Abstract
Background
COVID-19 is diagnosed using RT-PCR assays of samples from nasal and oropharyngeal swabs. People with negative RT-PCR often presented with clinical manifestations of COVID-19. The data on such patients are lacking. The present study aims to characterize the patients who were suspected COVID-19 cases and tested negative in RT-PCR compared to patients who had been tested RT-PCR positive.
Methods
This is a retrospective, observational study of adult suspected and confirmed patients of COVID-19 admitted to King Saud University Medical City, Riyadh, Saudi Arabia, from 1st March 2020 until 30th November 2020. Laboratory confirmation is done through nasal/pharyngeal swab specimens, tested positive in RT-PCR assay. Patients with initial negative RT-PCR test results were assessed again within 48−72 h to avoid false-negative results. Patient data were extracted from the electronic medical files of each included patient using a predesigned case report form.
Results
The study included 488 (80.93%) patients with RT-PCR swab results positive, and 115 (19.07%) patients who were negative. Respiratory rate and diastolic blood pressure were higher among the swab-positive cases. More number of swab-negative patients had comorbidities such as coronary heart disease, chronic kidney disease, and carcinoma. Fever, cough, and shortness of breath were reported higher among the swab-positive cases. ALT and AST, and LDH levels were found higher among RT-PCR-positive patients. Serum creatinine, blood urea nitrogen and troponin were more elevated in RT-PCR-negative patients. Antibiotics, anticoagulants, and corticosteroids were used more by swab-positive patients. Significantly higher number of RT-PCR-positive patients required proning, high-flow nasal cannula, non-invasive mechanical ventilation, and invasive mechanical ventilation. Acute cardiac ischemia and death were found to be similar among the patients. However, deaths occurred significantly earlier among the swab-positive cases when compared to the swab-negative group.
Conclusion
Distinctive symptoms and markers of COVID-19 are more frequent among patients who had RT-PCR-positive results.
Abbreviations: ACE-2, angiotensin-converting enzyme-2; ALP, alkaline phosphatase; ALT, alanine aminotransferase; ARDS, acute respiratory distress syndrome; AST, aspartate aminotransferase; BMI, body mass index; BUN, blood urea nitrogen; CK, creatine kinase; CKD, chronic kidney disease; COVID-19, coronavirus disease-2019; CRP, C-reactive protein; GGT, gamma-glutamyl transferase; ICU, intensive care unit; INR, international normalised ratio; LDH, lactate dehydrogenase; PT, prothrombin time; PTT, partial thromboplastin time; RT-PCR, reverse-transcriptase polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome-coronavirus-2; SPSS, Statistical Package for the Social Sciences
Keywords: COVID-19, Clinical characteristics, Negative RT-PCR, Suspected COVID-19
Introduction
The coronavirus disease-2019 (COVID-19) caused by severe acute respiratory syndrome-coronavirus 2 (SARS-CoV-2) became a global pandemic in March 2020. There have been ∼187 million cases and ∼4 million deaths as of 13th July 2021 [1]. The clinical characteristics of COVID-19 patients ranged from asymptomatic in mild cases to acute respiratory distress syndrome and death in severe cases. The most common symptoms of COVID-19 are headache, loss of smell, nasal congestion, cough, asthenia, myalgia, rhinorrhea, sore throat, fever, shortness of breath, nausea or vomiting, and diarrhea [2,3]. Commonly reported comorbidities of COVID-19 are hypertension, obesity, diabetes, and cardiovascular disease [4]. Risk factors for mortality were age (≥60 years), male gender, smoking history, COPD, hypertension, diabetes, heart disease, and chronic kidney disease [5]. The first case of COVID-19 was reported on 2nd March 2019 in Saudi Arabia. As of 13th July 2021, Saudi Arabia has reported ∼500,000 cases and 8006 deaths with a case fatality rate of 1.6% [6].
Cough, fever, fatigue, dyspnea, and sore throat were the most common symptoms seen in COVID-19 patients in Saudi Arabia [7]. In addition, 52.5% of the hospitalized patients had comorbid conditions, with diabetes and hypertension being the most frequent comorbidities [8]. COVID-19 mainly affects the respiratory system, as the virus SARS-CoV-2 enters and replicates in the epithelial mucosa of the upper respiratory tract. The virus binds with angiotensin-converting enzyme-2 (ACE-2) receptor protein to enter the cells and downregulate it in the process. It leads to acute lung injury through the uncontrolled action of proinflammatory angiotensin II [9]. Rapid replication of the virus in the respiratory tract triggers an intense inflammatory response and inflammatory cytokine production named ‘cytokine storm.’ The acute inflammatory response leads to severe manifestations of COVID-19 such as tissue injury, coagulopathy, multiple organ dysfunction, and sepsis [10].
COVID-19 is diagnosed using real-time reverse-transcriptase polymerase chain reaction (RT-PCR) assays by detecting the SARS-CoV-2 from nasal and oropharyngeal swabs. The initial false positive RT-PCR may be as high as 54%, indicating the need for repeat testing and additional diagnostic tests. Apart from the analytical factors, the viral load, time of the sample, and quantity of specimen contribute to the false-positive cases of COVID-19 [11]. It has been even proposed to use deep tracheal aspirate instead of nasal or pharyngeal swabs to avoid false negative results in patients hospitalized with pneumonia [12]. People with negative RT-PCR reports often presented with clinical manifestations of COVID-19 [13]. These patients would require appropriate clinical assessment and treatment. The data on the clinical characteristics, laboratory profile, and outcomes of such patients are lacking.
The epidemiological and clinical characterizations mainly focused on the COVID-19 patients who tested positive in RT-PCR. There is a lack of data on clinical presentation and biochemical profiles of patients who tested negative RT-PCR. The present study aims to characterize the patients who were suspected COVID-19 cases and tested negative in RT-PCR compared to patients who had been tested RT-PCR positive.
Methods
Study design, setting and participants
This is a retrospective observational study that included adult suspected and confirmed cases of COVID-19 admitted to King Saud University Medical City, Riyadh, Saudi Arabia, from 1st March 2020 until 30th November 2020.
Case definition
The case definition for this study was based on the Ministry of Health, Saudi Arabia, and the Saudi Center for Disease Prevention and Control definition published in April 2020 [14]. Suspected COVID-19 case was defined as either patient with acute respiratory illness (sudden onset of at least one of the following: fever (measured or by history), cough, or shortness of breath) and in the 14 days prior to symptom onset, met at least one of the following epidemiological criteria with one or more of epidemiological link such as having a history of travel abroad or visiting or being a resident of the high-risk area for COVID-19 in the Kingdom or having a close physical contact prior to symptom onset with a confirmed COVID-19 case or working in a healthcare facility, or any admitted adult patient with unexplained severe acute respiratory illness (SARI), either Community-Acquired Pneumonia (CAP) or Hospital Acquired Pneumonia (HAP).
A confirmed case is defined as a suspected case with laboratory confirmation of COVID-19 infection [14]. Laboratory confirmation is done through nasal/pharyngeal swab specimens, tested positive for 2019-nCoV nucleic acid using RT-PCR assay. Patients with negative initial RT-PCR test results were assessed again within 48−72 h to avoid false-negative results. The result was defined as positive if it appeared positive once, and it was considered negative only if it remained negative consecutively.
Data collection
Data were extracted from the electronic medical files of each included patient using a predesigned case report form. Socio-demographic characteristics, vital signs, underlying comorbid conditions, clinical symptoms, and laboratory parameters on admission, treatment, and outcomes were retrieved. The study protocol was reviewed and approved by the institutional review board at the College of Medicine, King Saud University, Riyadh, Saudi Arabia.
Statistical analysis
Continuous data are represented as mean and standard deviation, or median and interquartile range and analyzed using Analysis of Variance (ANOVA) or independent t-test or Mann–Whitney U test. Proportions are expressed as frequencies and percentages and were compared using the Chi-square test. Association of demographic and clinical characteristics with swab-positive cases was assessed using logistic regression. Data analysis was conducted using the Statistical Package for Social Sciences (SPSS) version 26 (SPSS Inc, Armonk, New York, USA), and a p value of <0.05 was considered statistically significant.
Results
A total of six hundred and three patients were included in the study; among them, 488 (80.93%) patients had RT-PCR swab results positive, and 115 (19.07%) patients had RT-PCR swab results negative for SARS-CoV-2. The demographic profile of the study participants is presented in Table 1 . The gender ratio was similar in both swab-negative and swab-positive group. However, the swab-negative cases were significantly older than the swab-positive patients, with 48.6% of cases among the swab-positive group are less than 50 years of age. The vital signs did not differ significantly between the two groups, except for the respiratory rate and diastolic blood pressure, which were significantly higher among the swab-positive cases (p = 0.012 and p = 0.037, respectively). Although the mean body mass index (BMI) was similar between the groups, the percentage of overweight and obese patients was higher among the swab-positive patients (p < 0.001).
Table 1.
Demographic, anthropometric and vital signs of the study subjects.
| Variables | All patients | RT-PCR positive result | RT-PCR negative result | p Values |
|---|---|---|---|---|
| Mean ± SD or N (%) | Mean ± SD or N (%) | Mean ± SD or N (%) | (RT-PCR positive vs RT-PCR negative) | |
| N = 603 | N = 488 | N = 115 | ||
| Demography | ||||
| Gender | ||||
| Male | 317 (52.7%) | 256/488 52.5%) | 61/115 (53.0%) | 0.927 |
| Female | 285 (47.3%) | 231/488 (47.3%) | 54/115 (47.0%) | |
| Age, in years | ||||
| Overall | 51.60 ± 18.11 | 50.06 ± 17.21 | 57.86 ± 20.35 | |
| 18−29 | 74 (12.3%) | 60 (12.3%) | 14 (12.2%) | <0.001* |
| 30−39 | 109 (18.1%) | 96 (19.7%) | 13 (11.3%) | |
| 40−49 | 90 (14.9%) | 81 (16.6%) | 9 (7.8%) | |
| 50−64 | 182 (30.2%) | 148 (30.3%) | 34 (30.0%) | |
| ≥65 | 148 (24.5%) | 103 (21.1%) | 45 (39.1%) | |
| Marital status | ||||
| Single | 103 (17.1%) | 81/488 (16.6%) | 22/114 (19.3%) | |
| Married | 381 (63.3%) | 311/488 (63.7%) | 70/114 (61.4%) | 0.787 |
| Widowed/Divorced | 118 (19.6%) | 96/488 (19.7%) | 22/114 (19.3%) | |
| Respiratory rate (per min) | 21.84 ± 4.98 | 22.12 ± 5.20 | 20.81 ± 3.91 | 0.012* |
| Pulse rate (per min) | 90.19 ± 15.47 | 90.29 ± 15.00 | 90.08 ± 17.46 | 0.897 |
| SBP, in mmHg | 125.50 ± 18.35 | 125.82 ± 17.65 | 124.32 ± 21.35 | 0.436 |
| DBP, in mmHg | 72.79 ± 12.62 | 73.32 ± 11.97 | 70.59 ± 14.88 | 0.037* |
| BMI, kg/m2 | ||||
| Overall mean | 29.75 ± 7.33 | 30.05 ± 7.02 | 28.59 ± 8.51 | 0.063 |
| <18.5 | 14 (2.5%) | 8/462 (1.7%) | 6/108 (5.6%) | |
| 18.5−24.9 | 129 (22.6%) | 97/462 (21.0%) | 32/108 (30.0%) | 0.018* |
| 25−29.9 | 192 (33.7%) | 159/462 (34.4%) | 33/108 (30.1%) | |
| ≥30 | 235 (41.2%) | 198/462 (43.0%) | 37/108 (34.3%) | |
Significant p value between RT-PCR positive and negative groups.
The frequency of comorbidities such as diabetes mellitus, hypertension and immunosuppressive disease did not differ significantly between the groups. On the other hand, the swab-negative cases had significantly higher rates of coronary heart disease, chronic kidney disease, and carcinoma (p < 0.001, p = 0.001 and p = 0.015, respectively) compared to the swab-positive cases (Table 2 ). Although the history of chronic obstructive pulmonary disease among the RT-PCR-positive patients was almost twice the rate among the RT-PCR-negative patients, the difference did not reach a statistically significant level (p = 0.057). The most common symptoms among both groups were fever, cough, and shortness of breath. These symptoms were reported significantly higher among the swab-positive cases compared to the swab-negative group (p < 0.001, p < 0.001 and p = 0.023, respectively). Other symptoms such as sore throat, tiredness, diarrhea, runny nose, loss of sense of odor, and loss of sense of taste were reported by fewer participants; however, they were significantly higher among the swab-positive cases.
Table 2.
Comorbidities and clinical presentation of the study subjects.
| Variables | All patients | RT-PCR positive result | RT-PCR negative result | p Values |
|---|---|---|---|---|
| N (%) | N (%) | N (%) | (RT-PCR positive vs RT-PCR negative) | |
| N = 603 | N = 488 | N = 115 | ||
| Comorbidities | ||||
| Diabetes | 267 (44.4%) | 215/488 (44.1%) | 52/114 (45.6%) | 0.763 |
| Hypertension | 242 (40.3%) | 188/486 (38.7%) | 54/115 (47.0%) | 0.104 |
| Coronary heart disease | 91 (15.1%) | 60/488 (12.3%) | 31/115 (27.0%) | <0.001* |
| COPD | 35 (5.8%) | 31/487 (6.4%) | 4/114 (3.5%) | 0.057 |
| CKD | 62 (10.3%) | 40/485 (8.2%) | 22/114 (19.3%) | 0.001* |
| Carcinoma | 27 (4.5%) | 17/488 (3.5%) | 10/115 (8.7%) | 0.015* |
| Immunosuppressive disease | 9 (1.5%) | 7/488 (1.4%) | 2/115 (1.7%) | 0.808 |
| Clinical presentation | ||||
| Loss of sense of odour | 34/599 (5.7%) | 33/488 (6.7%) | 1/115 (0.9%) | 0.013* |
| Loss of sense of taste | 43/599 (7.2%) | 42/488 (8.6%) | 1/115 (0.9%) | 0.004* |
| Loss of appetite | 70/600 (11.7%) | 59/485 (12.2%) | 11/115(9.6%) | 0.435 |
| Fever | 302/598 (50.5%) | 275/484 (56.8%) | 27/114 (23.7%) | <0.001* |
| Cough | 318/599 (53.1%) | 283/484 (58.5%) | 35/115 (30.4%) | <0.001* |
| Tiredness | 108/600 (18.0%) | 96/485 (19.8%) | 12/115 (10.4%) | 0.019* |
| Shortness of breath | 334/600 (55.7%) | 283/485(58.3%) | 51/115 (44.3%) | 0.023* |
| Runny nose | 45/598 (7.5%) | 43/483 (8.9%) | 2/115 (1.7%) | 0.009* |
| Sore throat | 111/599 (18.5%) | 105/484 (21.7%) | 6/115 (5.2%) | <0.001* |
| Body aches | 155/600 (25.8%) | 132/485 (27.2%) | 23/115 (20.0%) | 0.112 |
| Diarrhoea | 85/597 (14.2%) | 83/482 (17.2%) | 2/115 (1.7%) | <0.001* |
| CNS presentation | 50/594 (8.4%) | 44/481 (9.1%) | 6/113 (5.3%) | 0.186 |
| Headache | 38/593 (6.4%) | 34/481 (7.1%) | 4/112 (3.6%) | 0.173 |
| Confusion | 14/592 (2.4%) | 11/480 (2.3%) | 3/112 (2.7%) | 0.808 |
Significant p value between RT-PCR positive and negative groups.
Comparison of the hematological and biochemical parameters of the study participants revealed interesting findings. Unlike the clinical characteristics, which were suggestive of clinical COVID-19 in the majority of the RT-PCR positive cases, the biochemical profile of the study participants presented a mixed scenario (Table 3 ). Basophils and eosinophils were significantly lower among swab-positive cases (p = 0.017 and p = 0.010, respectively). Hemoglobin level and hematocrit were significantly lower in swab-negative patients (p < 0.001 and p < 0.001, respectively), whereas platelet count was significantly lower in swab-positive cases (p = 0.001). Liver enzymes ALT and AST, and LDH levels were found to be significantly higher among RT-PCR-positive patients (p < 0.001, p = 0.001, and p < 0.001, respectively). Renal function markers serum creatinine and blood urea nitrogen and troponin, a cardiac injury marker, were significantly more elevated in RT-PCR-negative patients (p = 0.014, p = 0.008, and p = 0.001, respectively). On the other hand, the coagulation profile (INR and PT) was significantly higher among RT-PCR-positive cases (p < 0.001 and p < 0.001, respectively). Even though the fibrinogen levels were higher in RT-PCR-positive patients, it was not statistically significant (p = 0.055). RT-PCR-positive patients reported a significantly higher level of ferritin when compared to swab-negative cases (p < 0.001). The inflammatory marker CRP was significantly higher in RT-PCR-positive patients (p < 0.001). Although the fasting blood glucose levels were higher among RT-PCR-positive patients, it was not statistically significant (p = 0.341).
Table 3.
Hematology and biochemical parameters of the study subjects.
| Variables | All patients | RT-PCR positive | RT-PCR negative | p Values |
|---|---|---|---|---|
| N = 603 | N = 488 | N = 115 | (RT-PCR positive vs RT- PCR negative) | |
| Hematology | ||||
| Granulocyte, (×109/L) | 6.71 ± 9.05 (0.7−79.0) | 6.72 ± 9.97 | 6.58 ± 3.39 | 0.936 |
| Lymphocytes, (×109/L) | 1.32 ± 0.77 (0.10−4.90) | 1.29 ± 0.75 | 1.43 ± 0.81 | 0.165 |
| Monocytes, (×109/L) | 0.60 ± 0.42 (0.1−4.1) | 0.58 ± 0.43 | 0.67 ± 0.34 | 0.085 |
| Eosinophils, (×109/L) | 0.09 ± 0.23 (0−2.0) | 0.08 ± 0.22 | 0.16 ± 0.26 | 0.010* |
| Basophils, (×109/L) | 0.03 ± 0.06 (0−0.70) | 0.03 ± 0.07 | 0.05 ± 0.05 | 0.017* |
| MCV, (fL) | 84.45 ± 7.22 (52.3−113.1) | 84.17 ± 7.24 | 85.61 ± 7.15 | 0.063 |
| MCH, (pg) | 28.53 ± 2.91 (17−38) | 28.48 ± 2.92 | 28.71 ± 2.92 | 0.468 |
| MCHC, (g/L) | 337.01 ± 12.43 (287−384) | 337.34 ± 12.37 | 335.73 ± 12.73 | 0.229 |
| Hgb, (g/L) | 125.89 ± 22.56 (46−173) | 127.77 ± 21.14 | 119.24 ± 25.66 | <0.001* |
| Hct, (%) | 37.36 ± 6.36 (13.6−54.3) | 37.90 ± 5.90 | 35.49 ± 7.38 | <0.001* |
| Platelets, (×109/L) | 252.32 ± 107.04 (17−732) | 244.31 ± 103.85 | 281.41 ± 110.69 | 0.001* |
| MPV, (fL) | 8.37 ± 1.09 (3.3−12.7) | 8.41 ± 1.06 | 8.23 ± 1.24 | 0.141 |
| Blood biochemistry | ||||
| AST, (U/L) | 32.00 (19.0–53.0) | 34.0 (6.0−494.0) | 25.5 (6.0−606.0) | 0.001* |
| ALT, (U/L) | 34.00 (22.0−53.0) | 37.0 (8.0−395.8) | 25.0 (5.0−1000) | <0.001* |
| ALP, (U/L) | 78.00 (61.0−116.0) | 76.0 (30.0−670.0) | 89.0 (24.0−975.0) | 0.079 |
| GGT, (U/L) | 54.00 (27.25−107.75) | 54.00 (3.0−1501) | 44.00 (13.0−748.0) | 0.606 |
| LDH, (U/L) | 335.5 (236.75−452.25) | 342.0 (101−1979) | 167.00 (99−1059) | <0.001* |
| Calcium, (mmol/L) | 2.18 ± 0.26 | 2.17 ± 0.24 | 2.22 ± 0.26 | 0.156 |
| Phosphates, (mmol/L) | 1.08 ± 0.42 | 1.11 ± 0.35 | 1.30 ± 0.63 | <0.001* |
| Creatinine, (μmol/L) | 78.0 (20.8−56.0) | 76.0 (15.0−1268) | 86.50 (19.0−1520) | 0.041* |
| BUN, (mmol/L) | 4.70 (3.2−7.45) | 4.60 (1.20−41.20) | 6.25 (1.33−78.4) | 0.008* |
| Troponin, (ng/L) | 8.10 (1.50−23.25) | 7.00 (0.001−28,389) | 14.70 (1.5−15,039) | 0.001* |
| Creatine kinase, (U/L) | 117.50 (58.2−242.5) | 118.0 (8−3062) | 111.0 (15−753) | 0.852 |
| D-dimer, (μg/L) | 1.02 (0.64−1.73) | 1.01 (0.27−20.0) | 1.28 (0.27−20.0) | 0.163 |
| Ferritin, (μg/L) | 435.60 (137.63−1106.6) | 500.0 (5−12,961) | 191.0 (21−2263) | <0.001* |
| Fibrinogen, (g/L) | 5.53 (3.9−7.01) | 5.53 (2.13−9.60) | 3.57 (3.27−6.08) | 0.055 |
| INR | 1.05 (0.97−1.15) | 0.99 (0.86−1.53) | 1.08 (0.94−1.28) | <0.001* |
| PT, (s) | 14.20 (13.5−15.4) | 13.60 (12.0−19.2) | 15.30 (13.0−16.7) | <0.001* |
| PTT, (s) | 38.5 (34.7−44.2) | 38.93 (28.7−51.5) | 34.6 (31.2−47.0) | 0.864 |
| CRP, (mg/L) | 70.3 (24.7−123.0) | 79.3 (0.84−233) | 63.40 (27.4−128.0) | <0.001* |
| FBS, (mmol/L) | 6.6 (5.2−9.25) | 7.0 (4.0−35.0) | 5.47 (4.0−7.0) | 0.341 |
Values are expressed as mean ± standard deviation with range given in the parenthesis or median and interquartile range in the parenthesis.
Significant p value between RT-PCR positive and negative groups.
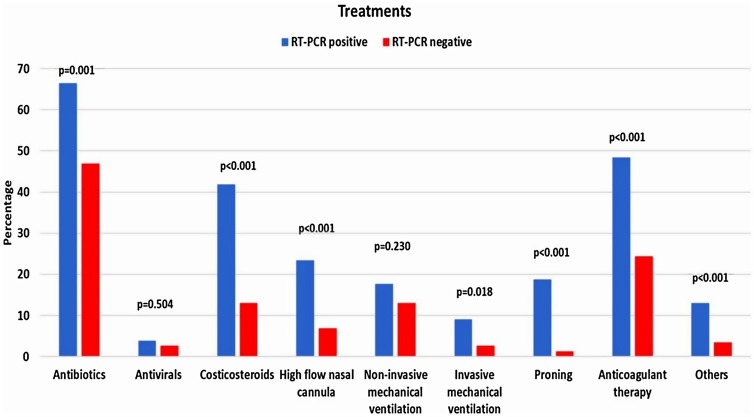
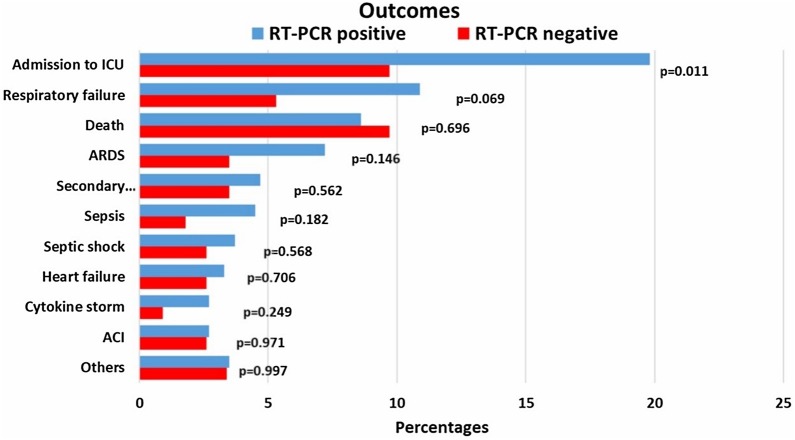
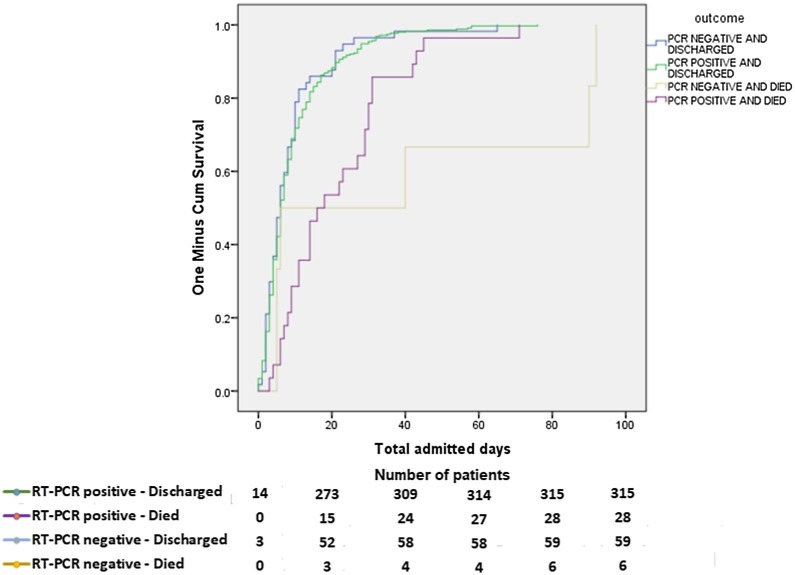
The most used drugs were antibiotics, anticoagulants, and corticosteroids in both groups, in which swab-positive cases were significantly higher in receiving those treatments (Fig. 1 ) (p = 0.001, p < 0.001 and p < 0.001, respectively). Only a minor proportion of patients received antiviral therapy, and it was similar between the groups. A significantly higher number of RT-PCR-positive patients required proning, high-flow nasal cannula, non-invasive mechanical ventilation, and invasive mechanical ventilation (p < 0.05). Fig. 2 reports the outcomes of the patients. Admission to the intensive care unit was found significantly higher among swab-positive cases (p = 0.011). Respiratory failure, acute respiratory distress syndrome, secondary infections, sepsis, septic shock, heart failure, and cytokine storm were found higher among RT-PCR-positive patients but not statistically significant. Acute cardiac ischemia and death were found to be similar among the patients. The rate of discharge and death among the groups are presented as the Kaplan Meier curve in Fig. 3 . The curve was constructed after adjustment (exclusion) of patients with comorbid conditions such as coronary heart disease, chronic kidney disease and carcinoma which were found to be significant confounders for mortality and discharge. The number of days for being discharged from the hospital between the two groups was not different. However, deaths occurred significantly earlier among the swab-positive cases when compared to the swab-negative group (19.56 ± 2.44 days vs. 39.67 ± 17.14 days, p = 0.037).
Fig. 1.
Treatments provided to the patients categorized according to the RT-PCR status.
Fig. 2.
Clinical outcomes of the study patients by RT-PCR status.
Fig. 3.
Cumulative hospitalization outcomes of patients by RT-PCR status.
Discussion
This study describes the clinical characteristics, treatments, and outcomes of SARS-CoV-19 RT-PCR positive patients and RT-PCR negative patients. All these patients had either exposure history and or symptoms suggestive of COVID-19 and required hospitalization. RT-PCR-negative patients were older and had a higher rate of comorbidities such as coronary heart disease, chronic kidney disease, and carcinoma. Even though the two groups showed a similar pattern of clinical presentation, the patients in RT-PCR positive group are characterized by a higher rate of typical COVID-19 symptoms. Lower lymphocyte count on admission is one characteristic of COVID-19 observed in earlier studies [15]. However, in the present study, lymphocyte count in the swab-positive group is only slightly less than in the RT-PCR-negative group and not significant. Previous studies comparing RT-PCR positive and RT-PCR negative cases showed a significantly lower lymphocyte count among the positive patients [13,16]. On the other hand, eosinophils and basophils were found lower among the RT-PCR-positive patients. Lower levels of eosinophils and basophils are reported to be associated with delayed recovery in COVID-19 patients [17]. Contrastingly, we observed a similar time to discharge among the survivors in both groups.
Hemoglobin and hematocrit levels are significantly higher in RT-PCR-positive patients than in RT-PCR-negative patients. A progressive decrease in hemoglobin levels with the increasing disease severity was reported in COVID-19 patients [18]. The laboratory parameters of the present study were assessed at the time of admission to the hospital. The lower hemoglobin levels among the RT-PCR-negative cases might be attributed to higher chronic kidney disease and coronary disease patients. It may be noted that the serums creatinine, phosphate, and BUN levels are significantly more elevated among RT-PCR negative cases indicating poor renal function. Ferritin, the other hemoglobin-related marker, was found to be very high among RT-PCR-positive patients. This is in agreement with the previous reports [19]. Increased circulating ferritin level is considered as a marker of an uncontrolled and heightened immune response. The inflammatory cytokines produced during the COVID-19 infection induce hepatocytes, Kupffer cells, and macrophages to secrete ferritin [20]. Ferritin has been implicated as an immune mediator and is associated with the hyperinflammatory state of COVID-19. Another inflammatory marker, C-reactive protein (CRP), was also found to be significant in the RT-PCR positive cases.
Significantly higher levels of serum LDH among RT-PCR positive cases indicate COVID-19 had already progressed to severe levels. Elevated liver enzymes were observed among both groups. But the RT-PCR positive group showed significantly higher serum ALT and AST levels than the RT-PCR negative group. SARS-CoV-19 infection is known to increase liver enzymes by affecting the liver in multiple ways. Direct injury to ACE2 expressing liver epithelial cells, inflammatory response mediated damage to liver cells, hypoxia-induced cell damage, and hepatotoxicity arising from multiple drug treatments are the major causes of elevated liver enzymes observed among COVID-19 patients [21]. The other liver enzymes ALP and GGT, were not different between the groups. However, they were abnormally high among many patients in both groups. This contrasts with the serum AST and ALT levels, which were too abnormal in both groups but significantly higher among RT-PCR positive cases. Such a difference was not observed in serum ALP and GGT levels.
Serum creatine kinase (CK), an indicator of muscle damage, has been found to be associated with poor outcomes among COVID-19 patients [22]. We found abnormally high CK levels in both groups in the present study, although the range has been broader among RT-PCR-positive patients. Elevated CK levels are also implicated in cardiovascular and renal diseases. Therefore, individual assessment of the patients will be more appropriate to identify the causes of elevated CK levels. Troponin, a marker of myocardial inflammation, was found higher among RT-PCR-negative patients, though the RT-PCR-positive group had a more comprehensive range. Troponin levels are often associated with COVID-19 indicating myocardial injury mediated by inflammatory cytokines. Acute respiratory infections and sepsis also increase troponin levels. As sepsis and secondary infections were reported less frequently among the RT-PCR-negative group, the higher level of serum troponin among this group may be ascribed to the presence of more patients with acute myocardial injury. Coagulopathy in COVID-19 is defined by decreased platelet counts, elevated D-dimer, fibrinogen levels, and prolongation of prothrombin time/partial thromboplastin time [23]. With almost all the coagulation parameters in the abnormal range, coagulopathy seems to be present in both groups in the present study. D-dimer, fibrinogen, and PTT levels were similar between the RT-PCR-positive and RT-PC-negative groups. It should be kept in mind that chronic kidney disease can independently increase the coagulation markers [24]. Therefore, its influence on the coagulopathy observed among RT-PCR negative cases cannot be ruled out.
Even though the admission to ICU was higher in RT-PCR positive cases, mortality has been similar between the groups. Except for ICU admission, respiratory failure, and ARDS, all other severe outcomes were reported in similar frequency between RT-PCR-positive and RT-PCR-negative groups. In addition, the presence of comorbidities was higher among RT-PCR-negative patients. Death among RT-PCR positive patients occurred earlier than RT-PCR negative patients. With more patients admitted to ICU, experienced respiratory failure and ARDS, the RT-PCR-positive group, appears to have a severe manifestation of COVID-19, which shortened the time to death. Comparable time to discharge rates between groups indicates that both groups’ survivors had a similar disease severity.
Clinical and biochemical characteristics of RT-PCR-negative patients overlap with RT-PCR-positive patients to some extent. A substantial number of RT-PCR-negative patients were symptomatic and required hospital admission. In addition, many of them had coronary heart disease and chronic kidney disease. Even though RT-PCR-positive patients presented with many of the distinctive markers of COVID-19, few markers were observed equally between the two groups. While some of them could be attributed to underlying disease conditions, it may not rule out clinical COVID-19 in RT-PCR negative cases. The RT-PCR-positive group had a severe manifestation of COVID-19 whereas, mortality and time to discharge were similar between the groups. It may be possible that the RT-PCR positive group had a higher viral load that resulted in the severe disease and shorter time to death. Therefore, patients with an RT-PCR-negative report should not be discharged without clinical assessment, especially in patients with comorbidities. RT-PCR-negative patients with clinical manifestations should be dealt with a high degree of suspicion and treated appropriately. Imaging could provide additional information about the presence of a SARS-CoV-2 infection. Serological testing also can be used to detect exposure to SARS-CoV-2. Therefore, in RT-PCR negative cases, clinical presentation, imaging, and serology should be evaluated depending on the availability and level of suspicion. Identifying the false-negative cases will lead to isolation and prevent clusters of nosocomial infections. Protocols should be developed to deal with highly suspected false-negative cases, particularly those with comorbidities.
The strength of this study is that we included RT-PCR-negative patients that were negative in two consecutive tests. We were able to report the outcomes of the RT-PCR negative patients as we included only patients who were hospitalized. At the same time, it is also one of the limitations of this study, as we do not know the outcomes of RT-PCR negative cases who have COVID-19 like symptoms but were not hospitalized. The hospitalized RT-PCR-negative patients are more likely to have had clinical COVID-19 symptoms. Besides, the RT-PCR-negative patients presented with more frequent comorbidities. This could have been one of the reasons for the hospitalization of those RT-PCR-negative patients. Therefore, the results of this study cannot be generalized to all RT-PCR negative cases. Further, we did not investigate the imaging reports of the patients. We do not know how many of the RT-PCR negative patients had radiology image assessments.
Conclusion
The clinical and biochemical characteristics of RT-PCR-negative patients have certain similarities to the RT-PCR-positive patients. Distinctive symptoms and markers of COVID-19 are more frequent among patients who had RT-PCR-positive results. Comorbidities were higher among RT-PCR-negative patients. Patients in the RT-PCR-positive group had severe outcomes of COVID-19. However, mortality was similar between groups, although the RT-PCR-positive group had a shorter time to death. Both groups had comparable time to discharge among survivors. Clinical presentation, imaging, and serology should be evaluated in RT-PCR negative cases depending on the availability and level of suspicion.
Funding
No funding sources.
Competing interests
None declared.
Ethical approval
Not required.
Acknowledgments
The authors acknowledge Ms. Deema Althagabi and Dr. Mohammad Al Wetidy from College of Medicine Research Center, King Saud University, Riyadh, Saudi Arabia, for patient consenting and clinical data collection, and coordination with clinical laboratory, Ms. Heba Mohammed, Ms. Salini Scaria Joy, and Teena George, Ms. Nourhan Ourhan, Ms. Tahany Mossa Edrees and Ms. Faiza Abood from the Strategic Center for Diabetes Research, College of Medicine, King Saud University, Riyadh, Saudi Arabia for data collection and encoding respectively. The services of Mr. Saud Alanazi, from the Strategic Center for Diabetes Research, and Ms. Amina Fallata, and Mr. Kenneth Domero from the Obesity Research Center, College of Medicine, King Saud University, Riyadh, Saudi Arabia for assisting in phlebotomy is appreciated. We acknowledge Mr. Adnan Mahmood Usmani from the Strategic Center for Diabetes Research, College of Medicine, King Saud University, Riyadh, Saudi Arabia, for assisting with the English language editing. We also acknowledge the partial funding and services provided by the College of Medicine Research Center, King Saud University, Riyadh, Saudi Arabia, in conducting the study.
References
- 1.WHO . 2021. WHO coronavirus (COVID-19) dashboard.https://covid19.who.int/ [Google Scholar]
- 2.ECDC . European Centre for Disease Prevention and Control; 2020. Clinical characteristics of COVID-19.https://www.ecdc.europa.eu/en/covid-19/latest-evidence/clinical [Google Scholar]
- 3.CDC . Centers for Disease Control and Prevention; 2021. Interim clinical guidance for management of patients with confirmed coronavirus disease (COVID-19)https://www.cdc.gov/coronavirus/2019-ncov/hcp/clinical-guidance-management-patients.html# [Google Scholar]
- 4.Thakur B., Dubey P., Benitez J., Torres J.P., Reddy S., Shokar N. A systematic review and meta-analysis of geographic differences in comorbidities and associated severity and mortality among individuals with COVID-19. Sci Rep. 2021;11:8562. doi: 10.1038/s41598-021-88130-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dorjee K., Kim H., Bonomo E., Dolma R. Prevalence and predictors of death and severe disease in patients hospitalized due to COVID-19: a comprehensive systematic review and meta-analysis of 77 studies and 38,000 patients. PLoS One. 2020;15 doi: 10.1371/journal.pone.0243191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ministry of Health . 2021. COVID-19 dashboard.https://covid19.moh.gov.sa/ [Google Scholar]
- 7.Al-Omari A., Alhuqbani W.N., Zaidi A.R.Z., Al-Subaie M.F., AlHindi A.M., Abogosh A.K. Clinical characteristics of non-intensive care unit COVID-19 patients in Saudi Arabia: a descriptive cross-sectional study. J Infect Public Health. 2020;13:1639–1644. doi: 10.1016/j.jiph.2020.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barry M., AlMohaya A., AlHijji A., Akkielah L., AlRajhi A., Almajid F. Clinical characteristics and outcome of hospitalized COVID-19 patients in a MERS-CoV endemic area. J Epidemiol Glob Health. 2020;10:214–221. doi: 10.2991/jegh.k.200806.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heyman S.N., Kinaneh S., Abassi Z. The duplicitous nature of ACE2 in COVID-19 disease. EBioMedicine. 2021;67 doi: 10.1016/j.ebiom.2021.103356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang Y., Liu J., Zhang D., Xu Z., Ji J., Wen C. Cytokine storm in COVID-19: the current evidence and treatment strategies. Front Immunol. 2020;11:1708. doi: 10.3389/fimmu.2020.01708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arevalo-Rodriguez I., Buitrago-Garcia D., Simancas-Racines D., Zambrano-Achig P., Del Campo R., Ciapponi A. False-negative results of initial RT-PCR assays for COVID-19: a systematic review. PLoS One. 2020;15 doi: 10.1371/journal.pone.0242958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chirico F., Nucera G., Magnavita N. Hospital infection and COVID-19: do not put all your eggs on the “swab” tests. Infect Control Hosp Epidemiol. 2021;42:372–373. doi: 10.1017/ice.2020.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Middleton P., Perez-Guzman P.N., Cheng A., Kumar N., Kont M.D., Daunt A. Characteristics and outcomes of clinically diagnosed RT-PCR swab negative COVID-19: a retrospective cohort study. Sci Rep. 2021;11:1–7. doi: 10.1038/s41598-021-81930-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saudi Center for Disease Prevention and Control . 2020. Coronavirus disease COVID-19 guidelines. [Google Scholar]
- 15.Terpos E., Ntanasis-Stathopoulos I., Elalamy I., Kastritis E., Sergentanis T.N., Politou M. Hematological findings and complications of COVID-19. Am J Hematol. 2020;95:834–847. doi: 10.1002/ajh.25829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karimi F., Vaezi A.A., Qorbani M., Moghadasi F., Gelsfid S.H., Maghoul A. Clinical and laboratory findings in COVID-19 adult hospitalized patients from Alborz province/Iran: comparison of rRT-PCR positive and negative. BMC Infect Dis. 2021;21:1–8. doi: 10.1186/s12879-021-05948-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mao J., Dai R., Du R.-C., Zhu Y., Shui L.-P., Luo X.-H. Hematologic changes predict clinical outcome in recovered patients with COVID-19. Ann Hematol. 2021;100:675–689. doi: 10.1007/s00277-021-04426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan B.E., Chong V.C.L., Chan S.S.W., Lim G.H., Lim K.G.E., Tan G.B. Hematologic parameters in patients with COVID-19 infection. Am J Hematol. 2020;95:E131–E134. doi: 10.1002/ajh.25774. [DOI] [PubMed] [Google Scholar]
- 19.Gómez-Pastora J., Weigand M., Kim J., Wu X., Strayer J., Palmer A.F. Hyperferritinemia in critically ill COVID-19 patients – is ferritin the product of inflammation or a pathogenic mediator? Clin Chim Acta. 2020;509:249–251. doi: 10.1016/j.cca.2020.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng L., Li H., Li L., Liu C., Yan S., Chen H. Ferritin in the coronavirus disease 2019 (COVID-19): a systematic review and meta-analysis. J Clin Lab Anal. 2020;34 doi: 10.1002/jcla.23618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clark R., Waters B., Stanfill A.G. Elevated liver function tests in COVID-19: causes, clinical evidence, and potential treatments. Nurse Pract. 2021;46:21–26. doi: 10.1097/01.NPR.0000722316.63824.f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akbar M.R., Pranata R., Wibowo A., Lim M.A., Sihite T.A., Martha J.W. The prognostic value of elevated creatine kinase to predict poor outcome in patients with COVID-19 – a systematic review and meta-analysis. Diabetes Metab Syndr. 2021;15:529–534. doi: 10.1016/j.dsx.2021.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mucha S.R., Dugar S., McCrae K., Joseph D., Bartholomew J., Sacha G.L. Coagulopathy in COVID-19: manifestations and management. Cleve Clin J Med. 2020;87:461–468. doi: 10.3949/ccjm.87a.ccc024. [DOI] [PubMed] [Google Scholar]
- 24.Huang M.-J., Wei R.-B., Wang Y., Su T.-Y., Di P., Li Q.-P. Blood coagulation system in patients with chronic kidney disease: a prospective observational study. BMJ Open. 2017;7:e014294. doi: 10.1136/bmjopen-2016-014294. [DOI] [PMC free article] [PubMed] [Google Scholar]