Sir,
During the COVID-19 pandemic, there has been substantial interest in the ability to rapidly diagnose infection serologically, in particular to clear individuals to attend their workplace or for travel. Currently, a negative test result for severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2)-specific IgM immediately prior to travel has been mandated for some destinations in addition to nucleic acid testing (NAT).1 While pathogen-specific IgM is an early immune marker for many infectious diseases, there are exceptions. Many respiratory viruses, including SARS-CoV-2, demonstrate delayed IgM seroconversion often greater than 10 days from onset of illness, and this follows the appearance of specific IgG antibodies.2 We explored the utility of SARS-CoV-2-specific IgM test as a marker of infective risk and thus as a guide for international traveller screening.
We performed a retrospective cross-sectional study on all SARS-CoV-2-specific serology performed by NSW Health Pathology's Institute of Clinical Pathology and Medical Research, Australia before 10 November 2020. Sera were tested on a validated in-house quantitative indirect immunofluorescence assay (IFA) for SARS-CoV-2-specific IgM, IgA and IgG detection with a sensitivity of 91.3% and specificity of 98.9% for any antibody class and a sensitivity of 62.2% and specificity of 99.7% for IgM for NAT-positive patients.3 Clinical information was obtained from pathology request forms.
Individuals were classified as infectious if they had confirmed or probable SARS-CoV-2 infection (using Australian Public Health definitions)4 and were within 14 days from illness onset (the estimated maximum duration of infectivity adopted by the Communicable Diseases Network of Australia in their guidance on release from isolation).4 A confirmed case was defined by laboratory confirmation using a positive SARS-CoV-2 NAT, isolation of SARS-CoV-2 in cell culture or SARS-CoV-2-specific IgG seroconversion or a four-fold increase in IgG titres. A probable case was defined as someone who had a positive SARS-CoV-2-specific IgG with either clinical or epidemiological history compatible with SARS-CoV-2 infection.4
Individuals with confirmed or probable infection were considered non-infectious once they were >14 days after illness onset, as were individuals in whom SARS-CoV-2 infection was excluded with a negative NAT. As the current study aimed to determine the utility of IgM as a screening test prior to travel and the timing of IgM positivity relative to infective course could not be determined, individuals with confirmed or probable infection in whom the date of illness onset was unknown, or who were asymptomatic, were not included in the evaluation. Similarly, as the COVID-19 status could not be confirmed, individuals with a positive SARS-CoV-2-specific IgG but not meeting public health definitions for a confirmed or probable infection were excluded. Several individuals had multiple serology tests at different time points in their illness and these were assessed individually. The sensitivity, specificity, positive predictive value and negative predictive value of a positive SARS-CoV-2-specific IgM being able to identify an infectious individual were calculated.
Ethics was approved by Western Sydney Local Health District Human Research Ethics Committee (2008–10 QA).
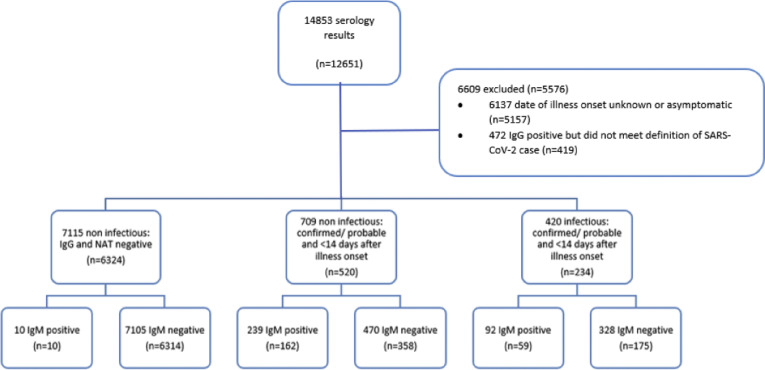
The study included 7075 individuals, after excluding confirmed and probable cases without a known illness onset date (n=419) and cases with positive SARS-CoV-2-specific IgG who did not meet the criteria for confirmed or probable infection (n=5157) (Fig. 1 ). In total, 420 samples were taken while an individual was infectious and 7824 were taken while an individual was non-infectious. SARS-CoV-2-specific IgM results by infectious or non-infectious status are indicated in Table 1 . Mean SARS-CoV-2-specific IgM titres peaked in the third week and were undetectable by approximately 10 weeks, on average.
Fig. 1.
Breakdown of serology results by IgM results and infectiousness.
Table 1.
IgM detection between infectious and non-infectious periods
| Infectious | Non-infectious | Total | |
|---|---|---|---|
| IGM positive | 92 | 249 | 341 |
| IGM negative | 328 | 7575 | 7903 |
| Total | 420 | 7824 | 8244 |
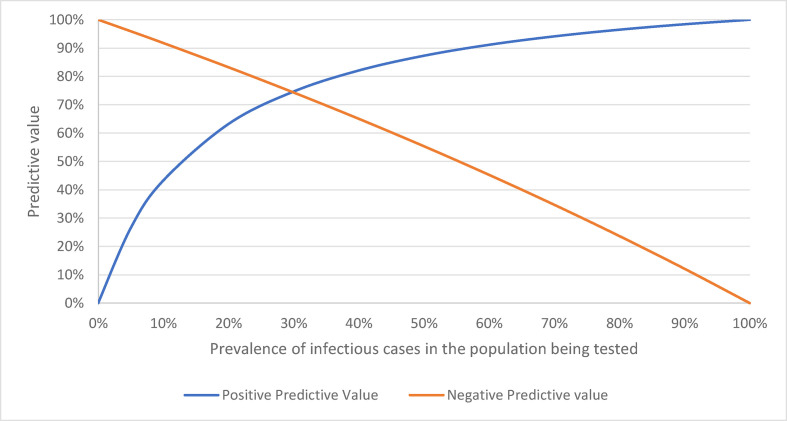
The specificity of SARS-CoV-2-specific IgM detection within the presumed 14-day infectious period was 96.8% and sensitivity was 21.9%. Figure 2 summarises the positive and negative predictive value over a range of prevalence. At the end of December 2020 there were only 220 active SARS-CoV-2 infections in Australia.5 Assuming an estimated infection to case ratio of 3.5,6 this would suggest a prevalence of infectious individuals in Australia (population 25 million) of 0.003%. At this prevalence, the positive predictive value of IgM to predict an individual is infectious would be 0.007%. Even at the peak of the second wave in Australia, when there were 8195 active cases on 12 August 2020, the positive predictive value of IgM to determine infectious state would be only 0.07%.
Fig. 2.
Change in positive and negative predictive value according to prevalence.
While serology is important in understanding the extent of antibody response and sero-prevalence, its role in diagnosis of infection in the acute, infectious period remains limited. This brief review reinforces that serology, in particular SARS-CoV-2-specific IgM, is not useful in identifying infectious individuals. This has implications for the use of serology in determining suitability for travel.
This study demonstrates very low positive predictive value for a positive SARS-CoV-2-specific IgM to potentially identify an infectious individual. In Australia, where the prevalence of infection is low (<1%),6 the positive predictive value of a SARS-CoV-2-specific IgM to detect an infectious individual is less than 1% and a positive result may exclude an individual who may have already recovered from travelling. There is evidence that development of antibodies to SARS-CoV-2 may directly coincide with a loss of infectivity.7 Even in a high prevalence setting, neither a negative nor a positive SARS-CoV-2-specific IgM would be able to appropriately exclude infectious individuals from travelling. The variable accuracy of serological testing using various commercial and rapid SARS-CoV-2-specific IgM, which have differing sensitivity for IgM,8 further makes interpretation difficult, particularly in the context of a negative SARS-CoV-2 NAT. A study looking at different commercial SARS-CoV-2 assays found the specificity of IgM to be high (96.3–99.7%), however the sensitivity varied between assays (42.0–82.7%).8 Assays with a lower sensitivity would perform poorer in predicting the infectivity of an infectious traveller.
Asymptomatic individuals may be less likely to develop SARS-CoV-2-specific IgM,9 although asymptomatic individuals can pose a risk of transmission,10 which further reduces the utility of SARS-CoV-2-specific IgM for pre-travel screening. The rollout of COVID-19 vaccines will also impact the utility of SARS-CoV-2-specific IgM to identify infectious individuals, depending on administered vaccine components and the antigens utilised in serological assays; in particular, the COVID-19 vaccine can elicit an IgM response in a subset of individuals.11 In one study, all participants demonstrated an IgM response after receiving the second but not first dose of the BNT162b2 mRNA COVID-19 vaccine.12
In conclusion, SARS-CoV-2-specific IgM measurement is not a useful screening test as an addition to NAT to identify infectious individuals prior to travel.
References
- 1.Embassy of the People’s Republic of China in the United States of America. Notice on the requirement adjustment of the nucleic acid rt-PCR and IgM serum antibody tests. Dec 2020. www.china-embassy.org/eng/notices/t1841416.htm cited Jul 2021. [Google Scholar]
- 2.Hsueh P.R., Huang L.M., Chen P.J., et al. Chronological evolution of IgM, IgA, IgG and neutralisation antibodies after infection with SARS-associated coronavirus. Clin Microbiol Infect. 2004;10:1062–1066. doi: 10.1111/j.1469-0691.2004.01009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hueston L., Kok J., Guibone A., et al. The antibody response to SARS-CoV-2 infection. Open Forum Infect Dis. 2020;7:ofaa387. doi: 10.1093/ofid/ofaa387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Australian Government Department of Health. Coronavirus disease 2019 (COVID-19): CDNA National guidelines for public health units. Cited Jul 2021. https://www1.health.gov.au/internet/main/publishing.nsf/Content/cdna-song-novel-coronavirus.htm
- 5.Live C.O.V.I.D. Nov 2020. Daily active cases.https://covidlive.com.au/report/daily-active-cases/aus cited Jul 2021. [Google Scholar]
- 6.Gidding H., Machalek D., Hendry A., et al. Seroprevalence of SARS-CoV-2-specific antibodies in Sydney, Australia following the first epidemic wave in 2020. Med J Aust. 2021;214:179–185. doi: 10.5694/mja2.50940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwon J.S., Kim J.Y., Kim M.C., et al. Factors of severity in patients with COVID-19: cytokine/chemokine concentrations, viral load, and antibody responses. Am J Trop Med Hyg. 2020;103:2412–2418. doi: 10.4269/ajtmh.20-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harritshøj L.H., Gybel-Brask M., Afzal S., et al. Comparison of 16 serological SARS-CoV-2 immunoassays in 16 clinical laboratories. J Clin Microbiol. 2021;59 doi: 10.1128/JCM.02596-20. e02596-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shirin T., Bhuiyan T.R., Charles R.C., et al. Antibody responses after COVID-19 infection in patients who are mildly symptomatic or asymptomatic in Bangladesh. Int J Infect Dis. 2020;101:220–225. doi: 10.1016/j.ijid.2020.09.1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Letizia A.G., Ramos I., Obla A., et al. SARS-CoV-2 transmission among marine recruits during quarantine. N Engl J Med. 2020;383:2407–2416. doi: 10.1056/NEJMoa2029717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jalkanen P., Kolehmainen P., Häkkinen H.K., et al. COVID-19 mRNA vaccine induced antibody responses against three SARS-CoV-2 variants. Nat Commun. 2021;12:3991. doi: 10.1038/s41467-021-24285-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Izumo T., Kuse N., Awano N., et al. Side effects and antibody titer transition of the BNT162b2 messenger ribonucleic acid coronavirus disease 2019 vaccine in Japan. Respir Investig. 2021;59:635–642. doi: 10.1016/j.resinv.2021.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]