Abstract
The optic tectum of Xenopus receives innervation from retinal ganglion cells, which controls visual information processing and behavioral output. Several indicators can be used to evaluate the functional inputs/outputs of tectal neurons, such as spontaneous activity, visually evoked currents, temporal receptive fields and spatial receptive fields. Analysis of multiple functional properties in the same neurons allows increased understanding of mechanisms underlying visual system function and plasticity. Patch-clamp recordings combined with gene expression or morpholino-mediated knockdown techniques have been especially powerful in the study of specific genes during development and circuit function. The protocol described here provides instructions for performing in vivo electrophysiological recordings from individual tectal neurons to study retinotectal circuitry in the developing Xenopus tectum.
MATERIALS
It is essential that you consult the appropriate Material Safety Data Sheets and your institution’s Environmental Health and Safety Office for proper handling of equipment and hazardous material used in this protocol.
RECIPES: Please see the end of this protocol for recipes indicated by <R>. Additional recipes can be found online at http://cshprotocols.cshlp.org/site/recipes.
Reagents
Deionized (DI) water
Extracellular bath saline <R>
Internal recording saline <R>
MS-222 (0.02%) (Tricane methanesulfonate; in 1 × Steinberg’s solution)
Steinberg’s solution <R>
Sylgard-184 (Electron Microscopy Sciences, cat. no. 24236-10)
Equipment
Back-projector screen
Clampex 10.2 (Molecular Devices)
Clampfit 10.2 (Molecular Devices)
Digidata 1440A data acquisition system (Molecular Devices)
Dissecting stereomicroscope (Nikon, model no. SMZ 1500 or similar)
Epifluorescence microscope (equipped with 10× or 20× objective, Nikon Instruments)
Forceps (Dumont no. 5; Fine Science Tools)
Insect pins
Kodak neutral density filters
MATLAB (The MathWorks)
Microforge (Narishige, MF-830)
Micromanipulator controller (e.g., MP-225, Sutter Instrument)
Micropipettes with filaments (Borosilicate glass capillaries, Warner Instruments)
Micropipette puller (P-97, Sutter Instrument)
Recording chamber
Patch-clamp amplifier (Multiclamp 700B amplifier, Molecular Devices)
Projector (Samsung SP-P310ME LED)
Screen luminance meter (SM208, M&A Instruments Inc)
METHOD
Recording Chamber Preparation
- Custom make a recording chamber with a back-projector screen to present stimulus to Xenopus retina.Present stimulus from a projector to one side of the chamber with a 4.5 × 4.5 cm2 viewing area. Insert a rotatable bar across the chamber and place a sylgard on the bar.
Remove the specimen holder and the light condenser so the recording chamber fits.
Mount the recording chamber on the stage of the inverted microscope.
Adjust the distance between the projector and screen until all of the stimulus images of 8×8 grid fit within the animal’s field of view.
Patch-Clamp System Preparation
-
5.
Seal the faraday cage with light-tight materials.
-
6.Silver new electrodes by fresh bleach and mount the Ag/AgCl electrode to the pipette holder.This will deposit a layer of AgCl on the recording and ground electrodes.
-
7.
Pull recording micropipettes with glass capillaries using a pipette puller and inspect the shape of micropipettes under a microforge.
-
8.Use a custom-made plastic syringe to add internal saline to the micropipettes and place the micropipettes in the pipette holder.Heat and melt the plastic tubing of a disposable sterile plastic syringe and manually pull a disposable sterile needle-like injector. Control the speed used to pull the syringe while heating to obtain a suitable size of the plastic tubing to fit into the micropipette. The amount of internal solution needs to submerge electrode.
-
9.
Turn on the amplifier, digitizer and Clampex software in the computer.
-
10.Move micropipettes with a micromanipulator to the chamber and check the resistances.The resistance of recording pipette should be in the range of 7–9 MΩ. This indicates a suitable tip size required to form a giga-ohm seal and maintain a long-term whole cell recording without the cell membrane resealing.
Tadpole Preparation for Patch-Clamp
-
11.
Set and maintain the room temperature at 20°C.
-
12.
Anesthetize tadpoles in MS-222 solution.
-
13.
Peel the dorsal skin back with forceps to expose the tectum.
-
14.
Rinse the tadpole with extracellular saline, and secure it on a sylgard cushion with pins.
-
15.
Attach the sylgard to the rotatable bar with grease in the recording chamber.
-
16.Adjust the angle of the bar so that the tadpole’s eyes are vertically facing the back-projector screen.The computer-generated image is projected to the retina via the screen.
Mapping Receptive Fields
-
17.Adjust the brightness of the projector with Kodak neutral density filters, and control the image size with a convex lens.The luminance determines the strength of visually evoked currents. Measure the luminance every time with a luminance detector before the recording. The range of the luminance should be 3 to 55 cd/m2.
-
18.Generate the stimulus image via MATLAB using a separate computer.The stimulus consists of an 8 × 8 grid of 0.5 × 0.5 cm2 nonoverlapping squares covering the visual field. White squares on a dark background or dark squares on a white background are presented for 1.5 s in random order, 64 times with 5 s intervals, until the entire visual field is mapped.
-
19.Move the stage to find a clear image of the optic tectum under the microscope.Record the neurons from the same area of the optic tectum to decrease the variation of responses due to differential maturation stages along the rostro-caudal tectal axis.
-
20.
Gently break a micropipette tip with a Kimwipe and apply a gentle negative pressure in the micropipette, then remove a piece of the membranes (dura mater and pia mater) overlying the tectum to expose the tectal cells.
-
21.
Use the syringe and valve to maintain a gentle positive pressure in the micropipette.
-
22.
Use the micromanipulator to move the patch pipette close to the cell.
-
23.
Slowly push the micropipette against the cell until the measured resistance rises.
-
24.
Remove positive pressure and apply negative pressure to obtain a giga-ohm seal.
-
25.Run the stimulation protocol and collect cell-attached recording for three repetitions per cell.The cell membrane current is held at 0 pA. Cell spikes are recorded for 20 min. The recording was repeated to eliminate possible contamination by spontaneous activity.SeeTroubleshooting.
-
26.
Apply a brief strong negative pressure to break the cell membrane and continue whole-cell voltage clamp recording.
-
27.Run the same stimuli to record excitatory and inhibitory currents by holding membrane potential at −60 mV and 0 mV respectively.Membrane potential is voltage clamped at −60 mV to record AMPAR-mediated currents or at 0 mV to record GABAAR-mediated currents. Signals are filtered at 2 kHz sampled at 10 kHz.SeeTroubleshooting.
Data analysis
-
28.
For cell-attached recording, determine the average number of total spikes per stimulus and map the spiking receptive field (sRF) or temporal receptive field (tRF) by MATLAB.
-
29.For whole-cell recording, compute and normalize total synaptic charge transfer over 600 ms from the onset of stimulus to map excitatory receptive field (eRF) and inhibitory receptive field (iRF) by MATLAB.All values that are larger than three times the standard deviation of spontaneous activity are included in the analysis of spatial receptive fields. Only neurons from which both eRF and iRF are successfully recorded are included in the correlation analysis (Shen et al. 2011).SeeTroubleshooting.
-
30.Represent spatial receptive fields for synaptic currents on gray-scale map, according to the normalized total charge transfer in response to the visual stimulus.SeeTroubleshooting.
TROUBLESHOOTING
Problem (step 25-27):
Baseline shifts and recordings (currents or spikes) are unstable over time.
Solution:
Wait 10 minutes and allow the cell to reach a stable condition after the formation of cell-attached or whole cell mode.
Problem (step 27):
Each stimulus presented to different grids evokes similar values of currents.
Solution:
Reduce the luminance intensity of white squares by applying neutral density filters to avoid the saturation of light sources. Measure the luminance intensity every time before performing the visual stimulation experiment with a luminance detector.
DISCUSSION
This protocol is based on our study showing that inhibition to excitation ratio regulates visual system responses and behavior in the developing Xenopus in vivo (Shen et al. 2011; He et al. 2018). This protocol provides an effective method to map spiking, excitatory and inhibitory receptive fields in individual neurons without physically removing the lens from the retina as shown before (Engert et al. 2002; Tao and Poo 2005). The custom-built recording chamber with a back-projector screen mimics the visual stimulus from the nature environment. The advantage of using the chamber is that multiple recordings (spontaneous activity, receptive field mapping, and visually evoked responses) can be performed in one cell without changing the stimulus equipment. An alternative way to present visual stimuli is to use a rather expensive piece of medical equipment, the multicore optical image fiber (FIGH-30–650S, Myriad Fiber) (Dong et al. 2009; Van Horn et al. 2017).
RECIPES
Steinberg’s solution
| Reagent | Final concentration | Amount to add (100 × stock) |
|---|---|---|
| NaCl | 58 mM | 34 g |
| KCl | 0.67 mM | 0.5 g |
| Ca(NO3)2 | 0.34 mM | 0.8 g |
| MgSO4 | 0.83 mM | 2.06 g |
| HEPES | 10 mM | 119 g |
To prepare 1 L of 100 × Steinberg’s solution, dissolve the reagents listed above in 800 mL of ddH2O. Adjust the pH to 7.5 with NaOH, and then add ddH2O to 1 L. Store the solution at 4°C. Pour 100mL stock and add ddH2O to 10 L to get the 1× solution. Store at room temperature.
Extracellular saline
| Reagent | Final concentration | Amount to add (1L solution) |
|---|---|---|
| NaCl | 115 mM | 6.72 g |
| KCl | 4 mM | 0.30 g |
| CaCl2-2H2O | 3 mM | 0.44 g |
| MgCl2-6H2O | 3 mM | 0.61 g |
| HEPES | 5 mM | 1.19 g |
| Glycine | 0.01 mM | 0.00075 g |
| Glucose | 10 mM | 1.80 g |
| Tubocurarine | 0.1 mM | 0.068 g |
To prepare 1 L extracellular saline, dissolve the reagents listed above in 800 mL of ddH2O. Adjust the pH to 7.2 with NaOH, and then add ddH2O to 1 L. The final Osmolarity is 255 mOsm. Store at 4°C.
Internal saline
| Reagent | Final concentration | Amount to add (20 ml solution) |
|---|---|---|
| K-gluconate | 110 mM | 515.4 mg |
| KCl | 8 mM | 11.9 mg |
| NaCl | 5 mM | 5.8 mg |
| MgCl2-6H2O | 1.5 mM | 6.1 mg |
| HEPES | 20 mM | 95.3 mg |
| EGTA | 0.5 mM | 3.8 mg |
| ATP | 4 mM | 44.1 mg |
| GTP | 0.3 mM | 3.1 mg |
To prepare 20 mL internal saline, dissolve the reagents listed above in 15 ml of ddH2O. Adjust the pH to 7.2 with KOH, and then add ddH2O to 20 mL. The final Osmolarity is 255 mOsm. Store at −20°C and thaw it on ice just before the experiment.
Figure 1.
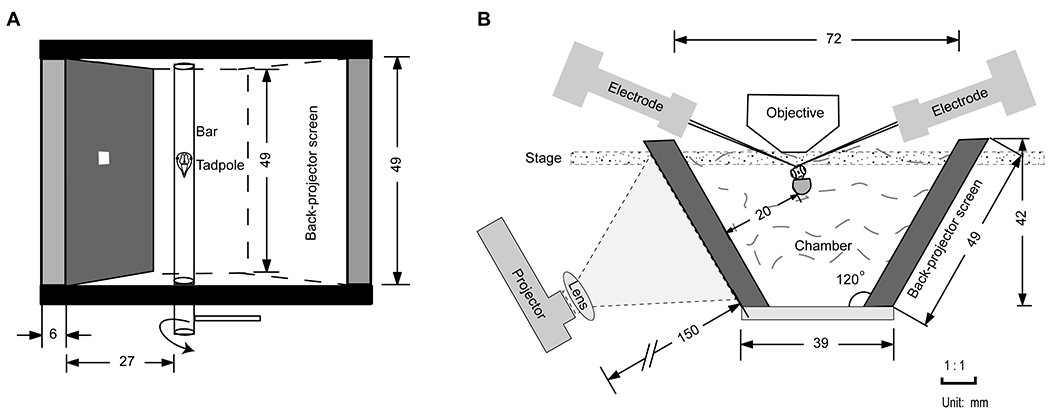
Schematic diagram of recording chamber. (A) Overview of the recording chamber. The grey screen is made of a back-projector screen, which presents the stimulus to the eye while the animal is in the chamber. Tadpole is secured on a sylgard cushion with insect pins and stuck to the rotatable bar with grease. Adjust the angle of the bar and face the retina vertical to the back-projector screen. The white square represents one stimulus grid on the black background. (B) Front view of the recording chamber. Place a convex lens in front of the projector to control the projected image size. Remove the condenser before placing the chamber to the stage.
Acknowledgements
This work was supported by the National Nature Sciences Foundation of China (NSFC 31871041 to WS) and the United States National Institutes of Health (EY011261 and EY027437 to HTC).
Footnotes
Declaration of competing interest
The authors declare no conflicts of interest with respect to the authorship and/or publication of this article.
REFERENCES
- Dong W, Lee RH, Xu H, Yang S, Pratt KG, Cao V, Song YK, Nurmikko A, Aizenman CD. 2009. Visual avoidance in Xenopus tadpoles is correlated with the maturation of visual responses in the optic tectum. J Neurophysiol 101: 803–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engert F, Tao HW, Zhang LI, Poo MM. 2002. Moving visual stimuli rapidly induce direction sensitivity of developing tectal neurons. Nature 419: 470–475. [DOI] [PubMed] [Google Scholar]
- He HY, Shen W, Zheng L, Guo X, Cline HT. 2018. Excitatory synaptic dysfunction cell-autonomously decreases inhibitory inputs and disrupts structural and functional plasticity. Nat Commun 9: 2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W, McKeown CR, Demas JA, Cline HT. 2011. Inhibition to excitation ratio regulates visual system responses and behavior in vivo. J Neurophysiol 106: 2285–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao HW, Poo MM. 2005. Activity-dependent matching of excitatory and inhibitory inputs during refinement of visual receptive fields. Neuron 45: 829–836. [DOI] [PubMed] [Google Scholar]
- Van Horn MR, Strasser A, Miraucourt LS, Pollegioni L, Ruthazer ES. 2017. The Gliotransmitter d-Serine Promotes Synapse Maturation and Axonal Stabilization In Vivo. J Neurosci 37: 6277–6288. [DOI] [PMC free article] [PubMed] [Google Scholar]


