Abstract
Clinical annotations are one of the most popular resources available on the Pharmacogenomics Knowledgebase (PharmGKB). Each clinical annotation summarizes the association between variant‐drug pairs, shows relevant findings from the curated literature, and is assigned a level of evidence (LOE) to indicate the strength of support for that association. Evidence from the pharmacogenomic literature is curated into PharmGKB as variant annotations, which can be used to create new clinical annotations or added to existing clinical annotations. This means that the same clinical annotation can be worked on by multiple curators over time. As more evidence is curated into PharmGKB, the task of maintaining consistency when assessing all the available evidence and assigning an LOE becomes increasingly difficult. To remedy this, a scoring system has been developed to automate LOE assignment to clinical annotations. Variant annotations are scored according to certain attributes, including study size, reported P value, and whether the variant annotation supports or fails to find an association. Clinical guidelines or US Food and Drug Administration (FDA)‐approved drug labels which give variant‐specific prescribing guidance are also scored. The scores of all annotations attached to a clinical annotation are summed together to give a total score for the clinical annotation, which is used to calculate an LOE. Overall, the system increases transparency, consistency, and reproducibility in LOE assignment to clinical annotations. In combination with increased standardization of how clinical annotations are written, use of this scoring system helps to ensure that PharmGKB clinical annotations continue to be a robust source of pharmacogenomic information.
Pharmacogenomics (PGx) is a key component of personalized medicine, putting the patient and their genome front and center in the search for the best medication and dose for that person. The Pharmacogenomics Knowledge Base (PharmGKB, https://www.pharmgkb.org) has been a leading source of manually curated, publicly accessible PGx knowledge for more than 2 decades and is a vital tool in both guiding PGx research as well as facilitating the implementation of PGx in the clinic.
The PharmGKB was launched in 2000 as part of the National Institutes of Health (NIH)‐funded Pharmacogenetics Research Network (PGRN). It was originally established to be the PGRN’s genotype and phenotype data repository but quickly grew to encompass manually curated knowledge about gene‐drug associations supported by peer‐reviewed literature. In 2003, PharmGKB began aggregating curated gene‐drug associations creating illustrative diagrams of pharmacokinetic and pharmacodynamic drug‐centered pathways, accompanied by text summaries, and publishing these as review articles.1 Shortly afterwards, PharmGKB began creating and publishing peer‐reviewed summaries of genes important for the metabolism of, or response to, one or more drugs as Very Important Pharmacogenes (VIPs). By 2007, PharmGKB focused on capturing variant‐drug relationships from the published literature as freeform text summaries. This was subsequently formalized by introducing webforms with standardized fields and vocabularies that PharmGKB curators could use to record and annotate specific characteristics of the associations and the studies reporting them, resulting in our machine‐readable variant annotations. Each variant annotation reports a single finding from a single study, along with details of the study cohort and any statistical analyses undertaken by the study authors.
Today, the foundation of PharmGKB remains the manual curation of the peer‐reviewed published literature. The annotation types and curation processes at PharmGKB are illustrated in Figure 1. Publications for curation are identified from a core set of PGx journals that are routinely curated, literature searches performed to create pathway, VIPs, guidelines or other reviews, and user recommendations. PharmGKB has also recently begun to use PGxMine,2 a natural language processing program to identify a wider range of articles for manual curation.
Figure 1.
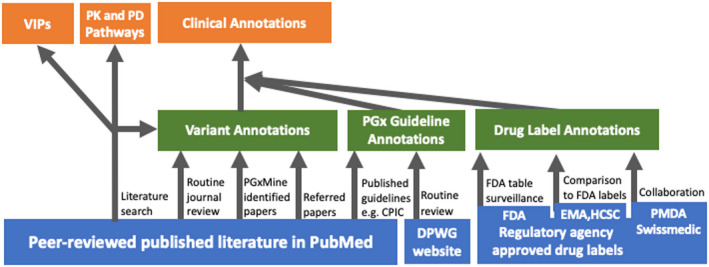
The PharmGKB curation process. Blue boxes represent primary literature and source documents that are curated. Green boxes represent basic PharmGKB annotations derived from curated literature and documents. Orange boxes represent PharmGKB annotations based on aggregated and evaluated basic annotations and curated literature. CPIC, Clinical Pharmacogenetics Implementation Consortium; DPWG, Royal Dutch Association for the Advancement of Pharmacy‐ Pharmacogenetics Working Group; EMA, European Medicines Agency; FDA, US Food and Drug Administration; HCSC, Health Canada (Santé Canada); PD, pharmacodynamics; PharmGKB, Pharmacogenomics Knowledgebase; PK, pharmacokinetics; PMDA, Pharmaceuticals and Medical Devices Agency, Japan; VIPs, Very Important Pharmacogenes.
Through the years, PharmGKB curators have read thousands of papers and created tens of thousands of variant annotations. As mentioned previously, papers are added to the PharmGKB curation queue via a number of different avenues. As we curated the PGx literature, it became apparent that many variant‐drug associations had been replicated in multiple studies and, in some cases, conflicting results had been reported. This emphasized the need for brief summaries of the curated evidence on variant‐drug associations. In 2010, PharmGKB began creating clinical annotations which summarized variant annotations. Variant annotations on the same drug, variant, and phenotype are brought together to support a clinical annotation. Clinical annotations summarize the curated evidence base of a variant‐drug pair with all relevant variant annotations, both supporting and contradictory, which are listed underneath.
A “level of evidence” (LOE) is assigned to each clinical annotation to indicate the amount of literature support annotated for the variant‐drug association.3 The clinical annotation LOEs were originally developed using qualitative criteria with input from collaborators.
Clinical annotations remain one of the most popular features on PharmGKB and are used by others to annotate the human genome,4, 5, 6, 7 develop PGx testing panels,8 and when reporting PGx testing results to clinicians and patients. The Clinical Pharmacogenetics Implementation Consortium (CPIC) considers PharmGKB clinical annotations when assigning CPIC levels to gene‐drug pairs. Over the past decade, we have applied the original LOE to thousands of clinical annotations based on thousands of publications. However, as with other biocuration efforts, we face a challenge in trying to maintain accurate and consistent assessment of LOE in the face of an ever‐increasing number of PGx publications.9 This challenge was further complicated by our use of qualitative criteria when assigning LOE. For example, the qualitative criteria for level 1A included PharmGKB knowledge of variant‐drug associations implemented in a clinical setting. The specifics of this information or how it was obtained by PharmGKB was not documented in the clinical annotation, leading to user confusion. However, PharmGKB annotates prescribing guidance from genotype‐based drug dosing guidelines and regulatory agency approved drug labels, as described in the next paragraphs. This information supplements the variant‐drug association knowledge PharmGKB curates from the primary literature and can add to the evidence summarized in, and LOE assigned to, clinical annotations in a transparent manner.
In 2009, PharmGKB partnered with the PGRN to create CPIC.10 CPIC produces genotype‐based drug‐prescribing guidelines established on extensive literature reviews and expert recommendations. These peer‐reviewed, published guidelines are available on the PharmGKB website, along with guideline summaries and functionality to enter specific genotypes to retrieve CPIC recommendations. In addition, PharmGKB partnered with the Royal Dutch Association for the Advancement of Pharmacy—Pharmacogenetics Working Group (DPWG) to host webpages with their PGx guideline recommendations, and we continue to curate DPWG guidelines today.
PharmGKB also curates US Food and Drug Administration (FDA)‐approved drug labels based on the information posted on the FDA’s Table of Pharmacogenomic Biomarkers in Drug Labeling11 and created a PGx level system for annotating labels with the type of PGx information found on them.12 Over the years, PharmGKB annotations of drug labels have expanded and we now curate labels containing PGx information from the European Medicines Agency and Health Canada (HCSC) as these labels come to our attention. We have also partnered with collaborators in one‐off projects to curate labels from Swissmedic13 and Japan’s Pharmaceuticals and Medical Devices Agency (PMDA).
Herein, we describe a formalized, quantitative LOE system where a score for each clinical annotation is calculated based on the supporting annotations, including variant annotations, guideline annotations, and drug label annotations, and used to determine the LOE. Variant annotations are assigned a score based on specific attributes and the scores for all variant annotations included in a clinical annotation are summed together to give an overall score for the clinical annotation. Drug label annotations and PGx guideline annotations can also contribute to the clinical annotation score. The score is then translated to an LOE.
FRAMEWORK FOR ANNOTATION SCORING AND LOE ASSIGNMENT
Variant annotation scoring system
Variant annotation scores are calculated in a five‐step process based on several annotation attributes, as shown in Table 1. These attributes are defined fields in the variant annotation tool that curators populate when extracting information from a publication. Attributes used in variant annotation scoring cover phenotype category, association P value, cohort size, effect size, study type, and association and significance. Some of these attributes, such as P value and cohort size, are stored in a section of the variant annotation called “study parameters.” A variant annotation may have more than one study parameters section if, for example, multiple analyses were carried out by the study authors.
Table 1.
VA scoring system
| VA attributes | Points | |
|---|---|---|
| Step 1 | Phenotype category: toxicity, efficacy or dosage | 1 |
| Phenotype category: metabolism/PK or PD | 0.5 | |
| All others | 0 | |
| Step 2 | Non‐GWAS with P value < 0.01 | 1 |
| Non‐GWAS with a P value ≤ 0.05 and ≥ 0.01 OR GWAS with a P value ≤ 5 × 10‐8 | 0.5 | |
| All others | 0 | |
| Non‐GWAS with at least 2 P values ≤ 0.05 receive additional points | 0.5 | |
| Step 3 | Cohort size > 500 | 1 |
| Cohort size 251–500 | 0.75 | |
| Cohort size 101–250 | 0.5 | |
| Cohort size 51–100 | 0.25 | |
| Cohort size ≤ 50 | 0 | |
| Step 4 | OR/HR/RR value < 0.5 or > 2 and has a 95% confidence interval which does not cross 1 and has a P value ≤ 0.05 | 0.5 |
| Step 5A | In vitro study | 0 |
| Meta‐analysis with metabolizer phenotype terms rather than genotypes | 0.25 | |
| Meta‐analysis OR phenotype terms rather than genotypes, not from a meta‐analysis | 0.5 | |
| All others | 1 | |
| Step 5B | Author reports no association; study cohort ≥ 50 | −1 |
| Author reports no association; study cohort is < 50 | −0.25 | |
| Association reported as “not significant” OR “significance not stated” OR P value > 0.05; study cohort is ≥ 50 | 0 | |
| From unreplicated GWASs (1 study parameter with P value reported) or GWAS P value > 5 × 10‐8 and replication P value > 0.05 | 0 | |
| Association reported as “not significant” OR “significance not stated” OR P value > 0.05; study cohort is < 50 | 0.25 | |
| All others | 1 | |
| Score | (Step 1 + Step 2 + Step 3 + Step 4) * (Step 5A * Step 5B) | |
GWAS, genomewide association study; HR, hazard ratio; OR, odds ratio; PD, pharmacodynamic; PK, pharmacokinetic; RR, risk ratio; VA, variant annotation.
The basic formula for variant annotation scoring is:
An overview of each step is given below. Further details of the scoring system, including the points awarded for specific attributes, can be found in Table 1 and on the PharmGKB website.14
Step 1—Phenotype category (0–1 point)
Variant annotation phenotype categories reflect the type of outcomes assessed in the annotated study and are added by curators during the curation process. Clinical outcomes (efficacy, toxicity, and drug dose) are important for determining the potential clinical value of a variant‐drug association and are therefore awarded more points. Pharmacokinetic or pharmacodynamic findings, such as drug metabolism studies or receptor binding studies, may have less clinical relevance than clinical outcome studies but still add to evidence to support a variant‐drug association, so are awarded fewer points.
Step 2—Association P value (0–1 point)
Points for this step are based on the variant annotation study parameters with the lowest, most significant P value. In the scoring system, findings from hypothesis‐driven association analyses with strong P values are awarded the highest score. The scoring system is also able to account for findings from genomewide association study (GWAS) analyses where genomewide significance is reported. Step 5B includes further assessment of GWAS findings.
Step 3—Cohort size (0–1 point)
Larger cohort sizes result in more accurate mean values, reduced impact of outliers, and increased power to detect weaker effects, which minimizes type II errors. As an acknowledgement of this, more points are awarded for larger reported cohort sizes. Some variants, however, are so rare that they are primarily reported in case studies or family studies and may never be studied in a large cohort. This is compensated for in Step 5B and by the use of a scoring range specifically for rare variants.
Step 4—Effect size (0–0.5 points)
Effect size of a variant‐drug association indicates the degree to which patients with the variant may be affected by drug efficacy or toxicity. Associations with statistically significant odds ratio, hazard ratio, or relative risk greater than twice that of the control group or less than half that of the control group are considered relevant and are given an additional score.
Step 5A—Study type (multiplication factor of 0–1)
Variant annotations from in vitro assays, meta‐analyses, or studies lacking genotype information are given reduced scores, decreasing the contribution of these annotations to the total clinical annotation score. Findings from in vitro studies are not necessarily translatable to PGx relationships in vivo. Meta‐analyses are given reduced scores as they may introduce complications through their comparison of secondary datasets. Additionally, PharmGKB cannot easily determine which datasets that are re‐evaluated in meta‐analyses that have already been annotated from the original study publication, so these datasets could potentially be given double weight when calculating a score for a clinical annotation. Publications that only provide an associated phenotype rather than genotype information (e.g., “poor metabolizer” or “slow acetylator” without providing mapping information to specific diplotypes or alleles) are also given reduced scores. Although these types of annotations can provide a general idea of the direction of a pharmacogenetic association, they lack the specificity required to make the genotype‐ or allele‐specific assertions found in a clinical annotation.
Step 5B—Association and significance (multiplication factor of −1–1)
Variant annotations which do not show an association or do not report a significant finding are given reduced scores, decreasing the contribution of these annotations to the total clinical annotation score. Although recognizing that a failure to find a significant association does not prove that there is no association between a drug and a variant, this system of scoring is in place because these findings do not corroborate findings which do claim an association. Findings from GWAS studies which were not replicated in the same publication also score less in this step. Variant annotations with cohorts < 50 are given a smaller weighting to reflect the difficulty of finding significant associations in small cohorts.
The following types of variant annotation are scored 0 in steps 5A or 5B and therefore do not contribute to a clinical annotation score:
Variant annotations from in vitro studies.
Variant annotation from studies with a cohort ≥ 50 where the association is not significant (either by author report or P > 0.05) or the association significance is not stated.
Variant annotations of unreplicated GWAS associations.
Voiding the variant annotation score
The scoring system allows a curator to void the calculated score for a variant annotation. Although the variant annotation score is not intended to be a judgment of the quality of the curated study, there may be issues with a variant annotation such that it should not be included in a clinical annotation’s score. For example, a variant annotation from a paper where the authors do not explicitly state which allele was associated or the direction of the effect. In such cases, the curator will manually set the variant annotation score to zero and add a publicly available written justification for the change. These variant annotations are still added to relevant clinical annotations to acknowledge the publication as part of the evidence base.
PGx guideline and drug label annotation scoring system
PharmGKB annotations of PGx guidelines and drug labels can be used as additional supporting evidence for clinical annotations. A clinical annotation must already be created based on one or more variant annotations based on published literature in order for PGx guideline or drug label annotations to be added.
To qualify for addition to a clinical annotation, a PGx guideline or drug label annotation must:
Be annotated in the PharmGKB database.
Provide recommendations or guidance based on the association (i.e., be “actionable”). If a PGx guideline or drug label mentions an association was evaluated or may exist, but does not provide a recommendation or guidance, it will not be added to the clinical annotation. If a guideline explicitly says no recommendation can be provided for a gene‐drug pair, the guideline is not added as support for the clinical annotation score. However, if the clinical annotation is at level 1B or 2, then a written statement acknowledging the “no recommendation” is added to the clinical annotation.
Refer to a specific allele or genotype. If a guideline or label refers to phenotypes or metabolizer status without explicit mapping to specific alleles or genotypes, it cannot be added to a clinical annotation. This avoids requiring curators to infer genotypes and allele function from the given phenotype or metabolizer status.
If these criteria are met, the respective drug label and PGx guideline annotations are added as evidence to the clinical annotation. One hundred points are added to the clinical annotation score for each, increasing the clinical annotation score so that level 1A can be automatically assigned.
Some PGx guidelines only provide recommendations for a subset of phenotype groups. For example, the CPIC guideline for ondansetron and tropisetron gives a recommendation for CYP2D6 ultrarapid metabolizers but has a “no recommendation” for CYP2D6 intermediate and poor metabolizers. In these cases, the guideline can still be used to support a level 1A clinical annotation if the clinical annotation includes alleles which can contribute to the phenotypes with recommendations. A note is added to each phenotype description to inform users that a “no recommendation” exists for certain phenotype groups.
Clinical annotation scoring system
The clinical annotation score is based on the sum of scores for each supporting annotation, illustrated in Figure 2. When selecting variant annotations for inclusion in a clinical annotation, curators can indicate if a variant annotation reports results that conflict with the others. Conflicting variant annotations report associations in an opposite direction to other variant annotations in the clinical annotation (e.g., a variant annotation that associates a variant with increased response to a particular drug, whereas other variant annotations associate the same variant with decreased response to the same drug). The scores of the variant annotations in each direction are summed; the direction with the largest score is considered the direction of the association and the variant annotations showing opposite direction of effect are marked as conflicting. The scores from the conflicting variant annotations are subtracted from the clinical annotation score.
Figure 2.
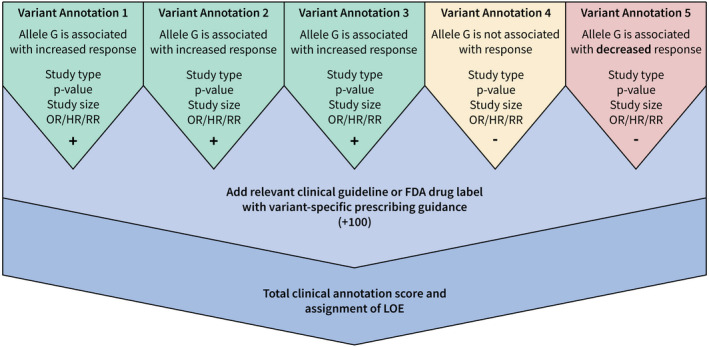
Illustrated example of clinical annotation scoring. FDA, US Food and Drug Administration; HR, hazard ratio; LOE, level of evidence; OR, odds ratio; RR, relative risk.
Level of evidence and clinical annotation scoring ranges
The LOE is automatically set within the web‐based curator tool according to the clinical annotation score. The scoring ranges for each level were set by evaluating the scores of all clinical annotations in PharmGKB and binning them based on the level descriptions (Table 2). Levels range from high (level 1) to associations unsupported by the evidence (level 4). The levels from the original LOE system and curators’ experience with the PGx literature were also taken into account in this process. As mentioned previously, there is a separate scoring range specifically for clinical annotations on rare variants as these are likely to only be reported in small studies, increasing the difficulty of detecting an underlying PGx association. Users will note in Table 2 that the upper bounds of the many levels end in 0.9375. This is because the lowest possible positive score of a variant annotation is 0.0625.
Table 2.
Levels of evidence
| Level of evidence | Standard scoring range | Rare variant scoring rangea | Description |
|---|---|---|---|
| 1A | ≥ 80 | ≥ 80 | High level of evidence for the association. Level 1A clinical annotations describe variant‐drug combinations that have variant‐specific prescribing guidance in a current clinical guideline annotation or an FDA‐approved drug label annotation. Annotations of drug labels or clinical guidelines must give prescribing guidance for specific variants (e.g., CYP2C9*3, HLA‐B*57:01) or provide mapping from defined allele functions to diplotypes and phenotypes to be used as supporting evidence for a level 1A clinical annotation. Level 1A clinical annotations must be supported by at least one variant annotation in addition to a clinical guideline or drug label annotation with variant‐specific prescribing guidance. |
| 1B | 25–79.9375 | 10–79.9375 | High level of evidence for the association. Level 1B clinical annotations describe variant‐drug combinations with a high level of evidence supporting the association but no variant‐specific prescribing guidance in an annotated clinical guideline or FDA drug label. Level 1B clinical annotations must be supported by variant annotations from at least two independent publications. |
| 2A | 8–24.9375 variant in a tier 1 VIP | 3–9.9375 variant in a tier 1 VIP | Moderate level of evidence for the association. Variants in level 2A clinical annotations are found in PharmGKB’s tier 1 VIPs. These variants are in known pharmacogenes, implying causation of drug phenotype is more likely. Level 2 clinical annotations describe variant‐drug combinations with a moderate level of evidence supporting the association. For example, the association may be found in multiple cohorts, but there may be a minority of variant annotations that do not support the majority assertion. Level 2 clinical annotations must be supported by variant annotations from at least two independent publications. |
| 2B | 8–24.9375 | 3–9.9375 | Moderate level of evidence for the association. Variants in level 2B clinical annotations are not in PharmGKB’s tier 1 VIPs. Level 2 clinical annotations describe variant‐drug combinations with a moderate level of evidence supporting the association. For example, the association may be found in multiple cohorts, but there may be a minority of variant annotations that do not support the majority assertion. Level 2 clinical annotations must be supported by variant annotations from at least two independent publications. |
| 3 | 0–7.9375 | 0–2.9375 | Low level of evidence for the association. Level 3 clinical annotations describe variant‐drug combinations with a low level of evidence supporting the association. This association may be based on a single publication annotated in PharmGKB, or there may be several variant annotations that failed to replicate the association. The annotation may also be based on preliminary evidence (e.g., a case report, nonsignificant study, or in vitro, molecular, or functional assay evidence), resulting in a lower calculated score. |
| 4 | < 0 | < 0 | Association is unsupported. Level 4 clinical annotations describe variant‐drug combinations where the total score is negative, and the evidence does not support an association between the variant and the drug phenotype. |
FDA, US Food and Drug Administration; PharmGKB, Pharmacogenomics Knowledgebase; VIP, very important pharmacogene.
A separate scoring range is used for clinical annotations on rare variants.20
In cases where the level assigned by the scoring system is not fully reflective of the evidence base supporting a clinical annotation, a curator will bring the issue to the PharmGKB curation team for discussion. If consensus is reached, the level of a clinical annotation will be manually overridden. A justification for the override will be displayed to users on the website and in the download files.
Clinical annotations creation: Standard operating procedures
In conjunction with the revision of the LOE definition and assignment, PharmGKB has updated and standardized how clinical annotations are structured and worded. New annotation structuring means that clinical annotations are now typically based on a single phenotype category and a single drug or drug class. This ensures that the LOE is a more accurate reflection of the underlying evidence.
PharmGKB offers two distinct types of clinical annotation: variant‐level and gene‐level, because associations of genetic variation with a drug phenotype can be documented for a variant at a particular genomic position (e.g., single‐nucleotide polymorphism (SNP) or indel) or with a “star” allele, which represents variation across the gene. Variant‐level annotations provide genotype‐based summaries for a specific rsID. For example, a variant‐level clinical annotation on the ABCB1 variant rs1045642 provides phenotype descriptions for the AA, AG, and GG genotypes. Gene‐level clinical annotations display summaries for one of more star alleles of a gene, as depicted in Figure 4 where descriptions for the CYP2C19*1, *2, *3, and *29 alleles are given. Star allele definitions are sourced from the Pharmacogene Variation Consortium (PharmVar)15 or other official nomenclature groups (e.g., TPMT Nomenclature Committee16). When available, CPIC allele functions are displayed on gene‐level annotations. These formats have always been part of our clinical annotations, but have now been formalized with templates to standardize annotation writing.
Figure 4.
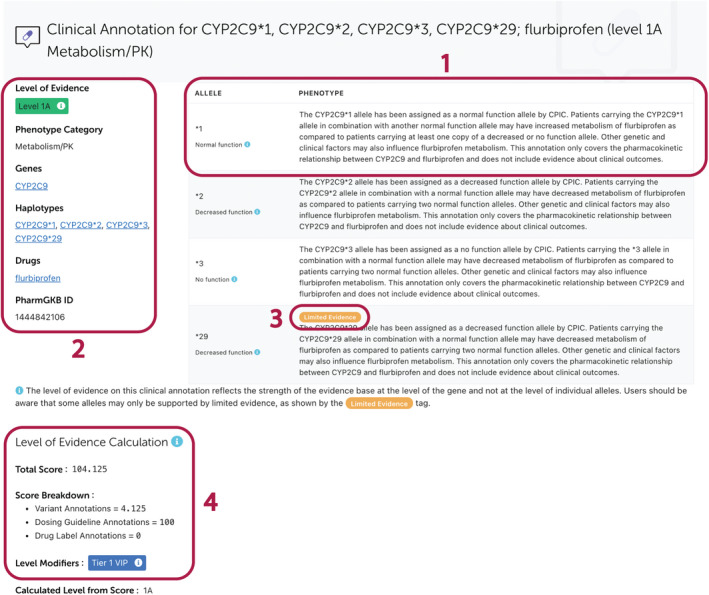
An example gene‐level clinical annotation. All clinical annotations provide genotype‐ or allele‐specific summaries of the curated evidence (Box 1) and display the LOE along with associated genes, haplotypes, drugs and phenotypes (Box 2). The Limited Evidence tag on gene‐level clinical annotations is added to alleles which are supported by substantially less evidence than other alleles in the same annotation (Box 3). The clinical annotation score and a scoring breakdown are displayed under the main annotation text (Box 4). This annotation can be viewed on the PharmGKB website at https://www.pharmgkb.org/clinicalAnnotation/1444842106. CPIC, Clinical Pharmacogenetics Implementation Consortium; LOE, level of evidence; PharmGKB, Pharmacogenomics Knowledgebase; PK, pharmacokinetic.
Variants are typically binary, a person either has the variant or the reference at that position. However, there are examples of tri‐allelic variation or multiple insertion/deletion/repeat variation at a genomic position. In these cases, the clinical annotation will reflect all variant possibilities that have been curated and the score will be the sum of all variant annotations for all variant possibilities. Likewise, when the association is reported as star allele variation across the gene, clinical annotations will reflect the reported star allele possibilities for that gene, and the score is the sum of all variant annotations for all reported allele possibilities. In some cases, certain alleles may be less well represented in the evidence base than others, typically because they are not frequently tested for. To address this, we have introduced a “Limited Evidence” tag that denotes individual alleles on gene‐level annotations which are supported by notably less evidence than other alleles. Use of the tag does not alter the assigned LOE of the clinical annotation.
Clinical annotations must now be independently reviewed by a second curator in the following cases:
Newly created level 1 or 2 clinical annotation.
Clinical annotation previously at level 3 or 4 becoming level 1 or 2 following the addition of new evidence.
Clinical annotation previously at level 1 or 2 becoming level 3 or 4 following the addition of new evidence.
Changes to clinical annotations are logged in the history section, which is displayed to users at the bottom of each annotation page.
All clinical annotations must be based on at least one variant annotation which shows an association; a clinical annotation is not written based only on a variant annotation which reports no association. Annotations of PGx guidelines or drug labels cannot be used as the basis for a new clinical annotation, they can only support existing annotations. Clinical annotations at level 1 or 2 must have supporting annotations from at least two sources. For both levels 1 and 2, one annotation must be a variant annotation from published literature. For levels 1B or 2, the other annotation will be a variant annotation from an independent publication, whereas for level 1A, it must be an annotated guideline publication or annotated drug label which supports the association. Clinical annotations which have a high enough score to rise above level 3 but are only supported by variant annotations from a single publication, or multiple publications from the same author group, will remain at level 3 until support from an independent publication (defined as no overlapping first or senior authors in each paper’s author list as determined by a curator) is curated and added to the clinical annotation.
As part of the clinical annotation update, all level 1 and 2 clinical annotations were updated to follow the revised standard operating procedure and have been independently reviewed by at least 2 curators.
Application of the framework to clinical annotations in PharmGKB
PharmGKB has 4,750 clinical annotations across 6 levels of evidence (from 1A to 4); a breakdown by level is shown in Table 3. Of the 4,750 clinical annotations, 4,203 are written at the variant‐level and the remaining 547 are written at the gene level. In total, the clinical annotations describe 2,451 variants and gene alleles from 1,039 genes, and 218 variants found in intergenic regions. There are 155 clinical annotations for 109 rare variants. The clinical annotations describe genetic associations with 346 drugs/chemicals. PharmGKB curators have overridden the calculated LOE on only nine clinical annotations (1.9%).
Table 3.
Number of clinical annotations at each LOE
| Number | LOE |
|---|---|
| 231 | 1A |
| 17 | 1B |
| 52 | 2A |
| 25 | 2B |
| 4,182 | 3 |
| 243 | 4 |
Data taken on March 30 2021.
LOE, level of evidence.
Level 1A clinical annotations have at least one supporting annotation from a CPIC or DPWG guideline or an FDA‐approved drug label. Figure 3 illustrates level 1A annotation support. Across all levels, 12,929 variant annotations from 5,427 publications, 44 CPIC guideline annotations, 13 DPWG guideline annotations, and 16 FDA‐approved drug label annotations are used as support for clinical annotations.
Figure 3.
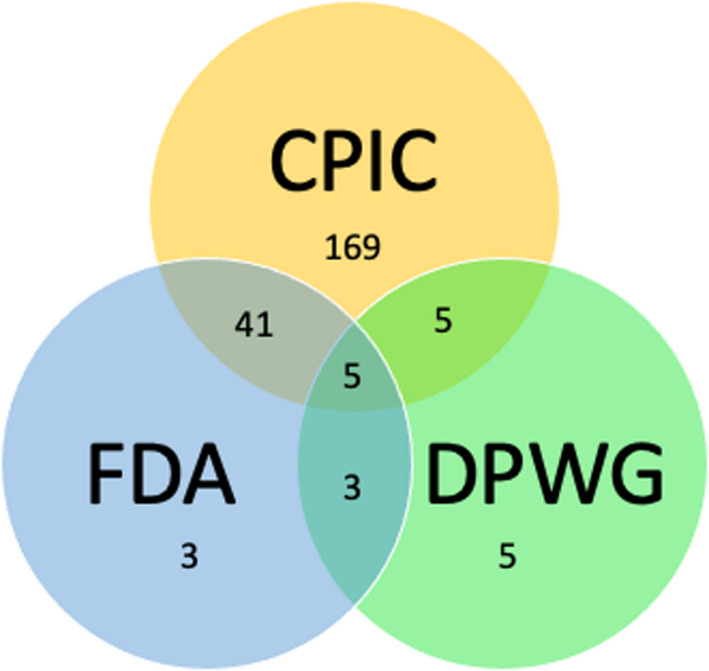
Venn diagram showing the number of level 1A clinical annotations with PGx guideline or drug label annotations as support. CPIC, Clinical Pharmacogenetics Implementation Consortium; DPWG, Royal Dutch Association for the Advancement of Pharmacy‐ Pharmacogenetics Working Group; FDA, US Food and Drug Administration; PGx, pharmacogenomics.
Clinical annotations are displayed individually on the PharmGKB website, available for bulk download as a set of tab‐separated value files, and can be queried through the PharmGKB application programming interface. The website display is seen in Figure 4 and the description of the set of download files is found on the PhamGKB website.17
DISCUSSION
PharmGKB was the first public resource to curate PGx literature and annotate human genomes with variant‐based PGx associations.4, 5, 6, 7 While carrying out this work, it became apparent that regularly maintained summaries of the total evidence for each variant would facilitate the genome annotation process, offering a viable alternative to repeatedly reviewing multiple publications and synthesizing the information from scratch for each genome project. This led PharmGKB to create clinical annotations and assign an LOE to indicate the strength of evidence contributing toward them.
As the number of variant and clinical annotations in PharmGKB grows and use of clinical annotations for PGx implementation expands, there is a need for a more objective system for assigning the LOE. Although specific for each level, the criteria in the original system were qualitative and therefore somewhat subjective. As new curated evidence was added to an existing clinical annotation, that annotation could be reassessed by multiple curators over time, leading to slight variance in evidence interpretation between curators. Whereas this redundancy was good for quality control, it could also lead to differences in how the supporting evidence was assessed and the LOE assigned.
We have addressed the issues with the original LOE system by developing a quantitative approach that systematically and consistently evaluates the supporting evidence of a clinical annotation and automatically assigns the LOE for the curator to review. Similar systems are used by other biological databases, most notably the Clinical Genome Resource, to facilitate curation and to maintain consistent standards.18, 19 Although curators can override the LOE assignment, curation standard operating procedures require the consensus of the curation team to do so, and a justification for the override to be publicly displayed with the clinical annotation for transparency.
Levels of evidence
The LOE definitions are largely similar to the original system, with the exception of level 4. Level 1A indicates a variant‐drug pair with actionable prescribing guidance available, whereas associations at level 1B are supported by the preponderance of annotated evidence. Level 2 is a moderate level of annotated evidence requiring replication of the association and level 2A still refers to variants in tier 1 VIP genes. Level 3 is a low level of annotated evidence that may be based on one study, or multiple studies with conflicting results leading to uncertainty, and has expanded to include case reports, studies that did not reach significance, and in vitro studies (previously level 4). The definition for level 4 has changed substantially. It previously applied to annotations based on case reports, studies that did not reach significance, or in vitro studies, but now applies to annotations where the preponderance of annotated evidence does not support an association. The new system can also accommodate situations where a curator disagrees with the level based on the clinical annotation score.
Variant annotation scoring
The scoring of variant annotations is not a judgment of study quality. It is a metric used by PharmGKB curators when comparing variant annotations against each other as part of the process of creating and updating clinical annotations. The variant annotation score is based on the information collected from the publication, without evaluation of the study design, which alleles were tested, the experimental approach, etc. The five‐step scoring system for variant annotations is based on criteria that curators already considered in the original LOE system, but the application of those criteria has now been formalized and automated.
Hypothesis driven association studies are weighted more than GWAS, meta‐analyses, or in vitro assays in this system. Findings from a GWAS tend to be preliminary and often require functional validation or at least independent replication to be clinically relevant. Although variant annotations of unreplicated GWAS score 0, replicated GWAS within the same publication have a positive score. Subsequent publications which replicate a GWAS are scored as hypothesis driven studies. Therefore, evidence (and points) can build to support GWAS findings and the LOE for the association can increase accordingly. Likewise, in vitro studies typically require additional evidence to show clinical relevance. Again, these studies alone score a 0, but subsequent in vivo studies of these associations can increase the LOE for the association. Meta‐analyses have the potential to discover new associations but can come with a range of problematic issues ranging from the inclusion criteria used to the analyses performed. As PharmGKB does not judge study quality, it takes a conservative approach to meta‐analyses by limiting the score. As with GWAS, evidence can build to support meta‐analysis findings and increase the LOE.
Drug label and PGx guideline annotation scoring
Although PharmGKB annotates clinical dosing guidelines and drug labels from multiple sources, the only guidelines and labels that are currently annotated in the PharmGKB database and capture the specific variants/alleles (or mappings to the variants/alleles) and prescribing guidance as described in the “PGx guideline and drug label annotation scoring system” section are PGx guidelines from CPIC or DPWG and FDA‐approved drug labels. CPIC guidelines do not require PharmGKB curators to infer the allele‐to‐phenotype mapping because CPIC allele function assignments and genotype‐to‐phenotype translations are already curated and stored in the PharmGKB database. For this first release of the scoring system, only DPWG guidelines that refer to specific alleles in the recommendation text are used to support clinical annotations. DPWG guidelines referring only to metabolizer status do not meet the criteria outlined in the PGx guideline and drug label annotation scoring system section. However, DPWG does provide some mapping materials and PharmGKB is currently curating and storing this information in the PharmGKB database for scoring use on a gene‐by‐gene basis. PharmGKB systematically captures both the specific variants referenced and prescribing guidance given in FDA‐approved drug labels, but not labels from other sources. Many FDA‐approved labels refer only to the metabolizer status and PharmGKB is not aware of mapping materials available for translation of FDA‐approved drug labels. Therefore, FDA‐approved labels with metabolizer status only are not used for scoring at this time because they do not meet the criteria outlined in the PGx guideline and drug label annotation scoring system section.
All guideline and label annotations are given equal weight without regard for what the recommended prescribing action is or for which group provided the guideline/label. It is up to the user to determine if the recommendations among supporting guidelines/labels agree or disagree and to what extent. The purpose of clinical annotations is to provide a summary of the association and the evidence annotated in PharmGKB supporting that association, not to declare clinical actionability or a recommended course of action. PharmGKB defers to the clinical guidelines and agency‐approved drug labels for such guidance and does not evaluate or compare organizations or agencies that provide genotype‐based drug prescribing guidance.
Guidelines/labels are given such a large score with the express purpose of assigning an LOE of level 1A to comply with the level definition. Prescribing guidance from an authoritative source, such as CPIC or the FDA, means that the evidence supporting the association between that variant and drug is so high that these groups consider the association clinically actionable. PharmGKB recognizes this evidence to be at a higher level than variant annotations of curated literature evidence by assigning level 1A. The large score and equal weight for each guideline/label may result in some clinical annotations with inflated scores if multiple guidelines/labels provide guidance for the same association. However, the clinical annotation score is only used to determine the LOE (in this case, level 1A) and should not be used to rank or compare clinical annotations within this or any other LOE.
CONCLUSION
The clinical annotation scoring system is able to account for conflicting evidence and multiple study types and is adjustable for use with rare variants. Public display of clinical annotation scores and supporting annotation scores help users to better understand why an LOE has been assigned. Overall, the scoring and LOE system, in combination with increased standardization of how clinical annotations are written, help ensure that PharmGKB clinical annotations continue to be a robust source of PGx information.
FUNDING
This work is supported by the NIH/NHGRI/NICHD grant U24 HG010615.
CONFLICT OF INTEREST
The authors report no conflict of interest.
ACKNOWLEDGMENTS
The authors sincerely appreciate the valuable feedback and advice from Andrea Gaedigk and Houda Hachad. We thank all members of the PharmGKB team for their contributions, including Clarissa Klein, Mark Woon, Reid Barber, Binglan Li, Rachel Dalton, and Russ Altman.
References
- 1.Eichelbaum, M., Altman, R.B., Ratain, M. & Klein, T.E. New feature: pathways and important genes from PharmGKB. Pharmacogenet. Genomics 19, 403 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lever, J.et al. PGxMine: text mining for curation of PharmGKB. Pac. Symp. Biocomput. 25, 611–622 (2020). [PMC free article] [PubMed] [Google Scholar]
- 3.Whirl‐Carrillo, M.et al. Pharmacogenomics knowledge for personalized medicine. Clin. Pharmacol. Ther. 92, 414–417 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashley, E.A.et al. Clinical assessment incorporating a personal genome. Lancet 375, 1525–1535 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dewey, F.E.et al. Clinical interpretation and implications of whole‐genome sequencing. JAMA 311, 1035–1045 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, R.et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell 148, 1293–1307 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dewey, F.E.et al. Phased whole‐genome genetic risk in a family quartet using a major allele reference sequence. PLoS Genet. 7, e1002280 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson, J.A., Burkley, B.M., Langaee, T.Y., Clare‐Salzler, M.J., Klein, T.E. & Altman, R.B. Implementing personalized medicine: development of a cost‐effective customized pharmacogenetics genotyping array. Clin. Pharmacol. Ther. 92, 437–439 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burge, S.et al. Biocurators and biocuration: surveying the 21st century challenges. Database 2012, bar059 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Relling, M.V. & Klein, T.E. CPIC: Clinical Pharmacogenetics Implementation Consortium of the pharmacogenomics research network. Clin. Pharmacol. Ther. 89, 464–467 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.U.S. Food and Drug Administration (FDA) Table of Pharmacogenomic Biomarkers in Drug Labeling <https://www.fda.gov/drugs/science‐and‐research‐drugs/table‐pharmacogenomic‐biomarkers‐drug‐labeling>. Accessed March 2021. [Google Scholar]
- 12.PharmGKB ‐ Drug Label Information and Legend <https://www.pharmgkb.org/page/drugLabelLegend>. Accessed March 2021. [Google Scholar]
- 13.Jeiziner, C.et al. Pharmacogenetic information in Swiss drug labels ‐ a systematic analysis. Pharmacogenomics J. 10.1038/s41397-020-00195-4. [e‐pub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.PharmGKB ‐ Variant Annotation Scoring page <https://www.pharmgkb.org/page/varAnnScoring>. Accessed March 2021. [Google Scholar]
- 15.Gaedigk, A., Ingelman‐Sundberg, M., Miller, N.A., Leeder, J.S., Whirl‐Carrillo, M. & Klein, T.E. The Pharmacogene Variation (PharmVar) Consortium: incorporation of the human cytochrome P450 (CYP) allele nomenclature database. Clin. Pharmacol. Ther. 103, 399–401 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Appell, M.L.et al. Nomenclature for alleles of the thiopurine methyltransferase gene. Pharmacogenet. Genomics 23, 242–248 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.PharmGKB ‐ Clinical Annotations Help File page <https://www.pharmgkb.org/page/downloadClinicalAnnotationsHelp>. Accessed March 2021. [Google Scholar]
- 18.Strande, N.T.et al. Evaluating the clinical validity of gene‐disease associations: an evidence‐based framework developed by the clinical genome resource. Am. J. Hum. Genet. 100, 895–906 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buza, T.J., McCarthy, F.M., Wang, N., Bridges, S.M. & Burgess, S.C. Gene Ontology annotation quality analysis in model eukaryotes. Nucleic Acids Res. 36, e12 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.PharmGKB ‐ Rare Variants <https://www.pharmgkb.org/page/rareVariant>. Accessed July 2021. [Google Scholar]


