Abstract
When different preparations of Zymolyase were included in the pretreatment protocol of a panfungal PCR assay using a primer system for the 18S rRNA gene, an amplification product occurred in negative controls. The amplified fragment showed 100.0% sequence identity to the Saccharomyces sensu stricto complex and Kluyveromyces lodderae. Lyticase, lysing enzymes, and proteinase K appeared to be free from fungal DNA.
Early diagnosis of invasive fungal infection is a major concern in modern medicine, because of the high morbidity and mortality in high-risk patients. Conventional methods, including microscopy, culture, and antigen detection, lack sufficient sensitivity and/or specificity. Therefore, sensitive PCR assays have been devised for detection of fungal DNA in blood and bronchoalveolar lavage fluid, often with panfungal primers (2, 16). The sample pretreatment often includes the use of enzymes for digestion of yeast cell walls in order to extract fungal DNA (1, 3, 4, 6, 9, 13–15, 17). We consistently amplified DNA from the enzyme Zymolyase by using a universal fungal primer system for the 18S rRNA gene (10). To determine the origin of the amplified DNA, we sequenced this fragment.
Universal precautions to prevent PCR assay contamination, including the use of separate rooms and plasticware supplies for PCR setup and products, aliquoted reagents, and aerosol-resistant pipette tips, have been observed (11).
Several different enzymes used for sample pretreatment for fungal PCR assays, including three different batches of Zymolyase-20T from two companies, lyticase, lysing enzymes (from Trichoderma harzianum), and proteinase K, were examined (Table 1). The lyophilized enzymes were dissolved in sterile, pyrogen- and DNA-free water (Aqua ad injectabilia; Braun, Melsungen, Germany) to result in a final concentration of 300 μg/ml. One hundred microliters of this solution were heated for 10 min at 95°C and then processed by a purification and concentration procedure for DNA by using the GeneClean II kit (Bio 101, La Jolla, Calif.) according to the recommendations of the manufacturer. The final DNA preparation was dissolved in 25 μl of water. Sterile water as a negative sample control was processed the same way each time a sample preparation was done. Control DNA was prepared from Candida albicans and Aspergillus fumigatus from the University of Heidelberg strain collection.
TABLE 1.
Sensitive fungal PCR assays with fungi and commercial enzymes frequently used for pretreatment
| Sample | Source | Amplification product result with primer pair:
|
|
|---|---|---|---|
| S1-CUF1 | F5-F6 | ||
| C. albicans | DNA | Positive | Positive |
| A. fumigatus | DNA | Positive | Positive |
| Zymolyase-20T | Seikagaku, Tokyo, Japan | Positivea | Positivea |
| Zymolyase-20T | ICN, Aurora, Ohio | Positivea | Positivea |
| Lyticase | Sigma, St. Louis, Mo. | Negative | Negative |
| Lysing enzymes (T. harzianum) | Sigma, St. Louis, Mo. | Negative | Negative |
| Proteinase K | Amresco, Solon, Ohio | Negative | Negative |
| Proteinase K | Boehringer, Mannheim, Germany | Negative | Negative |
| Negative control | Sterile water | Negative | Negative |
Identified by sequencing as DNA from a member of the Saccharomyces sensu stricto complex or K. lodderae.
PCR amplifications were carried out in 100-μl volumes with 10 μl of sample added as described previously (10). Mixtures were subjected to an initial denaturation of 2 min at 95°C and 35 cycles of 95°C for 0.5 min, 53°C for 1 min, and 72°C for 1 min and a final extension at 72°C for 3 min and refrigeration. Two panfungal primer pairs complementary to fungal 18S rRNA sequences that are highly conserved were used: S1 and CUF1 (10), amplifying a 194-bp fragment (bp 86 to 279), including the variable region V2; and F5 (5′-AGTCTTAACCATAAACTATG) and F6 (5′-AGACAAATCACTCCACCA), amplifying a 295-bp fragment (bp 1012 to 1304), including the variable region V5.
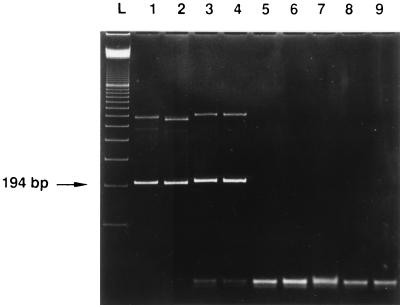
Both primer pairs consistently yielded amplification products when the Zymolyase preparation was added to the master mix. The amplicons had exactly the sizes of the fragments which were amplified from fungal DNA (194 and 295 bp, respectively). No products occurred when water or the preparations of the other enzymes were added (Fig. 1 and Table 1). To identify the contaminating DNA, both PCR fragments were sequenced as described previously (5). Briefly, the resulting PCR products were purified by using the Qiaquick PCR purification kit (Qiagen, Hilden, Germany), according to the recommendations of the manufacturer. About 200 ng of purified PCR product was submitted to cycle sequencing with biotinylated dideoxynucleoside triphosphates (GATC-Biocycle sequencing kit; GATC, Konstanz, Germany) and Thermo sequenase (Amersham Buchler, Braunschweig, Germany). The primers S1, CUF1, F5, and F6 were chosen for sequencing. DNA fragments were transferred to a nylon membrane during gel electrophoresis with a GATC 1500 direct blotting electrophoresis sequencer and visualized as blue bands with streptavidin alkaline phosphatase and a nitroblue tetrazolium chloride–X-phosphate mix (Boehringer, Mannheim, Germany). The results of sequence analysis of the two amplified fragments of the contaminating fungal DNA from the three tested batches of Zymolyase were 100% identical among the three batches with both primer pairs.
FIG. 1.
Ethidium bromide-stained polyacrylamide gel of PCR products obtained from enzymes used for pretreatment of samples for fungal PCR and controls by using primers S1 and CUF1. Lanes: L, 100-bp ladder; 1, 100 pg of C. albicans DNA; 2, 100 pg of A. fumigatus DNA; 3, Zymolyase (Seikagaku); 4, Zymolyase (ICN); 5, lysing enzymes (Sigma); 6, lyticase (Sigma); 7, proteinase K (Amresco); 8, proteinase K (Boehringer); 9, negative control.
DNA sequence similarity searches were performed by using the EMBL and GenBank sequence data banks. These searches revealed that both nucleotide sequences had 100% identity with 18S rRNA gene sequences of the following species: Saccharomyces cerevisiae (EMBL database accession no. Z75578), Saccharomyces pastorianus (accession no. X97805), Saccharomyces paradoxus (accession no. X97806), Saccharomyces bayanus (accession no. X97777), and Kluyveromyces lodderae (accession no. X83824). The four Saccharomyces species mentioned above display ≥99.9% sequence similarity in the 18S rRNA gene and form the Saccharomyces sensu stricto complex. K. lodderae is also a closely related yeast (8). Therefore, any of the five species could be the source of the contaminating fungal DNA in Zymolyase.
Thus, our finding raises concerns about possible false-positive results, if universal fungal primers are being used in sensitive PCR assays in combination with a Zymolyase pretreatment. Zymolyase is a registered trademark of the Kirin Brewery Co., Ltd., Tokyo, Japan. According to the distributors, Zymolyase-20T is manufactured by a submerged culture of Arthrobacter luteus by Yeast Related Business, Development Dept. (Kirin Brewery Co., Ltd.). Our data show that Zymolyase-20T is contaminated with DNA from the Saccharomyces sensu stricto complex or K. lodderae. This contamination is still present after partial purification of the Zymolyase by affinity chromatography, which was done by ICN Biomedicals, Inc.
Other enzymes, which are also widely used for pretreatment of samples for fungal PCR assays, including lysing enzymes from T. harzianum and lyticase (both from Sigma, Munich, Germany) and proteinase K (Amresco, Solon, Ohio, and Boehringer, Mannheim, Germany), did not yield amplification products when the panfungal primer pairs S1-CUF1 (Fig. 1) and F5-F6 (Table 1) were used; thus, they appeared to be largely free from fungal DNA.
The contamination of Taq polymerase with bacterial DNA, which is of relevance for PCR with universal bacterial primers, has been described previously (7, 12). This is now the first documented report of fungal DNA being present in an enzyme used in the PCR assay process. We conclude that if Zymolyase pretreatment is used for fungal PCR, primers and probes must not cross-react with Saccharomyces DNA, or enzymes other than Zymolyase should be used, in order to avoid false-positive results.
Acknowledgments
This work was supported in part by the Deutscher Akademischer Austauschdienst (DAAD) (A. P. Garg) and by DFG grant Ha 1921/3-1/2.
We thank Matthias Maiwald for critical reading of the manuscript.
REFERENCES
- 1.Burgener-Kairuz P, Zuber J-P, Jaunin P, Buchman T G, Bille J, Rossier M. Rapid detection and identification of Candida albicans and Torulopsis (Candida) glabrata in clinical specimens by species-specific nested PCR amplification of a cytochrome P-450 lanosterol-α-demethylase (L1A1) gene fragment. J Clin Microbiol. 1994;32:1902–1907. doi: 10.1128/jcm.32.8.1902-1907.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Einsele H, Hebart H, Roller G, Löffler J, Rothenhöfer I, Müller C A, Bowden R A, van Burik J-A, Engelhard D, Kanz L, Schumacher U. Detection and identification of fungal pathogens in blood by using molecular probes. J Clin Microbiol. 1997;35:1353–1360. doi: 10.1128/jcm.35.6.1353-1360.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fujita S-I, Lasker B A, Lott T J, Reiss E, Morrison C J. Microtitration plate enzyme immunoassay to detect PCR-amplified DNA from Candida species in blood. J Clin Microbiol. 1995;33:962–967. doi: 10.1128/jcm.33.4.962-967.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glee P M, Russell P J, Welsch J A, Pratt J C, Cutler J E. Methods for DNA extraction from Candida albicans. Anal Biochem. 1987;164:207–213. doi: 10.1016/0003-2697(87)90387-3. [DOI] [PubMed] [Google Scholar]
- 5.Haas W H, Schilke K, Brand J, Amthor B, Weyer K, Fourie P B, Bretzel G, Sticht-Groh V, Bremer H J. Molecular analysis of katG gene mutations in strains of Mycobacterium tuberculosis complex from Africa. Antimicrob Agents Chemother. 1997;41:1601–1603. doi: 10.1128/aac.41.7.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holmes A R, Cannon R D, Shepherd M G, Jenkinson H F. Detection of Candida albicans and other yeasts in blood by PCR. J Clin Microbiol. 1994;32:228–231. doi: 10.1128/jcm.32.1.228-231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hughes M S, Beck L-A, Skuce R A. Identification and elimination of DNA sequences in Taq DNA polymerase. J Clin Microbiol. 1994;32:2007–2008. doi: 10.1128/jcm.32.8.2007-2008.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.James S A, Cai J, Roberts I N, Collins M D. A phylogenetic analysis of the genus Saccharomyces based on 18S rRNA gene sequences: description of Saccharomyces kunashirensis sp. nov. and Saccharomyces martiniae sp. nov. Int J Syst Bacteriol. 1997;47:453–460. doi: 10.1099/00207713-47-2-453. [DOI] [PubMed] [Google Scholar]
- 9.Kan V L. Polymerase chain reaction for the diagnosis of candidemia. J Infect Dis. 1993;168:779–783. doi: 10.1093/infdis/168.3.779. [DOI] [PubMed] [Google Scholar]
- 10.Kappe R, Okeke C N, Fauser C, Maiwald M, Sonntag H-G. Molecular probes for the detection of pathogenic fungi in the presence of human tissue. J Med Microbiol. 1998;47:811–820. doi: 10.1099/00222615-47-9-811. [DOI] [PubMed] [Google Scholar]
- 11.Kwok S, Higuchi R. Avoiding false positives with PCR. Nature. 1989;339:237–238. doi: 10.1038/339237a0. [DOI] [PubMed] [Google Scholar]
- 12.Maiwald M, Ditton H-J, Sonntag H-G, von Knebel-Doeberitz M. Characterization of contaminating DNA in Taq polymerase which occurs during amplification with a primer set for Legionella 5S ribosomal RNA. Mol Cell Probes. 1994;8:11–14. doi: 10.1006/mcpr.1994.1002. [DOI] [PubMed] [Google Scholar]
- 13.Miyakawa Y, Mabuchi T, Fukazawa Y. New method for detection of Candida albicans in human blood by polymerase chain reaction. J Clin Microbiol. 1993;31:3344–3347. doi: 10.1128/jcm.31.12.3344-3347.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Müller F-M C, Werner K E, Kasai M, Francesconi A, Chanock S J, Walsh T J. Rapid extraction of genomic DNA from medically important yeasts and filamentous fungi by high-speed cell disruption. J Clin Microbiol. 1998;36:1625–1629. doi: 10.1128/jcm.36.6.1625-1629.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rand K H, Houck H, Wolff M. Detection of candidemia by polymerase chain reaction. Mol Cell Probes. 1994;8:215–222. doi: 10.1006/mcpr.1994.1030. [DOI] [PubMed] [Google Scholar]
- 16.van Burik J-A, Myerson D, Schreckhise R W, Bowden R A. Panfungal PCR assay for detection of fungal infection in human blood specimens. J Clin Microbiol. 1998;36:1169–1175. doi: 10.1128/jcm.36.5.1169-1175.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Deventer A J M, Goessens W H F, van Belkum A, van Vliet H J A, van Etten E W M, Verbrugh H A. Improved detection of Candida albicans by PCR in blood of neutropenic mice with systemic candidiasis. J Clin Microbiol. 1995;33:625–628. doi: 10.1128/jcm.33.3.625-628.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]