Abstract
The QIAstat-Dx SARS-CoV-2 panel is a multiplex cartridge based assay based on real time PCR which can detect 17 respiratory viruses, including the novel coronavirus SARS-CoV-2. A syndromic approach is the need of the hour for COVID-19 diagnostics among patients presenting with respiratory symptoms. The present study was done to evaluate 120 archived respiratory clinical specimens for SARS-CoV-2 on the SARS-CoV-2 panel. Further, 27 specimens were tested for other respiratory viruses, in comparison with the BioFire RP1.7 platform. The sensitivity and specificity for SARS-CoV-2 on SARS panel was found to be 90.00 % and 100 % respectively, indicating good diagnostic accuracy. The positive predictive value was found to be 100 %, negative predictive value was found to be 99.93 % and accuracy was 99.93 %. Detection of other respiratory viruses observed a concordance of 77.7 %. Despite advantages of speed, minimal expertise and accurate results; significant costs and discrepancies at Ct >35 remain important limitations of the SARS panel.
Keywords: Point of care, COVID-19, CBNAAT, Clinical evaluation, Diagnostics
1. Introduction
The introduction of multiplex, point-of-care tests (POC) for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) diagnostics are game changing when deciding triaging, isolation and therapy of patients in populous countries like India. At the height of the coronavirus disease 2019 (covid-19) pandemic, standard testing methods included real time PCR for detection of pan β - Coronavirus (target E gene) and SARS-CoV-2 (target S gene, N gene, RdRp gene, Orf1b gene).(Group ICS et al., 2020) These assays are time, labour intensive and requiring advanced laboratory infrastructure. Further, common respiratory viruses like influenza, rhinovirus and coronavirus 229E are widespread in the community and are often difficult to clinically differentiate from SARS-CoV-2. The recently introduced, QIAstat-Dx Respiratory SARS-CoV-2 panel (SARS-CoV-2 Panel, Qiagen, Hilden, Germany) is a cartridge based nucleic acid amplification test which can be operated in the field, requiring minimal technical expertise with a run time of 70 min. (Boers et al., 2020) It can detect SARS-CoV-2 along with 17 additional respiratory viruses including Influenza A, Influenza B, Parainfluenza, Respiratory syncytial viruses, Coronavirus, Metapneumoviruses, Adenovirus and Rhino/Enterovirus similar to the QIAstat-Dx Respiratory Panel. The new SARS-CoV-2 Panel detects two genes of the SARS-CoV-2 virus genome (Orf1b/RdRp gene and E genes) with the same fluorescence channel and amplification of either or both target regions leads to a single fluorescence signal. (Leber et al., 2020; Visseaux et al., 2020) The aim of the present study was to evaluate the clinical performance of this assay against the standard Real Time PCR assay for SARS-CoV-2 (RealStar® SARS-CoV-2 RT-PCR Kit, Altona Diagnostics, Hamburg, Germany) and other respiratory viruses with Respiratory panel of multiplex assay, Biofire FilmArray RP1.7. (FilmArray, BioMérieux, France).
2. Methods
A retrospective study was conducted over 2 months on archived respiratory specimens (throat and nasopharyngeal swabs, NPS) from 1 November 2020 to 31 December 2020. The study was approved by Institutional Ethics committee and performed as per the Helsinki code. All clinical information were obtained from the hospital information system (HIS).
2.1. Specimen selection
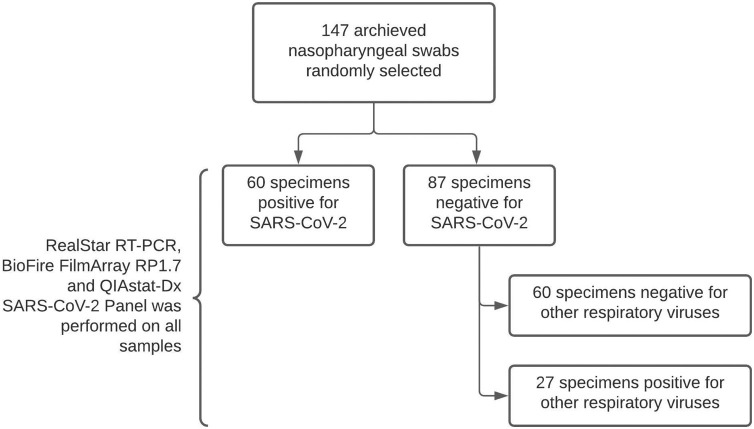
The study was done on archived samples kept at −80 °C in the Department repository. The study involved a total of 147 samples that were randomly selected from the database. Group 1 included 120 samples which were selected as per the SARS-CoV-2 RT-PCR results (60 positive and 60 negative) and they all were negative for any other respiratory virus, group 2 included 27 samples that were all negative for SARS-CoV-2 but positive for other respiratory viruses. (Fig. 1 ) This division was done to evaluate the QIAstat-Dx assay across all the respiratory viral pathogens. All the retrieved samples were tested, in a single freeze thaw cycle, in parallel on QIAstat-Dx and re tested on both RealStar RT-PCR as well as the Biofire FilmArray RP1.7 to reconfirm the earlier lab results.
Fig. 1.
Flow chart depicting sample selection.
2.1.1. Real time PCR (RT-PCR) for SARS CoV-2
The standard test used in our study for the detection of SARS-CoV-2 was RealStar® SARS-CoV-2 RT-PCR. The RealStar RT-PCR targets E gene and S gene for the detection of SARS-CoV-2. RNA extraction was done by QIAsymphony DSP Virus/pathogen Mini kit (Qiagen, Germany) and 10 μL of RNA elute, from an original 500 μL of VTM specimen, was required for the PCR. The assay requires 40 cycles of amplification and amplification for both the genes were considered as positive during the inclusion.
2.1.2. QIAstat-dx assay
All 147 specimens were tested in parallel on the SARS-CoV-2 panel. Manufacturers protocol was followed for testing on the QIAStat-Dx Analyzer 1.0. Briefly, 300 μL of clinical sample was added to the main port of the SARS-CoV-2 Panel cartridge and placed within the cartridge port. QIAstat-Dx Analyzer 1.0 Software processes controls, interprets the sample data including CT values and provides a final report.
2.1.3. BioFire film array respiratory panel RP 1.7
Detection of additional respiratory viruses was done on the automated multiplex system BioFire Film Array respiratory panel RP1.7. The BioFire is a closed system that performs sample preparation, reverse transcription and PCR in order to detect nucleic acid from multiple respiratory pathogens from a single nasopharyngeal swab.
All samples were processed and handled as per the recommended bio-safety guidelines. Any discrepant results were retested on both detection platforms (RealStar RT-PCR & QIAStat-Dx or BioFire FilmArray & QIAStat-Dx) for confirmation. Statistical analysis was performed using SPSS Ver 22 (IBM Corp., Armonk, NY, USA).
3. Results
We evaluated a total of 147 specimens on the SARS-CoV-2 Panel. The median age of the study group was 38 (IQR:15 – 68) years with Male: Female ratio was 3.1:1.
-
a)
Evaluation of SARS-CoV-2 Panel while using RealStar RT-PCR as the reference method for detection of SARS-CoV-2.
Out of 120 tested samples concordant results were seen in 114, 54 were positive and 66 were negative on the QIAStat-Dx. Taking prevalence of 0.7 % (as reported in India(Group ICS et al., 2020)) into consideration, the sensitivity of SARS-CoV-2 Panel was found to be 90.00 % (95 % CI:79.49%–96.24%) the specificity was found to be 100 % (95 % CI:94.04%–100.00%). (Table 1 ) Overall agreement of SARS-CoV-2 Panel with RT PCR was 95.0 %. There were 6 discordant samples which were positive on RealStar RT-PCR but negative on SARS-CoV-2 Panel. These discrepant cases were retested on RealStar RT-PCR and it was seen that there was amplification in both the genes but with higher CT values (>35).
Table 1.
Performance evaluation of QIAstat-Dx SARS-CoV-2 Panel when compared to RT-PCR.
| Value | 95 % CI | |
|---|---|---|
| Sensitivity | 90.00 % | 79.49%–96.24% |
| Specificity | 100.00 % | 94.04%–100.00% |
| Positive Likelihood Ratio | – | – |
| Negative Likelihood Ratio | 0.10 | 0.05 to 0.21 |
| Positive Predictive Value | 100.00 % | – |
| Negative Predictive Value | 99.93 % | 99.85%–99.97% |
| Accuracy | 99.93 % | – |
* Disease prevalence.0.70 %.
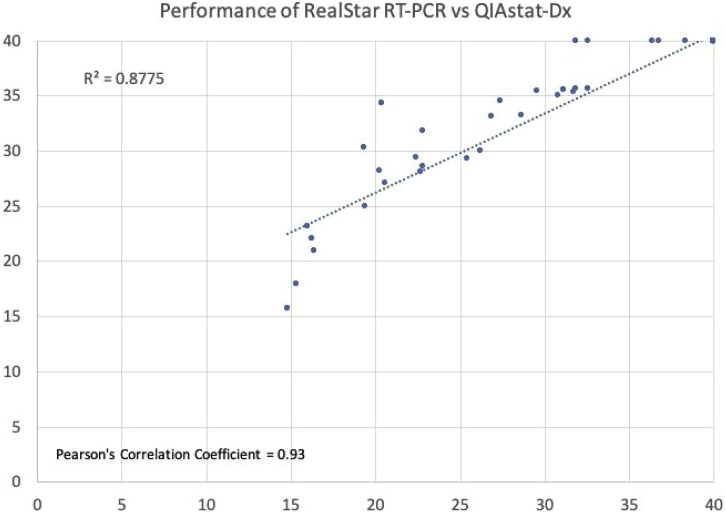
The performance of the assay was further evaluated and correlation was assessed. Pearson’s correlation coefficient was found to be 0.93 and R2 coefficient of determination was found to be 0.88 (Fig. 2 ). No other respiratory viruses were detected on the SARS-CoV-2 Panel among these samples.
-
b)
Evaluation of SARS-CoV-2 Panel while using BioFire RP1.7 for non-SARS-CoV-2 respiratory viruses.
Fig. 2.
Correlation between the cycle threshold (CT) values obtained by SARS-CoV-2 Panel on the QIAStat-Dx and by RealStar® RT-PCR for detection of SARS-CoV-2 with Pearson’s correlation coefficient was found to be 0.93. Negative samples have been illustrated as CT value of 40.
The detection of 17 other respiratory viruses performed on 27 known positive clinical samples with various respiratory pathogens. There was 77.7 % concordance between viruses detected on SARS-CoV-2 Panel and BioFire RP1.7. Discrepant results were observed in 6 specimens (Table 2 ). Among these, 4 specimens were negative on SARS-CoV-2 Panel but detected Human Rhinovirus/Enterovirus on BioFire RP1.7. Further, 2 specimens were found to show infection with dual respiratory viruses on BioFire RP1.7 (Cornonavirus 229E + Human Rhinovirus/Enterovirus and Coronavirus 229E + human metapneumovirus A/B). SARS-CoV-2 Panel could not detect Coronavirus 229E in both the specimens.
Table 2.
Results description of BioFire RP 1.7 vs SARS-CoV-2 Panel (QIAStat-Dx) along with concordance.
| Specimen No. | BioFire RP 1.7 | SARS-CoV-2 Panel | Concordant |
|---|---|---|---|
| 1 | Human Rhinovirus/Enterovirus | Human Rhinovirus/Enterovirus | Yes |
| 2 | Coronavirus HKU 1 | Coronavirus HKU 1 | Yes |
| 3 | Influenza AH1 2009 | Influenza AH1 2009 | Yes |
| 4 | Influenza AH3 | Influenza AH3 | Yes |
| 5 | Human Rhinovirus/Enterovirus | Human Rhinovirus/Enterovirus | Yes |
| 6 | Influenza AH1 2009 | Influenza AH1 2009 | Yes |
| 7 | Influenza AH1 2009 | Influenza AH1 2009 | Yes |
| 8 | Influenza AH1 2009 | Influenza AH1 2009 | Yes |
| 9 | Coronavirus 229E | Coronavirus 229E | Yes |
| 10 | Human Rhinovirus/Enterovirus | Human Rhinovirus/Enterovirus | Yes |
| 11 | Human Rhinovirus/Enterovirus | Human Rhinovirus/Enterovirus | Yes |
| 12 | Human Rhinovirus/Enterovirus | Human Rhinovirus/Enterovirus | Yes |
| 13 | Human Rhinovirus/Enterovirus | Human Rhinovirus/Enterovirus | Yes |
| 14 | Human Rhinovirus/Enterovirus | Human Rhinovirus/Enterovirus | Yes |
| 15 | Human Rhinovirus/Enterovirus | Human Rhinovirus/Enterovirus | Yes |
| 16 | Human Rhinovirus/Enterovirus | Human Rhinovirus/Enterovirus | Yes |
| 17 | Human Rhinovirus/Enterovirus | TND | No |
| 18 | Human Rhinovirus/Enterovirus | TND | No |
| 19 | Coronavirus 229E | Coronavirus 229E | Yes |
| 20 | Human Rhinovirus/Enterovirus | Human Rhinovirus/Enterovirus | Yes |
| 21 | Cornonavirus 229E + Human Rhinovirus/Enterovirus | Human Rhinovirus/Enterovirus | No |
| 22 | Influenza AH3 | Influenza A + influenza A H3 | Yes |
| 23 | Human Rhinovirus/Enterovirus | Human Rhinovirus/Enterovirus | Yes |
| 24 | Human Rhinovirus/Enterovirus | TND | No |
| 25 | Human Rhinovirus/Enterovirus | TND | No |
| 26 | Coronavirus 229E + human metapneumovirus A/B | Human metapneumovirus A/B | No |
| 27 | Influenza AH3 | Influenza A + influenza A H3 | Yes |
TND – Target Not Detected.
4. Discussion
The arrival of POC tests for SARS-CoV-2 diagnostics are game changing when deciding triaging, isolation implementation and therapy of patients.(Boers et al., 2020) The SARS-CoV-2 Panel provides a novel sensitive and specific method to diagnose multiple viral infections in at risk patients in the midst of a pandemic. Delay in reporting due to centralised laboratory PCR testing is a significant challenge and SARS-CoV-2 Panel represents an important step to improve detection potential co-infections or superinfections.(Audi et al., 2020; Brendish et al., 2020) Our study found good performance characteristics, similar to other published studies on the QIAstat-Dx, where a positive percent agreement and negative percent agreement with RT-PCR as the reference standard was more than 90 %.(Visseaux et al. (2020); Lebourgeois et al. (2021)) Further, correlation between the CT values on the RealStar and QIAstat-Dx platforms were observed. We observed discrepant results on SARS-CoV-2 Panel in 6 cases with CT values >35 on the Qiastat-Dx. The higher CT values of these cases may indicate presence of degraded viral RNA and viral clearance or possible false positive on RT PCR. (Drew et al., 2020) Now even Indian council of medical research (ICMR) has stated in their guidelines that samples on RT-PCR should be given positive only with Ct values < 35. (Aranha et al., 2021) However, earlier all amplifications in both the genes even if > 35 Ct value were considered as positive while reporting. The SARS-CoV-2 Panel was also evaluated for detection of respiratory viruses other than SARS-CoV-2 using the BioFire RP1.7 as the reference standard. Concordance was found to be 77.7 % between both platforms. This could be due to different primer targets for various viruses on BioFire RP1.7 and SARS-CoV-2 Panel. Thus, a larger prospective study is required to assess the sensitivity of these platforms in the Indian setting. Sensitivity for detection of Human Rhinovirus/Enterovirus and coronavirus 229E on the SARS-CoV-2 Panel appears to be poor in comparison to the Biofire RP 1.7. However, present study could not accurately assess detection of non-SARS-CoV-2 respiratory viruses during the ongoing pandemic as due to the ongoing pandemic and lockdown situation, requests for assessing other respiratory virus were drastically reduced. Present study was further unable to evaluate detection of SARS-CoV-2 by the newer BioFire RP2.1 panel which includes targets for COVID-19 due to non-availability of this revised version of assay at our laboratory. The study is further limited by its small sample size and lack of direct NP/OP swab assessment in the SARS-CoV-2 Panel cartridge. Ease of specimen loading, rapid results and minimal need of technical expertise are in favour of use of the SARS-CoV-2 Panel. However, the decreased sensitivity among SARS-CoV-2 low viral load specimens remains an important limitation of this assay, although with limited clinical relevance.
In conclusion, performance and costs of implementation need to be further evaluated with a larger number of subjects before application in routine investigations in India. Furthermore, evaluation of this point-of-care test in critical care settings is desirable as SARS-CoV-2 becomes a regular differential among respiratory viral infections.
Author Contribution
AG retrieved samples, performed QIAStat-Dx assay, data analysis and drafted the manuscript, AS performed QIAStat-Dx assay and edited manuscript, SR assisted in retrieving samples and performed QIAStat-Dx assay, DP edited the manuscript, RA designed the study and edited manuscript, EG conceived/designed the study and edited/approved final manuscript.
Funding
None.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
We are thankful to Mr. Keshaw Singh and Mr. Jagat Singh Solanki, along with all technical staff for their assistance during sample collection, storage and processing.
References
- Aranha C., Patel V., Bhor V., Gogoi D. Cycle threshold values in RT-PCR to determine dynamics of SARS-CoV-2 viral load: an approach to reduce the isolation period for COVID-19 patients. J. Med. Virol. [Internet]. 2021;15(July) doi: 10.1002/jmv.27206. [cited 2021 Aug 9];n/a(n/a). Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audi A., AlIbrahim M., Kaddoura M., Hijazi G., Yassine H.M., Zaraket H. Seasonality of Respiratory Viral Infections: Will COVID-19 Follow Suit? Front. Public Health. 2020;8:576. doi: 10.3389/fpubh.2020.567184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boers S.A., Melchers W.J.G., Peters C.J.A., Toonen M., McHugh M.P., Templeton K.E., et al. Multicenter Evaluation of QIAstat-Dx Respiratory Panel V2 for Detection of Viral and Bacterial Respiratory Pathogens. Carroll KC, editor. J. Clin. Microbiol. 2020;58(6 May):e01793–19. doi: 10.1128/JCM.01793-19. 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendish N.J., Poole S., Naidu V.V., Mansbridge C.T., Norton N.J., Wheeler H., et al. Clinical impact of molecular point-of-care testing for suspected COVID-19 in hospital (COV-19POC): a prospective, interventional, non-randomised, controlled study. Lancet Respir. Med. 2020;8(12 December):1192–1200. doi: 10.1016/S2213-2600(20)30454-9. 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew R.J., O’Donnell S., LeBlanc D., McMahon M., Natin D. The importance of cycle threshold values in interpreting molecular tests for SARS-CoV-2. Diagn. Microbiol. Infect. Dis. 2020;98(3 November) doi: 10.1016/j.diagmicrobio.2020.115130. 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Group ICS, Team CE& DM, Team C.L., Team V. Laboratory surveillance for SARS-CoV-2 in India: performance of testing & descriptive epidemiology of detected COVID-19, January 22 - April 30, 2020. Indian J. Med. Res. 2020;151(5 May):424. doi: 10.4103/ijmr.IJMR_1896_20. 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leber A.L., Lisby J.G., Hansen G., Relich R.F., Schneider U.V., Granato P., et al. Multicenter Evaluation of the QIAstat-Dx Respiratory Panel for Detection of Viruses and Bacteria in Nasopharyngeal Swab Specimens. Tang Y-W, editor. J. Clin. Microbiol. 2020;58(5 April):e00155–20. doi: 10.1128/JCM.00155-20. 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebourgeois S., Storto A., Gout B., Le Hingrat Q., Ardila Tjader G., Cerdan M del C., et al. Performance evaluation of the QIAstat-Dx® respiratory SARS-CoV-2 panel. Int. J. Infect. Dis. 2021;107(June):179–181. doi: 10.1016/j.ijid.2021.04.066. 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visseaux B., Le Hingrat Q., Collin G., Bouzid D., Lebourgeois S., Pluart D., et al. Evaluation of the QIAstat-Dx Respiratory SARS-CoV-2 Panel, the first rapid multiplex PCR commercial assay for SARS-CoV-2 detection. J. Clin. Microbiol. 2020;27(April):58. doi: 10.1128/JCM.00630-20. [DOI] [PMC free article] [PubMed] [Google Scholar]