Abstract
Here, we report the synthesis, structure-activity relationship studies, enzyme inhibition, antiviral activity and X-ray crystallographic studies of 5-chloropyridinyl indole carboxylate derivatives as a potent class of SARS-CoV-2 chymotrypsin-like protease inhibitors. Compound 1 exhibited SARS-CoV-2 3CLpro inhibitory IC50 value of 250 nM and antiviral EC50 value of 2.8 μM in VeroE6 cells. Remdesivir, an RNA-dependent RNA polymerase inhibitor has shown antiviral EC50 value of 1.2 μM in the same assay. Compound 1 showed comparable antiviral activity with remdesivir in immunocytochemistry assays. Compound 7d with N-allyl derivative showed the most potent enzyme inhibitory IC50 value of 73 nM. To obtain molecular insight into the binding properties of these molecules, we have determined X-ray crystal structures of compounds 2-bound SARS-CoV-2 3CLpro, 7b-bound SARS-CoV 3CLpro, and 9d-bound SARS-CoV-2–3CLpro and compared their binding properties.
Keywords: Antiviral activity, covalent inhibitors, COVID-19, drug design, protein X-ray structures, SARS-CoV-2 3CLpro
Graphical Abstract
INTRODUCTION
The human coronavirus disease 2019 (COVID-19), is an infectious disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a newly discovered coronavirus.1,2 The disease first originated in Wuhan, China and then spread rapidly around the world. The outbreak escalated into an ongoing pandemic, leading to a catastrophic public health crisis and incredible uncertainty around the globe.3,4 As of July 5, 2021, more than 185 million patients were infected with COVID-19 and there were more than 4 million deaths worldwide.5 Currently, there is no specific efficacious drug treatment available for COVID-19, except remdesivir, which has been granted an emergency-use authorization.6,7 Research suggests remdesivir may provide only modest benefit to patients with COVID-19 infection.8,9 While current large-scale vaccination efforts are showing benefits, the hope that COVID-19 will be under control with ‘herd immunity’ is quite uncertain.10,11 Furthermore, there is no conclusive evidence yet that COVID-19 convalescent people with SARS-CoV-2 antibodies are immune to reinfection.12,13 Therefore, it is an urgent priority to develop potent antiviral drugs that can effectively mitigate the lethal consequences of cytokine storm in COVID-19 patients.
SARS-CoV-2 RNA genome sequence provided an excellent starting point for drug discovery and development of effective therapeutics against COVID-19.14,15 SARS-CoV-2 belongs to a family of beta coronaviruses, including SARS-CoV and MERS-CoV, responsible for outbreaks of SARS and MERS in 2003 and 2012, respectively.16,17 SARS-CoV-2 genome has overall 80% nucleotide identity with SARS-CoV and the main proteases of these viruses feature more than 90% amino acid sequence identity.18,19 Earlier medicinal chemistry efforts for the control of SARS-CoV and MERS-CoV infections have laid important early groundwork for design, synthesis and development of small molecule lead inhibitors of the main proteases, mainly chymotrypsin-like protease (3CLpro and Mpro) and papain-like protease (PLpro).20–22 Both of these proteases are essential for viral replication and transcription of the genome, and they represent very important targets for developing antiviral therapeutics against COVID-19. The structure, activity, and active sites of these proteases have been elucidated.23 Early and recent drug development efforts, including development of covalent and noncovalent inhibitors of these proteases have been recently reviewed.23,24 Among small molecule drug development targets for treatment of COVID-19, there is considerable interest and activity in SARS-CoV-2 3CLpro inhibitors.23–25 Recently, we have reported that combination of a peptidomimetic SARS-CoV-2 3CLpro inhibitor with remdesivir exhibits synergism against SARS-CoV-2.26 Oral or intraperitoneal administration of dipeptidyl 3CLpro inhibitors have been shown to reduce lung viral loads and lung lesions in a transgenic mouse model of SARS-CoV-2 infection.27 A small molecule inhibitor of SARS-CoV-2 3CLpro has been reported as entering into phase I clinical trials.28
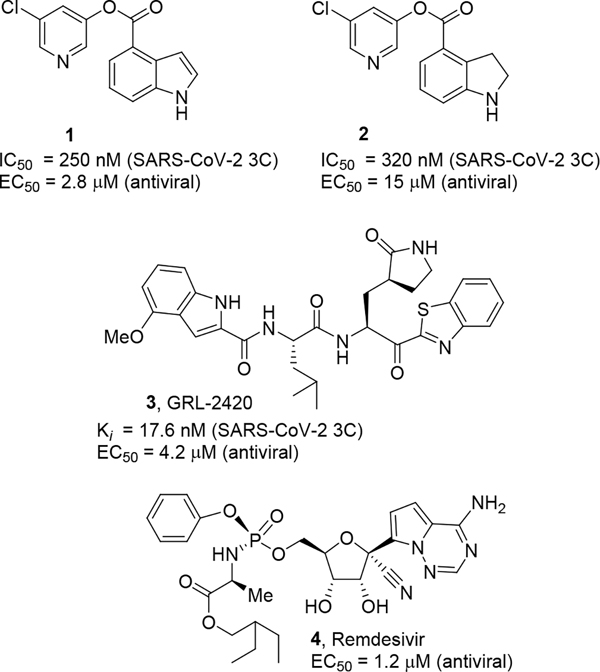
In our continuing efforts towards the treatment of SARS-CoV and SARS-CoV-2, we have developed a variety of potent peptidomimetic and nonpeptide-derived small molecule protease inhibitors.26,29–34 The SARS-CoV and SARS-CoV-2 3CLpro active sites possess a catalytic dyad where Cys145 acts as a nucleophile and His41 acts as the general acid-base. We and others have reported a series of covalent 3CLpro inhibitors against SARS-CoV and SARS-CoV-2 where the mode of inhibition involves acylation of the active site cysteine, forming a covalent bond with the inhibitor.35–38 In particular, ester derivatives of 5-chloropyridin-3-ol with various indole carboxylic acids (compounds 1, 2, Figure 1) displayed potent 3CLpro inhibitory activity and exerted potent antiviral activity against SARS-CoV and SARS-CoV-2 in VeroE6 cell-based assays.26,29 Keto-benzothiazole derivative 3 exhibited potent enzyme inhibitory and antiviral activity.26 Remdesivir (4), an RNA-dependent RNA-polymerase inhibitor, displayed comparable antiviral activity of inhibitors 1 and 2.6,26 We have further investigated various indole carboxylic acid-derived inhibitors. It appears that the position of the carboxylic acids, as well as substituents on the indole ring, are important to SARS-CoV-2 3CLpro inhibitory potency and antiviral activity. Presumably, the indole carboxylate scaffold plays a key role in binding to the 3CLpro active site. For an understanding of active-site interactions, we determined X-ray crystal structures of inhibitor-bound SARS-CoV-2 and SARS-CoV-1 3CLpro. Herein, we report the design, synthesis, structure-activity studies, enzyme inhibition studies, antiviral activity, and X-ray structural studies of 3-chloropyridinyl ester-derived SARS-CoV-2 3CLpro inhibitors.
Figure 1.
Structures of SARS-CoV-2 3CLpro inhibitors 1–3 and RdRp inhibitor, remdesivir, 4.
Results and Discussion
The general synthesis of various 3-chloropyridinyl ester-derived SARS-CoV-2 3CLpro inhibitors is shown in Scheme 1. Inhibitors 1–2, 7a-m were synthesized by esterification of respective carboxylic acids. The majority of the requisite carboxylic acids are commercially available and a few modified derivatives were readily prepared. Carboxylic acids 5a-l and 5-chloro-3-pyridinol were esterified using EDC in the presence of DMAP in CH2Cl2 at 23 °C for 12 h. The resulting esters were purified by silica gel chromatography to provide air stable ester derivatives in good to excellent yields (60–80%). Similarly, commercially available 3,4-dihydro-2H-benzo[b][1,4]oxazine-8-carboxylic acid 5m was converted to ester derivative 7m. The structures of the various 3-chloropyridinyl esters are shown in Table 1. The synthesis of active ester derivatives 9a-f, containing methyl-substituted 3-chloropyridinol was achieved by reaction of indole-4-carboxylic acid 5j and 5n with the respective commercially available substituted pyridinol derivatives 8a-f with EDC in the presence of DMAP under similar conditions as stated above. The synthesis of ester derivatives 7a, 7b, 7d, and 7e, is shown in Scheme 2. Commercially available methyl-1H-indole-5-carboxylate 10 was saponified with 1M LiOH in aqueous THF solution at 23 °C for 30 min. The resulting acid was esterified with 5-chloro-3-pyridinol 6 to provide ester derivative 7a. For the synthesis of ester 7b, methyl ester 10 was treated with NaH in THF at 23 °C for 1 h. After this period, 3-nitrophenylsulfonyl chloride was added and the resulting mixture was stirred for 23 h to provide sulfonamide derivative 11.39 Saponification of ester 11 with aqueous LiOH provided the corresponding carboxylic acid which was converted to 3-chloro-pyridinyl ester 7b as described above.
Scheme 1.
Synthesis of inhibitors 7a-m and 9a-f. Reagents and chemicals. (a) EDC, DMAP, CH2Cl2, 23 °C (60–80%).
Table 1.
Structures and activity of 3-chloropyridinyl ester-derived SARS-CoV-2 3CLpro inhibitors.
| Entry | Entry Compound Structure | SARS CoV-2 3CLpro IC50 (μM) | SARS-CoV-2 EC50 (μM)a |
|---|---|---|---|
|
| |||
| 1 |

|
0.25 | 2.8 |
| 2 |

|
0.32 | 15 |
| 3 |
|
0.31 | 43.7 |
| 4 |
|
0.12 | 69.8 |
| 5 |
|
0.90 | 8.1 |
| 6 |
|
0.073 | 15 |
| 7 |
|
0.38 | 11.5 |
| 8 |
|
0.47 | 56.7 |
| 9 |
|
10.3 | >100 |
| 10 |
|
0.59 | 3.1 |
| 11 |
|
0.87 | 14 |
| 12 |
|
0.34 | >100 |
| 13 |
|
0.49 | 57.1 |
| 14 |

|
1.19 | >100 |
| 15 |
|
0.42 | >100 |
Compounds 3 and 4 exhibited antiviral EC50 values 3.4 μM and 3.2 μM, respectively.
Scheme 2.
Synthesis of inhibitors 7a, 7b, 7d, and 7e. Reagents and chemicals. (a) LiOH, THF-H2O, 23 °C, 30 min; (b) 5-chloro-3-pyridinol 6, EDC, DMAP, CH2Cl2, 23 °C, 12 h; (c) NaH, m-NO2-PhSO2Cl, THF, 23 °C, 24 h; (d) NaH, allylbromide, THF, 23 °C, 12 h; (e) 1N NaOH, EtOH-H2O, 85 °C, 3 h.
For synthesis of active esters 7d, and 7e, commercially available methyl-1H-indole-4-carboxylate 12 was treated with NaH in THF at 0 °C to 23 °C for 1 h. Then, allyl bromide was added and the resulting reaction was stirred for 12 h at 23 °C to provide monoallyl derivative 13 and diallyl derivative 14 as a 7.5:1 mixture.40 These isomers were separated by silica gel chromatography. The respective ester was saponified with 1N NaOH in EtOH at 85 °C for 3 h to provide the corresponding acid. The resulting carboxylic acid derivatives were esterified as described above to provide esters 7d and 7e in good yields.
Our SARS-CoV-2 3CLpro inhibition assays of active ester derivatives were carried out using the authentic SARS-CoV-2 3CLpro enzyme that would be genenerated during viral replication, i.e no additional amino acids on the N- or C-termini. The details of expression and purification of fully active SARS-CoV-2 3CLpro construct have been published recently.41 Inhibitory activity (IC50 values) of compounds were assessed using a continuous fluorescence assay and the FRET-based substrate UIVT3 (HiLyte Fluor488™-ESATLQSGLRKAK-QXL520™-NH2) (Anaspec, Fremont, CA) as described previously.31,42
We resynthesized and assessed activity of 4-chloropyridinyl indole-4-carboxylate 1, which was previously identified by us as a potent and irreversible inhibitor of SARS-CoV (IC50 = 30 nM; antiviral EC50 = 6.9 μM in VeroE6 cells).35 The structure and activity of various new compounds are shown in Tables 1 and 2. Compound 1 exhibited SARS-CoV-2 3CLpro enzyme IC50 value of 250 nM. Compound 1 also displayed potent antiviral activity which was assessed using quantitative VeroE6 cell-based assay with RNA-qPCR.The details of the assay have been published recently.29
Table 2.
Structures and activity of substituted active ester-derived SARS-CoV-2 3CLpro inhibitors.
| Entry | Compound Structure | SARS-CoV-2 3CLpro IC50(μM) | SARS-CoV-2 EC50 (μM)a |
|---|---|---|---|
|
| |||
| 1 |
|
100 | >100 |
| 2 |
|
100 | >100 |
| 3 |
|
>100 | >100 |
| 4 |
|
2.2 | 19.3 |
| 5 |
|
15.3 | 30 |
| 6 |
|
4.4 | 57 |
Compounds 3 and 4 exhibited antiviral EC50 values 3.4 μM and 3.2 μM, respectively.
Compound 1 displayed an EC50 value of 2.8 μM. The high ratio of antiviral EC50 and enzyme IC50 may be due to expression of the efflux transporter P-glycoprotein in VeroE6 cells. Such discrepencies have been documented recently.43,44 This compound exhibited cytotoxicity (CC50) value >100 μM. Remdesivir (4), a nucleotide derivative that reportedly blocks the infectivity of SARS-CoV-2 by inhibiting viral RNA-dependent RNA polymerase (RdRp), was also assayed in the same assay to compare antiviral activity. Remdesivir exhibited antiviral EC50 value of 1.2 μM. Furthermore, we investigated antiviral activity of compound 1 and remdesivir at 1, 10, and 100 μM to assess if these compounds exert antiviral activity without significant cytostatic or cytotoxic effects. As reported recently, both compounds completely blocked the infectivity and cytopathic effect of SARS-CoV-2wk-521 in VeroE6 cells.26,29
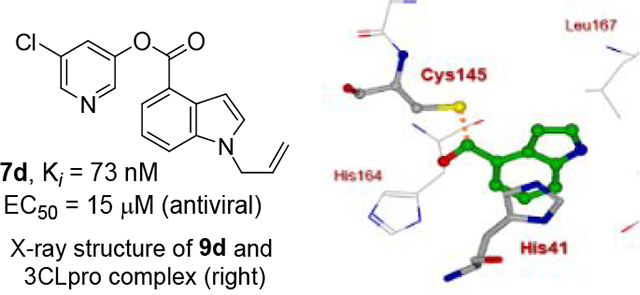
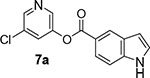
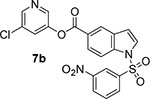
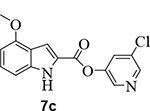
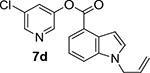
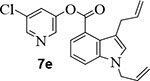
We then investigated the effect of various substitutions on the indole ring as well as on the chloropyridine ring of the ester. As can be seen, indoline derivative 2 showed only a slight reduction in inhibitory potency against SARS-CoV-2 3CLpro enzyme (IC50 = 320 nM) compared to indole derivative 1. It displayed SARS-CoV 3CLpro inhibitory IC50 value of 190 nM. Also, it showed a 5-fold reduction in antiviral activity (EC50 = 15 μM) compared to 1. Substitution of chloropyridinyl ester at the 5-position of the indole ring provided derivative 7a (entry 3) which displayed a comparable IC50 value to compounds 1 and 2 but it was significantly less potent than 1 or 2 against the virus (EC50 = 43.7 μM). Compound 7a exhibited comparable SARS-CoV 3CLpro activity (IC50 = 405 nM). Incorporation of a 3-nitro sulfonamide functionality on the indole nitrogen provided compound 7b. This compound showed improvement in SARS-CoV-2 3CLpro inhibitory activity (entry 4). Substitution of chloropyridinyl ester at the 4-position of indole ring resulted compound 7c. Interestingly, it showed reduction in enzyme inhibitory activity, but showed an antiviral EC50 value of 8 μM (entry 5). Both compounds 7b and 7c were less potent against SARS-CoV 3CLpro (IC50 185 nM and 253 nM, respectively). We then examined the structure-activity relationships associated with compound 1. Incorporation of N-allyl substituent resulted in compound 7d which exhibited over 5-fold improvement in inhibitory potency over 1. However, compound 7d displayed an antiviral EC50 value of 15 μM, a nearly 5-fold reduction over compound 1 (entry 6). The bis-allyl derivative 7e showed a 5-fold reduction in enzyme inhibitory activity but had a slight improvement in antiviral activity over 7d (entry 7).
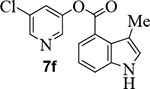
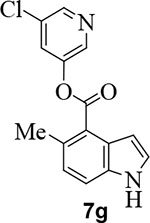
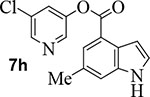
We further investigated the effect of methyl substitution on the indole ring in an attempt to facilitate the bioactive conformation of the pyridyl ester. Accordingly, incorporation of methyl group on the indole at positions 3, 5, 6 resulted in compounds 7f, 7g, and 7h, respectively (entries 8–10). Inhibitor 7g with 5-methyl indole resulted in significant loss of SARS-CoV-2 3CLpro inhibitory activity and this compound did not exhibit any appreciable antiviral activity (EC50 >100 μM). Compound 7h containing 6-methyl indole showed a slight reduction in enzyme inhibitory activity, however, this compound maintained potent antiviral activity (EC50 = 3.1 μM), comparable to inhibitor 1 (entry 10). The incorporation of the methyl group at position 5 on the indole ring most likely disrupts or reduces the available angle of attack of the Cys145 nucleophile on the ester group thereby reducing its potency.45
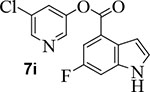
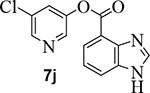
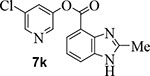
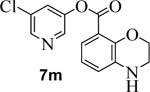
Incorporation of fluorine at position 6 provided inhibitor 7i with loss of enzyme inhibitory and antiviral activity. We then investigated the effect of incorporation of heteroatoms and the importance of the 5-membered pyrrole ring on indole. Compound 7j with a benzoimidazole aromatic heterocyclic group showed an enzyme IC50 value of 340 nM, but no appreciable antiviral activity (entry 12). Further incorporation of a 2-methyl group on compound 7j provided derivative 7k which displayed an enzyme IC50 value of 490 nM and antiviral EC50 value of 57 μM (entry 13). Compound 7l with a benzooxazole heterocyclic group showed a reduction in potency (entry 14). Compound 7m with a dihydrobenzooxazine heterocycle exhibited good enzyme inhibitory activity, however, antiviral activity was also poor (EC50 >100 μM).
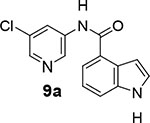
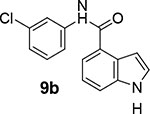
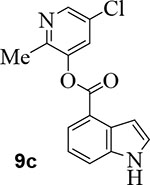
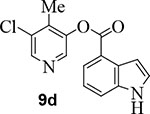
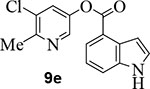
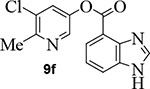
We then examined the importance of the 5-chloropyridinyl ester in compound 1. The structure and activity of various derivatives are shown in Table 2. As shown, we examined the importance of ester over its amide derivative. The 5-chloropyridyl indole carboxamide derivative 9a did not exhibit any appreciable enzyme inhibitory or antiviral activity. We also prepared a 3-chlorophenyl indole derivative 9b but this compound did not exhibit any activity either, showing the importance of the pyridinyl ester functionality (entries 1 and 2). We then incorporated a methyl group on the pyridine ring to examine the effect of the alkyl group on potency. Substitution of 2-methyl, 4-methyl, or 6-methyl group resulted in compounds 9c, 9d, and 9e. Interestingly, 2-methyl substitution provided inhibitor 9c with no appreciable activity (entry 3). Substitutions at 4 and 6-postions also resulted in significant reductions in potency (entries 4 and 5). Benzimidazole derivative 9f with a 6-methyl-5-chloropyridynyl ester showed a significant reduction in protease inhibitory activity compared to its desmethyl derivative 7j in Table 1 (entry 6).
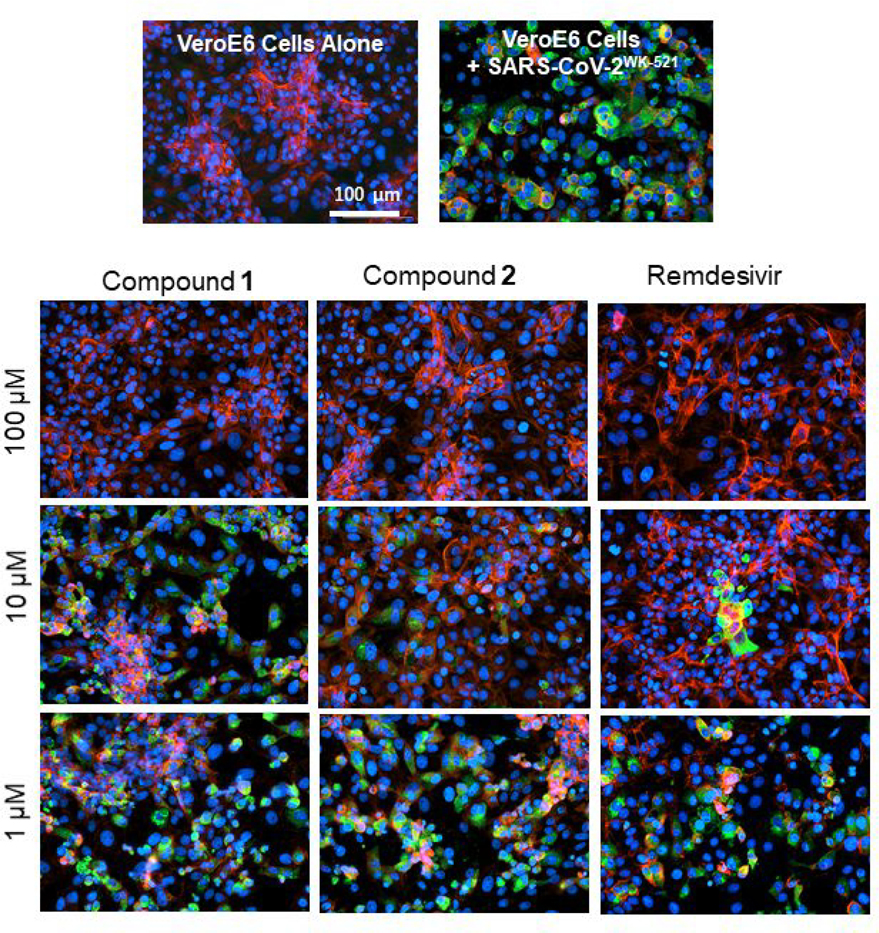
As mentioned earlier, our antiviral activity assays were performed using the quantitative RNA-qPCR assay in VeroE6 cells.26,29 In the present study, in order to confirm and corroborate the anti-SARS-CoV-2 activity (EC50, μM) shown in Tables 1 and 2, which often fails to differentiate the actual antiviral activity from the misleading and distractive reduction in RNA copy numbers caused by the cytostatic effect or cytotoxicity of test compounds, we have employed immunocytochemistry, which allows us to examine the antiviral activity of test compounds at cellular level. In the immunocytochemistry, the IgG fraction isolated from a COVID-19-convalescent patient, who had high titer SARS-CoV-2-binding IgG,46 and a green fluorescence-conjugated goat polyclonal anti-human-IgG-Alexa Fluor 488 Fab fragment were employed as the primary and secondary antibodies, respectively, together with Texas Red-X dye-conjugated Phalloidin and DAPI for visualization of F-actin and cell nuclei, respectively.26, 29 As shown in Figure 2, when VeroE6 cells were cultured alone, robust cellular cytoskeleton filamentous actin (F-actin) was seen as mesh-like structures in red and a number of nuclei (in blue) were identified, signifying that those VeroE6 cells were healthy and replicating (top left in Figure 2). However, when VeroE6 cells were exposed to SARS-CoV-2wk-521 and cultured in the absence of test compound, the F-actin structure was lost and a number of cells had been infected and destroyed by the virus and stained in green (top right in Figure 2). In contrast, when the SARS-CoV-2wk-521-exposed VeroE6 cells were cultured in the presence of 10 μM Compounds 1 and 2, there was significant reduction in the number of SARS-CoV-2wk-521-infected cells and there were essentially no infected cells when the cells were cultured in the presence of 100 μM of each compound. Remdesivir, the only FDA-approved antiviral therapeutic as of writing, also significantly reduced the number of infected cells at 10 and 100 μM, while there was viral breakthrough in the culture with 10 μM. Of particular note, among various active esters examined, compound 1 exerted potent antiviral activity (EC50 = 2.8 μM) and significantly reduced the infectivity, replication, and cytopathic effect of SARS-CoV-2 without significant toxicity.
Figure 2.
Inhibitors 1, 2 and remdesivir potently blocked the infectivity and cytopathic effect of SARS-CoV-2WK-521 in VeroE6 cells. VeroE6 cells were exposed to SARS-CoV-2WK-521 at an MOI of 0.05 and cultured in the presence or absence of test compounds. After 3 days, the cells were fixed with 4% paraformaldehyde and immunocytochemistry was conducted using the IgG fraction from serum of a COVID-19-convalescent patient, who had high-titer SARS-CoV-2-binding IgG,44 and a green fluorescence-conjugated goat polyclonal anti-human-IgG-Alexa Fluor 488 Fab fragment as the primary and secondary antibodies, respectively. SARS-CoV-2 antigens, filamentous actin, and nuclei are shown in green, red, and blue, respectively.
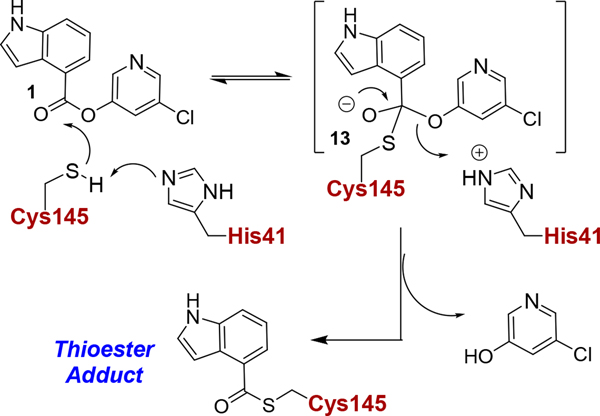
The mechanism of inhibition of SARS protease has been studied by us and others previously.35,36,47 Indole chloropyridinyl ester 1 covalently modifies SARS-CoV-2 3CLpro forming a thioester bond with the catalytic Cys145 and indole carbonyl group. The presence of the covalent bond was verified by electrospray ionization, quadruple time-of-flight mass spectrometry (ESI-QTOF/MS). The proposed inhibition of SARS-CoV-2 3CLpro is shown in Figure 3. As shown, the catalytic dyad of 3CLpro, His41 and Cys145, is involved in the nucleophilic attack on the 5-chloropyridinyl ester of inhibitor 1 to form a tetrahedral intermediate 13 which then expels the chloropyridinyl group and forms a covalent bond with Cys145 of SARS-CoV-2 3CLpro. In essence, 5-chloropyridinyl indole ester acylated Cys145 in the active site. In mouse hepatitis virus (MHV) coronavirus 3CLpro, this thioester bond can be slowly hydrolyzed by bulk water releasing active enzyme, and the rates of release are affected by drug-resistant mutants.48
Figure 3.
The mechanism of inhibition of SARS-CoV-2 3CLpro by compound 1. Structure 13 represents the presumed tetrahedral intermediate formed prior to the thioester intermediate.
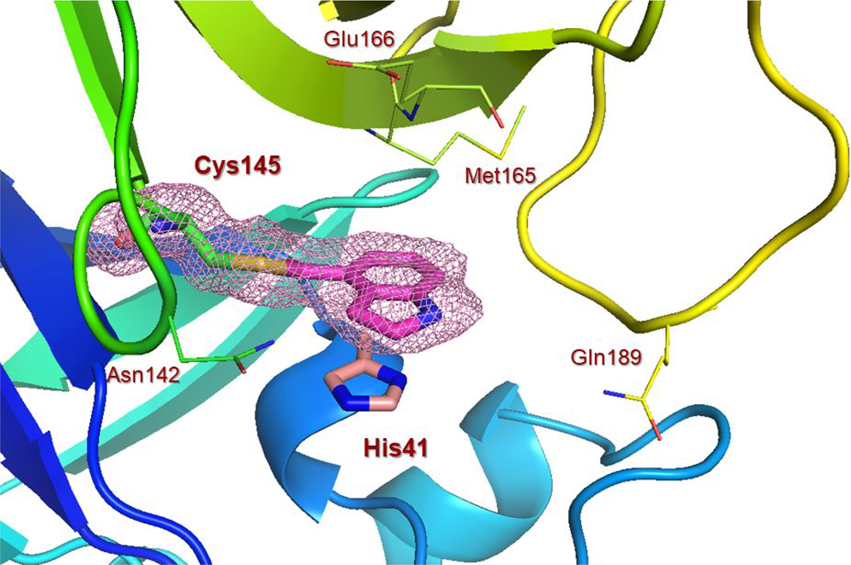
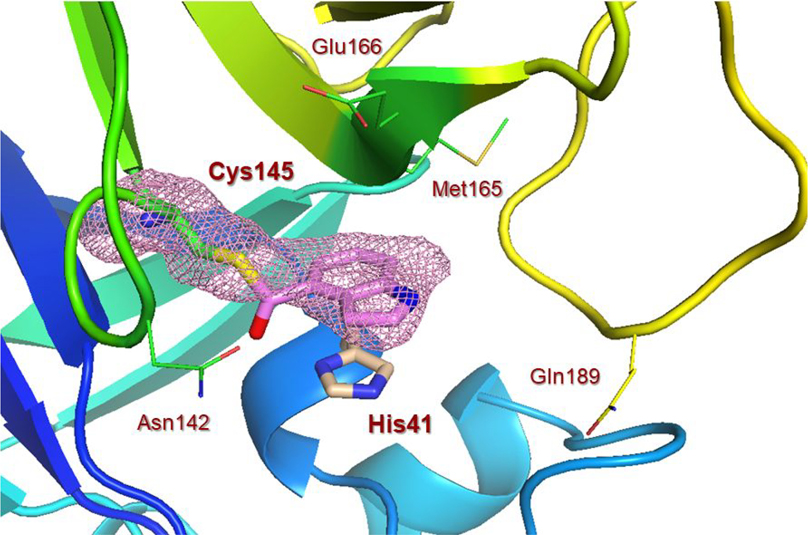
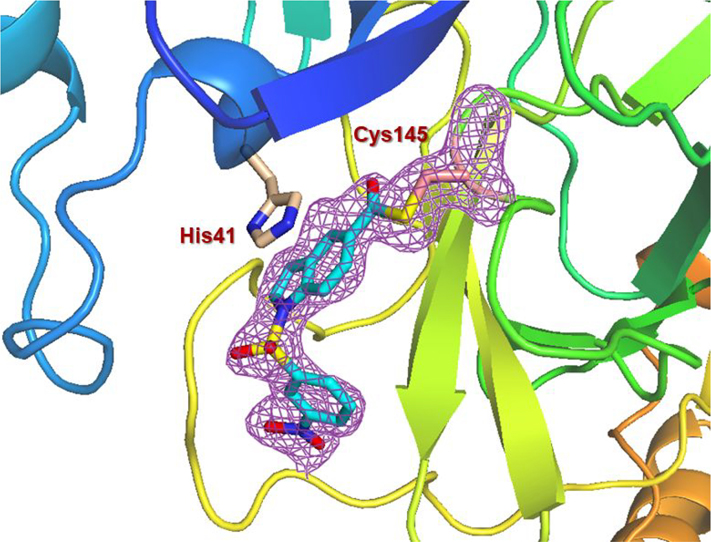
To obtain further molecular insight into the inhibition of SARS-CoV-2 3CLpro by the active esters, we determined the X-ray structures of SARS-CoV-2 3CLpro bound with inhibitor 2 and 9d at 1.65 Å resolution. Also, we determined the X-ray structure of SARS-CoV 3CLpro in complex with inhibitor 7b at 1.63 Å resolution. The X-ray data collection and refinement statistics for all three complexes are summarized in Table S1 (Please see Supporting Information). The electron density maps for the reaction product of inhibitor 2 and SARS-CoV-2 3CLpro and inhibitor 9d and SARS-CoV-2 3CLpro are shown in Figures 4 and 5, respectively. As can be seen, Cys145 is covalently attached to the indoline carbonyl group of 2 and indole carbonyl group of 9d in the S1 pocket. It is important to note that after X-ray structural refinement that included occupancy refinement of the ligand and His41, the thioester intermediate of compound 9d was bound at 80% occupancy along with His41 at about 80% occupancy in one conformation (inhibitor bound) and about 20% occupancy which represents the unbound structure. This is important because His41 needs to rotate out of the way to form an interaction with the indole group. Also important is that in this conformation, the carbonyl group is more protected from solvent keeping it stable from hydrolysis and this is observable in the maps. The indole ring forms π-π stacking with the shifted imidazole ring of His41 in both structures. There is approximately 3.5 Å distance between the His41 imidazole ring and phenyl ring of structure 2 and indole of structure 9d. Other key residues such as Asn142, Met165, Glu166, and Gln189 are within the hydrophobic pocket surrounding these rings in both structures.
Figure 4.
X-ray structure of SARS-CoV-2 3CLpro bound with compound 2 at 1.65 Å. Polder electron density omit maps (mFobs – DFmodel) surrounding compound 2 and Cys145. Electron density is contoured at 3.5σ. (+) density is colored pink and (−) density is colored green. Final Rwork =16.7% and Rfree = 18.9%. The thioester bond between Cys145 and compound 2 is clearly visible. (PDB code:7RBZ)
Figure 5.
X-ray structure of SARS-CoV-2 3CLpro bound with compound 9d at 1.65 Å. Polder electron density omit maps (mFobs – DFmodel ) surrounding 9d and Cys145. Electron density is contoured at 3.5 σ. (+) density is colored pink and (−) density is colored green but is not visible. Final Rwork = 12.6% and Rfree = 17.4%. The thioester bond between Cys145 and 9d is clearly visible. (PDB code:7RC0).
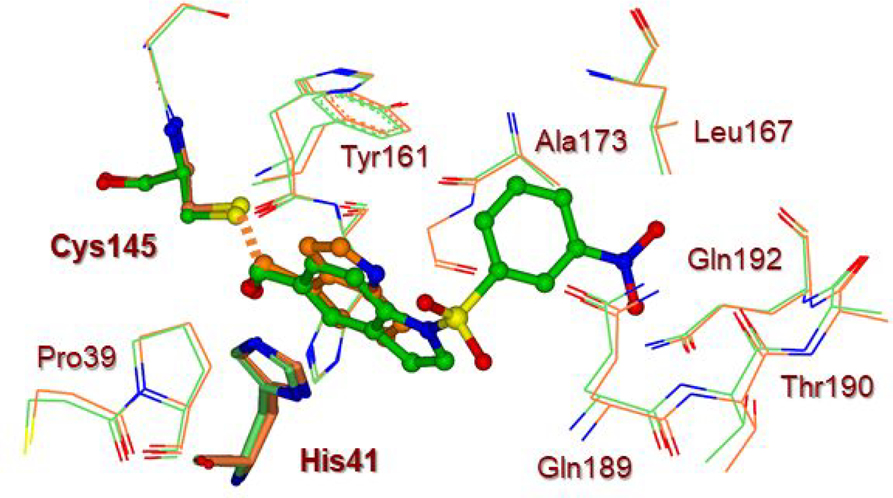
We have also determined the X-ray structure of SARS-CoV 3CLpro bound with inhibitor 7b at 1.63 Å resolution. The electron density map for the reaction product of inhibitor 7b and SARS-CoV 3CLpro is shown in Figure 6. The structure is very similar to compound 9d-bound SARS-CoV-2 3CLpro structure. The indole ring of 7b also forms π-π stacking with the shifted imidazole ring of His41 and the distance between the His41 imidazole ring and indole ring is about 3.5 Å. We compared the interactions of 7b-bound SARS-CoV 3CLpro with compound 9d-bound SARS-CoV-2 3CLpro structure. The superimposed structures are shown in Figure 7. As can be seen, the inhibitors overlapped nicely. The 3-nitrobenzenesulfonamide of compound 7b is positioned between the Gln189 and Gln192 after reaction with the cysteine. The structures of the proteins SARS-CoV 3CLpro and SARS-CoV-2 3Clpro have excellent overlap in many areas, particularly in the active sites of these enzymes.
Figure 6.
X-ray structure of SARS-CoV 3CLpro bound with compound 7b at 1.63 Å. Polder electron density omit maps (mFobs – DFmodel) surrounding 7b and Cys145. Electron density is contoured at 3.5 σ. (+) density is colored violet and (−) density is colored green. Final Rwork = 19.1% and Rfree = 22.2%. The thioester bond between Cys145 and 7b is clearly visible. (PDB code:7RC1).
Figure 7.
Superposition of SARS-CoV-2 3CLpro bound to compound 9d (orange carbon) and SARS-CoV 3CLpro bound to compound 7b (green carbon).
Conclusions
In summary, we have designed, synthesized, and evaluated a series of potent indole 5-chloropyridinyl ester-derived SARS-CoV-2 3CLpro inhibitors for the treatment of COVID-19. A number of compounds exhibited low nanomolar 3CLpro inhibitory activity. The mode of inhibition involves nucleophilic attack of catalytic Cys145 on the inhibitor’s ester carbonyl group and forming a covalent bond between Cys145 and the carbonyl group of the active ester. Our SAR studies show that the position of the carboxylic acid on the indole ring is important for activity. A number of compounds show potent antiviral activity in VeroE6 cell-based assays with RNA-qPCR and immunocytochemistry assays. In particular, compounds 1 and 7h exhibited potent antiviral activity (EC50 ~ 3 μM) comparable to compound 4 and FDA approved therapy, remdesivir. Furthermore, we demonstrate that the antiviral activity of compounds is not because of misleading cytostatic effects or cytotoxic effects, but due to an apparent destructive ‘antiviral effect.’ We determined high resolution X-ray structures of compound-bound to SARS-CoV 3CLpro and SARS-CoV-2 3CLpro enzymes. The structures revealed that catalytic Cys145 formed covalent bond with indole carbonyl group. Furthermore, the indole rings of inhibitors form π-π stacking interactions with the imidazole ring of the His41, a residue that must move substantially before, during and after reaction with the inhibitors. The present studies provided a number of lead compounds with potent antiviral activity. Our X-ray strutural studies offer important molecular insight into the active site interactions of these small molecules for further improvement of molcular features and activity. Further design and optimization are in progress in our laboratories.
Experimental Section.
General Methods.
All reactions were carried out under an argon atmosphere in either flame or oven-dried (120 °C) glassware. All reagents and chemicals were purchased from commercial suppliers and used without further purification unless otherwise noted. Anhydrous solvents were obtained as follows: Dichloromethane from calcium hydride, diethyl ether and tetrahydrofuran from Na/Benzophenone, methanol and ethanol from activated magnesium under argon. All purification procedures were carried out with reagent grade solvents (purchased form VWR) in air. TLC analysis was conducted using glass-backed Thin-Layer Silica Gel Chromatography Plates (60 Å, 250 μm thickness, F-254 indicator). Column chromatography was performed using 230–400 mesh, 60 Å pore diameter silica gel. 1H, 13C NMR spectra were recorded at room temperature on a Bruker AV-III-400 and AV-III-800. Chemical shifts (δ values) are reported in parts per million, and are referenced to the deuterated residual solvent peak. NMR data is reported as: δ value (chemical shift, J-value (Hz), integration, where s = singlet, d = doublet, t = triplet, q = quartet, brs = broad singlet). LRMS and HRMS spectra were recorded at the Purdue University Department of Chemistry Mass Spectrometry Center. HPLC analysis was done an on Agilent 1260 series instrument using a YMC Pack ODS-A column of 4.6 mm ID for analysis. The purity of all test compounds was determined by HPLC analysis to be ≥90% pure.
5-chloropyridin-3-yl 1H-indole-4-carboxylate (1):
To a stirred solution of commercially available methyl-1H-indole-4-carboxylate 12 (100 mg, 0.57 mmol) in a mixture of ethanol (3 mL) and water (3 mL), sodium hydroxide (46 mg, 1.14 mmol) was added. The resulting reaction mixture was stirred at 85 °C for 3 h. After this period, the reaction mixture was concentrated under reduced pressure. The residue was cooled at 0 °C and the solution was acidified to pH 4.5 using 1 M aqueous HCl. The mixture was extracted with ethyl acetate. The organic layer was dried over anhydrous Na2SO4 and concentrated under reduced pressure to give carboxylic acid (100 mg).
To a stirred solution of above acid (100 mg, 0.57 mmol) in CH2Cl2 (3 mL), 5-chloropyridin-3-ol (97 mg, 0.74 mmol), EDC (178 mg, 0.93 mmol) and DMAP (38 mg, 0.3 mmol) were added. The resulting reaction mixture was stirred at 23 °C for 12 h. After this period, the reaction mixture was washed with saturated aqueous NaHCO3. The organic layer was dried over anhydrous Na2SO4 and concentrated under reduced pressure to give a residue. The residue was purified via silica gel column chromatography (40% ethyl acetate in hexanes) to afford the title ester 1 (110 mg, 71% in 2-steps) as an amorphous solid. 1H NMR (400 MHz, CDCl3) δ 8.73 (s, 1H), 8.56 – 8.48 (m, 2H), 8.10 (d, J = 7.6 Hz, 1H), 7.78 – 7.67 (m, 2H), 7.42 (t, J = 2.9 Hz, 1H), 7.32 (t, J = 7.8 Hz, 1H), 7.27 – 7.19 (m, 1H); 13C NMR (100 MHz, CDCl3) δ 164.91, 147.68, 145.62, 141.61, 136.59, 131.73, 129.85, 127.91, 127.11, 124.43, 121.17, 119.17, 117.32, 103.70.; LRMS-ESI (m/z): 273.0 [M+H]+. HRMS (ESI/LTQ) m/z. [M+H]+ calcd for C14H10ClN2O2 273.0425; found 273.0431.
5-chloropyridin-3-yl indoline-4-carboxylate (2):
To a stirred solution of commercially available methyl-1H-indole-4-carboxylate 12 (100 mg, 0.57 mmol) in acetic acid (3 mL) at 10 °C, sodium cyanoborohydride (130 mg, 2.85 mmol) was added. The resulting reaction mixture was allowed to warm to 23 °C and stirred at that temperature for 12 h. After this period, the reaction mixture was concentrated under reduced pressure. The residue was diluted with aqueous NaHCO3 solution. The mixture was extracted with ethyl acetate. The organic layer was dried over anhydrous Na2SO4 and concentrated under reduced pressure to give crude indoline derivative. The product was purified by silica gel chromatography to give the corresponding indoline derivative (70 mg, 69%).
To a stirred solution of above indoline methyl ester (50 mg, 0.28 mmol) in a mixture of ethanol (1 mL) and water (1 mL), sodium hydroxide (23 mg, 0.56 mmol) was added. The resulting reaction mixture was stirred at 85 °C for 3 h. After this period, the reaction mixture was concentrated under reduced pressure. The residue was cooled at 0 °C and the solution was acidified to pH 4.5 using 1 M aqueous HCl. The mixture was extracted with ethyl acetate. The organic layer was dried over anhydrous Na2SO4 and concentrated under reduced pressure to give carboxylic acid (50 mg).
To a stirred soloution of indoline carboxylic acid (120 mg) in CH2Cl2 (3 mL), 5-chloropyridin-3-ol (115 mg, 0.88 mmol), EDC (211 mg, 1.1 mmol) and DMAP (45 mg, 0.5 mmol) were added. The resulting reaction mixture was stirred at 23 °C for 12 h. After this period, the reaction mixture was washed with saturated aqueous NaHCO3. The organic layer was dried over anhydrous Na2SO4 and concentrated under reduced pressure to give a residue. The residue was purified via silica gel column chromatography (40% ethyl acetate in hexanes) to afford the title ester 2 (160 mg, 55% over 3-steps) as an amorphous solid. 1H NMR (400 MHz, CDCl3) δ 8.47 (dd, J = 19.6, 2.2 Hz, 2H), 7.66 (t, J = 2.2 Hz, 1H), 7.49 (dd, J = 7.9, 1.0 Hz, 1H), 7.15 (t, J = 7.8 Hz, 1H), 6.87 (dd, J = 7.8, 1.0 Hz, 1H), 3.68 – 3.62 (m, 2H), 3.45 (t, J = 8.5 Hz, 2H); 13C NMR (100 MHz, CDCl3) δ 164.3, 145.7, 141.4, 129.6, 127.7, 127.0, 124.4, 121.2, 120.5, 117.3, 114.3, 103.7, 46.8, 30.9; LRMS-ESI (m/z): 275.0 [M+H]+. HRMS (ESI/LTQ) m/z. [M+H]+ calcd for C14H12ClN2O2 275.0582; found 275.0580.
5-chloropyridin-3-yl 1H-indole-5-carboxylate (7a):
To a stirred solution of methyl-1H-indole-5-carboxylate 10 (92 mg, 0.52 mmol), aqueous solution of 1 M lithium hydroxide (2 mL) and THF (0.5 mL) were added. The resulting reaction mixture was stirred at 23 °C for 24 h. After this period, solvent was evaporated under reduced pressure to give a residue. The residue was cooled at 0 °C and the solution was acidified to pH 3 using 10% citric acid. The mixture was extracted with ethyl acetate. The organic layer was dried over anhydrous Na2SO4 and concentrated under reduced pressure to give carboxylic acid (5a).
To a stirred solution of above acid (69.5 mg, 0.4 mmol) in CH2Cl2 (3 mL), 5-chloropyridin-3-ol (67 mg, 0.5 mmol), EDC (124 mg, 0.6 mmol) and DMAP (53 mg, 0.4 mmol) were added. The resulting reaction mixture was stirred at 23 °C for 4 h. After this period, the reaction mixture was washed with saturated aqueous NaHCO3. The organic layer was dried over anhydrous Na2SO4 and concentrated under reduced pressure to give a residue. The residue was purified via silica gel column chromatography (30% ethyl acetate in hexanes) to afford the title ester 7a (55 mg, 47% in 2-steps) as an amorphous solid. 1H NMR (400 MHz, CDCl3) δ 9.02 (s, 1H), 8.58 (dt, J = 1.6, 0.8 Hz, 1H), 8.50 (dd, J = 5.1, 2.2 Hz, 2H), 8.01 (dd, J = 8.6, 1.7 Hz, 1H), 7.71 (t, J = 2.2 Hz, 1H), 7.45 (dt, J = 8.6, 0.8 Hz, 1H), 7.31 (dd, J = 3.3, 2.3 Hz, 1H), 6.70 (ddd, J = 3.1, 2.0, 0.9 Hz, 1H); 13C NMR (100 MHz, CDCl3) δ 165.42, 147.89, 145.44, 141.54, 139.09, 131.77, 129.88, 127.62, 126.17, 124.88, 123.72, 119.45, 111.20, 104.09; LRMS-ESI (m/z): 273.0 [M+H]+. HRMS (ESI/LTQ) m/z. [M+H]+ calcd for C14H10ClN2O2 273.0425; found 273.0433.
5-chloropyridin-3-yl 1-((3-nitrophenyl)sulfonyl)-1H-indole-5-carboxylate (7b):
To a stirred soloution of indole methyl ester 11 (200 mg, 1.1 mmol) in THF (5 mL), sodium hydride (41 mg, 60% suspension in oil, 1.7 mmol) was added. The resulting reaction mixture was stirred at 0 °C for 1 h. After this period, 3-nitrobenzenesulfonyl chloride (380 mg, 1.7 mmol) was added and and the reaction mixture was warmed to 23 °C and stirred for 9 h. After completion of the reaction, the reaction mixture cooled to 0 °C and saturated aqueous NaCl was added. The reaction mixture was evaporated under reduced pressure to give a crude residue. The residue was extracted with ethyl acetate and washed with brine. The organic layer was dried over anhydrous Na2SO4 and concentrated under reduced pressure. The crude compound was purified via silica gel column chromatography (15% ethyl acetate in hexanes) to afford sulfonamide derivative 10. (170 mg, 41%). 1H NMR (400 MHz, CDCl3) δ 8.72 – 8.70 (m, 1H), 8.40 (ddd, J = 8.2, 2.2, 1.0 Hz, 1H), 8.27 (t, J = 1.2 Hz, 1H), 8.19 (ddd, J = 7.9, 1.8, 1.0 Hz, 1H), 8.04 (s, 2H), 7.71 – 7.66 (m, 1H), 7.63 (d, J = 3.7 Hz, 1H), 6.80 (dd, J = 3.7, 0.6 Hz, 1H), 3.92 (s, 3H); 13C NMR (100 MHz, CDCl3) δ 166.74, 148.20, 139.68, 137.06, 131.96, 130.84, 130.61, 128.45, 127.11, 126.37, 126.13, 123.96, 121.98, 112.98, 110.79, 52.15; LRMS-ESI (m/z): 391.2 [M+H]+.
To a stirred solution of above ester (63.6 mg, 0.18 mmol), aqueous 1M lithium hydroxide (2 mL) and THF (0.5 mL) were added. The reaction mixture was stirred at 23 °C for 9 h. After completion of the reaction, the reaction mixture was concentrated under reduced pressure. The resulting mixture was cooled at 0 °C and acidified to pH 3 by using 10% citric acid. The mixture was extracted with ethyl acetate. The organic layer was dried over anhydrous Na2SO4 and concentrated under reduced pressure to provide crude acid which was used for the next reaction without further purification.
To a stirred soloution of above acid (24 mg, 0.07 mmol) in CH2Cl2 (3 mL), 5-chloropyridin-3-ol (11 mg, 0.08 mmol), EDC (20 mg, 0.10 mmol) and DMAP (9 mg, 0.07 mmol) were added. The resulting reaction mixture was stirred at 23 °C for 5 h. After completion of the reaction, the reaction mixture was washed with saturated aqueous NaHCO3. The organic layer was dried over anhydrous Na2SO4 and concentrated under reduced pressure. The crude compound was purified via silica gel column chromatography (30% ethyl acetate in hexanes) to afford the title ester 7b (20.4 mg, 64% from 10) as an amorphous solid. 1H NMR (400 MHz, CDCl3) δ 8.75 – 8.70 (m, 1H), 8.58 (dt, J = 1.6, 0.8 Hz, 1H), 8.50 (t, J = 1.9 Hz, 3H), 8.02 (dd, J = 8.6, 1.7 Hz, 2H), 7.71 (t, J = 2.2 Hz, 1H), 7.48 (dt, J = 8.6, 0.8 Hz, 2H), 7.33 (dd, J = 3.3, 2.3 Hz, 2H), 6.71 (ddd, J = 3.1, 2.0, 1.0 Hz, 1H); 13C NMR (100 MHz, CDCl3) δ 165.30, 147.84, 145.50, 141.60, 141.57, 138.99, 131.69, 129.79, 127.60, 126.12, 126.02, 125.00, 124.88, 123.85, 123.81, 119.63, 111.13, 111.12, 104.22, 104.20; LRMS-ESI (m/z): 391.2 [M+H]+. HRMS (ESI/LTQ) m/z. [M+-NOS] calcd for C14H10ClN2O2 273.0425; found 273.0428.
5-chloropyridin-3-yl 4-methoxy-1H-indole-2-carboxylate (7c):
Commercially available 4-methoxy-1H-indole-2-carboxylic acid (50 mg, 0.26 mmol) was esterified with 5-chloropyridin-3-ol (41 mg, 0.31 mmol) by following the procedure for ester 7a to provide the title ester 7c (78 mg, 99%) as an amorphous solid.
1H NMR (400 MHz, CDCl3) δ 9.19 (s, 1H), 8.51 (dd, J = 4.9, 2.3 Hz, 2H), 7.72 (t, J = 2.1 Hz, 1H), 7.62 – 7.58 (m, 1H), 7.30 (t, J = 8.1 Hz, 1H), 7.03 (d, J = 8.3 Hz, 1H), 6.54 (d, J = 7.8 Hz, 1H), 3.98 (s, 3H); 13C NMR (100 MHz, CDCl3) δ 159.22, 154.75, 147.07, 145.88, 141.26, 138.99, 131.74, 129.42, 127.65, 123.62, 119.00, 109.15, 104.76, 99.97, 55.30; LRMS-ESI (m/z): 303.0 [M+H]+. HRMS (ESI/LTQ) m/z. [M+H]+ calcd for C15H12ClN2O3 303.0531; found 303.0538.
5-chloropyridin-3-yl 1-allyl-1H-indole-4-carboxylate (7d):
1-Allyl indole-4-carboxylic acid (40 mg, 0.20 mmol) was esterified with 5-chloropyridin-3-ol (31 mg, 0.23 mmol) by following the procedure for ester 7b to provide the title ester 7d (50 mg, 80%) as an amorphous solid. 1H NMR (400 MHz, CDCl3) δ 8.52 (dd, J = 5.7, 2.2 Hz, 2H), 8.08 (dd, J = 7.6, 0.9 Hz, 1H), 7.74 (t, J = 2.2 Hz, 1H), 7.63 (dt, J = 8.2, 0.9 Hz, 1H), 7.36 – 7.28 (m, 2H), 7.18 (dd, J = 3.1, 0.9 Hz, 1H), 6.01 (ddt, J = 17.1, 10.4, 5.3 Hz, 1H), 5.24 (dd, J = 10.3, 1.3 Hz, 1H), 5.07 (dd, J = 17.1, 1.1 Hz, 1H), 4.81 (d, J = 5.3 Hz, 2H); 13C NMR (100 MHz, CDCl3) δ 164.81, 147.65, 145.63, 141.65, 136.83, 132.80, 131.64, 130.83, 129.75, 128.64, 124.12, 120.74, 119.30, 117.59, 115.78, 102.51, 48.95; LRMS-ESI (m/z): 313.0 [M+H]+. HRMS (ESI/LTQ) m/z. [M+H]+ calcd for C17H14ClN2O2 313.0738; found 313.0745.
5-chloropyridin-3-yl 1,3-diallyl-1H-indole-4-carboxylate (7e):
1,3-Diallyl indole-4-carboxylic acid (20 mg, 0.1 mmol) was esterified with 5-chloropyridin-3-ol (13 mg, 0.1 mmol) by following the procedure for ester 7b to provide the title ester 7e (27 mg, 96%) as an amorphous solid. 1H NMR (400 MHz, CDCl3) δ 8.51 (s, 2H), 7.88 (dd, J = 7.5, 1.0 Hz, 1H), 7.71 (t, J = 2.2 Hz, 1H), 7.57 (dd, J = 8.3, 1.0 Hz, 1H), 7.31 – 7.24 (m, 1H), 7.10 (d, J = 0.9 Hz, 1H), 6.12 – 5.93 (m, 2H), 5.22 (dt, J = 10.3, 1.4 Hz, 1H), 5.12 – 5.01 (m, 2H), 4.93 (dd, J = 17.1, 1.9 Hz, 1H), 4.75 (dt, J = 5.3, 1.7 Hz, 2H), 3.70 (dq, J = 6.1, 1.4 Hz, 2H).; 13C NMR (100 MHz, CDCl3) δ 165.17, 145.64, 141.60, 138.06, 137.85, 132.85, 131.68, 129.84, 129.66, 125.52, 123.35, 123.20, 121.86, 120.36, 117.51, 115.07, 114.83, 113.82, 48.72, 31.64; LRMS-ESI (m/z): 353.0 [M+H]+. HRMS (ESI/LTQ) m/z. [M+H]+ calcd for C20H18ClN2O2 353.10513; found 353.10442.
5-chloropyridin-3-yl 3-methyl-1H-indole-4-carboxylate (7f):
Commercially available 3-methyl indole-4-carboxylic acid (20 mg, 0.11 mmol) was esterified with 5-chloropyridin-3-ol (18 mg, 0.14 mmol) by following the procedure for ester 7b to provide the title ester 7f (30 mg, 92%) as an amorphous solid. 1H NMR (400 MHz, CDCl3) δ 8.51 (t, J = 2.3 Hz, 2H), 8.37 (s, 1H), 7.92 (dd, J = 7.5, 1.0 Hz, 1H), 7.73 (t, J = 2.2 Hz, 1H), 7.61 (dd, J = 8.1, 1.0 Hz, 1H), 7.29 – 7.23 (m, 1H), 7.16 (dt, J = 2.2, 1.1 Hz, 1H), 2.47 (d, J = 1.0 Hz, 3H).; 13C NMR (100 MHz, CDCl3) δ 165.23, 147.70, 145.64, 141.56, 137.87, 131.77, 129.67, 125.97, 123.96, 121.34, 120.90, 120.73, 116.77, 112.62, 13.79; LRMS-ESI (m/z): 287.0 [M+H]+. HRMS (ESI/LTQ) m/z. [M+H]+ calcd for C15H12ClN2O2 287.05818; found 287.05871.
5-chloropyridin-3-yl 5-methyl-1H-indole-4-carboxylate (7g):
Commercially available 5-methyl indole 4-carboxylic acid (20 mg, 0.11 mmol) was esterified with 5-chloropyridin-3-ol (18 mg, 0.14 mmol) by following the procedure for ester 7b to provide the title ester 7g (32 mg, 98%) as an amorphous solid. 1H NMR (400 MHz, CDCl3) δ 8.53 (dd, J = 6.9, 2.2 Hz, 3H), 7.76 (t, J = 2.2 Hz, 1H), 7.52 (dd, J = 8.2, 0.9 Hz, 1H), 7.34 (dd, J = 3.2, 2.5 Hz, 1H), 7.15 (d, J = 8.3 Hz, 1H), 7.00 (ddd, J = 3.2, 2.1, 1.0 Hz, 1H), 2.75 (s, 3H); 13C NMR (100 MHz, CDCl3) δ 165.69, 147.52, 145.64, 141.61, 134.93, 134.60, 131.77, 129.84, 128.03, 126.30, 125.86, 118.54, 115.91, 103.81, 22.00; LRMS-ESI (m/z): 287.0 [M+H]+. HRMS (ESI/LTQ) m/z. [M+H]+ calcd for C15H12ClN2O2 287.05818; found 287.05867.
5-chloropyridin-3-yl 6-methyl-1H-indole-4-carboxylate (7h):
Commercially available 6-methyl-indole-4-carboxylic acid (25 mg, 0.14 mmol) was esterified with 5-chloropyridin-3-ol (22 mg, 0.17 mmol) by following the procedure for ester 7b to provide the title ester 7h (34 mg, 83%) as an amorphous solid. 1H NMR (400 MHz, CDCl3) δ 8.52 (dd, J = 7.0, 2.2 Hz, 3H), 7.93 (dd, J = 1.4, 0.7 Hz, 1H), 7.74 (t, J = 2.2 Hz, 1H), 7.49 (dt, J = 1.7, 0.9 Hz, 1H), 7.34 (dd, J = 3.2, 2.4 Hz, 1H), 7.14 (ddd, J = 3.1, 2.1, 1.0 Hz, 1H), 2.54 (s, 3H); 13C NMR (100 MHz, CDCl3) δ 164.99, 147.70, 145.60, 141.63, 137.07, 131.72, 131.15, 129.83, 126.39, 125.90, 125.83, 118.76, 117.47, 103.45, 21.32; LRMS-ESI (m/z): 287.0 [M+H]+. HRMS (ESI/LTQ) m/z. [M+H]+ calcd for C15H12ClN2O2 287.05818; found 287.05881.
5-chloropyridin-3-yl 6-fluoro-1H-indole-4-carboxylate (7i):
Commercially available 6-fluoro-indole-4-carboxylic acid (20 mg, 0.11 mmol) was esterified with 5-chloropyridin-3-ol (17 mg, 0.13 mmol) by following the procedure for ester 7b to provide the title ester 7i (30 mg, 93%) as an amorphous solid. 1H NMR (400 MHz, CDCl3) δ 8.61 – 8.50 (m, 3H), 7.83 (dd, J = 9.9, 2.3 Hz, 1H), 7.74 (t, J = 2.2 Hz, 1H), 7.44 – 7.39 (m, 2H), 7.19 (ddd, J = 3.2, 2.1, 0.9 Hz, 1H); 13C NMR (100 MHz, CDCl3) δ 159.61, 157.23, 145.86, 141.44, 136.64, 129.70, 127.43, 124.87, 119.70, 112.34, 112.08, 103.97, 103.75, 103.71; LRMS-ESI (m/z): 291.0 [M+H]+. HRMS (ESI/LTQ) m/z. [M+H]+ calcd for C14H9ClFN2O2 291.03311; found 291.03273.
5-chloropyridin-3-yl 1H-benzo[d]imidazole-4-carboxylate (7j):
Commercially available 1-benzoimidazole-4-carboxylic acid (50 mg, 0.30 mmol) was esterified with 5-chloropyridin-3-ol (60 mg, 0.5 mmol) by following the procedure for ester 7b to provide the title ester 7j (75 mg, 91%) as an amorphous solid. 1H NMR (400 MHz, MeOD) δ 8.47 (dd, J = 5.0, 2.2 Hz, 2H), 8.19 (s, 1H), 8.13 (dd, J = 7.7, 1.0 Hz, 1H), 8.03 (dd, J = 8.0, 1.0 Hz, 1H), 7.79 (t, J = 2.2 Hz, 1H), 7.43 – 7.37 (m, 2H); 13C NMR (100 MHz, MeOD) δ 163.62, 145.56, 142.81, 141.20, 132.04, 130.25, 130.21, 126.200, 126.17, 125.68, 122.00, 121.89, 112.43; LRMS-ESI (m/z): 274.0.[M+H]+. HRMS (ESI/LTQ) m/z. [M+H]+ calcd for C13H9ClN3O2 274.03778; found 274.03732.
5-chloropyridin-3-yl 2-methyl-1H-benzo[d]imidazole-4-carboxylate (7k):
Commercially available 2-methyl-benzoimidazole-4-carboxylic acid (15 mg, 0.1 mmol) was esterified with 5-chloropyridin-3-ol (13 mg, 0.1 mmol) by following the procedure for ester 7b to provide the title ester 7k (20 mg, 87%) as an amorphous solid. 1H NMR (400 MHz, CDCl3) δ 10.41 (s, 1H), 8.52 (dd, J = 8.0, 2.2 Hz, 2H), 8.01 (dd, J = 27.0, 7.8 Hz, 2H), 7.68 (t, J = 2.2 Hz, 1H), 7.36 (t, J = 7.9 Hz, 1H), 2.69 (s, 3H); 13C NMR (100 MHz, CDCl3) δ 164.19, 152.70, 147.07, 146.08, 141.37, 136.46, 135.13, 131.84, 129.54, 125.39, 124.80, 121.80, 111.01, 14.95; LRMS-ESI (m/z): 288.0 [M+H]+. HRMS (ESI/LTQ) m/z. [M+H]+ calcd for C14H11ClN3O2 288.05343; found 288.05302.
5-chloropyridin-3-yl benzo[d]oxazole-7-carboxylate (7l):
Commercially available benzoxazole-7-carboxylic acid (30.0 mg, 0.18 mmol) was esterified with 5-chloropyridin-3-ol (29 mg, 0.22 mmol) by following the procedure for ester 7b to provide the title ester 7l (50 mg, 99%) as an amorphous solid. 1H NMR (400 MHz, CDCl3) δ 8.51 (d, J = 2.1 Hz, 1H), 8.49 (d, J = 2.3 Hz, 1H), 8.43 (dd, J = 1.6, 0.7 Hz, 1H), 8.29 (s, 1H), 8.24 (dd, J = 8.4, 1.6 Hz, 1H), 7.92 (dd, J = 8.4, 0.7 Hz, 1H), 7.70 (t, J = 2.2 Hz, 1H); 13C NMR (100 MHz, CDCl3) δ 13C NMR (100 MHz, CDCl3) δ 163.65, 155.41, 149.64, 147.27, 146.09, 144.87, 141.28, 131.76, 129.46, 126.88, 125.78, 120.78, 113.52; LRMS-ESI (m/z): 275.0 [M+H]+. HRMS (ESI/LTQ) m/z. [M+H]+ calcd for C13H8ClN2O3 275.02180; found 275.02129.
5-chloropyridin-3-yl 3,4-dihydro-2H-benzo[b][1,4]oxazine-8-carboxylate (7m):
Commercially available 3,4-dihydro-benzoxazine-8-carboxylic acid (15 mg, 0.1 mmol) was esterified with 5-chloropyridin-3-ol (13 mg, 0.1 mmol) by following the procedure for ester 7b to provide the title ester 7m (20 mg, 83%) as an amorphous solid. 1H NMR (400 MHz, CDCl3) δ 8.45 (dd, J = 8.9, 2.2 Hz, 2H), 7.68 (t, J = 2.2 Hz, 1H), 7.36 (dd, J = 7.1, 2.4 Hz, 1H), 6.87 – 6.77 (m, 2H), 4.38 (dd, J = 5.1, 3.7 Hz, 2H), 4.00 (s, 1H), 3.55 – 3.41 (m, 2H); 13C NMR (100 MHz, CDCl3) δ 163.13, 147.50, 145.53, 145.29, 141.55, 134.61, 131.55, 129.73, 120.97, 120.45, 120.05, 116.90, 65.52, 40.10; LRMS-ESI (m/z): 291.0 [M+H]+. HRMS (ESI/LTQ) m/z. [M+H]+ calcd for C14H12ClN2O3 291.05310; found 291.05275.
N-(5-chloropyridin-3-yl)-1H-indole-4-carboxamide (9a):
Commercially available indole-4-carboxylic acid (50 mg, 0.31 mmol) was coupled with 5-chloro-3-aminopyridin (48 mg, 0.37 mmol) by following the procedure of EDC and DMAP for ester 7b to provide the title ester 9a (40 mg, 47%) as an amorphous solid. 1H NMR (400 MHz, MeOD) δ 8.81 (d, J = 2.2 Hz, 1H), 8.47 (t, J = 2.2 Hz, 1H), 8.29 (d, J = 2.2 Hz, 1H), 7.66 – 7.54 (m, 2H), 7.40 (d, J = 3.2 Hz, 1H), 7.23 (t, J = 7.8 Hz, 1H), 6.91 (dd, J = 3.2, 1.0 Hz, 1H); 13C NMR (100 MHz, DMSO) δ 167.81, 142.54, 140.14, 137.55, 136.96, 130.83, 127.56, 126.51, 126.30, 125.88, 120.50, 119.68, 115.66, 102.00; LRMS-ESI (m/z): 272.0 [M+H]+. HRMS (ESI/LTQ) m/z. [M+H]+ calcd for C14H11ClN3O 272.0585; found 272.0591.
N-(3-chlorophenyl)-1H-indole-4-carboxamide (9b):
Indole-4-carboxylic acid (20 mg, 0.12 mmol) was coupled with 3-chloroaniline (19 mg, 0.15 mmol) by following the procedure of EDC and DMAP for ester 7b to provide the title amide derivative 9b (32 mg, 95%) as an amorphous solid. 1H NMR (400 MHz, MeOD) δ 7.83 (t, J = 2.0 Hz, 1H), 7.57 – 7.48 (m, 3H), 7.31 (d, J = 3.2 Hz, 1H), 7.25 (t, J = 8.1 Hz, 1H), 7.17 (dd, J = 8.1, 7.4 Hz, 1H), 7.07 (ddd, J = 8.0, 2.1, 1.0 Hz, 1H), 6.86 (dd, J = 3.2, 1.0 Hz, 1H); 13C NMR (100 MHz, MeOD) δ 169.09, 140.20, 136.91, 133.89, 129.55, 129.50, 126.05, 123.48, 120.15, 120.11, 118.85, 118.72, 118.38, 114.46, 101.01; LRMS-ESI (m/z): 271.0 [M+H]+. HRMS (ESI/LTQ) m/z. [M+H]+ calcd for C15H12ClN2O 271.0633; found 271.0639.
5-chloro-2-methylpyridin-3-yl 1H-indole-4-carboxylate (9c):
Indole-4-carboxylic acid (50 mg, 0.31 mmol) was esterified with 5-chloro-2-methyl-pyridin-3-ol (54 mg, 0.37 mmol) by following the procedure for ester 7b to provide the title ester 9c (70.0 mg, 79%) as an amorphous solid. 1H NMR (400 MHz, CDCl3) δ 8.64 (s, 1H), 8.42 (d, J = 2.1 Hz, 1H), 8.12 (d, J = 7.5 Hz, 1H), 7.72 (d, J = 8.1 Hz, 1H), 7.62 (d, J = 2.2 Hz, 1H), 7.43 (t, J = 2.9 Hz, 1H), 7.33 (t, J = 7.8 Hz, 1H), 7.24 (dd, J = 7.3, 4.5 Hz, 1H), 2.52 (s, 3H); 13C NMR (100 MHz, CDCl3) δ 164.76, 150.04, 145.82, 145.01, 136.59, 130.05, 129.12, 127.94, 127.02, 124.37, 121.23, 119.36, 117.20, 103.79, 19.14; LRMS-ESI (m/z): 287.0 [M+H]+. HRMS (ESI/LTQ) m/z. [M+H]+ calcd for C15H12ClN2O2 287.05818; found 287.05766
5-chloro-4-methylpyridin-3-yl 1H-indole-4-carboxylate (9d):
Indole-4-carboxylic acid (50 mg, 0.31 mmol) was esterified with 5-chloro-4-methyl pyridin-3-ol (45 mg, 0.31 mmol) by following the procedure for ester 7b to provide the title ester 9d (70 mg, 79%) as an amorphous solid. 1H NMR (400 MHz, CDCl3) δ 8.71 (s, 1H), 8.50 (s, 1H), 8.41 (s, 1H), 8.16 – 8.10 (m, 1H), 7.72 (d, J = 8.1 Hz, 1H), 7.42 (t, J = 2.9 Hz, 1H), 7.33 (t, J = 7.8 Hz, 1H), 7.27 – 7.20 (m, 1H), 2.34 (s, 3H); 13C NMR (100 MHz, CDCl3) δ 164.95, 146.95, 146.11, 142.06, 138.82, 136.62, 132.83, 127.94, 127.07, 124.42, 121.21, 119.17, 117.25, 103.76, 13.35; LRMS-ESI (m/z): 287.0 [M+H]+. HRMS (ESI/LTQ) m/z. [M+H]+ calcd for C15H12ClN2O2 287.05818; found 287.05774.
5-chloro-6-methylpyridin-3-yl 1H-indole-4-carboxylate (9e):
Indole-4-carboxylic acid (25 mg, 0.15 mmol) was esterified with 5-chloro-6-methyl-pyridin-3-ol (22 mg, 0.15 mmol) by following the procedure for ester 7b to provide the title ester 9e (32 mg, 72%) as an amorphous solid. 1H NMR (400 MHz, CDCl3) δ 8.66 (s, 1H), 8.42 (d, J = 2.4 Hz, 1H), 8.09 (dd, J = 7.6, 0.9 Hz, 1H), 7.72 – 7.67 (m, 2H), 7.41 (dd, J = 3.2, 2.5 Hz, 1H), 7.31 (t, J = 7.8 Hz, 1H), 7.23 (ddd, J = 3.2, 2.1, 1.0 Hz, 1H), 2.66 (s, 3H); 13C NMR (100 MHz, CDCl3) δ 165.15, 153.13, 145.85, 140.65, 136.57, 130.76, 130.30, 127.87, 126.97, 124.38, 121.18, 119.40, 117.15, 103.76, 22.12; LRMS-ESI (m/z): 287.0 [M+H]+. HRMS (ESI/LTQ) m/z. [M+H]+ calcd for C15H12ClN2O2 287.05818; found 287.05768.
5-chloro-6-methylpyridin-3-yl 1H-benzo[d]imidazole-4-carboxylate (9f):
Commercially available benzoimidazole-4-carboxylic acid (20 mg, 0.12 mmol) was esterified with 5-chloro-6-methyl-pyridin-3-ol (21 mg, 0.15 mmol) by following the procedure for ester 7b to provide the title ester 9f (22 mg, 62%) as an amorphous solid. 1H NMR (400 MHz, CDCl3) δ 8.40 (d, J = 2.4 Hz, 1H), 8.20 (s, 1H), 8.17 – 8.09 (m, 3H), 7.64 (d, J = 2.4 Hz, 1H), 7.42 (t, J = 7.9 Hz, 1H), 2.65 (s, 3H); 13C NMR (100 MHz, CDCl3) δ 164.21, 153.77, 145.21, 143.50, 141.95, 140.38, 133.61, 130.91, 129.98, 126.62, 125.98, 122.11, 112.13, 22.11; LRMS-ESI (m/z): 288.0 [M+H]+. HRMS (ESI/LTQ) m/z. [M+H]+ calcd for C14H11ClN3O2 288.0534; found 288.0540.
Purification and Co-Crystallization of SARS-CoV-2 and SARS-CoV with inhibitors
SARS-CoV-2 with its authentic N- and C-termini was purified according to our recently published methods.25,38 Crystals grew at 4 °C from 3 μL droplets prepared by adding 1 μL of 110 μM SARS-CoV-2 3CLpro in 25 mM HEPES pH 7.50, 2.5 mM DTT, and 1% (v/v) DMSO containing inhibitor stocks to 2 μL of reservoir solution. Inhibitors were dissolved in DMSO and added to both crystallization droplets and cryo-protectant at concentrations of 400 μM for compound 2 (GRL-01720) and compound 9d (GRL-09120). The reservoir solution for the cocrystal complexes had constant concentrations of 3 mM DTT, 50 mM MES pH 6.0, 1% MPD, and varying concentrations of PEG-10,000 and KCl. The reservoir solution for compound 2 had a PEG-10,000 concentration of 22%, and a KCl concentration of 160 mM. The reservoir solution for compound 9d had a PEG-10,000 concentration of 18% and a KCl concentration of 120 mM. 3 μL of a cryo-solution containing an equivalent concentration of inhibitor prepared in DMSO, 25 mM HEPES pH 7.50, and 30% MPD was added to the crystal droplets. Crystals were soaked in the resulting solution for 30 minutes, then flash-frozen in liquid nitrogen in 0.05–0.2 μm nylon loops. SARS-CoV with its authentic N- and C-termini was purified according our previously published methods.48
The SARS-CoV 3Clpro-7b (GRL-686) inhibitor complex was crystallized using methods as previously described.49 Briefly, purified SARS-CoV 3CLpro was concentrated to 16 mg/mL in a buffer composed of 20 mM HEPES, pH 7.5, and 5 mM 2-mercaptoethanol. The enzyme was incubated on ice with a final concentration of 1 mM inhibitor. A 1:1 enzyme to crystallization solution ration was used, and the crystallization solution consisted of 14.5% PEG 20000, 50 mM MES, pH 6.0, 50 mM potassium chloride, and 1% MPD. Crystallization trials were set up at room temperature using the method of hanging drop vapor diffusion.
Data collection and structure refinement of SARS-CoV-2 3CLpro inhibitor complexes
X-ray diffraction data were collected on crystals of enzyme-inhibitor complexes using Life Sciences Collaborative Access Team (LS-CAT) beamlines 21-ID-F and 21-ID-G at the Argonne National Laboratory Advanced Photon Source, Argonne, Illinois, USA. X-ray data were indexed, integrated and scaled using the HKL2000 software package.50 The Phaser-MR module in the PHENIX software suite was used for molecular replacement.51 The search model used for molecular replacement was PDB: 6WNP41 with ligands and solvent molecules removed. Inhibitor coordinates and restraints were generated using eLBOW (PHENIX).52 Manual modeling building was performed using WinCoot.53 Automated structural refinement was performed using the Refine module available in PHENIX. Near the end of refinement, TLS restraints were generated and used in refinement and were left in only if it made a significant impact on Rfree, i.e. better than a drop of 1%. Anisotropic B-factors for individual atoms were automatically added by Phenix if appropriate. The X-ray data collection and refinement statistics for each 3CLpro-inhibitor complex are summarized in Table 3 in the supporting information.
IC50 value determination.
IC50 values were determined for compounds that covalently inhibit SARS-CoV-2 3CLpro using our recently described assay25 and data fitting methods that were derived from our previous work on SARS-CoV 3CLpro and inhibition by chloropyridyl esters.34 The only differences were that pre-incubation of the enzyme with the compounds was 10 minutes instead of 20 minutes. In addition, the Morrison Equation was only used to determine the IC50 value when they were below 1 μM.
Supplementary Material
ACKNOWLEDGEMENTS
The research was supported in part by grants from the National Institute of Allergy and Infectious Diseases, National Institutes of Health (AI158649 and AI150466, and A.D.M., contract No. HHSN272201700060C). The present work was also supported by a grant for Development of Novel Drugs for Treating COVID-19 from the Intramural Research Program of National Center for Global Health and Medicine (H.M., 19A3001 and S.H., 20A2001D), in part by Japan Agency for Medical Research and Development (AMED) (H.M., 20fk0108257). The authors acknowledge support from the Purdue Center for Cancer Research, NIH grant P30 CA023168 for use of the shared NMR and mass spectrometry facilities. This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817).
ABBREVIATIONS USED:
- 3CLpro
3-chymotrypsin-like protease
- DCC
dicyclohexylcarbodiimade
- DMAP
dimethylaminopyridine
- EDC
1-ethyl-3-(3-dimethylaminopropyl)carbodiimide
- F-actin
filamentors actin
- MERS-CoV
Middle east respiratory syndrome coronavirus
- MHV coronavirus
Mouse Hepatitis Virus coronavirus
- SARS-CoV-2
severe acute respiratory syndrome coronavirus 2
Footnotes
ASSOCIATED CONTENT
Supporting Information. The Supporting Information is available free of charge on the ACS Publication website at http://pubs.acs.org.
Full NMR spectroscopic data for all final compounds
X-ray structural data for inhibitors 2-bound SARS-CoV-2 3CLpro, 7b-bound SARS-CoV 3CLpro, and 9d-bound SARS-CoV-2 3CLpro
Molecular formula strings and some data (CSV)
PDB ID Codes. Inhibitors 2-bound SARS-CoV-2 3CLpro, 7b-SARS-CoV 3CLpro, and 9d-bound SARS-CoV-2 3CLpro X-ray structures are: 7RBZ, 7RC1, and 7RC0, respectively. Authors will release the atomic coordinates upon article publication.
The PDB accession codes for X-ray structures of inhibitors 2-bound SARS-CoV-2 3CLpro, 7b-SARS-CoV 3CLpro, and 9d-bound SARS-CoV-2 3CLpro are: 7RBZ, 7RC1, and 7RC0, respectively.
REFERENCES
- [1].Li Q; Guan X; Wu P; Wang X; Zhou L; Tong Y; Ren R; Leung KSM; Lau EHY; Wong JY; Xing X; Xiang N; Wu Y; Li C; Chen Q; Li D; Liu T; Zhao J; Liu M; Tu W; Chen C; Jin L; Yang R; Wang Q; Zhou S; Wang R; Liu H; Luo Y; Liu Y; Shao G; Li H; Tao Z; Yang Y; Deng Z; Liu B; Ma Z; Zhang Y; Shi G; Lam TTY; Wu JT; Gao GF; Cowling BJ; Yang B; Leung GM; Feng Z Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N. Engl. J. Med. 2020, 382, 1199–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Zhou P; Yang X-L; Wang X-G; Hu B; Zhang L; Zhang W; Si H-R; Zhu Y; Li B; Huang C-L; Chen H-D; Chen J; Luo Y; Guo H; Jiang R-D; Liu M-Q; Chen Y; Shen X-R; Wang X; Zheng X-S; Zhao K; Chen Q-J; Deng F; Liu L-L; Yan B; Zhan F-X; Wang Y-Y; Xiao G-F; Shi Z-L A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 2020, 579, 270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Fauci AS; Lane HC; Redfield RR Covid-19 — Navigating the uncharted. N. Engl.Med. 2020, 382, 1268–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Mitsuya H; Kokudo N Sustaining containment of COVID-19: Global sharing for pandemic response. Glob. Health Med. 2020, 2, 53–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].World Health Organization. Coronavirus disease (COVID-19) Situation Report, https://www.who.int/emergencies/diseases/novel-coronavirus-2019
- [6].Scavone C; Brusco S; Bertini M; Sportiello L; Rafaniello C; Zoccoli A; Berrino L; Racagni G; Rossi F; Capuano A Current pharmacological treatments for COVID-19: What’s next? Brit. J. Pharmacol. 2020, 177, 4813–4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Wang M; Cao R; Zhang L; Liu J; Xu M; Shi Z; Hu Z; Zhong W; Xiao G Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2019, 30, 269–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].McCreary EK; Angus DC Efficacy of remdesivir in COVID-19. JAMA 2020, 324, 1041–1042. [DOI] [PubMed] [Google Scholar]
- [9].Spinner CD; Gottlieb RL; Criner GJ; López JRA; Cattelan AM; Viladomiu AS; Ogbuagu O; Malhotra P; Mullane KM; Castagna A; Chai LYA; Roestenberg M; Tsang OTY; Bernasconi E; Le Turnier P; Chang S-C; SenGupta D; Hyland RH; Osinusi AO; Cao H; Blair C; Wang H; Gaggar A; Brainard DM; McPhail MJ; Bhagani S; Ahn MY; Sanyal AJ; Huhn G; Marty FM Effect of remdesivir vs standard care on clinical status at 11 days in patients with moderate COVID-19. A randomized clinical trial. JAMA 2020, 324, 1048–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kaur SP; Gupta V COVID-19 vaccine: A comprehensive status report. Virus Res. 2020, 288, 198114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Forni G; Mantovani A COVID-19 vaccines: Where we stand and challenges ahead. Cell Death & Differ. 2021, 28, 626–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Spellberg B; Nielsen TB; Casadevall A Antibodies, immunity, and COVID-19. JAMA Intern. Med. 2021, 181, 460–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Poland GA; Ovsyannikova IG; Kennedy RB SARS-CoV-2 immunity: Review and applications to phase 3 vaccine candidates. The Lancet. 2020, 396, 1595–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Wan Y; Shang J; Graham R; Baric RS; Li F Receptor recognition by the novel coronavirus from Wuhan: An analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020, 94, e00127–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Tse LV; Meganck RM; Graham RL; Baric RS The current and future state of vaccines, antivirals and gene therapies against emerging coronaviruses. Front. Microbiol. 2020, 98, 495–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kaul D An overview of coronaviruses including the SARS-2 coronavirus – Molecular biology, epidemiology and clinical implications. Curr. Med. Res. Pract. 2020, 10, 54–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Fehr AR; Perlman S (2015) Coronaviruses: An overview of their replication and pathogenesis. In: Maier H, Bickerton E, Britton P. (eds) Coronaviruses. Methods in Molecular Biology, vol 1282. Humana Press, New York, NY. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Coronaviridae Study Group of the International Committee on Taxonomy of Viruses, The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2019, 5, 536–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Forni D; Cagliani R; Clerici M; Sironi M Molecular evolution of human coronavirus genomes. Trends Microbiol. 2017, 25, 35–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ghosh AK; Xi K; Johnson ME; Baker SC; Mesecar AD Progress in anti-SARS coronavirus chemistry, biology and chemotherapy. Annu. Rep. Med. Chem. 2007, 41, 183–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Pillaiyar T; Manickam M; Namasivayam V; Hayashi Y; Jung S-H An overview of severe acute respiratory syndrome–coronavirus (SARS-CoV) 3CL protease inhibitors: Peptidomimetics and small molecule chemotherapy. J. Med. Chem. 2016, 59, 6595−6628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Pillaiyar T; Meenakshisundaram S; Manickam M Recent discovery and development of inhibitors targeting coronaviruses. Drug Discov. Today 2020, 25, 668–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ghosh AK; Brindisi M; Shahabi D; Chapman ME; Mesecar AD Drug development and medicinal chemistry efforts toward SARS-coronavirus and Covid-19 therapeutics. ChemMedChem, 2020, 15, 907–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Roe MK; Junod NA; Young AR; Beachboard DC; Stobart CC Targeting novel structural and functional features of coronavirus protease nsp5 (3CLpro, Mpro) in the age of COVID-19. J. Gen. Virol. 2021, 102, 001558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Han SH; Goins CM; Arya T; Shin W-J; Maw J; Hooper A; Sonawane DP; Porter MR; Bannister BE; Crouch RD; Lindsey AA; Lakatos G; Martinez SR; Alvarado J; Akers WS; Wang NS; Jung JU; Macdonald JD; Stauffer SR Structure-based optimization of ML300-derived, noncovalent inhibitors targeting the severe acute respiratory syndrome coronavirus 3CL protease (SARS-CoV-2 3CLpro). J. Med. Chem. 2021, DOI: 10.1021/acs.jmedchem.1c00598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hattori S.-i.; Higashi-Kuwata N; Hayashi H; Allu SR; Raghavaiah J; Bulut H; Das D; Anson BJ; Lendy EK; Takamatsu Y; Takamune N; Kishimoto N; Murayama K; Hasegawa K; Li M; Davis DA; Kodama EN; Yarchoan R; Wlodawer A; Misumi S; Mesecar AD; Ghosh AK; Mitsuya H A small molecule compound with an indole moiety inhibits the main protease of SARS-CoV-2 and blocks virus replication. Nat. Commun. 2021, 12, 668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Qiao J; Li Y-S; Zeng R; Liu F-L; Luo R-H; Huang C; Wang Y-F; Zhang J; Quan B; Shen C; Mao X; Liu X; Sun W; Yang W; Ni X; Wang K; Xu L; Duan Z-L; Zou Q-C; Zhang H-L; Qu W; Long Y-H-P; Li M-H; Yang R-C; Liu X; You J; Zhou Y; Yao R; Li W-P; Liu J-M; Chen P; Liu Y; Lin G-F; Yang X; Zou J; Li L; Hu Y; Lu G-W; Li W-M; Wei Y-Q; Zheng Y-T; Lei J; Yang S; SARS-CoV-2 Mpro inhibitors with antiviral activity in a transgenic mouse model. Science 2021, 371, 1374–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Halford B Pfizer unveils its oral SARS-CoV-2 inhibitor. Chem. Eng. News, 2021, 99, 7–7. [Google Scholar]
- [29].Hattori S.-i.; Higshi-Kuwata N; Raghavaiah J; Das D; Bulut H; Davis DA; Takamatsu Y; Matsuda K; Takamune N; Kishimoto N; Okamura T; Misumi S; Yarchoan R; Maeda K; Ghosh AK; Mitsuya H GRL-0920, an indole chloropyridinyl ester, completely blocks SARS-CoV-2 infection. mBio 2020, 11, e01833–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Ghosh AK; Xi K; Ratia K; Santarsiero BD; Fu W; Harcourt BH; Rota PA; Baker SC; Johnson ME; Mesecar AD Design and synthesis of peptidomimetic severe acute respiratory syndrome chymotrypsin-like protease inhibitors. J. Med. Chem. 2005, 48, 6767–6771. [DOI] [PubMed] [Google Scholar]
- [31].Ghosh AK; Xi K; Grum-Tokars V; Xu X; Ratia K; Fu W; Houser KV; Baker SC; Johnson ME; Mesecar AD Structure-based design, synthesis, and biological evaluation of peptidomimetic SARS-CoV 3CLpro inhibitors. Bioorg. Med. Chem. Lett. 2007, 17, 5876–5880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ratia K; Pegan S; Takayama J; Sleeman K; Coughlin M; Baliji S; Chaudhuri R; Fu W; Prabhakar BS; Johnson ME; Baker SC; Ghosh AK; Mesecar AD A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication. Proc. Natl. Acad. Sci. USA 2008, 105, 16119–16124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Ghosh AK; Takayama J; Aubin Y; Ratia K; Chaudhuri R; Baez Y; Sleeman K; Coughlin M; Nichols DB; Mulhearn DC; Prabhakar BS; Baker SC; Johnson ME; Mesecar AD Design, synthesis, and evaluation of novel fluoroquinolone–aminoglycoside hybrid antibiotics. J. Med. Chem. 2009, 52, 228–5240. [DOI] [PubMed] [Google Scholar]
- [34].Ghosh AK; Takayama J; Rao KV; Ratia K; Chaudhuri R; Mulhearn DC; Lee H; Nichols DB; Baliji S; Baker SC; Johnson ME; Mesecar AD Severe acute respiratory syndrome coronavirus papain-like novel protease inhibitors: Design, synthesis, protein−ligand x-ray structure and biological evaluation. J. Med. Chem. 2010, 53, 4968–4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Ghosh AK; Gong G; Grum-Tokars V; Mulhearn DC; Baker SC; Coughlin M; Prabhakar BS; Sleeman K; Johnson ME; Mesecar AD Design, synthesis and antiviral efficacy of a series of potent chloropyridyl ester-derived SARS-CoV 3CLpro inhibitors. Bioorg. Med. Chem. Lett. 2008, 18, 5684–5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Wu CY; King KY; Kuo CJ; Fang JM; Wu YT; Ho MY; Liao CL; Shie JJ; Liang PH; Wong CH Stable benzotriazole esters as mechanism-based inactivators of the severe acute respiratory syndrome 3CL protease. Chem. Biol. 2006, 13, 261–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Blanchard JE; Elowe NH; Huitema C; Fortin PD; Cechetto JD; Eltis LD; Brown ED High-throughput screening identifies inhibitors of the SARS coronavirus main proteinase. Chem. Biol. 2004, 11, 1445–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Zhang J; Pettersson HI; Huitema C; Niu C; Yin J; James MN; Eltis LD; Vederas JC Design, synthesis, and evaluation of inhibitors for severe acute respiratory syndrome 3C-like protease based on phthalhydrazide ketones or heteroaromatic esters. J. Med. Chem. 2007, 50, 1850–1864. [DOI] [PubMed] [Google Scholar]
- [39].Yao C-H; Song J-S; Chen C-T; Yeh T-K; Hsieh T-C; Wu S-H; Huang C-Y; Huang Y-L; Wang M-H; Liu Y-W; Tsai C-H; Kumar CR; Lee J-C Synthesis and biological evaluation of novel C-indolylxylosides as sodium-dependent glucose co-transporter 2 inhibitors. Eur. J. Med. Chem. 2012, 55, 32–38. [DOI] [PubMed] [Google Scholar]
- [40].Cardillo B; Casnati G; Pochini A; Ricca A Alkylation of indole sodium salt as ambifunctional nucleophilic system. Tetrahedron. 1967, 23, 3771–3783. [Google Scholar]
- [41].Anson BJ; Chapman ME; Lendy EK; Pshenychnyi S; D’Aquila RT; Satchell KJF; Mesecar AD, Broad-spectrum inhibition of coronavirus main and papain-like proteases by HCV drugs. Res. Square, preprint. 10.21203/rs.3.rs-26344/v1 (2020). [DOI] [Google Scholar]
- [42].Grum-Tokars V; Ratia K; Begaye A; Baker SC; Mesecar AD Evaluating the 3C-like protease activity of SARS-Coronavirus: recommendations for standardized assays for drug discovery. Virus. Res. 2008, 133, 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Hoffman RL; Kania RS; Brothers MA; Davies JF; Ferre RA; Gajiwala KS; He M; Hogan RJ; Kozminski K; Li LY; Lockner JW; Lou J; Marra MT; Mitchell LJ Jr.; Murray BW; Nieman JA; Noell S; Planken SP; Rowe T; Ryan K; Smith III GJ; Solowiej JE; Steppan CM; Taggart B; Discovery of ketone-based covalent inhibitors of coronavirus 3CL proteases for the potential therapeutic treatment of COVID-19. J. Med. Chem. 2020, 63, 12725–12747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].de Vries M; Mohamed AS; Prescott RA; Valero-Jimenez AM; Desvignes L; O’Connor R; Steppan C; Devlin JC; Ivanova E; Herrera A; Schinlever A; Loose P; Ruggles K; Koralov SB; Anderson AS; Binder J; Dittmann M A comparative analysis of SARS-CoV-2 antivirals characterizes 3CLpro inhibitor PF-00835231 as a potential new treatment for COVID-19. J. Virol 2021, 95, e01819–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Mesecar AD; Stoddard BL; Koshland DE Jr. Orbital steering in the catalytic power of enzymes: Small structural changes with large catalytic consequences. Science. 1997, 277, 202–206. [DOI] [PubMed] [Google Scholar]
- [46].Maeda K, Higashi-Kuwata N, Kinoshita N, Kutsuna S, Tsuchiya K, Hattori S-I, Matsuda K, Takamatsu Y, Gatanaga H, Oka S, Sugiyama H, Ohmagari N, and Mitsuya H. Neutralization of SARS-CoV-2 with IgG from COVID-19-convalescent plasma. Sci. Reports, 2021, 11, 5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Verschueren KHG; Pumpor K; Anemuller S; Chen S; Mesters JR; Hilgenfeld R A structural view of the inactivation of the SARS coronavirus main proteinase by benzotriazole esters. Chem. Biol. 2008, 15, 597–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Deng X; StJohn SE; Osswald HL; O’Brien A; Banach BS; Sleeman K; Ghosh AK; Mesecar AD; Baker SC Coronaviruses resistant to a 3C-like protease inhibitor are attenuated for replication and pathogenesis, revealing a low genetic barrier but high fitness cost of resistance. J. Virol. 2014, 88, 11886–11898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Jacobs J; Grum-Tokars V; Zhou Y; Turlington M; Saldanha SA; Chase P; Eggler A; Dawson ES; Baez-Santos YM; Tomar S; Mielech AM; Baker SC; Lindsley CW; Hodder P; Mesecar A; Stauffer SR Discovery, synthesis, and structure-based optimization of a series of N-(tert-butyl)-2-(N-arylamido)-2-(pyridin-3-yl) acetamides (ML188) as potent noncovalent small molecule inhibitors of the severe acute respiratory syndrome coronavirus (SARS-CoV) 3CL protease. J. Med. Chem. 2013, 56, 534–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Otwinowski Z; Minor W Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997, 276, 307–326. [DOI] [PubMed] [Google Scholar]
- [51].Adams PD; Afonine PV; Bunkozi G; Chen VB; Davis IW; Echols N; Headd JJ; Hung L-W; Kapral GJ; Grosse-Kunstleve RW; McCoy AJ; Moriarty NW; Oeffner R; Read RJ; Richardson DC; Richardson JS; Terwilliger TC; Zwart PH PHENIX: A comprehensive python-based system for macromolecular structure solution. Acta. Crystallogr. D Biol. Crystallogr. 2010, 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Moriarty NW; Grosse-Kunstleve RW; Adams PD electronic ligand builder and optimization workbench (eLBOW): A tool for ligand coordinate and restraint generation. Acta. Crystallogr. D Biol. Crystallogr. 2009, 65, 1074–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Emsley P; Lohkamp B; Scott WG; Cowtan K Features and development of coot. Acta. Crystallogr. D. Biol. Crystallogr. 2010, 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.