Abstract
Originally relatively ignored in forensic investigations because its genetic analysis lacks inference of individual identification, the value of Y chromosome analysis has been proven in cases of sexual assault, particularly where the amount of material left by a male assailant is limited in comparison with female DNA. All routine analysis of autosomal DNA, however, targets a gene (AMELY) on the Y chromosome in order to identify the sex of the DNA source and this is discussed in the context of the genetic structure of this male-specific chromosome. Short-tandem repeat markers on the chromosome are tested in dedicated multiplexes that have developed over time and these are described alongside international guidance as to their use in a forensic setting. As a marker of lineage, the Y chromosome provides additional tools to assist in the inference of ancestry, both geographical and familial and the value of Y chromosome testing is illustrated through descriptions of cases of criminal and historical interest. A decision to analyse the Y chromosome has to be considered in the context, not only of the circumstances of the case, but also with regard to the ethical questions it might raise, and these are discussed in relation to the cases that have been described in more detail in the accompanying online supplementary material.
Keywords: familial linkage, forensic, genetic intelligence, Y chromosome
Introduction
Genetic characteristics on the human Y chromosome provide a lineage marker in the form of a single haplotype transferred directly from father to son [1]. A haplotype is the set of short-tandem repeat (STR) alleles typed on a single Y chromosome. Analysis of the Y chromosome has been relatively ignored for some time in forensic casework because it does not provide the near certainty of identification that autosomal DNA can offer. The uniparental nature of this marker can be useful in certain circumstances: sexual assault; missing persons; disaster victim identification; complex kinship analysis; and population inference [2].
The Y chromosome is one of a pair, with the X chromosome, of the human sex chromsomes located in the nucleus of human cells along with 22 pairs of autosomal chromosomes. Individuals with an X and Y chromosome normally are phenotypically male, and those with two X chromosomes, female, although there are both rare differences in the numbers of these chromosomes, or other distinct mutations that can influence that phenotype [3].
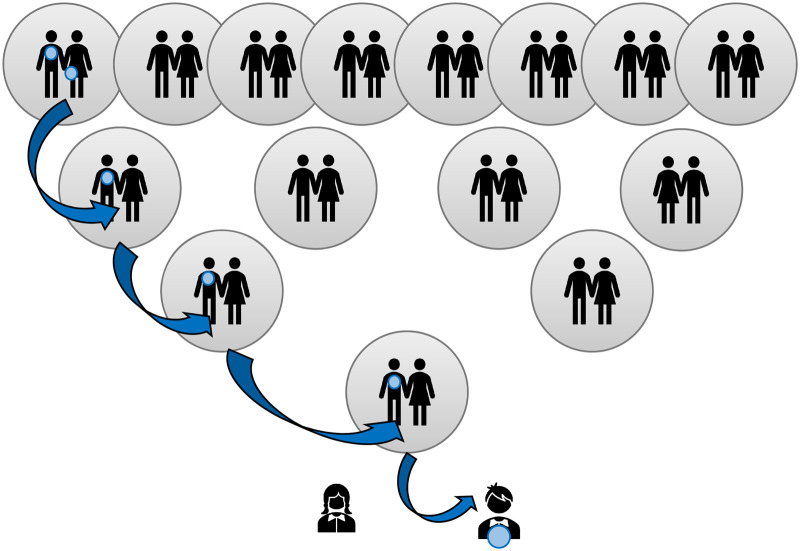
Figure 1 illustrates the inheritance typical of males in the population in which the single Y chromosome is passed down virtually intact (barring mutations) from father to son.
Figure 1. Illustration of Y chromosome inheritance over four generations  indicates ancestral males that share the same Y chromosome haplotype as the child at the base.
indicates ancestral males that share the same Y chromosome haplotype as the child at the base.
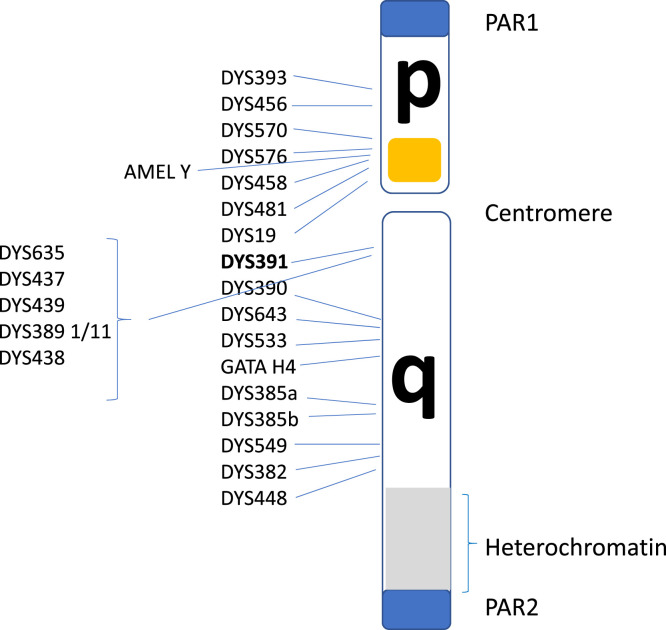
The human Y chromosome has two pseudoautosomal regions (PARs) with a short arm Yp and long arm Yq, separated by a centromere (Figure 2). The PARs, the short arm and the proximal part of the long arm consist of lightly packed chromatin enriched with genes PAR1 and PAR2, both recombine with areas on the X chromosome; the remaining regions are non-recombining and contain about 70 genes in the male-specific region (MSY). Of particular interest in forensic DNA typing is the detection of markers associated with the amelogenin (AMEL). The locus has two homologous genes on the X and Y chromosomes [4]: AMELY [5] encodes a protein involved in the demineralisation of tooth enamel and has a paralogue on the X chromosome (AMELX). The first intron of AMELX is six base-pairs (bp) [6] shorter and primers that target both genes are included in many forensic DNA kits in order to reveal the sex of the donor. As early as 1998 [7] there were reports in the literature of 2/24 males in a Sri Lankan population that lacked AMELY (AMELYnull) and others since have reported different classes of deletions involving the gene [8], seen predominantly in South Asians with a frequency of between 1% and 6.5% in other larger studies of this population group [9–13]. Sometimes the deletion will encompass a larger area resulting in incomplete Y chromosome STR loci in profiles routinely used in Y chromosome forensic analysis. This has led to other markers, such as DYS391 (shown in bold in Figure 2), being included in standard autosomal DNA analysis kits as this marker is on the long arm of the Y chromosome and not involved in the above-reported deletion event.
Figure 2. Structural diagram of the human Y chromosomes identifying the relative locations of PowerPlex Y23short tandem repeat (STR) loci used routinely in forensic DNA casework.
The approximate region  identified in the diagram is sometimes the subject of variable size deletions.
identified in the diagram is sometimes the subject of variable size deletions.
The inability of most of the Y chromosome to recombine with the X chromosome is a feature that has developed over time [14] and the sequenced chromosome is full of small repeats, inversions, palindromic sequences (where nucleotide sequences are identical with that on the complementary strand) [15]. The latter, in particular, can facilitate gene conversion and increase independent deletion and mutation events in certain areas [16,17]. Where these impact on genes involved in spermatogenesis this can impact on male fertility, but the chromosome can tolerate quite large deletions without consequence.
STRs on the Y chromosome are no more likely to mutate than those selected for forensic analysis on autosomes but, not only are they are individually selected based on significant diversity measures [18], the more regular appearance of mutations between father and son is likely to reflect the high number of mitotic events in spermatogenesis. Because of this Y-STR haplotypes will be much more variable than seen in a single autosomal STR and Y-STR haplotype reference databases are required to be significantly larger than autosomal STR allele reference databases in order to provide reliable frequency estimates.
Mutations are not randomly distributed along the chromosome; some regions are more vulnerable to a mutational event than others. Those that mutate more frequently on average (often referred to as rapidly mutating STRs) have some advantages if they are included in forensic multiplexes, particularly in the distinction of closely related males.
In 2014, in a worldwide collaborative effort [19], many contributors typed almost 20 thousand samples in total from their respective populations using the Y23 loci, confirming high diversity and revealing many null (missing), duplicate (repeated) and off-ladder (not coincident with expected size) alleles. The number of alleles at any one locus varied from 10 to 31, with a median of 16, not including DYS385 which had 146 different unordered variants; 92.9% of the 19 630 haplotypes observed were unique.
In 2018, Aliferi et al. [20] published a collection of over 3000 Y23-STRs from the U.K. and Ireland. In addition to white British (n = 1062) and Irish (n = 720) the population set included black British (n = 977) and British Asians (n = 369) providing 97.9% different haplotypes with shared haplotypes being seen within the different population sets, or between the Irish and white British. Adding the five additional loci from Y filer Plus increased that to 99.5%.
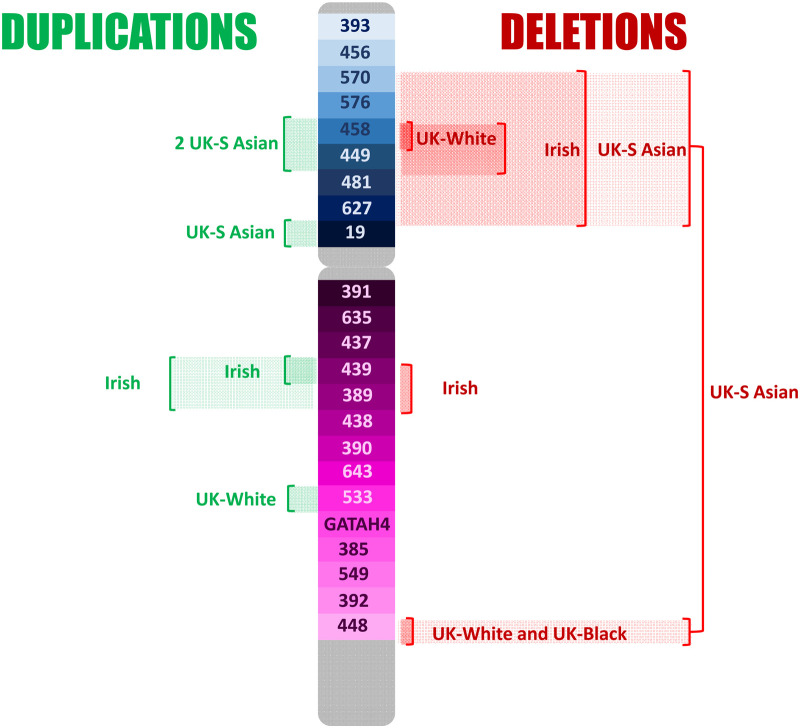
Duplications and deletions were observed in eleven and eight loci, respectively, in the U.K. and Irish set. These are shown in Figure 3. The duplication of DYS448 in 11 black British suggests a population frequency of around 1% for this variation in the black population and has previously been described in the study by Purps et al. [19].
Figure 3. Duplications and deletions reported in a U.K. (three ancestries) and Irish population set of 3654 individuals typed for the Y23 set of Y-STRs [20].
Deletions were seen predominantly associated with the short arm and in the British (south) Asian population group, with two individuals demonstrating a large deletion of most of the short arm and DYS448 on the distant end of the long arm in addition. The observed deletions in this dataset are typical of those including AMELY that are seen at higher frequency in south Asians reported in other studies. These findings suggest a single origin for these variants clustered on the short arm of the chromosome that has gained prominence due to genetic drift. In YHRD deletions are seen in 1/9200 worldwide, but 1/148 of individuals in the Indian metapopulation. In contrast, deletions of DYS448 and DYS458, seen in 17/37 and 10/37 of the total observed deletions, suggest a form of instability in these areas of the chromosome. Both areas show multiple different forms of structural variation occurring in different populations.
Y single nucleotide polymorphisms (SNPs) were also seen in the Aliferi study at two loci (DYS392 and DYS576) resulting in null alleles. The latter is a recognised SNP (rs757752030) with a frequency of around 1.4% in the South Asian 1000 genomes study [21]. Additionally, mutations upstream of DYS481 produced mobility shifts in the electropherogram that led to false off-ladder allele reports. The G > A SNP, which is 16 bp upstream of the CTT locus repeat, is hypothesised to enlarge the local secondary structure [22] and appears to be population specific, being seen in 7.3% of British South Asians, predominantly associated with allele 25. A further shift was seen associated with this locus in another population, but involving a G > T SNP further upstream, suggesting that any mutation in the flanking region of this locus may produce a change in the secondary structure, resulting in a mobility shift.
Forensic Y chromosome DNA analysis
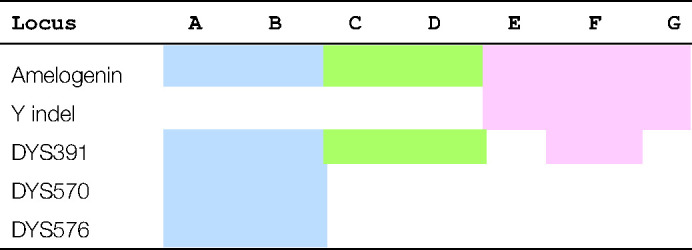
Routine autosomal DNA analysis makes use of STR loci that comprise a standard set. In Europe, these are the twelve European Standard Set (ESS) loci, last defined in 2008, and in 2015 the United States increased their standard set to twenty in the combined DNA index system (CODIS) [23]. The main manufacturers of autosomal multiplex kits have always included AMEL as an indicator of sex but are now adding additional markers in order to avoid incidences of AMELYnull in casework. These additional markers are usually Y-STRs but a Y-specific insertion/deletion (Yindel) rs2032678 locus is used within the kits from one manufacturer [24]. Where Y-STRs have been included, all include DYS391 which is distant from the common deletion site. Table 1 summarises the current set of autosomal multiplex kits that include additional Y chromosome markers.
Table 1. Autosomal STR kits supplied by Promega (blue), Qiagen (green) and ThermoFisher (pink) that include additional Y loci in their kits.
|
Multiplex keys: B, PowerPlex Fusion 6C; C, Versaplex 27PY; E, Investigator 24plex; F, Investigator 26plex; H, AmpFLSTR NGM Detect; I, Globalfiler; J, Verifiler Plus.
Y chromosome STR multiplexes
Two manufacturers produce the main Y-STR multiplex kits used in forensic casework today employing up to 23 and 27 loci, respectively. Table 2 shows the loci included.
Table 2. Y STR markers currently used in forensic DNA analysis from multiplex kit manufacturers – Promega (blue), ThermoFisher (pink) and a non-commercial set (grey). Loci in bold are ‘rapidly mutating’.
a Kayser, M., Caglia, A., Copach, D. et al. (1997). Evaluation of Y-chromosomal STRs: a mulicenter study. Int. J. Leg. Med. 110, 141–149.
Y-STRs may or may not be tested routinely by forensic providers worldwide and the ability to compare with reference profiles will depend on the retention policies of individual jurisdictions. While their usefulness will be discussed below, because the inheritance of these loci is not independent, the product rule used for autosomal STRs cannot be employed and haplotype frequencies must be used instead. Because of the high variability of haplotypes, so the reference haplotypes used to provide reliable frequencies need to be extremely large and representative of the different populations, and that has relied on the collaboration of many scientists across the world.
YHRD
YHRD (www.yhrd.org) is an online Y chromosome haplotype reference database built from an initial collaboration with 31 forensic and anthropological institutions across Europe who contributed almost 5000 male minimal haplotypes to the collection initiated by academics at the Charité University in Berlin; the database went live in 2000 [25]. Since then, it has expanded enormously with additional contributors, worldwide, and is now in release 63 (September 2020). A similar database, set up in the US, was to be decommissioned in 2019 and all US haplotypes transferred to YHRD with appropriate guidance for SWGDAM compliant searches being provided for US subpopulations.
The collection of populations is classified into metapopulations based on both geographical and linguistic determinants and now consists of 33 different metapopulations, with about 50% being Eurasian (mainly Western and Eastern European), 25% Asian (mainly from China) and the other composed of mainly admixed populations and people from African, Native American, Aborigine and Inuit populations [26]. The database is essentially five separate databases, each with a defined set of Y-STR markers. Table 3 shows the current position of the database which now also includes a collection of SNPs.
Table 3. Composition of the Y-HRD database release 64 (May 2021) giving the number of haplotypes in each set. The minimal haplotype includes alleles at STRs DYS19, DYS389I, DYS389II, DYS390, DYS391, DYS392, DYS393 and DYS385. The maximal haplotype includes alleles at the 27 STRs typed across the different commercial multiplexes, including Yfiler Plus.
| Dataset | Number of haplotypes |
|---|---|
| Minimal | 337 449 |
| PowerPlex Y | 296 791 |
| Yfiler | 227 004 |
| PowerPlex Y23 | 93 998 |
| Maximal | 40 201 |
| Y-SNPs | 24 429 |
For whatever purpose a Y-haplotype is defined, it is likely that an estimate of its frequency in a relevant population will be needed. The YHRD database provides frequency estimates based on different statistical models.
Y-STR guidelines
Y-STR loci exhibit the same characteristics as autosomal loci in terms of stutter and repeat unit types, and interpretation rules need to be defined locally. The rules more obviously differ from autosomal guidance because Y-STRs are not observed in females and normally only present with a single allele unless the locus is duplicated. Because of its familial nature consideration must also be given to any relevant legal requirements of the jurisdiction and the prevailing population substructure.
Y chromosome STR loci variations are more common than seen in autosomal STRs, with multicopy loci such as DYS385 and DYF387S1 being observed routinely. DYS385 is a constitutively duplicated locus comprised of two linked STR sub-loci with a GAAA repeat structure in different locations [27]; the two alleles are frequently different sizes and when one primer pair is used both are amplified and these are considered together as a single haplotype. The parts are called ‘a’ and ‘b’, and by convention the one with the smallest number of repeats is defined as ‘a’, but this does not imply assignation to one or other of the two chromosomal locations.
SWGDAM guidance
The Scientific Working Group on DNA Analysis Methods (SWGDAM) published Y chromosome interpretation guidelines in 2014 [28], which superseded their 2009 guidance, outlining the principle of using Y-STR analysis as an additional tool to autosomal DNA, or instead of autosomal DNA when wanting to detect males in a mixture with an abundance of female DNA. The guidance also recommends the retention of samples for possible future Y-STR analysis.
The use of the counting method is suggested for reporting frequencies, preferably also with a confidence interval. This is calculated from n/(N + 1), where n is the number of times the haplotype is seen in a database of size N. They recommend reporting the match probability as:
where A is the haplotype of interest, Pr(A|A) is the probability of observing haplotype A given that is has already been seen once. pA is the upper bound of the confidence interval for the profile probability estimated by counting. Theta (θ), representing population substructure and broadly similar to FST [29], is likely to be very low (at a maximum of around 0.0007 for a 12-locus haplotype in all populations) and decreases as the number of loci typed increases and so will have little effect on the estimation.
Because the Y chromosome is inherited independently of other chromosomes, they remark that the haplotype frequency can be multiplied, applying the product rule, with frequencies of autosomal loci.
Finally, the guidance recommends that mixtures in which it has not been possible to determine a major or conditioned haplotype should only be used for exclusionary purposes.
ISFG guidance
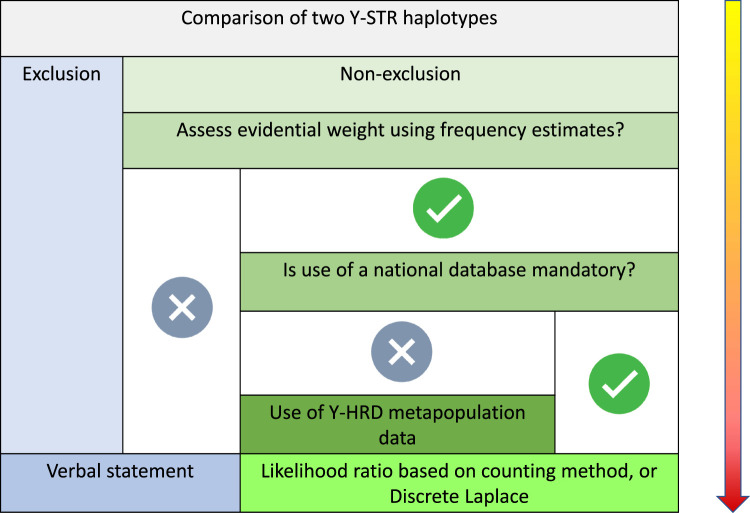
The DNA Commission of the International Society of Forensic Genetics (ISFG) has also provided recommendations on the interpretation of forensic Y-STRs [30]. While they recognise that comparisons of haplotypes can provide a simple qualitative statement in forensic reports: inconclusive; exclusion; or non-exclusion, they regard that as a minimum requirement and state that a quantitative statement is desirable. They have set out a conditional decision tree which is illustrated in Figure 4.
Figure 4. ISFG Y chromosome guidelines, illustrated here in a flow chart with successive decisions in Y-STR haplotype analysis [30].
The use of either the counting method or the Discrete Laplace formula [31] to estimate population frequencies is recommended. The former may be over conservative, especially when profiles are not represented in the database, which is commonly seen. The DNA Commission prefers the use of the Discrete Laplace model which ensures that rare haplotypes retain evidential power, providing that ancestral haplotype clusters are represented in the database. Some jurisdictions use the augmented counting method, estimating frequencies as (n + 1)/(N + 1) in contrast with the simpler n/(N + 1) used by SWGDAM.
When a quantitative inference is made, the quality of the data supporting it is relevant. The ISFG has set down fundamental requirements for reference Y-STR databases: full specific kit-haplotypes from randomly selected individuals; meta- and sub-population data; population datasets of sufficient size to ensure representation of the population and its extended lineage clusters; submitters of data to have engaged in national or international proficiency tests. Instead of using a match probability which will not account for extended lineage matches, Anderson and Balding [32] propose reporting the median and 95 percentile number of males expected to match others in a database of a particular size. Guidance also suggests that reports should use a standard format stating competing hypotheses:
Hypothesis 1: Person X is the source of the DNA
Hypothesis 2: A random man Y from the reference population Z is the source of the DNA, and Y is another man than X
To be followed by a statement akin to: The Y-STR profile detected in the crime stain is LR (where LR is the likelihood ratio) times more probable to observe under hypothesis 1 than under hypothesis 2. This notwithstanding, paternal relatives have a high probability to have the same Y-STR profile and will in that case have the same likelihood ratio.
To put this into context, paternally related individuals of 20 generations distant are 18% more likely to share the same Y23 haplotype and even if 27 markers are used, 5.5% will still share the same haplotype [32].
Uses of Y-STRs in the criminal justice system
Y-STRs have been used in criminal casework for almost 30 years [33], highlighting its main advantage in cases of alleged sexual assault in which the contribution of a male can be detected when present in a small amount in a mixture with female DNA. It may sometimes also reveal that more than one male has contributed to a mixture, which might not have been revealed in the standard autosomal analysis.
Routine analysis of sexual assault cases will normally employ chemical tests to suggests the presence of semen, confirmed with microscopic visualisation of sperm cells. This will often be followed by a differential extraction of epithelial and sperm cells to enable standard autosomal DNA profiling of the male specific-enriched contribution to a male–female mixture. Differential extraction, or differential lysis, was first described by Peter Gill in 1985 [34], at a time when DNA analysis was in its infancy and the polymerase chain reaction (PCR) that has transformed DNA typing had not yet been published [35], only just having been invented. Using a process that continues today, albeit with refinements, Gill described incubation of extracted DNA with an SDS/proteinase K/dithiothreitol mixture which tends to lyse epithelial cells, whereas sperm nuclei have a highly developed proteome, rich in protamine to allow the DNA to be tightly packed in the head, offering more protection in a lytic environment. The robust structure of sperm nuclei serves to protect these cells from damage which might otherwise significantly impact on the health of a fertilised oocyte [16].
Use of Y-STRs in routine casework is limited, however, particularly because of the inability to identify a person rather than a lineage, and a separate analysis for these markers could consume valuable case material. Massively parallel sequencing (MPS) techniques allow analysis of large numbers of DNA markers, including Y-STRs, in a single analysis, but until these techniques are introduced into routine forensic casework, the decision to undertake a Y-STR analysis will remain with the individual laboratory and associated guidance. Without an enrichment process for the male material, analysis of the male contribution in MPS multiplexes may still be compromised if present at low levels because of competitive amplification with more abundant DNA. In some cases, such as if semen has originated from an azoospermic male, or there has been contact or penetration of a female in which non-sperm DNA has been deposited at a low concentration, a separate analysis of Y-STRs can provide key evidence in a case.
Ethical issues associated with Y chromosome testing in criminal justice
There are several important cases involving Y-STR evidence, described more fully in the online supplementary material, that raise important ethical issues.
Albert DeSalva (the Boston Strangler)’s identity was confirmed after he had died years later through Y chromosome matching. DeSalva had confessed to the murder, yet no evidence had been found linking him to the crime. Investigators, determined to resolve the issue, violated the privacy of his nephew by testing a discarded water bottle. Although there are various laws in different US states, almost all allow for the non-consensual collection of human tissue for DNA [36]. This approach has been used in many cases, such as in the identification of the Golden State Killer. Clearly envisaged as a way to identify the potential guilty party, rather than a known innocent, the practice also raises issues about the identification of (untested) relatives of the individual and the potential for revealing unintentional secrets. In contrast, in 2006, the U.K. made DNA theft a criminal offence without consent [37] unless for law enforcement purposes, which would not fit these circumstances. Deliberate familial searching in the US is limited to using an offender DNA database in many states [38] and so there may have been an additional violation in this particular case.
After others were accused, the 1996 murderer of Angie Dodge was solved in 2019, but not without problems involving a very close Y-STR match impacting on an innocent that include a clear example of confirmation bias. Michael Usry Jnr, was required to provide a DNA sample because a close match was identified between the crime scene material and Usry's father. Usry had been in the state at the time of the murder. Although he was cleared of involvement after a month of waiting, nevertheless questions continue to be raised about his innocence in social media [39].
The murder of Marianne Vaastra in 1999 in The Netherlands raises additional ethical concerns. The racial prejudice exhibited by the local population led to the biased investigation of asylum seekers in the early stages of the investigation, unethical also because of wasting of time in catching the perpetrator, but also for the associated costs. The later illegal, albeit potentially justifiable, inference from analysis of the biogeographic ancestry (BGA) of the Y chromosome carried by the perpetrator, suggesting a male of Northwest European origin, has led to changes in the law to allow the process in future.
The prediction of BGA remains a highly sensitive issue, however, particularly in Germany, which continues to disallow its use; inference through the Y chromosome is, nevertheless, allowed [40]. Concerns have not diminished, however, as there was suspicion that haplotypes submitted to YHRD from minority groups, such as Uyghurs and Roma people, were collected without proper informed consent [41]. While there is a consensus that such profiles should be removed, this is contrasted with the possible negative impact on these minority groups when estimating the frequency of Y chromosome match.
Biogeographic ancestry
While a person's ancestry is rooted in concepts of language and culture associated with social constructs around the concepts of race and ethnicity, inference of a person's biogeographical ancestry is assessed through an understanding of genetic diversity in different population groups with respect to their distribution across world regions.
The distribution of polymorphisms that are not impacted by natural selection reflects the history of mobility of our species as populations have dispersed from Africa over many thousands of years. The influence of assortive mating, in which individuals are more likely to mate with each other, and genetic drift, in which the effect of random variability is more marked in smaller populations, plus the increase in population size over time, has produced genetic differentiation across the world [42]. Analysis of genetic markers, whether STRs, SNPs or other polymorphisms has shown that individuals living on the same continent are more similar in comparison with those in different continents.
Direct-to-consumer testing has led to a lay expectation of certainty in BGA but a broad continental ancestry from a broad view across an individual's genome is likely the best that can be done [43].
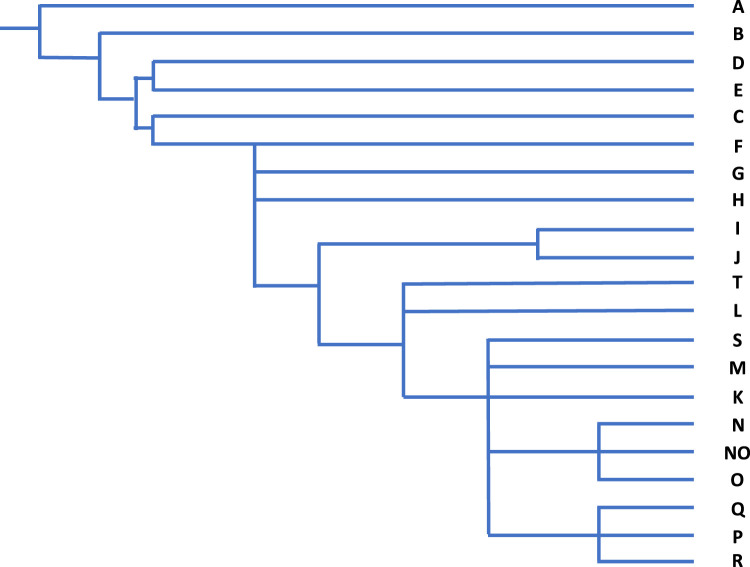
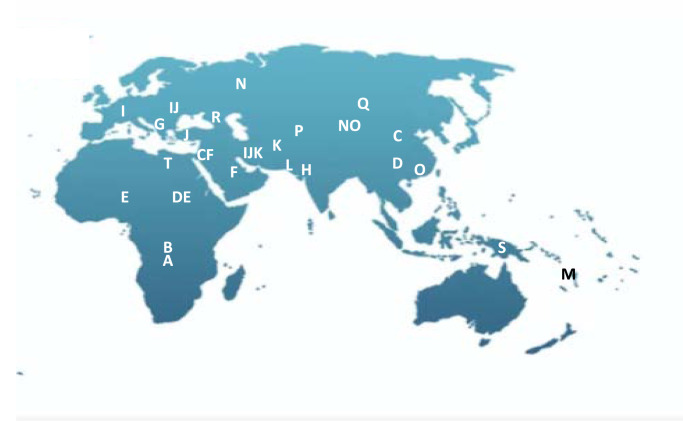
Examination of the Y chromosome can, however, only inform us about the BGA of our paternal ancestors, and only if we are male. While Y-STRs haplotypes can infer geographic ancestry, population origin and human migration routes can be best examined through examination of SNPs because of their low mutation rates. Phylogenetic trees of evolutionary relationships are modelled from characteristic SNPs in different population groups, defining core haplogroups [44,45]. Named alphabetically, the deepest branches in the phylogeny are haplogroups A and B that are essentially restricted to Africa, supporting a likely African ancestry for humans. Figure 5 illustrates the rooted phylogenetic tree with haplogroups being defined by characteristic SNPs and Figure 6 shows the core geographic locations.
Figure 5. Y chromosome core haplogroup phylogeny [46].
Figure 6. Geographic representation of likely origin of dominant Y-DNA haplogroups (data from [46]).
BGA made from the point of view of the Y chromosome may be very different from autosomal inference and its usefulness will depend on the ancestral groups within the relevant population. In the Vaastra case an inference was made of a north-western European origin for the perpetrator; this presumably was because more, or only, people from that region were represented within the YHRD database. That might have been a reasonable inference because the crime took place in a northern province that has not been impacted by immigration from other parts of the world, but that confidence may not be as strong in other parts of Europe. Such an inference may in truth be very deep-rooted as Figure 1 makes clear and the British Isles has a long history of the presence of Africans in its population which is not evident on a general population view. Haplogroup A is regarded as being African-specific, yet a population study of the British Isles revealed a collection of ‘indigenous’ Northern England residents with this same haplogroup, all, in this study, with the same surname, that was likely to have been introduced into the country some time before the late eighteen century [46]. This will, of course, not be the only example and highlights the potential danger of relying on BGA inference from the Y chromosome in populations with differing demographic histories.
Kinship
Y-STRs are frequently used in the examination of a paternal lineage, whether for criminal of civil investigations. The Y-STRs selected for including in forensic multiplexes are generally chosen for their population differentiation although early multiplex collection use, such as the STRs from the minimal haplotype collection, showed that this could be population dependent [47] and subsequent multiplexes have included markers that can offer better cross-population differentiation.
While it is expected that the Y chromosome passed from father to son will provide the same Y-STR haplotype, mutations are not infrequent. This is because male germ cells undergo more multiplication events than in female egg cells leading to such mutational events being more likely to be revealed in investigations of paternity than in maternity.
Rapidly mutating Y-STRs
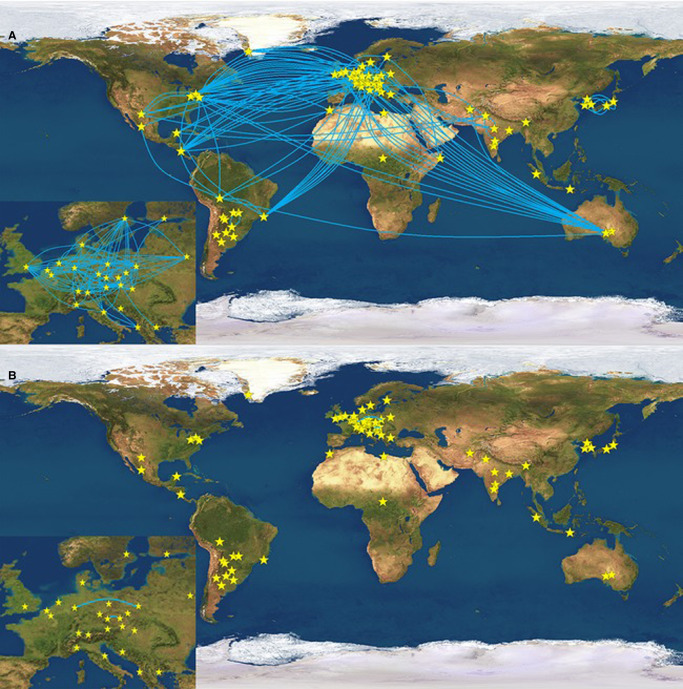
Analysis of a large number of Y-STRs in father-son pairs undertaken by Ballantyne et al. [48] added a further set of Y-STRs with high mutation rates that can be particularly useful in some instances, such as the increased potential for discriminating between people that otherwise share a Y-haplotype. Figure 7 illustrates the power of these 13 rapidly mutating (RM) YSTRs (DYF387S1, DYF399S1, DYF403S1, DYF404S1, DYS449, DYS518, DYS526, DYS547, DYS570, DYS576, DYS612, DYS626 and DYS627) to separate individuals living in different continents, or different counties, that otherwise share the same Yfiler haplotype. About half of these RM loci are now included in the Yfiler Plus multiplex. Potentially the set has the power to differentiate close relatives such as siblings, or even father and son. These markers also improve the ability to discriminate between closely related individuals with 26.9% of father–son relationships having at least one mutational difference, in comparison with Yfiler, and brothers being distinguished from each other in 56.3% of cases in comparison with 10% when using Yfiler. This distinction can be of particular importance in certain forensic cases.
Figure 7. Illustration of the power of rapidly mutating (RM) Y-STRs [48].
Blue lines link pairs that share the same Y-haplotype. A — testing using 17 Yfiler STRs; B-testing using 13 RM YSTRs. Links across Europe are magnified in the inset.
Civil uses of Y-STRs
Because of their paternal lineage, Y-STRs, while not being able to prove paternity, can be of assistance in certain cases. For example, when if the putative father is deceased or missing, a matching Y chromosome haplotype with another paternally related family member can help support the relationship. Y-STRs are frequently used in cases of inheritance where individuals may want to claim on an estate where the link may be disputed. In such circumstances, it is helpful to analyse as large many Y-STRs as possible, while avoiding the more rapidly mutating STRs.
Y-STRs can also be helpful in the triage of disaster victim identification, or in the identification of bodies in mass graves.
Use of Y-STRs in cases of historical interest
Y-STRs have been used in many cases of historical interest and but can raise other non-genetic matters that may need to be considered by living participants to these studies. In addition, it must always be foremost in the minds of investigators that these are lineage markers and there may be other explanations for their findings. Four examples are detailed in the supplementary material. These, far from providing definitive answers to the truth in some cases, have also raised other disappointments and unexpected findings, Three are historic, and one more recent.
The genetic investigation of the claims that Thomas Jefferson, one of the American Founding Fathers, fathered a child with one of his African slaves, has revealed several undesired consequences. Furthermore, the disproving of a theory that an American nineteenth-century mass murderer (The American Ripper) did not escape punishment and went on to commit similar crimes in the U.K. as Jack the Ripper, has disappointed the conspiracy theorists.
In the U.K., the discovery and identification of the skeleton of Richard III using mitochondrial DNA has, through analysis of the Y chromosome, revealed genetic inconsistencies in the line of succession of some members of the peerage and a more recent investigation in the Baronetcy of Stichill has forced transfer of a peerage from one side of a family to another which has raised issues of ‘informed consent’ as set down in data protection law.
Summary
Analysis of Y-STRs can be of high importance, particularly in sexual assault cases where female material in a female–male mixture may otherwise overwhelm the genetic evidence.
There are key circumstances where analysis of the Y chromosome can provide investigators with additional intelligence, such as ancestry, but this should always be viewed in the context of other available genetic and non-genetic information because it can only inform on lineages, rather than being a tool for identification.
As a lineage marker, the Y chromosome can provide a useful tool to triage remains in a mass disaster and can also provide historic evidence linking families over many generations.
Analysis of the Y chromosome can, however, raise important ethical issues that may adversely affect many people and should never be undertaken without that consideration.
Abbreviations
- CODIS
combined DNA index system
- MPS
massively parallel sequencing
- PARs
pseudoautosomal regions
- SNPs
Y-single nucleotide polymorph
- STR
short-tandem repeat
Competing Interests
The author declares that there are no competing interests associated with this manuscript.
Supplementary Material
References
- 1.Jobling, M.A. and Tyler-Smith, C. (1995) Fathers and sons: the Y chromosome and human evolution. Trends Genet. 11, 449–456 10.1016/S0168-9525(00)89144-1 [DOI] [PubMed] [Google Scholar]
- 2.Jobling, M.A., Pandya, A. and Tyler-Smith, C. (1997) The Y chromosome in forensic analysis and paternity testing. Int. J. Leg. Med. 110, 118–124 10.1007/s004140050050 [DOI] [PubMed] [Google Scholar]
- 3.Quintana-Murci, L. and Fellous, M. (2001) The human Y chromosome: the biological role of a “Functional Wasteland”. J. Biomed. Biotechnol. 1, 18–24 10.1155/S1110724301000080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sasaki, S. and Shimokawa, H. (1995) The amelogenin gene. Int. J. Dev. Biol. 39, 127–133 [PubMed] [Google Scholar]
- 5.Cooke, H. (1999) Y chromosome and male infertility. Rev. Reprod. 4, 5–10 10.1530/ror.0.0040005 [DOI] [PubMed] [Google Scholar]
- 6.Sullivan, K., Mannucci, A., Kimpton, C.P. and Gill, P. (1993) A rapid and quantitative DNA sex test: fluorescence-based PCR analysis of an X-Y homologous gene amelogenin. Biotechniques 15, 636–638. 640–641 PMID: [PubMed] [Google Scholar]
- 7.Santos, F.R., Pandya, A. and Tyler-Smith, C. (1999) Reliability of DNA based sex tests. Nat. Genet. 18, 103 10.1038/ng0298-103 [DOI] [PubMed] [Google Scholar]
- 8.Jobling, M.A., Lo, I.C., Turner, D.J., Bowden, G.R., Lee, A.C., Xue, Y.et al. (2007) Structural variation on the short arm of the human Y chromosome recurrent multigene deletions encompassing amelogenin Y. Hum. Mol. Genet. 16, 307e16 10.1093/hmg/ddl465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thangaraj, K., Reddy, A.G. and Singh, L. (2002) Is the amelogenin gene reliable for gender identification in forensic casework and prenatal diagnosis? Int. J. Leg. Med. 116, 121–123 10.1007/s00414-001-0262-y [DOI] [PubMed] [Google Scholar]
- 10.Parkin, E., Kraayenbrink, T., Opgenort, J.R.M.L., van Driem, G., Tuladhar, N.M., de Knijff, P.et al. (2007) Diversity of 26-locus Y-STR haplotypes in a Nepalese population sample: isolation and drift in the Himalayas. Forensic Sci. Int. 166, 176–181 10.1016/j.forsciint.2006.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yong, R.Y.Y., Gan, L.S.H., Chang, Y.M. and Yap, E.P.H. (2007) Molecular characterization of a polymorphic 3-Mb deletion at chromosome Yp11.2 containing the AMELY locus in Singapore and Malaysia populations. Hum. Genet. 122, 237–249 10.1007/s00439-007-0389-0 [DOI] [PubMed] [Google Scholar]
- 12.Cadenas, A.M., Regueiro, M., Gayden, T., Singh, N., Zhivotovsky, L.A., Underhill, P.A.et al. (2007) Male amelogenin dropouts: phylogenetic context, origins and implications. Forensic Sci. Int. 166, 155–163 10.1016/j.forsciint.2006.05.002 [DOI] [PubMed] [Google Scholar]
- 13.Chang, Y.M., Perumal, R., Keat, P.Y., Yong, R.Y., Kuehn, D.L., Burgoyne, L. (2007) A distinct Y-STR haplotype for amelogenin negative males characterized by a large Yp11.2 (DYS458-MSY1-AMEL-Y0 deletion. Forensic Sci. Int. 166, 115–120 10.1016/j.forsciint.2006.04.013 [DOI] [PubMed] [Google Scholar]
- 14.Charlesworth, B. and Charlesworth, D. (2000) The degeneration of Y chromosomes. Philos. Trans. R. Soc. Lond., B Biol. Sci. 355, 1563–1572 10.1098/rstb.2000.0717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carvalho, C.M., Zhang, F. and Lupski, J.R. (2011) Structural variation of the human genome: mechanisms, assays, and role in male infertility. Syst. Biol. Reprod. Med. 57, 3–16 10.3109/19396368.2010.527427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Champroux, A., Torres-Carreira, J., Gharagozloo, P., Drevet, J.R. and Kocer, A. (2016) Mammalian sperm nuclear organization: resiliencies and vulnerabilities. Basic Clin. Androl. 26, 17 10.1186/s12610-016-0044-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Balaresque, P., Bowden, G.R., Parkin, E.J., Omran, G.A., Heyer, E., Quintana-Murci, L.et al. (2008) Dynamic nature of the proximal AZFc region of the human Y chromosome: multiple independent events revealed by microsatellite analysis. Hum. Mutat. 29, 1171–1180 10.1002/humu.20757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kayser, M. and Sajantila, A. (2001) Mutations at Y-STR loci: implications for paternity testing and forensic analysis. Forensic Sci. Int. 118, 116–121 10.1016/S0379-0738(00)00480-1 [DOI] [PubMed] [Google Scholar]
- 19.Purps, J., Siegert, S., Willuweit, S., Nagy, M., Alves, C., Salazar, R.et al. (2014) The global analysis of Y chromosome diversity for 23 STR loci. Forensic Sci. Int. 12, 12–23 10.1016/j.fsigen.2014.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aliferi, A., Thomson, J., McDonald, A., Paynter, C.M., Ferguson, S., Vanhinsbergh, D.et al. (2018) UK and Irish Y-STR population data – a catalogue of variant alleles. Forensic Sci. Int.: Genet. 34, e1–e6 10.1016/j.fsigen.2018.02.018 [DOI] [PubMed] [Google Scholar]
- 21.1000 Genomes Project Consortium. (2015) A global reference for human genetic variation. Nature 526, 68–74 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee, E.Y., Lee, H.Y. and Shin, K. (2016) Off-ladder alleles due to a single nucleotide polymorphisms in the flanking region at DYS481 detected by the PowerPlex Y23 system. Forensic Sci. Int.: Genet. 24, 7–8 10.1016/j.fsigen.2016.06.019 [DOI] [PubMed] [Google Scholar]
- 23.National Institute of Standards and Technology (NIST) (2015) Core STR loci used in human identity testing – STRBase. https://strbase.nist.gov/coreSTRs.htm [accessed 1 January 2021]
- 24.Tao, R., Qi, W., Chen, C., Zhang, J., Yang, Z., Song, W.et al. (2019) Pilot study for forensic evaluations of the precision ID globalFiler™ NGS STR panel v2 with the Ion S5™ system. Forensic Sci. Int.: Genet. 43, 102147 10.1016/j.fsigen.2019.102147 [DOI] [PubMed] [Google Scholar]
- 25.Roewer, L., Krawczak, M., Willuweit, S., Nagy, M., Alves, C., Amorim, A.et al. (2001) Online reference database of European Y-chromosomal short tandem repeat (STR) haplotypes. Forensic Sci. Int. 118, 106–113 10.1016/S0379-0738(00)00478-3 [DOI] [PubMed] [Google Scholar]
- 26.Willuweit, S. and Roewer, L. (2015) The new Y chromosome haplotype reference database. Forensic Sci. Int.: Genet. 15, 43–48 10.1016/j.fsigen.2014.11.024 [DOI] [PubMed] [Google Scholar]
- 27.Unuvar, D., Baransel Isir, A., Cankus, G. and Pehlivan, S. (2008) Y chromosomal STR locus DYS385 in azoospermic and fertile men from the Aegean Region of Turkey: is there any forensic relevance? Balk. J. Med. Genet. 11, 39–44 10.2478/v10034-008-0027-8 [DOI] [Google Scholar]
- 28.SWGDAM (2014). Interpretation guidelines for Y chromosome STR typing. http://media.wix.com/ugd/4344b0_da25419ba2dd4363bc4e5e8fe7025882.pdf [accessed 1 January 2021]
- 29.Séré, M., Thévenon, S., Belem, A.M.G. and De Meeûs, T. (2017) Comparison of different genetic distances to test isolation by distance between populations. Heredity 119, 55–63 10.1038/hdy.2017.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DNA commission of the ISFG. (2020) Recommendations on the interpretation of Y-STR results in forensic analysis. Forensic Sci. Int.: Genet. 48, 202308. 10.1016/s0379-0738(01)00498-4 [DOI] [PubMed] [Google Scholar]
- 31.Anderson, M.M., Eriksen, P.S. and Morling, N. (2013) The discrete Laplace exponential family and estimation of Y-STR haplotype frequencies. J. Theor. Biol. 329, 39–51 10.1016/j.jtbi.2013.03.009 [DOI] [PubMed] [Google Scholar]
- 32.Anderson, M.M. and Balding, D.J. (2017) How convincing is a matching Y-chromosome profile? PLoS Genet. 13, e1007028 10.1371/journal.pgen.1007028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prinz, M. and Sansone, M. (2001) Y-chromosome specific short tandem repeats in forensic casework. Croat. Med. J. 42, 288–291 PMID: [PubMed] [Google Scholar]
- 34.Gill, P., Jeffreys, A. and Werrett, D. (1985) Forensic application of DNA “Fingerprints.”. Nature 318, 577–579 10.1038/318577a0 [DOI] [PubMed] [Google Scholar]
- 35.Mullis, K.F., Faloona, F., Scharf, S., Saiki, R., Horn, G. and Erlich, H. (1986) Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Cold Spring Harb. Symp. Quant. Biol. 51, 263–273 10.1101/SQB.1986.051.01.032 [DOI] [PubMed] [Google Scholar]
- 36.Lund, S. (2020) Ethical implications of forensic genealogy in criminal cases. J. Bus. Entrep. Law 13, 2 https://digitalcommons.pepperdine.edu/jbel/vol13/iss2/6 [Google Scholar]
- 37.Legislation.gov.uk. The Human Tissue Act (2004). https://www.legislation.gov.uk/ukpga/2004/30/contents [Accessed 1 January 2021]
- 38.Kim, J., Mammo, D., Siegel, M.B. and Kafsanis, S.H. (2011) Policy implications for familial searching. Investig. Genet. 2, 22 10.1186/2041-2223-2-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scudder, N. and McNevin, D. (2018). Is your DNA really your own? The public and forensic value of DNA. The conversation May 1. https://theconversation.com/is-your-genome-really-your-own-the-public-and-forensic-value-of-dna-95786 [Accessed 1 January 2021]
- 40.Editorial (2017). Forensics: Germany considers wider use of DNA evidence in criminal cases. Nature 543: 589-590 [DOI] [PubMed] [Google Scholar]
- 41.Schiermeier, Q. (2021) Crime-solving DNA database faces ethical scrutiny. Nature 594, 320–322 10.1038/d41586-021-01584-w [DOI] [PubMed] [Google Scholar]
- 42.Bamshad, M., Wooding, S., Salisbury, B.A. and Claiborne Stephens, J. (2004) Deconstructing the relationship between genetics and race. Nat. Rev. Genet. 5, 598–609 10.1038/nrg1401 [DOI] [PubMed] [Google Scholar]
- 43.McNevin, D. (2020) Forensic inference of biogeographical ancestry from genotype: the genetic ancestry lab. WIREs Forensic Sci. 2, e1356 10.1002/wfs2.1356 [DOI] [Google Scholar]
- 44.The Y Chromosome Consortium. (2002) A nomenclature system for the tree of human Y chromosomal binary haplgroups. Genome Res. 12, 339–348 10.1101/gr.217602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chiaroni, J., Underhill, P.A. and Cavalli-Sforza, L. (2009) Y chromosome diversity, human expansion, drift and cultural evolution. Proc. Natl Acad. Sci. USA 106, 20174–20179 10.1073/pnas.0910803106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.King, T.E., Parkin, E.J., Swinfield, G., Cruciani, F., Scozzari, R., Rosa, A.et al. (2007) Africans in Yorkshire? The deepest-rooting clade of the Y phylogeny within an English genealogy. Eur. J. Hum. Genet. 15, 288–293 10.1038/sj.ejhg.5201771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leat, N., Benjeddou, M. and Davison, S. (2004) Nine-locus Y chromosome STR profiling of Caucasian and Xhosa populations from Cape Town, South Africa. Forensic Sci. Int. 144, 73–75 10.1016/j.forsciint.2004.02.022 [DOI] [PubMed] [Google Scholar]
- 48.Ballantyne, K.N., Ralf, A., Aboukhalid, R., Achakzai, N.M., Anjos, M.J., Ayub, Q.et al. (2014) Toward male individualization with rapidly mutating Y-chromosomal short tandem repeats. Hum. Mutat. 35, 1021–1032 10.1002/humu.22599 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.