Abstract
We have investigated the in vivo and in vitro regulation of the human urokinase-type plasminogen activator (uPA) gene by interleukin-1 (IL-1) and analyzed the transcription factors and signalling pathways involved in the response of the −2.0-kb uPA enhancer to IL-1 induction and to tetradecanoyl phorbol acetate (TPA) induction. Mutational analysis showed the cooperative activity of the Ets-binding site (EBS) and the two AP-1 elements of the enhancer. The results reveal that the EBS is required for the response to both inducers mediated by Ets-2, which is regulated at a level subsequent to DNA binding, by an IL-1- and phorbol ester-inducible transactivation domain. Both the IL-1 and the TPA-mediated induction result in a drastic increase of AP-1 binding to the downstream site of the enhancer (uPA 3′ TPA-responsive element), while a mostly qualitative change, resulting from the interplay between ATF-2 homodimers and c-Jun–ATF-2 heterodimers, takes place at the upstream AP-1 element. The analysis of two distinct mitogen-activated protein kinase pathways shows that stress-activated protein kinase–Jun N-terminal kinase activation, resulting in the phosphorylation of ATF-2, c-Jun, and JunD, is required not only for the IL-1- but also for the TPA-dependent induction, while the extracellular signal-related kinase 1 (ERK-1) and ERK-2 activation is involved in the TPA- but not in the IL-1-dependent stimulation of the uPA enhancer.
The urokinase-type plasminogen activator (uPA) is a secreted serine protease involved in many biological processes requiring extracellular matrix degradation and cell migration, such as wound healing, mammary gland involution, macrophage migration, and tumor metastasis. The uPA activity, which results in the proteolytic cleavage of plasminogen to plasmin, is finely controlled at multiple levels. Urokinase can be rapidly inactivated by binding to specific plasminogen activator inhibitors (PAI-1 and PAI-2); in addition, the cell surface localization of the uPA proteolytic activity and the urokinase internalization are controlled by the membrane-bound uPA receptor (reviewed in references 1, 9, and 10).
The transcriptional control of the urokinase gene expression has been characterized in many experimental systems. The uPA gene transcription is modulated by a variety of signals, including cyclic AMP and polypeptide hormones (calcitonin), growth factors (epidermal growth factor, fibroblast growth factor 2 [FGF-2], and hepatocyte growth factor [HGF]), tumor promoters, several oncogene products, cytoskeletal reorganization, retinoic acid, glucocorticoids, etc. (reviewed in reference 7). Recently, the signalling pathways involved in uPA gene induction by different agents have been dissected in various experimental systems. The roles of individual components of the Ras/extracellular signal-regulated kinase (ERK) signalling pathway have been established for uPA gene induction by FGF-2 (5), cytoskeletal reorganization (28), and different transforming oncogenes, such as the polyomavirus middle-T antigen (6), and the activated c-Ha-ras (36) and the v-mos (35) oncoproteins.
We and others have shown that the growth factor- and phorbol ester-dependent transcriptional regulation of the human uPA gene is mediated by a complex enhancer element spanning a 120-bp region, localized 2 kb upstream of the transcription start site (44, 45, 50, 54).
The uPA enhancer activity results from the functional cooperation between an upstream inducible element (uPA 5′ tetradecanoyl phorbol acetate (TPA)-responsive element [TRE]) formed by an Ets-binding site (EBS) and a c-Jun–ATF-2 site (uPA 5′ AP-1) and a downstream AP-1 binding site (uPA 3′ TRE) (18, 44). The cooperation between the two inducible elements is mediated by a 74-bp protein-binding domain, the cooperation mediator (COM) element (44), localized between the two AP-1 sites and interacting with four distinct nuclear proteins (urokinase enhancer factors 1 to 4) (4, 16, 17). While the uPA 5′ TRE and 3′ TRE are able to function autonomously as TREs, the isolated COM region does not exhibit any transcriptional stimulatory activity but rather appears to play an architectural role for the uPA enhancer function. The tight interdependence between the adjacent EBS and the c-Jun–ATF-2 site, which are unable to function as independent inducible elements, confirms the general importance of the cooperation between the Ets members and the AP-1 factor, well documented by the analysis of several oncogene-responsive promoters (11, 26, 27, 38). The role of Ets–AP-1 cooperation in uPA gene induction has been further substantiated by the characterization of a second Ets–AP-1 element, localized further upstream (−6.9 kb in mouse and −5.3 kb in human) and cooperating with the downstream Ets–AP-1 element, in the response to TPA and FGF-2 induction (20).
Because of the various transduction pathways modulated by different agents and the multiple transcription factors interacting with the uPA regulatory region, studying the uPA gene regulation makes it possible to address the potential cross talk between different signalling pathways and the individual roles of distinct transcription factors as targets of growth-, transformation-, or stress-dependent regulatory cascades.
Interleukin-1 (IL-1) plays an essential function as an inflammation mediator responsible for complex local and systemic reactions in many different cell types. The stimulatory effect of IL-1 on uPA biosynthesis has been characterized in several cell systems, including normal human chondrocytes (13), pulmonary epithelial cell lines (41), and primary cultures of human hepatocytes (12). The effect observed in the hepatocytes is important because of the major role played by IL-1 and other inflammatory cytokines, such as IL-6 and tumor necrosis factor alpha, in the induction of the acute-phase response (3).
In this work, we have characterized the in vivo induction of the urokinase mRNA by IL-1α, and dissected the in vitro IL-1- and TPA-dependent regulation of the uPA enhancer, in the HepG2 hepatoma cell line. We have determined the activities, compositional changes, and phosphorylation states of several AP-1 components (c-Jun, JunD, and ATF-2) and shown the essential role played by Ets-2, likely mediated by its phosphorylation, as a target for each of the two uPA gene inducers. Finally, the comparative analysis of the effects of IL-1 and TPA allowed us to define the level of cross talk and the essential role played by the Jun N-terminal kinase (JNK) activation in response to both signalling agonists.
MATERIALS AND METHODS
Materials.
Human recombinant IL-1α and IL-1β were obtained from Boehringer Mannheim. TPA and anisomycin were obtained from Sigma. Cycloheximide was from Calbiochem; [γ-32P]ATP, [α-32P]dCTP, [α-32P]GTP, and ECL (enhanced chemiluminescence) reagent were obtained from Amersham. Antibodies were purchased from Santa Cruz Biotechnology.
Cell culture and transfection analysis.
HepG2 cells were grown in Dulbecco’s modified Eagle medium (DMEM; Life Technologies, Inc.) containing 5% fetal calf serum, 0.2 mg of streptomycin per ml, and 50 U of penicillin per ml. Approximately 1 × 106 to 2 × 106 cells were electroporated with the indicated amounts of the reporter constructs and expression vectors, in 0.5 ml of complete medium, at 250 V and 960 μF, with a Gene Pulser apparatus (Bio-Rad). Cells were plated in DMEM containing 5% fetal calf serum and allowed to adhere overnight. Medium was changed to DMEM containing 0.5% fetal calf serum and effectors as indicated (IL-1α [1 ng/ml] or TPA [100 ng/ml]), for 24 h prior to harvest. Chloramphenicol acetyltransferase (CAT) activity of cells extracts was assayed as previously described (18).
Probes and plasmids.
The −2345 uPA promoter-CAT fusion, the constructs containing site-specific mutations (mEBS, m5′ AP-1, m3′ AP-1, and mCOM), and the uPAenh-tkCAT construct, containing the uPA enhancer upstream of the heterologous thymidine kinase promoter, have been described previously (18, 44). The vector expressing the Ets-2 DNA-binding domain–LacZ fusion protein and neomycin resistance (pAPr-etsZ-neo) (34) and the reporter construct containing a palindromic high-affinity Ets-2 site upstream of the c-fos minimal promoter (E18-luc) (23) were obtained from M. C. Ostrowski, Ohio State University. The luciferase reporter construct containing five copies of the GAL4 binding site upstream of the c-fos minimal promoter (FR-luc) was obtained from Stratagene. The vector expressing the Ets-2 protein fused to the heterologous GAL4(1–147) DNA-binding domain (Gal4-Ets-2) (55) was obtained from B. Wasylyk, IGBMC, Strasbourg, France. The E-c-Jun (amino acid 1 to 253) and GAL4–ATF-2 (amino acid 1 to 109) fusion constructs were obtained from D. Bohmann (European Molecular Biology Laboratory, Heidelberg, Germany) and M. Green (University of Massachusetts), respectively. The vector expressing the catalytically inactive dominant-negative c-Jun N-terminal kinase (pCMV-JNK-APF) (19) was provided by R. J. Davis, University of Massachusetts. The vectors expressing the catalytically inactive derivatives of ERK-1 (pcDNA-ERK-1-T192A) (48) and ERK-2 (pCMV5-ERK-2-K→R) (33) were kindly provided by J. Pouysségur (Centre Nationale de la Recherche Scientifique, Nice, France) and P. Shaw (Max-Planck-Institut, Freiburg, Germany), respectively.
RNA extraction, RNase protection, and Northern analysis.
Total RNA was extracted by the guanidine thiocyanate method (15). RNA preparations were treated with RNase-free DNase I (Promega) and purified by phenol extraction and ethanol precipitation. RNA probes were uniformly labelled with [α-32P]GTP, with an in vitro transcription kit (Promega). RNA samples were analyzed by quantitative RNase protection (51). Fifty micrograms of total cellular RNA was incubated with 105 cpm of the indicated ribonucleotide probes at optimal annealing temperatures (54°C for uPA, 45°C for c-Jun, and 48°C for ATF-2) for 14 to 16 h. Following RNase treatment, protected fragments were separated on a denaturing 6% acrylamide gel. Northern blot hybridization was carried out as described previously (53). The murine uPA cDNA probe was labelled with [α-32P]dCTP with the random oligonucleotide primer (Ready-To-Go; Pharmacia) to a specific activity of about 108 cpm/μg.
Nuclear extracts and EMSA.
Nuclear extract preparation and electrophoretic mobility shift assays (EMSAs) were performed as described elsewhere (53). The oligonucleotides used were as follows: PEA3/ets, 5′-TTTGTCCAGGAGGAAAATGATCCG-3′; uPA EBS, 5′-TTTGTCCAGGAGGAAATGATCCG-3′; uPA mEBS, 5′-TTTGTCCAGGACCAAATGATCCG-3′; Py EBS, 5′-TCGAGCAGGAAGTTCGA-3′; uPA 5′ TRE mEBS, 5′-GATCGGAGGAAGTGAAGTCATCTGC-3′; uPA 5′ TRE, 5′-GATCGGACCAAATGAAGTCATCTGC-3′; and coll TRE, 5′-CGCTTGATGAGTCAGCCGGAA-3′.
For the antibody supershift analysis, the reactions were performed by preincubating nuclear extracts with 0.5 μg of antibody at 4°C for 3 h. After addition of the labelled oligonucleotide and 15 min of incubation at room temperature, the products were resolved by 8% polyacrylamide gel electrophoresis (PAGE). The anti-c-Jun/AP-1 and anti-ATF-2 antibodies were purchased from Santa Cruz Biotechnology, Inc.
Immunoblotting analysis.
Nuclear extracts were separated by sodium dodecyl sulfate (SDS)–9% PAGE as previously described (53). For mitogen-activated protein (MAP) kinase analysis, total extracts were prepared and resolved by SDS–10% PAGE, according to the method described by Besser et al. (5). Proteins were transferred to Immobilon-P membranes (Millipore), which were subsequently blocked with 5% nonfat milk proteins and incubated with the indicated antibodies at a concentration of 0.2 μg/ml. All antibodies were from Santa Cruz Biotechnology, except for the anti-phospho-c-Jun (Ser-73) and phospho-ATF-2 (Thr-71), which were purchased from New England Biolabs. Bound antibodies were detected by the appropriate horseradish peroxidase-conjugated secondary antibodies followed by enhanced chemiluminescence (ECL; Amersham).
RESULTS
In vitro and in vivo induction of uPA mRNA by IL-1: differential regulation by IL-1 and TPA.
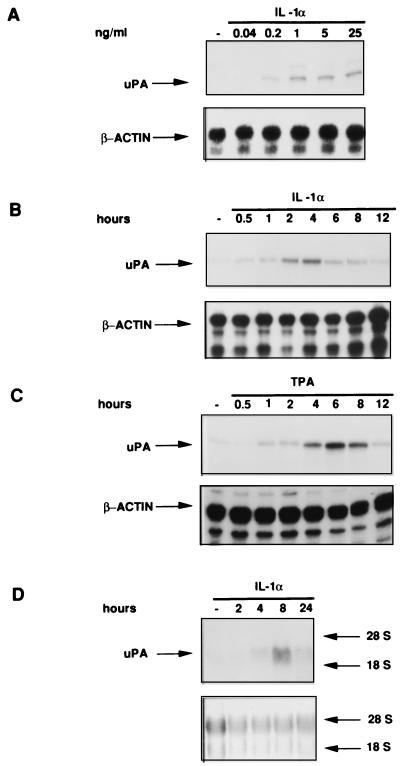
We have previously characterized the regulatory factors required for the induction of the uPA gene by phorbol esters, in the HepG2 cell line. To define the transduction pathways involved in responses to different inducers, we have now analyzed the mechanism of induction of the uPA gene by the inflammatory cytokine IL-1. The IL-1α treatment of HepG2 cells resulted in a dose-dependent increase of the uPA transcript (Fig. 1A), with a peak of mRNA accumulation between 2 and 4 h after the beginning of treatment (Fig. 1B). A similar result was obtained by treatment of the cells with IL-1β (data not shown). The phorbol ester-dependent induction exhibited a different kinetics, resulting in a rather sustained uPA mRNA accumulation with a peak at approximately 6 h (Fig. 1C).
FIG. 1.
In vitro and in vivo regulation of uPA mRNA by IL-1 and TPA. (A) Dose dependence of IL-1 induction. HepG2 cells were treated with the indicated concentrations of IL-1α for 8 h. Total RNA was extracted and quantitatively analyzed (30 μg/sample) by RNase protection with the two indicated ribonucleotide probes. The human β-actin ribonucleotide probe was utilized as a control for RNA loading. (B and C) Time course of the IL-1 and TPA induction. HepG2 cells were stimulated for the indicated times with IL-1α (1 ng/ml) (B) or TPA (100 ng/ml) (C), and total RNA was analyzed by RNase protection. (D) In vivo uPA mRNA regulation by IL-1 in rat liver. Rats were treated with IL-1α for the indicated times. Total RNA was extracted from rat livers and subjected to Northern blot analysis by hybridization to the radiolabelled mouse uPA cDNA probe. The equal loading of total RNA was confirmed by methylene blue staining of rRNAs (bottom panel).
To understand the in vivo role of IL-1-mediated uPA gene induction, we analyzed the time course of uPA mRNA expression, in livers of IL-1-treated rats. The uPA transcript was markedly induced by the cytokine treatment, with a peak at 8 h after stimulation (Fig. 1D). Therefore, the uPA mRNA is induced by IL-1 both in vitro and in intact animals.
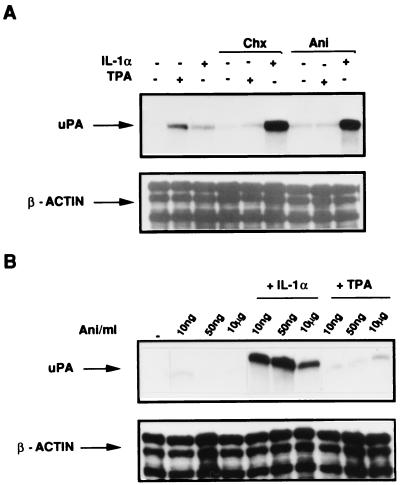
The results shown in Fig. 1 also suggest that different mechanisms underlie the effects of IL-1 and phorbol esters on uPA mRNA. To test whether de novo protein synthesis was required for the induction, we analyzed the effects of two protein synthesis inhibitors (cycloheximide and anisomycin), in combination with each of the two inducers. Both cycloheximide and anisomycin applied alone did not significantly affect the uPA mRNA level. When the two drugs were added in combination with each of the two inducers, we found that, while the TPA-mediated induction was slightly inhibited, the IL-1-mediated induction was dramatically increased (Fig. 2A). The uPA mRNA superinduction could arise from translational arrest or by an effect of the signalling activity of the two drugs. It has been shown elsewhere that it is possible to discriminate between these two different effects, since anisomycin signalling activity can be detected in the presence of drug concentrations (below 70 to 80 ng/ml) which do not inhibit protein synthesis (39). Therefore, to understand whether the observed uPA mRNA superinduction was independent of translational arrest, we tested the anisomycin dose response, in the presence of subinhibitory drug concentrations, which do not interfere with protein synthesis. IL-1-dependent superinduction was detected even at the lowest anisomycin concentration (10 ng/ml [Fig. 2B]), showing that the observed synergism was independent of the inhibition of protein synthesis.
FIG. 2.
Effect of protein synthesis inhibitors on IL-1α and TPA induction of uPA mRNA in HepG2 cells. (A) Effect of anisomycin and cycloheximide on IL-1α and TPA induction of uPA mRNA. Cells were preincubated with 10 μg of cycloheximide (Chx) per ml or 10 μg of anisomycin (Ani) per ml for 30 min before a 2-h treatment with IL-1α or TPA. Total RNA was analyzed by RNase protection as described for Fig. 1A. (B) Dose dependence of anisomycin effect. HepG2 cells were pretreated with the three indicated concentrations of anisomycin for 30 min before a 2-h incubation with IL-1α or TPA prior to harvest.
The response to both IL-1 and TPA is mediated by the cooperative activity of the Ets/AP-1, AP-1, and COM elements of the −2.0-kb uPA enhancer.
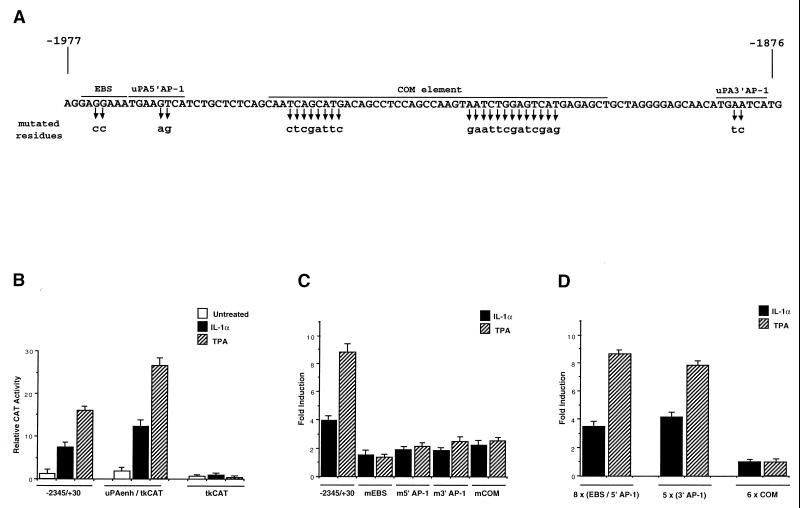
To identify the regulatory sequences involved in the IL-1-dependent induction of the uPA gene, we analyzed the previously described CAT constructs (18, 44) containing the phorbol ester-responsive enhancer, localized 2 kb upstream of the uPA transcription start site (shown in Fig. 3A). The results of transfection analysis showed that the −2.0-kb enhancer region is necessary and sufficient both for the IL-1- and for the TPA-mediated induction of the uPA promoter: we detected the same (about fivefold) induction for the −2345 to +30 uPA promoter-CAT construct and for the chimeric reporter gene containing the 120-bp enhancer (nucleotides −1977 to −1858) fused to the heterologous thymidine kinase promoter (Fig. 3B). The IL-1 induction of the CAT constructs was about one-half of the TPA induction, in agreement with the different levels of stimulation of the endogenous uPA mRNA level by the two different inducers (Fig. 1).
FIG. 3.
Identification of IL-1-responsive elements in the −2.0-kb uPA enhancer. (A) Nucleotide sequence and protein-binding domains of the −2.0-kb human uPA enhancer. The positions of the EBS, the 5′ AP-1, the COM, and the 3′ AP-1 element are overlined. The nucleotide residues replaced by the four site-specific mutations (mEBS, m5′ AP-1, mCOM, and m3′ AP-1) are indicated by the arrows. (B) IL-1-responsive activity of the uPA enhancer fused to a heterologous promoter. HepG2 cells were electroporated with 20 μg of a CAT reporter driven by the wild-type uPA promoter (−2345 to +30) or by the uPA enhancer region (−1995 to −1870) fused to the herpes simplex virus thymidine kinase minimal promoter (uPAenh/tkCAT). Transfected cells were incubated with IL-1α (1 ng/ml) or TPA for 24 h prior to harvesting, and CAT activity was assayed by thin-layer chromatography and PhosphorImager scanning. The values represent the means of four independent experiments with standard deviations indicated by bars. (C) IL-1 and TPA inducibility of uPA 5′ flanking region (−2345 to +30) derivatives containing the site-directed mutations described for panel A (mEBS, m5′ AP-1, m3′ AP-1, and mCOM). Fold induction represents the ratio of CAT enzyme activity in IL-1α- or TPA-stimulated cells to that in untreated controls. (D) IL-1 and TPA inducibility of uPA-tkCAT fusion constructs containing the indicated multimerized oligonucleotides fused to the thymidine kinase promoter. Fold induction was determined as described for panel C.
We have previously shown that the activity of the uPA enhancer depends on the cooperation between an upstream inducible element (uPA 5′ TRE) formed by an EBS and an AP-1 motif (uPA 5′ AP-1) and a downstream AP-1 binding site (uPA 3′ AP-1). The cooperation between the two inducible elements is mediated by a complex region, the COM, localized between the two AP-1 sites and formed by multiple protein-binding domains (Fig. 3A). First, we showed that the 120-bp enhancer region was sufficient to mediate the IL-1-dependent activation of the thymidine kinase heterologous promoter, with a fold induction comparable to that of the (−2345 to +30) uPA promoter construct (Fig. 3B). To determine the role of the individual sites in the response to IL-1, we tested the effect of site-directed mutations disrupting each of the described regulatory elements. The mutations in the AP-1 sites (m5′ AP-1 and m3′ AP-1), the mutation in the ets site (mEBS), and the mutation in the COM element (mCOM) affected both the IL-1 and the TPA inducibility of the uPA promoter (Fig. 3C). To test whether the individual protein-binding domains of the uPA enhancer could function as autonomous IL-1- and TPA-responsive elements, we analyzed the activities of the fusion constructs containing the multimerized EBS/5′ AP-1, 3′ AP-1, and COM element fused to the thymidine kinase heterologous promoter. The results (Fig. 3D) showed that, while the isolated EBS/5′ AP-1 and 3′ AP-1 were able to mediate induction in response to both signalling pathways, the COM element did not confer any activity on the heterologous promoter.
Role of the EBS in the uPA enhancer regulation: induction of Ets-2 transcriptional activity by both IL-1 and TPA.
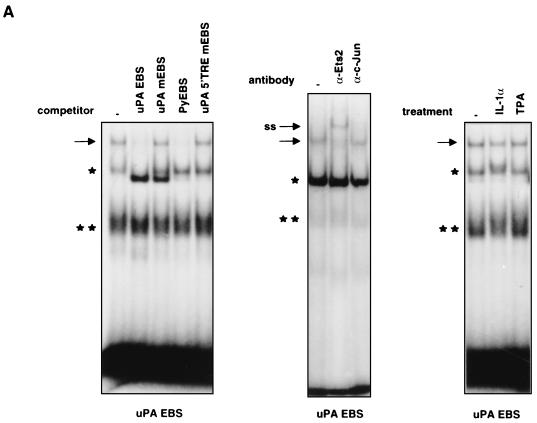
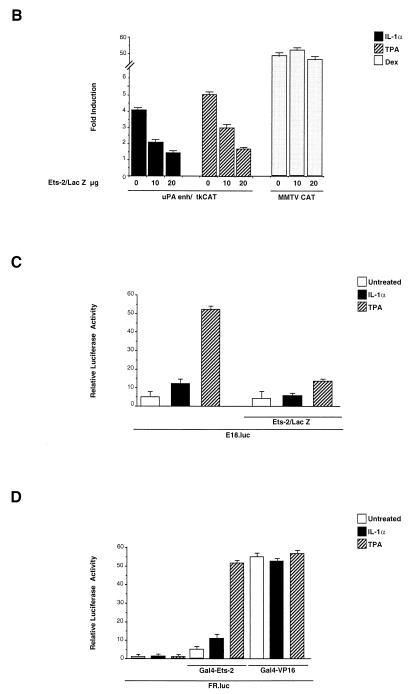
The effect of the EBS mutation showed that the EBS element plays a role in response to both IL-1 and TPA induction. To investigate the role of the ets family factors in the mechanism of IL-1 and TPA induction, we analyzed EBS binding activities in nuclear extracts from HepG2 cells treated with each of the two different uPA inducers. The binding to the EBS oligonucleotide of nuclear proteins from untreated cells resulted in the formation of multiple gel-retarded complexes; only one of the observed complexes was competed by the urokinase (uPA) and polyomavirus enhancer (Py) ets-binding sequences but not by the mutated derivatives, allowing the identification of the ets-specific complex (Fig. 4A). To identify the ets family member interacting with the EBS, we utilized an antibody selective for Ets-2, which has been previously implicated in the regulation of the uPA promoter (52). The results (Fig. 4A) showed that the gel-retarded complex was almost completely supershifted by the antibody, indicating Ets-2 as the major (if not the only) ets family member involved in the regulation of the uPA enhancer. The analysis of the binding activities of IL-1- and TPA-treated cells did not reveal any increase or modification of the preexisting complex or the appearance of new EBS-bound complexes (Fig. 4A), suggesting that the Ets-2 activity might be regulated at a level subsequent to EBS binding in HepG2 cells. To test the functional role of Ets-2 in the IL-1 and TPA induction of the uPA enhancer, we utilized an expression vector encoding the Ets-2–LacZ fusion protein, which has been characterized previously as a dominant-negative repressor of Ets activity (34). The results of the cotransfection of the uPA enhancer-CAT construct in the presence of increasing amounts of the Ets-2–LacZ expression vector showed that the coexpression of the Ets dominant-negative protein resulted in the almost complete inhibition of the uPA enhancer induction, both by IL-1 and by TPA, while the dexamethasone-induced activity of the mouse mammary tumor virus (MMTV)-CAT control construct was not affected (Fig. 4B). Therefore, the uPA EBS element is implicated not only in the TPA- but also in the IL-1-mediated induction.
FIG. 4.
Role of the uPA EBS and regulation of an EBS-driven minimal promoter and Ets-2 transcriptional activity in response to IL-1 and TPA. (A) In vitro binding to the uPA EBS oligonucleotide. (Left [binding competition assay]) Nuclear extracts (5 μg/lane) were preincubated for 15 min with a 200-fold molar excess of the indicated cold competitor oligonucleotides before the addition of the probe. The arrow shows the migration of the specifically bound complex, while the asterisks (single and double) indicate two nonspecific complexes formed with partial reproducibility by the uPA EBS. (Center [supershift assay]) Nuclear extracts were preincubated for 3 h with the indicated antibodies before the addition of the probe. (Right) HepG2 cells were treated with IL-1α or TPA for 2 h prior to nuclear extract preparation and binding to the uPA EBS oligonucleotide; ss indicates the supershifted complex. (B) Effect of the dominant-negative Ets-2–LacZ fusion protein on the IL-1 and TPA induction of the uPA enhancer. The uPAenh/tkCAT reporter (20 μg) was cotransfected with increasing amounts of the vector (pAPr-etsZ-neo) expressing the Ets-2–LacZ chimeric protein; the total amount of DNA was kept constant in each transfection assay, by addition of varying amounts of the empty expression vector. The MMTV-CAT control construct contains the dexamethasone (Dex)-inducible MMTV enhancer fused to the tkCAT reporter. The results represent the averages of five independent experiments. (C) IL-1 and TPA inducibility of an Ets-2 reporter construct. The E18-luc plasmid (10 μg), expressing the luciferase gene driven by a palindromic high-affinity Ets-2 binding site inserted upstream of the c-fos minimal promoter, was electroporated in the presence or absence of the pAPr-etsZ-neo vector (20 μg) into the HepG2 cells, which were subsequently treated with IL-1α or TPA for 24 h. The total amount of DNA in each transfection was kept constant as described for panel B. The results represent the averages of three independent experiments. (D) Modulation of Ets-2-mediated transactivation by IL-1 and TPA. The FR-luc reporter construct (10 μg), containing five copies of the GAL4 binding site upstream of the c-fos minimal promoter, was cotransfected with the Gal4-Ets-2 (0.25 μg) or Gal4-VP16 (0.25 μg) expression vector in the HepG2 cells, which were subsequently treated with IL-1α or TPA. The control was represented by the FR-luc reporter alone. The total amount of DNA in each transfection was kept constant as described for panel B. The results represent the averages of three independent experiments.
The ets factor could be constitutively active in cooperating with the adjacently bound IL-1- and TPA-inducible AP-1 dimer or could represent an independent target for one or both transduction pathways. To investigate this point, we analyzed the activity of a reporter construct containing a high-affinity palindromic Ets-2 binding site fused to a minimal promoter (23). This construct was chosen on the basis of the results of competition assays, showing that the palindromic Ets-2 site (E18) exhibited the same binding specificity as the uPA EBS (data not shown). The results revealed that the E18-luc reporter construct was inducible in response to both TPA and IL-1, with a stronger effect for the phorbol ester than for the inflammatory cytokine (about 10-fold compared with 2.5-fold); the induction was antagonized by coexpression of dominant-negative Ets-2–LacZ (Fig. 4C). Therefore, an isolated Ets-2 binding element can autonomously mediate a moderate transcriptional activation in response to the IL-1-dependent pathway and a significantly stronger induction in response to phorbol ester stimulation. The results of Fig. 4A (right panel) led us to postulate that the IL-1 and TPA regulation of Ets-2 activity might take place at a level subsequent to DNA binding. Therefore, we examined the regulation of the Ets-2 transactivating potential, by analyzing the activity of a protein fusion between Ets-2 and the GAL4 (amino acids 1 to 147) DNA-binding domain. The GAL4-Ets-2-dependent reporter gene activity was induced by both IL-1 and TPA, with a higher fold stimulation for the phorbol ester (about sixfold compared to threefold). The reporter gene activity induced by the control GAL4-VP16 chimeric protein was not affected by both treatments (Fig. 4D). Therefore, the transactivation activity of Ets-2 is regulated by the two different signalling pathways in HepG2 cells. In summary, the results of Fig. 4 indicate that the factor interacting with the uPA EBS is Ets-2 and that its transactivating activity is regulated in response to both IL-1 and TPA.
IL-1- and TPA-dependent compositional change of the AP-1 complex and ATF-2 dimers binding to the (3′ and 5′) sites of the uPA enhancer.
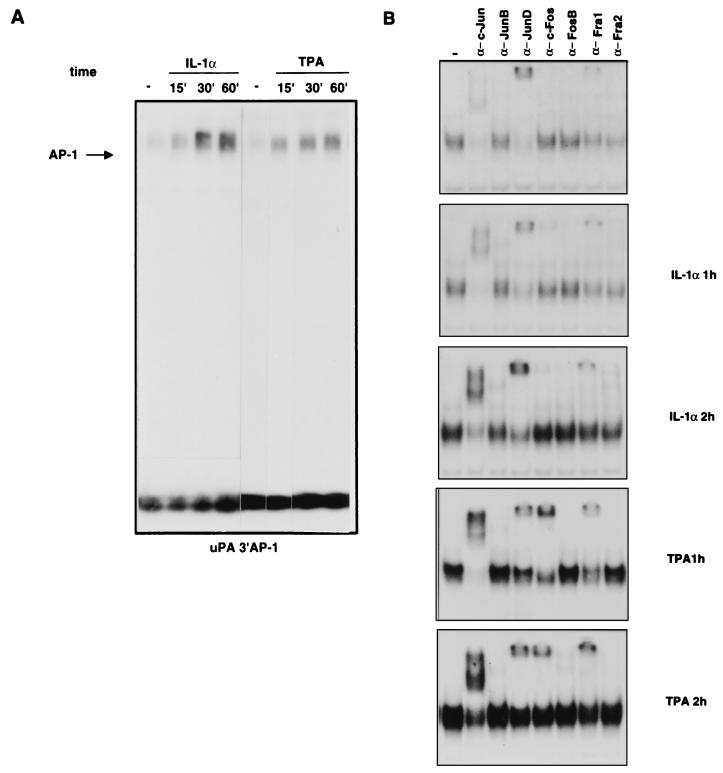
We then analyzed the IL-1- and TPA-dependent modifications of binding activity and composition of the dimers interacting with the AP-1 sites of the uPA enhancer. The binding to the uPA 3′ AP-1 site was strongly induced both by IL-1 and by TPA; the IL-1 induction resulted in a stronger accumulation of the DNA-bound complex than did the TPA induction, which was characterized by a slower accumulation during the analyzed time course (60 min following the addition of the drug [Fig. 5A]). The supershift analysis with antibodies selective for each Jun and Fos family member showed that the AP-1 complex in unstimulated HepG2 cells contains mostly JunD and a small amount of c-Jun and Fra-1. The complex detected in the IL-1-induced HepG2 cells (1 or 2 h of treatment) was characterized by a strong increase of c-Jun, along with the appearance of a smaller amount of JunB and c-Fos, while the level of JunD and Fra-1 supershifted complexes remained relatively constant. The compositional change induced by TPA was very similar except for the stronger accumulation of c-Fos, which started to decline between 1 and 2 h after the induction, and the increase of the Fra-1-containing complex (Fig. 5B). Immunoblotting analysis showed that the modifications detected by gel retardation were determined by variations of the protein levels, with a strong increase of c-Jun, a small induction of JunB, and a slight increase of the preexisting JunD in response to both modulators, while c-Fos and Fra-1 were differently induced (strongly by TPA and weakly by IL-1) by the two treatments (Fig. 6C and 7B and data not shown).
FIG. 5.
In vitro analysis of protein binding to the uPA 3′ AP-1 site during the course of IL-1 or TPA induction. (A) EMSA of IL-1α- and TPA-induced binding to the uPA 3′ AP-1 oligonucleotide. Nuclear proteins were extracted from HepG2 cells at different times (in minutes) after the induction and incubated with the labelled oligonucleotide before the gel retardation assay. (B) Antibody supershift analysis of the complex bound to the uPA 3′ AP-1 oligonucleotide. Nuclear extracts from uninduced, IL-1α-induced (1 and 2 h), or TPA-induced (1 and 2 h) HepG2 cells were preincubated for 3 h with the indicated antibodies, before the addition of the probe.
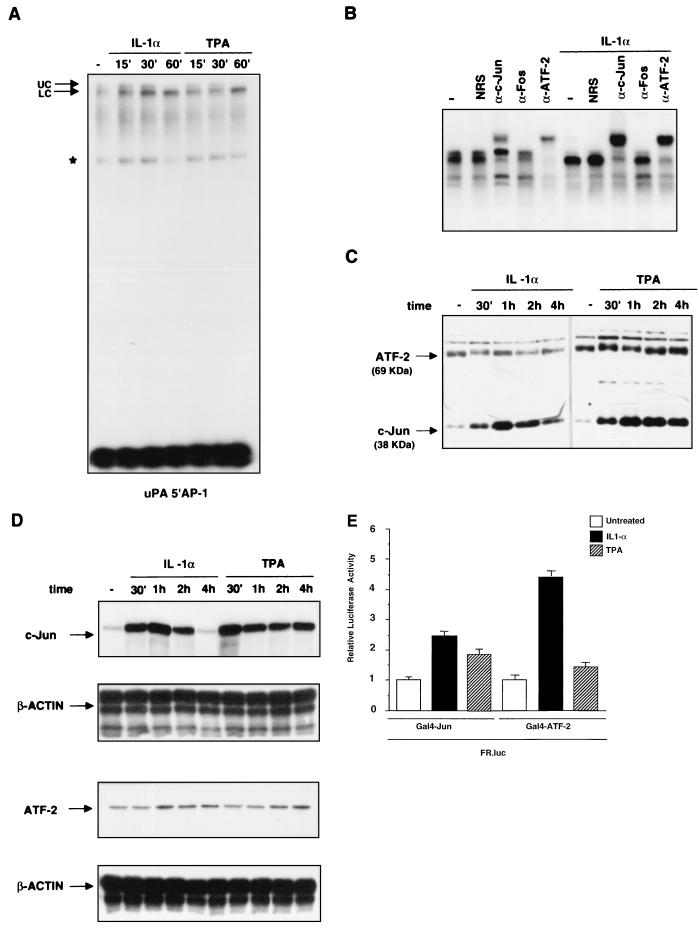
FIG. 6.
In vitro analysis of protein binding to the uPA 5′ AP-1 site during the course of IL-1 or TPA induction. (A) EMSA of IL-1α- and TPA-induced binding to the uPA 5′ AP-1 oligonucleotide. Nuclear proteins were extracted from HepG2 cells at different times (in minutes) after the induction and incubated with the labelled oligonucleotide before the gel retardation assay. (B) Antibody supershift analysis of the complex bound to the uPA 5′ AP-1 oligonucleotide. Nuclear extracts from uninduced or IL-1α- or TPA-induced (60 min) HepG2 cells were preincubated for 3 h with the indicated antibodies, before the addition of the probe (NRS, normal rabbit serum; α-Fos, antibody recognizing all Fos family members). (C) Immunoblotting analysis of ATF-2 and c-Jun expression. Nuclear proteins were extracted at the indicated time points from IL-1α- or TPA-induced cells, separated by SDS-PAGE (20 μg/lane), and transferred to polyvinylidene difluoride membranes. Western blots were first incubated with both anti-c-Jun and anti-ATF-2 antibodies and then with horseradish peroxidase-conjugated secondary antibodies followed by enhanced chemiluminescence. As a control for equal loadings, the blotted proteins were stained with Ponceau red. The arrows indicate the apparent molecular masses of c-Jun (39 kDa) and ATF-2 (69 kDa). (D) RNase protection analysis of c-Jun and ATF-2 mRNA in response to IL-1 induction. Total RNA was extracted at different times (minutes or hours) after induction as indicated and annealed to the human c-Jun or ATF-2 ribonucleotide probes along with the internal reference β-actin ribonucleotide probe, before RNase digestion and polyacrylamide-urea gel electrophoresis. (E) Modulation of c-Jun and ATF-2 transactivation activity by IL-1 and TPA. The FR-luc reporter construct was cotransfected with the pDB10 (containing the mouse c-Jun amino acid residues 1 to 253 fused to the GAL4 DNA-binding domain) or the GAL4-ATF-2 (containing the human ATF-2 amino acid residues 1 to 109 fused to the GAL4 DNA-binding domain) expression vector in the HepG2 cells, which were subsequently treated with IL-1α or TPA. The results represent the averages of three independent experiments.
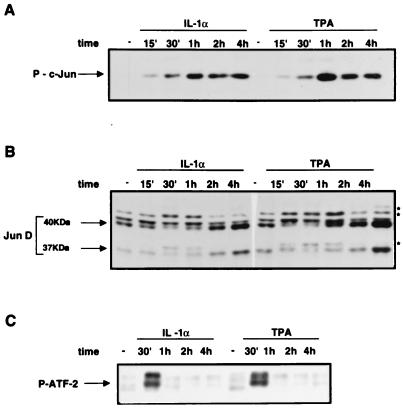
FIG. 7.
IL-1- and TPA-dependent induction of c-Jun, JunD, and ATF-2 phosphorylation. (A) IL-1- and TPA-dependent induction of c-Jun phosphorylation. Western blots were incubated with a phosphospecific polyclonal antibody, recognizing the Ser73-phosphorylated, but not the unphosphorylated, c-Jun. (B) Mobility shift of JunD isoforms during IL-1- or TPA-mediated induction. Immunoblotting was performed as described for Fig. 6C. The arrows indicate the two JunD translation products (40 and 37 kDa), while the asterisks show the JunD modified isoforms. The immunoblotting specificity was verified by preincubating the antibodies with the JunD immunizing peptide. (C) IL-1- and TPA-dependent induction of ATF-2 phosphorylation. Western blots were incubated with a phosphospecific polyclonal antibody, recognizing the Thr71-phosphorylated, but not the unphosphorylated, ATF-2. Primes indicate minutes.
The binding to the octameric uPA upstream AP-1 site (uPA 5′ AP-1) gave rise to a complex which was both quantitatively and qualitatively modified by the two inducers. The extended electrophoretic separation of the DNA-bound complexes allowed us to detect a gel-retarded doublet. The induction (both by IL-1 and by TPA) resulted in augmented binding, which was associated with a sharp increase of the faster-migrating component (lower complex) and with the disappearance of the slower component (upper complex) of the doublet (Fig. 6A). We then examined the composition of the doublet by antibody supershift analysis. Since we have shown previously that the cyclic AMP response element-like sequence of the uPA 5′ TRE is bound by c-Jun–ATF-2 heterodimers, we used anti-c-Jun and anti-ATF-2 antibodies, in addition to normal rabbit serum and anti-Fos antibodies, which were included as negative controls. The results showed that the faster-migrating complex was recognized by both the anti-c-Jun and anti-ATF-2 antibodies, while the upper component of the doublet was supershifted by the anti-ATF-2, but not the anti-c-Jun, antibodies (Fig. 6B). Therefore, we concluded that the faster component is represented by c-Jun–ATF-2 heterodimers and that the slower component is formed by ATF-2 homodimers. The only complex detected in the IL-1α-stimulated cells was supershifted by both the anti-c-Jun and anti-ATF-2 antibodies (Fig. 6B); an identical result was obtained with the TPA-induced complex (data not shown). These data indicate that, while in unstimulated cells both ATF-2 homodimers and c-Jun–ATF-2 heterodimers interact with the uPA 5′ AP-1 site, in IL-1- or TPA-stimulated cells only c-Jun–ATF-2 heterodimers bind to such an element.
To understand the mechanism of the transition between the different dimers, we analyzed the ATF-2 and c-Jun protein levels during the time course of stimulation. The results showed that, while the c-Jun level was strongly increased by both inducers, the ATF-2 protein level remained constant (Fig. 6C). The results also showed the different kinetics of accumulation of c-Jun during the course of the IL-1 treatment and TPA stimulation. While the IL-1-dependent induction resulted in the transient increase of the c-Jun level, which started to decline between 1 and 2 h following induction, the TPA stimulation resulted in a larger induction of c-Jun, which underwent a very small decrease during the 4-h course. The absence of quantitative changes of the 69-kDa ATF-2 polypeptide was associated with a qualitative change, represented by a small protein mobility shift, detected in both the IL-1- and the TPA-treated samples. Interestingly, while such a modification was apparent at only 30 min from the beginning of the IL-1 induction, the shift was still detectable after 1 h, during the course of TPA stimulation. These results likely indicate the actions of both inducers on ATF-2 phosphorylation, with a more sustained effect of TPA than of IL-1, in the absence of quantitative changes of ATF-2 protein level. In agreement with the observed variation in protein levels, we found that the c-Jun mRNA was dramatically induced by IL-1, with an increase already detectable at 15 min, while the ATF-2 mRNA was not affected by the induction. A similar result was obtained from the analysis of the TPA-treated samples, which revealed a sustained accumulation of c-Jun mRNA and no induction of the ATF-2 transcript, in agreement with the immunoblotting data (Fig. 6D). Therefore, the modified ratio of the expression level of c-Jun to that of ATF-2 is responsible for the compositional change leading to the prevalence of the c-Jun–ATF-2 heterodimer as the nuclear factor bound to the uPA 5′ TRE in induced cells.
To define the relative contribution of the transactivation domain of each of the two components of the c-Jun–ATF-2 heterodimer to the transcriptional response to IL-1 and TPA, we analyzed the inducibility of the chimeric proteins containing the amino-terminal c-Jun and ATF-2 transactivation domains (amino acid residues 1 to 253 and 1 to 109, respectively), fused to the GAL4 DNA-binding domain. The results showed that both IL-1 and TPA stimulated the transactivation activity of both c-Jun and ATF-2 in the HepG2 cell line (Fig. 6E).
IL-1 and TPA stimulate the c-Jun and JunD phosphorylation in HepG2 cells.
The transcriptional induction detected by the GAL4 chimeric proteins reflects the posttranslational modifications of the c-Jun and ATF-2 amino-terminal domains. In particular, the role of the JNK-dependent phosphorylation of multiple Ser and Thr residues (c-Jun Ser-63, Ser-73, Thr-91, and Thr-93 and ATF-2 Thr-69 and Thr-71) in response to various stress-inducing agents has been analyzed in detail. The role of in vivo phorbol ester-induced phosphorylation is less well characterized for both transcription factors: it has been shown elsewhere that ATF-2 constitutes a poor substrate and that c-Jun is almost completely nonphosphorylatable by the TPA-inducible Erk-type MAP kinases (37). To characterize the changes of c-Jun amino-terminal phosphorylation induced by the IL-1 and TPA treatment of HepG2 cells, we utilized a phosphospecific antibody, recognizing only the phosphorylated c-Jun Ser-73 residue. The results showed that the phospho-Ser-73 c-Jun, undetectable in unstimulated cells, was strongly induced both by IL-1 and by TPA, with a maximum about 1 h after treatment, with each of the two inducers (Fig. 7A). The kinetics of phospho-c-Jun induction were identical for the two inducers, in contrast with the pattern of c-Jun accumulation, which exhibited a different time course (Fig. 7A). The selectivity of the phosphospecific antibody was verified by the in situ acid phosphatase treatment of Western blots, which prevented the detection of the c-Jun polypeptide by the phosphospecific antibody but not by the phosphorylation-insensitive antibody (data not shown).
As described above, the other Jun family component detected in the AP-1 complex of (both uninduced and induced) HepG2 cells is represented by JunD. The results of the immunoblotting with the JunD-selective antibodies showed two major JunD polypeptides, of about 41 and 37 kDa (Fig. 7B). Both the IL-1- and the TPA-mediated induction resulted in a moderate accumulation of the JunD polypeptides and in the appearance of electrophoretically slower JunD isoforms. Therefore, both inducers do not significantly affect the JunD expression level but result in JunD modification, which likely reflects the phosphorylation of the protein. As described above for c-Jun, the effect of TPA was more evident than the effect of IL-1, with the JunD mobility shift detectable after 15 min of stimulation (compared to 30 min for IL-1).
To investigate the phosphorylation of ATF-2, we took advantage of the phosphospecific antibody recognizing the ATF-2 Thr-71, which represents (along with Thr-69) the substrate for the JNK-mediated phosphorylation of ATF-2. The immunoblotting result indicated that the ATF-2 Thr-71 was strongly phosphorylated in response to both signalling agonists, with a rapidly reversible effect, which disappeared between 30 min and 1 h. In summary, our results indicate that both IL-1 and phorbol esters can induce the phosphorylation of c-Jun, JunD, and ATF-2 in HepG2 cells.
Involvement of different MAP kinase pathways in uPA enhancer activity: IL-1 induction requires JNK but not ERK, while TPA induction requires both JNK and ERK activation.
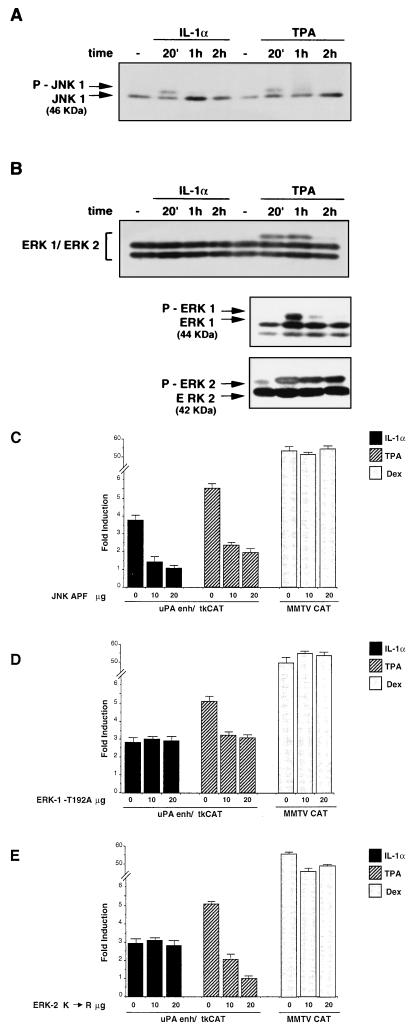
To characterize the MAP kinase pathways involved in the uPA enhancer induction, we analyzed the activity of the JNK- and ERK-type kinases, in response to the two different inducers. The changes of MAP kinase activity in response to the IL-1- or TPA-dependent induction were evaluated by analyzing the phosphorylation-dependent shift of the proteins by Western blotting. The results showed that the electrophoretic mobility shift of the 46-kDa JNK1 polypeptide took place in response to both IL-1 and TPA (Fig. 8A), while a mobility shift of the slower-migrating ERK polypeptide (44-kDa ERK-1) was detected following TPA but not IL-1 treatment, suggesting that ERK activity is not stimulated by IL-1 in HepG2 cells. The result obtained with the anti-ERK-1 and -2 antiserum did not allow us to analyze the mobility shift of ERK-2, because of the electrophoretic comigration of phosphorylated ERK-2 and unphosphorylated ERK-1. Therefore, we also used the (ERK-1 and ERK-2) selective antibodies. The result indicated a clear difference between the TPA-dependent activation of ERK-1 and that of ERK-2, showing that ERK-1 phosphorylation was transient, with an almost complete decline between 20 min and 1 h, while ERK-2 activity exhibited a sustained kinetics, with the phosphorylated protein fully detectable more than 2 h after the induction (Fig. 8B).
FIG. 8.
Role of JNK- and ERK-type MAP kinases in the IL-1- and TPA-mediated induction of the uPA enhancer. (A) IL-1- and TPA-mediated induction of JNK phosphorylation. Immunoblotting was performed as described for Fig. 7, except for the acrylamide-bisacrylamide cross-linking and the pH of the separation SDS gel (see Materials and Methods), to detect the phosphorylation-dependent mobility shift. The arrows indicate the unmodified and the shifted isoforms of the 46-kDa JNK1 polypeptide. (B) Phosphorylation-dependent mobility shift of ERK proteins in response to IL-1 or TPA induction. Immunoblotting was performed as described for panel A, with a polyclonal antibody recognizing both ERK-1 and ERK-2 (upper panel) or selective for ERK-1 and ERK-2 (lower panels). The arrows indicate the 42-kDa ERK-2, the 44-kDa ERK-1, and the phosphorylated isoform of ERK-1. (C) Effect of the transdominant-negative JNK-APF derivative on the IL-1- or TPA-mediated induction of the uPA enhancer. The uPAenh/tkCAT reporter or the MMTV CAT dexamethasone (Dex)-inducible control (20 μg) was cotransfected with increasing amounts of the pCMV-JNK-APF vector expressing the catalytically inactive JNK1 protein; the total amount of DNA was kept constant in each transfection assay by addition of varying amounts of the empty expression vector. The results represent the averages of three independent experiments. (D) Effect of the transdominant-negative ERK-1 on the IL-1- or TPA-mediated induction of the uPA enhancer. The uPAenh/tkCAT reporter or the MMTV CAT dexamethasone (Dex)-inducible control (20 μg) was cotransfected with increasing amounts of the pCMV-derived vector expressing the catalytically inactive ERK-1 (ERK-1 T192A); the experiments were performed as described for panel C. (E) Effect of the transdominant-negative ERK-2 derivatives on the IL-1- or TPA-mediated induction of the uPA enhancer. The uPAenh/tkCAT reporter or the MMTV CAT dexamethasone (Dex)-inducible control (20 μg) was cotransfected with increasing amounts of the pCMV-derived vector expressing the catalytically inactive ERK-2 (ERK-2 K→R); the experiments were performed as described for panel C.
To test the role of JNK and ERK activation in the response of the uPA enhancer to IL-1 or TPA, we tested the effect of the selective inhibition of each of the two MAP kinases. First, we analyzed the effect of a nonphosphorylatable JNK1 derivative, in which the phosphorylation site Thr-Pro-Tyr is changed to Ala-Pro-Phe (JNK-APF). The expression vector encoding the dominant-negative JNK-APF protein, which behaves as a competitive inhibitor of JNK, was coexpressed with the uPA enhancer reporter construct (uPA-tkCAT). The results showed that the stimulation of the uPA enhancer by both IL-1 and TPA was decreased in the presence of the dominant-negative JNK, with the strongest effect on the IL-1 induction, which was totally suppressed by the inhibition of JNK activity (Fig. 8C). Therefore, the observed increase of c-Jun phosphorylation is functionally relevant for not only the IL-1- but also the TPA-dependent induction of the uPA enhancer.
To assess the relative importance of the activation of the ERK-1 and -2 kinases, we tested the effect of the catalytically inactive dominant-negative derivatives of ERK-1 and ERK-2 on the TPA-dependent induction of the uPA enhancer. The dominant-negative ERKs exhibited a significantly different effect: while TPA induction was fully suppressed by ERK-2 inhibition, the dominant-negative ERK-1 resulted in a partial effect (less than 50% inhibition), which was not augmented by the addition of increasing amounts of expression vector (Fig. 8D). In addition, we showed that each of the dominant-negative ERKs did not affect the IL-1 induction of the uPA enhancer, as expected on the basis of the observed lack of IL-1-dependent induction of ERKs in HepG2 cells (data not shown). In summary, the results of Fig. 8 indicate that, while the IL-1-dependent induction of the uPA enhancer requires the JNK but not the ERK-type MAP kinases, the TPA-mediated induction is mediated by the activation of both the JNK- and the ERK-type MAP kinases.
DISCUSSION
IL-1, in combination with IL-6, tumor necrosis factor alpha, and other inflammatory cytokines, plays a major role in mediating complex regulatory changes of gene expression in the liver acute-phase response (3). In this paper, we describe the in vivo and in vitro regulation of the urokinase gene by IL-1 in liver cells and the regulatory elements and signalling pathways involved in the IL-1 induction of the uPA transcriptional enhancer. In particular, we show that the uPA enhancer regulation involves the increased expression and phosphorylation of multiple AP-1 components such as c-Jun, JunD, and ATF-2, along with the IL-1- and TPA-dependent activation of the ets family member Ets-2. In addition, we show that the IL-1 induction requires the JNK- but not the ERK-type MAP kinases, while the TPA induction requires the activation of both JNKs and ERKs, with a major role being played by ERK-2.
Mutational analysis of the human uPA promoter showed the essential role played by the evolutionally conserved −2.0-kb regulatory region (−2.4 kb in mouse) and confirmed the functional cooperation among the three elements previously characterized for their role in the growth factor- and phorbol ester-dependent induction of the uPA enhancer: the 5′ TRE, the COM, and the 3′ TRE (4, 17, 18, 44, 45).
The in vitro analysis revealed that IL-1 induction results in the increased binding and compositional change of different dimeric species bound to the uPA 5′ TRE and 3′ TRE. While the increased binding to the uPA 3′ TRE reflects the well-established induction of multiple AP-1 components (2) in response to both IL-1 and TPA, the modification of binding to the uPA 5′ TRE reflects the interplay between ATF-2 homodimers and c-Jun–ATF-2 heterodimers, resulting from variation of the ratio between the constitutively expressed ATF-2 and the highly inducible c-Jun. Both c-Jun and ATF-2 contain IL-1-inducible transactivation domains (25, 30), as confirmed by our analysis in the HepG2 cell line; therefore, both the ATF-2 homodimers and c-Jun–ATF-2 heterodimers appear to be equally important in mediating the transcriptional response to the inflammatory cytokine. The functional difference between the two dimeric species (ATF-2–ATF-2 and c-Jun–ATF-2) remains to be investigated. On the basis of recent reports showing that ATF-2, differing from c-Jun, represents a direct substrate for protein kinase Cα (PKCα)-dependent phosphorylation (31) and that c-Jun and ATF-2 are differentially phosphorylated by distinct JNK isoforms (24), it can be envisaged that the two distinct dimers (ATF-2–ATF-2 and c-Jun–ATF-2) might perform different regulatory tasks. A complete understanding of the functional role of ATF-2 in uPA enhancer activity will be made possible by studying uPA gene regulation in response to multiple stimuli in the recently described ATF-2-deficient mice (49).
The role of the ets family factors in the regulation of the urokinase promoter has been characterized in several cellular systems and in response to various agents, such as epidermal growth factor (50), granulocyte-monocyte colony-stimulating factor (GM-CSF) (52), and TPA (44). The role of the ets-2 gene product in in vivo uPA gene regulation has been recently substantiated by the analysis of the uPA expression in the skin of ets-2-knockout mice (57). The inhibition by the dominant-negative Ets-2–LacZ derivative shows the functional role of the uPA EBS; the gel retardation data identify Ets-2 as the ets family member interacting with the urokinase enhancer in the HepG2 cell line (Fig. 4A). In IL-1- or TPA-treated HepG2 cells, the Ets-2 activity is regulated at a level subsequent to DNA binding, indicating a different mechanism than that of the Ets-2-dependent regulation described in the GM-CSF- or phorbol myristate acetate-induced macrophage cell lines (52) and in the developing avian heart (40), in which the changes of urokinase gene expression are paralleled by changes of Ets-2 expression and DNA-binding activity. The inducible activity of the GAL4-Ets-2 chimeric protein strongly suggests a role for Ets-2 modification(s) in response to both IL-1 and phorbol esters. Phosphorylation of one Ets-2 residue (Thr-72) is essential for mediating the oncogenic activation of several ras-responsive promoters, suggesting a role for ras-dependent MAP kinases in Ets-2 phosphorylation (58). Recently, it has been shown that Ets-2 phosphorylation strongly correlates with the activation of p42 and p44 ERKs in the response to GM-CSF (22); in addition, it has been reported that a related component of the ets family, PEA3, can be activated by two distinct MAP kinase cascades, the ERKs and the stress-activated protein kinases (47). Interestingly, PEA3 activation by both pathways plays a role in the ras-mediated signalling triggered by the HER/Neu oncogenic tyrosine kinase (46). Therefore, the convergence of two MAP kinase pathways on the same transcription factor is not restricted to Elk-1 (56) but involves different ets family members. Our results suggest that Ets-2 phosphorylation might play a role in response to both signalling agonists (IL-1 and TPA). Since IL-1, differing from TPA, leads to the induction of JNK but not ERK activity in HepG2 cells, Ets-2 might represent a substrate of JNK (or other MAP kinases distinct from ERK-1 and -2) in the IL-1-treated cells, while it would be phosphorylated by ERK-1 and -2 (or ERK-1 and -2 and JNK) in TPA-treated cells.
The analysis of c-Jun amino-terminal phosphorylation and the effect of inhibition of the c-Jun NH2 kinase shows that the induction of JNK activity is required for uPA transcriptional enhancement both by IL-1 and by TPA. Our results confirm the role of c-Jun N-terminal kinase-mediated signalling in uPA enhancer induction, in agreement with the recently characterized mechanism of UV-dependent uPA gene induction in NIH 3T3 fibroblasts (42), but in contrast with a previous report showing that the UV-induced JNK activity is not associated with uPA promoter induction in pig kidney epithelial cells (28). To explain such a discrepancy, it can be speculated that some cell-specific component, lacking in the pig kidney cells, might be required for the induction of the uPA enhancer by the JNK-dependent pathway.
The stimulatory effect of JNK activity on the uPA enhancer in HepG2 cells is mediated by phosphorylation of multiple factors, including c-Jun, ATF-2, and possibly Ets-2. In addition, the observed modification by both IL-1 and TPA treatment likely represents the effect of JunD phosphorylation. The recently shown recruitment of JunD to JNK by c-Jun and JunB (29) shows the importance of the heterodimerization partner; the identification of JunD–ATF-2 (in addition to c-Jun–ATF-2) heterodimers bound to the uPA 5′ TRE (data not shown) suggests the possibility that the ATF-2-mediated recruitment of JNK might be responsible for JunD phosphorylation.
The TPA-dependent phosphorylation of c-Jun raises a question regarding the pathway linking PKC activation and the induction of JNK activity. Since the activation of the PKC substrate Raf-1 does not result in a significant activation of the JNK pathway, different levels of cross talk can be postulated. In particular, the recent report showing the PKC-dependent phosphorylation of Shc (21) suggests that the TPA signal can act at a level upstream of ras, therefore affecting not only the Raf- but also the MEKK1-dependent pathway and resulting in the induction of the c-Jun NH2 kinase activity, with quantitatively different effects in different cell lines.
In our system, the effect of IL-1 required the activation of the JNK- but not the ERK-type MAP kinases, in agreement with previous reports describing the induction by IL-1 of the p54 stress-activated protein kinase but not ERK-1 and -2 MAP kinases in HepG2 cells (8). The TPA-dependent stimulation of the ERKs is relevant for uPA enhancer induction, as shown by functional inhibition (Fig. 8). The more pronounced effect of ERK-2 than of ERK-1 inhibition confirms the observation reported for the renal epithelial cell system (28) and raises the question about the differential activation and/or distinct substrate specificities of ERK-1 and ERK-2.
The diversity of regulatory circuits involved in the differential induction of uPA mRNA by IL-1 or phorbol esters is further shown by the different interactions with protein synthesis inhibitors. Cycloheximide and anisomycin dramatically synergize with IL-1, but not with TPA, with an effect already detectable at a subinhibitory dosage for translational arrest (14). It will be interesting to investigate whether the observed synergism results from the combination of effects on both uPA transcription and mRNA stability, or if it takes place only at the transcriptional level, possibly affecting the activity of multiple factors interacting with the uPA enhancer.
The relevance of the regulation characterized in vitro in the hepatoma cell line is validated by the uPA induction detected in the livers of rats treated in vivo with IL-1. These findings raise questions regarding the physiological significance of the increased synthesis of liver urokinase during the acute-phase response. The previously characterized role of urokinase as a pro-HGF convertase, responsible for the proteolytic activation of the matrix-associated HGF (43), suggests that the IL-1-mediated regulation of uPA might functionally control HGF activity, responsible for hepatocyte mitogenesis and induction of acute-phase response genes (3). Furthermore, it is interesting to speculate that IL-1 induction may play a role in controlling the increase of plasminogen activator activity associated with the extracellular matrix remodeling which takes place during the early stages of liver regeneration (32).
ACKNOWLEDGMENTS
We thank Diego Di Lorenzo (Civic Hospital of Brescia) for the generous gift of samples from IL-1-treated rats and those persons mentioned in the text for kindly providing us with various expression vectors. We also thank Maria Terracciano for excellent technical assistance and Luigi Lania and Andrea Riccio for critical review of the manuscript.
The work was supported by grants from the Associazione Italiana per la Ricerca sul Cancro (AIRC) to P.V. G.C. and D.V. were supported by Fellowships from CNR; L.C. and A.C. were recipients of AIRC Fellowships.
G. Cirillo and L. Casalino contributed equally to this work.
REFERENCES
- 1.Andreasen P A, Kjoller L, Christensen L, Duffy M J. The urokinase-type plasminogen activator system in cancer metastasis: a review. Int J Cancer. 1997;72:1–22. doi: 10.1002/(sici)1097-0215(19970703)72:1<1::aid-ijc1>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 2.Angel P, Karin M. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim Biophys Acta. 1991;1072:129–157. doi: 10.1016/0304-419x(91)90011-9. [DOI] [PubMed] [Google Scholar]
- 3.Baumann H, Morella K K, Wong G H. TNF-alpha, IL-1 beta, and hepatocyte growth factor cooperate in stimulating specific acute phase plasma protein genes in rat hepatoma cells. J Immunol. 1993;151:4248–4257. [PubMed] [Google Scholar]
- 4.Berthelsen J, Vandekerkhove J, Blasi F. Purification and characterization of UEF3, a novel factor involved in the regulation of the urokinase and other AP-1 controlled promoters. J Biol Chem. 1996;271:3822–3830. doi: 10.1074/jbc.271.7.3822. [DOI] [PubMed] [Google Scholar]
- 5.Besser D, Presta M, Nagamine Y. Elucidation of a signaling pathway induced by FGF-2 leading to uPA gene expression in NIH 3T3 fibroblasts. Cell Growth Differ. 1995;6:1009–1107. [PubMed] [Google Scholar]
- 6.Besser D, Urich M, Sakaue M, Messerschmitt A, Ballmer-Hofer K, Nagamine Y. Urokinase-type plasminogen activator gene regulation by polyomavirus middle-T antigen. Oncogene. 1995;11:2383–2391. [PubMed] [Google Scholar]
- 7.Besser D, Verde P, Nagamine Y, Blasi F. Signal transduction and the uPA/uPAR system. Fibrinolysis. 1996;10:215–237. [Google Scholar]
- 8.Bird T A, Kyriakis J M, Tyshler L, Gayle M, Milne A, Virca G D. Interleukin-1 activates p54 mitogen-activated protein (MAP) kinase/stress-activated protein kinase by a pathway that is independent of p21ras, Raf-1, and MAP kinase kinase. J Biol Chem. 1994;269:31836–31844. [PubMed] [Google Scholar]
- 9.Blasi F. Urokinase and urokinase receptor: a paracrine/autocrine system regulating cell migration and invasiveness. Bioessays. 1993;15:105–111. doi: 10.1002/bies.950150206. [DOI] [PubMed] [Google Scholar]
- 10.Blasi F, Verde P. Urokinase-dependent cell surface proteolysis and cancer. Semin Cancer Biol. 1990;1:117–126. [PubMed] [Google Scholar]
- 11.Bruder J T, Heidecker G, Rapp U R. Serum-, TPA-, and Ras-induced expression from Ap-1/Ets-driven promoters requires Raf-1 kinase. Genes Dev. 1992;6:545–556. doi: 10.1101/gad.6.4.545. [DOI] [PubMed] [Google Scholar]
- 12.Busso N, Nicodeme E, Chesne C, Guillouzo A, Belin D, Hyafil F. Urokinase and type I plasminogen activator inhibitor production by normal human hepatocytes: modulation by inflammatory agents. Hepatology. 1994;20:186–190. doi: 10.1016/0270-9139(94)90152-x. [DOI] [PubMed] [Google Scholar]
- 13.Campbell I K, Piccoli D S, Butler D M, Singleton D K, Hamilton J A. Recombinant human interleukin-1 stimulates human articular cartilage to undergo resorption and human chondrocytes to produce both tissue- and urokinase-type plasminogen activator. Biochim Biophys Acta. 1988;967:183–194. doi: 10.1016/0304-4165(88)90008-6. [DOI] [PubMed] [Google Scholar]
- 14.Cano E, Hazzalin C A, Mahadevan L C. Anisomycin-activated protein kinases p45 and p55 but not mitogen-activated protein kinases ERK-1 and -2 are implicated in the induction of c-fos and c-jun. Mol Cell Biol. 1994;14:7352–7362. doi: 10.1128/mcb.14.11.7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 16.De Cesare D, Palazzolo M, Berthelsen J, Blasi F. Characterization of UEF-4, a DNA-binding protein required for transcriptional synergism between two AP-1 sites in the human urokinase enhancer. J Biol Chem. 1997;272:23921–23929. doi: 10.1074/jbc.272.38.23921. [DOI] [PubMed] [Google Scholar]
- 17.De Cesare D, Palazzolo M, Blasi F. Functional characterization of COM, a DNA region required for cooperation between AP-1 sites in urokinase gene transcription. Oncogene. 1996;13:2551–2562. [PubMed] [Google Scholar]
- 18.De Cesare D, Vallone D, Caracciolo A, Sassone-Corsi P, Nerlov C, Verde P. Heterodimerization of c-Jun with ATF-2 and c-Fos is required for positive and negative regulation of the human urokinase enhancer. Oncogene. 1995;11:365–376. [PubMed] [Google Scholar]
- 19.Derijard B, Hibi M, Wu I H, Barrett T, Su B, Deng T, Karin M, Davis R J. JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell. 1994;76:1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- 20.D’Orazio D, Besser D, Marksitzer R, Kunz C, Hume D A, Kiefer B, Nagamine Y. Cooperation of two PEA3/AP1 sites in uPA gene induction by TPA and FGF-2. Gene. 1997;201:179–187. doi: 10.1016/s0378-1119(97)00445-9. [DOI] [PubMed] [Google Scholar]
- 21.El-Shemerly M Y, Besser D, Nagasawa M, Nagamine Y. 12-O-Tetradecanoylphorbol-13-acetate activates the Ras/extracellular signal-regulated kinase (ERK) signaling pathway upstream of SOS involving serine phosphorylation of Shc in NIH3T3 cells. J Biol Chem. 1997;272:30599–30602. doi: 10.1074/jbc.272.49.30599. [DOI] [PubMed] [Google Scholar]
- 22.Fowles L F, Martin M L, Nelsen L, Stacey K J, Redd D, Clark Y M, Nagamine Y, McMahon M, Hume D A, Ostrowski M C. Persistent activation of mitogen-activated protein kinases p42 and p44 and ets-2 phosphorylation in response to colony-stimulating factor 1/c-fms signaling. Mol Cell Biol. 1998;18:5148–5156. doi: 10.1128/mcb.18.9.5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galang C K, Der C J, Hauser C A. Oncogenic Ras can induce transcriptional activation through a variety of promoter elements, including tandem c-Ets-2 binding sites. Oncogene. 1994;9:2913–2921. [PubMed] [Google Scholar]
- 24.Gupta S, Barrett T, Whitmarsh A J, Cavanagh J, Sluss H K, Derijard B, Davis R J. Selective interaction of JNK protein kinase isoforms with transcription factors. EMBO J. 1996;15:2760–2770. [PMC free article] [PubMed] [Google Scholar]
- 25.Gupta S, Campbell D, Derijard B, Davis R J. Transcription factor ATF2 regulation by the JNK signal transduction pathway. Science. 1995;267:389–393. doi: 10.1126/science.7824938. [DOI] [PubMed] [Google Scholar]
- 26.Gutman A, Wasylyk B. The collagenase gene promoter contains a TPA and oncogene-responsive unit encompassing the Pea3 and AP-1 binding sites. EMBO J. 1990;9:2241–2246. doi: 10.1002/j.1460-2075.1990.tb07394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gutman A, Wasylyk B. Nuclear targets for transcription regulation by oncogenes. Trends Genet. 1991;7:49–54. doi: 10.1016/0168-9525(91)90231-E. [DOI] [PubMed] [Google Scholar]
- 28.Irigoyen J P, Besser D, Nagamine Y. Cytoskeleton reorganization induces the urokinase-type plasminogen activator gene via the Ras/extracellular signal-regulated kinase (ERK) signaling pathway. J Biol Chem. 1997;272:1904–1909. doi: 10.1074/jbc.272.3.1904. [DOI] [PubMed] [Google Scholar]
- 29.Kallunki T, Deng T, Hibi M, Karin M. c-Jun can recruit JNK to phosphorylate dimerization partners via specific docking interactions. Cell. 1996;87:929–939. doi: 10.1016/s0092-8674(00)81999-6. [DOI] [PubMed] [Google Scholar]
- 30.Karin M. The regulation of AP-1 activity by mitogen-activated protein kinases. J Biol Chem. 1995;270:16483–16486. doi: 10.1074/jbc.270.28.16483. [DOI] [PubMed] [Google Scholar]
- 31.Kawasaki H, Song J, Eckner R, Ugai H, Chiu R, Taira K, Shi Y, Jones N, Yokoyama K K. p300 and ATF-2 are components of the DRF complex, which regulates retinoic acid- and E1A-mediated transcription of the c-jun gene in F9 cells. Genes Dev. 1998;12:233–245. doi: 10.1101/gad.12.2.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim T H, Mars W M, Stolz D B, Petersen B E, Michalopoulos G K. Extracellular matrix remodeling at the early stages of liver regeneration in the rat. Hepatology. 1997;26:896–904. doi: 10.1002/hep.510260415. [DOI] [PubMed] [Google Scholar]
- 33.Kortenjann M, Thomae O, Shaw P E. Inhibition of v-raf-dependent c-fos expression and transformation by a kinase-defective mutant of the mitogen-activated protein kinase Erk2. Mol Cell Biol. 1994;14:4815–4824. doi: 10.1128/mcb.14.7.4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langer S J, Bortner D M, Roussel M F, Sherr C J, Ostrowski M C. Mitogenic signaling by colony-stimulating factor 1 and ras is suppressed by the ets-2 DNA-binding domain and restored by myc overexpression. Mol Cell Biol. 1992;12:5355–5362. doi: 10.1128/mcb.12.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lengyel E, Singh B, Gum R, Nerlov C, Sabichi A, Birrer M, Boyd D. Regulation of urokinase-type plasminogen activator expression by the v-mos oncogene. Oncogene. 1995;11:2639–2648. [PubMed] [Google Scholar]
- 36.Lengyel E, Stepp E, Gum R, Boyd D. Involvement of a mitogen-activated protein kinase signaling pathway in the regulation of urokinase promoter activity by c-Ha-ras. J Biol Chem. 1995;270:23007–23012. doi: 10.1074/jbc.270.39.23007. [DOI] [PubMed] [Google Scholar]
- 37.Livingstone C, Patel G, Jones N. ATF-2 contains a phosphorylation-dependent transcriptional activation domain. EMBO J. 1995;14:1785–1797. doi: 10.1002/j.1460-2075.1995.tb07167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Logan S K, Garabedian M J, Campbell C E, Werb Z. Synergistic transcriptional activation of the tissue inhibitor of metalloproteinases-1 promoter via functional interaction of AP-1 and Ets-1 transcription factors. J Biol Chem. 1996;271:774–782. doi: 10.1074/jbc.271.2.774. [DOI] [PubMed] [Google Scholar]
- 39.Mahadevan L C, Edwards D R. Signalling and superinduction. Nature. 1991;349:747–748. doi: 10.1038/349747c0. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 40.Majka S M, McGuire P G. Regulation of urokinase expression in the developing avian heart: a role for the Ets-2 transcription factor. Mech Dev. 1997;68:127–137. doi: 10.1016/s0925-4773(97)00138-x. [DOI] [PubMed] [Google Scholar]
- 41.Marshall B C, Xu Q P, Rao N V, Brown B R, Hoidal J R. Pulmonary epithelial cell urokinase-type plasminogen activator. Induction by interleukin-1 beta and tumor necrosis factor-alpha. J Biol Chem. 1992;267:11462–11469. [PubMed] [Google Scholar]
- 42.Miralles F, Parra M, Caelles C, Nagamine Y, Felez J, Munoz-Canoves P. UV irradiation induces the murine urokinase-type plasminogen activator gene via the c-Jun N-terminal kinase signaling pathway: requirement of an AP1 enhancer element. Mol Cell Biol. 1998;18:4537–4547. doi: 10.1128/mcb.18.8.4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Naldini L, Tamagnone L, Vigna E, Sachs M, Hartmann G, Birchmeier W, Daikuhara Y, Tsubouchi H, Blasi F, Comoglio P M. Extracellular proteolytic cleavage by urokinase is required for activation of hepatocyte growth factor/scatter factor. EMBO J. 1992;11:4825–4833. doi: 10.1002/j.1460-2075.1992.tb05588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nerlov C, De Cesare D, Pergola F, Caracciolo A, Blasi F, Johnsen M, Verde P. A regulatory element that mediates co-operation between a PEA3-AP-1 element and an AP-1 site is required for phorbol ester induction of urokinase enhancer activity in HepG2 hepatoma cells. EMBO J. 1992;11:4573–4582. doi: 10.1002/j.1460-2075.1992.tb05559.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nerlov C, Rørth P, Blasi F, Johnsen M. Essential AP-1 and PEA3 binding elements in the human urokinase enhancer display cell type-specific activity. Oncogene. 1991;6:1583–1592. [PubMed] [Google Scholar]
- 46.O’Hagan R C, Hassell J A. The PEA3 Ets transcription factor is a downstream target of the HER2/Neu receptor tyrosine kinase. Oncogene. 1998;16:301–310. doi: 10.1038/sj.onc.1201547. [DOI] [PubMed] [Google Scholar]
- 47.O’Hagan R C, Tozer R G, Symons M, McCormick F, Hassell J A. The activity of the Ets transcription factor PEA3 is regulated by two distinct MAPK cascades. Oncogene. 1996;13:1323–1333. [PubMed] [Google Scholar]
- 48.Pages G, Lenormand P, L’Allemain G, Chambard J C, Meloche S, Pouyssegur J. Mitogen-activated protein kinases p42mapk and p44mapk are required for fibroblast proliferation. Proc Natl Acad Sci USA. 1993;90:8319–8323. doi: 10.1073/pnas.90.18.8319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reimold A M, Grusby M J, Kosaras B, Fries J W, Mori R, Maniwa S, Clauss I M, Collins T, Sidman R L, Glimcher M J, Glimcher L H. Chondrodysplasia and neurological abnormalities in ATF-2-deficient mice. Nature. 1996;379:262–265. doi: 10.1038/379262a0. [DOI] [PubMed] [Google Scholar]
- 50.Rørth P, Nerlov C, Blasi F, Johnsen M. Transcription factor PEA3 participates in the induction of urokinase plasminogen activator transcription in murine keratinocytes stimulated with epidermal growth factor or phorbol ester. Nucleic Acids Res. 1990;18:5009–5017. doi: 10.1093/nar/18.17.5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 52.Stacey K J, Fowles L F, Colman M S, Ostrowski M C, Hume D A. Regulation of urokinase-type plasminogen activator gene transcription by macrophage colony-stimulating factor. Mol Cell Biol. 1995;15:3430–3441. doi: 10.1128/mcb.15.6.3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vallone D, Battista S, Pierantoni G M, Fedele M, Casalino L, Santoro M, Viglietto G, Fusco A, Verde P. Neoplastic transformation of rat thyroid cells requires the junB and fra-1 gene induction which is dependent on the HMGI-C gene product. EMBO J. 1997;16:5310–5321. doi: 10.1093/emboj/16.17.5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Verde P, Boast S, Franze A, Robbiati F, Blasi F. An upstream enhancer and a negative element in the 5′ flanking region of the human urokinase plasminogen activator gene. Nucleic Acids Res. 1988;16:10699–10716. doi: 10.1093/nar/16.22.10699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wasylyk C, Bradford A P, Gutierrez-Hartmann A, Wasylyk B. Conserved mechanisms of Ras regulation of evolutionary related transcription factors, Ets1 and Pointed P2. Oncogene. 1997;14:899–913. doi: 10.1038/sj.onc.1200914. [DOI] [PubMed] [Google Scholar]
- 56.Whitmarsh A J, Shore P, Sharrocks A D, Davis R J. Integration of MAP kinase signal transduction pathways at the serum response element. Science. 1995;269:403–407. doi: 10.1126/science.7618106. [DOI] [PubMed] [Google Scholar]
- 57.Yamamoto H, Flannery M L, Kupriyanov S, Pearce J, McKercher S R, Henkel G W, Maki R A, Werb Z, Oshima R G. Defective trophoblast function in mice with a targeted mutation of Ets2. Genes Dev. 1998;12:1315–1326. doi: 10.1101/gad.12.9.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang B-S, Hauser C A, Henkel G, Colman M S, Van Beveren C, Stacey K J, Hume D A, Maki R A, Ostrowski M C. Ras-mediated phosphorylation of a conserved threonine residue enhances the transactivation activities of c-Ets1 and c-Ets2. Mol Cell Biol. 1996;16:538–547. doi: 10.1128/mcb.16.2.538. [DOI] [PMC free article] [PubMed] [Google Scholar]