Abstract
Biologically-derived nanoparticles such as extracellular vesicles are promising candidates for therapeutic applications. In vivo toxicity of biological nanoparticles can result in tissue or organ damage, immunological perturbations, or developmental effects, but cannot be readily predicted from in vitro studies. Therefore, an essential component of the preclinical assessment of these particles for their use as therapeutics requires screening for adverse effects and a detailed characterization of their toxicity in vivo. However, there are no standardized, comprehensive methods to evaluate the toxicity profile of nanoparticle treatment in a pre-clinical model. Here, we first describe a method to prepare bovine milk-derived nanovesicles (MNVs). These MNV are inexpensive to isolate, have a scalable production platform, and can be modified to achieve a desired biological effect. We also describe the use of two vertebrate animal models, mice and zebrafish, that can be employed as needed to evaluate the toxicity profile of biologically derived nanoparticles, using MNVs as an example. Treatment-induced organ toxicity and immunological effects can be assessed in mice receiving systemic injections of MNVs, and developmental toxicity can be assessed in zebrafish embryos exposed to MNVs in embryo water. Utilizing these animal models provides opportunities for the analysis of the toxicity profiles of therapeutic extracellular vesicles in vivo.
Keywords: biological nanoparticles, safety, nanotherapeutics, zebrafish, developmental toxicity
INTRODUCTION
Biologically-derived nanoparticles based on extracellular vesicles (EVs) are promising candidates for therapeutic applications and are garnering increasing attention for use in the treatment of many different conditions such as organ injury, cancer, and neurodegenerative diseases (A. Li et al., 2021; Muthu, Bapat, Jain, Jeyaraman, & Jeyaraman, 2021). These nanoparticles can be released by virtually all mammalian cells and are present in bodily fluids including urine, milk, and blood (Tomiyama et al., 2021; Zhang, Xiong, & Yang, 2021; Zonneveld et al., 2014). These nanoparticles are imparted with cargo from their host cells and are enclosed within a lipid bilayer, which enables the safe transfer of the cargo to cells nearby or at distances far away from the host cell. Some nanoparticles, such as EVs, generally exhibit surface expression of several tetraspannins and they may also be endowed with unique markers to enable them to home to certain tissues or organs (Cohen et al., 2021). Recent developments in the isolation, characterization, and functional adaptation of biological nanoparticles have resulted in an increase in the different types of nanotherapeutics that are being developed for their therapeutic applications (Gardiner et al., 2016; Royo, Théry, Falcón-Pérez, Nieuwland, & Witwer, 2020).
An assessment of the safety of such biological nanoparticles is essential to evaluate their potential use as therapeutics. Additionally, since the nanoparticles exhibit batch-to-batch differences due to cell sources, methods of isolation, and downstream processing techniques, the toxicity profile of each batch of nanoparticles should be studied. A required step in this assessment is the evaluation of both their in vitro and in vivo toxicity. Screening for in vitro toxicity can be performed by examining their effects on cell viability and cellular responses that could result in cell injury or DNA damage in human cells. Although the potential for genotoxic effects can be detected in vitro by DNA damage assays and may reflect the potential for developmental abnormalities, toxicity to whole organisms cannot be readily inferred or predicted from in vitro studies. In vivo toxicity of biological nanoparticles can result, for instance, in tissue or organ damage, immunological perturbations, or developmental effects and thus, preclinical assessment of in vivo toxicity is necessary. As such, there is a need for the implementation of rapid screening approaches to detect adverse effects as an essential step in the preclinical assessment of potential toxicity of biological nanoparticles being developed for therapeutic use.
Given the promising therapeutic potential of biological nanotherapeutics, we have developed methods to screen for in vivo effects of their application and evaluate their developmental toxicity. Here, we detail these approaches using biological nanoparticles derived from milk as an example. Milk-derived nanovesicles (MNVs) can be isolated at scale and can serve as delivery vehicles (Ishiguro, Yan, Lewis-Tuffin, & Patel, 2020). We describe a method for their isolation that can be used to generate a source of biological nanoparticles for therapeutic use (Basic Protocol 1: Preparation of milk derived nanovesicles). Then, to illustrate a systematic assessment of in vivo toxicity, and using MNVs as an example, we describe two protocols, in two different vertebrate animal models, that could be used depending on the needs and goals of the research study. The first, is a method for determining acute organ injury and immunological effects in mice following systemic administration of single or multiple doses of nanoparticles (Basic Protocol 2: Acute and subacute toxicity screening in mice). At various time-points after nanoparticle administration, tissues are harvested to evaluate immune response and histopathological changes. The second, is a protocol for determining teratogenicity and developmental effects, using zebrafish as a model system (Basic Protocol 3: Developmental toxicity screening in zebrafish). In this protocol, zebrafish embryos are exposed to nanoparticles and evaluated for survival, developmental delay, and malformations. MNVs are used in these two protocols as an example of one therapeutic biological nanoparticle to test, but Basic Protocols 2 and 3 could be adapted for use with therapeutic extracellular vesicles or any other nanoparticle of interest, isolated using methods different from those described in Basic Protocol 1.
The use of these protocols will allow users to implement a systematic approach to generate toxicological data on organ injury, immunological effects, or developmental effects of biological nanoparticles that are being developed for use as therapeutic agents.
BASIC PROTOCOL 1: Preparation of milk derived nanovesicles
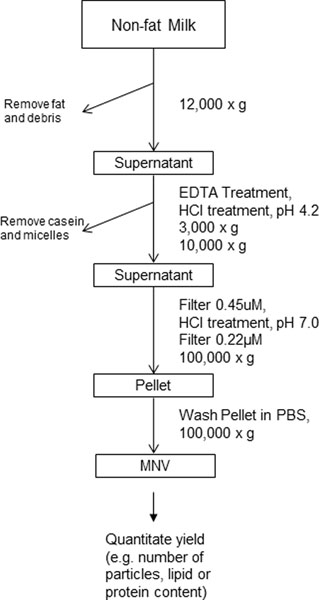
Recent studies have identified the use of biological nanoparticles such as extracellular vesicles as delivery vehicles for therapeutic agents. Biological nanoparticles derived from bovine milk, or MNVs, are particularly advantageous for this purpose due to the scalability and low cost of their isolation (Maji et al., 2017; Matsuda et al., 2020). MNVs can be isolated from commercially available non-fat bovine milk. Milk, however, contains casein proteins that arrange in micelles and can confound the isolation of MNVs. Different sources of milk will vary in the amount of casein micelles present and in their fat content. In the context of commercial or raw milk, the processing of homogenization and pasteurization may influence their cargo (Sanwlani, Fonseka, Chitti, & Mathivanan, 2020). This protocol combines approaches to disrupt casein micelles with sequential centrifugation steps, for purification (Figure 1). For quantitation of MNV size and yield, nanoparticle tracking analysis can then be performed, using, for instance, the NanoSight LM10, and such steps are also included.
Figure 1:
Flow chart of milk nanovesicle (MNV) isolation and purification as described in Basic Protocol 1.
Based on the specific research goals, the user can then proceed with toxicity profiling in mice (Basic Protocol 2) and/or in zebrafish (Basic Protocol 3).
NOTE:
commercially available fat-free milk is used for this protocol and should be used by the expiration date and kept at 4°C.
NOTE:
All centrifugations should be performed at 4°C.
NOTE:
If sterility is required for downstream assays, sterile centrifuge tubes should be used. All manipulations should be performed in a cell culture hood to reduce risk of contamination.
Materials
Reagents
Non-fat commercially available bovine milk
Ca2+ and Mg2+ free PBS (HyClone, cat no. SH30256.01)
0.5M Ca2+ free EDTA, pH 8.0 (Promega, cat no. V4321)
3.0M HCl (Fisher Scientific, cat no. SA56–500)
3.0M NaOH (Sigma, cat no. S5881)
Cold PBS (ThermoFisher, cat no. 14190144)
Maintain PBS at 4°C
Hardware and Instruments
Stainless steel spatula (Fisherbrand, cat no. 13–820-059)
2L glass beaker (Millipore sigma, cat no. CLS10032L)
Magnetic stir bar (Fisherbrand, cat no. 14–512-126)
Magnetic stir plate (Fisher scientific, cat no. 11–497-13)
Hexagonal weigh boat, large (Thomas Scientific, cat no. 1163Z61)
Polycarbonate Bottles with Screw-On Cap, 250ml (Beckman Coulter, cat no. 356013)
Polycarbonate Bottles with Cap Assembly, 26.3ml (Beckman Coulter, cat no. 355618)
Fixed Angle Rotor (Beckman Coulter, JLA-16.250)
High Speed High Capacity Centrifuge (Beckman Coulter, Avanti J-26XP)
Benchtop pH meter (Thermo Scientific, Orion 2-Star)
0.45uM PES vacuum filter system (Olympus, cat no. 25–228)
0.22uM PES vacuum filter system (Olympus, cat no. 25–227)
Fixed Angle Rotor, Type 70Ti (Beckman Coulter, cat no. 337922)
Ultracentrifuge (Beckman Coulter, XL-80)
Nanosight (Malvern Pananalytic, LM10)
Protocol Steps
Casein removal
-
1
Dilute 200 ml of non-fat milk with 200 ml of PBS. Mix gently and divide equally into two 250 ml polycarbonate bottles.
-
2
Centrifuge at 12,000 x g for 30 mins at 4°C.
-
3Carefully skim off the top white layer with a spatula. Transfer supernatant to a 2L beaker without disturbing the pellet.Be sure not to disturb the pellet, which may contain large debris. Using a pipet instead of pouring off the supernatant can help reduce contamination. Leave behind 5–10 ml of supernatant as a precaution.
-
4Add 0.5M Ca2+ free EDTA to the supernatant. Mix gently using a stir bar until the solution becomes yellowish and slightly clear.Add 1 ml of 0.5M Ca2+ free EDTA at a time and allow solution to mix. Depending on the starting material, a total of 8–12 ml of EDTA may be required. Solution should turn from opaque to slightly transparent.
-
5Adjust pH of the supernatant to 4.2 using 3.0M HCl.Add 1ml of 3.0M HCl at a time and allow the solution to mix. Visible precipitation will appear, and the solution will turn mostly transparent.
-
6
Cover the beaker with a plastic weigh boat and transfer to 4°C. Allow the solution to precipitate for 30 mins.
-
7
Stir gently using a magnetic stir plate and divide the entire mixture equally into two new 250 ml polycarbonate bottles.
-
8
Centrifuge at 3,000 x g for 10 mins at 4°C to remove large precipitates.
-
9
Transfer the supernatant to two new 250 ml polycarbonate bottles.
-
10
Centrifuge at 10,000 x g for 30 mins at 4°C.
-
11
Carefully transfer the supernatant and filter using a 0.45 μM vacuum filter.
-
12Adjust pH to 7.0 using 3.0M NaOH.Add 1 ml of 3.0M NaOH at a time and allow solution to mix. The amount of NaOH required should be similar to that used of HCl in step 5. The solution should remain transparent.
-
13
Transfer the solution and filter using a 0.22 μM vacuum filter. Store preparation at 4°C for up to 72hrs.
MNV purification
-
14
Divide the supernatant into separate polycarbonate bottles with cap assembly (add 24 mL to each) and spin at 100,000g for 70 mins at 4°C.
-
15
15. Discard the supernatant and resuspend the pellet with 24 mL of cold PBS and spin again at 100,000g for 70 mins at 4°C.
-
16Discard the supernatant and aspirate all remaining PBS. Add 250–500 μl of PBS to each tube and allow the pellet to soak for 3 hrs-overnight at 4°C.This volume may be adjusted to obtain a particular MNV concentration. However, pellet may remain as a precipitate if the volume used is too low.Make sure the pellet is submerged in PBS. Storing the tubes at a 45° angle can facilitate this.
-
17
Resuspend the MNV pellet by pipetting up and down. Transfer the suspension into a microcentrifuge tube and use immediately for preparation of therapeutic delivery vehicles or store at 4°C for up to 48hrs. If quantification is required, proceed to next step.
MNV quantification
Following MNV isolation, particle concentration must be quantitated prior to conducting any in vivo studies. There are several approaches available (Hartjes, Mytnyk, Jenster, van Steijn, & van Royen, 2019) and one of them, using NanoSight, is described below.
-
18Prepare the following dilutions of the MNVs from step 17 in PBS:
- First, prepare a 1:1,000 dilution: transfer 1 μL of MNVs to 1000 μL of PBS. Mix by pipetting
- Prepare the following serial dilutions using the 1:1,000 dilution that you prepared above as a starting point: 1:10,000, 1:100,000, and 1:1,000,000. Mix by pipetting.
-
19
Load 450 μL of the 1:1,000,000 dilution of MNVs into the NanoSight sample holder. Turn on the laser and place the holder underneath the microscope objective
-
20
Capture a single one-minute video. Export the data to a spreadsheet and record the concentration of MNVs. Calculate the average particle concentration from at least three separate assessments.
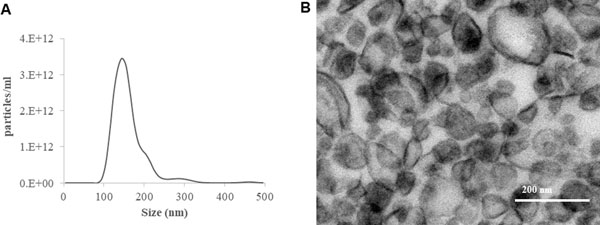
Particles isolated following this protocol range in size from 100 to 250nm, with a yield of up to 1 × 1013 particles/ml on nanoparticle tracking analysis. See Figure 2 for results and analysis of a representative MNV sample. Note that size and count may vary with the use of other techniques for quantitation.
At this point, the MNVs can be subjected to surface modifications or be loaded with therapeutic molecules of interest. The preparation of these therapeutic delivery vehicles is out of the scope of this paper, and readers interested in these methods are referred elsewhere (Han et al., 2021; Rayamajhi & Aryal, 2020). Such MNVs can then be subjected to in vivo toxicity assays (Basic Protocol 2 and 3).
Figure 2: Size and concentration of isolated MNVs.
(A) Assessment of size distribution of isolated MNVs by nanoparticle tracking analysis using NanoSight LM10. (B) Transmission electron microscopy image of MNV.
BASIC PROTOCOL 2: In vivo toxicology screening in mice
To evaluate the in vivo toxicity of therapeutic biological nanoparticles such as MNVs, these can be systematically administered to mice, and the effects on biochemical, immunological, or histological features of injury can be evaluated. The protocol described here can be used for empty therapeutic biological nanoparticles, to assess their inherent toxicity, but also for biological nanoparticles that are loaded with therapeutic cargo. This protocol can also be used to examine the toxicity of different concentrations or different formulations of therapeutic vesicles. Furthermore, screening can be performed with different batches to evaluate any variations that may be due to source, method of isolation, or post-isolation modifications.
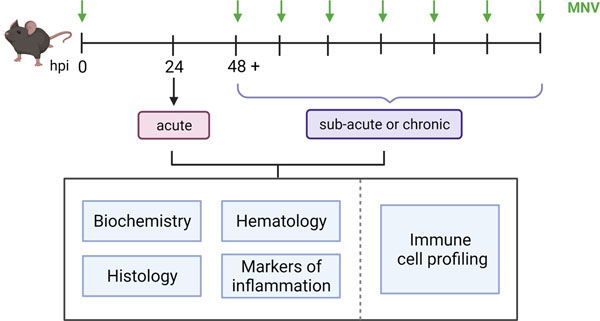
In this protocol, MNVs are used as an example of a type of therapeutic biological nanoparticle to test. Mice are administered a single dose of MNVs to assess acute toxicity, or multiple doses of MNVs to assess subacute toxicity. Mice can be euthanized 24 hours after treatment with a single dose of MNVs, or 2 weeks after treatment with a course of 8 doses of MNVs (Figure 3). At the treatment endpoints, serum is collected and used for biological assays. Tissue from solid organs such as the heart, lungs, kidneys, liver, or other organs of interest can be collected to assess treatment-induced tissue injury.
Figure 3. Evaluation of in vivo toxicity in mice.
The in vivo toxicity of biological nanoparticles can be evaluated in mice after a single administration (acute effects) or after repeated administration over a period of time (sub-acute or chronic effects). The dose of biological nanoparticles administered can be based on number of particles, or on protein or lipid content (see Background Information). At predetermined timepoints, blood and tissue are obtained for detailed systematic evaluation of toxicity. MNV, milk derived nanovesicles; hpi, hours post-injection.
If an assessment of treatment-induced effects on immune cells is desired, the spleen can be harvested, and single cell suspensions of splenocytes can be prepared and used to evaluate immune cell profiles by mass cytometry using CyTOF.
The type of therapeutic vesicles, the dosage schedule, and the duration of dose administration that are described here are illustrative and can be modified based on the specific needs and intended application. Same with the number of particles used.
Animal studies should be carried out in accordance with an approved institutional animal care and use protocol. Typically, mice are housed at 22°C with 40–70% humidity in a 12-hour light/dark cycle and fed food and water ad libitum.
Materials
Reagents
5 × 1011 particles of freshly prepared MNV therapeutic delivery vehicles or other vesicle of interest
A typical administration of MNVs may consist of 1 × 1010 particles to each mouse, but can be adjusted based on the goals of the study and type of vesicle.
Warm tap water
Ten 6-week-old male C57BL/6 mice (Jackson labs, cat no. 000634)
10% neutral buffered formalin (Fisherbrand, cat no. 032–059)
Sterile Ca2+- and Mg2+-free PBS (Hyclone, cat no. SH30256.01) supplemented with 0.1% BSA (Santa Cruz, cat no. sc-2323)
Sterile Ca2+- and Mg2+-free PBS (Hyclone, cat no. SH30256.01) supplemented with 0.1% w/v BSA (Santa Cruz, cat no. sc-2323) and 0.6% w/v sodium citrate (Sigma, cat no. S-4641).
Freezing media [FBS (Sciencell, cat no. 0025) plus 10% v/v DMSO (Sigma, cat no. D2650–5X5ML)]
DNase I (Sigma, cat no. 10104159001)
Trypan blue (Thomas Scientific, cat no. T8154–20m)
Tissue and Biopsy Cassettes (Fisher Scientific, cat no. 15–200-403D)
Ethanol (Fisher scientific, cat no. A405P-4)
Xylene (Millipore sigma, cat no. 534056)
Gill’s Hematoxylin (Fisher scientific, cat no. 6765010)
Eosin-Y (Fisher scientific, cat no. 71211)
Deionized (DI) water
Slides (Fisherbrand, cat no. 22–037-246)
Coverslips (Fisherbrand, cat no. 12–547)
Cytoseal XYL (Fisher scientific, cat no. 22–050-262)
For immune cell profiling (optional)
RPMI 1640 media (Invitrogen, cat no. 11875093)
MaxPar cell staining buffer (Fluidigm, cat no. 201068)
Cell-ID cisplatin (Fluidigm, cat no. 201064)
MaxPar PBS (Fluidigm, cat no. 2010580)
TruStain FcX PLUS (anti-mouse CD16/32) antibody (Biolegend, cat no. 156603)
Metal-conjugated antibody cocktail:
Refer to Table 1 for antibody volumes
Table 1.
Metal conjugated anti-mouse antibodies used for mass cytometry
| Antigen | Metal Tag | Clone | Volume (μL) | Fluidigm cat. No. |
|---|---|---|---|---|
| CD45 | 089Y | 30-F11 | 0.50 | 3089005C |
| TCRb | 143Nd | H57-597 | 0.50 | 3169002C |
| CD4 | 145Nd | RM4-5 | 0.50 | 3145002C |
| I-A/I-E | 150Nd | M5/114.15.2 | 0.20 | 3209006C |
| CD25 (IL-2R) | 151Eu | 1.00 | 3151007C | |
| CD3e | 152Sm | 145-2C11 | 0.25 | 3152004C |
| F4/80 | 159Tb | BM8 | 0.50 | 3159009C |
| CD62L | 160Gd | 1.00 | 3160008C | |
| Ly6G | 161Dy | 1A8 | 0.25 | 3141008C |
| CD44 | 162Dy | IM7 | 0.25 | 3171003C |
| CD19 | 166Er | 6D5 | 0.50 | 3149002C |
| CD8a | 168Er | 53-6.7 | 0.25 | 3146003C |
| CD161 (NK1.1) | 170Er | PK136 | 0.50 | 3170002C |
| CD11b (Mac-1) | 171Yb | M1/70 | 0.25 | 3148003C |
| CD45R (B220) | 176Yb | RA3-6B2 | 0.50 | 3176002C |
Anti-mouse 89Y-CD45 (Fluidigm, cat no. 3089005C, AB_2651152)
Anti-mouse 143Nd-TCRb (Fluidigm, cat no. 3169002C)
Anti-mouse 145Nd-CD4 (Fluidigm, cat no. 3145002C)
Anti-mouse 150Nd-I-A/I-E (Fluidigm, cat no. 3209006C)
Anti-mouse 151Eu-CD25 (Fluidigm, cat no. 3151007C)
Anti-mouse 152Sm-CD3e (Fluidigm, cat no. 3152004C)
Anti-mouse 159Tb-F4/80 (Fluidigm, cat no. 3159009C)
Anti-mouse 160Gd-CD62L (Fluidigm, cat no. 3160008C)
Anti-mouse 161Dy-Ly6g (Fluidigm, cat no. 3141008C)
Anti-human/mouse 162Dy-CD44 (Fluidigm, cat no. 3171003C)
Anti-mouse 166Er-CD19 (Fluidigm, cat no. 3149002C)
Anti-mouse 168Er-CD8a (Fluidigm, cat no. 3146003C)
Anti-mouse 170Er-CD161 (Fluidigm, cat no. 3170002C)
Anti-mouse 171Yb-CD11b (Fluidigm, cat no. 3148003C)
Anti-mouse 176Yb-CD45R (Fluidigm, cat no. 3176002C)
16% formaldehyde (w/v), methanol free (ThermoFisher Scientific, cat no. 28906)
MaxPar fix and perm buffer (Fluidigm, cat no. 201067)
Cell-ID Intercalator-Ir 125μM (Fluidigm, cat no. 201192A)
MaxPar water (Fluidigm, cat no. 201069)
MaxPar cell acquisition solution (Fluidigm, cat no. 201237)
EQ four element calibration beads (Fluidigm, cat no. 201078)
Protein block, serum free (Agilent, cat. no. X0909)
Hardware and Instruments
27-gauge insulin syringes
Mouse restrainer
25-gauge needles
1 mL syringes
Comprehensive blood chemistry panel (Idexx Radil, cat. no. 6006)
Alcohol wipes (Covidien, cat no. 6818)
70 μM cell strainers (MidSci, cat no. 2583668)
50 mL sterile conical tubes (Thermo Fisher, cat no. 339652)
Hemocytometer (Cardinal Healthcare, Cat no. B3192)
5 mL Eppendorf tubes (Olympus, cat no. 24–285)
Water bath
1.5 mL autoclaved Eppendorf tubes (Thomas scientific, cat no. 1148T71)
Surgical tape (ULINE, cat no. S-19225)
Polystyrene round-bottom tubes with 35 μM cell strainers (Thermal Scientific, cat no. BD352235)
For tissue processing:
Automated tissue processor e.g. Leica Biosystems
For immune cell profiling:
Mass cytometry system - Helios CyTOF (Fluidigm)
Protocol Steps
In vivo study
-
1
Place one mouse at a time in the restrainer and submerge the tail in warm water for approximately 15 seconds to dilate the tail veins.
-
2
As a vehicle control, inject 100 μL of PBS into the tail vein of one mouse, using a 27-guage insulin syringe.
-
3
Prepare single aliquots of 1 × 1010 MNV particles in 100 μL of PBS, one aliquot per each mouse.
-
4
Transfer the particle solution to a 27-guage insulin syringe
-
5Administer the MNVs by tail vein injection, as in Steps 1 and 2.To assess acute toxicity, administer a single dose of MNV to 4 mice, which will be euthanized at 24 hours post-treatment (Figure 3).To assess sub-acute toxicity, administer this dose of MNVs every 2 days, for a total of 8 doses. The mice will be euthanized two weeks after the first dose was administeredFor immune profiling, administer a single dose of MNV to 1 mouse, which will be euthanized at 4 hours post-treatment.
-
6
At the designated treatment endpoint, euthanize the mice by CO2 asphyxiation followed by cervical dislocation (See (Donovan & Brown, 2006)).
-
7Perform cardiac puncture using a 25-gauge needle to collect circulating blood from the mice used to assess acute and sub-acute toxicity. Transfer the blood to a 1.5 mL Eppendorf tube. This sample can be used for serum biochemistry analysis protocol (Step 22).Cardiac puncture does not need to be performed on a mouse that will be used for immune profiling studies.Prior to performing cardiac puncture, lie the mouse in a supine position and secure its arms and legs to the tabletop using surgical tape.To perform cardiac puncture, place the syringe bevel side up at a 40° angle slightly below and to the left of the sternum, pointed towards the head. Slowly withdraw the circulating blood. If performed correctly, approximately 500uL of blood can be collected from a single mouse.
-
8Harvest the heart, liver, kidney, and lungs and preserve them in 10% v/v formalin overnight.The volume of formalin added should be enough to completely submerge the tissues. Examine the gross appearance, presence of any hemorrhage or necrosis, and take note of the organ’s and total body weight.
-
9Transfer the fixed tissues from Step 8 to histology cassettes and submerge them in 70% v/v ethanol until further processing.Tissue processing and embedding in paraffin should be done using an available automated tissue processor and following their associated protocols. Processing steps will include a dehydration step to replace water in the cells with alcohol, a clearing step to dissolve the alcohol using an intermediate solvent such as xylene, and an infiltration step in which the specimen is infiltrated with paraffin. Specimens are then formed into a block and placed into a microtome for section cutting. Sections are cut and placed onto slides for further analysis.
-
1010. Harvest the spleen and place it in a tube with 5 mL of pre-warmed RPMI mediaPlace the media in a 37°C bead or water bath for 15 minutes prior to use.
At this point, users can proceed to downstream analyses that may be of interest to them based on the needs of their study. We have outlined below the steps for some common types of analyses typically used.
Histopathological analysis
-
11
Prepare tissue sections on microscope slides as in Step 9.
-
12
To de-paraffinize the slides, incubate them in a 60°C steamer for 1 hour.
-
13
To remove the paraffin from the slides, soak them in xylene for 4 minutes
-
14
Remove slides and soak slides in fresh xylene for 4 minutes.
-
15
Repeat step 14 three additional times.
-
16
Gradually rehydrate the slides with a graded series of ethanol solutions: first soak the slides in 100% ethanol for 3 minutes. Then, transfer the slides to a 95% ethanol solution and soak for 3 minutes. Then, transfer the slides to a 75% ethanol solution and soak for 3 minutes. Lastly, move the slides into fresh 75% ethanol and soak for 3 minutes.
-
17
To rehydrate the slides, rinse them with water
-
18
Dip the slides in H&E and soak for 1 minute
-
19
Wash the slides 3 times with DI water
-
20
Incubate the slides in 70% ethanol for 3 minutes and subsequently incubate them in 100% ethanol for 3 minutes.
-
21
Incubate the slides in xylene for 5 minutes
-
22Add two drops of Cytoseal to each slide, place a coverslip over the tissue, and allow the slides to air dry.Evaluate for evidence of tissue necrosis, inflammation, fibrosis or hemorrhage. See sample data in Figure 4.
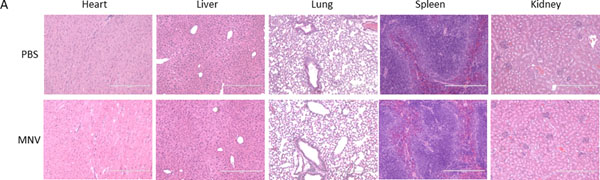
Figure 4. Histopathological analysis.
Empty, unmodified MNVs or PBS-vehicle controls were administered to mice, and the heart, liver, lung, spleen, and kidney were harvested after 24 hours. Hematoxylin and eosin (H&E) staining was performed on sections of the abovementioned tissues to assess histological damage. No visible damage was apparent in these studies.
Serum biochemistry analysis
-
23
Allow the mouse blood (from Step 7) to coagulate at room temperature for 30 minutes
-
24
Centrifuge the whole blood at 10,000 x g for 5 minutes
-
25
Collect the serum and transfer 250 μL of serum to 1.5 mL sterile Eppendorf tubes
-
26
Send serum samples to a clinical biochemistry analytical laboratory, e.g. Idexx Radil, to perform a comprehensive blood chemistry panel (e.g. Table 2)
Table 2.
Biochemical assessment of organ or tissue injury
| Biochemical tests | |
|---|---|
| Hepatic | Alanine aminotransferase, Aspartate aminotransferase, Alkaline phosphatase, Bilirubin, Albumin, Globulin |
| Renal and electrolytes | Creatinine, Blood urea nitrogen, Bicarbonate, Chloride Sodium, Potassium, Calcium |
| Cardiac and muscle | Creatine kinase |
| Pancreatic | Amylase |
| Inflammation | IL-6, C-reactive protein |
Immune response
-
27Visually inspect the spleen (Step 10) to evaluate growth in response to the treatment.An enlarged spleen indicates a potent immune response.
Immune cell profiling by mass cytometry
-
28
Place the harvested spleen (Step 10) in a 70 μM cell strainer over a 50 mL conical tube containing a small volume of cold PBS supplemented with 0.1% BSA and 0.6% sodium citrate.
-
29Using the plastic side of the syringe plunger, macerate the spleen to create a single-cell suspension.Do not use the plunger region of the syringe to macerate the tissue, as this region contains a large amount of barium, a common contaminant in CyTOF.
-
30
Rinse the cell strainer at regular intervals with PBS supplemented with 0.1% BSA and 0.6% sodium citrate
-
31
Centrifuge the splenocytes at 180 x g for 10 minutes at 4°C
-
32
Remove the supernatant and resuspend the pellet in 10 mL of PBS supplemented with 0.1% BSA and 0.6% sodium citrate, and centrifuge the splenocytes at 180 x g for 10 minutes at 4°C.
-
33
Repeat step 32
-
34
Remove the supernatant and resuspend the cells in 1 mL of PBS supplemented with 0.1% BSA
-
35Count the cells with a hemocytometer.The concentration of the splenocytes is typically very high, so it is best to prepare a 1/10 dilution of your cell suspension before counting
-
36
Centrifuge the cells at 180 x g for 5 minutes at 4°C
-
37
Discard the supernatant and resuspend the cells in cold freezing media (FBS supplemented with 10% DMSO) to achieve a final concentration of 1×107 cells/mL.
-
38Prepare 1 mL aliquots of cells and store them in a cool cell at −80°C. This is a safe stopping point.Splenocytes can be stored at −80 for up to two weeks
-
39
When ready to analyze, place a vial of frozen splenocytes in a 37°C water bath.
-
40
Prepare pre-warmed RPMI 1640 media supplemented with 10 units/mL DNase I.
-
41
Transfer the thawed splenocytes to a conical tube containing 10 mL of RPMI media supplemented with DNase I and return the pre-warmed RPMI 1640 media to the water bath.
-
42
Centrifuge the cells at 180 x g for 5 minutes at room temperature.
-
43
Remove the supernatant and resuspend the cells in 1 mL of pre-warmed RPMI 1640 media.
-
44
Count the cells with a hemocytometer.
-
45Centrifuge the cells at 180 x g for 5 minutes, then remove the supernatant and resuspend the cells in MaxPar cell staining buffer to achieve a concentration of 3.5×105 cells in 150 μLIf there are less than 3.5×105 cells, thaw an additional vial of splenocytes and repeat steps 40–44. Combine the thawed splenocytes to obtain the desired number of splenocytes.
-
46Transfer the cells to 5 mL Eppendorf tubesThe Eppendorf tubes used hereafter must not be autoclaved, as autoclaving can introduce barium which is a common CyTOF contaminant
-
47Prepare a 0.625μM solution of Cell-ID cisplatin in MaxPar PBSPrepare at least 150 μl per sample. This concentration of cisplatin was optimized for our samples.
-
48
Add 150 μL of 0.625μM cisplatin to each tube, vortex briefly to mix
-
49
Incubate the cells with cisplatin for 5 minutes at room temperature
-
50
Add 1 mL of MaxPar cell staining buffer to each tube and centrifuge the cells at 300 x g for 5 minutes at 4°C
-
51
Resuspend each cell pellet in 100 μL of MaxPar cell staining buffer and incubate the cells with 0.125 μg of TruStain FcX PLUS antibody for 5 minutes on ice to block the Fc receptors.
-
52
In the meantime, prepare the antibody cocktail, using the volumes listed in Table 1 and adding MaxPar cell staining buffer to achieve a final volume of 50 μL.
-
53
After the 5-minute incubation from step 51, add 50 μL of the antibody cocktail (Step 52) to each tube and pipette to mix
-
54
Incubate the cells for 15 minutes at room temperature
-
55
Wash the cells with 1 mL of MaxPar cell staining buffer. Centrifuge the cells at 300 x g for 5 minutes
-
56
Discard the supernatant and repeat step 55 once more.
-
57
Discard most of the supernatant but leave approximately 30 μL of supernatant behind. Briefly vortex
-
58
Resuspend the cell pellets with 500 μL of MaxPar PBS and pipette to mix
-
59Prepare a 3.2% v/v formaldehyde solution in MaxPar PBS in a total volume of 1 mL. Add 500 μL of 3.2% formaldehyde solution to each tubeIn order to minimize the number of doublets, the cells are first resuspended in PBS(Step 58) to create a single cell solution and then an equal volume of a 3.2% formaldehyde solution is added to the cells (this step), such that the final concentration of formaldehyde used to fix the cells is 1.6%
-
60
Vortex the cells and incubate for 10 minutes at room temperature
-
61
Centrifuge the cells at 800 x g for 5 minutes at 4°C
-
62
Prepare 1 mL per sample of a 125 μM solution of intercalator-Ir in MaxPar fix and perm buffer
-
63
Discard the supernatant and resuspend each pellet with 1 mL of 125 μM intercalator-Ir solution
-
64
Incubate the cells overnight at 4°C
-
65
Centrifuge the cells at 800 x g for 5 minutes at 4°C
-
66
Remove the supernatant and wash the cells once with 2 mL of MaxPar cell staining buffer
-
67
Repeat Step 65, remove supernatant, and resuspend the cells in 1 mL of MaxPar water
-
68
Count the cells with a hemocytometer.
-
69
Centrifuge the cells at 800 x g for 5 minutes at 4°C and aspirate most of the supernatant, leaving a small volume of supernatant behind (approximately 10 μL)
-
70
Prepare 5 mL of a 1/10 dilution of EQ equilibration beads in MaxPar water
-
71Add enough of the 1/10 EQ bead solution to each of the tubes to achieve a final cell concentration of 1 × 105 cells/mL.The volume of EQ bead solution needed will depend on the concentration of the cell pellet.
-
72
Transfer the cell suspensions to 5 mL FACS tubes. FACS will not be performed but the samples must be placed in FACS tubes for CyTOF.
-
73Acquire data and analyze using visualization tools as suitable to define the cell proportions.>Live CD45+ events should be identified. CD45 is a pan-lymphocyte marker, enabling the identification of any cells deriving from the lymphocyte lineage. A gating strategy for this is outlined in Figure 5. Once identified, these events can be analyzed by various dimensionality-reduction algorithms, reviewed in detail elsewhere (Kimball et al., 2018; Palit, Heuser, de Almeida, Theis, & Zielinski, 2019).
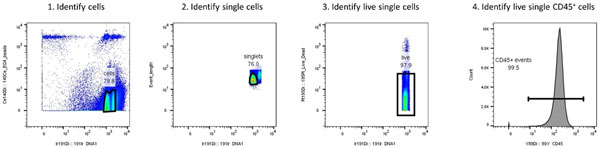
Figure 5: Gating strategy for mass cytometry data.
After systemic administration of biological nanoparticles such as MNVs, splenocytes were isolated and stained with a cocktail of metal-conjugated antibodies, and mass cytometry was performed. Live single CD45+ events were identified by drawing a gate around the events that had high 191Ir staining and low levels of the 140 Ce metal tag to discriminate the cells from beads, bead-cell doublets, and debris. Next, a daughter population of the cell gate was created to identify single cells, which had moderate 191Ir staining compared to the doublets. Then a daughter population of the single cells was created to identify the live cells, which had low to moderate levels of 191Pt. Lastly, a daughter plot of the live single cells was generated to identify CD45+ events, which are expressed by all lymphocyte subtypes.
BASIC PROTOCOL 3: In vivo developmental toxicology screening in zebrafish
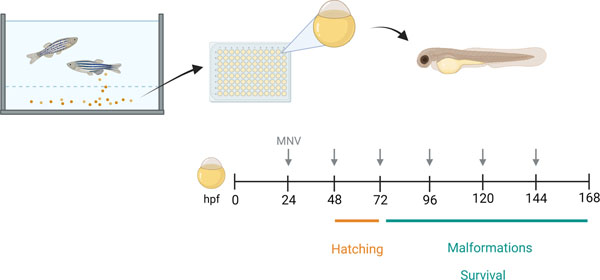
Assessment of developmental toxicity is an important requirement for the safety characterization of biological nanoparticles intended for in vivo use. Developmental toxicity can be examined in zebrafish embryos by monitoring survival, evaluating the hatching rate, and looking for gross malformations (Figure 6). Here, we use MNVs as an example test nanoparticle. By adding MNV therapeutic delivery vehicles into the embryo water, fish are exposed by direct surface contact and ingestion. The effects of different concentrations or formulations of MNVs on survival, hatching, and malformations can be systematically analyzed over a period spanning up to 144 hours post-fertilization (hpf). Mortality rate can be assessed using different treatment concentrations at a given time point. Developmental abnormalities may contribute to a delay in hatching. As zebrafish typically hatch between 48–72 hours, an assessment of hatching proportions can be performed at 45hpf, 48hpf, 51hpf, and 72hpf. In addition, teratogenicity and documentation of visible malformations can be determined by systematically evaluating for malformations at pre-defined time-points, up to 144 hpf. Zebrafish heart rate and pericardial edema should also be monitored, as MNV treatment-induced alterations may contribute to developmental toxicity.
Figure 6: Screening for developmental toxicity in zebrafish.
After fertilization, zebrafish embryos are collected and placed into individual wells of a 96-well plate in embryo water with or without the biological nanoparticles to test, such as milk derived nanovesicles (MNVs). The water is replaced every 24 hours, and the embryos monitored for up to 6 days. Hatching is monitored starting from 48 hours post fertilization (hpf) until all embryos have hatched. The zebrafish are examined daily, and any mortality noted. Survivors are examined daily for visible malformations using a checklist.
Details of zebrafish housing and husbandry will typically be available within zebrafish facilities or laboratories conducting zebrafish studies, and have been described elsewhere (Harper & Lawrence, 2011; Nusslein-Volhard & Dahm, 2002). All zebrafish studies should be performed under an approved institutional animal care and use protocol. Zebrafish (Danio rerio) should be maintained in a light/dark cycle of 14:10 hours at 28 ± 1°C in an automatic circulating water system. Zebrafish are fed twice a day with live brine shrimp (Artemia nauplii) and once a day with dry flakes . Collecting viable embryos and sorting them into proper vessels should be done in a delicate manner. This can be done using a pipet pump with a glass Pasteur pipette attached. Incubation at 28 ± 1°C is critical because temperature fluctuations can affect development and, thereby, obscure results.
Materials
Reagents
Male and female Danio rerio AB strain (ZIRC, cat no. ZL1)
Brine shrimp cysts (for hatching) (Reed mariculture). These are hatched as needed and fed as a live food source.
Dry food flakes (Ziegler larval diet) (Pentair, cat no. LD50-AQ)
E3 Embryo water (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4, and 10−5 w/v % Methylene Blue)
Freshly prepared MNVs (or particle of interest)
Each zebrafish embryo will be treated with 2 × 108 particles
Tricaine 0.015% v/v in embryo water (Pentair, cat no. MS-222)
Hardware and Instruments
96 well plate (ThermoFisher, cat no. 268200)
Inverted microscope with 4X objective (Olympus CKX53)
Glass Pasteur pipets (Fisher, 1367820A)
Pipet Pump (Bel-Art, 13683C)
Petri dishes (100 mm x 15 mm)
Protocol Steps
Embryo Collection
-
1
Cross the wild-type zebrafish by placing pairs or groups of fish into the crossing tank pre-filled with adequate amount of water.
-
2
In a Petri dish, collect embryos 30–60 minutes after fertilization and rinse in E3 embryo water.
-
3
Incubate the embryos at 28 ± 1°C for 24hrs.
-
4
Discard non-viable or unfertilized embryos and transfer the viable embryos onto a fresh Petri dish containing fresh E3 medium.
-
5
Using a glass Pasteur pipette and a manual pipet aid, transfer each fertilized embryo into a single well of a 96-well plate containing 100 μl of embryo water.
Treatment with MNVs
-
6Per each treatment well, prepare a mix of 2 × 108 MNVs in 100 μl of embryo water.For survival studies, use up to 24 embryos per group to determine the survival rate.
-
7
Replace the entire 100 μl of embryo water (Step 5) with the 100 μl MNV mix (Step 6) at 24 hours post-fertilization (hpf). Maintain 10 embryos (i.e. 10 wells) in embryo water only. These will serve as controls.
-
8
Incubate the plate at 28 ± 1°C.
-
9
Replace the media in the treatment wells with freshly prepared MNV mix (Step 6) every 24 hours for continuous exposure for 4–5 days. For control wells, replace the embryo water every 24 hours until the study endpoint.
At this point, users can proceed to the downstream analyses that may be of interest to them based on the needs of their study. We have outlined below some examples of some common types of analyses typically used.
Lethality
-
10
Determine the number of fish that remain viable at pre-determined timepoints (e.g. at 24, 48, 72 and 96 hpf). Inspect the MNV-treated and control zebrafish under a microscope and monitor the heartbeat to determine viability.
Developmental toxicity
-
11For development toxicity, determine the hatching rate and examine for malformations. Malformations are determined by examining the gross morphology of the embryos at 24 hpf and following a checklist (Table 3). As zebrafish embryos hatch between 48–72 hpf, begin assessing embryos at 45 hpf. Using a microscope at low magnification, determine when zebrafish are released out of the chorion at different time points (e.g. at 45 hpf, 48 hpf, 51 hpf, and 72 hpf).Larva may have increased movements after 72 hpf and may require the use of anesthesia to allow for detailed morphological assessments. This can be accomplished by using a 0.015% Tricaine solution.
Table 3:
Checklist for developmental malformations
| System | Morphological features | Count |
|---|---|---|
| Body growth | Eel-like body Emaciated body Stunted growth |
|
| Cardiac function and morphology | Acardia Bradycardia Tachycardia Cardiac Enlargement Tubular heart Pericardial edema |
|
| Brain and facial structure | Brachycephaly Dolichocephaly/beak face Microcephaly |
|
| Fin | Irregular fin(s) Missing fin(s) Stunted fin(s) |
|
| Ocular | Microphthalmia Ocular edema |
|
| Body shape | Kink in tail Lordosis Scoliosis |
|
| Yolk sac | Yolk sac edema | |
| Other | ||
Cardiotoxicity
-
12
At predetermined intervals after 48 hpf, assess cardiac toxicity by observing the cardiac chamber size and appearance, or the presence of pericardial edema, and assess cardiac function by measuring the heart rate using a microscope and counting the heart rate over 30 seconds.
COMMENTARY
Background information
The use of biological nanoparticles such as MNVs or others such as EVs derived from mesenchymal stem cells have garnered attention due to their intrinsic therapeutic properties as well as their potential to serve as drug delivery vehicles (Ishiguro et al., 2020). These nanotherapeutics offer several advantages over traditional drug therapies. For instance, the use of these biological nanoparticles as delivery vehicles could enhance the bioavailability and biocompatibility of their protein or nucleic acid-based therapeutic cargo by reducing their degradation (Lamichhane & Jay, 2018). Further, they could be modified to ensure targeted delivery of the cargo to a desired location, and thereby reduce off-target adverse effects.
In this article, a protocol for the isolation of MNVs using an ultracentrifugation-based method with a casein removal step is described. The casein micelles exhibit diameters similar in size to MNVs and, therefore, must be removed to prevent contamination of the final MNV product (Bijl, de Vries, van Valenberg, Huppertz, & van Hooijdonk, 2014). Given that casein is the major protein component in bovine milk, its removal is necessary to isolate pure MNVs, free of any protein contaminants (Davoodi et al., 2016). This method can be used to process a large volume of starting material in a cost-efficient manner. The resulting MNVs can be further modified through surface modifications or modulation of contents as desired to enhance their therapeutic potential (Ishiguro et al., 2020; Matsuda, Ishiguro, Yan, & Patel, 2019). Detailed protocols on potential MNV surface modifications or cargo manipulations can be found elsewhere (S. P. Li, Lin, Jiang, & Yu, 2018; O’Brien, Breyne, Ughetto, Laurent, & Breakefield, 2020; Yokoi et al., 2019). Other methods such as density-gradient ultracentrifugation, ultrafiltration, size-exclusion chromatography, and immunoaffinity can enable isolation of specific subtypes of biological nanoparticles of higher purity or from cellular sources, that may potentially be used to deliver therapeutic cargo.
It should be noted that the potential toxicity of biological nanoparticle- or EV-based therapeutics would vary with their sources, cargo, surface modifications, and methods of isolation, thus warranting individual assessment of every formulation. Here, we also describe two complimentary vertebrate models to evaluate in vivo toxicity of nanoparticles, using MNVs as an example. A mouse model is used to assess acute and sub-acute toxicity as well as immunological effects, whereas a zebrafish model is used to screen for developmental toxicity induced by MNV exposure. The mouse model is used to elucidate the effects of MNV treatment on adult tissues and to assess its impact on the immune system. Mouse models are ubiquitously used as a model organism for drug screening in pre-clinical studies. A mouse model is suitable for systemic administration of MNV and permits histological assessment of tissue injury concomitantly in solid organs such as liver, spleen, heart, kidneys, and lungs. In addition, blood samples can be obtained for biochemical analysis of markers of tissue response or injury. Previous biodistribution studies have suggested that bovine milk-derived MNVs preferentially home to and accumulate in the liver and spleen (Manca et al., 2018; Munagala, Aqil, Jeyabalan, & Gupta, 2016). The biodistribution of therapeutic biological nanoparticles can be influenced by particle size and route of administration. For these reasons, a mouse model is suitable to evaluate toxicity in vivo following acute or sub-acute exposure to MNV treatment. Furthermore, since bovine MNVs can be taken up within the spleen, the use of a mouse model allows for detailed assessment of treatment-related effects on immune cells. This can be done by mass cytometry to assess the immune profiles of mice treated with unloaded or therapeutic MNVs or vehicle only. While mice possess many of the same immune cell subtypes as humans, there are marked differences in the levels of circulating immune cells between mice and humans, with the former dominated by lymphocytes and the latter, dominated by neutrophils (Mestas & Hughes, 2004). Therefore, any conclusions drawn from the immune profiling of MNV treatment in mice should be further evaluated using human cells to mimic the immune environment found in humans.
There has been a growing interest in utilizing zebrafish as model organisms for genetic and drug screening studies (Nishimura et al., 2016). Zebrafish possess many advantages over traditional model organisms for such evaluations. Indeed, in comparison to rodent models, zebrafish are small and are kept in high density aquariums that are cost-effective to maintain. The high fecundity of zebrafish makes them especially appealing for drug screening studies, as one zebrafish mating can yield 50–300 embryos. A major advantage of using zebrafish for these studies is their rapid development time. At 72 hours post-fertilization, all internal organs are developed (van Wijk, Krekels, Hankemeier, Spaink, & van der Graaf, 2016). Therefore, developmental toxicity drug screening studies can be completed in a much shorter time frame and at a significantly reduced cost in comparison to using a rodent model. These embryos are transparent in their early developmental stage and, thus, it is easy to monitor any treatment-induced developmental changes. For drug screening studies, embryos can be placed in single wells of a 96 well plate, enabling high-throughput screening while also minimizing the amount of nanotherapeutic or drug needed for testing. Despite these benefits, there are some drawbacks to consider. For example, the use of these approaches may not be ideal for drugs that are not soluble in aqueous solutions. Similarly, drugs with poor absorption are not suitable for high-throughput screens as they need to be injected to get exposure to the drug. Also, the differences in anatomy and physiology will, overall, produce fewer translatable clinical toxicity data as compared to the mammalian system. For instance, zebrafish embryos lack fully functioning immune systems in the first few weeks of life and, thus, cannot be used for treatment-induced immunological or adult tissue effects (which is why we describe Basic Protocol 2 for those interested in these endpoints). Focusing on the interrogation of systems and chemicals with the highest likelihood of translatability will make the most of this zebrafish model. Furthermore, there may be differences in the genes involved in drug metabolism or effect between human and zebrafish (Howe et al., 2013). Other drawbacks include that some procedures, such as assessment of heart rate or malformations in individual embryos, can be time consuming and laborious. Lastly, other features, including the capacity of zebrafish to regenerate some tissues (including the fin, brain, retina, spinal cord, and heart) may impact tissue responses to toxic insults.
Critical Parameters
There are two aspects of MNV isolation that warrant special attention. Casein protein represents approximately 80% of the total protein found in bovine milk and form micelles that are of similar size to MNVs and, thus, can confound analyses of MNV yield. As such, it is imperative that casein is removed (Davoodi et al., 2016). This can be performed by the addition of EDTA to the MNV prep. Since the amount of casein protein can vary from one batch to another, we suggest adding between 8 to 12 mL of EDTA to the MNV prep. Upon the addition of EDTA, the MNV prep should become clear.
Another important consideration is to soak the MNV pellet overnight in PBS, as this will increase the recovery of MNVs. After washing the MNVs, the supernatant is removed, and a small volume of PBS is added to the tube. Due to the extremely high yield of MNVs, the pellets may be compact and, therefore, difficult to disperse in solution. We recommend soaking the pellet in PBS overnight at 4°C before resuspending the MNV pellet to maximize the yield and reduce the number of MNV aggregates.
To assess toxicity profiles of MNVs in mice (Basic Protocol 2), we describe the administration of a defined number of particles. Please note that approaches to count the number of particles may not accurately provide a true count of therapeutic vesicles used, as these are based on indirect measurements. An alternative approach would be to describe doses administered in terms of protein or lipid content. The protein and lipid content can be quantified using commercially available kits (i.e. BCA protein assay kit from Thermo Fisher Pierce and Phospholipid assay from Abcam). The amount of lipid may reflect the number of nanoparticles that are delivered, whereas the amount of protein may reflect the quantity of therapeutic protein that is delivered.
To ensure systemic delivery of the MNVs, the tail veins must be properly dilated, which can be accomplished by submerging the tail in warm water or by wrapping it in a heating pad. We recommend the former, as this method uniformly warms the tail. To ensure that the desired amount of MNVs are administered, syringes that do not have any dead space such as those used for insulin administration are recommended for tail vein injections.
For the zebrafish studies, it is important to ensure that only healthy embryos are used. For the first 48 hours post-fertilization, the zebrafish embryos are protected by the chorion. Hatching can occur between 48 and 72 hpf. For toxicity studies, it is vital that dechorionation occurs, as the chorion has limited permeability and can attenuate uptake of therapeutic candidates being tested (Henn & Braunbeck, 2011). While dechorionation is one of the parameters we monitor to assess developmental toxicity, it is important to exclude any zebrafish that have not hatched by 85 hpf. It is also important to replace E3 embryo water daily to avoid toxicity. Avoid the use of tissue culture treated plates for hatched embryos to minimize the effects of embryos adhering to the surfaces of the plates. If the protocols are being used for nanoparticles other than MNVs, the solubility of the solvent used for these should be first tested in embryo water.
Troubleshooting
MNV isolation issues
The protocol for the isolation of MNVs will typically yield up to 1 × 1013 particles/ml from fat-free milk. If the yield of isolated MNV is unexpectedly low, consider increasing the volume of starting material and ensure that fat-free milk is used as a source. If toxicity is observed during any in vitro studies, consider removing endotoxins prior to use for in vivo studies such as those in BP2/BP3. This could be done using an endotoxin removal kit (Pierce High Capacity Endotoxin Removal Resin; Thermo Fisher Scientific, Waltham, MA).
Biochemical and histopathological analyses issues
One of the most common issues encountered with biochemical analysis comes from having a serum volume that is too low. If the whole blood is collected several minutes after euthanasia, the blood may be coagulated, thereby limiting the amount of serum that can be isolated. To avoid this issue, one should perform cardiac puncture immediately after euthanasia.
For a detailed review of common H&E staining issues and tips for addressing them, refer to the National Society for Histotechnology guidelines for H&E staining (NSH).
Immune profiling issues
One common issue that may be encountered with mass cytometry is the presence of a large amount of debris in the splenocyte sample that can clog the mass cytometer and prevent the acquisition of data. This can arise from poor splenocyte isolation technique. To minimize the amount of debris, first inspect the excised spleen to ensure that all other tissues have been removed. After placing the spleen over a cell strainer, keep the spleen moist and gently macerate the tissue with the plastic end of a syringe plunger. If the spleen is macerated too harshly, debris can be introduced into your single cell suspension. Another common issue is barium contamination, which can be minimized by using non-autoclaved tubes for sample preparation and by working in a hood to minimize exposure of the samples to barium in the air. If a sample is contaminated with barium, a high signal will be detected in the Ba138 channel. Other issues include over- or undersaturation of the cisplatin and iridium staining. The concentrations recommended herein have been titrated according to our splenocyte sample; however, the user may need to perform a titration experiment to optimize the concentrations of cisplatin and iridium used for staining. The range of recommended concentrations used for titration are provided in the cisplatin and iridium data sheets published by Fluidigm and available from their website by searching for the appropriate catalog numbers provided in the reagents section of Basic Protocol 2.
There are several potential challenges with using zebrafish for screening studies. One issue is that some water-insoluble molecules cannot be administered to zebrafish because the amount of carrier solvents (such as ethanol or DMSO) required for their solubility is toxic. Moreover, toxicity and death can result from prolonged incubation of the zebrafish without any medium change. Furthermore, visualization may be difficult in fully developed zebrafish larvae (72 hpf) at higher magnifications and hamper the in vivo assessment of morphological, physiological, or genetic events on the cell- or tissue-specific level. Likewise, skin pigmentation can develop in response to the treatment and could hinder visualization of embryonic development.
Understanding results
The MNVs isolated by sequential ultracentrifugation exhibit a heterogenous size distribution, with an average particle diameter of 200 nm (Figure 2). These MNVs express several hallmark markers similar to those found on extracellular vesicles from cellular sources, such as TSG101, and tetraspannins CD81, CD63, and CD9, which can be assessed by western blotting or mass spectrometry (Kowal, Ter-Ovanesyan, Regev, & Church, 2017; Lötvall et al., 2014; Matsuda et al., 2020).
Systemic administration of 1 × 1010 MNVs without any modifications or cargo loading prepared as in Basic Protocol 1 does not elicit acute or sub-acute toxicity in mice. There should be no significant differences in the levels of the blood biochemical markers between MNV-treated mice and vehicle controls. Similarly, there should be no evidence of histological injury (i.e. fibrosis, necrosis, inflammation, or hemorrhages) (Figure 4). MNV treatment alters the immune profile in the spleen, resulting in an increase in the population of CD19+B220− B cells as well as a decrease in CD4+ naïve T cells.
Treatment with 2 × 108 MNV does not induce developmental toxicity in zebrafish, although they do exhibit accelerated hatching rates, compared to controls. Treatment with this dose of MNVs does not induce the formation of developmental abnormalities, nor does it affect mortality. Treatment with higher doses of MNVs, however, have been shown to result in developmental abnormalities, most commonly pericardial and yolk sac edema, that can contribute to mortality (Figure 7).
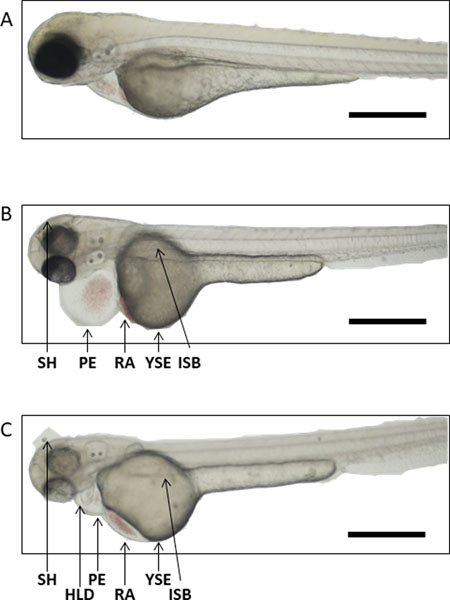
Figure 7. Developmental abnormalities in zebrafish.
(A) Representative image of a healthy zebrafish. Zebrafish can present with a host of different morphological abnormalities (B-C), including pericardial edema (PE), yolk sac edema (YSE), small head (SH), heart looping defect (HLD), red blood cell accumulation (RA), irregular swim bladder (ISB). Scale bars represent 500 μm.
If Basic Protocols 2 or 3 are used for other types of biological nanoparticles, they can be modified or expanded based upon the specific modes of administration and intended applications. For example, more detailed assessment of pulmonary, gastrointestinal, or neurological effects should be considered with the use of inhaled, oral, or intranasally administered nanotherapeutics, respectively. Similarly, specific characteristics such as size, tendency for aggregation of nanoparticles, as well as exposure dose may also influence their toxicity. Spontaneous occurrence of developmental changes can occur. Thus, a knowledge of the underlying background incidence of developmental abnormalities, and the use of appropriate concurrently used controls, is necessary. Developmental toxicity can be determined by comparing the concentrations of MNVs that cause toxicity (e.g. no observed adverse effect level or EC50) compared with those that cause lethality (e.g. as LC50).
Time considerations
The procedure of isolation of MNVs (Basic Protocol 1) can be completed in 1 day. Basic Protocol 2 can take up to 2 weeks depending on the goals and time points selected. In Basic Protocol 3, fertilization can take place within 48 hours, and the zebrafish studies can be completed within a week of fertilization.
Acknowledgments
This work was supported by the National Institutes of Health (NIH R01 CA217833). The contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Footnotes
Conflict of Interest Statement
The authors declare no conflicts of interest
Data Availability Statement
Data sharing is not applicable to this article as no new data resources were created or analyzed in this study
Literature Cited
- Bijl E, de Vries R, van Valenberg H, Huppertz T, & van Hooijdonk T (2014). Factors influencing casein micelle size in milk of individual cows: Genetic variants and glycosylation of κ-casein. International Dairy Journal, 34(1), 135–141. doi: 10.1016/j.idairyj.2013.08.001 [DOI] [Google Scholar]
- Cohen O, Betzer O, Elmaliach-Pnini N, Motiei M, Sadan T, Cohen-Berkman M, . . . Popovtzer R (2021). ‘Golden’ exosomes as delivery vehicles to target tumors and overcome intratumoral barriers: in vivo tracking in a model for head and neck cancer. Biomater Sci, 9(6), 2103–2114. doi: 10.1039/d0bm01735c [DOI] [PubMed] [Google Scholar]
- Davoodi SH, Shahbazi R, Esmaeili S, Sohrabvandi S, Mortazavian A, Jazayeri S, & Taslimi A (2016). Health-Related Aspects of Milk Proteins. Iran J Pharm Res, 15(3), 573–591. [PMC free article] [PubMed] [Google Scholar]
- Donovan J, & Brown P (2006). Euthanasia. Current Protocols in Immunology, 73(1), 1.8.1–1.8.4. doi: 10.1002/0471142735.im0108s73 [DOI] [PubMed] [Google Scholar]
- Gardiner C, Di Vizio D, Sahoo S, Théry C, Witwer KW, Wauben M, & Hill AF (2016). Techniques used for the isolation and characterization of extracellular vesicles: results of a worldwide survey. J Extracell Vesicles, 5, 32945. doi: 10.3402/jev.v5.32945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Jones TW, Dutta S, Zhu Y, Wang X, Narayanan SP, . . . Zhang D (2021). Overview and Update on Methods for Cargo Loading into Extracellular Vesicles. Processes (Basel), 9(2). doi: 10.3390/pr9020356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper C, & Lawrence C (2011). The Laboratory Zebrafish: CRC Press. [Google Scholar]
- Hartjes TA, Mytnyk S, Jenster GW, van Steijn V, & van Royen ME (2019). Extracellular Vesicle Quantification and Characterization: Common Methods and Emerging Approaches. Bioengineering (Basel), 6(1). doi: 10.3390/bioengineering6010007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn K, & Braunbeck T (2011). Dechorionation as a tool to improve the fish embryo toxicity test (FET) with the zebrafish (Danio rerio). Comp Biochem Physiol C Toxicol Pharmacol, 153(1), 91–98. doi: 10.1016/j.cbpc.2010.09.003 [DOI] [PubMed] [Google Scholar]
- Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, Muffato M, . . . Stemple DL (2013). The zebrafish reference genome sequence and its relationship to the human genome. Nature, 496(7446), 498–503. doi: 10.1038/nature12111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro K, Yan IK, Lewis-Tuffin L, & Patel T (2020). Targeting Liver Cancer Stem Cells Using Engineered Biological Nanoparticles for the Treatment of Hepatocellular Cancer. Hepatol Commun, 4(2), 298–313. doi: 10.1002/hep4.1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimball AK, Oko LM, Bullock BL, Nemenoff RA, van Dyk LF, & Clambey ET (2018). A Beginner’s Guide to Analyzing and Visualizing Mass Cytometry Data. J Immunol, 200(1), 3–22. doi: 10.4049/jimmunol.1701494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowal EJK, Ter-Ovanesyan D, Regev A, & Church GM (2017). Extracellular Vesicle Isolation and Analysis by Western Blotting. Methods Mol Biol, 1660, 143–152. doi: 10.1007/978-1-4939-7253-1_12 [DOI] [PubMed] [Google Scholar]
- Lamichhane TN, & Jay SM (2018). Production of Extracellular Vesicles Loaded with Therapeutic Cargo. Methods Mol Biol, 1831, 37–47. doi: 10.1007/978-1-4939-8661-3_4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A, Zhao Y, Li Y, Jiang L, Gu Y, & Liu J (2021). Cell-derived biomimetic nanocarriers for targeted cancer therapy: cell membranes and extracellular vesicles. Drug Deliv, 28(1), 1237–1255. doi: 10.1080/10717544.2021.1938757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SP, Lin ZX, Jiang XY, & Yu XY (2018). Exosomal cargo-loading and synthetic exosome-mimics as potential therapeutic tools. Acta Pharmacol Sin, 39(4), 542–551. doi: 10.1038/aps.2017.178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liangsupree T, Multia E, & Riekkola ML (2021). Modern isolation and separation techniques for extracellular vesicles. J Chromatogr A, 1636, 461773. doi: 10.1016/j.chroma.2020.461773 [DOI] [PubMed] [Google Scholar]
- Lötvall J, Hill AF, Hochberg F, Buzás EI, Di Vizio D, Gardiner C, . . . Théry C (2014). Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J Extracell Vesicles, 3, 26913. doi: 10.3402/jev.v3.26913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maji S, Yan IK, Parasramka M, Mohankumar S, Matsuda A, & Patel T (2017). In vitro toxicology studies of extracellular vesicles. J Appl Toxicol, 37(3), 310–318. doi: 10.1002/jat.3362 [DOI] [PubMed] [Google Scholar]
- Manca S, Upadhyaya B, Mutai E, Desaulniers AT, Cederberg RA, White BR, & Zempleni J (2018). Milk exosomes are bioavailable and distinct microRNA cargos have unique tissue distribution patterns. Sci Rep, 8(1), 11321. doi: 10.1038/s41598-018-29780-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda A, Ishiguro K, Yan IK, & Patel T (2019). Extracellular Vesicle-Based Therapeutic Targeting of β-Catenin to Modulate Anticancer Immune Responses in Hepatocellular Cancer. Hepatol Commun, 3(4), 525–541. doi: 10.1002/hep4.1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda A, Moirangthem A, Angom RS, Ishiguro K, Driscoll J, Yan IK, . . . Patel T (2020). Safety of bovine milk derived extracellular vesicles used for delivery of RNA therapeutics in zebrafish and mice. J Appl Toxicol, 40(5), 706–718. doi: 10.1002/jat.3938 [DOI] [PubMed] [Google Scholar]
- Mestas J, & Hughes CC (2004). Of mice and not men: differences between mouse and human immunology. J Immunol, 172(5), 2731–2738. doi: 10.4049/jimmunol.172.5.2731 [DOI] [PubMed] [Google Scholar]
- Munagala R, Aqil F, Jeyabalan J, & Gupta RC (2016). Bovine milk-derived exosomes for drug delivery. Cancer Lett, 371(1), 48–61. doi: 10.1016/j.canlet.2015.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthu S, Bapat A, Jain R, Jeyaraman N, & Jeyaraman M (2021). Exosomal therapy-a new frontier in regenerative medicine. Stem Cell Investig, 8, 7. doi: 10.21037/sci-2020-037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura Y, Inoue A, Sasagawa S, Koiwa J, Kawaguchi K, Kawase R, . . . Tanaka T (2016). Using zebrafish in systems toxicology for developmental toxicity testing. Congenit Anom (Kyoto), 56(1), 18–27. doi: 10.1111/cga.12142 [DOI] [PubMed] [Google Scholar]
- NSH. Guidelines for Hematoxylin & Eosin Staining. Retrieved from http://nsh.org/sites/default/files/Guidelines_For_Hematoxylin_and_Eosin_Staining.pdf
- Nusslein-Volhard C, & Dahm R (2002). Zebrafish: a practical approach: New York: Oxford University Press. [Google Scholar]
- O’Brien K, Breyne K, Ughetto S, Laurent LC, & Breakefield XO (2020). RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat Rev Mol Cell Biol, 21(10), 585–606. doi: 10.1038/s41580-020-0251-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palit S, Heuser C, de Almeida GP, Theis FJ, & Zielinski CE (2019). Meeting the Challenges of High-Dimensional Single-Cell Data Analysis in Immunology. Front Immunol, 10, 1515. doi: 10.3389/fimmu.2019.01515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayamajhi S, & Aryal S (2020). Surface functionalization strategies of extracellular vesicles. Journal of Materials Chemistry B, 8(21), 4552–4569. [DOI] [PubMed] [Google Scholar]
- Royo F, Théry C, Falcón-Pérez JM, Nieuwland R, & Witwer KW (2020). Methods for Separation and Characterization of Extracellular Vesicles: Results of a Worldwide Survey Performed by the ISEV Rigor and Standardization Subcommittee. Cells, 9(9). doi: 10.3390/cells9091955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanwlani R, Fonseka P, Chitti SV, & Mathivanan S (2020). Milk-Derived Extracellular Vesicles in Inter-Organism, Cross-Species Communication and Drug Delivery. Proteomes, 8(2). doi: 10.3390/proteomes8020011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomiyama E, Matsuzaki K, Fujita K, Shiromizu T, Narumi R, Jingushi K, . . . Nonomura N (2021). Proteomic analysis of urinary and tissue-exudative extracellular vesicles to discover novel bladder cancer biomarkers. Cancer Sci. doi: 10.1111/cas.14881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijk RC, Krekels EHJ, Hankemeier T, Spaink HP, & van der Graaf PH (2016). Systems pharmacology of hepatic metabolism in zebrafish larvae. Drug disovery today: disease models, 22, 27–34. [Google Scholar]
- Yokoi A, Villar-Prados A, Oliphint PA, Zhang J, Song X, De Hoff P, . . . Sood AK (2019). Mechanisms of nuclear content loading to exosomes. Sci Adv, 5(11), eaax8849. doi: 10.1126/sciadv.aax8849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LL, Xiong YY, & Yang YJ (2021). The vital roles of mesenchymal stem cells and the derived extracellular vesicles in promoting angiogenesis after acute myocardial infarction. Stem Cells Dev. doi: 10.1089/scd.2021.0006 [DOI] [PubMed] [Google Scholar]
- Zonneveld MI, Brisson AR, van Herwijnen MJ, Tan S, van de Lest CH, Redegeld FA, . . . Nolte-’t Hoen EN (2014). Recovery of extracellular vesicles from human breast milk is influenced by sample collection and vesicle isolation procedures. J Extracell Vesicles, 3. doi: 10.3402/jev.v3.24215 [DOI] [PMC free article] [PubMed] [Google Scholar]