC patients were at lower frequencies, and the production of IFN‐γ was remarkably decreased upon IL‐12 plus IL‐18 stimulation in vitro. Hence, their results indicated that tumor‐infiltrating ILC1s undergo functional conversion during CRC progression. Furthermore, tumor‐infiltrating ILC2s were shown to be heterogeneous in CRC tumors and were categorized into three groups, termed ILC2‐A (PD‐1low ILC2s), ILC2‐B, and ILC2‐C (PD‐1high ILC2s) [41]. Among these groups, ILC2‐Cs which highly express HS3ST1 and PD‐1, predominate in the late stage of CRC [41]. In vitro and in vivo experiments showed that deletion of HS3ST1 or PD‐1 in the ILC2‐C subset significantly suppressed tumor growth [41]. Previously, a regulatory subpopulation of ILCs expressing IL‐10 was identified as regulatory innate lymphoid cells (ILCregs) [82]. In CRC, ILC3s were reported to transdifferentiate into ILCregs upon TGF‐β stimulation, and deletion of IL‐10 in ILCregs suppressed tumor development, indicating a tumor‐promoting role of ILCregs during CRC progression [41].
In non‐muscle‐invasive bladder cancer (NMIBC), intravesical instillations with the Bacillus Calmette‐Guérin (BCG) were applied to prevent tumor recurrence and progression. Chevalier et al. reported that the BCG vaccine‐induced an infiltration of neutrophils, T cells, M‐MDSCs, and ILC2s in NMIBC patients (Fig. 1E) [83]. Notably, ILC2 frequency positively correlates with M‐MDSC frequency, and patients with a T cell‐to‐MDSC ratio less than one showed dramatically lower recurrence‐free survival than those with a ratio greater than one [83]. In vitro, upon BCG or tumor cell stimulation, ILC2s were able to produce IL‐13 which in turn recruited and induced a suppressive function in M‐MDSCs through the IL‐13 receptor α1. Hence, these results indicate an immunosuppressive function of the ILC2/IL‐13/MDSC axis in NMIBC patients receiving BCG therapy, which might be a reason for the failure of current BCG therapies [83].
Together with NK and ILC1 cells, both ILC2s and ILC3s can integrate antitumor responses, but the mode of ILC2s and ILC3s mainly depends on the stimuli generated by their resident tissue microenvironments. For example, the primary melanoma growth restriction is managed by the lung resident ILC2s subgroup with the release of IL‐5 [51]. Interestingly, the NCR+ILC3 population activated by IL‐12 was also reported to be involved in the control of this tumor type [39]. In contrast, the roles of ILCs were also shown to promote tumor growth in both in‐vivo and in‐vitro tumor models. Indeed, IL‐22 producing ILC3s promote tumor growth in the pancreas and gut [75, 78, 80], as do IL‐13 releasing ILC2s in breast cancer and leukemia [60, 65]. Aside from these examples, CCL21 mediated migration of RORγt+ILC3s at tumor sites can enhance LN metastasis by altering the local population of chemokines in the TME [66].
We have briefly summarized in this section the current knowledge of the involvement of ILCs in tumor progression or inhibition. Taken together, similar to T cells, which are divided into many subtypes and their dysregulation leads to tumor development, increasing evidence has proved that helper ILCs are a plastic population with multidimensional roles in tumor immunity (Table 1), but further investigations underlying the mechanisms used by these cells are still necessary. ILCs behaviors are modulated by their communication with the different residents of the TME [28]. Therefore, in the next section, we further reveal the interaction of ILCs with other immune cells (myeloid cells, DC cells, T and B cells), modulators (cytokines) of the TME, and tumor cells.
Table 1.
ILC phenotypes, functions and clinical correlations in different tumors
| ILC type | Tumor type | Phenotype | Clinical status/tumor progression | Function/mechanism | Organism | References |
|---|---|---|---|---|---|---|
| ILC1 | CLL | Lin−CRTH2–CD117− | Poor clinical outcome | Impaired function due to disturbed TNF‐α production. | Human | [13] |
| AML | Lin–CD127+ T‐bet+ | Poor clinical outcome | Impaired in the production of IFN‐γ or type 2 cytokines. | Human | [14] | |
| CD16−CD127+c‐Kit−CRTH2−CD56+ | Poor clinical outcome | Impaired cytotoxicity; the cytotoxicity of this subset is regulated by TRAIL, NKp30, NKp80, NKG2A and is KIR independent; TGF‐β1 and AhR ligands might impair their cytotoxicity in AML. | Human | [15] | ||
| NSCLC | CD45+Lin−c‐kit−CRTH2− CD127+CD56−T‐bet+Eomeslo | Associated with tumor progression both in human and mice | The cytotoxicity of this subset is positively associated with Eomes expression; impaired cytotoxicity due to less IFN‐γ production. | Human and mice | [40] | |
| Breast Cancer | TCR−NK1.1+CD49ahi | Inhibited tumor growth | Expresses high levels of GzmB, GzmC, and TRAIL and exhibit cytotoxicity towards tumor cells; constitutive IL‐15 overexpression is able to expand this subset. | Mice | [16] | |
| Melanoma | CD49a+CD49b−Eomesint | Unable to control local tumor growth and metastasis | Expresses higher level inhibitory immunological checkpoint receptors, such as CTLA‐4, CD96 and LAG‐3, and produces more myeloid growth factor GM‐CSF and TNF compared to tumor NK cells; TGF‐β drives the conversion of NK cells into ILC1s. | Mice | [67] | |
| CRC | Lin−CD45+CD127+NK1.1+NKp46+ | NA | ILC1s expresses high levels of activating receptors (Klrd1, Ncr1, Klrc2, Klrb1c), and inhibitory receptors (Klre1, Klra7) at the early and late stage of CRC, respectively, while the production of IFN‐γ in ILC1s from late stage of CRC is remarkably decreased. | Mice | [41] | |
| ILC2 | APL | Lin−CD127+CRTH2+cKit−/+ | Increases in the PBMC of APL patients; associated with increased APL mice mortality. | ILC2s are increased and hyperactivated through the interaction of CRTH2 and NKp30 with elevated tumor‐derived PGD2 and B7H6, respectively; in turn, ILC2s activates M‐MDSCs via producing IL‐13. | Human and mice | [60] |
| MM | Lin–CD127+CD25+KLRG1hi | Have no effect on the growth and dissemination of myeloma cells | IL‐33‐inducing circulating KLRG1hi ILC2s inhibits protective type 1 immune responses against MM. | Mice | [43] | |
| Metastatic lung cancer | Lin−ST2+CD127+CD90.2+ | Lacking of ILC2s associated with tumor growth and metastasis | IL‐33 induces ILC2s accumulation in tumor, which in turn mediate tumor immune‐surveillance by cooperating with DCs to promote adaptive cytolytic T‐cell responses. | Mice | [49] | |
| LLC | ICOS+ST+ | Promotes tumor growth | Enhances the type II cytokine levels; increases AMM. | Mice | [63] | |
| Melanoma | CD11b−CD11c−NK1.1+FcεRI+ CD25+CD45+CD90.2+ | Promotes tumor growth | IL‐33 promoted the expansion of ILC2s, which in turn inhibited NK activation and cytotoxicity through expressing the immunosuppressive ectoenzyme CD73. | Mice | [69] | |
| CD45+CD3–B220–NK1.1–Lin–CD127+RORγt–GATA3+ | Promotes lung metastases and mortality | IL‐33‐denpendent ILC2s activation suppresses the production of IFN‐γ and cytotoxicity of lung NK cells, which might be reliant on IL‐5‐induced lung eosinophilia. | Mice | [70] | ||
| PDAC | CD25+CD127+ST2+ GATA3+ | Correlated with longer survival in PDAC patients; ILC2s inhibits pancreas‐specific tumor growth in PDAC mice. | IL‐33 induces ILC2s activation to prime CD8+ T cells; ILC2s stimulates tissue‐specific cancer immunity by recruiting intratumoral dendritic cells and partially contribute to the efficacy of PD‐1 pathway blockade. | Human and mice | [58] | |
| Gastric Cancer | Lin−ICOS+IL‐17RB+ | Increased frequency in patients with gastric cancer. | Contributes to immunosuppressive microenvironment and closely related to the upregulation of MDSCs and M2 macrophages in gastric patients. | Human | [71] | |
| Breast Cancer | Lin−Sca‐1+ST+ | Promotes breast cancer progression | IL‐33 induces ILC2s activation, which might influence immunosuppressive functionality of MDSCs through producing IL‐13. | Mice | [65] | |
| CD45+Lin−CD56−CD127+CRTH2+cKit +/− | Enriched in human breast cancer tissue | ILC2s highly expressed PD‐1 in comparison to circulating ILCs in peripheral blood. | Human | [64] | ||
| NMIBC | Lin−CD127+CRTH2+ | Poor clinical outcome | ILC2s recruits immunosuppressive cells, including M‐MDSCs and monocytes, through producing IL‐13, especially in the presence of BCG or tumor cells. | Human | [83] | |
| CCA | Lin−ST2+ | Promote cholangiocyte hyperplasia and cholangiocarcinoma with liver metastases | Releases high levels of IL‐13 that promotes cholangiocyte hyperplasia; IL‐33/ILC2/IL‐13 circuit associated with constitutive activation of AKT and YAP in bile ducts lead to cholangiocarcinoma with liver metastases. | Mice | [72] | |
| CRC | Lin−CD45+CD127+ST2+KLRG1+PD1high | Promotes tumor growth | Expresses high levels of PD‐1 and HS3ST1; deletion PD‐1 or HS3ST1 suppressed tumor development and proliferation. | Mice | [41] | |
| ILC3 | NSCLC | Lin− CD127+CD117+NKp44+ | Accumulates in stage I/II NSCLC than in more advanced tumor stages and correlates with the density of intratumoral tertiary lymphoid structures. | Produces IL‐22, TNF‐α, IL‐8, and IL‐2, and activates endothelial cells; recognize lung tumor cells via NKp44; possesses lymphoid tissue inducing properties. | Human | [50] |
| SqCC | CD3−CD117+RORγt+ | Associated with short survival of patients with SqCC | Promotes IL17‐mediated tumor cell proliferation. | Human | [62] | |
| Melanoma | NKp46+RORγt+ | Represses subcutaneous tumor growth | Induces upregulation of adhesion molecules in the tumor vasculature and resulted in more leukocyte infiltration. | Mice | [52] | |
| Breast cancer | CD3−CD11c−B220−CD127+CD90.2+NKp46− | Correlates with lymphatic tumor cell invasion and draining LN metastasis | Stimulated the production of the CXCL13 by TME stromal cells, which in turn promoted ILC3‐stromal interactions and production of the cancer cell motile factor RANKL. | Human and mice | [66] | |
| PC | Lin–CD127+CRTH2–c‐Kit+ NKp44+ | Correlates with tumor distant metastasis and vascular invasion in PC patients | Promotes the proliferation, invasion, and migration of PC cell lines by secreting IL‐22 to activate AKT signaling | Human | [75] | |
| CRC | B220−CD3−NK1.1−CD45+CD90.2+ RORγt+ | Promotes tumor growth | Reduced IL‐22 production in ILC2s in Card9 deficient mice. | Mice | [81] | |
| Lin−CD45+CD127+RORγt− | Promotes tumor growth | ILC3s transdifferentiate into IL‐10‐producing ILCregs during CRC progression in the presence of TGF‐β. | Mice | [41] |
ALCL, anaplastic large cell lymphoma; AHR, aryl hydrocarbon receptor; AML, acute myeloid leukemia; APL, acute promyelocytic leukemia; CCA, Cholangiocarcinoma; CRC, colorectal cancer; DC, dendritic cell HCC, hepatocellular carcinoma; ILCregs, regulatory innate lymphoid cells; LLC, Lewis lung cancer; LN, lymph node; MDSCs, myeloid‐derived suppressor cells; MM, multiple myeloma; NA, not available; NK, natural killer cell; NMIBC, non‐muscle–invasive bladder cancer; NSCLC, non‐small cell lung cancer; PBMC, peripheral blood mononuclear cells; PC, pancreatic cancer; PDAC, pancreatic ductal adenocarcinoma; SqCC, Squamous Cell Carcinomas; ST2, IL‐33 receptor. TME, tumor microenvironment;
ILCs shape other immune cells in the TME
In cancer development, the immune system serves as a double‐edged sword. On one side, it inhibits tumor growth by restricting angiogenesis via the release of different cytokines, specifically by the release of IFN‐γ, antibodies, and natural cytotoxicity‐assisted phagocytosis, antibody‐dependent cell cytotoxicity, and complement activation [84, 85, 86]. Whereas, on the other side, signaling cascades leading to an immunosuppressive environment favor tumor growth [84, 87]. Here, we will discuss the potential interactions of ILCs with other immune cells and the resultant responses.
NK cells and ILC1s
Currently, there are few reports on the interaction of ILC1s and adaptive immunity [88]. In certain scenarios, the accumulation of ILC1s in the chronically inflamed gut after conversion from ILC3s in response to different cytokines, including IL‐12 or IL‐15, was reported [45, 89]. These ILC1s, in response, produce excess amounts of IFN‐γ that recruit macrophages and neutrophils to initiate tissue injuries [28, 90]. In a murine tumor model, NK cells were shown to be involved in employing classical type 1 DCs. The group revealed that the TME mediated compromised interaction among NK and DC cells via release of PGE2, leading to impaired cytotoxicity of NK cells towards tumor cells [91]. The polarization of Th1 in LNs was also reported to be facilitated by NK cells via the secretion of IFN‐γ [92, 93]. In lung carcinoma, activation of NK cells led to increased infiltration of highly activated T cells and resulted in tumor suppressive outcomes [94]. Association of LIGHT, a member of the TNF superfamily, with NK cells resulted in activation of CD8+ T cells in the TME [95].
ILC2s
In tumorigenesis, the anti‐ or protumor activity of ILC2s is attributed to the cytokines produced by ILC2s, their interaction with other immune cells, and molecules expressed by tumor cells [96]. For instance, ILC2s was significantly increase in the PBMC of gastric cancer patients and positively correlated with the mRNA expression of arginase‐1 (Arg‐1) and iNOS, indicating a correlation of increased ILC2s frequency with the upregulation of MDSCs [71]. Via the release of PGD2 and the expression of B7H6, APL blasts attract CRTH2+NKp30+ILC2s and induce their activation and IL‐13 release, which in turn drives the expansion and the immune suppressive function of IL‐13Rα+M‐MDSCs through upregulating Arg‐1 and iNOS in M‐MDSCs [60]. In addition, ILC2‐derived IL‐13 was able to suppress the proliferation of CD3+T cells in vitro as well [60]. In line with this findings, IL‐13 produced by ILC2s upon BCG stimulation in vitro recruited and induced a suppressive function in M‐MDSCs, indicating the potential involvement of an ILC2s/IL‐13/MDSC axis in BCG treatment failure in bladder cancer [83]. In melanoma, IL‐33‐induced ILC2s expressed immunosuppressive ectoenzyme CD73 that was able to facilitate tumor growth through suppressing the cytotoxicity of NK cells [69]. ILC2s infiltration was associated with low intratumoral T to Tregs ratio in both primary and metastatic tumors of lung and led to tumor immunosuppression initiated by Kras pathway and Treg cells [97]. In contrast, IL‐5 produced by ILC2s is necessary for the infiltration of eosinophil into the lung, preventing tumor metastasis [51]. In addition, it has been proposed that ILC2s might be capable of facilitating antigen presentation and the recognition of tumor cells by T cells [49, 96]. In PDAC, ILC2s expansion was accompanied by enhanced intratumoral CD8+ T‐cell cytokine capacity and CD103+DCs [58]. Saranchova et al. revealed that tumor‐infiltrating ILC2s cooperated with DCs to promote adaptive cytolytic T‐cell responses, limiting tumor metastasis [49].
ILC3s
Regarding ILC3s and adaptive immunity, in a mouse model of bacteria induced CRC, a subset of Lin–IL‐22+ILCs accumulated during cancer development and induced myeloid cell recruitment through the production of IL‐22 [78]. In another mouse model of melanoma highly expressing CCL21, CD3–CD4+RORgt+LTi cells were found to be associated with CD11b+CD11c–F4/80–Gr1highMDSCs. Nevertheless, whether this phenomenon was required for the CCL21‐enhanced tumor growth remains unclear [98]. Liu et al. showed that NCR−ILC3s highly expressing MHC I significantly inhibited the proliferation of CD8+T cells and promoted their apoptosis in in vitro coculture experiments [74]. Whereas, tumor‐infiltrating NKp44+NCR+ILC3s were proved to be a unique and innate source of IL‐2 which is a crucial growth factor for the clonal expansion of tumor‐specific lymphocytes [50]. In addition, a regulatory ILC3‐like group in humans was shown to restrict the activity of a tumor‐associated subgroup of T cells through expressing NKp46 [99]. In another in vitro experiment, a mutualism was reported between B cells and ILC3s. The production of IL‐15 by B cells was induced by tonsillar and circulating ILC3s in humans which promoted the expression of CD40L on ILC3s surface. This particular subset of ILC3s was found to be responsible for prolonged survival, proliferation, and the cytokine production ability of B cells [100]. Moreover, ILC3s are capable of receiving an activating signal by DCs through the activating receptor DNAX accessory molecule 1 (DNAM‐1) and in turn act as early and effective activator of DCs. Although the interaction between ILC3s and DCs in tumor immunity remains unclear, the results highlight an advance in understanding of ILC3‐activating signals as well as their interactions with other players of innate immunity [101].
In summary, ILCs modulate adaptive immune cells in the TME, particularly T‐cell responses through the release of cytokines or direct interactions with T cells or accessory cells [60, 61, 74, 95, 102]. ILCs also monitor tumor infiltration and downstream responses of B cells, MDSCs, DCs, and eosinophils [49, 51, 60, 98, 100].
Plasticity of ILCs and tumor immunotherapy
Plasticity of ILCs in the TME
In inflammatory conditions and the TME, the phenotypes and functional capacities of ILC subsets can change in response to external signals (Fig. 2) [48, 102–105]. For example, Bernink et al. demonstrated that ILC1s differentiate to ILC3s upon release of IL‐2, IL‐23, and IL‐1β, and dependent on the TF RORγt in inflamed intestinal tissue [106]. CD14+DCs were shown to promote polarization of ILC3s to ILC1s in vitro [106]. In an animal model of HCC, ILC1s were induced to differentiate into ILC3s phenotypes by expressing IL‐17 and RORγt in the presence of IL‐23 [74]. Similarly, SqCC tumor cells were shown to produce IL‐23 promoting IL17‐mediated tumor growth by converting ILC1s into ILC3s, thereby shortening patient survival [62]. In addition, ILC3s may convert into NK cells in the presence of IL‐15 and IL‐12. Intriguingly, the TF AHR was shown to prevent the conversion of ILC3s to NK cells [107, 108]. Aiolos and Ikaros are two TFs important for the regulation of lymphocyte function [103]. Selective degradation of Aiolos and Ikaros by lenalidomide inhibited ILC1s and NK cells differentiation characterized by suppressed expression of ILC1s‐ and NK cell‐related transcripts (LEF1, PRF1, GRZB, CD244, NCR3, and IRF8) and increased expression of the ILC3s‐related TF Helios and ILC3s transcripts (TNFSF13B, IL22, NRP1, and RORC) [103].
Figure 2.
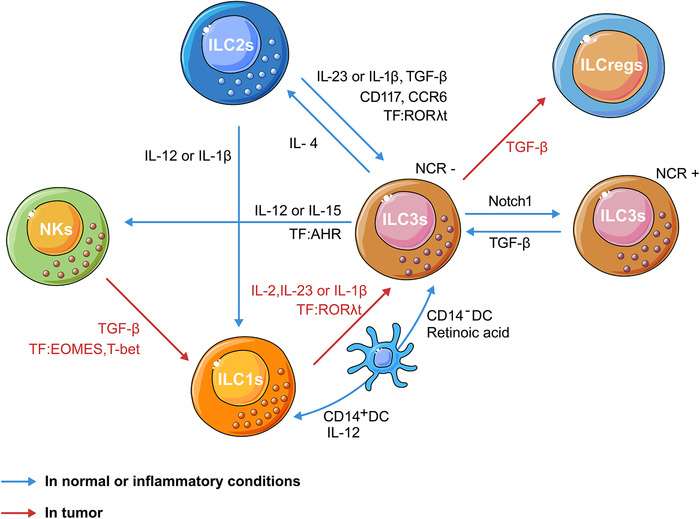
Schematic illustration of the plasticity of ILCs in inflammatory or tumor conditions. In inflammatory or tumor environments rich in TGF‐β, NK cells convert to an ILC1‐like phenotype through decreasing Eomes expression. In inflamed intestinal tissue, ILC1s differentiate to ILC3s upon release of IL‐2, IL‐23, and IL‐1β and dependent on RORγt [106]. In line with this finding, ILC1s were induced to differentiate into the ILC3s phenotype in the presence of IL‐23 in tumors [62, 74]. Moreover, TGF‐β has been implicated in the conversion of ILC3s into ILCregs in CRC [41]. In vitro, CD14+DCs promoted the polarization of ILC3s to ILC1s [106]. ILC3s may convert into NKs in the presence of IL‐15 and IL‐12; conversely, the transcription factor AHR prevented the conversion of ILC3s to NK cells [107, 108]. Notch signaling governs the conversion of NCR−ILC3s into NCR+ ILC3s by sustaining T‐bet expression; conversely, TGF‐β could convert NKp46+ILC3s into NKp46−ILC3s by opposing Notch signaling [105]. IL‐1β and IL‐12 are involved in the transdifferentiation of ILC2s into ILC1s [105]. ILC2s stimulated with IL‐1β, IL‐23, and TGF‐β acquired the phenotype of ILC3s; conversely, ILC3s can convert back to ILC2s in the presence of IL‐4 [48, 114, 115]. AHR, aryl hydrocarbon receptor; CCR, C‐C‐chemokine receptor; DC, dendritic cell; EOMES, eomesodermin; IL, interleukin; ILC, innate lymphoid cell; ILCregs, regulatory innate lymphoid cells; NCR, natural cytotoxicity receptor; NK, natural killer cell; TFs, transcription factors.
TGF‐β is known to be a potent immunosuppressive cytokine of the TME [109]. Several studies have demonstrated that NK cells could convert to an ILC1‐like phenotype in the presence of TGF‐β [67, 68, 110]. Previously, Pikovskaya et al. showed that ectopic expression of TF Eomes induces cNK‐like properties in ILC1s in mice [111]. It has also been reported that Notch signaling governs the conversion of NCR− ILC3 into NCR+ ILC3 by sustaining T‐bet expression. Conversely, TGF‐β could convert NKp46+ ILC3 into NKp46−ILC3 by opposing Notch signaling [105]. Moreover, TGF‐β has been implicated in the conversion of ILC3s into ILCregs in CRC [41]. The conversion of ILC2s to ILC1s is well documented in inflammatory conditions [17, 112, 113]. It has been proposed that IL‐1β and IL‐12 are two major cytokines that determine the transdifferentiation of ILC2s into ILC1s [105]. In addition, human ILC2s stimulated with IL‐1β, IL‐23, and TGF‐β acquired phenotype of ILC3s. Conversely, ILC3s can convert back to ILC2s in the presence of IL‐4 [48, 114, 115]. Collectively, these reports demonstrate broad‐range plasticity between nearly all ILC types, and the respective cytokines of the inflammation milieu and TME that serve as key regulators in governing this plasticity. Keeping in view the emerging concept of immunotherapy, it is imperative to investigate the impact of ILC plasticity when considering ILCs as a target for cancer immunotherapy.
ILCs and cancer immunotherapy
Strategies for improving cancer immunotherapy through targeting NK cells have been well reviewed and reported elsewhere, including modification of the expression of inhibitory and activating NK cell receptors, restoring NK cell antitumor activity, and transplantation of engineered NK cells with improved cytotoxic capacities [32, 35, 37, 39, 116]. However, approved or experimental cancer therapies targeting other helper ILCs are rarely reported [117]. Nevertheless, ILCs express many surface markers or immune checkpoints and are stimulated by different cytokines in common with T cells, NK cells, and other immune cells [47, 118]. Therefore, cytokine‐based immunotherapies and immune checkpoint inhibitors probably impact ILC responses in tumorigenesis.
Candidate cytokines that might be used for immunotherapies targeting ILCs include TGF‐β, IL‐33, IL‐23, and IL‐2. TGF‐β has been implicated in modulating the plasticity of ILCs, consequently affecting the status of tumor immunity [68, 109, 110]. In addition, the IL‐33/ILC2s cascade has been shown to play a pro‐ or antitumor role depending on the tumor type [49, 69, 72]. Moreover, IL‐23 secreted by HCC has been reported to induce ILC3s expansion, leading to an immunosuppressive environment and tumor progression [74]. However, the complexity of the TME may make it challenging to target cytokines for immunotherapies [47]. For instance, IL‐2‐based immunotherapy has been used in curing melanoma with success in a small fraction of patients due to unfavorable toxicity profiles of different immune cells [119]. As an activation marker, CD25, the α‐subunit of the IL‐2 receptor, was highly expressed by ILC2s and ILC3s in humans [120]. ILC2s and ILC3s were shown to either promote or inhibit melanoma progression depending on the tumor context [117]. Therefore, it would be of interest to further investigate the functions of these ILCs during the process of IL‐2‐based immunotherapies for melanoma.
The upregulation of immune checkpoints has been described as one of the hallmarks of T‐cell exhaustion and drugs targeting these immune checkpoints have been developed and used in clinical treatment to improve T‐cells antitumor function [34]. Similar to T cells, ILCs have been shown to overexpress T cells‐ related inhibitory receptors, including PD‐1, T‐cell immunoglobulin and mucin domain 3 (TIM‐3), lymphocyte‐activation gene 3 (LAG‐3), CTLA‐4, and T‐cell immunoglobulin and ITIM domain (TIGIT), which could be potential targets for immunotherapy through modulating antitumor function of ILCs (Table 2). PD‐1 has been identified as a marker of ILC committed progenitors, which can generate ILC1s, ILC2s, ILC3s, and a small fraction of circulating NK cells [121, 122]. Taylor et al. demonstrated that PD‐1 is a negative regulator of KLRG1+ILC2s in both mice and humans with helminth infections [123]. Their study showed that PD‐1 selectively inhibits the proliferation of KLRG1+ILC2s via modulating STAT5 function [123]. PD‐1‐ and ILC2s‐mediated tumor immunity has been reported in PC [58]. In an animal model of PDAC, combining rIL‐33 with αPD‐1 (a blocking antibody of PD‐1), dramatically promoted ILC2 expansion in tumors and enhanced tumor control [58]. These studies indicated that targeting PD‐1 could influence type 2 responses in cancer patients [124]. Moreover, PD‐1 is reported to be expressed by ILC3s and regulates the production of cytokines, including IL‐22, IL‐8, and TNF‐α, in ILC3s induced by IL‐23 in human decidua [125]. The results indicate that the expression or function of PD‐1 in ILC3s may play a role in maintaining immune tolerance during pregnancy [125]. However, the functional role of PD‐1 expression on ILC3s in tumor immunosurveillance modulation remains unclear and merits further investigation. TIM‐3 and LAG‐3 are also reported to be expressed on ILC1s but not on ILC2s and ILC3s; in addition, TIGIT is expressed on both ILC1s and ILC3s [126]. Blocking antibodies against TIM3, LAG3 and TIGIT have been in used in clinical trials and described elsewhere [47, 117]. In addition, KLRG1 has been detected on ILC1s, ILC2s, and ILC3s in breast and gastrointestinal cancer tissues [64]. Recently, NKG2A has been found to be highly expressed by ILC1s in AML patients [15]. Upon engagement with HLA‐E on leukemic targets, ILC1s expressing high levels of NKG2A are characterized with decreased degranulation and impaired cytotoxic functions [15]. CTLA‐4, another checkpoint molecule, has been reported to be expressed by ILC1s and ILC2s [41]. ILC1s with higher expression of CTLA‐4 resulted in impaired ability to secrete IFN‐γ but not TNF‐α. The excess of TNF‐α and VEGF secreted by ILC1s may be associated with protumoral and proangiogenic phenomena [67]. Hence, the use of therapeutic antibodies against these immune checkpoints expressed by ILCs would be of interest for developing novel immunotherapy methods against cancer.
Table 2.
Potential cytokines and immune checkpoints in cancer immunotherapy of targeting ILCs
| Cell type | Function | Cytokines, inhibitors, or mAbs available | Clinical status | |
|---|---|---|---|---|
| Immune checkpoints | ||||
| PD‐1 | ILC2s, ILC3s | A negative regulator of ILC2s; expressed on ILC3s [123, 125] | Pembrolizumab, nivolumab, etc. | Blocking antibody against PD‐1 approved by FDA in oncology. |
| PD‐L1 | ILC2s | Expressed on ILC2s [127] | Atezolizumab, avelumab, etc. | Blocking antibody against PD‐L1 approved by FDA in oncology. |
| TIM‐3 | ILC3s | Expressed on human decidual ILC3s [125] | Sym023, TSR‐022 | Blocking antibody against TIM‐3 in oncology are in clinical trials such as in adult primary liver cancer, NSCLC and Melanoma, etc. |
| LAG‐3 | ILC1s | Involved in the conversion of NKs to ILC1s in tumors [67] | Sym022, BMS 986016 | Blocking antibody against LAG‐3 in oncology are in clinical trials, such as in glioblastoma and melanoma. |
| TIGIT | ILC1s, ILC3s | Involved in the conversion of NKs to ILC1s in tumors [126] | BGB‐A1217, IBI939, COM902 | Blocking antibody against TIGIT in oncology are in clinical trials. |
| KLRG1 | ILC1s, ILC2s, ILC3s | Identified in ILC2s in lung and CRC and an activation marker on ILC1s, ILC2s, and ILC3s [43, 64] | ABC008 | Blocking antibody against KLRG1 is in clinical trial of inclusion body myositis. |
| NKG2A | ILC1s | Decreased degranulation and impairment cytotoxic functions [15] | Monalizumab | Blocking antibody against NKG2A in oncology is in clinical trials. |
| CTLA‐4 | ILC1s, ILC2s | Impaired in IFN‐γ secretion [41] | AK104 | Blocking antibody against CTLA‐4 in oncology is in clinical trials. |
| Cytokines | ||||
| IL‐2 | ILC2s, ILC3s | The α‐subunit of the IL‐2 receptor, CD25, is expressed by ILC2s and ILC3s [120] | Recombinant human IL‐2 | IL‐2 treatment approved by FDA in metastatic melanoma and renal cell carcinoma. |
| IL‐12 | ILC2s, ILC3s | Lead to immunodeficiency of ILC2s and tumor‐suppressive function of ILC3s [43, 52]; | Ustekinumab | Tested in clinical trials for inflammation diseases. |
| IL‐15 | ILC1s, ILC3s | Lead to ILC1s expansion and the conversion of ILC3s to NK cells [16, 104] | Recombinant human IL‐15 | Tested in clinical trials for cancer, such as refractory metastatic malignant melanoma and metastatic renal cell cancer, recurrent non‐small cell lung carcinoma, etc. |
| IL‐23 | ILC3s | Induces the expansion of NCR‐ILC3s [74] | Ustekinumab | Tested in clinical trials for inflammatory bowel diseases. |
| IL‐33 | ILC2s | Lead to activation of ILC2s and elevate the frequency of tumor‐infiltrating ILC2s in lung cancer; promote MM progression [43, 49, 58, 72] | IL‐33, vaccination against IL‐33, soluble ST2, Itepekimab etc. | Blocking antibodies against IL‐33/ST2 axis in allergic inflammatory diseases are in clinical trials. |
| TGF‐β | ILC1s, ILC2s, ILC3s | Convert NK cells to ILC1s, convert ILC3s into ILCregs, ILC2s acquire a phenotype of ILC3s [41, 67, 115] | Vactosertib, galunisertib, etc. | Inhibitors of the serine/threonine kinase TGF‐β receptor type 1 (TGFBR1) are currently in clinical trials. |
CTLA‐4, cytotoxic T‐lymphocyte antigen 4; KLRG1, killer cell lectin‐like receptor G1; LAG‐3, lymphocyte‐activation gene 3; PD‐1, Programmed cell death protein 1; PDL‐1, programmed cell death ligand 1; TIGIT, T‐cell immunoglobulin and ITIM domain; TIM‐3, T‐cell immunoglobulin and mucin domain 3.
Conclusions
The impact of ILCs on the immune microenvironment during tumor development, growth, and metastasis has been investigated in recent years. It has become clearer that different ILC subsets display either pro‐ or antitumor roles. These roles depend on the extensive communication between ILCs and the TME, including direct ILC‐tumor cell interactions, ILC‐mediated modulation of the vasculature, the stroma, and the ECM, innate‐adaptive lymphocyte crosstalk, and transdifferentiation of ILC members among themselves. The specific antitumor effects and mechanisms of different ILCs need to be confirmed by further indepth experiments in the future. Currently, the main problems that need to be addressed are that ILCs definition requires many combined markers and these markers are not exclusively expressed in ILCs but also in other cells. There is no decisive marker that specifically defines individual ILC subsets in humans and mice [105]. In the future, a key or exclusive indicator that defines each group of ILCs will help the development of ILC‐related immunotherapy. The use of novel techniques, such as single‐cell RNA sequencing and fluorescence live‐cell imaging, will empower us to specifically define individual ILC subsets in tumor tissues. Due to the characteristics of ILCs integrated in tissues, the analysis of the spatial transcriptome and integration analysis of the combination of the spatial transcriptome and single‐cell transcriptome can better describe the spatial location and role of ILCs in tumor tissues.
The modulation of the phenotypes and functions of ILCs in the TME is an emerging and promising strategy for cancer immunotherapies. ILCs are covered with surface makers or immune checkpoints common with those of T cells, indicating that immunotherapies targeting T cells may affect the phenotypes and functions of ILCs, which may lead to treatment failure or cancer regression and vice versa. Indeed, the use of specific genetic manipulations selectively targeting specific ILC subgroups without disturbing the adaptive immunity will support the idea to explore a clearer picture of their roles in tumorigenesis. Therefore, further investigations aided with improved experimental tools followed by clinical studies are obligatory to specifically address the value of ILCs in tumor immunity and therapy. Ultimately, the success to failure ratio of these efforts will deepen our understanding of ILC plasticity and dynamics in the TME. Although immunotherapies targeting helper‐like ILCs have just emerged and are being investigated in several trials on nontumor diseases, we anticipate that future studies will further drive new innovative treatments.
Conflict of interest
The authors declare no financial or commercial conflict of interest.
Abbreviations
- AAMs
alternatively activated macrophages
- ALCL
anaplastic large cell lymphoma
- AML
acute myeloid leukemia
- APL
acute promyelocytic leukemia
- Arg‐1
arginase‐1
- ATRA
all‐trans retinoic acid
- BCG
Bacillus Calmette‐Guérin
- CCA
cholangiocarcinoma
- CCR
C‐C‐chemokine receptor
- CLL
chronic lymphocyte leukemia
- CTCs
circulating tumor cells
- CTLA‐4
cytotoxic T‐lymphocyte antigen 4
- cILCs
colonic innate lymphoid cells
- CRC
colorectal cancer
- cNK
cytotoxic NK
- Eomes
eomesodermin
- GM‐CSF
granulocyte‐macrophage colony stimulating factor
- HCC
hepatocellular carcinoma
- ILCs
Innate lymphoid cells
- ILCregs
regulatory innate lymphoid cells
- iNOS
inducible nitric oxide synthase
- KLRG1
killer cell lectin‐like receptor G1
- LAG‐3
lymphocyte‐activation gene 3
- LLC
Lewis lung cancer
- LTis
lymphoid tissue inducer
- M‐MDSCs
monocytic myeloid‐derived suppressor cells
- MM
multiple myeloma
- NCR
natural cytotoxicity receptor
- NK
natural killer
- NMIBC
non‐muscle‐invasive bladder cancer
- NSCLC
non‐small cell lung cancer
- PBMC
peripheral blood mononuclear cells
- PC
pancreatic cancer
- PD‐1
Programmed cell death protein 1
- PDAC
pancreatic ductal adenocarcinoma
- PyMT
polyoma middle T oncoprotein
- RAs
retinoic acids
- ST2
IL‐33 receptor
- SqCC
squamous cell carcinomas
- TIGIT
T‐cell immunoglobulin and ITIM domain
- TIM‐3
T‐cell immunoglobulin and mucin domain 3
- TLSs
tertiary lymphoid structures
- TFs
transcription factors
- TME
tumor microenvironment
Acknowledgments
This work was supported by the National Natural Science Foundation of China (#8202290021, #91942310), Anhui Provincial Natural Science Foundation (#2008085J35), the Fundamental Research Funds for the Central Universities (#WK9110000055, WK9110000131).
References
- 1.Vivier, E., Artis, D., Colonna, M., Diefenbach, A., Di Santo, J. P., Eberl, G., Koyasu, S. et al., Innate lymphoid cells: 10 years on. Cell 2018. 174: 14–1066. [DOI] [PubMed] [Google Scholar]
- 2.Cherrier, M., Ramachandran, G. and Golub, R., The interplay between innate lymphoid cells and T cells. Mucosal. Immunol. 2020. 13: 732–742. [DOI] [PubMed] [Google Scholar]
- 3.Eberl, G., Colonna, M., Di Santo, J. P. and Mckenzie, A. N. J., Innate lymphoid cells. Innate lymphoid cells: a new paradigm in immunology. Science 2015. 348: aaa6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vivier, E., Raulet, D. H., Moretta, A., Caligiuri, M. A., Zitvogel, L., Lanier, L. L., Yokoyama, W. M. et al., Innate or adaptive immunity? The example of natural killer cells. Science 2011. 331: 44–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spits, H., Artis, D., Colonna, M., Diefenbach, A., Di Santo, J. P., Eberl, G., Koyasu, S. et al., Innate lymphoid cells–a proposal for uniform nomenclature. Nat. Rev. Immunol. 2013. 13: 145–149. [DOI] [PubMed] [Google Scholar]
- 6.Klose, C. S. N. and Artis, D., Innate lymphoid cells control signaling circuits to regulate tissue‐specific immunity. Cell Res. 2020. 30: 475–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuchs, A., ILC1s in tissue inflammation and infection. Front. Immunol., 2016. 7: 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peng, H., Jiang, X., Chen, Y., Sojka, D. K., Wei, H., Gao, X., Sun, R. et al., Liver‐resident NK cells confer adaptive immunity in skin‐contact inflammation. J. Clin. Invest. 2013. 123: 1444–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herberman, R. B., Nunn, M. E. and Lavrin, D. H., Natural cytotoxic reactivity of mouse lymphoid cells against syngeneic acid allogeneic tumors. I. Distribution of reactivity and specificity. Int. J. Cancer 1975. 16: 216–229. [DOI] [PubMed] [Google Scholar]
- 10.Gordon, S. M., Chaix, J., Rupp, L. J., Wu, J., Madera, S., Sun, J. C., Lindsten, T., et al., The transcription factors T‐bet and Eomes control key checkpoints of natural killer cell maturation. Immunity 2012. 36: 55–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abel, A. M., Yang, C., Thakar, M. S. and Malarkannan, S., Natural killer cells: development, maturation, and clinical utilization. Front. Immunol. 2018. 9: 1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paijens, S. T., Vledder, A., de Bruyn, M. and Nijman, H. W., Tumor‐infiltrating lymphocytes in the immunotherapy era. Cell Mol. Immunol. 2020. 18: 842‐859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Weerdt, I., Van Hoeven, V., Munneke, J. M, Endstra, S., Hofland, T., Hazenberg, M. D. and Kater, A. P., Innate lymphoid cells are expanded and functionally altered in chronic lymphocytic leukemia. Haematologica 2016. 101: e461–e464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trabanelli, S., Curti, A., Lecciso, M., Salome, B., Riether, C., Ochsenbein, A., Romero, P. et al., CD127+ innate lymphoid cells are dysregulated in treatment naive acute myeloid leukemia patients at diagnosis. Haematologica 2015. 100: e257–e260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salomé, B., Gomez‐Cadena, A., Loyon, R., Suffiotti, M., Salvestrini, V., Wyss, T., Vanoni, G. et al., CD56 as a marker of an ILC1‐like population with NK cell properties that is functionally impaired in AML. Blood Adv. 2019. 3: 3674–3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dadi, S., Chhangawala, S., Whitlock, B. M., Franklin, R. A., Luo, C. T., Oh, S. A., Toure, A. et al., Cancer immunosurveillance by tissue‐resident innate lymphoid cells and innate‐like T cells. Cell 2016. 164: 365–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bal, S. M., Bernink, J. H., Nagasawa, M., Groot, J., Shikhagaie, M. M., Golebski, K., Van Drunen, C. M. et al., IL‐1beta, IL‐4 and IL‐12 control the fate of group 2 innate lymphoid cells in human airway inflammation in the lungs. Nat. Immunol. 2016. 17: 636–645. [DOI] [PubMed] [Google Scholar]
- 18.Scanlon, S. T. and McKenzie, A. N., Type 2 innate lymphoid cells: new players in asthma and allergy. Curr. Opin. Immunol. 2012. 24: 707–712. [DOI] [PubMed] [Google Scholar]
- 19.Yagi, R., Zhong, C., Northrup, D. L., Yu, F., Bouladoux, N., Spencer, S., Hu, G. et al., The transcription factor GATA3 is critical for the development of all IL‐7Ralpha‐expressing innate lymphoid cells. Immunity 2014. 40: 378–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bouchery, T., Gros, G. Le. and Harris, N., ILC2s‐trailblazers in the host response against intestinal helminths. Front. Immunol. 2019. 10: 623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Withers, D. R. and Hepworth, M. R., Group 3 innate lymphoid cells: communications hubs of the intestinal immune system. Front. Immunol. 2017. 8: 1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Becker, M., Gnirck, A. C. and Turner, J. E., Innate lymphoid cells in renal inflammation. Front. Immunol. 2020. 11: 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Melo‐Gonzalez, F. and Hepworth, M. R., Functional and phenotypic heterogeneity of group 3 innate lymphoid cells. Immunology 2017. 150: 265–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abou‐Samra, E., Hickey, Z., Aguilar, O. A., Scur, M., Mahmoud, A. B., Pyatibrat, S., Tu, M. M. et al., NKR‐P1B expression in gut‐associated innate lymphoid cells is required for the control of gastrointestinal tract infections. Cell Mol. Immunol. 2019. 16: 868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Togni, P., Goellner, J., Ruddle, N., Streeter, P., Fick, A., Mariathasan, S., Smith, S. et al., Abnormal development of peripheral lymphoid organs in mice deficient in lymphotoxin. Science 1994. 264: 703–707. [DOI] [PubMed] [Google Scholar]
- 26.van de Pavert, S. A. and Vivier, E., Differentiation and function of group 3 innate lymphoid cells, from embryo to adult. Int. Immunol. 2016. 28: 35–42. [DOI] [PubMed] [Google Scholar]
- 27.Klose, C. S. and Artis, D., Innate lymphoid cells as regulators of immunity, inflammation and tissue homeostasis. Nat. Immunol. 2016. 17: 765–774. [DOI] [PubMed] [Google Scholar]
- 28.Ducimetiere, L., Vermeer, M. and Tugues, S., The interplay between innate lymphoid cells and the tumor microenvironment. Front. Immunol. 2019. 10: 2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohs, I., Van Den Broek, M., Nussbaum, K., Münz, C., Arnold, S. J., Quezada, S. A., Tugues, S. et al., Corrigendum: interleukin‐12 bypasses common gamma‐chain signalling in emergency natural killer cell lymphopoiesis. Nat. Commun. 2017. 8: 15185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krasnova, Y., Putz, E. M. and Smyth, M. J., Souza‐Fonseca‐Guimaraes, F., Bench to bedside: NK cells and control of metastasis. Clin. Immunol. 2017. 177: 50–59. [DOI] [PubMed] [Google Scholar]
- 31.Sun, C., Lan, P., Han, Q., Huang, M., Zhang, Z., Xu, G., Song, J. et al., Oncofetal gene SALL4 reactivation by hepatitis B virus counteracts miR‐200c in PD‐L1‐induced T cell exhaustion. Nat. Commun. 2018. 9: 1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun, H. and Sun, C., The rise of NK cell checkpoints as promising therapeutic targets in cancer immunotherapy. Front. Immunol. 2019. 10: 2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Golstein, P. and Griffiths, G. M., An early history of T cell‐mediated cytotoxicity. Nat. Rev. Immunol. 2018. 18: 527–535. [DOI] [PubMed] [Google Scholar]
- 34.Thommen, D. S. and Schumacher, T. N., T cell dysfunction in cancer. Cancer Cell 2018. 33: 547–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chiossone, L., Dumas, P. ‐ Y.Vienne, M. and Vivier, E., Natural killer cells and other innate lymphoid cells in cancer. Nat. Rev. Immunol. 2018. 18: 671–688. [DOI] [PubMed] [Google Scholar]
- 36.Chen, Y. and Tian, Z., Innate lymphocytes: pathogenesis and therapeutic targets of liver diseases and cancer. Cell Mol. Immunol. 2021. 18: 57–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruf, B., Heinrich, B. and Greten, T. F., Immunobiology and immunotherapy of HCC: spotlight on innate and innate‐like immune cells. Cell Mol. Immunol. 2021. 18: 112–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun, H., Liu, L., Huang, Q., Liu, H., Huang, M., Wang, J., Wen, H. et al., Accumulation of tumor‐infiltrating CD49a(+) NK cells correlates with poor prognosis for human hepatocellular carcinoma. Cancer Immunol. Res. 2019. 7: 1535–1546. [DOI] [PubMed] [Google Scholar]
- 39.Wu, S. ‐ Y., Fu, T., Jiang, Y. ‐ Z. and Shao, Z. ‐ M., Natural killer cells in cancer biology and therapy. Mol. Cancer 2020. 19: 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Verma, R., Er, J. Z., Pu, R. W., Sheik Mohamed, J., Soo, R. A., Muthiah, H. M., Tam, J. K. C. et al., Eomes expression defines group 1 innate lymphoid cells during metastasis in human and mouse. Front. Immunol. 2020. 11: 1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, S., Qu, Y., Xia, P., Chen, Y., Zhu, X., Zhang, J., Wang, G. et al., Transdifferentiation of tumor infiltrating innate lymphoid cells during progression of colorectal cancer. Cell Res. 2020. 30: 610–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun, C., Sun, H., Zhang, C. and Tian, Z., NK cell receptor imbalance and NK cell dysfunction in HBV infection and hepatocellular carcinoma. Cell Mol. Immunol. 2015. 12: 292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guillerey, C., Stannard, K., Chen, J., Krumeich, S., Miles, K., Nakamura, K., Smith, J. et al., Systemic administration of IL‐33 induces a population of circulating KLRG1(hi) type 2 innate lymphoid cells and inhibits type 1 innate immunity against multiple myeloma. Immunol. Cell Biol. 2021. 99: 65–83. [DOI] [PubMed] [Google Scholar]
- 44.Kini Bailur, J., Mehta, S., Zhang, L., Neparidze, N., Parker, T., Bar, N., Anderson, T. et al., Changes in bone marrow innate lymphoid cell subsets in monoclonal gammopathy: target for IMiD therapy. Blood Adv. 2017. 1: 2343–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bernink, J. H., Peters, C. P., Munneke, M., Te Velde, A. A., Meijer, S. L., Weijer, K., Hreggvidsdottir, H. S. et al., Human type 1 innate lymphoid cells accumulate in inflamed mucosal tissues. Nat. Immunol. 2013. 14: 221–229. [DOI] [PubMed] [Google Scholar]
- 46.Trabanelli, S., Chevalier, M. F., Derré, L. and Jandus, C., The pro‐ and anti‐tumor role of ILC2s. Semin. Immunol. 2019. 41: 101276. [DOI] [PubMed] [Google Scholar]
- 47.Crinier, A., Vivier, E. and Blery, M., Helper‐like innate lymphoid cells and cancer immunotherapy. Semin. Immunol. 2019. 41: 101274. [DOI] [PubMed] [Google Scholar]
- 48.Guia, S. and Narni‐Mancinelli, E., Helper‐like innate lymphoid cells in humans and mice. Trends Immunol. 2020. 41: 436–452. [DOI] [PubMed] [Google Scholar]
- 49.Saranchova, I., Han, J., Zaman, R., Arora, H., Huang, H., Fenninger, F., Choi, K. B. et al., Type 2 innate lymphocytes actuate immunity against tumours and limit cancer metastasis. Sci. Rep. 2018. 8: 2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carrega, P., Loiacono, F., Di Carlo, E., Scaramuccia, A., Mora, M., Conte, R., Benelli, R. et al., NCR(+)ILC3 concentrate in human lung cancer and associate with intratumoral lymphoid structures. Nat. Commun. 2015. 6: 8280. [DOI] [PubMed] [Google Scholar]
- 51.Ikutani, M., Yanagibashi, T., Ogasawara, M., Tsuneyama, K., Yamamoto, S., Hattori, Y., Kouro, T. et al., Identification of innate IL‐5‐producing cells and their role in lung eosinophil regulation and antitumor immunity. J. Immunol. 2012. 188: 703–713. [DOI] [PubMed] [Google Scholar]
- 52.Eisenring, M., Vom Berg, J., Kristiansen, G., Saller, E. and Becher, B., IL‐12 initiates tumor rejection via lymphoid tissue‐inducer cells bearing the natural cytotoxicity receptor NKp46. Nat. Immunol. 2010. 11: 1030–1038. [DOI] [PubMed] [Google Scholar]
- 53.Spits, H., Bernink, J. H. and Lanier, L., NK cells and type 1 innate lymphoid cells: partners in host defense. Nat. Immunol. 2016. 17: 758–764. [DOI] [PubMed] [Google Scholar]
- 54.Yang, S., Tian, Z., Wu, Y., Van Velkinburgh, J. C. and Ni, B., Pivotal roles of ILCs in hepatic diseases. Int. Rev. Immunol. 2015. 34: 509–522. [DOI] [PubMed] [Google Scholar]
- 55.Turchinovich, G., Ganter, S., Bärenwaldt, A. and Finke, D., NKp46 calibrates tumoricidal potential of type 1 innate lymphocytes by regulating TRAIL expression. J. Immunol. 2018. 200: 3762–3768. [DOI] [PubMed] [Google Scholar]
- 56.Smyth, M. J., Cretney, E., Takeda, K., Wiltrout, R. H., Sedger, L. M., Kayagaki, N., Yagita, H. et al., Tumor necrosis factor‐related apoptosis‐inducing ligand (TRAIL) contributes to interferon gamma‐dependent natural killer cell protection from tumor metastasis. J. Exp. Med. 2001. 193: 661–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kimberley, F. C. and Screaton, G. R., Following a TRAIL: update on a ligand and its five receptors. Cell Res. 2004. 14: 359–372. [DOI] [PubMed] [Google Scholar]
- 58.Moral, J. A., Leung, J., Rojas, L. A., Ruan, J., Zhao, J., Sethna, Z., Ramnarain, A. et al., ILC2s amplify PD‐1 blockade by activating tissue‐specific cancer immunity. Nature 2020. 579: 130–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Blom, B., van Hoeven, V. and Hazenberg, M. D., ILCs in hematologic malignancies: tumor cell killers and tissue healers. Semin. Immunol. 2019. 41: 101279. [DOI] [PubMed] [Google Scholar]
- 60.Trabanelli, S., Chevalier, M. F., Martinez‐Usatorre, A., Gomez‐Cadena, A., Salomé, B., Lecciso, M., Salvestrini, V. et al., Tumour‐derived PGD2 and NKp30‐B7H6 engagement drives an immunosuppressive ILC2‐MDSC axis. Nat. Commun. 2017. 8: 593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schleussner, N., Merkel, O., Costanza, M., Liang, H. ‐ C., Hummel, F., Romagnani, C., Durek, P. et al., The AP‐1‐BATF and ‐BATF3 module is essential for growth, survival and TH17/ILC3 skewing of anaplastic large cell lymphoma. Leukemia 2018. 32: 1994–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Koh, J., Kim, H. Y., Lee, Y., Park, I. K, Kang, C. H., Kim, Y. T., Kim, J. ‐ E. et al., IL23‐producing human lung cancer cells promote tumor growth via conversion of innate lymphoid cell 1 (ILC1) into ILC3. Clin. Cancer Res. 2019. 25: 4026–4037. [DOI] [PubMed] [Google Scholar]
- 63.Cui, W., Zhang, W., Yuan, X., Liu, S., Li, M., Niu, J., Zhang, P. et al., Vitamin A deficiency execrates Lewis lung carcinoma via induction of type 2 innate lymphoid cells and alternatively activates macrophages. Food Sci. Nutr. 2019. 7: 1288–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Salimi, M., Wang, R., Yao, X., Li, X., Wang, X., Hu, Y., Chang, X. et al., Activated innate lymphoid cell populations accumulate in human tumour tissues. BMC Cancer 2018. 18: 341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jovanovic, I. P., Pejnovic, N. N., Radosavljevic, G. D., Pantic, J. M., Milovanovic, M. Z., Arsenijevic, N. N. and Lukic, M. L., Interleukin‐33/ST2 axis promotes breast cancer growth and metastases by facilitating intratumoral accumulation of immunosuppressive and innate lymphoid cells. Int. J. Cancer 2014. 134: 1669–1682. [DOI] [PubMed] [Google Scholar]
- 66.Irshad, S., Flores‐Borja, F., Lawler, K., Monypenny, J., Evans, R., Male, V., Gordon, P. et al., RORgammat(+) innate lymphoid cells promote lymph node metastasis of breast cancers. Cancer Res., 2017. 77: 1083–1096. [DOI] [PubMed] [Google Scholar]
- 67.Gao, Y., Souza‐Fonseca‐Guimaraes, F., Bald, T., Ng, S. S., Young, A., Ngiow, S. F., Rautela, J. et al., Tumor immunoevasion by the conversion of effector NK cells into type 1 innate lymphoid cells. Nat. Immunol. 2017. 18: 1004–1015. [DOI] [PubMed] [Google Scholar]
- 68.Cortez, V. S., Ulland, T. K., Cervantes‐Barragan, L., Bando, J. K., Robinette, M. L., Wang, Q., White, A. J. et al., SMAD4 impedes the conversion of NK cells into ILC1‐like cells by curtailing non‐canonical TGF‐beta signaling. Nat. Immunol. 2017. 18: 995–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Long, A., Dominguez, D., Qin, L., Chen, S., Fan, J., Zhang, M., Fang, D. et al., Type 2 innate lymphoid cells impede IL‐33‐mediated tumor suppression. J. Immunol. 2018. 201: 3456–3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schuijs, M. J., Png, S., Richard, A. C., Tsyben, A., Hamm, G., Stockis, J., Garcia, C. et al., ILC2‐driven innate immune checkpoint mechanism antagonizes NK cell antimetastatic function in the lung. Nat. Immunol. 2020. 21: 998–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bie, Q., Zhang, P., Su, Z., Zheng, D., Ying, X., Wu, Y., Yang, H. et al., Polarization of ILC2s in peripheral blood might contribute to immunosuppressive microenvironment in patients with gastric cancer. J. Immunol. Res. 2014. 2014: 923135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li, J., Razumilava, N., Gores, G. J., Walters, S., Mizuochi, T., Mourya, R., Bessho, K. et al., Biliary repair and carcinogenesis are mediated by IL‐33‐dependent cholangiocyte proliferation. J. Clin. Invest. 2014. 124: 3241–3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Han, X., Huang, T. and Han, J., Cytokines derived from innate lymphoid cells assist Helicobacter hepaticus to aggravate hepatocellular tumorigenesis in viral transgenic mice. Gut Pathog. 2019. 11: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu, Y., Song, Y., Lin, D., Lei, L., Mei, Y., Jin, Z., Gong, H. et al., NCR(−) group 3 innate lymphoid cells orchestrate IL‐23/IL‐17 axis to promote hepatocellular carcinoma development. EBioMedicine 2019. 41: 333–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xuan, X., Zhou, J., Tian, Z., Lin, Y., Song, J., Ruan, Z., Ni, B. et al., ILC3 cells promote the proliferation and invasion of pancreatic cancer cells through IL‐22/AKT signaling. Clin. Transl. Oncol. 2020. 22: 563–575. [DOI] [PubMed] [Google Scholar]
- 76.Fuchs, A. and Colonna, M., Innate lymphoid cells in homeostasis, infection, chronic inflammation and tumors of the gastrointestinal tract. Curr. Opin. Gastroenterol. 2013. 29: 581–587. [DOI] [PubMed] [Google Scholar]
- 77.Man, S. M., Inflammasomes in the gastrointestinal tract: infection, cancer and gut microbiota homeostasis. Nat. Rev. Gastroenterol. Hepatol. 2018. 15: 721–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kirchberger, S., Royston, D. J., Boulard, O., Thornton, E., Franchini, F., Szabady, R. L., Harrison, O. et al., Innate lymphoid cells sustain colon cancer through production of interleukin‐22 in a mouse model. J. Exp. Med. 2013. 210: 917–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Geremia, A., Arancibia‐Cárcamo, C. V., Fleming, M. P. P., Rust, N., Singh, B., Mortensen, N. J., Travis, S. P. L. et al., IL‐23‐responsive innate lymphoid cells are increased in inflammatory bowel disease. J. Exp. Med. 2011. 208: 1127–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chan, I. H., Jain, R., Tessmer, M. S., Gorman, D., Mangadu, R., Sathe, M., Vives, F. et al., Interleukin‐23 is sufficient to induce rapid de novo gut tumorigenesis, independent of carcinogens, through activation of innate lymphoid cells. Mucosal Immunol. 2014. 7: 842–856. [DOI] [PubMed] [Google Scholar]
- 81.Bergmann, H., Roth, S., Pechloff, K., Kiss, E. A., Kuhn, S., Heikenwälder, M., Diefenbach, A. et al., Card9‐dependent IL‐1beta regulates IL‐22 production from group 3 innate lymphoid cells and promotes colitis‐associated cancer. Eur. J. Immunol. 2017. 47: 1342–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang, S., Xia, P., Chen, Yi, Qu, Y., Xiong, Z., Ye, B., Du, Y. et al., Regulatory innate lymphoid cells control innate intestinal inflammation. Cell 2017. 171: 201–216.e18. [DOI] [PubMed] [Google Scholar]
- 83.Chevalier, M. F., Trabanelli, S., Racle, J., Salomé, B., Cesson, V., Gharbi, D., Bohner, P. et al., ILC2‐modulated T cell‐to‐MDSC balance is associated with bladder cancer recurrence. J. Clin. Invest. 2017. 127: 2916–2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hosseini, S. H, Sharafkandi, N., Seyfizadeh, N., Hemmatzadeh, M., Marofi, F., Shomali, N., Karimi, M. et al., Progression or suppression: two sides of the innate lymphoid cells in cancer. J. Cell. Biochem. 2020. 121: 2739–2755. [DOI] [PubMed] [Google Scholar]
- 85.Mittal, D., Gubin, M. M., Schreiber, R. D. and Smyth, M. J., New insights into cancer immunoediting and its three component phases–elimination, equilibrium and escape. Curr. Opin. Immunol. 2014. 27: 16–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vesely, M. D., Kershaw, M. H., Schreiber, R. D. and Smyth, M. J., Natural innate and adaptive immunity to cancer. Annu. Rev. Immunol. 2011. 29: 235–271. [DOI] [PubMed] [Google Scholar]
- 87.Grivennikov, S. I., Greten, F. R. and Karin, M., Immunity, inflammation, and cancer. Cell 2010. 140: 883–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sonnenberg, G. F. and Hepworth, M. R., Functional interactions between innate lymphoid cells and adaptive immunity. Nat. Rev. Immunol. 2019. 19: 599–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fuchs, A., Vermi, W., Lee, J. S., Lonardi, S., Gilfillan, S., Newberry, R. D., Cella, M. et al., Intraepithelial type 1 innate lymphoid cells are a unique subset of IL‐12‐ and IL‐15‐responsive IFN‐gamma‐producing cells. Immunity 2013. 38: 769–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vonarbourg, C., Mortha, A., Bui, V. L., Hernandez, P. P., Kiss, E. A., Hoyler, T., Flach, M. et al., Regulated expression of nuclear receptor RORgammat confers distinct functional fates to NK cell receptor‐expressing RORgammat(+) innate lymphocytes. Immunity 2010. 33: 736–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Böttcher, J. P., Bonavita, E., Chakravarty, P., Blees, H., Cabeza‐Cabrerizo, M., Sammicheli, S., Rogers, N. C. et al., NK cells stimulate recruitment of cDC1 into the tumor microenvironment promoting cancer immune control. Cell 2018. 172: 1022–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Martín‐Fontecha, A., Thomsen, L. L., Brett, S., Gerard, C., Lipp, M., Lanzavecchia, A. and Sallusto, F., Induced recruitment of NK cells to lymph nodes provides IFN‐gamma for T(H)1 priming. Nat. Immunol. 2004. 5: 1260–1265. [DOI] [PubMed] [Google Scholar]
- 93.Paul, S., Kulkarni, N., Shilpi, . and Lal, G., Intratumoral natural killer cells show reduced effector and cytolytic properties and control the differentiation of effector Th1 cells. Oncoimmunology 2016. 5: e1235106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schmidt, L., Eskiocak, B., Kohn, R., Dang, C., Joshi, N. S., DuPage, M., Lee, D. ‐ Y. et al., Enhanced adaptive immune responses in lung adenocarcinoma through natural killer cell stimulation. Proc. Natl. Acad. Sci. USA. 2019. 116: 17460–17469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fan, Z., Yu, P., Wang, Y., Wang, Y., Fu, M. L., Liu, W., Sun, Y. et al., NK‐cell activation by LIGHT triggers tumor‐specific CD8+ T‐cell immunity to reject established tumors. Blood 2006. 107: 1342–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ercolano, G., Falquet, M., Vanoni, G., Trabanelli, S. and Jandus, C., ILC2s: new actors in tumor immunity. Front. Immunol. 2019. 10: 2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Domvri, K., Petanidis, S., Zarogoulidis, P., Anestakis, D., Tsavlis, D., Bai, C., Huang, H. et al., Treg‐dependent immunosuppression triggers effector T cell dysfunction via the STING/ILC2 axis. Clin. Immunol. 2021. 222: 108620. [DOI] [PubMed] [Google Scholar]
- 98.Shields, J. D., Kourtis, I. C., Tomei, A. A., Roberts, J. M. and Swartz, M. A., Induction of lymphoidlike stroma and immune escape by tumors that express the chemokine CCL21. Science 2010. 328: 749–752. [DOI] [PubMed] [Google Scholar]
- 99.Crome, S. Q., Nguyen, L. T., Lopez‐Verges, S., Yang, S. Y. C., Martin, B., Yam, J. Y., Johnson, D. J. et al., A distinct innate lymphoid cell population regulates tumor‐associated T cells. Nat. Med. 2017. 23: 368–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Komlósi, Z. I., Kovács, N., Van De Veen, W., Kirsch, A. I., Fahrner, H. B., Wawrzyniak, M., Rebane, A. et al., Human CD40 ligand‐expressing type 3 innate lymphoid cells induce IL‐10‐producing immature transitional regulatory B cells. J. Allerg. Clin. Immunol. 2018. 142: 178–194. [DOI] [PubMed] [Google Scholar]
- 101.Campana, S., Di Carlo, E., De Pasquale, C., Barberi, C., Oliveri, D., Migliore, G. S., Cannavò, S. P. et al., Dendritic cell recognition by group 3 innate lymphoid cells through DNAX accessory molecule 1 triggers proinflammatory reciprocal cell activation. J. Allerg. Clin. Immunol. 2019. 144: 1118–1122. [DOI] [PubMed] [Google Scholar]
- 102.Warner, K. and Ohashi, P. S., ILC regulation of T cell responses in inflammatory diseases and cancer. Semin. Immunol. 2019. 41: 101284. [DOI] [PubMed] [Google Scholar]
- 103.Mazzurana, L., Forkel, M., Rao, A., Van Acker, A., Kokkinou, E., Ichiya, T., Almer, S. et al., Suppression of Aiolos and Ikaros expression by lenalidomide reduces human ILC3‐ILC1/NK cell transdifferentiation. Eur. J. Immunol. 2019. 49: 1344–1355. [DOI] [PubMed] [Google Scholar]
- 104.Wagner, M., Moro, K. and Koyasu, S., Plastic heterogeneity of innate lymphoid cells in cancer. Trends Cancer 2017. 3: 326–335. [DOI] [PubMed] [Google Scholar]
- 105.Bald, T., Wagner, M., Gao, Y., Koyasu, S. and Smyth, M. J., Hide and seek: plasticity of innate lymphoid cells in cancer. Semin. Immunol. 2019. 41: 101273. [DOI] [PubMed] [Google Scholar]
- 106.Bernink, J. H., Krabbendam, L., Germar, K., De Jong, E., Gronke, K., Kofoed‐Nielsen, M., Munneke, J. M. et al., Interleukin‐12 and ‐23 control plasticity of CD127(+) group 1 and group 3 innate lymphoid cells in the intestinal lamina propria. Immunity 2015. 43: 146–160. [DOI] [PubMed] [Google Scholar]
- 107.Hughes, T., Briercheck, E. L., Freud, A. G., Trotta, R., Mcclory, S., Scoville, S. D., Keller, K. et al., The transcription factor AHR prevents the differentiation of a stage 3 innate lymphoid cell subset to natural killer cells. Cell Rep. 2014. 8: 150–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Raykova, A., Carrega, P., Lehmann, F. M., Ivanek, R., Landtwing, V., Quast, I., Lünemann, J. D. et al., Interleukins 12 and 15 induce cytotoxicity and early NK‐cell differentiation in type 3 innate lymphoid cells. Blood Adv. 2017. 1: 2679–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Larson, C., Oronsky, B., Carter, C. A., Oronsky, A., Knox, S. J., Sher, D. and Reid, T. R., TGF‐beta: a master immune regulator. Expert Opin. Ther. Targets 2020. 24: 427–438. [DOI] [PubMed] [Google Scholar]
- 110.Hawke, L. G., Mitchell, B. Z. and Ormiston, M. L., TGF‐beta and IL‐15 Synergize through MAPK pathways to drive the conversion of human NK cells to an innate lymphoid cell 1‐like phenotype. J. Immunol. 2020. 204: 3171–3181. [DOI] [PubMed] [Google Scholar]
- 111.Pikovskaya, O., Chaix, J., Rothman, N. J., Collins, A., Chen, Y.‐H., Scipioni, A. M., Vivier, E. et al., Cutting edge: eomesodermin is sufficient to direct type 1 innate lymphocyte development into the conventional NK lineage. J. Immunol. 2016. 196: 1449–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Silver, J. S., Kearley, J., Copenhaver, A. M., Sanden, C., Mori, M., Yu, L., Pritchard, G. H. et al., Inflammatory triggers associated with exacerbations of COPD orchestrate plasticity of group 2 innate lymphoid cells in the lungs. Nat. Immunol. 2016. 17: 626–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ohne, Y., Silver, J. S., Thompson‐Snipes, L., Collet, M. A., Blanck, J. P., Cantarel, B. L., Copenhaver, A. M. et al., IL‐1 is a critical regulator of group 2 innate lymphoid cell function and plasticity. Nat. Immunol. 2016. 17: 646–655. [DOI] [PubMed] [Google Scholar]
- 114.Bernink, J. H., Ohne, Y., Teunissen, M. B. M., Wang, J., Wu, J., Krabbendam, L., Guntermann, C. et al., c‐Kit‐positive ILC2s exhibit an ILC3‐like signature that may contribute to IL‐17‐mediated pathologies. Nat. Immunol. 2019. 20: 992–1003. [DOI] [PubMed] [Google Scholar]
- 115.Golebski, K., Ros, X. R., Nagasawa, M., Van Tol, S., Heesters, B. A., Aglmous, H., Kradolfer, C. M. A. et al., IL‐1beta, IL‐23, and TGF‐beta drive plasticity of human ILC2s towards IL‐17‐producing ILCs in nasal inflammation. Nat. Commun. 2019. 10: 2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sun, H., Huang, Q., Huang, M., Wen, H., Lin, R., Zheng, M., Qu, K. et al., Human CD96 correlates to natural killer cell exhaustion and predicts the prognosis of human hepatocellular carcinoma. Hepatology 2019. 70: 168–183. [DOI] [PubMed] [Google Scholar]
- 117.Pesce, S., Trabanelli, S., Di Vito, C., Greppi, M., Obino, V., Guolo, F., Minetto, P. et al., Cancer immunotherapy by blocking immune checkpoints on innate lymphocytes. Cancers (Basel) 2020. 12: 3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chiossone, L. and Vivier, E., Immune checkpoints on innate lymphoid cells. J. Exp. Med. 2017. 214: 1561–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pachella, L. A., Madsen, L. T. and Dains, J. E., The toxicity and benefit of various dosing strategies for interleukin‐2 in metastatic melanoma and renal cell carcinoma. J. Adv. Pract. Oncol. 2015. 6: 212–221. [PMC free article] [PubMed] [Google Scholar]
- 120.Simoni, Y., Fehlings, M., Kløverpris, H. N., Mcgovern, N., Koo, S.‐L., Loh, C. Y., Lim, S. et al., Human innate lymphoid cell subsets possess tissue‐type based heterogeneity in phenotype and frequency. Immunity 2017. 46: 148–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yu, Y., Tsang, J. C. H., Wang, C., Clare, S., Wang, J., Chen, Xi, Brandt, C. et al., Single‐cell RNA‐seq identifies a PD‐1(hi) ILC progenitor and defines its development pathway. Nature 2016. 539: 102–106. [DOI] [PubMed] [Google Scholar]
- 122.Seillet, C., Mielke, L. A., Amann‐Zalcenstein, D. B., Su, S., Gao, J., Almeida, F. F., Shi, W. et al., Deciphering the innate lymphoid cell transcriptional program. Cell Rep. 2016. 17: 436–447. [DOI] [PubMed] [Google Scholar]
- 123.Taylor, S., Huang, Y., Mallett, G., Stathopoulou, C., Felizardo, T. C., Sun, M‐An, Martin, E. L. et al., PD‐1 regulates KLRG1(+) group 2 innate lymphoid cells. J. Exp. Med. 2017. 214: 1663–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mariotti, F. R., Quatrini, L., Munari, E., Vacca, P. and Moretta, L., Innate lymphoid cells: expression of PD‐1 and other checkpoints in normal and pathological conditions. Front. Immunol. 2019. 10: 910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Vacca, P., Pesce, S., Greppi, M., Fulcheri, E., Munari, E., Olive, D., Mingari, M. C. et al., PD‐1 is expressed by and regulates human group 3 innate lymphoid cells in human decidua. Mucosal Immunol. 2019. 12: 624–631. [DOI] [PubMed] [Google Scholar]
- 126.Guia, S., Fenis, A., Vivier, E. and Narni‐Mancinelli, E., Activating and inhibitory receptors expressed on innate lymphoid cells. Semin. Immunopathol. 2018. 40: 331–341. [DOI] [PubMed] [Google Scholar]
- 127.Helou, D. G., Shafiei‐Jahani, P., Lo, R., Howard, E., Hurrell, B. P., Galle‐Treger, L., Painter, J. D. et al., PD‐1 pathway regulates ILC2 metabolism and PD‐1 agonist treatment ameliorates airway hyperreactivity. Nat. Commun. 2020. 11: 3998. [DOI] [PMC free article] [PubMed] [Google Scholar]


