Abstract
Fundamental asymmetries between the host and its microbiome in enzymatic activities and nutrient storage capabilities have promoted mutualistic adaptations on both sides. As a result, the enteric immune system has evolved so as not to cause a zero‐sum sterilization of non‐self, but rather achieve a non‐zero‐sum self‐reinforcing cooperation with its evolutionary partner the microbiome. In this review, we attempt to integrate the accumulated knowledge of immune—microbiome interactions into an evolutionary framework and trace the pattern of positive immune—microbiome feedback loops across epithelial, enteric nervous system, innate, and adaptive immune circuits. Indeed, the immune system requires commensal signals for its development and function, and reciprocally protects the microbiome from nutrient shortage and pathogen outgrowth. In turn, a healthy microbiome is the result of immune system curatorship as well as microbial ecology. The paradigms of host–microbiome asymmetry and the cooperative nature of their interactions identified in the gut are applicable across all tissues influenced by microbial activities. Incorporation of immune system influences into models of microbiome ecology will be a step forward toward defining what constitutes a healthy human microbiome and guide discoveries of novel host–microbiome mutualistic adaptations that may be harnessed for the promotion of human health.
Keywords: enteric nervous system, gut‐brain axis, immune system, microbiome, mutualism
Immune circuits of microbiome perception and response mediated by gut innate and adaptive immune systems, epithelial cells, and enteric nervous system have evolved to protect not only the host, but commensal microbiome as well. The immune curatorship together with the laws of microbial ecology defines the healthy microbiome.
Introduction
The host–microbiome mutualistic networks are fundamentally asymmetric in their interactions. The collective genomes of microbiome members harbor at least two orders of magnitude more genes than the human genome. The latest estimates of the size of microbial genomic space based on the analysis of 2182 gut microbiome metagenomes put it at 22,254,436 non‐redundant consensus genes, although about half of which are unique to a single sample [1]. In contrast, the size of the human genome is estimated to be 42,611 genes and only about half of them are protein‐coding [2]. Therefore, the enzymatic capacity of the microbiome is much greater than the host. Furthermore, while the genome of the host is stable throughout its life, the collective genome of microbiome adapts in response to external factors (e.g., diet change) and evolves under the influence of both positive [3] and negative [4] selections. The host‐microbiome genetic asymmetry has provided powerful evolutionary incentives for the host to harness the enzymatic potential of the microbiome to access otherwise unavailable small molecules and energy sources. In addition to the genetic host–microbiome asymmetry, the unique positioning of the microbiome at the interface between the host and environment creates additional evolutionary forces for the host to develop the capability to curate the microbiome's composition, both through positive reinforcement by supporting the beneficial members as well as through negative pressure by deselecting parasitic and pathogenic members of the microbiome. To achieve this goal, the host mucosal immune system employs a highly efficient, layered defense strategy featuring passive measures, such as the mucus barrier that creates a bacteria‐free zone separating the enteric epithelium from the microbiota [5, 6, 7], as well as active measures, such as the production of antimicrobial peptides (AMPs) and secretory IgA antibodies. The limited resources of the mucosal immune system are deployed in a coordinated and timely manner, notably through the involvement of intestinal epithelial cells (IECs), myeloid cells, innate lymphoid cells (ILCs), and the enteric nervous system (ENS), together with adaptive immune cells. For instance, in mice upon infection by pathogens, the ENS affects the development of IECs [8], activates ILCs [9, 10], and simultaneously engages damage‐control transcriptional programs in macrophages [11].
There is also an opposite direction of the asymmetric host–microbiome partnership: the host is capable of storing energy, whereas the microbiome is unable to do so. Taking advantage of this asymmetry, the host supports the beneficial members of the microbiome by supplying dietary polysaccharides or secreting sugar‐ and lipid‐decorated proteins into the lumen. In times of nutrient scarcity, the host uses its reserves to support the beneficial members of microbiome, for example, by fucosylation of the epithelium, thus protecting the commensals from starvation [12]. Such support of metabolically beneficial commensals indirectly leads to protection of the host from pathogenic infections through their occupation of the limited ecological niches—a concept known as colonization resistance [13].
At the fundamental level, the host–microbiome mutualism is based on cooperative behavior between two entities in which the actions of one depend on those of the other with the overall goal being homeostasis. Mathematically, given that an interaction between two entities has a defined goal and follows logical rules, such an interaction can be analyzed and an optimal strategy for each participant as well as important characteristics, such as boundary conditions where the interaction breaks down, may be calculated. The framework for the analysis of strategic interactions is termed “game theory” [14]. The theory was first developed from considerations of simplified strategic interactions namely zero‐sum games (e.g., poker) in which the sum of wins and losses of all players equals zero or in other words described strictly competitive interactions. Later the theory was expanded onto non‐zero‐sum interactions such as cooperation and it became clear that any kind of strategic interaction could be represented as a “game” and viewed in a framework of the theory [15]. The nature of immune system–microbiome interactions is not a zero‐sum sterilization of non‐self, but can be viewed as a non‐zero‐sum cooperation between interdependent evolutionary partners [16]. The non‐zero‐sum interaction with the microbiome mediates robustness and tunability in the immune network. This self‐reinforcing mutualistic host–microbiome coevolution is well captured by the concept of the microbiome as an extra organ of the human body [17]. A baseline understanding of their interdependent interactions also provides insights into how its collapse influences disease risk and what are potential targets for future lifestyle, drug, probiotic, prebiotic, and live biotherapeutic interventions. Herein, we summarize recent progress toward deciphering the principles of microbiome–immune bidirectional interdependent interactions in the intestine. As causality studies are predominantly only possible in animal models, unless stated otherwise research reviewed herein was performed in mice.
The innate immune system as the regulator of gut barrier function and immune homeostasis
During the past decade, there have been significant advances in our understanding of the gut microbiome and immune system bidirectional interactions. Molecular and cellular investigations have identified numerous cell types and molecules critical for microbial recognition and circuitry of signaling cascades to mount homeostatic and inflammatory immune responses. In particular, IECs, ENS, myeloid cells, and ILCs have been revealed to play critical roles as force multipliers to handle the otherwise uneven host‐microbiome standoff (Fig. 1, 2, 3).
Figure 1.
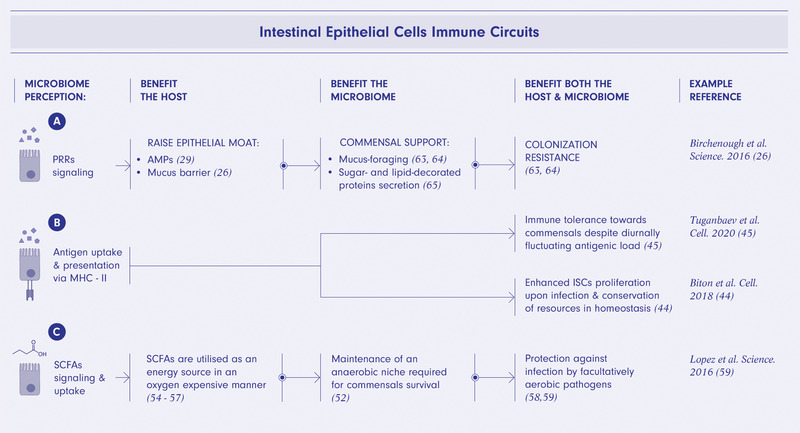
IECs immune programs are mutually beneficial for the host and microbiome. Perception of microbiome via pattern recognition receptors (PRRs) signaling promotes secretion of antimicrobial peptides (AMPs) and erection of a mucus barrier directly supporting beneficial mucus‐foraging bacteria and indirectly via colonization resistance to pathogens of other commensal bacteria (A). Microbiome sensing via luminal antigen uptake informs diurnal adjustment of intestinal barrier properties maintaining immunological tolerance towards microbiome around the clock and calibrates proliferative activity of intestinal stem cells (ISCs) promoting host‐microbiome homeostasis (B). Uptake of microbiome‐derived short‐chain fatty acids (SCFAs) promotes maintenance of an anaerobic environment required for homeostasis of commensal bacteria and protects against facultatively anaerobic pathogens (C). The figure was created using BioRender.com.
Figure 2.
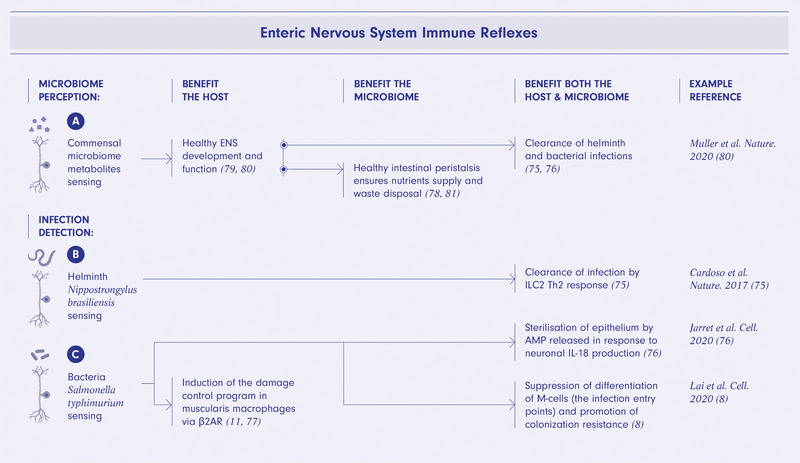
Enteric nervous system immune reflexes are mutually beneficial for the host and microbiome. Microbiome‐derived metabolites are required for healthy ENS development and function including peristalsis supplying nutrients to and disposing of waste from the microbiome community as well as benefiting the host and ultimately promoting clearance of parasite and bacterial infections by ENS‐immune reflexes (A). Sensing of Nippostrongylus brasiliensis infection by ENS neurons triggers innate lymphoid cells type 2 (ILC2) mediated Th2 immune response resulting in clearance of the helminth (B). Detection of Salmonella typhimurium infection by ENS leads to neuronal secretion of IL‐18 and a release of antimicrobial peptides (AMPs) by epithelial cells sterilizing the epithelium as well as suppression of M‐cells differentiation thus limiting infection entry points and the induction of neuronal damage control program in muscularis macrophages via β2AR (C). The figure was created using BioRender.com.
Figure 3.
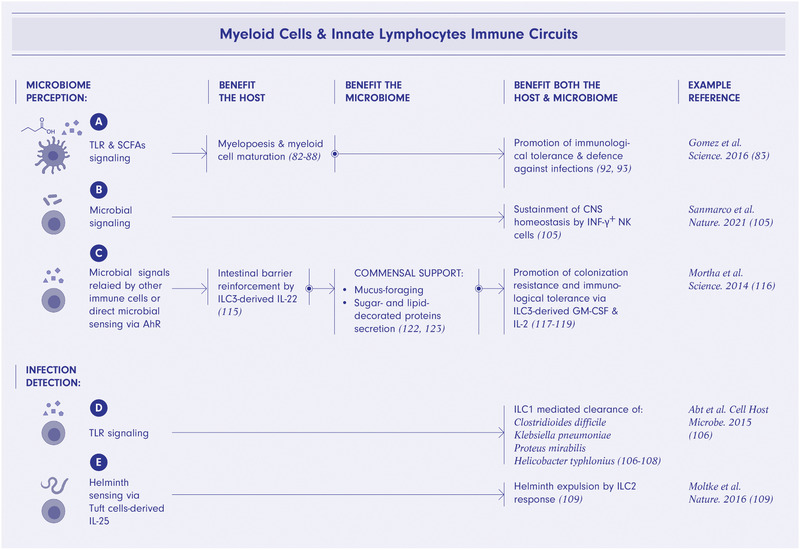
Mutually beneficial myeloid and innate lymphocytes ‐microbiome interactions. Microbiome perception via TLRs and short‐chain fatty acids (SCFAs) signaling promote myelopoiesis and myeloid cell maturation ultimately supporting immunological tolerance to commensal microbes and defense against infections (A). Microbial signaling in NK cells promotes CNS homeostasis (B). Microbial signals relayed by other immune cells or direct microbial sensing via aryl hydrocarbon receptor (AhR) leads to production of IL‐22, GM‐CSF and IL‐2 by innate lymphoid cells type 3 (ILC3) thus promoting colonization resistance and immunological tolerance as well as to secretion of mucus supporting mucus‐foraging commensals (C). TLR signaling in ILC1 triggers a bacterial infection clearing Th1 immune response benefiting commensal microbes as well as the host (D). Helminth sensing via Tuft cells‐derived IL‐25 by ILC2 leads to parasite expulsion by a Th2 immune response (E). The figure was created using BioRender.com.
Intestinal epithelial cells
Despite their non‐hematopoietic origin, IECs constitute a major part of the mucosal innate immune system in the context of both health and disease. A single cell layer of IECs forms the border that separates the host from the microbiome and the environment. Therefore, IECs need to simultaneously fulfill roles of a physical barrier guarding self from non‐self as well as a conduit facilitating the bidirectional interaction between the host's immune system and the microbiome. Despite the single‐cell layer organization, intestinal epithelium is functionally subdivided into distinct biogeographical niches spanning from the tip of villi to the bottom of the crypts. The bottom of the crypts serves as a niche for LGR5+ intestinal stem cells (ISCs), early progenies of ISCs, as well as Paneth cells that secrete AMPs into the lumen, preventing most bacteria from reaching the ISCs niche [18]. The villus body fulfills the absorptive, sensory, and secretory roles.
The epithelial layer is maintained by the proliferative activity of ISCs, which differentiate into six major IEC types. The absorptive lineage progenitors develop into enterocytes and microfold (M) cells, while the progenitors of the secretory lineage give rise to Paneth cells, goblet cells, enteroendocrine cells, and tuft cells [19, 20]. The enterocytes absorb nutrients and serve as a physical barrier. Enteroendocrine cells sense distinct dietary components, regulate intestinal motility, and modulate feeding behavior through the gut–brain axis [21]. Tuft cells detect parasite‐associated metabolites and elicit a type 2 immune response [22, 23, 24, 25]. Secretions from goblet cells form the mucus barrier precluding most bacteria from direct contact with the epithelium [26]. Located in Peyer's patches, M cells sample luminal antigens, helping promote IgA responses and thus curating the microbiome composition [27, 28]. The number of M cells is tightly regulated by gut‐innervating nociceptor sensory neurons and is reduced upon sensing by the neurons of Salmonella typhimurium that uses M cells as entry points for mucosal infection [8].
IECs perceive the microbiome composition directly via innate immune receptors as well as indirectly via recognition of microbiome‐associated metabolites. Microbial recognition by IECs via innate immune receptors is vital for maintaining mucosal health. Indeed, IEC‐specific deletion of elements of TLR signaling such as MyD88 [29], NEMO [30], and TRAF6 [31], as well as whole‐body knock‐out of NOD2 [32, 33], the DNA sensor AIM2 [34], or components of the NLRP3‐ or NLRP6‐dependent inflammasome [26, 35‐37] lead to either spontaneous severe pancolitis [30], spontaneous activation of small intestinal adaptive immune cells [29], or enhanced susceptibility in models of intestinal injury [26, 31‐37]. Mechanistically, the absence of innate immune signaling results in decreased Paneth cell numbers and consequently lower luminal concentration of AMPs as well as reduced mucin production by goblet cells, allowing more bacteria to come into direct contact with the epithelium and promote mucosal inflammation.
In addition to fortification of the mucosal barrier through the secretion of AMPs and mucin, IECs serve as non‐classical antigen presenting cells of microbial components. IECs in the small intestine take up luminal antigens [38, 39, 40], process [41] and present them via MHC class II (MHC‐II) to T cells [42, 43]. In the crypt niche, LGR5+ ISCs also express MHC‐II and present luminal antigen to CD4+ T cells, which reciprocally regulate differentiation rates of ISCs. CD4+ T cell‐derived proinflammatory cytokines such as IFN‐γ, IL‐17A, and IL‐13 promote active differentiation of ISCs, whereas regulatory IL‐10 attenuates differentiation and promotes self‐renewal [44]. Therefore, renewal of damaged IECs is accelerated upon detection of pathogen infection, whereas ISCs prioritize conservation of resources under homeostatic conditions. In the villus niche, MHC‐II‐mediated interactions between mature enterocytes and intraepithelial IL‐10+ CD4+ T cells regulate intestinal permeability diurnally to adapt barrier properties to fluctuating luminal antigenic load [45]. Diurnal nutrient intake, and consequently fluctuating microbiome composition, leads to dramatic differences in antigenic load at different times of the day [46, 47]. The mechanisms regulating intestinal permeability compensate for the antigenic fluctuations, preventing the mucosal sensory system from overloading at high antigen tide and maximizing its sensitivity at low tide. In addition to adapting regenerative or barrier properties of the epithelium, antigen sampling by IECs contributes to selecting inflammatory microbes to be targeted by humoral and cellular immune responses and thus actively curates the microbiome composition. Indeed, enterocytes sample antigens of mucosa‐attaching bacteria such as segmented filamentous bacteria (SFB) by employing clathrin‐independent, dynamin‐dependent cell division control protein 42 (CDC42) mediated endocytosis [40]. Such microbial adhesion‐triggered endocytosis (MATE) results in the induction of an SFB antigen‐specific mucosal Th17 response [40], suggesting a mucosa‐cleansing role of MATE. M cells, a specialized type of absorptive IECs, sample luminal antigens and simultaneously help to mediate the maturation of naïve B cells into IgA‐producing plasma cells [27] targeting inflammatory disease‐causing bacteria [48] as well as the induction of luminal pathogen‐specific Th1, Th17, and Th22 responses [49]. Similar to M cell‐mediated induction of cellular responses, goblet cells mediate goblet cell‐associated antigen passage (GAP), which supplies luminal antigens to tolerogenic CD103+ DCs in the lamina propria, thereby inducing differentiation of regulatory T (Treg) cells in the steadystate [50]. Successful clearance of pathogens requires a swift transition from a tolerant state of the immune system to an inflammatory one. In agreement with this paradigm, tolerogenic GAPs are inhibited during a Salmonella infection, thus allowing an inflammatory response to develop [51].
Complementary to the direct methods of microbiome perception, IECs sense the microbiome by detecting microbiome metabolites. In the colon, strictly anaerobic commensal bacteria digest dietary fiber to produce short‐chain fatty acids (SCFAs) as byproducts. SCFAs are sensed by IECs via GPR41, GPR43, GPR109A, and PPARγ [52, 53]. SCFAs can reinforce the epithelial barrier by modulating the mucus layer, epithelial cell survival, as well as expression of tight junction proteins [53]. SCFAs are also utilized by colonocytes as an energy source via the pathway of mitochondrial fatty acid β‐oxidation. This is an oxygen‐consuming process that reduces the amount of oxygen emanating from the epithelium into the lumen of the colon, thereby supporting the niche for anaerobic bacteria [54, 55, 56, 57]. SCFA‐mediated PPARγ‐signaling further limits the bioavailability of luminal oxygen and contributes to providing colonization resistance to pathogens [52]. In the absence of SCFAs producers due to antibiotic or dietary interventions, colonocyte metabolism shifts toward glycolysis, a process characterized by low oxygen consumption as well as activation of nitric oxide synthase. This results in luminal accumulation of oxygen and nitrate, thereby creating niches for facultative anaerobic pathogens such as Escherichia coli [55, 58] and Citrobacter rodentium [59]. Similar to regulation of oxygen metabolism by SCFAs in colonocytes, l‐lactate has recently been reported to promote lipid storage while conversely acetate is shown to trigger lipid oxidation via the AMPK/PGC‐1α/PPARα pathway in enterocytes in the small intestine [60]. Enterocytes are not the only cell type sensitive to lactate. Small intestinal CX3CR1+ mononuclear cells express the lactate G‐protein‐coupled receptor GPR31. GPR31 signaling promotes dendrite protrusion and enhances uptake of luminal antigens by monocytes [61]. Small intestinal LGR5+ ISCs express another lactate receptor GPR81; signaling through this receptor has been shown to stimulate ISCs expansion [62]. Concordantly, pre‐feeding of mice with lactate protected them from chemotherapy‐ and radiation‐mediated mucosal damage [62]. Metabolite sensing mechanisms are also utilized by tuft cells, which detect parasite‐ and microbiome‐derived succinate via the Sucnr1 receptor and initiate type 2 immune responses [22, 24, 25].
IECs employ additional strategies to actively assist commensals in competition with pathogens over an ecological niche in the intestine to reinforce colonization resistance [13]. The components of the mucus barrier secreted into the lumen by goblet cells are decorated with sugars and lipids conferring a fitness advantage to commensals such as Akkermansia muciniphila [63] and Ruminococcus gnavus [64], which possess specialized enzymatic systems for harvesting these resources. Furthermore, given that the host possesses the ability to store energy in reserve while beneficial prokaryotic symbionts do not, the host enterocytes upregulate fucosyltransferase 2 (Fut2) and shed fucose into the lumen to sustain the commensals during periods of fasting and nutrient unavailability [65]. Concordantly, human milk oligosaccharides promote colonization of the newborn gut by beneficial commensals such as Lactobacillus, thereby developing colonization resistance to pathogens [66].
Already in invertebrates, the epithelium features some of the simplest and therefore probably the most evolutionary ancient innate immune programs that are composed of just two steps: perception and reaction. Representative examples include production of AMPs upon microbial sensing [37] (Fig. 1). In contrast, mammalian epithelial programs additionally include integration with the adaptive arm and therefore represent a more recent innovation in immune‐microbiome co‐evolution. Representative examples include the program regulating epithelial proliferation based on the microbiome composition [44] and the program adjusting epithelial permeability in response to diurnally fluctuating luminal antigenic load [45]. Importantly, programs at all levels of complexity pursue not the one‐sided advantage for the host over the microbiome, but rather the collective benefits for the host and a diverse and stable microbiome (Fig. 1A‐C).
Enteric nervous system
Recent advances in studies of the ENS have firmly established it as a critical component of the mucosal immune system in health and disease [67, 68, 69]. Structurally, the ENS network is composed of two main plexuses—the myenteric plexus (also termed Auerbach's plexus), located within the muscularis layer between the longitudinal and the circular muscles; and the submucosal plexus (also termed Meissner's plexus), located between the mucosa and the circular layer of muscles. The ENS innervates all layers of the intestinal tissue from the epithelial layer through the lamina propria, submucosa, muscularis, serosa, and the mesentery throughout the gastrointestinal tract. The total number of neurons in ENS is estimated to be on par with that of the spinal cord [69]. The diverse aspects of intestinal homeostasis such as gastric acid secretion, intestinal motility, barrier integrity, fluid and nutrient absorption, regulation of local blood flow, and interactions with the gut immune and endocrine systems are all collectively managed by reflex arcs of local ENS, reflex arcs that pass through extrinsic sympathetic and sensory ganglia, as well as reflexes that circle through the CNS [69]. The bidirectional communication between the ENS and the CNS is established via three major routes: (1) parasympathetic vagal neurons that connect the ENS to the hindbrain, (2) sympathetic nerves that connect the ENS through the sympathetic visceral ganglia to the spinal cord, and (3) the pelvic pathways that connect the ENS directly with the spinal cord. Despite this interconnectivity, the ENS can manage the gastrointestinal functions even if the connection with the CNS is severely reduced (e.g., in the case of a vagotomy) [69]. In contrast, neuropathies involving failure of a part of the ENS, such as Hirschsprung disease [70] or Chagas disease [71], are potentially fatal.
A key player in ENS circuitry is the intrinsic sensory neuron (known as intrinsic primary afferent neurons, or IPANs) [72]. IPANs form ring‐shaped networks encircling the gut while sending projections deep into the mucosa [72]. Interneurons on the other hand form linear networks spanning in the oral and anal directions along the length of intestine. Ascending chains of interneurons synapse onto excitatory motor neurons while descending interneurons interconnect with inhibitory motor neurons [72]. Located in the myenteric plexus, excitatory and inhibitory motor neurons form mesh‐like networks interlaced by glial cells. In addition to classical subtyping of these ENS neurons based on morphology, electrochemical characteristics, neurotransmitters utilized for signal transduction and biogeographical location [73], recent single‐cell transcriptomics has revealed additional layers of complexity [68, 74]. Further investigation into how spatial distribution as well as morphological, electrochemical, and transcriptomic profiles combine to form functional identity of neurons is required.
It has been shown that mucosal ENS circuitry promotes group 2 innate lymphoid cell (ILC2) activation in response to helminth infections [75]. An infection with the helminth parasite Nippostrongylus brasiliensis leads to a release of the neuropeptide neuromedin U by lamina propria cholinergic neurons, which in turn induces rapid ILC2‐mediated responses, characterized by immediate and massive production of IL‐5, IL‐13, and amphiregulin (AREG) [75]. Similar to the ILC2‐inducing reflex, neurons of the myenteric and submucosal plexuses in the colon react to Salmonella typhimurium infection by mounting an IL‐18 response via the NLRP6‐ and Caspase 1/11‐dependent inflammasome pathway. IL‐18 then stimulates AMP production by goblet cells to sterilize the epithelium [76]. This pathway also induces pyroptosis of neurons, resulting in post‐infection neuronal damage and thereby possibly promoting chronic irritable bowel syndrome. To prevent post‐infection neuronal damage, sympathetic catecholaminergic neurons innervating the myenteric plexus rapidly respond to Salmonella infection by releasing norepinephrine, which stimulates local muscularis resident macrophages via the β2 adrenergic receptor (β2AR) [77]. In response, macrophages upregulate an M2‐like tissue protective and regenerative transcriptional program shielding neurons from damage [11]. In the steady state, myenteric neurons also secrete colony stimulatory factor 1 (CSF1), and muscularis macrophages in turn secrete bone morphogenetic protein 2 (BMP2). BMP2R signaling in myenteric neurons adjusts the pattern of smooth muscle contraction [78]. Thus, enteric macrophages and neurons cooperate to manage intestinal motility, homeostasis, and damage control in the steady state and during infection. Whereas ENS regulates the commensal microbiome composition by adjusting intestinal peristalsis [78], the number and function of ENS neurons are affected by the commensal microbiome [79]. This is exemplified by the downregulation of expression of neuropeptides associated with neuro‐immune crosstalk and ENS physiological function, such as neuromedin U, somatostatin, and Cocaine‐ and Amphetamine‐Regulated Transcript Protein (CARTP), in germ‐free (GF) or antibiotic‐treated mice. Concordantly, the numbers of intestinal ENS neurons are reduced in animals lacking a microbiome. Mechanistically, the microbiome promotes maintenance of ENS neurons via suppression of NLRP6‐ and caspase‐11‐mediated neuronal death [79].
In addition to gut intrinsic neurons, the intestine is innervated by different classes of extrinsic neurons, which detect and convey the information within the gut to the extrinsic ganglia and the CNS. Extrinsic ganglia, such as the sensory nodose ganglion and dorsal root ganglion, and the sympathetic coeliac‐superior mesenteric ganglia project neurons to the gut [80]. Complementary to the intrinsic ENS, gut‐extrinsic neurons are also involved in control of the mucosal immune system. For instance, transient receptor potential cation channel subfamily V member 1 (TRPV1)+ nociceptor neurons projected from the dorsal root ganglion respond to a Salmonella typhimurium infection by releasing a neuropeptide calcitonin gene‐related peptide (CGRP) and thus suppressing the differentiation of ileum M cells to block further entry of Salmonella [8]. The reduction in M cell density also leads to an increase in mucosal colonization by SFB, which competes with Salmonella for mucosal ecological niches, resulting in further limitation of Salmonella infection [8]. While extrinsic neurons regulate infections, their homeostasis is supported by the commensal microbiome. Indeed, the commensal microbiome plays an important role in preventing aberrant activation of the sympathetic neurons in the coeliac‐superior mesenteric ganglia [80]. In this preventive mechanism, nodose sensory neurons detect microbiome‐derived SCFAs and transmit suppressive signals to sympathetic neurons [80]. Epithelial cell subsets, particularly enteroendocrine cells, have also been implicated in transmitting luminal input to the intrinsic and extrinsic ENS. Mechanistically, enteroendocrine cells may communicate with the ENS directly via synapse‐like innervation [21] or via enteroendocrine hormone signaling in ENS neurons, as was recently predicted by a single cell survey of the mouse and human ENS [68]. This prediction was recently validated by the demonstration that glucagon‐like peptide 1 (GLP‐1) can activate gut sympathetic neurons [80]. Therefore, homeostatic communication in the intestine involves contributions by the gut microbiome, epithelium, ENS, CNS, and the immune system.
Epithelial and neuronal cells form the main sensory networks that perceive and interact with the microbiome. Similar to epithelial‐microbiome interaction programs, the immune reflexes of the ENS, both intrinsic and extrinsic, as well as of all levels of complexity function not solely for the benefit of the host but instead promote and rely on the health of the commensal microbiome. At steady state, intestinal peristalsis–crucial for both the microbiome and the host's homeostasis–requires sensing of the microbiome by enteric neurons [81]. During an infection that displaces commensals out of their ecological niches, ENS reflexes stimulate innate and adaptive immune responses to cleanse the pathogen, freeing the ecological niches for commensals to recolonize [76]. Commensals in turn produce metabolites vital for ENS homeostasis [80]. The commensal microbiome also promotes damage control programs within the ENS and CNS [11, 77], making the host more resilient to infections, reflecting the fact that the host's health is in the microbiome's best interest. Thus, the ENS and microbiome non‐zero‐sum interactions counter infections and promote intestinal homeostasis (Fig. 2A‐C).
Myeloid and innate lymphoid cells
The interaction between myeloid cells and the microbiome runs deep and starts before birth and continues throughout life. This is exemplified by the observation that antibiotic treatment of pregnant mice leads to a reduced number of neutrophils in their offspring, which could be prevented by microbial colonization of the pregnant mice [82]. Transitory colonization of pregnant female mice increases the number of ILC3s as well as F4/80+CD11c+ mononuclear cells in pups [83]. Furthermore, adult GF mice have a profound defect in myelopoiesis, characterized by a reduction of both yolk sac‐ and BM‐derived myeloid progenitors [84]. These defects can be restored by microbial recolonization, suggesting that healthy myelopoiesis requires continuous microbiome interaction throughout life rather than solely during the developmental stage [84]. Mechanistically, it has been proposed that the microbiome stimulates myelopoiesis via TLR signaling [85] and by producing metabolites such as SCFAs [84, 86]. In addition to influencing myelopoiesis, the microbiome drives maturation of neutrophils [87] and basophils [88]. The microbiome also affects the tissue‐specific adaptation of myeloid cells. The microglia of the brain of GF mice display an immature phenotype, which is phenocopied in SPF mice deficient for an SCFA receptor FFAR2 [89]. In the skin, the development of monocyte‐derived DCs depends on the presence of a skin microbiome [90]. The functional polarization of lung‐resident macrophages is modulated by the composition of the intestinal microbiome via prostaglandin E2 (PGE2) [91]. Similarly, the tolerogenic polarization of intestinal macrophages and DCs requires continuous SCFA‐mediated signaling [92, 93].
In contrast to the myeloid arm of the innate immune system, ILCs develop normally in number in the absence of the microbiota [94]. Instead, the microbiome regulates the maturation and tissue‐specific adaptation of ILCs [95, 96, 97]. ILCs consist of cytotoxic NK cells and non‐cytotoxic ILC1, ILC2, and ILC3 cells. ILC1s express the transcriptional factor T‐bet and produce IFN‐γ [98, 99, 100]. ILC2s are characterized by the expression of GATA3 and release IL‐5 and IL‐13 [101, 102, 103]. ILC3s express RORγt and secrete IL‐22 [104]. IFN‐γ expression by gut‐derived meningeal NK cells is induced by the gut microbiota [105]. Intestinal IFN‐γ+ NK cells migrate into the meninges and drive homeostatic expression of TRAIL by astrocytes, which limits inflammation in the CNS by inducing T cell apoptosis [105]. ILC1 cells are directly involved in mucosal defense against opportunistic pathogens such as Clostridioides difficile, as loss of ILC1, but not other ILC subsets or conventional T and B cells, rendered mice highly susceptible to infection with this organism [106]. Furthermore, the loss of T‐bet in Rag2‐deficient mice led to a spontaneous colitis that was mediated by pathobionts including Klebsiella pneumoniae, Proteus mirabilis and Helicobacter typhlonius [107, 108], suggesting that ILC1s are required for the management of specific members of the intestinal microbiome. ILC2s work in close coordination with tuft and goblet cells, promoting intestinal barrier function against bacteria and parasites. Following pathogen clearance, ILC2s‐mediated promotion of tuft and goblet cells expansion impairs future infections in a phenomenon termed concomitant immunity [25, 109]. Tuft cells sense parasites by detecting the helminth‐derived metabolite succinate and respond by producing IL‐25 [24, 25]. IL‐25 in turn activates ILC2s to produce the type 2 cytokine IL‐13, which helps to repel the infection and additionally stimulates tuft cells to promote feedforward production of IL‐25 [109]. ILC2s express TLR1, 4, and 6 [110], and the transcriptional profile of ILC2 cells changes upon microbial colonization [111]. However, further studies will be needed to clarify the issue of ILC2 plasticity and its dependency on the microbiome.
ILC3s are indispensable in integration of signals from the microbiome, IECs, ENS, as well as both the innate and adaptive arms of the immune system in order to promote mucosal homeostasis. ILC3s reinforce the intestinal barrier, promote immunological tolerance to commensals, and bolster colonization resistance to pathogens. Stimulated by the microbiome, myeloid and epithelial cells produce IL‐23 and IL‐1β [112, 113, 114]. In response, ILC3s release IL‐17 and IL‐22 in the steady state, and IFN‐γ and GM‐CSF under inflammatory conditions [115]. Cytokine release from ILC3s is drastically reduced in GF mice [95, 96, 116]. In addition to indirectly receiving microbial signals via myeloid and epithelial cells, ILC3s are capable of sensing the microbiome directly via the aryl hydrocarbon receptor (AhR) [117, 118, 119]. Regardless of the mode of ILC3 activation, ILC3‐derived IL‐22 has a profound reinforcing effect on the intestinal mucosal barrier. IL‐22 promotes ISC regeneration in the stem cell niche [120] and tight junction formation in the absorptive compartment of the epithelium [121]. IL‐22 also stimulates the release of AMPs, production of mucus [29, 122, 123], as well as fucosylation of the apical surface of IECs [12, 65]. Concordantly, mice deficient in ILC3s [124] or AhR [125] exhibit enhanced colonization of mucosal surfaces by SFB. In addition to AhR and IL‐23 signaling, IL‐22 production by ILC3s is stimulated by PGE2 [126]. Accordingly, PGE2 blockade results in an intestinal barrier failure and a systemic translocation by commensal microbes [126]. Furthermore, the ILC3‐mediated IL‐22 response can also be initiated by the ENS. The glial‐derived neurotrophic factor produced by enteric glial cells stimulates the RET tyrosine kinase receptor on ILC3s, resulting in the production of IL‐22 [127]. Moreover, ILC3s express vasoactive intestinal peptide receptor 2 and react to VIP produced by the ENS [9, 10]. As food consumption is a strong stimulator for the production of VIP from the ENS, the function of ILC3s is dynamically controlled in a coordinated manner and has a circadian pattern [9, 10]. On the other hand, microbiota‐stimulated ILC3s regulate the diurnal expression of the circadian transcription factor NFIL3 (also known as E4BP4) in IECs [128]. NFIL3 controls expression of a circadian lipid metabolic program and regulates lipid absorption in IECs [128]. In addition to their mucosal barrier reinforcing role via secretion of IL‐17 and IL‐22, ILC3s produce GM‐CSF and IL‐2, which contribute to the development of Treg cells and the maintenance of a tolerogenic state in the mucosal immune system [116, 129‐131]. Moreover, the CCR6+ subset of ILC3s is capable of antigen presentation to CD4 T cells [132] and iNKT cells [133]. MHC‐II expression by ILC3s leads to the control of mucosal IgA responses, thereby contributing to editing the intestinal microbial composition and sustaining its diversity [134]. The activity of ILC3s is also controlled by CD4 T cells. Without CD4 T cells, ILC3s exhibit extensive and persistent Stat3 activation and IL‐22 production, resulting in impaired host lipid metabolism by decreasing lipid transporter expression in the intestine [135]. Taken together, ILC populations interact with the microbiota and multiple host cells to contribute to mounting immune response against pathogens as well as shaping the mature commensal microbiome to help to maintain tissue homeostasis (Fig. 3A–E).
Bidirectional interactions between the adaptive immune system and the microbiome
It has long been known that the gut microbiome contributes both to the development and to set the tone of adaptive immune cells. This is exemplified by the overall diminished state of the adaptive immune system in GF mice, characterized by underdeveloped intestinal lymphoid tissue architecture [136], reductions in intraepithelial lymphocytes (IELs) [137], and in IgA‐producing B cells [138]. In addition, recent studies have shown that the microbiome directs the functional maturation of T cells, such as Th17, Treg, follicular helper T (Tfh), and CD8 T cells, and determines the gut antibody repertoire [139, 140, 141, 142]. The adaptive immune system in turn curates the composition of the microbiome, with the goal of sustaining maximal diversity thus promoting broad colonization resistance and preventing infections with pathogens. Therefore, the interaction between adaptive immune cells and the microbiome is bidirectional and mutualistic (Fig. 4).
Figure 4.
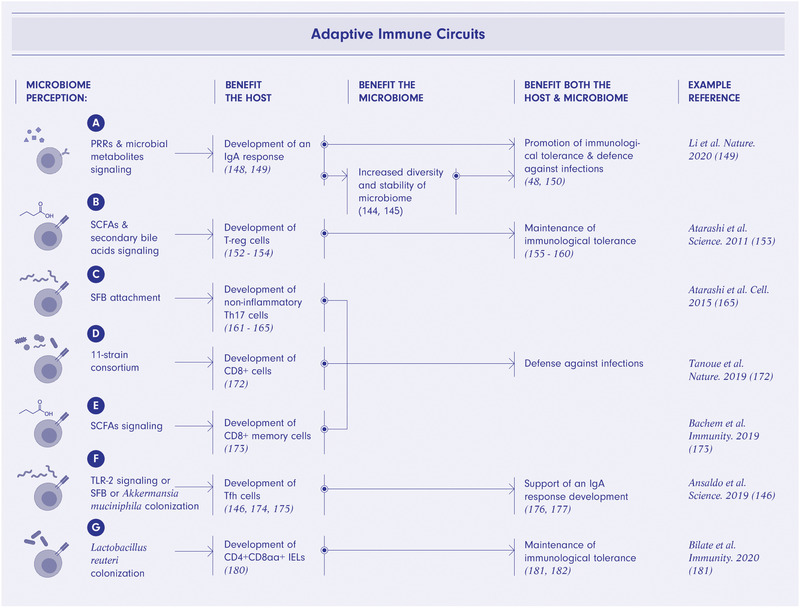
T and B cell‐microbiome mutually beneficial interactions. Microbial sensing via pattern recognition receptors signaling and microbial metabolites signaling promotes an IgA humoral response curating microbiome diversity, promoting immunological tolerance, and providing defense against infections (A). Short‐chain fatty acids (SCFAs) and secondary bile acids signaling promote differentiation of T regulatory cells ultimately bolstering immunological tolerance toward microbiome (B). Specific bacterial species such as segmented filamentous bacteria (SFB) or the 11‐strain consortium and SCFA signaling result in differentiation of Th17 (C), CD8+, (D) and memory CD8+ cells (E) respectively and provide defense of the host and microbiome from infections. TLR2 signaling or SFB or Akkermansia muciniphila colonization promotes the differentiation of Tfh cells that support development of an IgA response thus curating microbiome composition and benefiting the host and microbiome (F). Lactobacillus reuteri colonization promotes differentiation of tolerogenic CD4+CD8αα+ intraepithelial lymphocytes (IELs) thus enhancing immunological tolerance towards microbiome (G). The figure was created using BioRender.com.
B cells
B cells are curators of a healthy and diverse intestinal microbiome [143, 144]. Intestinal B lineage plasma cells produce several grams of a diverse array of secretory IgA per day [144, 145]. Although less effective as compared with IgA+ B cell responses, IgG1 responses specific to commensal species, such as Akkermansia muciniphila, can also be induced in the intestine [146]. In contrast to the T cell‐dependent IgG1 response, IgA‐producing cells develop in both T cell‐dependent and ‐independent ways. The diversity of the IgA repertoire, and antigen recognition possibilities, are increased by somatic hypermutation in germinal centers in a T cell‐dependent manner. The high diversity of the IgA repertoire helps to sustain high diversity of the microbial composition (Fig. 4A). Therefore, it has been considered that T cell‐dependent pathway is mostly responsible for the microbiome curatorship[144]. However, recent studies provide evidence that T cells do not substantially modulate the specificity of IgA [147, 148]. As IgA‐producing cells in tissues other than the small intestine are severely reduced in the absence of the microbiota and T cells, T cells are likely required more for the generation of extraintestinal IgA‐producing cell populations [148]. In any case, IgA‐producing cells in the intestinal and extraintestinal tissues typically show microbiota‐reactive and polyreactive specificities [148]. It has been shown that microbial exposures at the intestinal mucosa generate polyreactive but oligoclonal responses, different from the diverse IgG repertoire responses to systemic microbial exposures [149]. Moreover, priming thresholds differ depending on exposure routes; a higher dose exposure is needed to shape the mucosal IgA repertoire than that of systemic IgG [149]. As intestinal IgA induced by luminal microbes is predominantly specific to cell‐surface antigens [149], a broad, but distinct, subset of microbiota is bound by IgA antibodies in vivo. In particular, Enterobacteriaceae as well as mucus‐resident bacteria are heavily coated with IgA [48, 150]. IgA binds to commensal species and promotes their growth [144, 145]. On the other hand, IgA also induces agglutination, enchained growth, neutralization, and immune exclusion of microbes. Although it remains unclear what factors determine whether the IgA‐coating enhances or diminishes the fitness of microbes in the intestine, accumulating evidence indicates that IgA‐coating leads to alterations in microbial gene expression, motility, and/or spatial localization.
While microbes are required for induction of IgA, it appears that the opposite is true for IgE. IgE is elevated in the sera of GF mice and decreases upon colonization with commensal microbes [142]. GF mice begin to produce IgE shortly after weaning age to high levels and maintain these levels unless colonized with microbes within the first week of life.
T cells
Both circulating and tissue‐specific populations of CD4 T cells are influenced by the intestinal microbiome. This is exemplified by the observation that a significant proportion of circulating CD4 T cells are specific to intestinal microbes in the steady state [151]. Among gut‐resident CD4 T cells, several subsets depend on the presence of specific members of the microbiome. For instance, a subset of Treg cells depends on the metabolic activity of specific members of the gut microbiota [152, 153, 154]. In particular, microbes that ferment dietary fiber into SCFAs promote the development of Treg cells [153, 155‐157]. In addition, microbiome‐derived specific secondary bile acids can modulate the function and differentiation of Treg cells [158, 159, 160]. For example, isoallolithocholic acid (isoalloLCA) and isodeoxycholic acid (isoDCA) enhance the differentiation of naïve T cells into Treg cells [158, 159]. Mechanistically, vitamin D receptor and Nuclear Receptor Subfamily 1 Group H Member 4 [NR1H4, also known as Farnesoid X Receptor (FXR)] are implicated in recognition of secondary bile acids and regulation of gene expression in antigen presenting cells and/or T cells for Treg cell differentiation [158, 160].
Similarly, lamina propria resident Th17 cells are induced by specific bacteria [161, 162, 163, 164, 165]. In particular, Th17 cells are induced by intestinal epithelial adhesive microbes, such as SFB [162, 163, 164, 165], in a microbial antigen‐dependent manner [40, 166]. Interestingly, the functional profile of Th17 cells depends on bacterial stimuli. Th17 cells induced by SFB or other commensals are non‐inflammatory, while the opposite is true for the Th17 cells elicited by Citrobacter rodentium [167] or Helicobacter hepaticus in the condition of lacking IL‐10 [168]. Homeostatic Th17 cell differentiation is promoted by IL‐6 and TGF‐β produced by microbiome‐stimulated epithelial cells and gut resident DCs [162‐165, 169]. On the other hand, it has been shown that IEC‐derived serum amyloid A (SAA) proteins enhance differentiation of proinflammatory Th17 cells [170]. Epithelial cells express retinoic acid receptor β (RARβ), which activates vitamin A‐dependent expression of SAA proteins by binding directly to the Saa promoter [171]. Mirroring the development of CD4 T cells, IFNγ‐producing CD8 T cell differentiation is also promoted by specific commensal bacterial consortia [172]. Furthermore, the transition of antigen‐activated CD8 T cells into memory cells depends on microbiome‐derived SCFAs [173]. Another subset of T cells that depends on the microbiome is the Tfh cell, as its differentiation is impaired in GF mice but can be rescued by supplementation with TLR2 agonists [174] or colonization by SFB [175] or A. muciniphila [146]. In turn, Tfh cells indirectly influence the composition of the microbiome by participating in the induction of IgA‐mediated (and perhaps also IgG1‐mediated) microbiome selection [176, 177]. GF mice also feature an altered invariant natural killer T cell (iNKT) compartment. The iNKT cells from GF mice are hyporesponsive and have weaker effector functions after antigenic stimulation [178]. The microbiota also affects the distribution of iNKT cells. Whereas the frequency of iNKT cells in GF mice is lower in spleen and liver, it is increased in the intestines. The iNKT tissue distribution is established in early life and affects host sensitivity to Th2‐driven inflammation in later life [179]. Therefore, a time window exists within which the microbiota robustly affects the immune system. The development of CD4+CD8αα+ IELs is also affected by microbiota members, such as Lactobacillus reuteri [180]. The microbiota‐driven expression of MHC‐II and programmed death‐ligand 1 (PD‐L1) by IECs is required for the differentiation of CD4 T cells to become CD4+CD8αα+ IELs [181, 182].
The interactions of B and T cells with the microbiome provide perhaps the clearest example of non‐zero‐sum dynamics. These two compartments require microbial signals for development and activation and reciprocally promote microbiome homeostasis. In contrast to B cells, the contributions of the T cell compartment to microbiome homeostasis are indirect but broad. T cells cooperate with antigen‐presenting epithelial cells, myeloid cells, and ILCs to protect the host from infections and promote IgA curatorship for microbiome diversity (Fig. 4B‐G).
Conclusions
As the microbiome envelops all surfaces of the human body and mediates interactions with the environment, it is perhaps not surprising that it is involved in health and disease of virtually all organ systems. The microbiome participates in the maintenance of tissue homeostasis through its direct contact at the gastrointestinal [183, 184], respiratory [185], urogenital [186], and skin surfaces [187]. In addition, the microbiota produces a wide variety of molecules that can reach millimolar concentrations in the blood [188, 189, 190, 191, 192], thereby influencing remote organ systems such as the central and peripheral nervous systems [193], liver [194], adipose tissue [195, 196], skeletal muscles [197], and the cardio‐vascular system [198, 199]. Although the immune system no doubt serves as a major conduit of microbiome influences across multiple organs, major knowledge gaps still remain. The concept of microbiome‐immune non‐zero‐sum interactions based on studies of host–microbiome interactions at the intestine is likely to be applicable across multiple organ systems and therefore may help guide formulation of hypotheses to be tested in future studies. Innate and adaptive immunological programs as well as IECs and ENS that evolved to support host‐microbiome mutualism in the intestine may have counterparts in other organ systems that interact with microbiome directly and indirectly via immune system and microbiome‐derived metabolites.
Another avenue of promising future research lies in utilizing the concept of immune system–microbiome mutualism to better define what constitutes a healthy microbiome (Fig. 5). Defining a healthy microbiome is a long‐term goal of the field of microbiome research and will require to model the dynamics underpinning the seemingly endless compositional variability between individuals. Existing models of microbiome ecology successfully describe select aspects of bacteria‐to‐bacteria interactions by mapping limits of selfish behavior in a network [200], modeling an effect of the ratio of cooperative to competitive interactions on diversity and stability [201], applying game theory to metabolic interdependencies between microbes [15] and strategies of nutrients utilization [202], calculating an effect of cross‐feeding on colonization resistance [203], studying the relationship between nutrients availability and diversity [204], and defining rules of assembly of infant microbiome [205]. However, by design these models are limited to bacteria‐to‐bacteria interactions. Studies reviewed herein and elsewhere demonstrate that a healthy microbiome is the result of active host immune system curatorship as well as the outcome of microbial ecology. Incorporation of microbiome‐editing programs by innate and adaptive immune cells, IECs, as well as ENS will be an important step towards development of a model of healthy human microbiome (Fig. 5).
Figure 5.
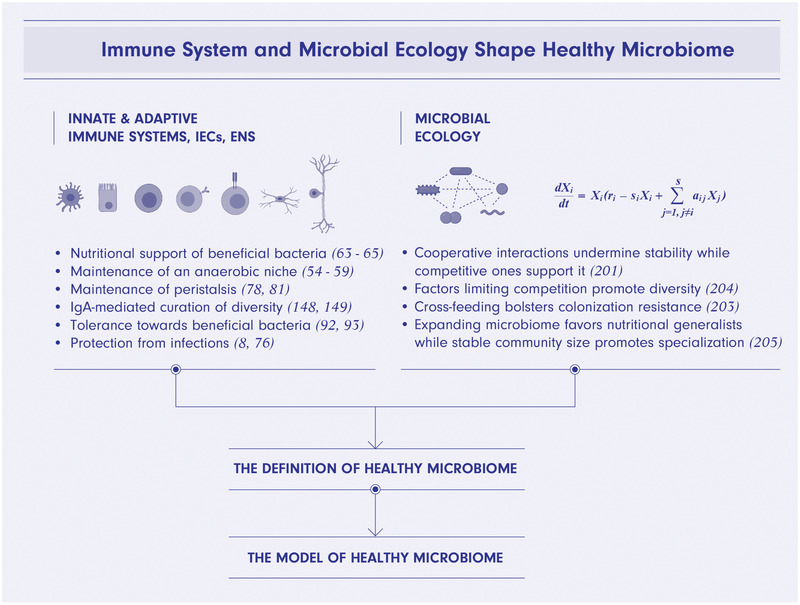
Immune system, IECs, ENS, and microbial ecology shape healthy microbiome. Integration of microbiome‐curating programs of immune system, intestinal epithelial cells (IECs) and enteric nervous system (ENS) and microbial ecology will be a step forward toward defining and modeling healthy microbiome. IECs provide nutritional support and maintain an anaerobic niche for beneficial bacteria. ENS sustains peristalsis providing microbiome with a nutrients flow and disposing of waste products. B‐cells secrete bacteria‐targeting IgA antibodies curating microbiome composition with the overall effect of increased diversity and stability. Myeloid cells, T regulatory cells as well as other cell types establish the state of immunological tolerance towards commensal microbes. Cells of both innate as well as adaptive arms of immune system protect the host and microbiome from infections. Known patterns of healthy microbiome's ecology include dominance of competitive interactions over cooperative, high diversity supported by external factors, cross‐feeding, and succession of nutrients utilization strategies from generalists in neonatal microbiome to mature communities dominated by specialists. The figure was created using BioRender.com.
Conflict of interest
K.H. is a scientific advisory board member of Vedanta Biosciences and 4BIO CAPITAL. T.T has no commercial or financial conflict of interests.
Abbreviations
- AhR
aryl hydrocarbon receptor
- AMP
antimicrobial peptide
- ENS
enteric nervous system
- IEC
intestinal epithelial cell
- IEL
intraepithelial lymphocyte
- ILC
innate lymphoid cell
- ISC
intestinal stem cell
- PGE2
prostaglandin E2
- SCFA
short‐chain fatty acid
- Tfh
follicular helper T cells
- Treg
regulatory T cell
Acknowledgments
We thank Peter D. Burrows for comments and Maria Eizner of EiznerDesign.com for graphic design. Kenya Honda was funded through AMED LEAP under grant number JP20gm0010003, Grant‐in‐Aid for Specially Promoted Research from the JSPS (No: 20H05627), the Public/Private R&D Investment Strategic Expansion Program (PRISM) from the Cabinet Office of the Government of Japan, the Naito Foundation, and the Takeda Science Foundation.
Contributor Information
Timur Tuganbaev, Email: tuganbaev.timur@keio.jp.
Kenya Honda, Email: kenya@keio.jp.
References
- 1.Tierney, B. T., Yang, Z., Luber, J. M., Beaudin, M., Wibowo, M. C., Baek, C., Mehlenbacher, E. et al., The landscape of genetic content in the gut and oral human microbiome. Cell Host Microbe. 2019. 26: 283–295 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pertea, M., Shumate, A., Pertea, G., Varabyou, A., Breitwieser, F. P., Chang, Y. C., Madugundu, A. K. et al., CHESS: a new human gene catalog curated from thousands of large‐scale RNA sequencing experiments reveals extensive transcriptional noise. Genome Biol.. 2018. 19: 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yilmaz, B., Mooser, C., Keller, I., Li, H., Zimmermann, J., Bosshard, L., Fuhrer, T. et al., Long‐term evolution and short‐term adaptation of microbiota strains and sub‐strains in mice. Cell Host Microbe. 2021. 29: 650–663 e9. [DOI] [PubMed] [Google Scholar]
- 4.Good, B. H., McDonald, M. J., Barrick, J. E., Lenski, R. E. and Desai, M. M., The dynamics of molecular evolution over 60,000 generations. Nature. 2017. 551: 45–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergstrom, K., Fu, J., Johansson, M. E., Liu, X., Gao, N., Wu, Q., Song, J. et al., Core 1‐ and 3‐derived O‐glycans collectively maintain the colonic mucus barrier and protect against spontaneous colitis in mice. Mucosal Immunol. 2017. 10: 91–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johansson, M. E., Phillipson, M., Petersson, J., Velcich, A., Holm, L. and Hansson, G. C., The inner of the two Muc2 mucin‐dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A. 2008. 105: 15064–15069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Post, S., Jabbar, K. S., Birchenough, G., Arike, L., Akhtar, N., Sjovall, H. et al., Structural weakening of the colonic mucus barrier is an early event in ulcerative colitis pathogenesis. Gut. 2019. 68: 2142–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lai, N. Y., Musser, M. A., Pinho‐Ribeiro, F. A., Baral, P., Jacobson, A., Ma, P., Potts, D. E. et al., Gut‐Innervating Nociceptor Neurons Regulate Peyer's Patch Microfold Cells and SFB Levels to Mediate Salmonella Host Defense. Cell. 2020. 180: 33–49 e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seillet, C., Luong, K., Tellier, J., Jacquelot, N., Shen, R. D., Hickey, P., Wimmer, V. C. et al., The neuropeptide VIP confers anticipatory mucosal immunity by regulating ILC3 activity. Nat. Immunol.. 2020. 21: 168–177. [DOI] [PubMed] [Google Scholar]
- 10.Talbot, J., Hahn, P., Kroehling, L., Nguyen, H., Li, D. and Littman, D. R., Feeding‐dependent VIP neuron‐ILC3 circuit regulates the intestinal barrier. Nature. 2020. 579: 575–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gabanyi, I., Muller, P. A., Feighery, L., Oliveira, T. Y., Costa‐Pinto, F. A. and Mucida, D., Neuro‐immune Interactions Drive Tissue Programming in Intestinal Macrophages. Cell. 2016. 164: 378–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pickard, J. M., Maurice, C. F., Kinnebrew, M. A., Abt, M. C., Schenten, D., Golovkina, T. V., Bogatyrev, S. R. et al., Rapid fucosylation of intestinal epithelium sustains host‐commensal symbiosis in sickness. Nature. 2014. 514: 638–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sorbara, M. T. and Pamer, E. G., Interbacterial mechanisms of colonization resistance and the strategies pathogens use to overcome them. Mucosal Immunol. 2019. 12: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Von Neumann, J. and Morgenstern, O., Theory of games and economic behavior (60th Anniversary Commemorative Edition). Princeton university press; 2007. [Google Scholar]
- 15.Zomorrodi, A. R. and Segre, D., Genome‐driven evolutionary game theory helps understand the rise of metabolic interdependencies in microbial communities. Nat. Commun.. 2017. 8: 1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wright, R., Nonzero: The logic of human destiny: Vintage; 2001.
- 17.O'Hara, A. M. and Shanahan, F., The gut flora as a forgotten organ. EMBO Rep.. 2006. 7: 688–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clevers, H. C. and Bevins, C. L., Paneth cells: maestros of the small intestinal crypts. Annu. Rev. Physiol.. 2013. 75: 289–311. [DOI] [PubMed] [Google Scholar]
- 19.Haber, A. L., Biton, M., Rogel, N., Herbst, R. H., Shekhar, K., Smillie, C., Burgin, G. et al., A single‐cell survey of the small intestinal epithelium. Nature. 2017. 551: 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gehart, H. and Clevers, H., Tales from the crypt: new insights into intestinal stem cells. Nat. Rev. Gastroenterol. Hepatol.. 2019. 16: 19–34. [DOI] [PubMed] [Google Scholar]
- 21.Latorre, R., Sternini, C., De Giorgio, R. and Greenwood‐Van Meerveld, B., Enteroendocrine cells: a review of their role in brain‐gut communication. Neurogastroenterol. Motil.. 2016. 28: 620–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lei, W., Ren, W., Ohmoto, M., Urban, J. F., Jr., Matsumoto, I., Margolskee, R. F. and Jiang, P., Activation of intestinal tuft cell‐expressed Sucnr1 triggers type 2 immunity in the mouse small intestine. Proc. Natl. Acad. Sci. U. S. A.. 2018. 115: 5552–5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, Q., Ma, L., Shen, S., Guo, Y., Cao, Q., Cai, X., Feng, J. et al., Intestinal dysbacteriosis‐induced IL‐25 promotes development of HCC via alternative activation of macrophages in tumor microenvironment. J. Exp. Clin. Cancer Res.. 2019. 38: 303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nadjsombati, M. S., McGinty, J. W., Lyons‐Cohen, M. R., Jaffe, J. B., DiPeso, L., Schneider, C., Miller, C. N. et al., Detection of Succinate by Intestinal Tuft Cells Triggers a Type 2 Innate Immune Circuit. Immunity. 2018. 49: 33–41 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schneider, C., O'Leary, C. E., von Moltke, J., Liang, H. E., Ang, Q. Y., Turnbaugh, P. J., Radhakrishnan, S. et al., A Metabolite‐Triggered Tuft Cell‐ILC2 Circuit Drives Small Intestinal Remodeling. Cell. 2018. 174: 271–284 e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Birchenough, G. M., Nystrom, E. E., Johansson, M. E. and Hansson, G. C., A sentinel goblet cell guards the colonic crypt by triggering Nlrp6‐dependent Muc2 secretion. Science. 2016. 352: 1535–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rios, D., Wood, M. B., Li, J., Chassaing, B., Gewirtz, A. T. and Williams, I. R., Antigen sampling by intestinal M cells is the principal pathway initiating mucosal IgA production to commensal enteric bacteria. Mucosal Immunol. 2016. 9: 907–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jung, C., Hugot, J. P., Barreau, F., Peyer's Patches: The Immune Sensors of the Intestine. Int J Inflam. 2010. 2010: 823710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vaishnava, S., Yamamoto, M., Severson, K. M., Ruhn, K. A., Yu, X., Koren, O., Ley, R. et al., The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011. 334: 255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nenci, A., Becker, C., Wullaert, A., Gareus, R., van Loo, G., Danese, S., Huth, M. et al., Epithelial NEMO links innate immunity to chronic intestinal inflammation. Nature. 2007. 446: 557–561. [DOI] [PubMed] [Google Scholar]
- 31.Vlantis, K., Polykratis, A., Welz, P. S., van Loo, G., Pasparakis, M. and Wullaert, A., TLR‐independent anti‐inflammatory function of intestinal epithelial TRAF6 signalling prevents DSS‐induced colitis in mice. Gut. 2016. 65: 935–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Couturier‐Maillard, A., Secher, T., Rehman, A., Normand, S., De Arcangelis, A., Haesler, R., Huot, L. et al., NOD2‐mediated dysbiosis predisposes mice to transmissible colitis and colorectal cancer. J. Clin. Invest.. 2013. 123: 700–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramanan, D., Tang, M. S., Bowcutt, R., Loke, P., Cadwell, K., Bacterial sensor Nod2 prevents inflammation of the small intestine by restricting the expansion of the commensal Bacteroides vulgatus. Immunity. 2014. 41: 311–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu, S., Peng, L., Kwak, Y. T., Tekippe, E. M., Pasare, C., Malter, J. S., Hooper, L. V. et al., The DNA Sensor AIM2 Maintains Intestinal Homeostasis via Regulation of Epithelial Antimicrobial Host Defense. Cell Rep.. 2015. 13: 1922–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song‐Zhao, G. X., Srinivasan, N., Pott, J., Baban, D., Frankel, G. and Maloy, K. J., Nlrp3 activation in the intestinal epithelium protects against a mucosal pathogen. Mucosal Immunol. 2014. 7: 763–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wlodarska, M., Thaiss, C. A., Nowarski, R., Henao‐Mejia, J., Zhang, J. P., Brown, E. M., Frankel, G. et al., NLRP6 inflammasome orchestrates the colonic host‐microbial interface by regulating goblet cell mucus secretion. Cell. 2014. 156: 1045–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levy, M., Thaiss, C. A., Zeevi, D., Dohnalova, L., Zilberman‐Schapira, G., Mahdi, J. A., David, E. et al., Microbiota‐Modulated Metabolites Shape the Intestinal Microenvironment by Regulating NLRP6 Inflammasome Signaling. Cell. 2015. 163: 1428–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buning, J., Schmitz, M., Repenning, B., Ludwig, D., Schmidt, M. A., Strobel, S. and Zimmer, K. P., Interferon‐gamma mediates antigen trafficking to MHC class II‐positive late endosomes of enterocytes. Eur. J. Immunol.. 2005. 35: 831–842. [DOI] [PubMed] [Google Scholar]
- 39.Gonnella, P. A. and Wilmore, D. W., Co‐localization of class II antigen and exogenous antigen in the rat enterocyte. J. Cell Sci.. 1993. 106: 937–940. [DOI] [PubMed] [Google Scholar]
- 40.Ladinsky, M. S., Araujo, L. P., Zhang, X., Veltri, J., Galan‐Diez, M., Soualhi, S., Lee, C. et al., Endocytosis of commensal antigens by intestinal epithelial cells regulates mucosal T cell homeostasis. Science. 2019. 363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hershberg, R. M., Framson, P. E., Cho, D. H., Lee, L. Y., Kovats, S., Beitz, J., Blum, J. S. et al., Intestinal epithelial cells use two distinct pathways for HLA class II antigen processing. J. Clin. Invest. 1997. 100: 204–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Westendorf, A. M., Fleissner, D., Groebe, L., Jung, S., Gruber, A. D., Hansen, W. and Buer, J., CD4+Foxp3+ regulatory T cell expansion induced by antigen‐driven interaction with intestinal epithelial cells independent of local dendritic cells. Gut. 2009. 58: 211–219. [DOI] [PubMed] [Google Scholar]
- 43.Koyama, M., Mukhopadhyay, P., Schuster, I. S., Henden, A. S., Hulsdunker, J., Varelias, A., Vetizou, M. et al., MHC Class II Antigen Presentation by the Intestinal Epithelium Initiates Graft‐versus‐Host Disease and Is Influenced by the Microbiota. Immunity. 2019. 51: 885–898 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Biton, M., Haber, A. L., Rogel, N., Burgin, G., Beyaz, S., Schnell, A., Ashenberg, O. et al., T Helper Cell Cytokines Modulate Intestinal Stem Cell Renewal and Differentiation. Cell. 2018. 175: 1307–1320 e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tuganbaev, T., Mor, U., Bashiardes, S., Liwinski, T., Nobs, S. P., Leshem, A., Dori‐Bachash, M. et al., Diet Diurnally Regulates Small Intestinal Microbiome‐Epithelial‐Immune Homeostasis and Enteritis. Cell. 2020. 182: 1441–1459 e21. [DOI] [PubMed] [Google Scholar]
- 46.Thaiss, C. A., Zeevi, D., Levy, M., Zilberman‐Schapira, G., Suez, J., Tengeler, A. C., Abramson, L. et al., Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell. 2014. 159: 514–529. [DOI] [PubMed] [Google Scholar]
- 47.Zarrinpar, A., Chaix, A., Yooseph, S. and Panda, S., Diet and feeding pattern affect the diurnal dynamics of the gut microbiome. Cell Metab. 2014. 20: 1006–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Palm, N. W., de Zoete, M. R., Cullen, T. W., Barry, N. A., Stefanowski, J., Hao, L., Degnan, P. H. et al., Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell. 2014. 158: 1000–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakamura, Y., Mimuro, H., Kunisawa, J., Furusawa, Y., Takahashi, D., Fujimura, Y., Kaisho, T. et al., Microfold cell‐dependent antigen transport alleviates infectious colitis by inducing antigen‐specific cellular immunity. Mucosal Immunol. 2020. 13: 679–690. [DOI] [PubMed] [Google Scholar]
- 50.McDole, J. R., Wheeler, L. W., McDonald, K. G., Wang, B., Konjufca, V., Knoop, K. A., Newberry, R. D. et al., Goblet cells deliver luminal antigen to CD103+ dendritic cells in the small intestine. Nature. 2012. 483: 345–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kulkarni, D. H., McDonald, K. G., Knoop, K. A., Gustafsson, J. K., Kozlowski, K. M., Hunstad, D. A., Miller, M. J. et al., Goblet cell associated antigen passages are inhibited during Salmonella typhimurium infection to prevent pathogen dissemination and limit responses to dietary antigens. Mucosal Immunol. 2018. 11: 1103–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Byndloss, M. X., Olsan, E. E., Rivera‐Chavez, F., Tiffany, C. R., Cevallos, S. A., Lokken, K. L., Torres, T. P. et al., Microbiota‐activated PPAR‐gamma signaling inhibits dysbiotic Enterobacteriaceae expansion. Science. 2017. 357: 570–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tan, J., McKenzie, C., Potamitis, M., Thorburn, A. N., Mackay, C. R. and Macia, L., The role of short‐chain fatty acids in health and disease. Adv. Immunol. 2014. 121: 91–119. [DOI] [PubMed] [Google Scholar]
- 54.Duszka, K., Oresic, M., Le May, C., Konig, J. and Wahli, W., PPARgamma Modulates Long Chain Fatty Acid Processing in the Intestinal Epithelium. Int. J. Mol. Sci. 2017. 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hughes, E. R., Winter, M. G., Duerkop, B. A., Spiga, L., Furtado de Carvalho, T., Zhu, W., Gillis, C. C. et al., Microbial Respiration and Formate Oxidation as Metabolic Signatures of Inflammation‐Associated Dysbiosis. Cell Host Microbe. 2017. 21: 208–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kelly, C. J., Zheng, L., Campbell, E. L., Saeedi, B., Scholz, C. C., Bayless, A. J., Wilson, K. E. et al., Crosstalk between Microbiota‐Derived Short‐Chain Fatty Acids and Intestinal Epithelial HIF Augments Tissue Barrier Function. Cell Host Microbe. 2015. 17: 662–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Litvak, Y., Byndloss, M. X. and Baumler, A. J., Colonocyte metabolism shapes the gut microbiota. Science. 2018. 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Winter, S. E., Winter, M. G., Xavier, M. N., Thiennimitr, P., Poon, V., Keestra, A. M., Laughlin, R. C. et al., Host‐derived nitrate boosts growth of E. coli in the inflamed gut. Science. 2013. 339: 708–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lopez, C. A., Miller, B. M., Rivera‐Chavez, F., Velazquez, E. M., Byndloss, M. X., Chavez‐Arroyo, A., Lokken, K. L. et al., Virulence factors enhance Citrobacter rodentium expansion through aerobic respiration. Science. 2016. 353: 1249–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Araujo, J. R., Tazi, A., Burlen‐Defranoux, O., Vichier‐Guerre, S., Nigro, G., Licandro, H., Demignot, S. et al., Fermentation Products of Commensal Bacteria Alter Enterocyte Lipid Metabolism. Cell Host Microbe. 2020. 27: 358–375 e7. [DOI] [PubMed] [Google Scholar]
- 61.Morita, N., Umemoto, E., Fujita, S., Hayashi, A., Kikuta, J., Kimura, I., Haneda, T. et al., GPR31‐dependent dendrite protrusion of intestinal CX3CR1(+) cells by bacterial metabolites. Nature. 2019. 566: 110–114. [DOI] [PubMed] [Google Scholar]
- 62.Lee, Y. S., Kim, T. Y., Kim, Y., Lee, S. H., Kim, S., Kang, S. W., Yang, J. Y. et al., Microbiota‐Derived Lactate Accelerates Intestinal Stem‐Cell‐Mediated Epithelial Development. Cell Host Microbe. 2018. 24: 833–846 e6. [DOI] [PubMed] [Google Scholar]
- 63.Derrien, M., Vaughan, E. E., Plugge, C. M. and de Vos, W. M., Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin‐degrading bacterium. Int. J. Syst. Evol. Microbiol. 2004. 54: 1469–1476. [DOI] [PubMed] [Google Scholar]
- 64.Bell, A., Brunt, J., Crost, E., Vaux, L., Nepravishta, R., Owen, C. D., Latousakis, D. et al., Elucidation of a sialic acid metabolism pathway in mucus‐foraging Ruminococcus gnavus unravels mechanisms of bacterial adaptation to the gut. Nat Microbiol. 2019. 4: 2393–2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goto, Y., Obata, T., Kunisawa, J., Sato, S., Ivanov, I. I., Lamichhane, A., Takeyama, N. et al., Innate lymphoid cells regulate intestinal epithelial cell glycosylation. Science. 2014. 345: 1254009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zuniga, M., Monedero, V. and Yebra, M. J., Utilization of Host‐Derived Glycans by Intestinal Lactobacillus and Bifidobacterium Species. Front Microbiol. 2018. 9: 1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yoo, B. B. and Mazmanian, S. K., The Enteric Network: Interactions between the Immune and Nervous Systems of the Gut. Immunity. 2017. 46: 910–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Drokhlyansky, E., Smillie, C. S., Van Wittenberghe, N., Ericsson, M., Griffin, G. K., Eraslan, G., Dionne, D. et al., The Human and Mouse Enteric Nervous System at Single‐Cell Resolution. Cell. 2020. 182: 1606–1622 e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Furness, J. B., The enteric nervous system and neurogastroenterology. Nat. Rev. Gastroenterol. Hepatol. 2012. 9: 286–294. [DOI] [PubMed] [Google Scholar]
- 70.Swenson, O., Hirschsprung's disease: a review. Pediatrics. 2002. 109: 914–918. [DOI] [PubMed] [Google Scholar]
- 71.Matsuda, N. M., Miller, S. M. and Evora, P. R., The chronic gastrointestinal manifestations of Chagas disease. Clinics (Sao Paulo). 2009. 64: 1219–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fung, C. and Vanden Berghe, P., Functional circuits and signal processing in the enteric nervous system. Cellular and Molecular Life Sciences. 2020. 77: 4505–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Furness, J. B., Callaghan, B. P., Rivera, L. R. and Cho, H. J., The enteric nervous system and gastrointestinal innervation: integrated local and central control. Adv. Exp. Med. Biol. 2014. 817: 39–71. [DOI] [PubMed] [Google Scholar]
- 74.Zeisel, A., Hochgerner, H., Lonnerberg, P., Johnsson, A., Memic, F., van der Zwan, J., Haring, M. et al., Molecular Architecture of the Mouse Nervous System. Cell. 2018. 174: 999–1014 e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cardoso, V., Chesne, J., Ribeiro, H., Garcia‐Cassani, B., Carvalho, T., Bouchery, T., Shah, K. et al., Neuronal regulation of type 2 innate lymphoid cells via neuromedin U. Nature. 2017. 549: 277–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jarret, A., Jackson, R., Duizer, C., Healy, M. E., Zhao, J., Rone, J. M., Bielecki, P. et al., Enteric Nervous System‐Derived IL‐18 Orchestrates Mucosal Barrier Immunity. Cell. 2020. 180: 50–63 e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Matheis, F., Muller, P. A., Graves, C. L., Gabanyi, I., Kerner, Z. J., Costa‐Borges, D., Ahrends, T. et al., Adrenergic Signaling in Muscularis Macrophages Limits Infection‐Induced Neuronal Loss. Cell. 2020. 180: 64–78 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Muller, P. A., Koscso, B., Rajani, G. M., Stevanovic, K., Berres, M. L., Hashimoto, D., Mortha, A. et al., Crosstalk between muscularis macrophages and enteric neurons regulates gastrointestinal motility. Cell. 2014. 158: 300–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Muller, P. A., Matheis, F., Schneeberger, M., Kerner, Z., Jove, V. and Mucida, D., Microbiota‐modulated CART(+) enteric neurons autonomously regulate blood glucose. Science. 2020. 370: 314–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Muller, P. A., Schneeberger, M., Matheis, F., Wang, P., Kerner, Z., Ilanges, A., Pellegrino, K. et al., Microbiota modulate sympathetic neurons via a gut‐brain circuit. Nature. 2020. 583: 441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Obata, Y., Á, C., Boeing, S., Bon‐Frauches, A. C., Fung, C., Fallesen, T., de Agüero, M. G. et al., Neuronal programming by microbiota regulates intestinal physiology. Nature. 2020. 578: 284–289. [DOI] [PubMed] [Google Scholar]
- 82.Deshmukh, H. S., Liu, Y., Menkiti, O. R., Mei, J., Dai, N., O'Leary, C. E., Oliver, P. M. et al., The microbiota regulates neutrophil homeostasis and host resistance to Escherichia coli K1 sepsis in neonatal mice. Nat. Med. 2014. 20: 524–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gomez de Agüero, M., Ganal‐Vonarburg, S. C., Fuhrer, T., Rupp, S., Uchimura, Y., Li, H., Steinert, A. et al., The maternal microbiota drives early postnatal innate immune development. Science. 2016. 351: 1296–1302. [DOI] [PubMed] [Google Scholar]
- 84.Khosravi, A., Yanez, A., Price, J. G., Chow, A., Merad, M., Goodridge, H. S. and Mazmanian, S. K., Gut microbiota promote hematopoiesis to control bacterial infection. Cell Host Microbe. 2014. 15: 374–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Balmer, M. L., Schurch, C. M., Saito, Y., Geuking, M. B., Li, H., Cuenca, M., Kovtonyuk, L. V. et al., Microbiota‐derived compounds drive steady‐state granulopoiesis via MyD88/TICAM signaling. J. Immunol. 2014. 193: 5273–5283. [DOI] [PubMed] [Google Scholar]
- 86.Trompette, A., Gollwitzer, E. S., Yadava, K., Sichelstiel, A. K., Sprenger, N., Ngom‐Bru, C., Blanchard, C. et al., Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 2014. 20: 159–166. [DOI] [PubMed] [Google Scholar]
- 87.Zhang, D., Chen, G., Manwani, D., Mortha, A., Xu, C., Faith, J. J., Burk, R. D. et al., Neutrophil ageing is regulated by the microbiome. Nature. 2015. 525: 528–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hill, D. A., Siracusa, M. C., Abt, M. C., Kim, B. S., Kobuley, D., Kubo, M., Kambayashi, T. et al., Commensal bacteria‐derived signals regulate basophil hematopoiesis and allergic inflammation. Nat. Med. 2012. 18: 538–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Erny, D., Hrabe de Angelis, A. L., Jaitin, D., Wieghofer, P., Staszewski, O., David, E., Keren‐Shaul, H. et al., Host microbiota constantly control maturation and function of microglia in the CNS. Nat. Neurosci. 2015. 18: 965–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tamoutounour, S., Guilliams, M., Montanana Sanchis, F., Liu, H., Terhorst, D., Malosse, C., Pollet, E. et al., Origins and functional specialization of macrophages and of conventional and monocyte‐derived dendritic cells in mouse skin. Immunity. 2013. 39: 925–938. [DOI] [PubMed] [Google Scholar]
- 91.Kim, Y. G., Udayanga, K. G., Totsuka, N., Weinberg, J. B., Nunez, G. and Shibuya, A., Gut dysbiosis promotes M2 macrophage polarization and allergic airway inflammation via fungi‐induced PGE(2). Cell Host Microbe. 2014. 15: 95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chang, P. V., Hao, L., Offermanns, S. and Medzhitov, R., The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci U S A. 2014. 111: 2247–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Singh, N., Gurav, A., Sivaprakasam, S., Brady, E., Padia, R., Shi, H., Thangaraju, M. et al., Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity. 2014. 40: 128–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sawa, S., Cherrier, M., Lochner, M., Satoh‐Takayama, N., Fehling, H. J., Langa, F., Di Santo, J. P. et al., Lineage relationship analysis of RORgammat+ innate lymphoid cells. Science. 2010. 330: 665–669. [DOI] [PubMed] [Google Scholar]
- 95.Sanos, S. L., Bui, V. L., Mortha, A., Oberle, K., Heners, C., Johner, C. and Diefenbach, A., RORgammat and commensal microflora are required for the differentiation of mucosal interleukin 22‐producing NKp46+ cells. Nat. Immunol. 2009. 10: 83–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Satoh‐Takayama, N., Vosshenrich, C. A., Lesjean‐Pottier, S., Sawa, S., Lochner, M., Rattis, F., Mention, J. J. et al., Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity. 2008. 29: 958–970. [DOI] [PubMed] [Google Scholar]
- 97.Sawa, S., Lochner, M., Satoh‐Takayama, N., Dulauroy, S., Berard, M., Kleinschek, M., Cua, D. et al., RORgammat+ innate lymphoid cells regulate intestinal homeostasis by integrating negative signals from the symbiotic microbiota. Nat. Immunol. 2011. 12: 320–326. [DOI] [PubMed] [Google Scholar]
- 98.Klose, C. S. N., Flach, M., Mohle, L., Rogell, L., Hoyler, T., Ebert, K., Fabiunke, C. et al., Differentiation of type 1 ILCs from a common progenitor to all helper‐like innate lymphoid cell lineages. Cell. 2014. 157: 340–356. [DOI] [PubMed] [Google Scholar]
- 99.Bernink, J. H., Peters, C. P., Munneke, M., te Velde, A. A., Meijer, S. L., Weijer, K., Hreggvidsdottir, H. S. et al., Human type 1 innate lymphoid cells accumulate in inflamed mucosal tissues. Nat. Immunol. 2013. 14: 221–229. [DOI] [PubMed] [Google Scholar]
- 100.Fuchs, A., Vermi, W., Lee, J. S., Lonardi, S., Gilfillan, S., Newberry, R. D., Cella, M. et al., Intraepithelial type 1 innate lymphoid cells are a unique subset of IL‐12‐ and IL‐15‐responsive IFN‐gamma‐producing cells. Immunity. 2013. 38: 769–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mjosberg, J., Bernink, J., Golebski, K., Karrich, J. J., Peters, C. P., Blom, B., te Velde, A. A. et al., The transcription factor GATA3 is essential for the function of human type 2 innate lymphoid cells. Immunity. 2012. 37: 649–659. [DOI] [PubMed] [Google Scholar]
- 102.Roediger, B., Kyle, R., Yip, K. H., Sumaria, N., Guy, T. V., Kim, B. S., Mitchell, A. J. et al., Cutaneous immunosurveillance and regulation of inflammation by group 2 innate lymphoid cells. Nat. Immunol. 2013. 14: 564–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hoyler, T., Klose, C. S., Souabni, A., Turqueti‐Neves, A., Pfeifer, D., Rawlins, E. L., Voehringer, D. et al., The transcription factor GATA‐3 controls cell fate and maintenance of type 2 innate lymphoid cells. Immunity. 2012. 37: 634–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cording, S., Medvedovic, J., Cherrier, M. and Eberl, G., Development and regulation of RORgammat(+) innate lymphoid cells. FEBS Lett. 2014. 588: 4176–4181. [DOI] [PubMed] [Google Scholar]
- 105.Sanmarco, L. M., Wheeler, M. A., Gutierrez‐Vazquez, C., Polonio, C. M., Linnerbauer, M., Pinho‐Ribeiro, F. A., Li, Z. et al., Gut‐licensed IFNgamma(+) NK cells drive LAMP1(+)TRAIL(+) anti‐inflammatory astrocytes. Nature. 2021. 590: 473–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Abt, M. C., Lewis, B. B., Caballero, S., Xiong, H., Carter, R. A., Susac, B., Ling, L. et al., Innate Immune Defenses Mediated by Two ILC Subsets Are Critical for Protection against Acute Clostridium difficile Infection. Cell Host Microbe. 2015. 18: 27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Garrett, W. S., Gallini, C. A., Yatsunenko, T., Michaud, M., DuBois, A., Delaney, M. L., Punit, S. et al., Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010. 8: 292–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Powell, N., Walker, A. W., Stolarczyk, E., Canavan, J. B., Gokmen, M. R., Marks, E., Jackson, I. et al., The transcription factor T‐bet regulates intestinal inflammation mediated by interleukin‐7 receptor+ innate lymphoid cells. Immunity. 2012. 37: 674–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.von Moltke, J., Ji, M., Liang, H. E. and Locksley, R. M., Tuft‐cell‐derived IL‐25 regulates an intestinal ILC2‐epithelial response circuit. Nature. 2016. 529: 221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Maggi, L., Montaini, G., Mazzoni, A., Rossettini, B., Capone, M., Rossi, M. C., Santarlasci, V. et al., Human circulating group 2 innate lymphoid cells can express CD154 and promote IgE production. J. Allergy Clin. Immunol. 2017. 139: 964–976 e4. [DOI] [PubMed] [Google Scholar]
- 111.Gury‐BenAri, M., Thaiss, C. A., Serafini, N., Winter, D. R., Giladi, A., Lara‐Astiaso, D., Levy, M. et al., The Spectrum and Regulatory Landscape of Intestinal Innate Lymphoid Cells Are Shaped by the Microbiome. Cell. 2016. 166: 1231–1246 e13. [DOI] [PubMed] [Google Scholar]
- 112.Buonocore, S., Ahern, P. P., Uhlig, H. H., Ivanov, I. I., Littman, D. R., Maloy, K. J. and Powrie, F., Innate lymphoid cells drive interleukin‐23‐dependent innate intestinal pathology. Nature. 2010. 464: 1371–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Peeters, P. M., Wouters, E. F. and Reynaert, N. L., Immune Homeostasis in Epithelial Cells: Evidence and Role of Inflammasome Signaling Reviewed. J Immunol Res. 2015. 2015: 828264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Shaw, M. H., Kamada, N., Kim, Y. G. and Nunez, G., Microbiota‐induced IL‐1beta, but not IL‐6, is critical for the development of steady‐state TH17 cells in the intestine. J. Exp. Med. 2012. 209: 251–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Melo‐Gonzalez, F. and Hepworth, M. R., Functional and phenotypic heterogeneity of group 3 innate lymphoid cells. Immunology. 2017. 150: 265–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mortha, A., Chudnovskiy, A., Hashimoto, D., Bogunovic, M., Spencer, S. P., Belkaid, Y. and Merad, M., Microbiota‐dependent crosstalk between macrophages and ILC3 promotes intestinal homeostasis. Science. 2014. 343: 1249288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kiss, E. A., Vonarbourg, C., Kopfmann, S., Hobeika, E., Finke, D., Esser, C. and Diefenbach, A., Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles. Science. 2011. 334: 1561–1565. [DOI] [PubMed] [Google Scholar]
- 118.Qiu, J., Heller, J. J., Guo, X., Chen, Z. M., Fish, K., Fu, Y. X. and Zhou, L., The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity. 2012. 36: 92–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lee, J. S., Cella, M., McDonald, K. G., Garlanda, C., Kennedy, G. D., Nukaya, M., Mantovani, A. et al., AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nat. Immunol. 2011. 13: 144–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gronke, K., Hernandez, P. P., Zimmermann, J., Klose, C. S. N., Kofoed‐Branzk, M., Guendel, F., Witkowski, M. et al., Interleukin‐22 protects intestinal stem cells against genotoxic stress. Nature. 2019. 566: 249–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wang, Y., Mumm, J. B., Herbst, R., Kolbeck, R. and Wang, Y., IL‐22 Increases Permeability of Intestinal Epithelial Tight Junctions by Enhancing Claudin‐2 Expression. J. Immunol. 2017. 199: 3316–3325. [DOI] [PubMed] [Google Scholar]
- 122.Sugimoto, K., Ogawa, A., Mizoguchi, E., Shimomura, Y., Andoh, A., Bhan, A. K., Blumberg, R. S., Xavier, R. J. et al., IL‐22 ameliorates intestinal inflammation in a mouse model of ulcerative colitis. J. Clin. Invest. 2008. 118: 534–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Dixon, B. R., Radin, J. N., Piazuelo, M. B., Contreras, D. C., Algood, H. M., IL‐17a and IL‐22 Induce Expression of Antimicrobials in Gastrointestinal Epithelial Cells and May Contribute to Epithelial Cell Defense against Helicobacter pylori. PLoS One. 2016. 11: e0148514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Guo, X., Liang, Y., Zhang, Y., Lasorella, A., Kee, B. L. and Fu, Y. X., Innate Lymphoid Cells Control Early Colonization Resistance against Intestinal Pathogens through ID2‐Dependent Regulation of the Microbiota. Immunity. 2015. 42: 731–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Qiu, J., Guo, X., Chen, Z. M., He, L., Sonnenberg, G. F., Artis, D. et al., Group 3 innate lymphoid cells inhibit T‐cell‐mediated intestinal inflammation through aryl hydrocarbon receptor signaling and regulation of microflora. Immunity. 2013. 39: 386–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Duffin, R., O'Connor, R. A., Crittenden, S., Forster, T., Yu, C., Zheng, X., Smyth, D. et al., Prostaglandin E(2) constrains systemic inflammation through an innate lymphoid cell‐IL‐22 axis. Science. 2016. 351: 1333–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ibiza, S., Garcia‐Cassani, B., Ribeiro, H., Carvalho, T., Almeida, L., Marques, R., Misic, A. M. et al., Veiga‐Fernandes H. Glial‐cell‐derived neuroregulators control type 3 innate lymphoid cells and gut defence. Nature. 2016. 535: 440–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wang, Y., Kuang, Z., Yu, X., Ruhn, K. A., Kubo, M. and Hooper, L. V., The intestinal microbiota regulates body composition through NFIL3 and the circadian clock. Science. 2017. 357: 912–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hoshi, N., Schenten, D., Nish, S. A., Walther, Z., Gagliani, N., Flavell, R. A., Reizis, B. et al., MyD88 signalling in colonic mononuclear phagocytes drives colitis in IL‐10‐deficient mice. Nat. Commun. 2012. 3: 1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Vicente‐Suarez, I., Larange, A., Reardon, C., Matho, M., Feau, S., Chodaczek, G., Park, Y. et al., Unique lamina propria stromal cells imprint the functional phenotype of mucosal dendritic cells. Mucosal Immunol. 2015. 8: 141–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhou, L., Chu, C., Teng, F., Bessman, N. J., Goc, J., Santosa, E. K., Putzel, G. G. et al., Innate lymphoid cells support regulatory T cells in the intestine through interleukin‐2. Nature. 2019. 568: 405–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hepworth, M. R., Fung, T. C., Masur, S. H., Kelsen, J. R., McConnell, F. M., Dubrot, J., Withers, D. R. et al., Immune tolerance. Group 3 innate lymphoid cells mediate intestinal selection of commensal bacteria‐specific CD4(+) T cells. Science. 2015. 348: 1031–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Saez de Guinoa, J., Jimeno, R., Farhadi, N., Jervis, P. J., Cox, L. R., Besra, G. S. and Barral, P., CD1d‐mediated activation of group 3 innate lymphoid cells drives IL‐22 production. EMBO Rep. 2017. 18: 39–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Melo‐Gonzalez, F., Kammoun, H., Evren, E., Dutton, E. E., Papadopoulou, M., Bradford, B. M., Tanes, C. et al., Antigen‐presenting ILC3 regulate T cell‐dependent IgA responses to colonic mucosal bacteria. J. Exp. Med. 2019. 216: 728–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mao, K., Baptista, A. P., Tamoutounour, S., Zhuang, L., Bouladoux, N., Martins, A. J., Huang, Y. et al., Innate and adaptive lymphocytes sequentially shape the gut microbiota and lipid metabolism. Nature. 2018. 554: 255–259. [DOI] [PubMed] [Google Scholar]
- 136.Bauer, H., Horowitz, R. E., Levenson, S. M. and Popper, H., The response of the lymphatic tissue to the microbial flora. Studies on germfree mice. Am. J. Pathol. 1963. 42: 471–483. [PMC free article] [PubMed] [Google Scholar]
- 137.Umesaki, Y., Setoyama, H., Matsumoto, S. and Okada, Y., Expansion of alpha beta T‐cell receptor‐bearing intestinal intraepithelial lymphocytes after microbial colonization in germ‐free mice and its independence from thymus. Immunology. 1993. 79: 32–37. [PMC free article] [PubMed] [Google Scholar]
- 138.Hapfelmeier, S., Lawson, M. A., Slack, E., Kirundi, J. K., Stoel, M., Heikenwalder, M., Cahenzli, J. et al., Reversible microbial colonization of germ‐free mice reveals the dynamics of IgA immune responses. Science. 2010. 328: 1705–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Round, J. L. and Mazmanian, S. K., The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 2009. 9: 313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Honda, K. and Littman, D. R., The microbiota in adaptive immune homeostasis and disease. Nature. 2016. 535: 75–84. [DOI] [PubMed] [Google Scholar]
- 141.Wesemann, D. R., Portuguese, A. J., Meyers, R. M., Gallagher, M. P., Cluff‐Jones, K., Magee, J. M., Panchakshari, R. A. et al., Microbial colonization influences early B‐lineage development in the gut lamina propria. Nature. 2013. 501: 112–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Cahenzli, J., Koller, Y., Wyss, M., Geuking, M. B. and McCoy, K. D., Intestinal microbial diversity during early‐life colonization shapes long‐term IgE levels. Cell Host Microbe. 2013. 14: 559–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Peterson, D. A., McNulty, N. P., Guruge, J. L. and Gordon, J. I., IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe. 2007. 2: 328–339. [DOI] [PubMed] [Google Scholar]
- 144.Sutherland, D. B., Suzuki, K. and Fagarasan, S., Fostering of advanced mutualism with gut microbiota by Immunoglobulin A. Immunol. Rev. 2016. 270: 20–31. [DOI] [PubMed] [Google Scholar]
- 145.Sterlin, D., Fadlallah, J., Adams, O., Fieschi, C., Parizot, C., Dorgham, K., Rajkumar, A. et al., Human IgA binds a diverse array of commensal bacteria. J. Exp. Med. 2020. 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ansaldo, E., Slayden, L. C., Ching, K. L., Koch, M. A., Wolf, N. K., Plichta, D. R., Brown, E. M. et al., Akkermansia muciniphila induces intestinal adaptive immune responses during homeostasis. Science. 2019. 364: 1179–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Bunker, J. J., Flynn, T. M., Koval, J. C., Shaw, D. G., Meisel, M., McDonald, B. D., Ishizuka, I. E. et al., Innate and Adaptive Humoral Responses Coat Distinct Commensal Bacteria with Immunoglobulin A. Immunity. 2015. 43: 541–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Bunker, J. J., Erickson, S. A., Flynn, T. M., Henry, C., Koval, J. C., Meisel, M., Jabri, B. et al., Natural polyreactive IgA antibodies coat the intestinal microbiota. Science. 2017. 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Li, H., Limenitakis, J. P., Greiff, V., Yilmaz, B., Scharen, O., Urbaniak, C., Zund, M. et al., Mucosal or systemic microbiota exposures shape the B cell repertoire. Nature. 2020. 584: 274–278. [DOI] [PubMed] [Google Scholar]
- 150.Nakajima, A., Vogelzang, A., Maruya, M., Miyajima, M., Murata, M., Son, A., Kuwahara, T. et al., IgA regulates the composition and metabolic function of gut microbiota by promoting symbiosis between bacteria. J. Exp. Med. 2018. 215: 2019–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hegazy, A. N., West, N. R., Stubbington, M. J. T., Wendt, E., Suijker, K. I. M., Datsi, A., This, S. et al., Circulating and Tissue‐Resident CD4(+) T Cells With Reactivity to Intestinal Microbiota Are Abundant in Healthy Individuals and Function Is Altered During Inflammation. Gastroenterology. 2017. 153: 1320–1337 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Geuking, M. B., Cahenzli, J., Lawson, M. A., Ng, D. C., Slack, E., Hapfelmeier, S., McCoy, K. D. et al., Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 2011. 34: 794–806. [DOI] [PubMed] [Google Scholar]
- 153.Atarashi, K., Tanoue, T., Shima, T., Imaoka, A., Kuwahara, T., Momose, Y., Cheng, G. et al., Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011. 331: 337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sefik, E., Geva‐Zatorsky, N., Oh, S., Konnikova, L., Zemmour, D., McGuire, A. M., Burzyn, D. et al., MUCOSAL IMMUNOLOGY. Individual intestinal symbionts induce a distinct population of RORgamma(+) regulatory T cells. Science. 2015. 349: 993–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Arpaia, N., Campbell, C., Fan, X., Dikiy, S., van der Veeken, J., deRoos, P., Liu, H. et al., Metabolites produced by commensal bacteria promote peripheral regulatory T‐cell generation. Nature. 2013. 504: 451–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Smith, P. M., Howitt, M. R., Panikov, N., Michaud, M., Gallini, C. A., Bohlooly, Y. M., Glickman, J. N. et al., The microbial metabolites, short‐chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013. 341: 569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Furusawa, Y., Obata, Y., Fukuda, S., Endo, T. A., Nakato, G., Takahashi, D., Nakanishi, Y. et al., Commensal microbe‐derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013. 504: 446–450. [DOI] [PubMed] [Google Scholar]
- 158.Campbell, C., McKenney, P. T., Konstantinovsky, D., Isaeva, O. I., Schizas, M., Verter, J., Mai, C. et al., Bacterial metabolism of bile acids promotes generation of peripheral regulatory T cells. Nature. 2020. 581: 475–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Hang, S., Paik, D., Yao, L., Kim, E., Trinath, J., Lu, J., Ha, S. et al., Bile acid metabolites control TH17 and Treg cell differentiation. Nature. 2019. 576: 143–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Song, X., Sun, X., Oh, S. F., Wu, M., Zhang, Y., Zheng, W., Geva‐Zatorsky, N. et al., Microbial bile acid metabolites modulate gut RORgamma(+) regulatory T cell homeostasis. Nature. 2020. 577: 410–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Gaboriau‐Routhiau, V., Rakotobe, S., Lecuyer, E., Mulder, I., Lan, A., Bridonneau, C., Rochet, V. et al., The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009. 31: 677–689. [DOI] [PubMed] [Google Scholar]
- 162.Ivanov, I. I., Frutos, R. L., Manel, N., Yoshinaga, K., Rifkin, D. B., Sartor, R. B., Finlay, B. B. et al., Specific microbiota direct the differentiation of IL‐17‐producing T‐helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008. 4: 337–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ivanov, I. I., Atarashi, K., Manel, N., Brodie, E. L., Shima, T., Karaoz, U., Wei, D. et al., Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009. 139: 485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Tan, T. G., Sefik, E., Geva‐Zatorsky, N., Kua, L., Naskar, D., Teng, F., Pasman, L. et al., Identifying species of symbiont bacteria from the human gut that, alone, can induce intestinal Th17 cells in mice. Proc Natl Acad Sci U S A. 2016. 113: E8141–E50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Atarashi, K., Tanoue, T., Ando, M., Kamada, N., Nagano, Y., Narushima, S., Suda, W. et al., Th17 Cell Induction by Adhesion of Microbes to Intestinal Epithelial Cells. Cell. 2015. 163: 367–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Yang, Y., Torchinsky, M. B., Gobert, M., Xiong, H., Xu, M., Linehan, J. L., Alonzo, F. et al., Focused specificity of intestinal TH17 cells towards commensal bacterial antigens. Nature. 2014. 510: 152–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Omenetti, S., Bussi, C., Metidji, A., Iseppon, A., Lee, S., Tolaini, M., Li, Y. et al., The Intestine Harbors Functionally Distinct Homeostatic Tissue‐Resident and Inflammatory Th17 Cells. Immunity. 2019. 51: 77–89 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Xu, M., Pokrovskii, M., Ding, Y., Yi, R., Au, C., Harrison, O. J., Galan, C. et al., c‐MAF‐dependent regulatory T cells mediate immunological tolerance to a gut pathobiont. Nature. 2018. 554: 373–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Persson, E. K., Uronen‐Hansson, H., Semmrich, M., Rivollier, A., Hagerbrand, K., Marsal, J., Gudjonsson, S. et al., IRF4 transcription‐factor‐dependent CD103(+)CD11b(+) dendritic cells drive mucosal T helper 17 cell differentiation. Immunity. 2013. 38: 958–969. [DOI] [PubMed] [Google Scholar]
- 170.Lee, J. Y., Hall, J. A., Kroehling, L., Wu, L., Najar, T., Nguyen, H. H., Lin, W. Y. et al., Serum Amyloid A Proteins Induce Pathogenic Th17 Cells and Promote Inflammatory Disease. Cell. 2020. 180: 79–91 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Gattu, S., Bang, Y. J., Pendse, M., Dende, C., Chara, A. L., Harris, T. A., Wang, Y. et al., Epithelial retinoic acid receptor beta regulates serum amyloid A expression and vitamin A‐dependent intestinal immunity. Proc Natl Acad Sci U S A. 2019. 116: 10911–10916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Tanoue, T., Morita, S., Plichta, D. R., Skelly, A. N., Suda, W., Sugiura, Y., Narushima, S. et al., A defined commensal consortium elicits CD8 T cells and anti‐cancer immunity. Nature. 2019. 565: 600–605. [DOI] [PubMed] [Google Scholar]
- 173.Bachem, A., Makhlouf, C., Binger, K. J., de Souza, D. P., Tull, D., Hochheiser, K., Whitney, P. G. et al., Microbiota‐Derived Short‐Chain Fatty Acids Promote the Memory Potential of Antigen‐Activated CD8(+) T Cells. Immunity. 2019. 51: 285–297 e5. [DOI] [PubMed] [Google Scholar]
- 174.Kubinak, J. L., Petersen, C., Stephens, W. Z., Soto, R., Bake, E., O'Connell, R. M. and Round, J. L., MyD88 signaling in T cells directs IgA‐mediated control of the microbiota to promote health. Cell Host Microbe. 2015. 17: 153–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Teng, F., Klinger, C. N., Felix, K. M., Bradley, C. P., Wu, E., Tran, N. L., Umesaki, Y. et al., Gut Microbiota Drive Autoimmune Arthritis by Promoting Differentiation and Migration of Peyer's Patch T Follicular Helper Cells. Immunity. 2016. 44: 875–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kawamoto, S., Tran, T. H., Maruya, M., Suzuki, K., Doi, Y., Tsutsui, Y., Kato, L. M. et al., The inhibitory receptor PD‐1 regulates IgA selection and bacterial composition in the gut. Science. 2012. 336: 485–489. [DOI] [PubMed] [Google Scholar]
- 177.Proietti, M., Cornacchione, V., Rezzonico Jost, T., Romagnani, A., Faliti, C. E., Perruzza, L., Rigoni, R. et al., ATP‐gated ionotropic P2X7 receptor controls follicular T helper cell numbers in Peyer's patches to promote host‐microbiota mutualism. Immunity. 2014. 41: 789–801. [DOI] [PubMed] [Google Scholar]
- 178.Wingender, G., Stepniak, D., Krebs, P., Lin, L., McBride, S., Wei, B., Braun, J. et al., Intestinal microbes affect phenotypes and functions of invariant natural killer T cells in mice. Gastroenterology. 2012. 143: 418–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Olszak, T., An, D., Zeissig, S., Vera, M. P., Richter, J., Franke, A., Glickman, J. N. et al., Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012. 336: 489–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Cervantes‐Barragan, L., Chai, J. N., Tianero, M. D., Di Luccia, B., Ahern, P. P., Merriman, J., Cortez, V. S. et al., Lactobacillus reuteri induces gut intraepithelial CD4(+)CD8alphaalpha(+) T cells. Science. 2017. 357: 806–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Bilate, A. M., London, M., Castro, T. B. R., Mesin, L., Bortolatto, J., Kongthong, S., Harnagel, A. et al., T Cell Receptor Is Required for Differentiation, but Not Maintenance, of Intestinal CD4(+) Intraepithelial Lymphocytes. Immunity. 2020. 53: 1001–1014 e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Moon, S., Park, Y., Hyeon, S., Kim, Y. M., Kim, J. H., Kim, H., Park, S. et al., Niche‐specific MHC II and PD‐L1 regulate CD4+CD8alphaalpha+ intraepithelial lymphocyte differentiation. J. Exp. Med. 2021. 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Lynch, S. V. and Pedersen, O., The Human Intestinal Microbiome in Health and Disease. N. Engl. J. Med. 2016. 375: 2369–2379. [DOI] [PubMed] [Google Scholar]
- 184.Dupont, H. L., Jiang, Z. D., Dupont, A. W. and Utay, N. S., The Intestinal Microbiome in Human Health and Disease. Trans. Am. Clin. Climatol Assoc. 2020. 131: 178–197. [PMC free article] [PubMed] [Google Scholar]
- 185.Mendez, R., Banerjee, S., Bhattacharya, S. K. and Banerjee, S., Lung inflammation and disease: A perspective on microbial homeostasis and metabolism. IUBMB Life. 2019. 71: 152–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Neugent, M. L., Hulyalkar, N. V., Nguyen, V. H., Zimmern, P. E. and De Nisco, N. J., Advances in Understanding the Human Urinary Microbiome and Its Potential Role in Urinary Tract Infection. mBio. 2020. 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Byrd, A. L., Belkaid, Y. and Segre, J. A., The human skin microbiome. Nat. Rev. Microbiol. 2018. 16: 143–155. [DOI] [PubMed] [Google Scholar]
- 188.Busnelli, M., Manzini, S. and Chiesa, G., The Gut Microbiota Affects Host Pathophysiology as an Endocrine Organ: A Focus on Cardiovascular Disease. Nutrients. 2019. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Donia, M. S. and Fischbach, MA. HUMAN MICROBIOTA. Small molecules from the human microbiota. Science. 2015. 349: 1254766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Fischbach, M. A., Microbiome: Focus on Causation and Mechanism. Cell. 2018. 174: 785–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Pedersen, H. K., Gudmundsdottir, V., Nielsen, H. B., Hyotylainen, T., Nielsen, T., Jensen, B. A., Forslund, K. et al., Human gut microbes impact host serum metabolome and insulin sensitivity. Nature. 2016. 535: 376–381. [DOI] [PubMed] [Google Scholar]
- 192.Perry, R. J., Peng, L., Barry, N. A., Cline, G. W., Zhang, D., Cardone, R. L., Petersen, K. F. et al., Acetate mediates a microbiome‐brain‐beta‐cell axis to promote metabolic syndrome. Nature. 2016. 534: 213–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Dinan, T. G. and Cryan, J. F., The Microbiome‐Gut‐Brain Axis in Health and Disease. Gastroenterol. Clin. North Am. 2017. 46: 77–89. [DOI] [PubMed] [Google Scholar]
- 194.Wang, R., Tang, R., Li, B., Ma, X., Schnabl, B. and Tilg, H., Gut microbiome, liver immunology, and liver diseases. Cell Mol Immunol. 2021. 18: 4–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Xiao, H. and Kang, S., The Role of the Gut Microbiome in Energy Balance With a Focus on the Gut‐Adipose Tissue Axis. Front Genet. 2020. 11: 297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Lundgren, P. and Thaiss, C. A., The microbiome‐adipose tissue axis in systemic metabolism. Am. J. Physiol. Gastrointest. Liver Physiol. 2020. 318: G717–G24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Ticinesi, A., Nouvenne, A., Cerundolo, N., Catania, P., Prati, B., Tana, C. and Meschi, T., Gut Microbiota, Muscle Mass and Function in Aging: A Focus on Physical Frailty and Sarcopenia. Nutrients. 2019. 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Kazemian, N., Mahmoudi, M., Halperin, F., Wu, J. C. and Pakpour, S., Gut microbiota and cardiovascular disease: opportunities and challenges. Microbiome. 2020. 8: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Tang, W. H. W., Li, D. Y. and Hazen, S. L., Dietary metabolism, the gut microbiome, and heart failure. Nat. Rev. Cardiol. 2019. 16: 137–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Corbo, J.andParkes, D., editors. The price of selfish behavior in bilateral network formation. Proceedings of the twenty‐fourth annual ACM symposium on Principles of distributed computing; 2005.
- 201.Coyte, K. Z., Schluter, J. and Foster, K. R., The ecology of the microbiome: Networks, competition, and stability. Science. 2015. 350: 663–666. [DOI] [PubMed] [Google Scholar]
- 202.Angell, I. L. and Rudi, K., A game theory model for gut bacterial nutrient utilization strategies during human infancy. Proc Biol Sci. 2020. 287: 20200824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Herren, C. M., Disruption of cross‐feeding interactions by invading taxa can cause invasional meltdown in microbial communities. Proc Biol Sci. 2020. 287: 20192945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Laan, A. and de Polavieja, G. G., Species diversity rises exponentially with the number of available resources in a multi‐trait competition model. Proc Biol Sci. 2018. 285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Rao, C., Coyte, K. Z., Bainter, W., Geha, R. S. and Martin, C. R., Rakoff‐Nahoum S. Multi‐kingdom ecological drivers of microbiota assembly in preterm infants. Nature. 2021. 591: 633–638. [DOI] [PMC free article] [PubMed] [Google Scholar]



