The inner workings of the clock system rely on communicating signals between distal tissues to maintain daily metabolism.
Abstract
The mammalian circadian clock, expressed throughout the brain and body, controls daily metabolic homeostasis. Clock function in peripheral tissues is required, but not sufficient, for this task. Because of the lack of specialized animal models, it is unclear how tissue clocks interact with extrinsic signals to drive molecular oscillations. Here, we isolated the interaction between feeding and the liver clock by reconstituting Bmal1 exclusively in hepatocytes (Liver-RE), in otherwise clock-less mice, and controlling timing of food intake. We found that the cooperative action of BMAL1 and the transcription factor CEBPB regulates daily liver metabolic transcriptional programs. Functionally, the liver clock and feeding rhythm are sufficient to drive temporal carbohydrate homeostasis. By contrast, liver rhythms tied to redox and lipid metabolism required communication with the skeletal muscle clock, demonstrating peripheral clock cross-talk. Our results highlight how the inner workings of the clock system rely on communicating signals to maintain daily metabolism.
INTRODUCTION
In mammals, the circadian clock system executes daily fluctuations of homeostasis, enabling organisms to temporally coordinate physiology and to align it with geophysical time (1). Models of clock disruption (including clock-mutant mice, jet lag paradigms, nutritional challenges, and aging) produce behavioral, cognitive, and metabolic phenotypes, placing circadian rhythms at the center of homeostatic control (2–4). Notably, circadian misalignment is linked to disease and aging and is reminiscent of mutant clock phenotypes, underscoring the importance of uncovering the connectivity of the clock system (2, 5).
Organizing systemic metabolism across time and space is a massive biological challenge. As molecular clocks are in virtually all cells of the body, a multilayered system is required for temporal coordination of metabolism (6, 7). While the suprachiasmatic nucleus (SCN) and other hypothalamic neurons set the pace of peripheral oscillators through behavioral, neuronal, and hormonal signals (8), numerous tissue-specific clock mutants demonstrate the physiological importance and necessity of local clocks in peripheral organs (9–12). We recently demonstrated, however, that the hepatic and epidermal clocks are sufficient only for a small fraction of rhythmic functions, which means that communication between distal clocks is paramount for the system (13, 14).
Undoubtedly, the brain is a major source of circadian regulation for peripheral tissues, with myriad signals emanating from it (8). In setting the sleep-wake cycle, the SCN evokes the feeding-fasting cycle, a major zeitgeber (synchronizer) for the periphery (15–18). Numerous studies demonstrate that both the liver clock and feeding rhythms are required to drive molecular oscillations (17–23), yet the interaction between the two has been poorly defined because of the lack of appropriate animal models. Here, we present a model in which the interaction between feeding and the liver clock can be isolated in vivo. With this approach, the molecular underpinnings of this interaction can be functionally dissected, and the contribution of extrahepatic clocks can be assessed.
The transcription factor (TF) BMAL1 (brain and muscle Arnt-like protein-1) is a key component of the molecular clock underlying circadian rhythmicity in mammals (24). We have previously generated a mouse model in which BMAL1 is expressed exclusively in hepatocytes (Liver-RE) (13). Devoid of all other clocks and their signals, the Liver-RE model is well suited to test the interaction between the tissue-autonomous clock and feeding rhythm, as well as to identify the contribution of extrahepatic clocks. This approach allowed us to demonstrate a mechanism through which BMAL1 and CEBPB (CCAAT/enhancer binding protein-β) coordinate daily metabolic transcriptional programs. By probing genes and metabolites that were not restored in the presence of the liver clock and a feeding rhythm through additional functional experiments, the muscle clock was identified as a regulator of liver oscillations.
RESULTS
Effects of imposed feeding-fasting rhythms on systemic metabolism and liver clock function
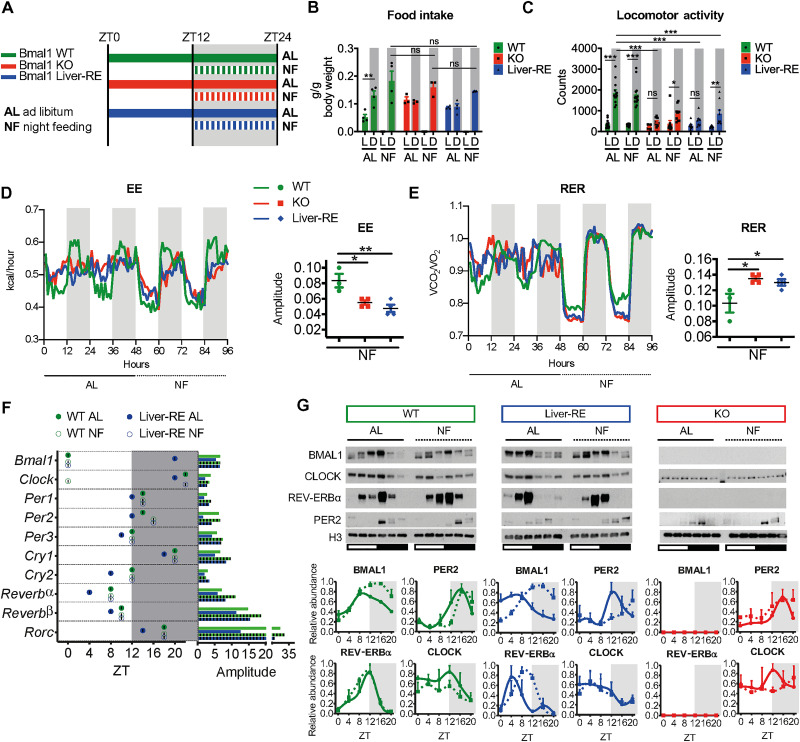
To reveal the impact of feeding-fasting rhythms on the autonomous liver clock, we housed 8- to 10-week-old male wild-type (WT), Bmal1–KO (knockout) (Bmal1 stop-Fl; KO), or Bmal1 Liver-RE–constituted (Bmal1 stop-Fl, AlfP-cre+/−; Liver-RE) mice under 12:12 hour light:dark conditions and provided either ad libitum (AL) access to normal chow or restricted food availability to the 12-hour dark phase for 2 weeks [night feeding (NF)] (Fig. 1A). Neither KO nor Liver-RE mice exhibited a detectable feeding-fasting rhythm under AL conditions, consistent with previous findings (Fig. 1B and fig. S1, A and B) (13). Consistent with the described accelerated aging phenotype of Bmal1 KO mice (13, 25), KO and Liver-RE mice weigh less than WT (fig. S1C). However, no differences were observed between NF- and AL-fed groups for total food consumption or weight for any genotype (fig. S1, A to C). NF induced a de novo feeding-fasting rhythm in KO and Liver-RE mice (Fig. 1B and fig. S1, A and B). While NF did not alter locomotor activity in WT mice, it increased the dark-phase ambulatory activity in Liver-RE and KO mice, leading to the appearance of detectable rhythms in locomotion with total activity levels remaining lower in KO and Liver-RE than WT (Fig. 1C and fig. S1D). For WT mice, NF had small effects on energy expenditure (EE) yet markedly increased the amplitude of respiratory exchange ratio (RER) (Fig. 1, D and E, and fig. S1E). NF in KO and Liver-RE mice induced metabolic rhythms [in line with a previous study on Bmal1-KO mice (26)] and strong rhythms of RER, indicative of a de novo switch from using lipids to carbohydrates between day and night (Fig. 1E, and fig. S1E). Compared to WT, the amplitude of RER rhythms under NF was higher in Liver-RE and KO, while the amplitude of EE was lower (Fig. 1, D and E). In addition, the rhythm of EE was phase-delayed in Liver-RE and KO compared to WT, an observation that may be tied to the timing of food intake during the dark phase (fig. S1A). Serum hormone and metabolite measurements also revealed similar metabolic states in Liver-RE and KO under NF. NF induced a day-night [zeitgeber time 4 (ZT4) versus ZT16] difference in insulin levels (fig. S1F) and tended to lower free fatty acids at ZT16, while levels at ZT4 remained significantly lower than WT (fig. S1G). Triglycerides were elevated in KO AL at ZT16 compared to both WT and Liver-RE but were restored by NF (fig. S1H). In addition, blood glucose tended to be higher at all time points in KO and Liver-RE mice under AL, whereas NF induced a rhythm in blood glucose tied to feeding and fasting in all genotypes (ZT8 versus ZT16) (fig. S1I). Thus, imposing a feeding rhythm induced a comparable behavioral and systemic metabolic state in KO and Liver-RE mice.
Fig. 1. Effects of an imposed feeding rhythm on systemic metabolism and the autonomous liver clock.
(A) Summary of experimental design. Horizontal bars indicate timing of food availability, and shaded area represents dark phase. (B) Food intake from metabolic cages (means ± SEM, n = 3 to 4 per group). L, light phase; D, dark phase. Two-way analysis of variance (ANOVA) with Bonferroni post hoc test; **P < 0.01. (C) Ambulatory locomotor activity measured from home cage (means ± SEM; WT, n = 13; KO, n = 10; Liver-RE, n = 9). Two-way ANOVA with Bonferroni post hoc test; *P < 0.05, **P < 0.01, and ***P < 0.001. ns, not significant. (D and E) Metabolic cage assessment of mice in light-dark. Traces show group averages. BioDare2 eJTK_CYCLE amplitude is shown to the right; P values for each parameter of individual mice are shown in fig. S1D. n = 3 to 4 per group; one-way ANOVA with Tukey post hoc tests; *P < 0.05 and **P < 0.01. (F) JTK_CYCLE phase and amplitude from total RNA sequencing (RNA-seq) (n = 3). (G) Western blot analysis of core clock components from liver chromatin fractions. Representative blots from three independent experiments are shown. Densitometry analysis displayed as means ± SEM, n = 3 per group, per time point, normalized to H3, except BMAL1 signal that is expressed as ratio of upper to lower band.
Next, we assessed the impact of NF on the activity of the core clock transcriptional loop in the liver by performing RNA sequencing (RNA-seq) generated over the daily cycle. While NF did not markedly change oscillations of core clock gene expression in WT mice (Fig. 1F and fig. S1J), it corrected the phase of core clock genes and modestly enhanced their amplitude in Liver-RE mice (Fig. 1F and fig. S1J). Realignment of core clock functions in Liver-RE mice was also found at the protein level, as evidenced by the timing of BMAL1, REV-ERBα, and CLOCK accumulation at chromatin (Fig. 1G). As reported previously, PER2 protein oscillated in KO mice under light-dark conditions (13, 27). In contrast, NF did not substantially affect protein levels of core clock components in either WT or KO mice (Fig. 1G). Hence, subjecting Liver-RE mice to a feeding-fasting rhythm corrects the phases of the core clock.
Daily liver transcriptome reveals drivers of nonautonomous oscillations
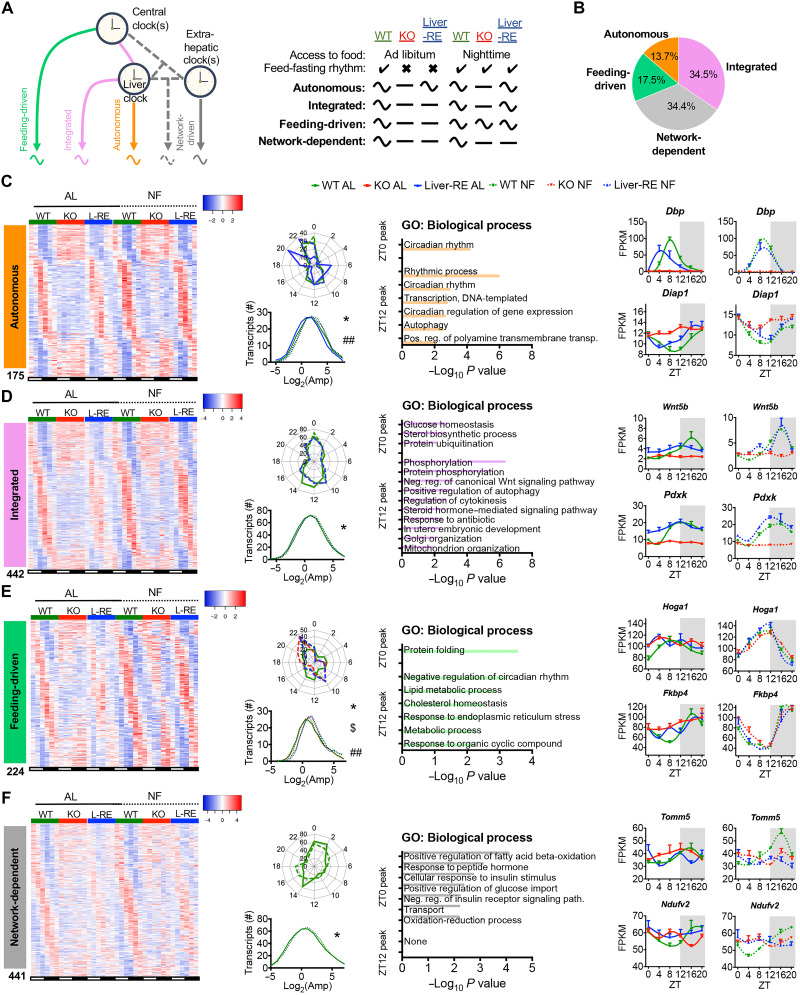
We next studied how feeding-fasting rhythms, local clocks, and the clock network interact to drive circadian gene expression. Principal components analyses (PCAs) of RNA-seq over 24 hours revealed that transcriptomic profiles of WT and Liver-RE clustered more closely to each other than to KO profiles under both AL and NF (fig. S2A). Compared to AL, NF profiles for all genotypes were more distinct between ZTs, while samples within ZTs were more clustered, illustrating coherence of gene expression over 24 hours (fig. S2A). Next, we used JTK_CYCLE (28) (fig. S2B and data S1) and BioCycle (29) (fig. S2C) to identify genes that oscillate with a 24-hour period. NF greatly increased the number of oscillatory genes in all genotypes, although Liver-RE and WT mice still displayed more oscillatory genes than KO mice (fig. S2D). We also detected 248 genes that exhibited de novo oscillations upon NF in all genotypes, oscillating with the same phase and amplitude (fig. S2, E to H); as these oscillations appeared to be driven by clock-independent mechanisms of NF, we removed them from downstream analysis. To classify drivers of gene oscillations in the liver, we defined four main classes of rhythmic genes: (i) autonomous (only liver clock required), (ii) integrated (both liver clock and feeding rhythm), (iii) feeding-driven (only feeding rhythm), and (iv) network-dependent (not sufficient with both liver clock and feeding rhythm) (Fig. 2, A and B). Similar to our previous reports (13, 14), 13.7% of oscillating genes were autonomous (Fig. 2, B and C). NF increased the amplitude and corrected the phase of autonomous genes, in line with its effect on the core clock (Figs. 1G and 2C). Autonomous genes included core clock genes and were mainly enriched for circadian processes (Fig. 2C). Of the 1236 nonautonomous genes, 666 (53.9%) gained oscillation under NF in Liver-RE. Of those, 442 (66.4%) were integrated by hepatic BMAL1, while only 224 (33.6%) oscillated in KO and were thus feeding-driven (Fig. 2, D and E). Phase and amplitude of integrated genes were similar in WT and Liver-RE (Fig. 2D). Moreover, the amplitude of integrated genes in WT was enhanced by NF as compared to AL, indicating that oscillation of these genes is cooperatively controlled by the hepatic clock and feeding (Fig. 2D). Feeding-driven genes displayed similar phases in Liver-RE and KO mice, yet amplitude was significantly lower in KO, suggesting reliance on the hepatic clock for robustness (Fig. 2E). Gene ontology (GO) analysis revealed that integrated and feeding-driven genes are functionally distinct. Integrated genes were enriched for pathways involving phosphorylation, steroid hormone signaling, and glucose homeostasis, among others (Fig. 2D), whereas feeding-driven genes were enriched for proteostasis-type pathways, such as protein folding, and for metabolic processes, including lipid and cholesterol metabolism (Fig. 2E). Unexpectedly, a large portion of nonautonomous genes (441, 34.4%) failed to oscillate in Liver-RE mice under NF (Fig. 2F); these genes were mostly enriched for oxidation-reduction and fatty acid β-oxidation metabolic processes and are likely driven by the clock network as a whole or by specific extrahepatic clocks (Fig. 2F).
Fig. 2. Defining drivers of gene oscillations in the liver.
(A to F) Data presented are liver transcriptome at six time points over the diurnal cycle (ZT0, ZT4, ZT8, ZT12, ZT16, and ZT20) by RNA-seq (n = 3) and JTK_CYCLE rhythmicity detection (P < 0.01). (A) Gene classification scheme used to determine the drivers of oscillating genes in the liver. Curved and flat lines represent oscillating and nonoscillating genes, respectively. (B) Breakdown of oscillating genes by mechanism, displayed as the percentage of total oscillating in WT under either AL or NF. (C to F) Features of each set of oscillating genes. Left: Phase-sorted heatmap. Middle left: Polar histogram of peak phase and amplitude distribution (one-way ANOVA with Newman-Keuls post hoc tests: autonomous, *WT AL versus Liver-RE AL and ##Liver-RE AL versus NF; integrated, *WT AL versus NF; feeding-driven, *WT NF versus KO NF, $KO NF versus Liver-RE NF, and ##WT AL versus NF; network-dependent, *WT AL versus NF; *P < 0.05. Middle right: Pathway enrichment analysis for the two main peaks of gene expression detected (ZT0 peak = ZT20 to ZT4, ZT12 peak = ZT8 to ZT16, P < 0.01). Right: Example genes (means ± SEM, n = 3 per group, per time point).
To add additional statistical power in support of each gene class, we performed differential rhythmicity analysis using LimoRhyde (30). Whereas JTK_CYCLE detects rhythmicity within each group individually, LimoRhyde analysis enables rhythmicity to be statistically compared between groups. To this end, we identified sets of genes that oscillate under one condition and not the other (P value for differential rhythmicity < 0.01). The sets of genes generated by LimoRhyde—which correspond to the autonomous, integrated, feeding-driven, and network-dependent gene classes—returned similar pathway enrichments to JTK_CYCLE analysis (fig. S3, A to E). For example, oxidation-reduction is an enriched pathway for both network-dependent genes and LimoRhyde WT NF versus Liver-RE NF (fig. S3D).
CEBPB co-occupies chromatin with BMAL1 in liver and supports rhythmic transcription
The feeding-fasting rhythm affected hepatic gene oscillations in several ways: It corrected the phase, increased the amplitude of autonomous genes, and induced de novo oscillation of genes in a hepatic clock–dependent manner (Fig. 2). To further dissect the transcriptional interplay, we measured RNA-seq coverage along intronic and exonic regions. This approach allows transcriptional and posttranscriptional contributions to be distinguished (31). A large percentage of integrated genes oscillated at both the intronic and exonic levels (67% intron/exon versus 33% exon only; fig. S4A), indicating that transcriptional mechanisms primarily regulate oscillation of this gene class.
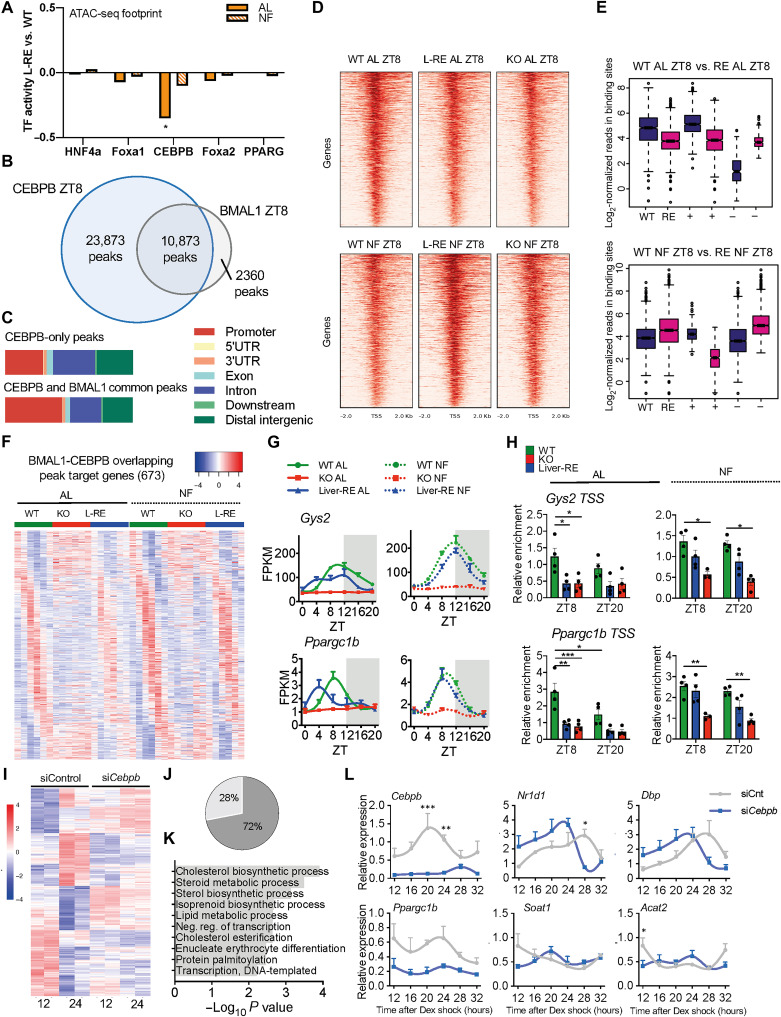
To explore the transcriptional mechanisms underlying rhythmicity of integrated genes, we used MotifMap (32, 33) to perform TF binding site (TFBS) enrichment analysis. CLOCK:BMAL1 was among the top enriched DNA binding motifs, confirming that the core clock is needed for rhythmic expression of these genes (fig. S4B). In addition, there was significant enrichment for several known liver metabolic regulators, such as HNF4α, CEBPB, and FOXA (fig. S4B). To investigate whether activities of these TFs differed between WT, KO, and Liver-RE mice, we determined chromatin accessibility by ATAC-seq (assay for transposase-accessible chromatin sequencing) at ZT8, which coincides with maximal DNA binding of BMAL1. We next merged our ATAC-seq dataset with our previously published BMAL1 chromatin immunoprecipitation followed by sequencing (ChIP-seq) (13) to identify regions of accessible chromatin bound by BMAL1. Consistent with a previous study (34), footprint analysis (35, 36) detected enrichment for HNF4a, FOXA1, and CEBPB at BMAL1-bound sites (data S3). Quantification of footprint signal in Liver-RE and KO revealed a significant reduction for CEBPB under AL compared to WT. Conversely, NF restored CEBPB signal in both Liver-RE and KO (Fig. 3A and fig. S4C), indicating that appropriate binding of CEBPB to chromatin may require a feeding rhythm. MotifMap analysis of the four gene classes revealed that CEBPB displayed the highest enrichment at integrated genes, followed by autonomous, and then feeding-driven genes (integrated q = 1.01 × 10−25; autonomous q = 1.35 × 10−17; feeding-driven q = 2.19 × 10−15; network-dependent q = 1.11 × 10−4; fig. S4D). This indicates that CEBPB is present at genes regulated by both the core clock and feeding. Although mRNA levels of Cebpb appear to oscillate, we did not observe significant time-dependent changes at the protein level (fig. S4, E and F). However, coimmunoprecipitation from liver chromatin fractions showed that CEBPB interacts with BMAL1 at ZT8 (fig. S4G), suggesting that CEBPB and BMAL1 may cooperate at chromatin.
Fig. 3. CEBPB cooperates with hepatic BMAL1 to drive oscillations in gene expression.
(A) Footprint of TF binding within regions of accessible chromatin at BMAL1 sites [n = 2 per group; unpaired t test; *P < 0.05, Liver-RE (L-RE) versus WT]. PPARG, peroxisome proliferator–activated receptor gamma. (B) Venn diagram of CEBPB and BMAL1 binding sites in livers of WT AL. (C) CEBPB ChIP peak distribution at ZT8. UTR, untranslated region. (D) Heatmap of CEBPB ChIP-seq binding profiles at CEBPB and BMAL1 common sites. (E) Boxplot of read distribution at CEBPB-BMAL1 common sites. + and − indicate sites with higher enrichment in WT or Liver-RE, respectively (DiffBind, P < 0.05; two-sided Wilcoxon-Mann-Whitney test; n = 2 per group). (F) Heatmap of oscillating target genes bound by CEBPB and BMAL1. (G) BMAL1/CEBPB target gene examples (means ± SEM, n = 3 per time point, per group). (H) CEBPB ChIP at Gys2 and Ppargc1b promoters (means ± SEM, n = 4 per time point, per group; two-way ANOVA with Holm-Sidak post hoc tests; *P < 0.05, **P < 0.01, and ***P < 0.001). (I) Hierarchical clustering of genes differentially expressed at 12 versus 24 hours after DEX in siControl (siCnt) AML12 cells (n = 2 per time point per group; FDR < 0.05). (J) Effect of siCebpb on time-regulated genes. (K) Biological processes of genes that lose time dependency on siCebpb. (L) Examples of genes modulated by siCebpb (means ± SEM, n = 3 per time point, per group; two-way ANOVA with Holm-Sidak post hoc tests; *P < 0.05, **P < 0.01, and ***P < 0.001).
To investigate genomic interplay of CEBPB and BMAL1, we performed ChIP-seq on liver from WT mice at ZT8 and ZT20. We identified 45,547 and 43,035 replicate-concordant CEBPB peaks at ZT8 and ZT20, respectively. Using our previously published BMAL1 ChIP-seq dataset (13), we calculated the peak overlap with BMAL1. This analysis revealed extensive overlap, as 82% of BMAL1 peaks overlapped with CEBPB (10,837 peaks at ZT8; Fig. 3B and data S4) and the majority of CEBPB sites (97%) did not change between ZT8 and ZT20 (fig. S5, A and B). Thus, recruitment of CEBPB to DNA does not appear to be rhythmic. CEBPB binding at BMAL1 common sites displayed a greater occupancy at promoter regions compared to noncommon sites (Fig. 3C). In line with the footprint results (Fig. 3A and fig. S4C), binding strength of CEBPB was decreased in Liver-RE and KO mice compared to WT under AL and increased in NF (Fig. 3D). Whereas the genomic binding of CEBPB was unaffected by genotype and feeding (fig. S5C), differential binding showed that overall binding was reduced in Liver-RE and KO fed AL, with only a minority of regions displaying increased binding affinity (Fig. 3E and fig. S5D). Notably, introducing a feeding rhythm was sufficient to rescue overall binding strength of CEBPB to DNA in Liver-RE and, to a lesser extent, in KO mice (Fig. 3E and fig. S5D).
BMAL1 binding alone is not sufficient to drive rhythmic transcription, raising the possibility that tissue-specific TFs located at common loci may contribute to rhythmic output (34, 37–40). We thus asked whether CEBPB cooperates with BMAL1 to drive rhythmic oscillation of target genes. The promoter region of ~46% of oscillating genes in WT mice were cobound by CEBPB and BMAL1. Under AL, BMAL1-CEBPB targets oscillated predominantly in WT, while NF induced strong oscillation in Liver-RE but not KO [121 genes oscillating in Liver-RE AL (18%); genes that recovered oscillation were 409 (61%) in Liver-RE NF and 178 (26%) in KO NF; Fig. 3F].
Moreover, phase was similar between NF WT and Liver-RE mice, with most genes peaking during the early night (fig. S5E). GO analysis revealed that shared BMAL1 and CEBPB targets are involved in rhythmic process (e.g., Rev-erbα and Dbp), lipid metabolic processes (e.g., Ppargc1b), and glucose metabolic processes (e.g., Gys2) (Fig. 3G and fig. S5F). ChIP analyses at Gys2 and Ppargc1b promoters confirmed the reduction of CEBPB binding at these sites in KO and Liver-RE mice under AL. NF significantly restored binding in Liver-RE mice (Fig. 3H). Additional transcriptional mechanisms are likely required for complete output of hepatic oscillations. HNF4a, recently implicated in hepatic circadian regulation (41), and FOXA1 were also highly enriched at BMAL1-bound sites; however, our footprint analysis failed to detect any significant difference in their transcriptional activity, suggesting that the genomic localization of these TFs is regulated by divergent mechanisms.
CEBPB modulates rhythmic gene expression in hepatocytes
To examine whether CEBPB is involved in rhythmic gene expression in hepatocytes, we used RNA interference to acutely knock down Cebpb expression in dexamethasone (DEX)–synchronized alpha mouse liver 12 (AML12) cells, a hepatocyte cell line (fig. S6A). RNA-seq analysis at two time points after DEX synchronization (12 and 24 hours) identified 502 genes with altered expression [false discovery rate (FDR) < 0.05; data S5] between the two time points in control cells (siControl). A total of 72% of these genes (358 of 502 genes) lost the time-dependent variation upon knockdown (KD) of Cebpb (Fig. 3, I and J). Consistent with in vivo results, affected genes showed significant enrichment for lipid metabolic processes (Fig. 3K). Moreover, ChIP enrichment analysis identified CEBPB as the top regulator of the 358 genes deregulated in KD cells (fig. S6B), indicating that a substantial percentage of these genes are direct targets of CEBPB. Multiple genes bound by CEBPB and BMAL1 displayed deregulated rhythmicity in CEBPB KD cells (Fig. 3L). Results obtained in AML12 cells were further confirmed in primary hepatocytes (fig. S6, C and D). In addition, CEBPB KD in unsynchronized cells led to deregulation of BMAL1-CEBPB target genes (fig. S6E). Together, these data demonstrate that DNA binding of CEBPB is dependent on a feeding-fasting rhythm and establish CEBPB as a liver TF that coregulates rhythmic expression of BMAL1 target genes.
Metabolome analysis reveals interplay of liver clock and feeding in energy metabolism
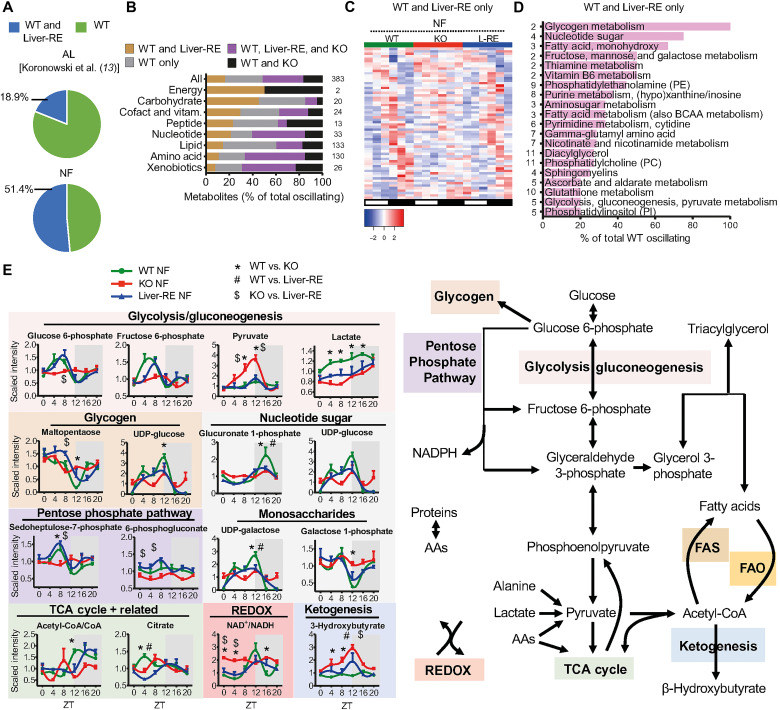
Since we observed restoration of genes involved in carbohydrate and lipid metabolism in NF Liver-RE mice, we sought to determine their functional status by probing daily metabolite oscillations. Liver profiles of 894 metabolites were generated over the daily cycle under NF by ultraperformance liquid chromatography–tandem mass spectroscopy (LC-MS/MS) (data S6). PCA and analysis of variance (ANOVA) comparisons at each ZT show that WT and Liver-RE profiles were more similar to each other than to KO profiles globally (13.24% of variation; fig. S7, A and B). Differences were notable at the most fasted time point (ZT12) and at the end of the feeding period (ZT0); for instance, at ZT12, >30% of carbohydrates, peptides, and cofactors/vitamins were altered in KO, while most were unaltered in Liver-RE (fig. S7B). In contrast, a similar number of altered metabolites were observed in KO and Liver-RE at ZT4 and ZT16, the time points that coincide with the observed phase shift of EE in KO and Liver-RE (Fig. 1D and fig. S7B).
JTK_CYCLE and BIO_CYCLE algorithms yielded similar results for the number of oscillating metabolites (fig. S8A). From JTK_CYCLE, 51.4% of WT oscillating metabolites also oscillated in Liver-RE, up from 18.9% in our previous study under AL (Fig. 4A and fig. S8, A and B) (13). A total of 135 metabolites (35% of WT) oscillated in all genotypes (Fig. 4B and fig. S8, A to E). Considering that we detected 40% more oscillating metabolites in WT with NF than AL (13), some of the commonly oscillating metabolites are gained by the strengthening of the endogenous feeding rhythm, while others are driven by the endogenous feeding rhythm (as observed in transcriptome analysis). This set of metabolites, which is enriched for amino acids, nucleotides, and xenobiotics, has a similar phase and amplitude in all groups, further supporting this notion (Fig. 4B and fig. S8, C to E). Oscillation of orotate is proportional to food intake for each genotype, demonstrating a clock-independent effect of NF (fig. S8E) (13). Alternatively, the nonautonomous oscillations of urea cycle and some tryptophan metabolites were restored in Liver-RE and KO, hence likely driven by the endogenous feeding rhythm (fig. S8E) (13).
Fig. 4. Influence of local BMAL1 on liver metabolite oscillations under NF.
(A to E) Metabolite profiles from six time points (ZT0, ZT4, ZT8, ZT12, ZT16, and ZT20) over the diurnal cycle in the liver under NF (n = 4, per group, per time point) and JTK_CYCLE (P < 0.05) rhythmicity detection. (A) Pie chart showing the percentage of metabolites oscillating in both WT and Liver-RE under AL (AL, previous report) or NF conditions. (B) Overlap of oscillating metabolites broken down into chemical class (with the number in each class shown to the right). Data show percentages of total oscillating metabolites for that class. (C) Phase-sorted heatmap of metabolites oscillating in WT and Liver-RE only. (D) Pathways of WT-only and Liver-RE–only oscillating metabolites (numbers in each class are shown to the left). BCAA, branched chain amino acid. (E) Example metabolites involved in macronutrient and energy metabolism (means ± SEM, n = 4 per group, per time point; two-way ANOVA with Bonferroni post hoc tests; P < 0.05; *WT versus KO, #WT versus Liver-RE, and $KO versus Liver-RE). AAs, amino acids; NADPH, reduced form of NAD phosphate.
In total, 62 (31.5%) WT and Liver-RE oscillating metabolites do not oscillate in KO and thus require the liver clock (Fig. 4C). This set of metabolites has a similar phase and amplitude between WT and Liver-RE and is primarily enriched for carbohydrates, cofactors, and vitamins (Fig. 4, B to D, and fig. S8F). A heatmap of all WT oscillating carbohydrates illustrates the similar phase and amplitude in Liver-RE and lack of oscillation in KO (fig. S8G). Autonomously oscillating carbohydrates are limited to glycogen metabolites and select monosaccharides (13). Here, in the presence of a feeding rhythm, we find a more expansive control of carbohydrate metabolism and oscillating glucose pathways in Liver-RE (Fig. 4, D and E, and fig. S9A), in line with pathway enrichment for integrated genes (Fig. 2D) and CEBPB-BMAL1–regulated pathways (fig. S5F). Glucose 6-phosphate (G6P) is a key intersection of glucose metabolism, by which glucose can be directed into glycogen synthesis, the pentose phosphate pathway, glycolysis, and the uronic acid pathway (42, 43). WT and Liver-RE have tight control over G6P levels, which peak during fasting and recover during feeding (Fig. 4E). G6P is a precursor of fructose 6-phosphate (F6P), which oscillates similarly and can feed the hexosamine biosynthetic pathway. Nearly all metabolites in these pathways oscillate in WT and Liver-RE and not KO, demonstrating the dependence of these fluctuations on the liver clock (Fig. 4E and fig. S9A).
Next, we asked whether a gain of metabolite rhythmicity in Liver-RE and KO translates into improved energy homeostasis, by examining the main hepatic macronutrient pathways (Fig. 4E) (4, 43, 44). During feeding (ZT12 to ZT24), liver activities include glycogenesis, glycolysis, amino acid anabolism, and fatty acid and triglyceride syntheses. During fasting (ZT0 to ZT12), activities switch to glycogenolysis, gluconeogenesis, amino acid catabolism, and fatty acid oxidation. In WT mice, this daily switch is evidenced by elevated glycogen breakdown products at ZT0 to ZT8 (maltopentaose), peaks of glycolytic/gluconeogenic metabolites at ZT8 (G6P and F6P), brief and modest peaks of tricarboxylic acid (TCA) cycle intermediates around ZT4 (citrate), and the inverse relationship between palmitate, which has a trough at ZT12, and long-chain acyl carnitines (myristoyl carnitine and pentadecanoyl carnitine) that peak at ZT12 (Figs. 4E and 5D and figs. S9A and S10A). The state of energy metabolism is reflected in the NAD+ (nicotinamide adenine dinucleotide)/NADH (reduced form of NAD+) ratio [as a readout of oxidation-reduction (redox) potential] (peak at ZT12), as well as a lack of activation of ketogenesis, with stable, low levels of β-hydroxybutyrate (Fig. 4E) (45, 46). KO fails to recapitulate any of these features of macronutrient pathways, consistent with static NAD+/NADH ratio and β-hydroxybutyrate levels that markedly rise and fall with fasting and feeding, respectively (Fig. 4E and fig. S9A). In contrast, Liver-RE mice displayed restored and nonrestored functions; in line with extensive carbohydrate oscillations, Liver-RE mice fully recapitulated the WT pattern of glycogen and glycolysis/gluconeogenesis with the exception of the gluconeogenic substrates lactate and alanine. In addition, Liver-RE mice displayed a delayed and blunted activation of ketogenesis and moderate oscillation of the NAD+/NADH ratio (Fig. 4E and fig. S9A). Still, several alterations were apparent, for example, a trough rather than a peak of citrate at ZT4 (and similar pattern of other TCA cycle intermediates), elevated palmitate at ZT12, and no elevation of long-chain acyl carnitines at ZT12 (Figs. 4E and 5D and figs. S9A and S10A). These results are consistent with improved systemic insulin sensitivity under AL and improved 24-hour fasting blood glucose under NF in Liver-RE compared to KO (fig. S9, B and C). Together, these data demonstrate the importance of a local clock in the presence of a feeding-fasting rhythm for glucose homeostasis, while other facets of energy homeostasis such as key lipid species remain deregulated despite transcriptional restoration of related genes.
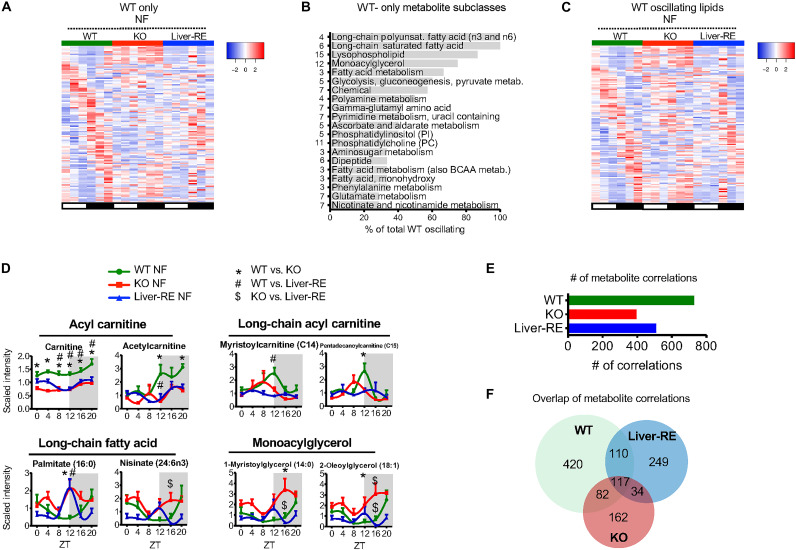
Fig. 5. A large portion of daily hepatic metabolism requires extrahepatic BMAL1 even in the presence of a feeding-fasting rhythm.
(A to F) Analysis of diurnal metabolite profiles under NF as in Fig. 4 (n = 4). (A) Phase-sorted heatmap of WT NF oscillating metabolites that failed to oscillate in Liver-RE NF, thereby showing a dependence on the rest of the clock network (“network-dependent”). (B) Pathways containing metabolites that oscillate in WT NF-only (JTK_CYCLE, P < 0.05), ranked by percentage of metabolites oscillating (numbers in each class are shown to the left of bars). (C) Phase-sorted heatmap showing all WT oscillating lipid metabolites. (D) Examples of WT NF-only oscillating and other informative lipid metabolites (means ± SEM, n = 4 per group, per time point; two-way ANOVA with Bonferroni post hoc tests; P < 0.05; *WT versus KO, #WT versus Liver-RE, and $KO versus Liver-RE). (E) Number of pairwise metabolite correlations under NF. (F) Overlap of metabolite correlation pairs between genotypes.
Requirement of extrahepatic BMAL1 for liver lipid metabolism
Daily metabolome analyses identified a substantial number of metabolites that only oscillate in WT, meaning they depend on extrahepatic clocks (32.1% of WT) (Fig. 5A). These metabolites were predominantly enriched for lipid species, such as long-chain polyunsaturated and saturated fatty acids, lysophospholipids, and monoacylglycerols (Fig. 5, B to D). Several metabolites are linked to β-oxidation; for instance, Liver-RE and KO have lower levels of carnitine and different patterns of long-chain acyl carnitines, which peak at ZT12 in WT and ZT8 in KO and do not peak in Liver-RE (Fig. 5D and fig. S10A) (47, 48). The general lack of lipid organization in KO and Liver-RE can be appreciated from a heatmap of all WT oscillating lipids (Fig. 5C). Joint enrichment analysis considering both genes and metabolites that depend on extrahepatic clocks suggested that mitochondrial derangements could contribute the disordered nature of lipids in Liver-RE. TCA cycle, respiratory electron transport, and biological oxidations were among the top gene-metabolite–enriched pathways (fig. S10B). This included genes encoding electron transport chain complex subunits, TOM (translocase of the outer membrane) and TIM (translocase of the inner membrane) complex subunits, and membrane transport proteins, among others (fig. S10C). Metabolites included mitochondria-associated cofactors and substrates such as FMN, FAD+, adenosine 5′-diphosphate, and thiamine diphosphate (fig. S10D). Together, these data show that extrahepatic clocks contribute to the interplay between mitochondrial function and lipid metabolism in the liver.
In addition to rhythmic analysis, we performed metabolite correlations over the daily cycle (fig. S11, A to C), reasoning that hepatic metabolism hinges on large-scale organization among specific pathways to form functional metabolic networks. Illustrating this point, WT displayed 729 significant correlations considering both intraclass (e.g. lipid-lipid) and interclass (e.g. lipid-carbohydrate) comparisons; in contrast, Liver-RE had 510, and KO had 395 (Fig. 5E). Overlapping the correlations of the three genotypes revealed that most correlations are unique to each genotype, with 420 (57.6%) correlations only present in WT livers (Fig. 5F). These data suggest that, in addition to contributing to rhythmicity of metabolic pathways, extrahepatic clocks are important for higher-order temporal coordination of metabolism.
The skeletal muscle clock regulates oscillations in the liver
A substantial number of genes (34%) were nonoscillatory in Liver-RE mice even in the presence of a feeding rhythm (termed network-dependent), suggesting that extrahepatic BMAL1 regulates oscillation of these genes. To hone in on which other tissue clocks may provide this regulation, we used the Quantitative Endocrine Network Interaction Estimation (QENIE) bioinformatic framework (49–51) to probe cross-tissue correlations of the Hybrid Mouse Diversity Panel (52) (Fig. 6A and data S7). Using our network-dependent genes as bait, we found that they were significantly more correlated with rhythmic genes than nonrhythmic genes (fig. S12A). Muscle had the highest number of significant correlations, which included several clock genes (Fig. 6, B and C, and fig. S12, B and C). In addition, we used a published dataset in which liver metabolites were correlated with metabolites in other metabolic tissues over the daily cycle (53) to identify connections. Metabolites that failed to oscillate in Liver-RE even under NF correlated mostly with muscle, followed closely by serum (fig. S12, D to F). Together, these analyses point to a potential role of the muscle BMAL1 in the regulation of daily liver metabolism.
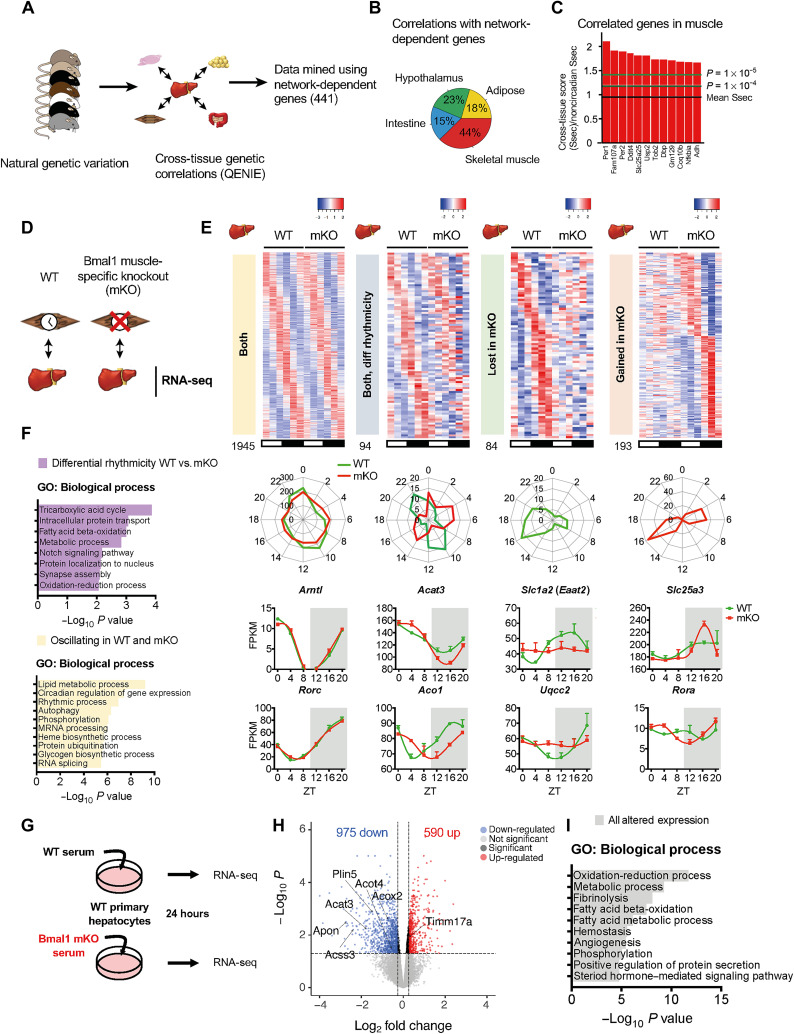
Fig. 6. Identification of the muscle clock as a key node supporting network-dependent circadian function in the liver.
(A) Scheme showing the QENIE approach used to identify genes in nonhepatic tissues correlated with network-dependent genes in the liver. (B) Tissue distribution of significant correlations related to network-dependent genes in the liver. (C) Top significant correlations in muscle with network-dependent genes in the liver; arrows indicate circadian clock genes. (D) Scheme of experimental setup; livers from WT or Bmal1 mKO mice were harvested over circadian time and subjected to RNA-seq. n = 3 animals of each genotype per time point (ZT0, ZT4, ZT8, ZT12, ZT16, and ZT20). (E) Phase-aligned heatmaps of circadian genes identified by JTK_CYCLE then subjected to LimoRhyde analysis to reveal differentially regulated transcripts. Both, genes oscillating in livers from WT and mKO; both differential rhythmicity, genes oscillating in livers from both genotypes but with different phases and/or amplitudes; lost in mKO, genes that lose oscillation in livers from mKO; gained in mKO, genes that oscillate in mKO exclusively. Phase maps of oscillating genes and example genes from each classification displayed below corresponding heatmap. (F) GO analysis from genes designated as either unaltered or differentially regulated by LimoRhyde analysis. (G) Experimental setup for serum treatment of primary hepatocytes and individual serum samples from WT and mKO mice were assessed by RNA-seq for their impact on gene expression in cultured hepatocytes (n = 5 per genotype). (H) Volcano plot of genes significantly regulated by treatment with mKO serum versus WT (FDR < 0.05). (I) GO analysis of significantly altered transcripts in primary hepatocytes treated with mKO versus WT serum.
To assess the role of the muscle clock in supporting liver transcriptional rhythms, we performed RNA-seq of livers harvested over the daily cycle from either WT or muscle-specific Bmal1-KO mice (mKO) (Fig. 6D and data S8). mKO mice do not exhibit behavioral phenotypes tied to feeding-fasting (9, 54). Liver core clock function at the protein and transcript levels was unaltered in and mKO (fig. S13, A and B). JTK_CYCLE found that 60% of oscillating genes in WT were maintained in mKO livers (fig. S13, C to E). Using JTK_CYCLE output, LimoRhyde revealed 371 liver transcripts (16% of total oscillating) displaying differential rhythmicity in the absence of the muscle clock (fig. S13F). These genes were enriched for TCA cycle, fatty acid β-oxidation, and oxidation-reduction pathways, among others (Fig. 6, E and F), similar to network-dependent transcripts and WT-only oscillating metabolite pathways. Of these, 94 (25%) oscillated in both genotypes but with different amplitudes or phases, 84 (23%) lost oscillation in mKO, and 193 (52%) gained oscillation in mKO (Fig. 6E and fig. S13F). Moreover, a clear separation of peak phase is observed in a subset of genes oscillating in both genotypes (Fig. 6E), revealing that the muscle clock is required for their proper phase alignment. Commonly oscillating genes had comparable amplitude (fig. S13G). Overlap of network-dependent genes with those lost in mKO (by JTK_CYCLE, P < 0.01) tied 30% of network-dependent genes to muscle BMAL1 (fig. S13, H to J). Together, these data demonstrate that the muscle clock regulates a subset of daily transcript rhythms in the liver.
Next, we next sought to reveal whether the serum represents a communication route between the muscle clock and the liver. Cultured primary hepatocytes were treated with serum harvested at ZT16 from either WT or mKO mice (serum from 1 mouse = 1 biological replicate, n = 5), and transcriptional changes were assessed by RNA-seq (Fig. 6G). A total of 1565 genes were differentially regulated (FDR < 0.05) in mKO serum-treated hepatocytes versus WT (975 genes down-regulated and 590 up-regulated) (Fig. 6H, fig. S14A, and data S9). Several enriched pathways matched those detected as altered in mKO livers, such as fatty acid β-oxidation and oxidation-reduction (Fig. 6I). Furthermore, many genes that identified as either differentially rhythmic in livers of mKO (fig. S14B) or network-dependent (fig. S14C) were differentially expressed (e.g., lipid metabolism: Acat3, Acss3, Acot4, Plin5, and Apon; and oxidation-reduction and mitochondrial function: Acox2 and Timm17a). The regulation of 13 network- and muscle clock–regulated genes was recapitulated in serum-treated hepatocytes (Dusp10, Efna1, Kank1, Snx33, Irs2, Iigp1, Stat3, Klkb1, Eaf1, Fez2, Akr1c14, Pah, and Timm9), suggesting that these genes are critical responders to muscle clock–dependent signaling in the liver. Our data overall highlight soluble factors as a communication route for BMAL1 signaling between muscle and liver.
DISCUSSION
Tissue- and organism-wide metabolism is remarkably coordinated across time and space (4, 53, 55). By restoring BMAL1 specifically in peripheral tissues (in RE lines) of mice otherwise devoid of clocks, we have previously demonstrated that peripheral clocks have limited functional autonomy (13, 14, 56). Pondering these nonautonomous functions, what tissue-extrinsic signals drive and direct the local node of the clock? By reinstating a feeding-fasting rhythm in Liver-RE mice, we showed that temporal regulation is mediated by the interaction of feeding behavior, the autonomous clock, and CEBPB. Probing the nonrestored genes and metabolites identified under these conditions, we also revealed muscle BMAL1 as a regulator of nonautonomous oscillations in the liver.
Imposed arrhythmic feeding on WT mice (17) and liver-specific clock disruption (10, 21, 27) have the same effect—eliminating most of the oscillating transcripts; thus, an interaction of the two components drives these oscillations. All genetic mouse models, including the Liver-RE model, have limitations in assessing this mechanism, but together, a clear picture is formed. Studying how the liver clock responds to an inverted (daytime) feeding regimen in WT mice has the caveat that conflicting signals from the central clock are present, since the SCN remains entrained to the light-dark cycle (16, 19, 21). Liver-specific mutants retain endogenous feeding-fasting rhythms and central clock signals, yet Bmal1 is not present in the liver and thus cannot be interrogated (3, 10, 21). Although the limitation of Liver-RE is that it also features an altered metabolic state due to loss of BMAL1 in other tissues, its strength is that it is possible to have both components in isolation. Using this approach, we were able to identify CEBPB as an important cooperating TF of BMAL1 in regulating daily transcription in hepatocytes.
The CEBP family of TFs has been identified as a possible regulator of circadian hepatic rhythms by several computational analyses (34, 37), and CEBPB regulates several metabolic processes in the liver, including daily autophagy (57). A circadian proteome study identified CEBPB as a candidate TF for diurnal regulation of glucose metabolism (58). Similar to our findings, an independent study showed that the transcriptional activity of CEBPB is reduced in the liver of Bmal1-KO mice (34). Our ATAC-seq and ChIP-seq results reveal that defective activity of CEBPB in Bmal1-KO mice is due to the lack of a feed-fast rhythm, as reinstating this rhythm restores its binding to target genomic regions. The presence of both CEBPB and BMAL1 is seemingly necessary for rhythmic output, as oscillation of genes associated with common loci was rescued preferentially in Liver-RE mice. Moreover, our data add to the known mechanisms by which BMAL1 regulates the transcription of output genes in the liver in concert with feeding rhythms (34, 37–40). In the liver, CEBPB cooperates with the glucocorticoid receptor (GR) (59), which plays a prominent role in circadian biology. During the day, GR binds to promoter regions in proximity of metabolic genes involved in glucose and lipid homeostasis at sites overlapping with CLOCK:BMAL1 (60). Since glucocorticoids can activate transcriptional activity and interact at chromatin with CEBPB (59), proper glucocorticoid signaling could provide a mechanistic link between systemic signals, CEBPB, and BMAL1. Another link could be through insulin signaling, which was recently identified as a systemic entrainer of hepatic rhythms to feeding (61) and was rescued in Liver-RE and KO upon NF. Insulin can trigger phosphorylation of CEBPB and, in turn, modify its DNA binding properties and its interaction with other transcriptional regulators (62).
We also found that NF was able to recruit a larger portion of oscillating metabolites than genes, in a clock-independent manner, as evidenced by a large overlap of all genotypes (63). However, we find that these oscillations do not correct functional defects of energy metabolism in KO and that Liver-RE mice display improvements under AL feeding conditions (13). One explanation stems from the observation that the liver clock amplifies and aligns even the feeding-driven metabolite oscillations, a plausibly critical regulatory step in transducing inputs into functional outputs. Still, in Liver-RE mice, energy metabolism was incompletely improved by NF. Intratissue correlation analysis revealed that in this incomplete state, temporal coherence between metabolic pathways remained disrupted, pointing to extrahepatic clocks as substantial organizers of large-scale hepatic metabolic networks (53). Considering genes and metabolites, our data divulge several levels of regulation derived from BMAL1 functions in nonliver tissues, including the aforementioned transcriptional control, with potentially feeding-sensitive translational sculpting of mRNA abundance (20, 22, 64), and posttranslational mechanisms that control metabolite levels.
Our metabolomic analysis shows that an interaction between the hepatic clock and feeding-fasting rhythm is not sufficient to drive oscillation of a large portion of lipids. In keeping with our results, most lipid species fail to oscillate in Per1/2 null mice subjected to NF (65). Moreover, oscillation of hepatic de novo lipid metabolism is still rhythmic in liver-specific Rev-Erbα and Rev-Erbβ double-KO mice (21). Notably, both adipose and muscle clocks have been implicated in systemic lipid homeostasis (11, 53, 66), although direct links to liver circadian function were yet to be uncovered. Here, our correlation analysis using the identified network-dependent genes pointed us to the muscle; in-depth characterization of livers from muscle-specific Bmal1-KO showed that the liver relies on muscle BMAL1 for daily regulation of transcripts involving oxidation-reduction, fatty acid, and TCA cycle metabolic processes. In addition, soluble factors in the serum tied to muscle BMAL1, whose identities remain unknown, were found to regulate many of the same genes in hepatocytes. Several muscle-secreted factors (myokines) are rhythmically released from synchronized human skeletal myotubes, in a manner dependent on the local clock (67). Myokines are demonstrated to mediate cross-talk between the muscle and liver (68, 69) and thus may coordinate daily metabolism between the two tissues.
One potential limitation of our study is that the exogenous feeding rhythm created by NF is more robust than the endogenous rhythm. This point is clear from its effect on WT mice, in which a subset of genes oscillated de novo upon NF. To circumvent this issue, we limited comparisons to WT features that were reproduced in both AL and NF regimes. Ultimately, this approach was chosen to directly and specifically test the interactions between the feeding-fasting rhythm and the autonomous clock. Brain reconstitution of BMAL1, another viable approach, would presumably reinstate other routes of top-down central clock regulation, such as temperature, autonomic nervous system activity, and neuropeptide release, among others, which would render it difficult to tease apart the contribution of each of these variables independently. Thus, we took advantage of the BMAL1-reconstituted model to specifically investigate feeding connections.
Myriad zeitgebers are sufficient but not necessary to synchronize peripheral oscillators (2, 13, 14, 17, 26, 70–75). Our data revealed the absolute minimum requirements: A light-dark cycle and feeding-fasting rhythm can propel an autonomous liver clock, making it indistinguishable from that of normal WT conditions. Many of the same zeitgebers are, however, necessary for oscillation of specific output (74, 76–80). We find that cyclic output in this minimal state was limited to ~50 to 65%. Thus, it appears that the array of circadian signals redundantly reinforces the stability of the core clock while providing unique driving inputs to couple the clock to specific output (13, 14). Our results highlight the critical importance of cooperating TFs and clocks in other tissues.
Reconstructing the clock system by simultaneously restoring multiple tissue clocks will shed light on intertissue communication. As desynchrony in the clock system is implicated in metabolic disease and aging (14, 81), mechanisms of intertissue clock communication may be an avenue to promote metabolic synchrony for therapeutic gain. Delineating these mechanisms will be the focus of our future studies.
METHODS
Animals
Mice were bred and housed in the animal facilities at Barcelona Science Park, Spain, in accordance with European Union and Spanish regulations. Animal care and experimental use were approved by the government of Catalonia, Spain, in line with national and local legislation. An additional cohort of mice was housed and used at the University of California, Irvine (UCI) vivarium, in accordance with the guidelines of the Institutional Animal Care and Use Committee at UCI. Animal experiments were designed and conducted with consideration of the ARRIVE (Animal Research: Reporting of In Vivo Experiments) guidelines, the details of which are as follows. Identical experimental conditions were in place at both institutions, and all mice were derived from the same founder line. Bmal1-stop-FL mice were generated as previously described (13, 14). All experiments used 8- to 12-week-old male mice that were on a 12:12 hour light:dark cycle and either fed AL or given AL access to food between ZT12 and ZT24 (NF group). Experimental mice were WT Bmal1wt/wt, Alfp-cre−/tg; KO Bmal1stopFL/stopFL, Alfp-cre−/−; Liver-RE Bmal1stopFL/stopFL, Alfp-cre−/tg. Muscle-specific Bmal1 KO mice were used as previously described (9, 54). Skeletal muscle–specific KO is achieved with a Myosin light chain 1f (Mlc1f) cre driver. Three- to 6-month-old mice were fed standard chow AL under a 12:12 hour light:dark cycle. WT littermates were used as controls.
Metabolic cage data acquisition
Indirect calorimetry was performed with negative flow Oxymax-CLAMS (Columbus Instruments, Columbus, OH) hardware system cages. Mice were given a 24-hour acclimation period to the metabolic cage. Measurements of VO2, VCO2, food intake, water intake, and feeding activity were taken every 10 min for up to five consecutive days at room temperature. The RER (RER = VCO2/VO2) was calculated by the accompanying Oxymax software.
Locomotor activity
Locomotor activity of individually housed mice was measured using optical beam motion detection (Starr Life Sciences). Data were collected using Mini Mitter VitalView 5.0 data acquisition software and analyzed using ClockLab (Actimetrics).
Insulin tolerance test
Mice were fasted from ZT0 until ZT4. At ZT4, mice were administered an intraperitoneal injection of insulin (0.3 U/kg). Blood glucose was measured via the tail with a standard glucose monitoring system at 0-, 15-, 30-, 60-, and 120-min post. Data are presented as the percentage of baseline to compare the relative drop in blood glucose for each genotype. The dose (0.3 U/kg) was determined through a pilot experiment, which showed that this was the lowest dose tested that consistently and substantially lowered blood glucose levels in WT mice.
Cell culture
AML12 cells (American Type Culture Collection) were cultured in Dulbecco’s modified Eagle’s medium:F12 (DMEM:F12; Lonza) supplemented with 10% fetal bovine serum (Gibco), 1% penicillin/streptomycin, insulin-transferring-selenium (ITS) liquid media supplement (Sigma-Aldrich, I3146), and DEX (40 ng/ml; Sigma-Aldrich). Primary hepatocytes were cultured in DMEM, low glucose (Thermo Fisher Scientific, 11885084), supplemented with 10% fetal bovine serum (Gibco) and 1% penicillin/streptomycin. Cells were maintained at 37°C, 5% CO2, and 95% air. For circadian synchronization, AML12 cells were plated in complete media without DEX. Cells were synchronized by addition of 100 nM DEX (Sigma-Aldrich; from 10mM in ethanol stock) for 1 hour, which induces synchronization through activation of GR signaling (82, 83). DEX was washed out, and cells were placed in fresh medium for the indicated time.
Primary hepatocyte isolation and culture
Mouse primary hepatocytes were isolated as previously described (84). Briefly, livers of 10- to 16-week-old male C57BL/6J mice were perfused through the hepatic vein with EGTA containing buffer A (115 mM NaCl, 5 mM KCl, 1 mM KH2PO4, 2.5 mM MgSO4, 25 mM Hepes, and 0.5 mM EGTA) and subsequent perfusion with buffer B (115 mM NaCl, 5 mM KCl, 1 mM KH2PO4, 25 mM Hepes, and 2 mM CaCl2) supplemented with collagenase (0.1 mg/ml). Livers were gently dispersed in buffer C (115 mM NaCl, 5 mM KCI, 1 mM KH2PO4, 25 mM Hepes, 2.5 mM MgSO4, and 4 mM CaCl2) containing 1% bovine serum albumin (BSA) to release hepatocytes. The cell suspension was filtered through 70 μm cell strainer and centrifuged at 30g to purify viable hepatocytes. Cell viability was determined by trypan blue exclusion, and only isolations with greater than 90% viability were used for experiments. Isolated hepatocytes were cultured on collagen-coated (Sigma-Aldrich, C7661) cell culture plates with DMEM low-glucose (Thermo Fisher Scientific, 11885) media supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. Cells were maintained at 37°C, 5% CO2, and 95% air.
Small interfering RNA transfection
Cells were plated in standard medium. The next day, cells were transfected with the following Silencer Select siRNA (small interfering RNA) (Thermo Fisher Scientific) against mouse Cebpb (ID s63859) or with a negative control (Thermo Fisher Scientific, 4390843) using Lipofectamine RNAiMAX Transfection Reagent (Thermo Fisher Scientific, 13-778-030), resuspended in Opti-MEM Reduced Serum Medium (Thermo Fisher Scientific, 31-985). The following day, cells were synchronized with DEX.
Serum treatment of primary hepatocytes
Serum from WT and mKO mice was collected at ZT16 and sterile-filtered. Serum from one mouse was used as one biological replicate, n = 5. Cultures were generated as described in the “Primary hepatocyte isolation and culture” section. The day after plating, cultures were washed 1× with phosphate-buffered saline and treated with 25% serum (in fetal bovine serum–free media). Twenty-four hours later, cells were collected for RNA-seq.
Blood glucose and serum measurements
Blood glucose was measured via the tail using the Accu-Chek Aviva Plus Blood Glucose Monitoring System. Blood serum was collected after centrifugation at 3000 rpm for 10 min at 4°C and stored at −80°C. The following commercially available kits were used to measure metabolic parameters in the serum, according to the manufacturer’s instructions: triglycerides, Triglyceride Quantification Colorimetric/Fluorometric Kit (Sigma-Aldrich, MAK266-1KT); free fatty acids, Free Fatty Acid Quantitation Kit (Sigma-Aldrich, MAK044-1KT); insulin, Ultra Sensitive Mouse Insulin ELISA Kit (Crystal Chem, 90080).
Chromatin fractionation, coimmunoprecipitation, and Western blot analysis
Chromatin fractions from mouse liver were prepared as previously described (85). Frozen liver was cut into small pieces, placed in STM buffer [50 mM tris-HCl (pH 7.4), 5 mM MgCl2, 250 mM sucrose, and 1 mM dithiothreitol (DTT)], homogenized by motorized tissue grinder with eight strokes, and incubated on ice for 10 min. All buffers were supplemented with 500 μM phenylmethylsulfonyl fluoride (PMSF; serine protease inhibitor), 20 mM NaF (phosphatase inhibitor), protease inhibitor cocktail (Roche, Basel, Switzerland), 10 mM nicotinamide (sirtuin inhibitor), and 330 nM trichostatin A (class I and II histone deacetylase inhibitor). After passage through a 100 μm filter, nuclei were pelleted by centrifugation at 3000 rpm for 10 min at 4°C. The nuclear pellet was washed by resuspension in Cyto buffer {1× buffer H [10 mM Hepes-NaOH (pH 8), 25 mM KCl, 650 μM spermidine, 1 mM EDTA, and 1 mM EGTA], 340 mM sucrose, 1% NP-40, and 1 mM DTT}, then incubated on ice for 10 min, and centrifuged at 3000 rpm for 10 min at 4°C. Nuclei were washed again in Nuc Low Salt buffer (1× buffer H, 20% glycerol, and 1 mM DTT) and collected by centrifugation at 3000 rpm for 10 min at 4°C. Nuclei were resuspended in Nuc High Salt buffer (1× buffer H, 400 mM NaCl, 20% glycerol, 1 mM DTT, and 500 μM PMSF), incubated on ice for 30 min, and centrifuged for 10 min at 14,000 rpm at 4°C. The resulting chromatin pellet was resuspended in radioimmunoprecipitation assay (RIPA) buffer [50 mM tris-HCl (pH 8), 150 mM NaCl, 5 mM EDTA, 15 mM MgCl2, and 1% NP-40] and sonicated at 60% amplitude, 5-s on, and 5-s off for four cycles. Following centrifugation for 10 min at 14,000 rpm at 4°C, the supernatant was used for coimmunoprecipitation or prepared for Western blot. For coimmunoprecipitation experiments, protein extracts were precleared with Dynabeads (protein G, Thermo Fisher Scientific) for 1 hour at 4°C and incubated with the indicated antibodies overnight at 4°C. The following day, samples were incubated with Dynabeads for 2 hours at 4°C. Beads were washed four times with RIPA buffer [50 mM tris-HCl (pH 8.0), 150 mM NaCl, 5 mM EDTA, 15 mM MgCl2, 1% NP-40, 0.5% deoxycholate, 1× protease inhibitor cocktail (Roche), and 1 mM PMSF], and samples were eluted with SDS loading buffer. For western blot, protein concentration was determined by the Bradford protein assay (Bio-Rad), and 5 to 10 μg of protein from each sample were separated on 6 to 10% gels by SDS–polyacrylamide gel electrophoresis. Proteins were transferred to a nitrocellulose membrane and blocked with 5% instant nonfat dry milk in TBS-T (0.1% Tween 20 and tris-buffered saline) for 2 hours at room temperature. Primary antibodies were diluted in 5% milk or 5% BSA TBS-T and incubated with membranes overnight at 4°C (BMAL1, Abcam, ab93806; Rev-Erbα, Cell Signaling, 13418; Per2, Alpha Diagnostic, PER21-A; CLOCK, Bethyl, A302-618A; H3, Abcam, ab1791; CEBPB, Abcam, ab32358; P84, GeneTex, GTX70220). Following horseradish peroxidase (HRP)–conjugated secondary antibody incubation [mouse immunoglobulin G (IgG)–HRP conjugate, EMD Millipore, AP160P; rabbit IgG-HRP linked, EMD Millipore, 12-348] for 1 hour at room temperature, blots were visualized with Immobilon Western chemiluminescent HRP substrate (Millipore, Burlington, MA) and developed on HyBlot CL autoradiography film (Denville Scientific, Holliston, MA).
RNA extraction and quantification of gene expression
Total RNA was extracted from liver tissue by homogenization in TRIzol (Invitrogen, Carlsbad, CA). Following RNA, DNA, and protein layer separation with chloroform, RNA was precipitated with a standard isopropanol and ethanol procedure. Final pelleted and washed RNA was resuspended in ribonuclease-free water and quantified using the NanoDrop (Thermo Fisher Scientific, Waltham, MA). RNA from AML12 cells and primary hepatocytes was extracted using the Direct-zol RNA Kit (Zymo Research, R2061). Complementary DNA (cDNA) was obtained by retrotranscription of 500 ng of RNA with the Maxima First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). Quantitative real-time polymerase chain reaction (qRT-PCR) analysis was performed using QuantStudio 3 (Applied Biosystems) with PowerUp SYBR Green Master Mix (Applied Biosystems). 18S was used as an endogenous control for AML12 cells, and Tbp was used for primary hepatocytes. The sequences of primer used are as follows: mouse 18S, 5′-CGCCGCTAGAGGTGAAATTC-3′ (forward) and 5′-CGAACCTCCGACTTTCGTTCT-3′ (reverse); mouse Tbp, 5′-CCCTTGTACCCTTCACCAAT-3′ (forward) and 5′-TTGAAGCTGCGGTACAATTC-3′ (reverse); mouse Cebpb, 5′-GGGGTTGTTGATGTTTTTGGT-3′ (forward) and 5′-TCGAAACGGAAAAGGTTCTCA-3′ (reverse); mouse Nr1d1, 5′-AGGCTGCTCAGTTGGTTGTT-3′ (forward) and 5′-CTCCATCGTTCGCATCAATC-3′ (reverse); mouse Nr1d2, 5′-GGGCACAAGCAACATTACCA-3′ (forward) and 5′-CACGTCCCCACACACCTTAC-3′ (reverse); mouse Dbp, 5′-AATGACCTTTGAACCTGATCCCGCT-3′ (forward) and 5′-GCTCCAGTACTTCTCATCCTTCTGT-3′ (reverse); mouse Ppargc1b, 5′-CTCCAGTTCCGGCTCCTC-3′ (forward) and 5′-CCCTCTGCTCTCACGTCTG-3′ (reverse); mouse Soat1, 5′-TCACTCTCCGCTAGGACCA-3′ (forward) and 5′-CCCAGCAGTGGTCAATAGGT-3′ (reverse); mouse Acat2, 5′-ACTACCACATGGGCATCACA-3′ (forward) and 5′-CTGGCACAATCTCCTTGTCA-3′ (reverse); mouse Hmgcs1, 5′-TATGGTTCCCTGGCTTCTGT-3′ (forward) and 5′-TGTGGCGTCTTGTGTGACTT-3′ (reverse); mouse Arntl, 5′-GCAGTGCCACTGACTACCAAGA-3′ (forward) and 5′-TTGCAATCTTACCCCAGACA-3′ (reverse); mouse Clock, 5′-ACCACAGCAACAGCAACAAC-3′ (forward) and 5′-GGCTGCTGAACTGAAGGAAG-3′ (reverse); mouse Per1, 5′-ACCAGCGTGTCATGATGACATA-3′ (forward) and 5′-GTGCACAGCACCCAGTTCCC-3′ (reverse); mouse Per2, 5′-CGCCTAGAATCCCTCCTGAGA-3′ (forward) and 5′-CCACCGGCCTGTAGGATCT-3′ (reverse); mouse Per3, 5′-CATACCAGGTGCCCGAGA-3′ (forward) and 5′-GCTGCTGTTCCATGCTCTG-3′ (reverse); mouse Cry1, 5′-CAGACTCACTCACTCAAGCAAGG-3′ (forward) and 5′-TCAGTTACTGCTCTGCCGCTGGAC-3′ (reverse); mouse Cry2, 5′-GGGACTCTGTCTATTGGCATCTG-3′ (forward) and 5′-GTCACTCTAGCCCGCTTGGT-3′ (reverse).
Chromatin immunoprecipitation
ChIP was performed on liver tissue from four biological replicates using the ChIP-IT High Sensitivity Kit (Active Motif, #53040), according to the manufacturer’s instructions using 6 μg of CEBPB antibody (Abcam, ab32358). Purified DNA was used for qRT-PCR analysis using QuantStudio 3 (Applied Biosystems) with PowerUp SYBR Green Master Mix (Applied Biosystems). The sequences of primer used are as follows: mouse ChIP Nr1d1, 5′-CTGCTTTGCAAAAGTGCTTG-3′ (forward) and 5′-CACCCTGACTCTTCAGAAAACC-3′ (reverse); mouse ChIP Gys, 5′-CACGGTGGTGCCTAGAGTC-3′ (forward) and 5′-AGAGGCAGAGGGAAGAGCA-3′ (reverse).
Next-generation sequencing sample preparation and analysis
RNA sequencing
For liver samples from WT, KO, and Liver-RE AL and NF, total RNA was monitored for quality control using the Agilent Bioanalyzer Nano RNA chip and NanoDrop absorbance ratios for 260/280 nm and 260/230 nm. Library construction was performed according to the Illumina TruSeq Total RNA stranded protocol (three biological replicates for all ZT points with the exception of ZT8 Liver-RE NF that has two biological replicates). The input quantity for total RNA was 1 μg, and ribosomal RNA (rRNA) was depleted using the Ribo-Zero rRNA Gold Removal Kit (human/mouse/rat). The rRNA-depleted RNA was chemically fragmented for 3 min. First-strand synthesis used random primers and reverse transcriptase to make cDNA. After second-strand synthesis, the double-stranded cDNA was cleaned using AMPure XP beads, the cDNA was end repaired, and then the 3′ ends were adenylated. Partial stub oligos for Illumina adapters were ligated on the ends, and the adapter-ligated fragments were enriched and barcoded by nine cycles of PCR using unique dual-indexing primers. The resulting libraries were validated by qPCR and sized by Agilent Bioanalyzer DNA high-sensitivity chip. The concentrations for the libraries were normalized and then multiplexed together. The pooled library concentration for denaturation was 2 nM, and the final concentration for loading was 400 pM. The multiplexed libraries were sequenced on the S2 flow cell using paired-end 100 cycles chemistry for the Illumina NovaSeq 6000. The version of NovaSeq control software was NVCS 1.6.0 with real-time analysis software, RTA 3.4.4. Postprocessing of the run to generate the FASTQ files was performed at the Institute for Genomics and Bioinformatics (UCI IGB).
The paired-end reads from each replicate were separately aligned to the reference genome assembly mm10 and corresponding transcriptome using the Tuxedo protocol (Illumina) (86). Reads uniquely aligned to known exons or splice junctions extracted with no more than two mismatches were included in the transcriptome. Reads uniquely aligned but with more than two mismatches or reads matching several locations in the reference genome were removed.
For time series datasets, rhythmic transcripts were detected with the nonparametric JTK_CYCLE algorithm (28) incorporating a period of 24 hours. Genes were considered rhythmic over the circadian cycle if their permutation-based, adjusted P value was <0.01. Metabolites were evaluated with an adjusted P < 0.05 given the inherent variability of metabolite quantification across mice and the n value of this experiment. Additional detection of rhythmicity for transcripts was conducted using a different, independently generated algorithm, BIO_CYCLE (29). Differential rhythmicity analysis was carried out using LimoRhyde (30). Starting rhythmicity detection was JTK_CYCLE; P < 0.01. Transcripts were considered differentially rhythmic at pDR < 0.01.
GO biological process (DIRECT) enrichment analysis was conducted using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) version 6.8 (87). TFBSs were analyzed in the promoter regions [−10,000 to +2000 base pairs (bp) of transcription starting site (TSS)] of circadian genes. Binding sites were determined from experimental ChIP-seq data and MotifMap (32) results for the mouse genome build mm10 [BBLS (Bayesian branch length score) > 1, FDR < 0.25]. Fisher’s exact test was performed between the circadian genes and the whole genome to establish enrichment.
For RNA-seq of AML12 cells and primary hepatocytes, library preparation and sequencing were performed by Novogene following standard procedures. Libraries were prepared using the NEBNext Ultra II RNA Library Prep (Illumina). Libraries were run on a NovaSeq 6000. Raw paired (150 × 150)–end reads were first quality checked with FastQC v0.11.8 and aligned to the to the reference genome assembly mm10 with the following settings (STAR--outFilterMultimapNmax 20 --alignSJoverhangMin 8 –alignSJDBoverhangMin 1 --outFilterMismatchNmax 999 --outFilterMismatchNoverReadLmax 0.04 --alignIntronMin 20--alignIntronMax 1000000 --alignMatesGapMax 1000000 --outSAMattributes NH HI NM MD). RNA-seq quality assessment was performed using RSeQC v3.0.1 tool (88). Differential expression analysis was assessed with edgeR v3.30.0 R Bioconductor package (89), and only genes with a cutoff expression of >1 CPM (counts per million reads mapped) in at least two samples were included in the analysis. TMM method was applied to normalize gene counts, and glmQLFTest and decideTestsDGE functions were used to perform differential analysis, with a minimum cutoff of an absolute fold change of ≥1.2 and an FDR of ≤0.05. Complete list of differentially expressed genes and statistical information (P value and FDR) for each condition are available at Gene Expression Omnibus (GEO: GSE158600, GSE171432, and GSE171184).
Exon-intron analysis
For intron-exon analysis, reads were mapped to the mouse genome (mm9) using STAR v2.6.0c (90) with the option “--quantMode TranscriptomeSAM,” and then gene expression level in fragments per kilobase million (FPKM) was quantified using RSEM v1.3.1 (91). The gene annotation file was downloaded from GENCODE (www.gencodegenes.org/mouse/release_M1.html), and the coordinates of exons and introns were generated from it. To avoid assigning the exon-junction reads to introns (92), introns were shrank by 10 bp at both 5′ and 3′ ends. The “coverageBed” module in bedtools v2.25.0 (93) was used to generate read count in each intron/exon. The option of “-F 0.5” was used to make sure a particular read was not counted in both exon and intron. The read count per kilobase per million reads (RPKM) value was calculated as the expression level of each intron/exon normalized by both intron/exon length and the total intronic/exonic read count of each sample. As the exonic-to-intronic ratio varies from sample to sample, exons and introns were processed separately.
Cross-tissue gene correlation
All datasets, analysis, and walk-through are provided at https://github.com/marcus-seldin/endocrine_communication-nonrestored-clocks. Briefly, genes identified as network-dependent (this study) were taken from liver gene expression in the Hybrid Mouse Diversity Panel (94, 95). All peripheral genes (adipose, muscle, hypothalamus, and intestine) from the same mice were then correlated across the network-dependent genes in the liver, where the global levels of cross-tissue correlation were quantified as a strength of cross-tissue predictions (Ssec) score (49). The Ssec score for each peripheral gene was normalized to the average correlation for each given tissue and scaled according to fold change relative to the same Ssec for noncircadian genes. Collectively, this enabled us to pinpoint peripheral genes that correlated specifically with network-dependent liver genes. Pathway enrichment for muscle genes ranked by Ssec score was calculated and visualized using WebGestalt (96).
ChIP sequencing
Library construction was performed from two biological replicates using the NEBNext Ultra DNA Library Prep for Illumina kit (E7370) according to the manufacturer’s protocol. Briefly, input and ChIP-enriched DNA were subjected to end repair and addition of “A” bases to 3′ ends, ligation of adapters, and USER excision. All purification steps were performed using AgenCourt AMPure XP beads (Beckman Coulter, A63882). Library amplification was performed by PCR using NEBNext Multiplex Oligos for Illumina (Index Primers Set 1, ref. E7335; Index Primers Set 2, ref. E7500; Index Primers Set 3, ref. E7710; and Index Primers Set 4, ref. E7730). Final libraries were analyzed using Agilent Bioanalyzer or Fragment Analyzer High Sensitivity assay (ref. 5067-4626 or DNF-474) to estimate the quantity and to check size distribution and were then quantified by qPCR using the KAPA Library Quantification Kit (Kapa Biosystems, ref. KK4835) before amplification with Illumina’s cBot. Libraries were sequenced 1 * 50 + 8 bp on Illumina’s HiSeq2500. To profile CEBPB-enriched regions, raw single-end reads were first quality checked with FastQC v0.11.8 and aligned to the UCSC Mus musculus release mm9, available at https://hgdownload.soe.ucsc.edu/goldenPath/mm9/bigZips/ using Bowtie2 v2.4.2 (97) with the following settings (bowtie2 -q –local). Mapped reads were then sorted using SAMtools 1.9 (98) and filtered with Sambamba v0.7.1 (99) with the following settings (sambamba view -h -t 2 -f bam -F “[XS] == null and not unmapped and not duplicate”). ChIP-seq quality assessment was performed using ChIPQC v1.26.0 R Bioconductor package (100). CEBPB-enriched regions relative to input DNA were assessed using the ENCODE recommended Irreproducibility Discovery Rate (IDR) framework. Peaks in both replicates were detected with MACS2 v2.2.7.1 (101) with the following settings (macs2 callpeak -f BAM -g 1.87e9 -B -p 1e-3). Peak consistency between true replicates was detected with IDR v2.0.4.2 (https://projecteuclid.org/journals/annals-of-applied-statistics/volume-5/issue-3/Measuring-reproducibility-of-high-throughput/10.1214/11-AOAS466.full) with the following settings (--input-file-type narrowPeak --rank p.value). Complete list of peak coordinates and statistical information (P value and FDR) in each sample and between replicates are available at Gene Expression Omnibus (GEO: awaiting reference number). To compute the overlap between CEBPB-enriched regions and previously published BMAL1-enriched regions (13), BMAL1 raw reads were retrieved from GEO:GSE132659 and were mapped, filtered, and peaks detected using the IDR framework. We then intersect CEBPB and BMAL1 peak coordinates with bedtools v2.30.0 with the following settings (bedtools intersect -b -a -u). Peak annotation was assessed with ChIPseeker v1.26.0 R Bioconductor package (102) using the prebuild annotation database TxDb.Mmusculus.UCSC.mm9.knownGene [tssRegion=c(-2000, 2000)]. Coverage tracks for each sample were computed with bedtools bamCoverage option, with the following settings (bamCoverage --binSize 20 --normalizeUsing BPM --smoothLength 60--extendReads 150 –centerReads). To obtain a global evaluation of enrichment around the TSS, we used both the computeMatrix and plotHeatmap options of bedtools package, with the following settings (computeMatrix reference-point --referencePoint TSS --beforeRegionStartLength 2000--afterRegionStartLength 2000 –skipZeros). Differential enrichment analysis was assessed with DiffBind v3.0.3 R Bioconductor package (103) using default system settings, providing prebuild DBA_BLACKLIST_MM9 black list coordinates and exporting reads with the following settings (bUsePval = TRUE,th = 0.05). Statistical analysis and visualization of functional profiles for genes and gene clusters associated with TSS region were computed with clusterProfiler R Bioconductor package v3.16.1 (104), using org.Mm.eg.db R Bioconductor annotation database, with a minimum threshold of an FDR of ≤0.05.
Assay for transposase-accessible chromatin sequencing
The Omni-ATAC protocol for isolation of nuclei and library preparation (105) was followed. Livers were homogenized by motorized tissue grinder at medium setting with 15 strokes in homogenization buffer [5 mM CaCl2, 3 mM Mg(Ac)2, 10 mM tris (pH 7.8), 16.7 μM PMSF, 166.7 μM β-mercaptoethanol, 320 mM sucrose, 100 μM EDTA, and 0.1% NP-40]. Samples were passed through 100-μm nylon mesh filters. Debris was removed by centrifugation for 1 min at 100 RCF (relative centrifugal force) at 4°C. One volume of 50% iodixanol solution [5 mM CaCl2, 3 mM Mg(Ac)2, 10 mM tris (pH 7.8), 16.7 μM PMSF, 166.7 μM β-mercaptoethanol, and 50% iodixanol] was added to the supernatant and mixed by gentle pipetting to get a 25% solution. A 29% iodixanol solution (as before plus 160 mM sucrose) was layered underneath, and a 35% iodixanol solution (plus sucrose) was layered underneath the 29% solution. Samples were centrifuged for 20 min at 3000 RCF at 4°C with the brake off in a swinging bucket centrifuge. The nuclei band was aspirated, placed into a fresh tube, and quantified using trypan blue staining and a manual cell counter. About 50,000 nuclei were washed in ATAC-RSB buffer [10 mM tris-HCl (pH 7.4), 10 mM NaCl, 3 mM MgCl2, and 0.1% Tween 20], centrifuged for 10 min at 500 RCF at 4°C, resuspended in Omni-ATAC Reaction Mix, and proceeded to the Optimized Transposition Reaction. Illumina Nextera transposase was used for the transposition reaction (106, 107). The reaction was cleaned up using the Zymo DNA Clean and Concentrator Kit. The transposed DNA was preamplified using Nextera index primers for five cycles of PCR. Additional cycles of PCR were determined by qPCR using 5 μl of partially amplified library. The resulting library was cleaned with AMPure XP beads and quantified by qPCR with KAPA SYBR Fast universal for the Illumina Genome Analyzer Kit. The library size was determined by analysis using the Bioanalyzer 2100 DNA High Sensitivity Chip. The library was sequenced on the Illumina NovaSeq 6000 using 150 cycles and paired-end dual-index read chemistry. The version of NovaSeq control software was NVCS 1.6.0 with real-time analysis software, RTA 3.4.4. Postprocessing of the run to generate the FASTQ files was performed at the UCI IGB.
ATAC-seq reads were aligned and processed following the ENCODE pipeline (www.encodeproject.org/atac-seq/) for paired-end sequencing reads. Reads were aligned to the mouse genome (mm9) using BowTie2 with the options of “-k 4 -X 2000.” Only properly paired reads were retained for analysis. The Picard tool (http://broadinstitute.github.io/picard/) was used to mark and remove duplicates. All samples were randomly down-sampled to 56,000,000 reads to exclude library size effects. Read coordinates were then transformed into the “BEDPE” format and fed to MACS2 for peak calling. The P value cutoff for MACS2 was 0.001. Peaks that overlapped with any ENCODE mm9 “blacklist” regions were excluded. The reads density in “bedGraph” format was generated by MACS2 and then transformed to bigWig using the “bedGraphToBigWig” tool in the UCSC binary utility directory (http://hgdownload.soe.ucsc.edu/admin/exe/). TF footprint detection was conducted on two biological replicates using an HMM-based software “rgt-hint” that is able to distinguish members of the same TF family (35) with the option of “--atac-seq.” The binding difference was determined by the “differential” module in rgt-hint.
Metabolomics
Sample preparation was carried out on four biological replicates at Metabolon Inc., in a manner similar to a previous study (108). Briefly, individual samples were subjected to methanol extraction and then split into aliquots for analysis by ultrahigh-performance LC-MS. The global biochemical profiling analysis composed of four unique arms consisting of reverse-phase chromatography positive ionization methods optimized for hydrophilic compounds (LC-MS Pos Polar) and hydrophobic compounds (LC-MS Pos Lipid), reverse-phase chromatography with negative ionization conditions (LC-MS Neg), and a hydrophilic interaction LC method coupled to negative (LC-MS Polar). All methods alternated between full-scan MS and data-dependent MSn scans. The scan range varied slightly between methods but generally covered 70 to 1000 m/z (mass/charge ratio). Metabolites were identified by automated comparison of the ion features in the experimental samples to a reference library of chemical standard entries that included retention time, molecular weight (m/z), preferred adducts, and in-source fragments as well as associated MS spectra and curated by visual inspection for quality control using software developed at Metabolon. Identification of known chemical entities was based on comparison to metabolomic library entries of purified standards (109).
General metabolite chemical classification (lipid, carbohydrate, etc.) and subpathways (fatty acid synthesis, polyunsaturated fatty acid, etc.) were determined by a combination of literature evidence and cross-reference to Metabolon’s internal database. Statistical analyses, including PCA and ANOVA, were carried out with MetaboAnalyst 3.0 (33) using prenormalized peak area under the curve values for each metabolite, which were then log-transformed and autoscaled. At each ZT, ANOVA with Fisher’s least significant difference (FDR < 0.1) determined metabolites altered in KO (WT versus KO, P < 0.05) and restored in Liver-RE (KO versus Liver-RE, P < 0.05; WT versus Liver-RE, not significant). Outliers were removed using the Grubbs’ test. Two types of statistical analyses were performed: (i) significance tests and (ii) classification analysis. Standard statistical analyses were performed in ArrayStudio on log-transformed data. For analyses not standard in ArrayStudio, the R program (http://cran.r-project.org/) was used. Following log transformation and imputation of missing values, if any, with the minimum observed value for each compound, Welch’s two-sample t test was used as a significance test to identify biochemicals that differed significantly (P < 0.05) between experimental groups. An estimate of the FDR (q value) was calculated to take into account the multiple comparisons that normally occur in metabolomic-based studies. Classification analyses used included PCA, hierarchical clustering, and random forest. For the scaled intensity graphics, each biochemical in original scale (raw area count) was rescaled to set the median across all animals and time points equal to 1.
Integrated pathway and enzyme-metabolite pair analysis
Pathway analysis integrating gene and metabolite enrichment was performed using the Integrated Molecular Pathway Level Analysis tool (110). This analysis returned a joint enrichment P value considering both genes and metabolites. Pathways were considered significantly coherent if enriched with ≥4 genes and ≥2 metabolites, P < 0.001.
Metabolite correlations
For the correlation analysis, xenobiotic and partially characterized metabolites were removed from further analysis. Correlations were estimated using Pearson’s correlation coefficient. Significant correlations P < 0.01 were selected after an FDR (Benjamini-Hochberg) correction. Distance maps were calculated by subtracting the condition-wise correlation coefficients (KO and RE) from the control (WT). Overall distance was estimated as the 1-norm of the distance map.
Statistical analysis
For each experiment, the number of biological replicates, statistical test, and significance threshold can be found in the figure legends or relevant sections of the Results and Methods. All data are shown as means ± SEM unless otherwise noted. Complex statistical analyses are described within their corresponding section in Methods. Unless otherwise stated, data were analyzed in Prism 6.0 (GraphPad). Sample size was determined by referencing literature standards recently reported for circadian studies (111).
Acknowledgments
We dedicate this paper to the memory of Paolo, who was an extremely supportive and enthusiastic scientist and who will remain an enduring influence for us all. We also extend special thanks to our animal technician S. Sato and laboratory manager W. Orquiz for valuable help, as well as to all members of the P.S.C. laboratory for support. We would like to thank the Genomics Unit at the CRG for assistance with ChIP-seq. Funding: C.M.G. was supported by the National Cancer Institute of the National Institutes of Health (NIH) under award number T32CA009054 and by the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement 749869. K.B.K. was supported by NIH-NINDS F32DK121425, and J.G.S. was supported by the Zymo-CEM Postdoctoral Fellowship (Zymo Research). P.P. was supported by a scholarship from the Wenner-Gren Foundations. C.V. and M.M.S. are supported by NIH grants DK20AU4084 and HL138193.The work of S.C., M.S., and P.B. was in part supported by NIH grant GM123558 to P.B. Work in the W.L. laboratory was supported by NIH grants R01HG007538, R01CA193466, and R01CA228140. Work in the P.S.-C. laboratory is supported by NIH grants R21DK114652 and R21AG053592, a Challenge Grant from the Novo Nordisk Foundation (NNF-202585), and through access to the Genomics High Throughput Facility Shared Resource of the Cancer Center Support Grant (CA-62203) at the UCI and NIH-shared instrumentation grants 1S10RR025496-01, 1S10OD010794-01, and 1S10OD021718-01. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. P.-S.W. is supported by a “Ramon y Cajal” contract (RYC2019-026661-I) from the Spanish Ministry of Science and Innovation (MICINN). Research in the P.M.-C. laboratory is supported by MINECO-Spain (RTI2018-096068), ERC-2016-AdG-741966, AFM, MDA-USA, La Marató/TV3 Foundation, LaCaixa-HEALTH-HR17-00040, and UPGRADE-H2020-825825 and María-de-Maeztu-Program for Units of Excellence to UPF (MDM-2014-0370) and Severo-Ochoa-Program for Centers of Excellence to CNIC (SEV-2015-0505). Research in the S.A.B. laboratory is supported by the European Research Council (ERC), the Government of Cataluña (SGR grant), the Government of Spain (MINECO), the La Marató/TV3 Foundation, and The Worldwide Cancer Research Foundation (WCRF). Author contributions: C.M.G., K.B.K., J.G.S., P.B., S.M., S.A.B., P.M.-C., and P.S.-C. conceived and designed the study. C.M.G., K.B.K., J.G.S., S.A.B., P.M.-C., and P.S.-C. wrote and edited the manuscript. C.M.G., K.B.K., J.G.S., P.-S.W., V.M.Z., T.M., K.S., T.S., P.P., S.K.C., and K.A.D. performed experiments. O.D., A.K. and M.V.-D. provided technical support. D.L., J.M.M.K., C.V., and M.M.S. performed bioinformatic correlation analyses. S.C., M.S., P.K., R.C., J.S., and W.L. performed sequencing analysis. Competing interests: The authors declare that they have no competing interests. Data and materials availability: The RNA-seq, ATAC-seq, and ChIP-seq data have been deposited in the GEO with accession numbers GSE158600, GSE171432, and GSE171184 (RNA-seq); GSE154902 (ATAC-seq); and GSE169483 (ChIP-seq). Metabolomic data are provided as data S6. For cross-tissue gene correlation, all datasets, analysis, and walk-through are provided at https://github.com/marcus-seldin/endocrine_communication-nonrestored-clocks. All transcriptomic data for this project are publicly available on the CircadiOmics web portal (112, 113) (http://circadiomics.igb.uci.edu/biocycle).
Supplementary Materials
This PDF file includes:
Supplementary Materials and Methods
Figs. S1 to S14
Legends for data S1 to S9
Other Supplementary Material for this manuscript includes the following:
Data S1 to S9
REFERENCES AND NOTES
- 1.Bass J., Lazar M. A., Circadian time signatures of fitness and disease. Science 354, 994–999 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Benitah S. A., Welz P. S., Circadian regulation of adult stem cell homeostasis and aging. Cell Stem Cell 26, 817–831 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Chaix A., Lin T., Le H. D., Chang M. W., Panda S., Time-restricted feeding prevents obesity and metabolic syndrome in mice lacking a circadian clock. Cell Metab. 29, 303–319.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Panda S., Circadian physiology of metabolism. Science 354, 1008–1015 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cederroth C. R., Albrecht U., Bass J., Brown S. A., Dyhrfjeld-Johnsen J., Gachon F., Green C. B., Hastings M. H., Helfrich-Förster C., Hogenesch J. B., Lévi F., Loudon A., Lundkvist G. B., Meijer J. H., Rosbash M., Takahashi J. S., Young M., Canlon B., Medicine in the fourth dimension. Cell Metab. 30, 238–250 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schibler U., Sassone-Corsi P., A web of circadian pacemakers. Cell 111, 919–922 (2002). [DOI] [PubMed] [Google Scholar]
- 7.Astiz M., Heyde I., Oster H., Mechanisms of communication in the mammalian circadian timing system. Int. J. Mol. Sci. 20, 343 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hastings M. H., Maywood E. S., Brancaccio M., The mammalian circadian timing system and the suprachiasmatic nucleus as its pacemaker. Biology 8, 13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dyar K. A., Hubert M. J., Mir A. A., Ciciliot S., Lutter D., Greulich F., Quagliarini F., Kleinert M., Fischer K., Eichmann T. O., Wright L. E., Peña Paz M. I., Casarin A., Pertegato V., Romanello V., Albiero M., Mazzucco S., Rizzuto R., Salviati L., Biolo G., Blaauw B., Schiaffino S., Uhlenhaut N. H., Transcriptional programming of lipid and amino acid metabolism by the skeletal muscle circadian clock. PLOS Biol. 16, e2005886 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lamia K. A., Storch K.-F., Weitz C. J., Physiological significance of a peripheral tissue circadian clock. Proc. Natl. Acad. Sci. U.S.A. 105, 15172–15177 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paschos G. K., Ibrahim S., Song W. L., Kunieda T., Grant G., Reyes T. M., Bradfield C. A., Vaughan C. H., Eiden M., Masoodi M., Griffin J. L., Wang F., Lawson J. A., FitzGerald G. A., Obesity in mice with adipocyte-specific deletion of clock component Arntl. Nat. Med. 18, 1768–1777 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sadacca L. A., Lamia K. A., deLemos A. S., Blum B., Weitz C. J., An intrinsic circadian clock of the pancreas is required for normal insulin release and glucose homeostasis in mice. Diabetologia 54, 120–124 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koronowski K. B., Kinouchi K., Welz P.-S., Smith J. G., Zinna V. M., Shi J., Samad M., Chen S., Magnan C. N., Kinchen J. M., Li W., Baldi P., Benitah S. A., Sassone-Corsi P., Defining the independence of the liver circadian clock. Cell 177, 1448–1462.e14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Welz P. S., Zinna V. M., Symeonidi A., Koronowski K. B., Kinouchi K., Smith J. G., Guillén I. M., Castellanos A., Furrow S., Aragón F., Crainiciuc G., Prats N., Caballero J. M., Hidalgo A., Sassone-Corsi P., Benitah S. A., BMAL1-driven tissue clocks respond independently to light to maintain homeostasis. Cell 178, 1029 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barber A. F., Erion R., Holmes T. C., Sehgal A., Circadian and feeding cues integrate to drive rhythms of physiology in Drosophila insulin-producing cells. Genes Dev. 30, 2596–2606 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Damiola F., le Minh N., Preitner N., Kornmann B., Fleury-Olela F., Schibler U., Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 14, 2950–2961 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greenwell B. J., Trott A. J., Beytebiere J. R., Pao S., Bosley A., Beach E., Finegan P., Hernandez C., Menet J. S., Rhythmic food intake drives rhythmic gene expression more potently than the hepatic circadian clock in mice. Cell Rep. 27, 649–657.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Stokkan K. A., Yamazaki S., Tei H., Sakaki Y., Menaker M., Entrainment of the circadian clock in the liver by feeding. Science 291, 490–493 (2001). [DOI] [PubMed] [Google Scholar]
- 19.Vollmers C., Gill S., DiTacchio L., Pulivarthy S. R., le H. D., Panda S., Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc. Natl. Acad. Sci. U.S.A. 106, 21453–21458 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J., Symul L., Yeung J., Gobet C., Sobel J., Lück S., Westermark P. O., Molina N., Naef F., Circadian clock-dependent and -independent posttranscriptional regulation underlies temporal mRNA accumulation in mouse liver. Proc. Natl. Acad. Sci. U.S.A. 115, E1916–E1925 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guan D., Xiong Y., Trinh T. M., Xiao Y., Hu W., Jiang C., Dierickx P., Jang C., Rabinowitz J. D., Lazar M. A., The hepatocyte clock and feeding control chronophysiology of multiple liver cell types. Science 369, 1388–1394 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Atger F., Gobet C., Marquis J., Martin E., Wang J., Weger B., Lefebvre G., Descombes P., Naef F., Gachon F., Circadian and feeding rhythms differentially affect rhythmic mRNA transcription and translation in mouse liver. Proc. Natl. Acad. Sci. U.S.A. 112, E6579–E6588 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weger B. D., Gobet C., David F. P. A., Atger F., Martin E., Phillips N. E., Charpagne A., Weger M., Naef F., Gachon F., Systematic analysis of differential rhythmic liver gene expression mediated by the circadian clock and feeding rhythms. Proc. Natl. Acad. Sci. U.S.A. 118, e2015803118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bunger M. K., Wilsbacher L. D., Moran S. M., Clendenin C., Radcliffe L. A., Hogenesch J. B., Simon M. C., Takahashi J. S., Bradfield C. A., Mop3 is an essential component of the master circadian pacemaker in mammals. Cell 103, 1009–1017 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kondratov R. V., Kondratova A. A., Gorbacheva V. Y., Vykhovanets O. V., Antoch M. P., Early aging and age-related pathologies in mice deficient in BMAL1, the core componentof the circadian clock. Genes Dev. 20, 1868–1873 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adamovich Y., Ladeuix B., Sobel J., Manella G., Neufeld-Cohen A., Assadi M. H., Golik M., Kuperman Y., Tarasiuk A., Koeners M. P., Asher G., Oxygen and carbon dioxide rhythms are circadian clock controlled and differentially directed by behavioral signals. Cell Metab. 29, 1092–1103.e3 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Kornmann B., Schaad O., Bujard H., Takahashi J. S., Schibler U., System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLOS Biol. 5, e34 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hughes M. E., Hogenesch J. B., Kornacker K., JTK_CYCLE: An efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J. Biol. Rhythms 25, 372–380 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Agostinelli F., Ceglia N., Shahbaba B., Sassone-Corsi P., Baldi P., What time is it? Deep learning approaches for circadian rhythms. Bioinformatics 32, 3051 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singer J. M., Hughey J. J., LimoRhyde: A flexible approach for differential analysis of rhythmic transcriptome data. J. Biol. Rhythms 34, 5–18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koike N., Yoo S.-H., Huang H.-C., Kumar V., Lee C., Kim T.-K., Takahashi J. S., Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338, 349–354 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daily K., Patel V. R., Rigor P., Xie X., Baldi P., MotifMap: Integrative genome-wide maps of regulatory motif sites for model species. BMC Bioinformatics 12, 495 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xia J., Sinelnikov I. V., Han B., Wishart D. S., MetaboAnalyst 3.0––Making metabolomics more meaningful. Nucleic Acids Res. 43, W251–W257 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beytebiere J. R., Trott A. J., Greenwell B. J., Osborne C. A., Vitet H., Spence J., Yoo S.-H., Chen Z., Takahashi J. S., Ghaffari N., Menet J. S., Tissue-specific BMAL1 cistromes reveal that rhythmic transcription is associated with rhythmic enhancer-enhancer interactions. Genes Dev. 33, 294–309 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Z., Schulz M. H., Look T., Begemann M., Zenke M., Costa I. G., Identification of transcription factor binding sites using ATAC-seq. Genome Biol. 20, 45 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vierstra J., Stamatoyannopoulos J. A., Genomic footprinting. Nat. Methods 13, 213–221 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Trott A. J., Menet J. S., Regulation of circadian clock transcriptional output by CLOCK:BMAL1. PLOS Genet. 14, e1007156 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fang B., Everett L. J., Jager J., Briggs E., Armour S. M., Feng D., Roy A., Gerhart-Hines Z., Sun Z., Lazar M. A., Circadian enhancers coordinate multiple phases of rhythmic gene transcription in vivo. Cell 159, 1140–1152 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sobel J. A., Krier I., Andersin T., Raghav S., Canella D., Gilardi F., Kalantzi A. S., Rey G., Weger B., Gachon F., Dal Peraro M., Hernandez N., Schibler U., Deplancke B., Naef F.; CycliX Consortium , Transcriptional regulatory logic of the diurnal cycle in the mouse liver. PLOS Biol. 15, e2001069 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J., Mauvoisin D., Martin E., Atger F., Galindo A. N., Dayon L., Sizzano F., Palini A., Kussmann M., Waridel P., Quadroni M., Dulić V., Naef F., Gachon F., Nuclear proteomics uncovers diurnal regulatory landscapes in mouse liver. Cell Metab. 25, 102–117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qu M., Duffy T., Hirota T., Kay S. A., Nuclear receptor HNF4A transrepresses CLOCK:BMAL1 and modulates tissue-specific circadian networks. Proc. Natl. Acad. Sci. U.S.A. 115, E12305–E12312 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bouché C., Serdy S., Kahn C. R., Goldfine A. B., The cellular fate of glucose and its relevance in type 2 diabetes. Endocr. Rev. 25, 807–830 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Rui L., Energy metabolism in the liver. Compr. Physiol. 4, 177–197 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reinke H., Asher G., Circadian clock control of liver metabolic functions. Gastroenterology 150, 574–580 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Puchalska P., Crawford P. A., Multi-dimensional roles of ketone bodies in fuel metabolism, signaling, and therapeutics. Cell Metab. 25, 262–284 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nocito L., Kleckner A. S., Yoo E. J., Jones IV A. R., Liesa M., Corkey B. E., The extracellular redox state modulates mitochondrial function, gluconeogenesis, and glycogen synthesis in murine hepatocytes. PLOS ONE 10, e0122818 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Knottnerus S. J. G., Bleeker J. C., Wüst R. C. I., Ferdinandusse S., IJlst L., Wijburg F. A., Wanders R. J. A., Visser G., Houtkooper R. H., Disorders of mitochondrial long-chain fatty acid oxidation and the carnitine shuttle. Rev. Endocr. Metab. Disord. 19, 93–106 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qu Q., Zeng F., Liu X., Wang Q. J., Deng F., Fatty acid oxidation and carnitine palmitoyltransferase I: Emerging therapeutic targets in cancer. Cell Death Dis. 7, e2226 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seldin M. M., Koplev S., Rajbhandari P., Vergnes L., Rosenberg G. M., Meng Y., Pan C., Phuong T. M. N., Gharakhanian R., Che N., Mäkinen S., Shih D. M., Civelek M., Parks B. W., Kim E. D., Norheim F., Krishnan K. C., Hasin-Brumshtein Y., Mehrabian M., Laakso M., Drevon C. A., Koistinen H. A., Tontonoz P., Reue K., Cantor R. M., Björkegren J. L. M., Lusis A. J., A strategy for discovery of endocrine interactions with application to whole-body metabolism. Cell Metab. 27, 1138–1155.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seldin M., Yang X., Lusis A. J., Systems genetics applications in metabolism research. Nat. Metab. 1, 1038–1050 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seldin M. M., Lusis A. J., Systems-based approaches for investigation of inter-tissue communication. J. Lipid Res. 60, 450–455 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lusis A. J., Seldin M. M., Allayee H., Bennett B. J., Civelek M., Davis R. C., Eskin E., Farber C. R., Hui S., Mehrabian M., Norheim F., Pan C., Parks B., Rau C. D., Smith D. J., Vallim T., Wang Y., Wang J., The hybrid mouse diversity panel: A resource for systems genetics analyses of metabolic and cardiovascular traits. J. Lipid Res. 57, 925–942 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dyar K. A., Lutter D., Artati A., Ceglia N. J., Liu Y., Armenta D., Jastroch M., Schneider S., Mateo S., Cervantes M., Abbondante S., Tognini P., Orozco-Solis R., Kinouchi K., Wang C., Swerdloff R., Nadeef S., Masri S., Magistretti P., Orlando V., Borrelli E., Uhlenhaut N. H., Baldi P., Adamski J., Tschöp M. H., Eckel-Mahan K., Sassone-Corsi P., Atlas of circadian metabolism reveals system-wide coordination and communication between clocks. Cell 174, 1571–1585.e11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dyar K. A., Ciciliot S., Wright L. E., Biensø R. S., Tagliazucchi G. M., Patel V. R., Forcato M., Paz M. I. P., Gudiksen A., Solagna F., Albiero M., Moretti I., Eckel-Mahan K. L., Baldi P., Sassone-Corsi P., Rizzuto R., Bicciato S., Pilegaard H., Blaauw B., Schiaffino S., Muscle insulin sensitivity and glucose metabolism are controlled by the intrinsic muscle clock. Mol. Metab. 3, 29–41 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dyar K. A., Eckel-Mahan K. L., Circadian metabolomics in time and space. Front. Neurosci. 11, 369 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krishnaiah S. Y., Wu G., Altman B. J., Growe J., Rhoades S. D., Coldren F., Venkataraman A., Olarerin-George A. O., Francey L. J., Mukherjee S., Girish S., Selby C. P., Cal S., Er U., Sianati B., Sengupta A., Anafi R. C., Kavakli I. H., Sancar A., Baur J. A., Dang C. V., Hogenesch J. B., Weljie A. M., Clock regulation of metabolites reveals coupling between transcription and metabolism. Cell Metab. 25, 961–974.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ma D., Panda S., Lin J. D., Temporal orchestration of circadian autophagy rhythm by C/EBPβ. EMBO J. 30, 4642–4651 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Y., Song L., Liu M., Ge R., Zhou Q., Liu W., Li R., Qie J., Zhen B., Wang Y., He F., Qin J., Ding C., A proteomics landscape of circadian clock in mouse liver. Nat. Commun. 9, 1553 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grøntved L., John S., Baek S., Liu Y., Buckley J. R., Vinson C., Aguilera G., Hager G. L., C/EBP maintains chromatin accessibility in liver and facilitates glucocorticoid receptor recruitment to steroid response elements. EMBO J. 32, 1568–1583 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Quagliarini F., Mir A. A., Balazs K., Wierer M., Dyar K. A., Jouffe C., Makris K., Hawe J., Heinig M., Filipp F. V., Barish G. D., Uhlenhaut N. H., Cistromic reprogramming of the diurnal glucocorticoid hormone response by high-fat diet. Mol. Cell 76, 531–545.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Crosby P., Hamnett R., Putker M., Hoyle N. P., Reed M., Karam C. J., Maywood E. S., Stangherlin A., Chesham J. E., Hayter E. A., Rosenbrier-Ribeiro L., Newham P., Clevers H., Bechtold D. A., O’Neill J. S., Insulin/IGF-1 drives PERIOD synthesis to entrain circadian rhythms with feeding time. Cell 177, 896–909.e20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tian J., Goldstein J. L., Brown M. S., Insulin induction of SREBP-1c in rodent liver requires LXRα-C/EBPβ complex. Proc. Natl. Acad. Sci. U.S.A. 113, 8182–8187 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Adamovich Y., Rousso-Noori L., Zwighaft Z., Neufeld-Cohen A., Golik M., Kraut-Cohen J., Wang M., Han X., Asher G., Circadian clocks and feeding time regulate the oscillations and levels of hepatic triglycerides. Cell Metab. 19, 319–330 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Benegiamo G., Mure L. S., Erikson G., Le H. D., Moriggi E., Brown S. A., Panda S., The RNA-binding protein NONO coordinates hepatic adaptation to feeding. Cell Metab. 27, 404–418.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aviram R., Manella G., Kopelman N., Neufeld-Cohen A., Zwighaft Z., Elimelech M., Adamovich Y., Golik M., Wang C., Han X., Asher G., Lipidomics analyses reveal temporal and spatial lipid organization and uncover daily oscillations in intracellular organelles. Mol. Cell 62, 636–648 (2016). [DOI] [PubMed] [Google Scholar]
- 66.Wada T., Ichihashi Y., Suzuki E., Kosuge Y., Ishige K., Uchiyama T., Makishima M., Nakao R., Oishi K., Shimba S., Deletion of Bmal1 prevents diet-induced ectopic fat accumulation by controlling oxidative capacity in the skeletal muscle. Int. J. Mol. Sci. 19, 2813 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Perrin L., Loizides-Mangold U., Skarupelova S., Pulimeno P., Chanon S., Robert M., Bouzakri K., Modoux C., Roux-Lombard P., Vidal H., Lefai E., Dibner C., Human skeletal myotubes display a cell-autonomous circadian clock implicated in basal myokine secretion. Mol. Metab. 4, 834–845 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen W., Wang L., You W., Shan T., Myokines mediate the cross talk between skeletal muscle and other organs. J. Cell. Physiol. 236, 2393–2412 (2021). [DOI] [PubMed] [Google Scholar]
- 69.Priest C., Tontonoz P., Inter-organ cross-talk in metabolic syndrome. Nat. Metab. 1, 1177–1188 (2019). [DOI] [PubMed] [Google Scholar]
- 70.Hamaguchi Y., Tahara Y., Hitosugi M., Shibata S., Impairment of circadian rhythms in peripheral clocks by constant light is partially reversed by scheduled feeding or exercise. J. Biol. Rhythms 30, 533–542 (2015). [DOI] [PubMed] [Google Scholar]
- 71.Husse J., Leliavski A., Tsang A. H., Oster H., Eichele G., The light-dark cycle controls peripheral rhythmicity in mice with a genetically ablated suprachiasmatic nucleus clock. FASEB J. 28, 4950–4960 (2014). [DOI] [PubMed] [Google Scholar]
- 72.Izumo M., Pejchal M., Schook A. C., Lange R. P., Walisser J. A., Sato T. R., Wang X., Bradfield C. A., Takahashi J. S., Differential effects of light and feeding on circadian organization of peripheral clocks in a forebrain Bmal1 mutant. eLife 3, e04617 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Saini C., Morf J., Stratmann M., Gos P., Schibler U., Simulated body temperature rhythms reveal the phase-shifting behavior and plasticity of mammalian circadian oscillators. Genes Dev. 26, 567–580 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Soták M., Bryndová J., Ergang P., Vagnerová K., Kvapilová P., Vodička M., Pácha J., Sumová A., Peripheral circadian clocks are diversely affected by adrenalectomy. Chronobiol. Int. 33, 520–529 (2016). [DOI] [PubMed] [Google Scholar]
- 75.Vujović N., Davidson A. J., Menaker M., Sympathetic input modulates, but does not determine, phase of peripheral circadian oscillators. Am. J. Physiol. Regul. Integr. Comp. Physiol. 295, R355–R360 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.de Goede P., Sen S., Oosterman J. E., Foppen E., Jansen R., la Fleur S. E., Challet E., Kalsbeek A., Differential effects of diet composition and timing of feeding behavior on rat brown adipose tissue and skeletal muscle peripheral clocks. Neurobiol. Sleep Circadian Rhythms 4, 24–33 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gerber A., Esnault C., Aubert G., Treisman R., Pralong F., Schibler U., Blood-borne circadian signal stimulates daily oscillations in actin dynamics and SRF activity. Cell 152, 492–503 (2013). [DOI] [PubMed] [Google Scholar]
- 78.Oishi K., Amagai N., Shirai H., Kadota K., Ohkura N., Ishida N., Genome-wide expression analysis reveals 100 adrenal gland-dependent circadian genes in the mouse liver. DNA Res. 12, 191–202 (2005). [DOI] [PubMed] [Google Scholar]
- 79.Preußner M., Goldammer G., Neumann A., Haltenhof T., Rautenstrauch P., Müller-McNicoll M., Heyd F., Body temperature cycles control rhythmic alternative splicing in mammals. Mol. Cell 67, 433–446.e4 (2017). [DOI] [PubMed] [Google Scholar]
- 80.Su Y., van der Spek R., Foppen E., Kwakkel J., Fliers E., Kalsbeek A., Effects of adrenalectomy on daily gene expression rhythms in the rat suprachiasmatic and paraventricular hypothalamic nuclei and in white adipose tissue. Chronobiol. Int. 32, 211–224 (2015). [DOI] [PubMed] [Google Scholar]
- 81.Greco C. M., Sassone-Corsi P., Circadian blueprint of metabolic pathways in the brain. Nat. Rev. Neurosci. 20, 71–82 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Balsalobre A., Brown S. A., Marcacci L., Tronche F., Kellendonk C., Reichardt H. M., Schütz G., Schibler U., Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science 289, 2344–2347 (2000). [DOI] [PubMed] [Google Scholar]
- 83.Reddy A. B., Maywood E. S., Karp N. A., King V. M., Inoue Y., Gonzalez F. J., Lilley K. S., Kyriacou C. P., Hastings M. H., Glucocorticoid signaling synchronizes the liver circadian transcriptome. Hepatology 45, 1478–1488 (2007). [DOI] [PubMed] [Google Scholar]
- 84.Chun S. K., Lee S., Flores-Toro J., U R. Y., Yang M.-J., Go K. L., Biel T. G., Miney C. E., Pierre Louis S., Law B. K., Law M. E., Thomas E. M., Behrns K. E., Leeuwenburgh C., Kim J.-S., Loss of sirtuin 1 and mitofusin 2 contributes to enhanced ischemia/reperfusion injury in aged livers. Aging Cell 17, e12761 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Greco C. M., Cervantes M., Fustin J.-M., Ito K., Ceglia N., Samad M., Shi J., Koronowski K. B., Forne I., Ranjit S., Gaucher J., Kinouchi K., Kojima R., Gratton E., Li W., Baldi P., Imhof A., Okamura H., Sassone-Corsi P., S-adenosyl-L-homocysteine hydrolase links methionine metabolism to the circadian clock and chromatin remodeling. Sci. Adv. 6, eabc5629 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D. R., Pimentel H., Salzberg S. L., Rinn J. L., Pachter L., Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Huang D. W., Sherman B. T., Lempicki R. A., Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- 88.Wang L., Wang S., Li W., RSeQC: Quality control of RNA-seq experiments. Bioinformatics 28, 2184–2185 (2012). [DOI] [PubMed] [Google Scholar]
- 89.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li B., Dewey C. N., RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gaidatzis D., Burger L., Florescu M., Stadler M. B., Analysis of intronic and exonic reads in RNA-seq data characterizes transcriptional and post-transcriptional regulation. Nat. Biotechnol. 33, 722–729 (2015). [DOI] [PubMed] [Google Scholar]
- 93.Quinlan A. R., Hall I. M., BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Parks B. W., Sallam T., Mehrabian M., Psychogios N., Hui S. T., Norheim F., Castellani L. W., Rau C. D., Pan C., Phun J., Zhou Z., Yang W.-P., Neuhaus I., Gargalovic P. S., Kirchgessner T. G., Graham M., Lee R., Tontonoz P., Gerszten R. E., Hevener A. L., Lusis A. J., Genetic architecture of insulin resistance in the mouse. Cell Metab. 21, 334–347 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hasin-Brumshtein Y., Khan A. H., Hormozdiari F., Pan C., Parks B. W., Petyuk V. A., Piehowski P. D., Brümmer A., Pellegrini M., Xiao X., Eskin E., Smith R. D., Lusis A. J., Smith D. J., Hypothalamic transcriptomes of 99 mouse strains reveal trans eQTL hotspots, splicing QTLs and novel non-coding genes. eLife 5, e15614 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liao Y., Wang J., Jaehnig E. J., Shi Z., Zhang B., WebGestalt 2019: Gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 47, W199–W205 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup , The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tarasov A., Vilella A. J., Cuppen E., Nijman I. J., Prins P., Sambamba: Fast processing of NGS alignment formats. Bioinformatics 31, 2032–2034 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Carroll T. S., Liang Z., Salama R., Stark R., de Santiago I., Impact of artifact removal on ChIP quality metrics in ChIP-seq and ChIP-exo data. Front. Genet. 5, 75 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nussbaum C., Myers R. M., Brown M., Li W., Liu X. S., Model-based analysis of ChIP-seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yu G., Wang L.-G., He Q.-Y., ChIPseeker: An R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31, 2382–2383 (2015). [DOI] [PubMed] [Google Scholar]
- 103.Ross-Innes C. S., Stark R., Teschendorff A. E., Holmes K. A., Ali H. R., Dunning M. J., Brown G. D., Gojis O., Ellis I. O., Green A. R., Ali S., Chin S.-F., Palmieri C., Caldas C., Carroll J. S., Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 481, 389–393 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yu G., Wang L.-G., Han Y., He Q.-Y., clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Corces M. R., Trevino A. E., Hamilton E. G., Greenside P. G., Sinnott-Armstrong N. A., Vesuna S., Satpathy A. T., Rubin A. J., Montine K. S., Wu B., Kathiria A., Cho S. W., Mumbach M. R., Carter A. C., Kasowski M., Orloff L. A., Risca V. I., Kundaje A., Khavari P. A., Montine T. J., Greenleaf W. J., Chang H. Y., An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 14, 959–962 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Buenrostro J. D., Wu B., Chang H. Y., Greenleaf W. J., ATAC-seq: A method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 109, 21.29.21–21.29.29 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Buenrostro J. D., Giresi P.-G., Zaba L.-C., Chang H.-Y., Greenleaf W.-J., Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Collet T. H., Sonoyama T., Henning E., Keogh J. M., Ingram B., Kelway S., Guo L., Farooqi I. S., A metabolomic signature of acute caloric restriction. J. Clin. Endocrinol. Metab. 102, 4486–4495 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dehaven C. D., Evans A. M., Dai H., Lawton K. A., Organization of GC/MS and LC/MS metabolomics data into chemical libraries. J. Chem. 2, 9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kamburov A., Cavill R., Ebbels T. M. D., Herwig R., Keun H. C., Integrated pathway-level analysis of transcriptomics and metabolomics data with IMPaLA. Bioinformatics 27, 2917–2918 (2011). [DOI] [PubMed] [Google Scholar]
- 111.Hughes M. E., Abruzzi K. C., Allada R., Anafi R., Arpat A. B., Asher G., Baldi P., de Bekker C., Bell-Pedersen D., Blau J., Brown S., Ceriani M. F., Chen Z., Chiu J. C., Cox J., Crowell A. M., DeBruyne J. P., Dijk D.-J., DiTacchio L., Doyle F. J., Duffield G. E., Dunlap J. C., Eckel-Mahan K., Esser K. A., FitzGerald G. A., Forger D. B., Francey L. J., Fu Y.-H., Gachon F., Gatfield D., de Goede P., Golden S. S., Green C., Harer J., Harmer S., Haspel J., Hastings M. H., Herzel H., Herzog E. D., Hoffmann C., Hong C., Hughey J. J., Hurley J. M., de la Iglesia H. O., Johnson C., Kay S. A., Koike N., Kornacker K., Kramer A., Lamia K., Leise T., Lewis S. A., Li J., Li X., Liu A. C., Loros J. J., Martino T. A., Menet J. S., Merrow M., Millar A. J., Mockler T., Naef F., Nagoshi E., Nitabach M. N., Olmedo M., Nusinow D. A., Ptáček L. J., Rand D., Reddy A. B., Robles M. S., Roenneberg T., Rosbash M., Ruben M. D., Rund S. S.-C., Sancar A., Sassone-Corsi P., Sehgal A., Sherrill-Mix S., Skene D. J., Storch K.-F., Takahashi J. S., Ueda H. R., Wang H., Weitz C., Westermark P. O., Wijnen H., Xu Y., Wu G., Yoo S.-H., Young M., Zhang E. E., Zielinski T., Hogenesch J. B., Guidelines for genome-scale analysis of biological rhythms. J. Biol. Rhythms 32, 380–393 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ceglia N., Liu Y., Chen S., Agostinelli F., Eckel-Mahan K., Sassone-Corsi P., Baldi P., CircadiOmics: Circadian omic web portal. Nucleic Acids Res. 46, W157–W162 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Patel V. R., Eckel-Mahan K., Sassone-Corsi P., Baldi P., CircadiOmics: Integrating circadian genomics, transcriptomics, proteomics and metabolomics. Nat. Methods 9, 772–773 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials and Methods
Figs. S1 to S14
Legends for data S1 to S9
Data S1 to S9