Abstract
Objective
Systemic inflammatory factors have been implicated in symptomatic hand osteoarthritis (OA). Gut microbiome dysbiosis promotes systemic inflammation. The aim of this study was to examine the association between the gut microbiome and the presence of symptomatic hand OA in a population‐based study.
Methods
Study participants were subjects of the Xiangya Osteoarthritis Study, a community‐based observational study conducted in the Hunan Province of China. Symptomatic hand OA was defined as the presence of both symptoms and radiographic OA in the same hand. The gut microbiome was analyzed using 16S ribosomal RNA gene sequencing in stool samples. We examined the relation of α‐diversity, β‐diversity, relative abundance of taxa, and potential bacterial functional pathways to symptomatic hand OA.
Results
A total of 1,388 participants (mean age 61.3 years, 57.4% women) were included in the study, of whom 72 had symptomatic hand OA (prevalence of symptomatic hand OA 5.2%). Beta‐diversity of the gut microbiome, but not α‐diversity, was significantly associated with the presence of symptomatic hand OA (P = 0.003). Higher relative abundance of the genera Bilophila and Desulfovibrio as well as lower relative abundance of the genus Roseburia was associated with symptomatic hand OA. Most functional pathways (i.e., those annotated in the KEGG Ortholog hierarchy) that were observed to be altered in participants with symptomatic hand OA belonged to the amino acid, carbohydrate, and lipid metabolic pathways.
Conclusion
This large, population‐based study provides the first evidence that alterations in the composition of the gut microbiome were observed among study participants who had symptomatic hand OA, and a low relative abundance of Roseburia but high relative abundance of Bilophila and Desulfovibrio at the genus level were associated with prevalent symptomatic hand OA. These findings may help investigators understand the role of the microbiome in the development of symptomatic hand OA and could contribute to potential translational opportunities.
INTRODUCTION
Hand osteoarthritis (OA) is highly prevalent within middle‐aged and older populations (1). Individuals with hand OA may experience pain and stiffness and develop structural joint damage, which can impair their ability to undertake activities of daily living (1). Results from previous studies have shown that symptomatic hand OA can have a clinical burden comparable to that of rheumatoid arthritis (2). Although the pathogenesis of hand OA remains largely unknown, systemic factors, including systemic inflammation, have been implicated as a potential risk factor for symptomatic hand OA (1, 3).
Gut microbiome dysbiosis can lead to the dysregulation of various important physiologic functions, such as production of small molecules that interact with the host, synthesis of essential amino acids, and regulation of fat metabolism, which can in turn contribute to the development of systemic inflammation (4). Over recent decades, many studies have found that the gut microbiome and its metabolites play important roles in the pathologic development and progression of several systemic inflammatory diseases, including inflammatory bowel disease, inflammatory arthritis, multiple sclerosis, and systemic lupus erythematosus (4). However, to the best of our knowledge, no study has examined the association between the gut microbiome and hand OA. Elucidating this association would help clarify the role of the microbiome in the development of hand OA and contribute to potential translational opportunities for the prevention and treatment of this common condition.
To help fill this knowledge gap, we examined the association between the gut microbiome and prevalent symptomatic hand OA, using data collected from the Xiangya Osteoarthritis (XO) Study, a population‐based observational study conducted among the residents of rural areas of China.
PATIENTS AND METHODS
Study participants
The XO Study is a population‐based longitudinal study of the natural history of and risk factors for the development of OA in a rural area of China (ClinicalTrials.gov identifier: NCT04033757) (5). Participants in the XO Study were a randomly selected sample of residents age ≥50 years from rural mountainous villages in Longshan County in the Hunan Province. Specifically, for selection of the initial 14 communities, we first adopted a sampling method of probability proportionate to population size. All of the villages in the selected communities were then listed in a random order. The village‐to‐village recruitment began in the first village in the first community until the number of participants in that community met the predetermined quota. Eventually, a total of 25 rural mountainous villages in Longshan County were included in the XO Study. Of note, the XO Study includes 3 subcohorts (i.e., subcohorts I, II, and III), comprising participants recruited in 2015, 2018, and 2019, respectively.
The XO Study was approved by the Research Ethical Committee of Xiangya Hospital, Central South University (approval no. 201510506). All participants gave their written informed consent to participate in the studies.
Assessment of hand OA
Posteroanterior radiographs of both hands were obtained from each participant in the XO Study. Radiographs of the bilateral second to fifth distal interphalangeal joints, second to fifth proximal interphalangeal joints, first to fifth metacarpophalangeal joints, thumb interphalangeal joints, and thumb base (carpometacarpal) joints were graded using a modified Kellgren/Lawrence (K/L) scale for radiographic hand OA (6). All hand radiographs were read by a single musculoskeletal researcher (TY; an orthopedic surgeon who was the primary reader). With each new batch of radiographs (n = 50 films), we commingled 5 previously read radiographs to test intrarater reliability. For assessment of interrater reliability, another reader (ADO; a musculoskeletal imaging specialist) scored a selected subset of 30 films independently. Intra‐ and interrater reliabilities were assessed using kappa statistics with 95% confidence intervals (95% CIs). Radiographic hand OA was defined as the presence of a K/L radiographic severity grade of ≥2 in any of the joints of each hand (i.e., those joints listed above). The intrarater reliability for the identification of radiographic hand OA (presence versus absence) was a kappa of 0.91 (95% CI 0.83–0.99), and the interrater reliability was a kappa of 0.71 (95% CI 0.45–0.96).
Presence of hand symptoms was ascertained by noting the patient’s response to the question, “On most days, do you have pain, aching, or stiffness in your left/right hand?” (7). Symptomatic hand OA was defined as the presence of both self‐reported symptoms and radiographic OA in the same hand. Participants were defined as having symptomatic hand OA if they had symptomatic hand OA in at least one hand (7).
Stool sample collection and DNA extraction
Stool samples collected from the participants at the recruitment site were immediately frozen and transported on dry ice within 20 minutes. Samples were stored in freezers at a temperature of −80°C until analyzed. For DNA assessment, DNA was extracted from 200 mg of each stool sample using a Magen Hipure Soil DNA kit, according to the manufacturer’s protocol. DNA was quantified using a Qubit version 2.0 fluorometer.
Gut microbiome analysis using 16S ribosomal RNA (rRNA) gene sequencing
The 16S rRNA gene was amplified using a 341F/806R primer set targeting the V3–V4 hypervariable region. DNA was sequenced using an Illumina MiSeq platform. Bioinformatics analysis of the gut microbiome was performed using a QIIME 2 2019.10 platform (https://qiime2.org). Raw sequence data were demultiplexed and quality filtered using a q2‐demux plugin, followed by denoising with DADA2 (via q2‐dada2).
All amplicon sequence variants (ASVs) were aligned with mafft (via q2‐alignment) and used to construct a phylogeny with fasttree2 (via q2‐phylogeny). Metrics for α‐diversity and β‐diversity and data from principal coordinates analysis (PCoA) were estimated using q2‐diversity after samples were rarefied (subsampled without replacement) to the minimal number of reads per sample. Taxonomy was assigned to ASVs using a q2‐feature‐classifier, classify‐sklearn naive Bayes taxonomy classifier, against the Greengenes 13_8 99% operational taxonomic units (OTUs) from reference sequences. All 16S rRNA sequencing data obtained in this study are available for download from the European Nucleotide Archive (project no. PRJEB33926; https://www.ebi.ac.uk/ena/browser/home).
Statistical analysis
Similarities in the composition of the gut microbiome between participants with symptomatic hand OA (i.e., the symptomatic hand OA group) and those without symptomatic hand OA (i.e., the control group) were compared using α‐diversity as measured by the Shannon diversity index, and β‐diversity as measured by the unweighted Unifrac distance. We used a Wilcoxon’s rank sum test to determine the differences in α‐diversity between groups, and a permutation multivariate analysis of variance test to determine the differences in β‐diversity between groups.
To gain more insight into which gut microbiome taxonomies drive the association with symptomatic hand OA, we performed multivariate linear regression analyses at the phylum, family, and genus levels, with adjustments for age, sex, body mass index (BMI), alcohol consumption, and frequency of dietary intake of meat/eggs, dairy, and vegetables. Specifically, we removed from the analyses any microbiome taxa that were present in <20% of samples, and then compared the difference in relative abundance of taxa at the phylum, family, and genus levels between the symptomatic hand OA and control groups. Bacterial metagenomes were imputed from 16S rRNA sequencing–based microbial DNA data using the Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) software package. Functional annotation was applied using the annotated pathways from the KEGG catalog (http://www.genome.ad.jp/kegg). We performed multivariate linear regression analyses to assess differences in KEGG level 3 pathways (i.e., those present in more than 20% of samples) between participants with and those without symptomatic hand OA, with adjustments for age, sex, BMI, alcohol consumption, and frequency of dietary intake of meat/eggs, dairy, and vegetables. Significant differences were assessed using P values corrected for multiple testing with the Benjamin and Hochberg false discovery rate method; corrected P values (and Q values) less than 0.1 were considered statistically significant.
In addition, 3 sensitivity analyses were performed to assess the robustness of our study findings. First, to minimize the potential residual effect of antibiotic use, we compared the differences in relative abundance of microbiota at the genus level between the symptomatic hand OA group and the control group, after exclusion of individuals who reported having received antibiotics within 2 or 3 months prior to the stool sample collection. Second, we conducted a matched case–control study in which up to 4 controls were matched to each case by age, sex, and BMI. We compared the difference in relative abundance of the microbiome genera and KEGG level 3 pathways between the case and control groups using multivariate linear regression analyses adjusted for alcohol consumption and frequency of dietary intake of meat/eggs, dairy, and vegetables. Third, we performed a sex‐specific analysis to explore the potential sex interaction between the microbiome and symptomatic hand OA. Detailed information on the methods and analysis codes used are provided in the Supplementary Methods (available on the Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.41729/abstract).
RESULTS
A flow chart depicting the participant selection process is shown in Figure 1A. Baseline characteristics of the 1,388 included study participants are shown in Supplementary Table 1 (available on the Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.41729/abstract). Participants with symptomatic hand OA (n = 72) were older than participants without symptomatic hand OA (n = 1,316) (age 70.9 years versus 62.8 years; P < 0.001), and those with symptomatic hand OA were more likely to be women (75% versus 57%; P = 0.003).
Figure 1.
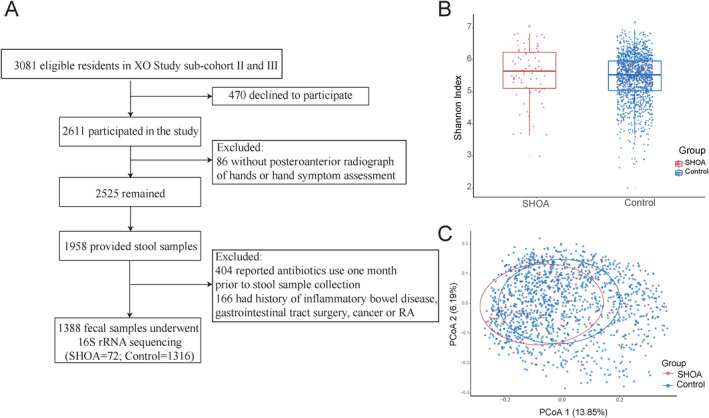
Profiling of the gut microbiome using 16S ribosomal RNA (rRNA) sequencing in subjects from the Xiangya Osteoarthritis (XO) Study cohort. A, Selection process of study subjects with symptomatic hand osteoarthritis (SHOA) and those without symptomatic hand OA (controls). B, Shannon index of microbial diversity (α‐diversity) in each group. Results are shown as box plots, in which symbols represent individual subjects, the horizontal line inside the box represents the median, each box represents the 25th to 75th percentiles, and the lines outside the box represent the smallest and largest value of 1.5 × the interquartile range. C, Principle coordinates analysis (PCoA) plots of β‐diversity in each group, constructed using the unweighted Unifrac distance. RA = rheumatoid arthritis.
A total of 90,608,388 raw sequence reads (mean reads per sample 65,279) were generated from all stool samples obtained from eligible participants. After quality filtering and removal of contaminants, there were 61,294,662 high‐quality reads that were used for analysis (mean reads per sample 44,160). Among all samples, 31,355 different ASVs were discovered.
The Shannon index of microbial α‐diversity was not significantly different between patients with symptomatic hand OA and controls without symptomatic hand OA (P = 0.095) (Figure 1B). However, the PCoA plot constructed using unweighted UniFrac distances showed that the structure and composition of the gut microbiome differed significantly between the 2 groups (P = 0.003) (Figure 1C). In both groups, the profile of the gut microbiome appeared to be dominated by Firmicutes and Bacteroidetes at the phylum level, by Lachnospiraceae, Bacteroidaceae, Prevotellaceae, and Ruminococcaceae at the family level, and by Bacteroides, Prevotella, Faecalibacterium, and Roseburia at the genus level (for details, see Supplementary Figures 1, 2, and 3, available on the Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.41729/abstract), consistent with the usual composition of the human gut microbiome.
The associations between microbiome taxonomies and symptomatic hand OA are shown in Figure 2. After adjustment for age, sex, BMI, alcohol consumption, and frequency of dietary intake of meat/eggs, dairy, and vegetables, there was no apparent difference in microbiome taxa at the phylum level between subjects with and those without symptomatic hand OA. However, at the family level, individuals with symptomatic hand OA had a higher relative abundance of Christensenellaceae (P < 0.001, Q < 0.001) (Figure 2A), Desulfovibrionaceae (P = 0.001, Q = 0.008) (Figure 2B), and Mogibacteriaceae (P = 0.01, Q = 0.053) (Figure 2C), but a lower relative abundance of Lachnospiraceae (P = 0.02, Q = 0.092) (Figure 2D) compared to those without symptomatic hand OA. Statistically significant differences in the gut microbiome were also observed at the genus level. Individuals with symptomatic hand OA had higher relative abundances of Bilophila (P = 0.001, Q = 0.006) (Figure 2E) and Desulfovibrio (P = 0.012, Q = 0.064) (Figure 2F) but a lower relative abundance of Roseburia (P = 0.011, Q = 0.062) (Figure 2G) compared to those without symptomatic hand OA. Full summary statistics of the associations of microbiome taxonomies with symptomatic hand OA adjusted for age, sex, BMI, alcohol consumption, and frequency of dietary intake of meat/eggs, dairy, and vegetables, determined using multivariate linear regression analyses, are presented in Supplementary Tables 2–4 (available on the Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.41729/abstract). After exclusion of individuals who reported having received antibiotic treatment within 2 months or 3 months prior to the stool sample collection, the associations remained statistically significant (Supplementary Tables 5 and 6; http://onlinelibrary.wiley.com/doi/10.1002/art.41729/abstract).
Figure 2.
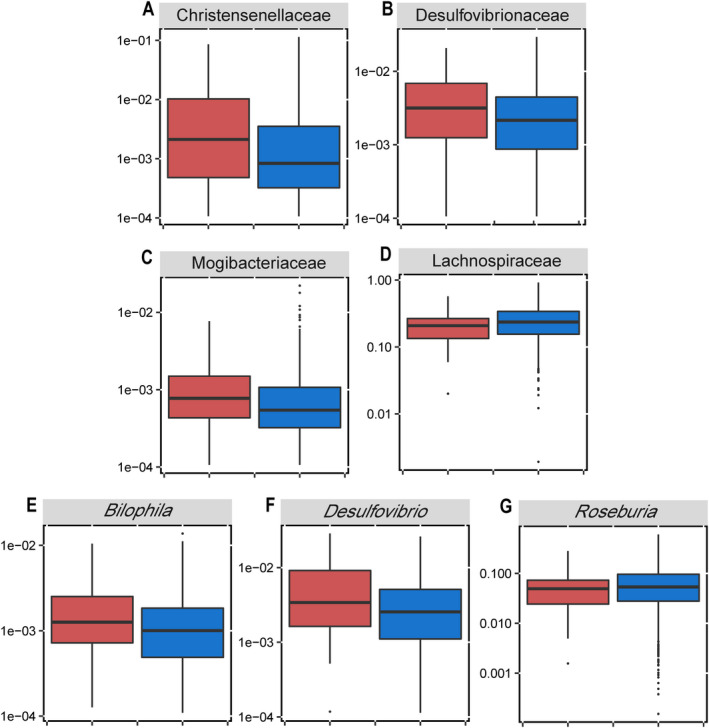
Differences in the composition (relative abundance) of the gut microbiota at the family level (A–D) and genus level (E–G) between individuals with symptomatic hand osteoarthritis (OA) (n = 72) (red) and individuals without symptomatic hand OA (controls) (n = 1,316) (blue), after adjustments for age, sex, body mass index, alcohol consumption, and frequency of dietary intake of meat/eggs, dairy, and vegetables. Data are shown as box plots, in which the horizontal line inside the box represents the median, each box represents the 25th to 75th percentiles, and the lines outside the box represent the smallest and largest value of 1.5 × the interquartile range. Circles represent individual outliers.
Sensitivity analyses conducted in case–control studies, with matching for age, sex, and BMI (68 participants with symptomatic hand OA versus 234 matched controls without symptomatic hand OA) showed similar results (Supplementary Table 7; http://onlinelibrary.wiley.com/doi/10.1002/art.41729/abstract). The sex‐specific analysis undertaken in the 54 women with symptomatic hand OA yielded results consistent with the primary analysis in terms of taxonomic associations with symptomatic hand OA (i.e., for Bilophila, β = 0.008, P = 0.003, Q = 0.031; for Desulfovibrio, β = 0.011, P = 0.026, Q = 0.148; for Roseburia, β = −0.046, P = 0.006, Q = 0.049). However, taxonomic associations in the 18 men with symptomatic hand OA were not statistically significant, although the findings were similar to that in women (i.e., relative abundance of the genera Bilophila and Desulfovibrio increased, and relative abundance of the genus Roseburia decreased in men with symptomatic hand OA).
The functional analysis, performed by reconstructing metagenomes using PICRUSt software, identified 15 KEGG level 3 pathways that were altered in association with symptomatic hand OA (Figure 3). Most KEGG pathways that were related to amino acids (metabolism of amino acids, tyrosine and lysine degradation, and cyano–amino acid metabolism), carbohydrates (metabolism of starch, sucrose, amino sugar and nucleotide sugar, and butanoate and propanoate), and lipids (sphingolipid metabolism) were significantly altered in individuals with symptomatic hand OA compared to controls. Full summary statistics of the associations of KEGG level 3 pathways with symptomatic hand OA adjusted for age, sex, BMI, alcohol consumption, and frequency of dietary intake of meat/eggs, dairy, and vegetables, determined using multivariate linear regression analyses, are presented in Supplementary Table 8 (available on the Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.41729/abstract). Similar results were observed in the case–control study with matching for age, sex, and BMI (Supplementary Table 9; http://onlinelibrary.wiley.com/doi/10.1002/art.41729/abstract).
Figure 3.
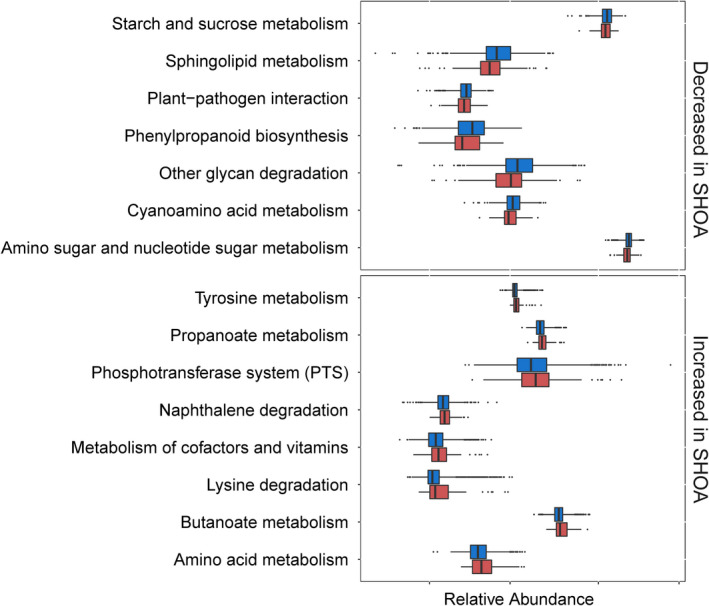
Differences in the relative abundance of predicted functions (third level of the KEGG Ortholog hierarchy), expressed as either a decrease or increase in the relative abundance of each level 3 functional pathway in individuals with symptomatic hand osteoarthritis (SHOA) (n = 72) (red) compared to individuals without symptomatic hand OA (controls) (n = 1,316) (blue), based on the Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) data set. Data are shown as box plots, in which the line inside the box represents the median, each box represents the 25th to 75th percentiles, and the lines outside the box represent the smallest and largest value of 1.5 × the interquartile range. Circles represent individual outliers.
DISCUSSION
Several studies have demonstrated that patients with inflammatory arthritis have a decreased relative abundance of the genus Roseburia (8), and there was a strong positive correlation between the relative abundance of the genus Desulfovibrio and inflammatory blood biomarkers in the general population (9). In accord with those findings, our results suggest that a high relative abundance of the genus Desulfovibrio but low relative abundance of the genus Roseburia may play a role in symptomatic hand OA. Furthermore, previous work has shown that metabolic pathways, namely those affecting metabolism of the branched‐chain amino acids, from arginine and phosphatidylcholine to lysophosphatidylcholine, were significantly associated with the pathogenesis of OA (10). Similarly, our results based on reconstruction of metagenomes using PICRUSt software also identified altered KEGG pathways related to metabolism of amino acids, lipids, and carbohydrates in association with symptomatic hand OA. Taken together, our findings suggest that metabolic dysfunction of the gut microbiome may play a key role in the state of systemic inflammation in patients with symptomatic hand OA by affecting the host metabolite levels, a concept that warrants further investigation.
Several biologic mechanisms linking the gut microbiome to systemic inflammation have been proposed. Bilophila member species have been shown to produce lipopolysaccharides that promote intestinal barrier dysfunction, bile acid dysmetabolism, and inflammation in mouse models (11). In addition, in in vitro experiments, a species belonging to Bilophila was able to convert taurine to the toxic metabolite hydrogen sulfide (H2S), which plays an important role in systemic inflammation (12). OTUs in Desulfovibrio have been shown to have a strong correlation with systemic and chronic inflammation in models of mice fed a high‐fat diet, suggesting that OTUs in Desulfovibrio might influence chronic inflammation in the host in a way that relates to weight gain and glucose tolerance (13). Several species included in Roseburia have been reported to serve an antiinflammatory function by producing butyrate, which is the main source of energy for colonic epithelial cells and which inhibits messenger RNA expression of proinflammatory cytokines in the mucosa by inhibiting NF‐κB activation (14).
Several strengths of our study are noteworthy. This was a population‐based study, and thus the findings may be generalizable to the entire Chinese population, among subjects with similar characteristics. This is supported by the prevalence of symptomatic hand OA in our study (5.2%), which was similar to that reported in other parts of China (15). In addition, our results provided novel evidence linking the gut microbiome composition to the prevalence of symptomatic hand OA. The significant associations of several genera with symptomatic hand OA parallel previous observations in inflammatory arthritis studies, supporting the validity of our findings. Furthermore, we demonstrated that several altered KEGG metabolism pathways were associated with symptomatic hand OA. This information may contribute to translational opportunities for the identification and treatment of individuals with symptomatic hand OA, and warrants further studies in independent populations.
There are several limitations of our study. First, the gut microbiomes were profiled by 16S rRNA gene sequencing. Although this technology can identify microbial taxonomies and composition, it has limitations in identifying genetically specific species and strains. Future studies using metagenomic approaches are needed to evaluate the relationship of a specific bacterial gene(s) and its function to symptomatic hand OA. Second, the current study was a cross‐sectional study; thus, we could not assess the temporal sequence between the gut microbiome and occurrence of symptomatic hand OA. Third, the present results were not validated and reproduced in an independent cohort. Changes in specific microbiome genera may not be replicable in other populations, given the heterogeneity of the gut microbiome in different geographic locations. Finally, although the main findings were shown to be similar in women with symptomatic hand OA, there may have been insufficient power to detect an association in sex‐specific analyses due to the relatively small number of men in the study, particularly in terms of fully exploring the potential sex interaction between the gut microbiome and symptomatic hand OA; this warrants further study.
This large population‐based study provides the first evidence that alterations in the gut microbiome composition are present in individuals with symptomatic hand OA, and a low relative abundance of Roseburia but high relative abundance of Bilophila and Desulfovibrio at the genus level were associated with prevalent symptomatic hand OA in this population. Our findings may help investigators understand the role of the microbiome in the development of symptomatic hand OA, and could contribute to potential translational opportunities.
AUTHOR CONTRIBUTIONS
All authors were involved in drafting the article or revising it critically for important intellectual content, and all authors approved the final version to be published. Drs. Lei and Zeng had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Study conception and design
Wei, C. Zhang, Y. Zhang, W. Zhang, Doherty, Zeng, Lei.
Acquisition of data
Wei, Yang.
Analysis and interpretation of data
Wei, Y. Zhang, W. Zhang, Zhai, Obotiba, Lyu, Zeng.
Supporting information
Supplementary Material
ACKNOWLEDGMENTS
We thank Sally A. Doherty and Piran Aliabadi for their assistance with radiograph readings, and we acknowledge Yilun Wang, Jiatian Li, Xiaoxiao Li, Dongxing Xie, Jing Wu, Bei Xu, Jian Tian, Yuqing Wang, Zhenglei Zhu, Fan Wu, Ke He, Haochen Wang, Hongyi He, Xing Huang, Ning Wang, Ziying Wu, Hui Li, Xiang Ding, Bin Zhou, Ye Yang, Ruijun Bai, Zhichen Liu, Kun Li, Junyu Zhu, Xinjia Deng, Manli Chen, Li Zhang, Ying Pan, Zikun Xie, and Renpeng Fang for their contribution to this study.
Supported by the National Natural Science Foundation of China (grants 81772413, 81702207, 81702206, 81930071, 81902265, and 82072502), the National Key Research and Development Project from the Ministry of Science and Technology of China (grant 2018YFB1105705), the Hunan Province Key Research and Development Program (grants 2018SK2070 and 2018SK2071), the Project Program of the National Clinical Research Center for Geriatric Disorders (grant 2020LNJJ03 to Xiangya Hospital), and the Hunan Province Science and Technology Program (grant 2019RS2010).
No potential conflicts of interest relevant to this article were reported.
Contributor Information
Chao Zeng, Email: zengchao@csu.edu.cn.
Guanghua Lei, Email: lei_guanghua@csu.edu.cn.
References
- 1.Marshall M, Watt FE, Vincent TL, Dziedzic K. Hand osteoarthritis: clinical phenotypes, molecular mechanisms and disease management [review]. Nat Rev Rheumatol 2018;14:641–56. [DOI] [PubMed] [Google Scholar]
- 2.Michon M, Maheu E, Berenbaum F. Assessing health‐related quality of life in hand osteoarthritis: a literature review. Ann Rheum Dis 2011;70:921–8. [DOI] [PubMed] [Google Scholar]
- 3.Conaghan PG, Cook AD, Hamilton JA, Tak PP. Therapeutic options for targeting inflammatory osteoarthritis pain [review]. Nat Rev Rheumatol 2019;15:355–63. [DOI] [PubMed] [Google Scholar]
- 4.Clemente JC, Manasson J, Scher JU. The role of the gut microbiome in systemic inflammatory disease [review]. BMJ 2018;360:j5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zeng C, Wei J, Terkeltaub R, Yang T, Choi HK, Wang YL, et al. Dose‐response relationship between lower serum magnesium level and higher prevalence of knee chondrocalcinosis. Arthritis Res Ther 2017;19:236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kellgren JH, Lawrence JS. Radiological assessment of osteo‐arthrosis. Ann Rheum Dis 1957;16:494–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Snyder EA, Alvarez C, Golightly YM, Renner JB, Jordan JM, Nelson AE. Incidence and progression of hand osteoarthritis in a large community‐based cohort: the Johnston County Osteoarthritis Project. Osteoarthritis Cartilage 2020;28:446–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forbes JD, Chen CY, Knox NC, Marrie RA, El‐Gabalawy H, de Kievit T , et al. A comparative study of the gut microbiota in immune‐mediated inflammatory diseases—does a common dysbiosis exist? Microbiome 2018;6:221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu BC, Hullar MA, Randolph TW, Franke AA, Monroe KR, Cheng I, et al. Associations of plasma trimethylamine N‐oxide, choline, carnitine, and betaine with inflammatory and cardiometabolic risk biomarkers and the fecal microbiome in the Multiethnic Cohort Adiposity Phenotype Study. Am J Clin Nutr 2020;111:1226–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhai G, Randell EW, Rahman P. Metabolomics of osteoarthritis: emerging novel markers and their potential clinical utility. Rheumatology (Oxford) 2018;57:2087–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Natividad JM, Lamas B, Pham HP, Michel ML, Rainteau D, Bridonneau C, et al. Bilophila wadsworthia aggravates high fat diet induced metabolic dysfunctions in mice. Nat Commun 2018;9:2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peck SC, Denger K, Burrichter A, Irwin SM, Balskus EP, Schleheck D. A glycyl radical enzyme enables hydrogen sulfide production by the human intestinal bacterium Bilophila wadsworthia. Proc Natl Acad Sci U S A 2019;116:3171–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang C, Zhang M, Pang X, Zhao Y, Wang L, Zhao L. Structural resilience of the gut microbiota in adult mice under high‐fat dietary perturbations. ISME J 2012;6:1848–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tamanai‐Shacoori Z, Smida I, Bousarghin L, Loreal O, Meuric V, Fong SB, et al. Roseburia spp.: a marker of health? [review]. Future Microbiol 2017;12:157–70. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, Xu L, Nevitt MC, Niu J, Goggins JP, Aliabadi P, et al. Lower prevalence of hand osteoarthritis among Chinese subjects in Beijing compared with white subjects in the United States: the Beijing Osteoarthritis Study. Arthritis Rheum 2003;48:1034–40. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


