Abstract
Liver transplantation is currently the only curative treatment for several liver diseases such as acute liver failure, end‐stage liver disorders, primary liver cancers, and certain genetic conditions. Unfortunately, despite improvements to transplantation techniques, including live donor transplantation, the number of organs available remains insufficient to meet patient needs. Hepatocyte transplantation has enabled some encouraging results as an alternative to organ transplantation, but primary hepatocytes are little available and cannot be amplified using traditional two‐dimensional culture systems. Indeed, although recent studies have tended to show that three‐dimensional culture enables long‐term hepatocyte culture, it is still agreed that, like most adult primary cell types, hepatocytes remain refractory to in vitro expansion. Because of their exceptional properties, human pluripotent stem cells (hPSCs) can be amplified indefinitely and differentiated into any cell type, including liver cells. While many teams have worked on hepatocyte differentiation, there has been a consensus that cells obtained after hPSC differentiation have more fetal than adult hepatocyte characteristics. New technologies have been used to improve the differentiation process in recent years. This review discusses the technical improvements made to hepatocyte differentiation protocols and the clinical approaches developed to date and anticipated in the near future.
Abbreviations
- ALF
acute liver failure
- BAL
bioartificial liver
- 2D/3D
two‐dimensional/three‐dimensional
- GMP
good manufacturing practices
- hESC
human embryonic stem cell
- hiPSC
human induced pluripotent stem cell
- HLC
hepatocyte‐like cell
- hPSC
human pluripotent stem cell
- MSC
mesenchymal stem cell
- OLT
orthotopic liver transplantation
- PHH
primary human hepatocyte
- PSC
pluripotent stem cell
Liver disorders have a variety of origins (metabolic, alcohol, viral, hereditary, cancer, immune, toxic, or drug‐related) and can lead to hepatic insufficiency, causing irreparable damage to the organ and becoming life‐threatening. In these cases, the only curative treatment is orthotopic liver transplantation (OLT),( 1 ) but some contraindications such as cardiac or respiratory failure may prevent this approach. Posttransplant immunosuppression is used to overcome any immune conflict between the recipient and the graft and prevent organ rejection. However, this lifelong treatment can also increase the risks of infection, malignancies, and other adverse effects. Operative or postoperative complications may also occur, such as failure of the newly transplanted organ. Finally, the limited number of donor livers remains a key issue; in 2018, 128 out of 1,000 patients died while on the waiting list for liver transplants in France (French Biomedicine Agency, https://www.agence‐biomedecine.fr/).
Artificial livers have already been proposed as an alternative or bridge to OLT. However, they have two important drawbacks that prevent their prolonged use: components in the system need to be replaced when they wear out and, more importantly, the patient plasma generated is markedly depleted in important biological substances. Proposals to include a bioreactor containing hepatocytes that might fulfill their detoxifying and synthetic functions have emerged, and such bioartificial livers (BALs) are now under development.
Hepatocyte transplantation might also offer an alternative to OLT, particularly when just liver function is deficient but the structure of the organ has remained unaffected (e.g., in some monogenic hereditary diseases) or as a temporary solution in patients waiting for a graft. However, whatever its type, cell therapy for liver diseases remains poorly developed.
Current Pitfalls Affecting Liver Cell Therapy
Until now, the principal obstacle to developing liver cell therapy was the lack of an unlimited and reliable source of hepatocytes. Several cell types can be used for this approach,( 2 ) summarized in Table 1; Supporting Table S1. Although primary human hepatocytes (PHHs) remain the gold standard, their poor availability still limits their use in clinical applications. To overcome this problem, culture techniques are currently being developed, including supporting matrices and three‐dimensional (3D) culture systems. Attempts have also been made to immortalize PHHs, but the results obtained so far have not been completely successful because of their tumorigenic tendency.( 3 )
TABLE 1.
Alternative Hepatocyte Source for Cell Transplantation and Therapeutic Applications. Additional Data to Table 1 are Available in Supplemental Table S1
| Cell Types | Definition and Clinical Applications | Drawbacks for Transplantation |
|---|---|---|
| Adult hepatocytes | Loss of functions in vitro | |
| Very low proliferation capacity under 2D culture conditions | ||
| Cell viability after cryopreservation | ||
| Clinic: transplantation in patients with liver diseases | Limited number of donor livers | |
| Limited engraftment into the liver | ||
| Fetal liver progenitors | Bipotent cells | Fetal origin |
| Highly proliferative in vitro | Need of cell purification | |
| Preclinic: can differentiate into mature hepatocytes after transplantation in animals | ||
| Less apoptotic and less immunogenic than adult hepatocytes | ||
| Clinic: transplanted in two patients with advanced liver cirrhosis | ||
| MSCs | Found in bone marrow and adipose tissue | |
| Able to differentiate into hepatocytes | ||
| Enhanced liver regeneration in animals | Phenotypic stability and contribution to long‐term tissue homeostasis to be demonstrated | |
| Embryonic stem cells and induced PSCs | Can be differentiated into hepatocytes | Need for GMP‐compatible protocols |
| Engraftment and rescue of ALF in animal | Careful examination of their genomic integrity needed |
Human pluripotent stem cells (hPSCs) can be differentiated in vitro into hepatocytes by the sequential addition of growth factors to the culture medium (Fig. 1) in order to mimic the principal stages of embryonic development and liver organogenesis (Table 2; Supporting Table S2). Human embryonic stem cells (hESCs) were initially proposed as a potential source for cell therapy, but their use is restricted by ethical issues. In 2007, Takahashi and Yamanaka( 5 ) were able to reprogram human adult somatic cells into embryo‐like cells called human induced pluripotent stem cells (hiPSCs). Since then, several reprogramming techniques and somatic cell types have been used to obtain hiPSCs (Table 3; Supporting Table S3).
FIG. 1.
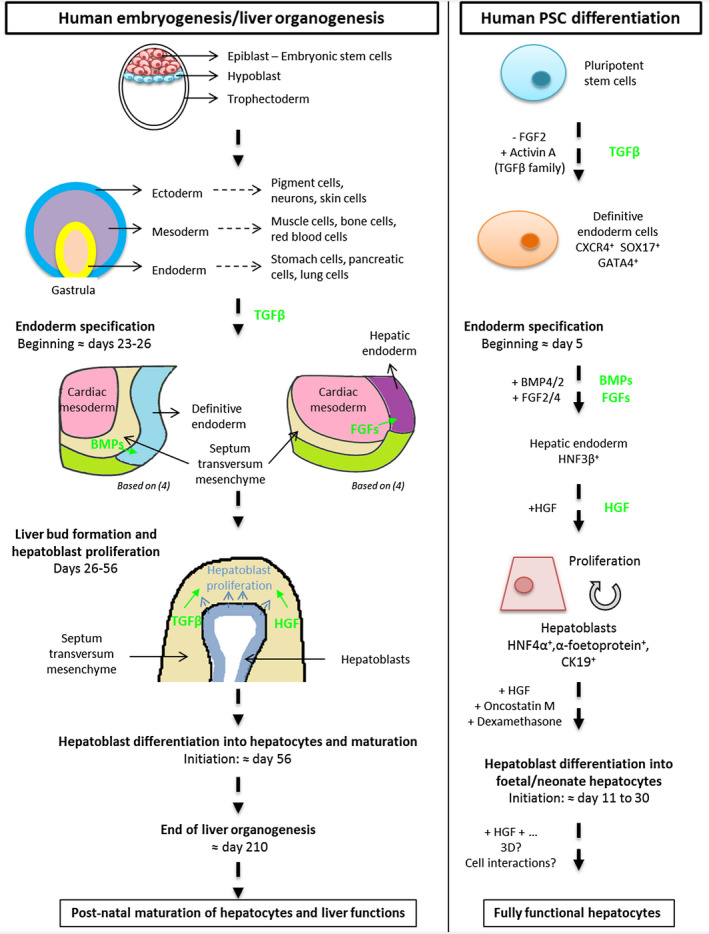
Main stages of human embryogenesis/liver organogenesis (left panel) mimicked and recapitulated in protocols for hPSCs into hepatocytes (right panel). Pathways involved during human liver embryogenesis are indicated in green. Abbreviations: BMP, bone morphogenic protein; CK19, cytokeratin 19; CXCR4, chemokine (C‐X‐C motif) receptor 4; GATA4, GATA binding protein 4; HGF, hepatocyte growth factor; HNF, hepatocyte nuclear factor; SOX17, SRY (sex determining region Y)‐box 17.
TABLE 2.
A Nonexhaustive Overview of the Principal Steps in the Liver Embryonic Development and Their Equivalents During the Hepatocyte Differentiation of PSCs
| Steps | In Vivo | In Vitro | |
|---|---|---|---|
| Embryonic Development | Growth Factors | Markers | |
| Definitive endoderm | Activin/Notch pathway | Activin A (TGFβ family) | CXCR4, SOX17, HNF3β, GATA4 |
| Endoderm specification | Factor secretion by the septum transversum | Bone morphogenetic protein 4/2 | Alpha‐fetoprotein, hepatocyte nuclear factor 4α |
| Factor secretion by the developing heart | FGF2/4 | ||
| Hepatoblast differentiation into hepatocytes and maturation | Members of the hepatocyte nuclear factor family | Hepatocyte growth factor | Hepatocyte nuclear factor 4α, alpha‐1‐anti‐trypsin, CYP3A7 |
| Inhibition of Notch and TGFβ pathways | Oncostatin M | Hepatocyte functions: albumin or urea secretion, energy metabolism, detoxification | |
| Dexamethasone | |||
Additional data are available in Supporting Table S2.
Abbreviations: CXCR4, chemokine (C‐X‐C motif) receptor 4; GATA4, GATA binding protein 4; HNF, hepatocyte nuclear factor; SOX17, SRY (sex determining region Y)‐box 17.
TABLE 3.
Reprogramming Strategies Developed to Generate Induced PSCs
| Reprogramming Methods | Advantages | Drawbacks | Induced PSC Generation (%) | Example of Somatic Cell Types Used | ||
|---|---|---|---|---|---|---|
| Integrative | Viral | Retrovirus | Highly efficient, stable, easy to use, efficient on a large number of cell types | Genomic integration (Cre/Lox system possible) | ≈0.1 | Fibroblasts, urine cells, MSCs, hepatocytes, GECs, NPCs |
| Lentivirus | Efficient, stable, easy to use, efficient on a large number of cell types | Genomic integration | ≈0.1 | Fibroblasts, PBMCs, adipose stem cells | ||
| Excisable lentiviral vectors | Excisable, relatively efficient | Genomic integration | ND | Fibroblasts | ||
| Inducible lentiviral systems | Controlled expression of reprogramming factors | Genomic integration | ND | Fibroblasts, melanocytes | ||
| Nonviral | PiggyBac | Excisable, relatively efficient | Genomic integration, efficiency? | ND | Fibroblasts | |
| Zinc finger nucleases | Targeted integration, excisable | Genomic integration | ND | Fibroblasts | ||
| Nonintegrative | Recombinant virus | Adenovirus | Very low genomic integration, easy‐to‐use | Time‐consuming, low efficiency, only in permissive cell types | ≈0.0002 | Fibroblasts, fetal liver cells |
| Sendai | No genomic integration, efficient on a large number of cell types, easily eliminated | Viral elimination not immediate (several passages required) | ≈0.1 | Fibroblasts, T cells, CD34+ cells | ||
| Episomal | Episomal vectors | Low risk of integration, easy to perform, controllably removable | Very low efficiency, time‐consuming | ≈0.0003‐0.0006 | Fibroblasts, CD34+ cells, dental pulp cells, PBMCs | |
| Others | mRNA | No genomic integration, very safe for clinical application | Daily transfection, efficiency dependent on cell type | ≈0.001‐2 | Fibroblasts, ADSCs, keratinocytes | |
| Proteins | No genomic integration, very safe for clinical application | Difficult to perform, low efficiency | ≈0.0005‐0.001 | Fibroblasts | ||
| Plasmids | Low risk of integration, easy to perform | Transient expression factors | ≈0.002‐0.01 | Fibroblasts, ADSCs, CD34+ cells, PBMCs | ||
| Small molecules | Highly efficient | Long to perform | ≈0.2 | Fibroblasts | ||
| miRNA | No genomic integration, very safe for clinical application | Can be time‐consuming, efficiency dependent on cell type | ≈0.03‐10 | Fibroblasts, hair follicle cells, ASCs | ||
Additional data are available in Supporting Table S3.
Abbreviations: ADSC, adipose‐derived stem cell; ASC, adipose stromal cell; CD34, cluster of differentiation 34; GEC, gastric epithelial cell; NPC, neural progenitor cell; PBMC, peripheral blood mononuclear cell.
During reprogramming, all epigenetic marks related to somatic cell identity need to be erased. Several studies have demonstrated differences in the epigenetic status of hiPSCs when compared to hESCs, probably due to incomplete erasure of the epigenetic marks during reprogramming. However, there is no clear consensus concerning the potential impact of this epigenetic memory on hiPSC‐derived cells. Nishizawa et al. showed that hiPSC lines with epigenetic variations displayed differing abilities to differentiate into blood cells.( 6 ) By contrast, 28 hiPSC lines differentiated into hepatocytes did not display any impact of tissue‐specific donor memory.( 7 ) These contrasting results may have been due to “transitory” epigenetic memory, which would be observed at early passages but lost after a few passages, because some culture parameters such as the passage number or culture time after cell thawing are rarely taken into account in such studies.( 8 )
In any case, the hESC( 9 ) and hiPSC( 10 ) differentiation approach enables a virtually inexhaustible source of hepatocytes, compatible with research and therapeutic applications. It is generally agreed that the hepatocytes obtained after the differentiation of pluripotent stem cells (PSCs) are better at mimicking fetal/neonatal hepatocytes than adult hepatocytes, as shown by their expression of the fetal form of albumin and limited cytochrome activities.( 11 ) However, they have been used effectively for transplantation in animal models of chronic liver disease or acute liver failure (ALF), thus indicating that the cells have continued to mature in vivo.( 12 ) These results tend to show that a lack of maturation of the differentiated cells might not necessarily be an obstacle to their use in transplantation. By contrast, drug‐screening approaches, toxicological studies, and BALs are reliant on hepatocyte functionality and require fully mature hepatocytes. New strategies have thus been developed, such as the addition of small molecules, coculture, and 3D culture systems (Fig. 2), which were initially developed to improve the culture of functional PHHs.
FIG. 2.
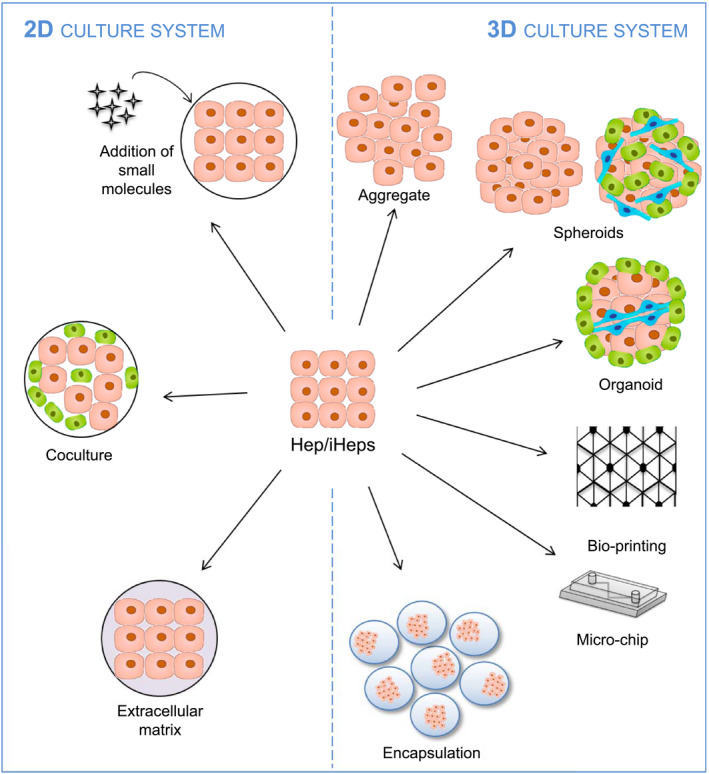
Improvements to hepatocyte or PSC‐derived hepatocyte culture systems. Abbreviations: Hep, hepatocyte; iHep, PSC‐derived hepatocyte.
Improvements to hPSC Differentiation into Hepatocytes Using 2D Culture Systems
Addition of Small Molecules
Although most of the protocols achieve hepatocyte differentiation using the growth factors or cytokines mentioned in Table 2, the addition of small molecules appears to enhance the efficiency of differentiation (Fig. 3). Examples of such molecules are CHIR99021, a small molecule that activates the WNT/β‐catenin pathway,( 13 ) and Ly294002, a phosphatidylinositol 3‐kinase inhibitor( 14 ) that synergizes with the Activin/Nodal pathway during the first days of differentiation.( 15 )
FIG. 3.
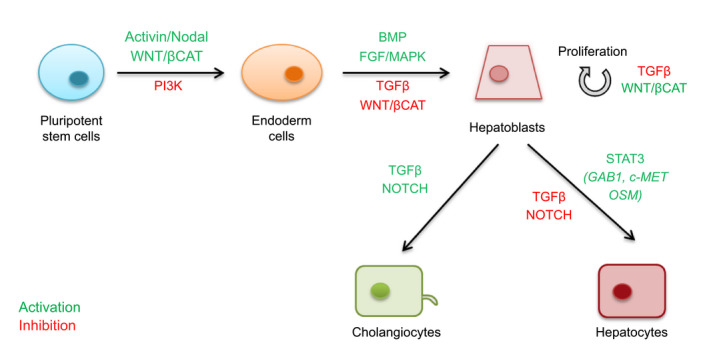
Pathways involved in PSC differentiation into hepatocytes and cholangiocytes. Abbreviations: βCAT, beta‐catenin; BMP, bone morphogenic protein; GAB1, growth factor receptor bound protein 2–associated protein 1; MAPK, mitogen‐activated protein kinase; OSM, oncostatin M; PI3K, phosphatidylinositol 3‐kinase; STAT3, signal transducer and activator of transcription 3.
During endoderm specification, the addition of retinoic acid,( 16 ) the chromatin modifier dimethyl sulfoxide,( 17 ) or the WNT/β‐catenin pathway inhibitor of Wnt response IWR‐1( 18 ) also enhances differentiation toward the hepatic lineage.
Finally, inhibition of the NOTCH pathway by compound E( 18 ) or of the TGFβ pathway by SB431542( 19 ) can prevent the cholangiocyte differentiation of hepatoblasts and thus favor the hepatocyte lineage.
Their principal value as chemicals is that they are less prone to batch variability and, unlike growth factors, are easily acceptable in good manufacturing practice (GMP) grade protocols.
Coculture of hPSC‐Derived Hepatocytes
Although the functions of PHHs rapidly decline in vitro, their coculture with mesenchymal stem cells (MSCs)( 20 ) or liver sinusoidal endothelial cells( 21 ) appears to improve their albumin production and cytochrome activity.
Coculture systems have thus been tested during PSC differentiation, in the presence of fibroblasts,( 22 ) liver stromal cells,( 23 ) and nonparenchymal human hepatic stellate cells.( 24 ) Interestingly, it has been shown that the improvement to the properties of hepatocyte‐like cells (HLCs) was mostly due to the coculture medium rather than the presence of human umbilical vein endothelial cells (HUVECs), although they are not liver endothelial cells.( 25 )
Extracellular Matrix
Because they are epithelial cells with complex polarization and strong adhesion properties, hepatocytes require a complex interplay of various factors to maintain their physiological characteristics and liver functions in vitro. Indeed, cell‐to‐cell interactions are largely responsible for retention of their xenobiotic metabolism capacity, whereas hepatic transport is mainly dependent on hepatocyte polarization, which is also related to cell–matrix interactions. Artificial constructs that can mimic the cell niche (such as nanofibers, films, or hydrogels) have thus been developed to enhance hepatocyte adhesion, migration, and proliferation. These scaffolds are generally made of natural polymers such as collagen, chitosan, gelatin, alginate, and agarose, or synthetic substances such as poly(ε‐caprolactone) and poly(L‐lactic acid).( 26 )
Culture of PHHs between two layers of collagen I or Matrigel, a widely used matrix composed of numerous basement membrane proteins, enables the formation of cell–cell and cell–matrix contacts similar to those observed in vivo.( 27 ) These results have been applied to hESCs and enabled better differentiation into definitive endoderm( 28 ) and a marked improvement in HLC polarization.( 29 )
Finally, several studies have reported the use of decellularized human liver extracellular matrix for hiPSC differentiation, causing the up‐regulation of hepatic functions when compared with standard differentiation.( 30 )
Fluidized Microchips
Microchips enable the miniaturization of traditional culture systems. They are generally molded in a transparent, hydrophobic, and biocompatible material such as polydimethylsiloxane that can be treated to modify their surface properties.
To best mimic the in vivo cell environment, they also include a blood flow equivalent, supplied by microfluidic systems which deliver oxygen and nutrients and drain waste. This approach was used with PHHs( 31 ) and later during hiPSC differentiation,( 32 ) thus confirming that biochips offer a favorable microenvironment for hepatocyte differentiation. Furthermore, the reported creation of a gradient of oxygen concentrations ranging from normoxia to severe hypoxia on a chip made it possible to mimic the physiological oxygen gradient generated in the liver.( 33 ) This could be of considerable interest for the study of liver zonation.
3D Culture Systems for hPSC Differentiation into Hepatocytes
Physical Properties of Cell Adhesion and Aggregation
Organ formation results from the ability of different cell populations to migrate and rearrange themselves. Regulation of these rearrangements has been well established through the study of adhesion molecules and cadherin expression. If a single cell type is involved in aggregate formation without any externally applied force, the homogeneous distribution of adhesion molecules leads to cells grouping like liquid droplets, sticking together to minimize their surface free energy.( 34 ) When several cell populations are mixed, the differences in their adhesion and chemotactic properties will produce a structure where less cohesive cells envelop those that are more cohesive because of their lower surface tension (Fig. 4).
FIG. 4.
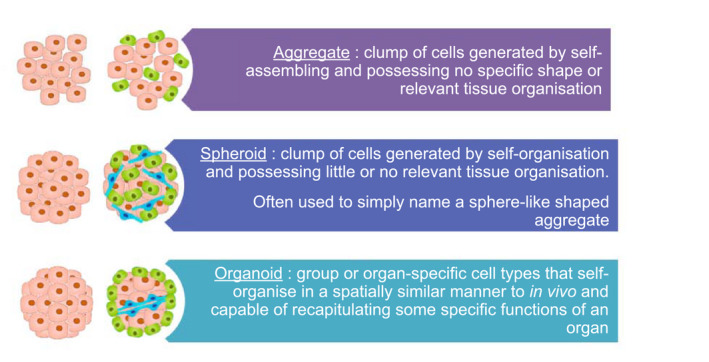
Three‐dimensional cell aggregation and organization in scaffold‐free culture systems.
Aggregates and Spheroids
The 3D culture systems have been tested on PHHs to define parameters such as the maximum size of spheroids to prevent necrosis or the number of cells to permit their optimal compaction. Culture of PHHs in aggregates enables prolongation of their culture in vitro while maintaining the expression of specific markers and CYP activity.( 35 ) The same results have been obtained during hESC( 36 ) and hiPSC( 37 ) hepatocyte differentiation.
Organoids
Organoids can be defined as self‐organizing 3D structures that mimic some of the in vivo functions of an organ.( 38 ) Because they self‐organize in vitro from stem cells into tissues, they can mimic many of the cellular interactions of the organ and be used to reconstruct microstructures that more or less resemble liver tissues.( 39 )
Takebe et al. were the first to report the formation of a vascularized hepatic bud composed initially of hiPSC‐derived endoderm cells, HUVECs, and human MSCs( 40 ) and 4 years later was made entirely of hiPSC‐derived cells.( 41 ) However, the lack of a biliary network in these coculture systems may prevent the long‐term culture of these organoids. Since then, several coculture systems have enabled improvements in terms of protein production when cocultured with stellate cells or cholangiocytes.( 42 )
Bioprinting
Bioprinting uses computer‐controlled printing technology to build, layer by layer or point by point, tissues and organs with cells or cell aggregates as the building blocks.( 43 ) hESC‐HLCs have been bioprinted in an alginate hydrogel,( 44 ) or hiPSC‐HLCs in gelatin, leading to improved albumin secretion and urea production as well as a higher expression of several cytochrome P450s when compared to a 2D culture system.( 45 ) Bioprinting approaches have also been used with spheroids as building blocks( 46 ) or associating different cell types, for example, the bioprinting of liver spheroids derived from hiPSCs with nonparenchymal cells.( 47 )
Microchips
Microchips can mimic the in vivo cell environment, including a blood flow equivalent. Proof of concept of the long‐term perfusion of 3D HepG2 spheroids showed that their use enabled a significant improvement in liver‐specific functions and metabolic activity compared to conventional perfusion methods.( 48 ) hiPSC‐HLC organoids have been included in a 3D fluidized chip and displayed a marked enhancement of liver‐specific functions, including albumin and urea production as well as metabolic capabilities, when compared to static systems.( 49 )
Bioprinting approaches can also be used in combination with microchips, using bioprinted HepG2 spheroids instead of single cells.( 50 )
Despite these promising results, bioprinting and microchips are designed, engineered, and human‐made, thus confining them to the limits of our own knowledge of the organ.
Encapsulation Systems
Encapsulation consists in entrapping cells in semipermeable spheres made of hydrogels that can be chemically modified to adapt their porosity as necessary. This technology enables the exchange of nutrients, oxygen, CO2, and signals through the bead while permitting the diffusion of growth factors, metabolites, and waste but preventing antibody penetration. Encapsulation in alginate poly‐l‐lysine beads maintained cell viability and allowed the differentiation of hPSCs into hepatocytes, using spheroids or organoids.
In vivo, the successful engraftment of encapsulated aggregates of hiPSC‐HLCs led to the secretion of human albumin in mouse blood at levels similar to those attained with PHHs.( 51 )
These encapsulated structures can also be used in BAL applications, for example, the use of HepG2 in alginate beads in a porcine model of liver failure( 52 ) and differentiated HepaRG cells self‐organized as spheroids.( 53 )
Applications for hPSC Differentiation into Hepatocytes
Disease Modeling
The differentiation of hiPSCs obtained from patient biopsies into a cell type of interest enables the in vitro modeling of genetic diseases (Fig. 5). For example, our team reported on the use of hiPSCs to model familial hypercholesterolemia and showed that hiPSC‐HLCs could be used to study the regulation of cholesterol metabolism.( 54 ) However, the viability of these models is heavily reliant on the characteristics and functions of the differentiated cells. In the case of hepatocytes, this feature may be limited by incomplete cell maturation.
FIG. 5.
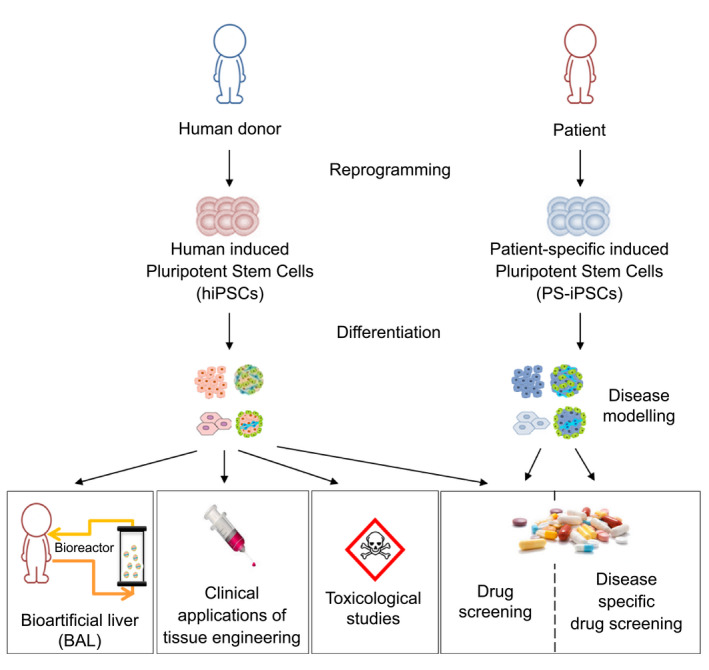
Applications for hPSC differentiation into hepatocytes.
Cystic fibrosis( 55 ) and alpha‐1 antitrypsin deficiency( 56 ) are examples of genetic diseases modeled using human organoids. However, in the majority of cases, the focus has mostly been on the effects of a given mutation by studying the expression of the protein involved, while downstream networks affected by the mutation are often poorly documented.
More than monogenic diseases, Ouchi et al. showed that treatment of iPSC‐HLC organoids with free fatty acids could mimic the lipid accumulation and fibrosis of nonalcoholic fatty liver disease.( 57 )
Toxicological Studies
In 2019, drug‐induced liver injury had an estimated annual incidence of between 1 per 10,000 and 1 per 100,000 people exposed to medications. In view of the important role of the liver in xenobiotic processing, 3D liver structures have been used during pharmacological studies.
PHHs offer a robust system for long‐term studies of drug‐induced hepatotoxicity,( 58 ) but 3D coculture systems using hepatic cell lines have also been used and revealed relevant differences between spheroids of HepG2( 59 ) and HepaRG( 60 ) cells.
In view of the shortage of PHHs and the controversial evidence regarding cell lines, liver tissue made from hESC‐HLCs( 61 ) and hiPSC‐HLCs( 62 ) was used and displayed a better response to apoptotic drugs than cell lines.( 63 ) However, Godoy and colleagues showed that monohepatocyte cultures have limited abilities to reproduce drug‐induced hepatotoxic effects because the toxic response observed in vivo is mediated by a complex interplay between different cell types.( 64 )
Drug Screening
Drug screening is used to develop drugs that can selectively interact with genes and gene products or can interfere with a specific molecular mechanism so that they can be used in human clinical trials. However, they do not always ensure safe and effective treatment in humans. One of the key factors that guides the success or failure of drug screening is a lack of pharmacokinetic and pharmacodynamic knowledge. In the context of conventional 2D screening, determining a “dose and drug effect” may be hampered by phenomena such as adsorption on plastic.( 65 ) Spheroids, organoids, and organs‐on‐chips seem more appropriate to investigate the pharmacokinetic profiles of drugs.( 66 )
BALs
Because the creation of an artificial device that can fully sustain liver functions remains problematic, considerable attention is being paid at present to BAL systems, where hepatocytes or liver organoids constitute the biological components that are missing from current artificial systems. BALs including bioreactors that host alginate‐encapsulated hepatocyte spheroids have thus been developed.( 67 ) Several systems have been evaluated in patients, but only two have produced promising results: the extracorporeal liver assist device and Hepat‐Assist, where the biological components are hepatoma cell lines and porcine hepatocytes, respectively. In 2017, the first GMP clinical‐scale BAL containing HepG2 organoids was developed on a porcine model of severe liver failure.( 52 )
As a functional BAL requires large quantities of hepatocytes to sustain hemodialysis and hepatic functions in the patient,( 68 ) PSC‐HLCs are emerging as attractive biological components; but their long‐term survival and functionality, as well as the high costs of their production, are the most serious drawbacks affecting their rapid application.
Clinical Applications of Tissue Engineering
Regenerative medicine is one of the principal challenges in terms of replacing damaged and/or nonfunctional tissue. The limited supply of healthy donor tissue and the inherent risks of tissue rejection that restrict this application can be overcome using isogenic or human leukocyte antigen (HLA)–compatible hiPSC‐HLCs. Two different strategies can be followed. The first consists in using a decellularized liver as a 3D matrix in which differentiated cells are seeded,( 69 ) and the second consists in using PSC‐derived organoids. However, regarding transplantation, a final challenge remains in terms of scalability and reproducibility of protocols to produce large and stable organoids following cryopreservation.( 70 )
In this context, a recent study reported the development of hESC‐derived organoids that could be expanded for 20 passages while stably maintaining the phenotypic features of bipotential progenitor cells.( 71 ) After transplantation in mice, they could differentiate into functional hepatocytes or cholangiocytes and displayed a remarkable capacity for repopulation. This large‐scale expansion of progenitor cells could overcome the scalability problem for clinical applications.
Moreover, Takebe et al. demonstrated the possibility of transplanting liver buds entirely composed of hiPSC‐derived cells,( 41 ) including endothelial cells that developed vascularization once transplanted in the animal, thus confirming the huge therapeutic potential of this approach. The technology has also been used in an ALF mouse model where animal survival was probably due to the early function of transplanted cells bridging the native recovery of the liver, highlighting the importance of the bridge function potentially assumed by transplanted cells, whatever their subsequent function after transplantation.
Limits to the Therapeutic Use of hPSC‐Derived Organoids
Practical Aspects
The autologous transplantation of differentiated cells requires not only reprogramming of the patient’s cells into hiPSCs but their characterization, their genetic correction in the case of a patient suffering from a monogenic disease, and their differentiation into hepatocytes. These steps can take from 3 to 4 months, thus limiting this approach to moderate or less severe cases. Moreover, the production cost of clinical‐grade hiPSC‐HLCs with all safety controls is estimated to be $200,000/patient, so this must also be taken into account. For these reasons, allogenic strategies are mostly privileged, with the development of banks of frozen HLA‐characterized and ready‐to‐use therapeutic products. The development of cryopreservation methods that sustain prefreezing hepatocyte functions will therefore be necessary. Moreover, the production of organoids at a very large scale is currently difficult as it first requires the mass culture of PSCs. Indeed, for transplantation, 108 cells/kg would be necessary, which represents 6 to 12 × 109 cells/transplantation.( 72 ) BALs would require even more cells, from 1010 to 2010 hepatocytes. For these reasons, the standardization and at least partial automation of organoid production will be necessary.( 73 )
Finally, the route of hepatocyte administration to the patient also needs to be determined. Four main strategies are currently under investigation and have been reviewed by Anderson and Zarrinpar( 74 ) Intraportal injection seems to be preferred as it can deliver a large number of cells into the hepatic sinusoids and is well tolerated in patients without fibrosis. However, it can cause elevated portal pressure and a risk of portal vein thrombosis. In any case, the cell anchorage could be improved by encapsulating hepatocytes in alginate beads, which would also protect them from the patient’s immune response. This strategy has produced promising results during preclinical studies.( 51 ) Other strategies such as cell sheets have been developed and have already been reviewed in depth.( 75 )
Safety of hPSC‐Derived Hepatocytes
Although 3D organoids are more physiologically relevant than monolayer cultures, there are some limitations to their therapeutic use. Considerable efforts have been made to develop appropriate GMP guidelines, in terms of culture media, matrix components, and growth factors, as well as reproducible and validated protocols to culture and differentiate hPSCs. Three GMP PSC lines have already been differentiated into hepatocytes and displayed highly reproducible phenotypes and functionality.( 76 ) Optimized GMP‐grade alginate encapsulation protocols have also been established for the transplantation of human hepatocytes.( 77 ) Quality controls on the final product, in terms not only of function but also of safety, also need to be defined.
Another major concern inherent to the use of hPSC‐derived cells is linked to the potential genetic instability of the cells. The genomic integrity of PSCs must be ensured, although not all mutations will result in undesirable effects or tumors. Recurrent characteristic abnormalities have been reported in hPSCs but have not been found in corresponding somatic cell samples.( 78 ) However, the preservation of genomic integrity during differentiation into hiPSC‐HLCs remains poorly documented, and no link has yet been established between the differentiation protocol and their genetic integrity. Our experiments evidenced the absence of de novo copy number variations during differentiation( 79 ) and showed that safety mainly depended on hiPSC clone genomic integrity.( 80 ) However, further studies are required in this area.
The potential presence of residual hPSCs in the final therapeutic product must also be taken into account, even if several studies have already reported the transplantation of differentiated hepatocytes, under either 2D( 81 ) or 3D( 71 ) conditions, without any formation of teratoma or tumors in mice.
Finally, many groups have investigated the immunogenicity of hPSC‐derived cells. Some of them showed that they were well tolerated by the immune system,( 82 ) but others reported different immune responses depending on the cell line used.( 83 ) However, several cellular therapies have been tested using hiPSC‐derived cells according to an autologous( 84 ) or allogenic( 85 ) strategy and caused no signs of immune rejection or tumor formation.
Ethical Concerns Relative to the Clinical Use of hPSC‐HLCs
The use of hPSC‐HLCs in cell therapy involves ethical issues regarding both their human origin and the potential use of hESCs. In this regard, the guidelines applied worldwide range from total prohibition to regulated authorization. However, hiPSCs bypass the ethical concerns of embryo destruction because they are produced from somatic cells. Several guidelines are currently available regarding the use of cells, tissues, and PSC products to treat patients, most of them issued by the American and European authorities (the US Food and Drug Administration and the European Medicines Agency, respectively).
In all cases, cell therapy, gene therapy, and tissue engineering require rapid changes to the legislation in line with the advancement of technologies in the fields of biomedicine or regenerative medicine and the emergence of new areas such as PSC research.
Conclusion
Because of the rapid loss of hepatic functions by PHHs in vitro, due in part to their poor spatial organization in a monolayer culture, numerous approaches have been successfully developed to improve cell–cell or cell–matrix interactions in 3D culture systems; but the problem of cell shortage persists. The 3D culture systems have been adapted to PSC differentiation and revealed clear improvements in the efficiency of hESC( 86 ) and hiPSC( 62 ) differentiation into hepatocytes. The use of liver organoids for drug screening and toxicity analyses could reduce the use of animal testing as animals would mainly be used for studies that require whole‐organism readouts. The potential of these new culture techniques is thus exciting, but the analysis of 3D structures requires the development of specific protocols and equipment; for example, microscopic analyses need to be redesigned to generate sufficiently informative images whose processing can rapidly become highly complex.
Therapeutic applications for clinical‐grade PSC‐HLCs are starting to emerge, and the problem of incomplete HLC maturation appears to be surmountable thanks to the development of 3D culture techniques. Interdisciplinary collaborations are necessary across cell biology, clinical care, bioengineering, and the science of biocompatible materials to enable further advances in the future.
Author Contributions
E.L. was involved in the conceptualization, investigation and visualization of the review as well as the lead writing of the original draft. A.M. participated to the investigation and the supporting writing of the review. J.‐C.D.‐V. and A.D.‐K. acquired the financial support and all authors revised the review and approved the final version.
Supporting information
Supporting Table T1–T3
Acknowledgment
We thank Dr. Anne Weber for her scientific expertise as well as her careful reading and revision of the manuscript.
Supported by the RHU program “iLite” on “Innovations for Liver Tissue Engineering” granted by PAI2 through ANR‐16‐RHUS‐0005.
Potential conflict of interest: Nothing to report.
References
Author names in bold designate shared co‐first authorship.
- 1.Starzl TE, Putnam CW, Koep LJ. Current status of liver transplantation. South Med J 1977;70:389‐390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Messina A, Luce E, Hussein M, Dubart‐Kupperschmitt A. Pluripotent‐stem‐cell‐derived hepatic cells: hepatocytes and organoids for liver therapy and regeneration. Cells 2020;9:420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hang H‐L, Liu X‐Y, Wang H‐T, Xu N, Bian J‐M, Zhang J‐J, et al. Hepatocyte nuclear factor 4A improves hepatic differentiation of immortalized adult human hepatocytes and improves liver function and survival. Exp Cell Res 2017;360:81‐93. [DOI] [PubMed] [Google Scholar]
- 4.Bort R. Hex homeobox gene‐dependent tissue positioning is required for organogenesis of the ventral pancreas. Development 2004;131:797‐806. [DOI] [PubMed] [Google Scholar]
- 5.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007;131:861‐872. [DOI] [PubMed] [Google Scholar]
- 6.Nishizawa M, Chonabayashi K, Nomura M, Tanaka A, Nakamura M, Inagaki A, et al. Epigenetic variation between human induced pluripotent stem cell lines is an indicator of differentiation capacity. Cell Stem Cell 2016;19:341‐354. [DOI] [PubMed] [Google Scholar]
- 7.Kajiwara M, Aoi T, Okita K, Takahashi R, Inoue H, Takayama N, et al. Donor‐dependent variations in hepatic differentiation from human‐induced pluripotent stem cells. Proc Natl Acad Sci U S A 2012;109:12538‐12543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vaskova EA, Stekleneva AE, Medvedev SP, Zakian SM. “Epigenetic memory” phenomenon in induced pluripotent stem cells. Acta Naturae 2013;5:15‐21. [PMC free article] [PubMed] [Google Scholar]
- 9.Touboul T, Hannan NRF, Corbineau S, Martinez A, Martinet C, Branchereau S, et al. Generation of functional hepatocytes from human embryonic stem cells under chemically defined conditions that recapitulate liver development. Hepatology 2010;51:1754‐1765. [DOI] [PubMed] [Google Scholar]
- 10.Si‐Tayeb K, Noto FK, Nagaoka M, Li J, Battle MA, Duris C, et al. Highly efficient generation of human hepatocyte‐like cells from induced pluripotent stem cells. Hepatology 2010;51:297‐305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baxter M, Withey S, Harrison S, Segeritz C‐P, Zhang F, Atkinson‐Dell R, et al. Phenotypic and functional analyses show stem cell–derived hepatocyte‐like cells better mimic fetal rather than adult hepatocytes. J Hepatol 2015;62:581‐589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fourrier A, Delbos F, Menoret S, Collet C, Thi Thuy LT, Myara A, et al. Regenerative cell therapy for the treatment of hyperbilirubinemic Gunn rats with fresh and frozen human induced pluripotent stem cells–derived hepatic stem cells. Xenotransplantation 2020;27:e12544. [DOI] [PubMed] [Google Scholar]
- 13.Mfopou JK, Geeraerts M, Dejene R, Van Langenhoven S, Aberkane A, Van Grunsven LA, et al. Efficient definitive endoderm induction from mouse embryonic stem cell adherent cultures: a rapid screening model for differentiation studies. Stem Cell Res 2014;12:166‐177. [DOI] [PubMed] [Google Scholar]
- 14.Mathew S, Jaramillo M, Zhang X, Zhang L, Soto‐Gutiérrez A, Banerjee I. Analysis of alternative signaling pathways of endoderm induction of human embryonic stem cells identifies context specific differences. BMC Syst Biol 2012;6:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hay DC, Fletcher J, Payne C, Terrace JD, Gallagher RCJ, Snoeys J, et al. Highly efficient differentiation of hESCs to functional hepatic endoderm requires ActivinA and Wnt3a signaling. Proc Natl Acad Sci U S A 2008;105:12301‐12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Negishi T, Nagai Y, Asaoka Y, Ohno M, Namae M, Mitani H, et al. Retinoic acid signaling positively regulates liver specification by inducing wnt2bb gene expression in medaka. Hepatology 2010;51:1037‐1045. [DOI] [PubMed] [Google Scholar]
- 17.Czysz K, Minger S, Thomas N. DMSO efficiently down regulates pluripotency genes in human embryonic stem cells during definitive endoderm derivation and increases the proficiency of hepatic differentiation. PLoS One 2015;10:e0117689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Touboul T, Chen S, To CC, Mora‐Castilla S, Sabatini K, Tukey RH, et al. Stage‐specific regulation of the WNT/β‐catenin pathway enhances differentiation of hESCs into hepatocytes. J Hepatol 2016;64:1315‐1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lv L, Han Q, Chu Y, Zhang M, Sun L, Wei W, et al. Self‐renewal of hepatoblasts under chemically defined conditions by iterative growth factor and chemical screening. Hepatology 2015;61:337‐347. [DOI] [PubMed] [Google Scholar]
- 20.Fitzpatrick E, Wu Y, Dhadda P, Hughes RD, Mitry RR, Qin H, et al. Coculture with mesenchymal stem cells results in improved viability and function of human hepatocytes. Cell Transplant 2015;24:73‐83. [DOI] [PubMed] [Google Scholar]
- 21.Bale SS, Golberg I, Jindal R, McCarty WJ, Luitje M, Hegde M, et al. Long‐term coculture strategies for primary hepatocytes and liver sinusoidal endothelial cells. Tissue Eng Part C Methods 2015;21:413‐422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu Y‐D, Kim K‐H, Lee S‐G, Choi S‐Y, Kim Y‐C, Byun K‐S, et al. Hepatic differentiation from human embryonic stem cells using stromal cells. J Surg Res 2011;170:e253‐e261. [DOI] [PubMed] [Google Scholar]
- 23.Elham H, Fardin F, Mahmod H. The roles of the co‐culture of mEScs with pancreatic islets and liver stromal cells in the differentiation of definitive endoderm cells. Biologicals 2017;45:9‐14. [DOI] [PubMed] [Google Scholar]
- 24.Javed MS, Yaqoob N, Iwamuro M, Kobayashi N, Fujiwara T. Generation of hepatocyte‐like cells from human induced pluripotent stem (iPS) cells by co‐culturing embryoid body cells with liver non‐parenchymal cell line TWNT‐1. J Coll Physicians Surg Pak 2014;24:91‐96. [PubMed] [Google Scholar]
- 25.Freyer N, Greuel S, Knöspel F, Strahl N, Amini L, Jacobs F, et al. Effects of co‐culture media on hepatic differentiation of hiPSC with or without HUVEC co‐culture. Int J Mol Sci 2017;18:1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eltom A, Zhong G, Muhammad A. Scaffold techniques and designs in tissue engineering functions and purposes: a review. Adv Mater Sci Eng 2019;2019:1‐13. [Google Scholar]
- 27.Deharde D, Schneider C, Hiller T, Fischer N, Kegel V, Lübberstedt M, et al. Bile canaliculi formation and biliary transport in 3D sandwich‐cultured hepatocytes in dependence of the extracellular matrix composition. Arch Toxicol 2016;90:2497‐2511. [DOI] [PubMed] [Google Scholar]
- 28.Lawton BR, Sosa JA, Roman S, Krause DS. Effect of a Matrigel sandwich on endodermal differentiation of human embryonic stem cells. Stem Cell Rev 2013;9:578‐585. [DOI] [PubMed] [Google Scholar]
- 29.Palakkan A, Drummond R, Anderson R, Greenhough S, Tv K, Hay D, et al. Polarisation and functional characterisation of hepatocytes derived from human embryonic and mesenchymal stem cells. Biomed Rep 2015;3:626‐636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jaramillo M, Yeh H, Yarmush ML, Uygun BE. Decellularized human liver extracellular matrix (hDLM)–mediated hepatic differentiation of human induced pluripotent stem cells (hIPSCs). J Tissue Eng Regen Med 2018;12:e1962‐e1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vinci B, Duret C, Klieber S, Gerbal‐Chaloin S, Sa‐Cunha A, Laporte S, et al. Modular bioreactor for primary human hepatocyte culture: medium flow stimulates expression and activity of detoxification genes. Biotechnol J 2011;6:554‐564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Danoy M, Bernier ML, Kimura K, Poulain S, Kato S, Mori D, et al. Optimized protocol for the hepatic differentiation of induced pluripotent stem cells in a fluidic microenvironment. Biotechnol Bioeng 2019;116:1762‐1776. [DOI] [PubMed] [Google Scholar]
- 33.Kang YBA, Eo J, Bulutoglu B, Yarmush ML, Usta OB. Progressive hypoxia‐on‐a‐chip: an in vitro oxygen gradient model for capturing the effects of hypoxia on primary hepatocytes in health and disease. Biotechnol Bioeng 2020;117:763‐775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Foty RA, Pfleger CM, Forgacs G, Steinberg MS. Surface tensions of embryonic tissues predict their mutual envelopment behavior. Dev Camb Engl 1996;122:1611‐1620. [DOI] [PubMed] [Google Scholar]
- 35.Bell CC, Hendriks DFG, Moro SML, Ellis E, Walsh J, Renblom A, et al. Characterization of primary human hepatocyte spheroids as a model system for drug‐induced liver injury, liver function and disease. Sci Rep 2016;6:25187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim JH, Jang YJ, An SY, Son J, Lee J, Lee G, et al. Enhanced metabolizing activity of human ES cell‐derived hepatocytes using a 3D culture system with repeated exposures to xenobiotics. Toxicol Sci 2015;147:190‐206. [DOI] [PubMed] [Google Scholar]
- 37.Pettinato G, Ramanathan R, Fisher RA, Mangino MJ, Zhang N, Wen X. Scalable differentiation of human iPSCs in a multicellular spheroid‐based 3D culture into hepatocyte‐like cells through direct Wnt/β‐catenin pathway inhibition. Sci Rep 2016;6:32888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lancaster MA, Knoblich JA. Organogenesis in a dish: modeling development and disease using organoid technologies. Science 2014;345:1247125. [DOI] [PubMed] [Google Scholar]
- 39.Rossi G, Manfrin A, Lutolf MP. Progress and potential in organoid research. Nat Rev Genet 2018;19:671‐687. [DOI] [PubMed] [Google Scholar]
- 40.Takebe T, Sekine K, Enomura M, Koike H, Kimura M, Ogaeri T, et al. Vascularized and functional human liver from an iPSC‐derived organ bud transplant. Nature 2013;499:481‐484. [DOI] [PubMed] [Google Scholar]
- 41.Takebe T, Sekine K, Kimura M, Yoshizawa E, Ayano S, Koido M, et al. Massive and reproducible production of liver buds entirely from human pluripotent stem cells. Cell Rep 2017;21:2661‐2670. [DOI] [PubMed] [Google Scholar]
- 42.Vyas D, Baptista PM, Brovold M, Moran E, Gaston B, Booth C, et al. Self‐assembled liver organoids recapitulate hepatobiliary organogenesis in vitro. Hepatology 2018;67:750‐761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bishop ES, Mostafa S, Pakvasa M, Luu HH, Lee MJ, Wolf JM, et al. 3‐D bioprinting technologies in tissue engineering and regenerative medicine: current and future trends. Genes Dis 2017;4:185‐195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Faulkner‐Jones A, Fyfe C, Cornelissen D‐J, Gardner J, King J, Courtney A, et al. Bioprinting of human pluripotent stem cells and their directed differentiation into hepatocyte‐like cells for the generation of mini‐livers in 3D. Biofabrication 2015;7:044102. [DOI] [PubMed] [Google Scholar]
- 45.Ma X, Qu X, Zhu W, Li Y‐S, Yuan S, Zhang H, et al. Deterministically patterned biomimetic human iPSC‐derived hepatic model via rapid 3D bioprinting. Proc Natl Acad Sci U S A 2016;113:2206‐2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kizawa H, Nagao E, Shimamura M, Zhang G, Torii H. Scaffold‐free 3D bio‐printed human liver tissue stably maintains metabolic functions useful for drug discovery. Biochem Biophys Rep 2017;10:186‐191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goulart E, de Caires‐Junior LC, Telles‐Silva KA, Araujo BHS, Rocco SA, Sforca M, et al. 3D bioprinting of liver spheroids derived from human induced pluripotent stem cells sustain liver function and viability in vitro. Biofabrication 2019;12:015010. [DOI] [PubMed] [Google Scholar]
- 48.Ma L‐D, Wang Y‐T, Wang J‐R, Wu J‐L, Meng X‐S, Hu P, et al. Design and fabrication of a liver‐on‐a‐chip platform for convenient, highly efficient, and safe in situ perfusion culture of 3D hepatic spheroids. Lab Chip 2018;18:2547‐2562. [DOI] [PubMed] [Google Scholar]
- 49.Wang Y, Wang H, Deng P, Chen W, Guo Y, Tao T, et al. In situ differentiation and generation of functional liver organoids from human iPSCs in a 3D perfusable chip system. Lab Chip 2018;18:3606‐3616. [DOI] [PubMed] [Google Scholar]
- 50.Bhise NS, Manoharan V, Massa S, Tamayol A, Ghaderi M, Miscuglio M, et al. A liver‐on‐a‐chip platform with bioprinted hepatic spheroids. Biofabrication 2016;8:014101. [DOI] [PubMed] [Google Scholar]
- 51.Song W, Lu Y‐C, Frankel AS, An D, Schwartz RE, Ma M. Engraftment of human induced pluripotent stem cell–derived hepatocytes in immunocompetent mice via 3D co‐aggregation and encapsulation. Sci Rep 2015;5:16884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Selden C, Bundy J, Erro E, Puschmann E, Miller M, Kahn D, et al. A clinical‐scale bioartificial liver, developed for GMP, improved clinical parameters of liver function in porcine liver failure. Sci Rep 2017;7:14518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pasqua M, Pereira U, Messina A, de Lartigue C, Vigneron P, Dubart‐Kupperschmitt A, et al. HepaRG self‐assembled spheroids in alginate beads meet the clinical needs for bioartificial liver. Tissue Eng Part A 2020;26:613‐622. [DOI] [PubMed] [Google Scholar]
- 54.Caron J, Pène V, Tolosa L, Villaret M, Luce E, Fourrier A, et al. Low‐density lipoprotein receptor–deficient hepatocytes differentiated from induced pluripotent stem cells allow familial hypercholesterolemia modeling, CRISPR/Cas‐mediated genetic correction, and productive hepatitis C virus infection. Stem Cell Res Ther 2019;10:221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sampaziotis F, Cardoso de Brito M, Madrigal P, Bertero A, Saeb‐Parsy K, Soares FAC, et al. Cholangiocytes derived from human induced pluripotent stem cells for disease modeling and drug validation. Nat Biotechnol 2015;33:845‐852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gómez‐Mariano G, Matamala N, Martínez S, Justo I, Marcacuzco A, Jimenez C, et al. Liver organoids reproduce alpha‐1 antitrypsin deficiency–related liver disease. Hepatol Int 2020;14:127‐137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ouchi R, Togo S, Kimura M, Shinozawa T, Koido M, Koike H, et al. Modeling steatohepatitis in humans with pluripotent stem cell–derived organoids. Cell Metab 2019;30:374‐384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bell CC, Dankers ACA, Lauschke VM, Sison‐Young R, Jenkins R, Rowe C, et al. Comparison of hepatic 2D sandwich cultures and 3D spheroids for long‐term toxicity applications: a multicenter study. Toxicol Sci 2018;162:655‐666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ramaiahgari SC, den Braver MW, Herpers B, Terpstra V, Commandeur JNM, van de Water B, et al. A 3D in vitro model of differentiated HepG2 cell spheroids with improved liver‐like properties for repeated dose high‐throughput toxicity studies. Arch Toxicol 2014;88:1083‐1095. [DOI] [PubMed] [Google Scholar]
- 60.Mueller D, Krämer L, Hoffmann E, Klein S, Noor F. 3D organotypic HepaRG cultures as in vitro model for acute and repeated dose toxicity studies. Toxicol In Vitro 2014;28:104‐112. [DOI] [PubMed] [Google Scholar]
- 61.Kim JH, Jang YJ, An SY, Son J, Lee J, Lee G, et al. Enhanced metabolizing activity of human ES cell–derived hepatocytes using a 3D culture system with repeated exposures to xenobiotics. Toxicol Sci 2016;149:269. [DOI] [PubMed] [Google Scholar]
- 62.Gieseck RL III, Hannan NRF, Bort R, Hanley NA, Drake RAL, Cameron GWW, et al. Maturation of induced pluripotent stem cell derived hepatocytes by 3D‐culture. PLoS One 2014;9:e86372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sjogren A‐KM, Liljevald M, Glinghammar B, Sagemark J, Li X‐Q, Jonebring A, et al. Critical differences in toxicity mechanisms in induced pluripotent stem cell–derived hepatocytes, hepatic cell lines and primary hepatocytes. Arch Toxicol 2014;88:1427‐1437. [DOI] [PubMed] [Google Scholar]
- 64.Godoy P, Hewitt NJ, Albrecht U, Andersen ME, Ansari N, Bhattacharya S, et al. Recent advances in 2D and 3D in vitro systems using primary hepatocytes, alternative hepatocyte sources and non‐parenchymal liver cells and their use in investigating mechanisms of hepatotoxicity, cell signaling and ADME. Arch Toxicol 2013;87:1315‐1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Groothuis FA, Heringa MB, Nicol B, Hermens JLM, Blaauboer BJ, Kramer NI. Dose metric considerations in in vitro assays to improve quantitative in vitro–in vivo dose extrapolations. Toxicology 2015;332:30‐40. [DOI] [PubMed] [Google Scholar]
- 66.Underhill GH, Khetani SR. Advances in engineered human liver platforms for drug metabolism studies. Drug Metab Dispos 2018;46:1626‐1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Figaro S, Pereira U, Rada H, Semenzato N, Pouchoulin D, Paullier P, et al. Optimizing the fluidized bed bioreactor as an external bioartificial liver. Int J Artif Organs. 2017;40:196‐203. [DOI] [PubMed] [Google Scholar]
- 68.Nibourg GA, Chamuleau RA, van Gulik TM, Hoekstra R. Proliferative human cell sources applied as biocomponent in bioartificial livers: a review. Expert Opin Biol Ther 2012;12:905‐921. [DOI] [PubMed] [Google Scholar]
- 69.Takeishi K, Collin de l’Hortet A, Wang Y, Handa K, Guzman‐Lepe J, Matsubara K, et al. Assembly and function of a bioengineered human liver for transplantation generated solely from induced pluripotent stem cells. Cell Rep 2020;31:107711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ding Q, Cowan CA. Liver in a dish. Cell Res 2013;23:1242‐1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang S, Wang X, Tan Z, Su Y, Liu J, Chang M, et al. Human ESC‐derived expandable hepatic organoids enable therapeutic liver repopulation and pathophysiological modeling of alcoholic liver injury. Cell Res 2019;29:1009‐1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hughes RD, Mitry RR, Dhawan A. Current status of hepatocyte transplantation. Transplantation 2012;93:342‐347. [DOI] [PubMed] [Google Scholar]
- 73.Yamashita T, Takayama K, Sakurai F, Mizuguchi H. Billion‐scale production of hepatocyte‐like cells from human induced pluripotent stem cells. Biochem Biophys Res Commun 2018;496:1269‐1275. [DOI] [PubMed] [Google Scholar]
- 74.Anderson TN, Zarrinpar A. Hepatocyte transplantation: past efforts, current technology, and future expansion of therapeutic potential. J Surg Res 2018;226:48‐55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tatsumi K, Okano T. Hepatocyte transplantation: cell sheet technology for liver cell transplantation. Curr Transplant Rep 2017;4:184‐192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Blackford SJI, Ng SS, Segal JM, King AJF, Austin AL, Kent D, et al. Validation of current good manufacturing practice compliant human pluripotent stem cell–derived hepatocytes for cell‐based therapy: validation of cGMP hPSCs for liver therapy. Stem Cells Transl Med 2019;8:124‐137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jitraruch S, Dhawan A, Hughes RD, Filippi C, Soong D, Philippeos C, et al. Alginate microencapsulated hepatocytes optimised for transplantation in acute liver failure. PLoS One 2014;9:e113609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Laurent LC, Ulitsky I, Slavin I, Tran H, Schork A, Morey R, et al. Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ESCs and iPSCs during reprogramming and time in culture. Cell Stem Cell 2011;8:106‐118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Steichen C, Luce E, Maluenda J, Tosca L, Moreno‐Gimeno I, Desterke C, et al. Messenger RNA– versus retrovirus‐based induced pluripotent stem cell reprogramming strategies: analysis of genomic integrity. Stem Cells Transl Med 2014;3:686‐691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Steichen C, Hannoun Z, Luce E, Hauet T, Dubart‐Kupperschmitt A. Genomic integrity of human induced pluripotent stem cells: reprogramming, differentiation and applications. World J Stem Cells 2019;11:729‐747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Takayama K, Akita N, Mimura N, Akahira R, Taniguchi Y, Ikeda M, et al. Generation of safe and therapeutically effective human induced pluripotent stem cell–derived hepatocyte‐like cells for regenerative medicine. Hepatol Commun 2017;1:1058‐1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.de Almeida PE, Meyer EH, Kooreman NG, Diecke S, Dey D, Sanchez‐Freire V, et al. Transplanted terminally differentiated induced pluripotent stem cells are accepted by immune mechanisms similar to self‐tolerance. Nat Commun 2014;5:3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu X, Li W, Fu X, Xu Y. The immunogenicity and immune tolerance of pluripotent stem cell derivatives. Front Immunol 2017;8:645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mandai M, Watanabe A, Kurimoto Y, Hirami Y, Morinaga C, Daimon T, et al. Autologous induced stem‐cell‐derived retinal cells for macular degeneration. N Engl J Med 2017;376:1038‐1046. [DOI] [PubMed] [Google Scholar]
- 85.Sugita S, Mandai M, Hirami Y, Takagi S, Maeda T, Fujihara M, et al. HLA‐matched allogeneic iPS cells‐derived RPE transplantation for macular degeneration. J Clin Med 2020;9:2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Subramanian K, Owens DJ, Raju R, Firpo M, O'Brien TD, Verfaillie CM, et al. Spheroid culture for enhanced differentiation of human embryonic stem cells to hepatocyte‐like cells. Stem Cells Dev 2014;23:124‐131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Table T1–T3


