Abstract
In this global pandemic situation of coronavirus disease (COVID-19), it is of foremost priority to look up efficient and faster diagnosis methods for reducing the transmission rate of the virus severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Recent research has indicated that radio-logical images carry essential information about the COVID-19 virus. Therefore, artificial intelligence (AI) assisted automated detection of lung infections may serve as a potential diagnostic tool. It can be augmented with conventional medical tests for tackling COVID-19. In this paper, we propose a new method for detecting COVID-19 and pneumonia using chest X-ray images. The proposed method can be described as a three-step process. The first step includes the segmentation of the raw X-ray images using the conditional generative adversarial network (C-GAN) for obtaining the lung images. In the second step, we feed the segmented lung images into a novel pipeline combining key points extraction methods and trained deep neural networks (DNN) for extraction of discriminatory features. Several machine learning (ML) models are employed to classify COVID-19, pneumonia, and normal lung images in the final step. A comparative analysis of the classification performance is carried out among the different proposed architectures combining DNNs, key point extraction methods, and ML models. We have achieved the highest testing classification accuracy of 96.6% using the VGG-19 model associated with the binary robust invariant scalable key-points (BRISK) algorithm. The proposed method can be efficiently used for screening of COVID-19 infected patients.
Keywords: COVID-19, Pneumonia, Image segmentation, Conditional generative adversarial network (C-GAN), Key point extraction, Deep neural networks (DNN), Classification
1. Introduction
Coronavirus disease (COVID-19) is caused by a severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), responsible for this ongoing pandemic. COVID-19 can be classified as a respiratory disease as the common symptoms include myalgia, sore throat, headache, fever, chest pain, and dry cough [1] also, an infected person can show complete signs in around 14 days. Up to July of 2021, 190 million COVID-19 cases have been reported in more than 200 countries and territories, eventuating approximately 4 million deaths [2]. This has resulted in the international community a significant cause of public health concern. The outbreak on January 30, 2020, was declared as public health emergency of international concern (PHEIC) by the world health organization (WHO) and recognized as a pandemic on March 11, 2020 [2]. At present, we have some vaccine options, but now it would take a long time for the vaccine to reach the corner of the world. Hence, for the rapid screening of infected patients, the alternative method that can be used is visual indicators. The most common symptom of this virus is a lung infection, for which the widely considered visual indicator is chest radiography images (chest X-ray/computed tomography (CT) images) [3]. The radiologists manually perform these images’ interpretation to find some visual patterns for confirming the COVID-19 infection.
Since the conventional diagnosis process has become more accurate with time, it is still prone to medical staff risks. Also, it is more costly because there is a need for diagnostic test kits for every patient. In comparison, medical imaging techniques, i.e., X-ray and CT scans, can be used for screening, which are relatively faster, safer, and easily accessible. The X-ray image screening is preferred over the CT scans for COVID-19 screening because it is widely available, and it relatively costs less to obtain [4], [5]. However, the manual diagnosis of the virus using X-ray images can be a time-consuming process. It can lead to many inaccuracies and human-based errors if there is no or less prior experience and knowledge about the virus and its symptoms. Hence, there is a solid need to automate such procedures widely, and it should be accessible for everyone so that the diagnosis can become more efficient, accurate, and fast. The recent works [6], [7], [8], [9], [10], [11], [12], use computer vision (CV) and artificial intelligence (AI) technologies that include the use of deep learning (DL) models, in particular, convolutional neural networks (CNNs) have been proven as practical approach for examining medical images. Recently, the use of CNN successfully helped to detect pneumonia in a patient’s chest X-ray image [13], [14], [15]. Many studies have been conducted for the diagnosis of COVID-19 through X-ray images using the DL models. For instance, Ozturk et al. [16] worked for automated diagnosis of COVID-19 based on X-ray images using a DL network which is named as DarkCovidNet. For multi-class classification (COVID-19, normal, and pneumonia), the model achieved an accuracy of 87.02%, and for two-class classification (COVID-19 and normal), the accuracy of 98.08% was achieved. Hemdan et al. [17] worked with X-ray images and developed a network named COVIDX-Net. This network was trained using seven different CNN models, and the validation dataset for the model consists of 50 X-ray images (25 normal and 25 COVID-19 cases). Their model’s achieved accuracy is 90.00%, which is very effective considering the amount of validation data used for testing the model. Wang et al. [18] achieved a testing accuracy of 93.3% with their designed COVID-Net, which is a deep CNN model. Sethy and Behera [19] used the combination of ResNet50 and support vector machine (SVM) classifier, which on 50 samples (25 normal and 25 COVID-19 cases) achieved the accuracy of 95.38%. Recently, Nayak et al. [20] proposed a method for screening the COVID-19 chest X-ray images using DL. They used a transfer learning concept with the most successful eight pre-trained CNN models, which are MobileNet-V2, AlexNet, VGG-16, GoogleNet, ResNet-34, SqueezeNet, Inception-V3, and ResNet-50. Their work’s major contributions include comparing the effectiveness of these eight pre-trained models and the impact of several hyper-parameters such as batch size, optimization techniques, learning rate, and the number of epochs.
The previous works in COVID-19 detection mainly DL algorithm-specific and did not focus on the regions of interest (ROI) in the X-ray images that reveal the particular patterns for the specific disease. Hence, there is a research scope in this field that deals with the exploitation of only the ROI in the provided X-ray images. It can lead to a much if not accurate but precise classification of images based on the actual medical terminologies. Here in our work, a new and comprehensive study focuses only on the X-ray images’ ROIs. The method is the composition of image segmentation, feature extraction, and classification processes for discriminating COVID-19, pneumonia, and normal X-ray images. The image segmentation process effectively uses a conditional generative adversarial network (C-GAN) [21] to obtain the lung images from the pre-processed input X-ray images. The feature extraction network computes the discriminatory features from the segmented lung images by combining deep neural networks (DNNs) and key point extraction algorithms such as scale-invariant feature transform (SIFT) [22] and binary robust invariant scalable key-points (BRISK) [23]. The DNNs considered in this work for feature extraction include four transfer learning models, namely DenseNet169, DenseNet201 [24] and VGG16, VGG19 [25], and a self-customized simple CNN (sCNN). Finally, various machine learning (ML) algorithms are employed to classify the extracted features into COVID-19, pneumonia, and normal classes. The best performing combinations of these selected models are evaluated and compared to determine the best-case scenario for the proposed method. The main contributions of this work can be stated as follows:
-
1.
The use of C-GAN network for segmentation of the chest X-ray images.
-
2.
Development of a feature extraction framework for fusing DNN and key-point features.
-
3.
Classification of COVID-19, pneumonia, and normal X-ray images using ML techniques.
-
4.
Performance evaluation of different combinations of the feature extraction network and ML methods.
The rest of the paper is organized as follows: Section 2 describes the datasets used for the segmentation and classification process. Section 3 illustrates the methods used for image segmentation, feature extraction, and classification. Sections 4, 5 present the experimental results and discusses the effectiveness of the proposed method, respectively. Finally, Section 6 concludes the paper.
2. Dataset of chest X-ray images and preprocessing
In this work, chest X-ray images have been used for the discrimination of COVID-19, pneumonia, and normal subjects. The images are obtained from different sources, which are publicly available online. The segmentation and classification datasets used in this work are described as follows:
2.0.1. Segmentation dataset
For training the segmentation network, we have used segmentation in chest radiographs (SCR) dataset X-ray images [26], available in [27]. This is a publicly available database containing 247 frontal viewed posterior-anterior (PA) chest radiographs to facilitate the segmentation of lung fields, heart, and clavicles. We split the entire dataset images into training, validation, and testing subsets. The testing subset included 13 images, which is around 5% of the total available images in the main dataset. The remaining 234 images are further bifurcated into training and validation subsets with a percentage split of around 90% and 10%, respectively. Considering that the number of images is low, we augmented the number of images in each subset by a factor of four using simple affine transformations. Finally, the segmentation network has been trained, validated, and tested for generating the lung masks from the chest radiographs.
2.0.2. Classification dataset
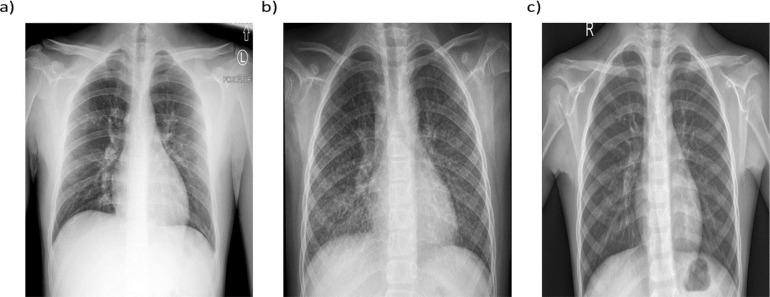
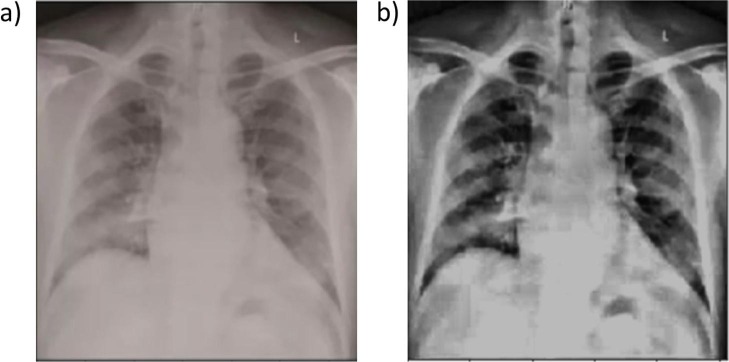
For the classification of chest X-ray images, two different public datasets are used. The first dataset contains the COVID-19 chest X-ray images provided by Cohen et al. [28]. This dataset is developed using images from different open access sources and gets regularly updated. Presently, the database contains 930 chest X-ray images of COVID-19 patients. Out of these 930 images, we selected 342 front viewed images labeled as COVID-19 affected chest X-rays with PA and anterior-posterior (AP) views in the metadata. Pneumonia and normal chest X-ray images are obtained from the second dataset, which is the pneumonia dataset [29], containing 1583 normal and 4273 pneumonia front viewed X-ray images. It should be noted that similar to segmentation dataset, all chest radiographs taken into consideration for classification are front viewed. For class balancing, we have randomly selected 341 normal and 347 pneumonia X-ray images, resulting into total 1030 images for classification. In this work, 73% of the total 1030 X-ray images (250 images from each class) is used for training the DNN, while the rest of the 27% images are kept for testing. As the number of training images was not adequate, we augmented the training images using basic affine transformations, which increased the number of training images to 3000, which is four times the original 750 training images. Further, the training images were split into train and validation datasets by a percentage division of 80% and 20%. Fig. 1 presents the sample X-ray images from COVID-19, pneumonia, and normal classes. All the images were visually inspected before generating the lung masks using the segmentation network. The low contrast images were identified and preprocessed using histogram equalization and thresholding operation (see Fig. 2 ). It is worth noting that the image preprocessing techniques have been found effective in literature for better training of the DNNs [30].
Fig. 1.
Class specific sample chest X-ray images (a) COVID-19 X-ray (b) Pneumonia X-ray c) Normal X-ray.
Fig. 2.
Preprocessing of low contrast image: (a) Original low contrast chest X-ray image (b) Contrast enhanced image.
3. Method
3.1. Segmentation of lung images using conditional-GAN
Goodfellow et al. in [31] proposed GAN models which have been vigorously employed in the image processing domain for translating an input image into its corresponding output image. The GAN network consists of two networks: a generator that generates convincing photo-realistic images and a discriminator that classifies the synthesized/generated image and the original image from the training dataset into fake and real categories. Formally, provided a set of noise samples z with distribution , the generator G transforms these samples to real-world data with distribution by implementing a min–max method along with the discriminator. During the training phase, the discriminator network tries to discriminate real data samples y having probability distribution from the transformed data samples with distribution . The min-imax GAN objective function can be mathematically expressed as,
| (1) |
Where E and stand for expectation and logarithmic operations, respectively. C-GAN extends GAN’s concept, where generator G synthesizes output data x from the real data y and random noise vector . On the other hand, the C-GAN discriminator D accepts both the synthesized and actual data (x and y) as inputs and tries to discriminate them. The objective function for C-GAN can be expressed as,
| (2) |
It should be noted that the task of the generator G is not only confusing the discriminator; at the same time, the output of it should match the actual data in an sense as,
| (3) |
Thus, the final objective function becomes as follows:
| (4) |
The parameter is the regularization weight. The Eqn. (4) can be solved by alternative training of generator G and discriminator D, respectively.
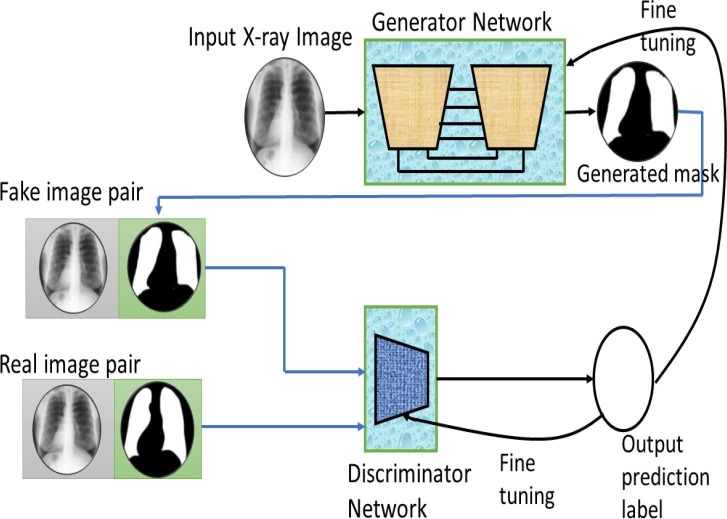
In this paper, we have used C-GAN for segmenting the COVID-19 chest X-ray images. We feed the chest X-rays as the generator’s input and expect the corresponding lung mask to be output from the generator. The discriminator has two pairs of inputs: real pair (the original chest X-ray and its ground truth lung mask) and fake pair (the original chest X-ray and the generated lung mask). The generator and discriminator both are trained in an adversarial way, as can be noticed in Fig. 3 . It can be noticed that G strives to generate increasingly realistic lung mask images to deceive D, whereas D aims to distinguish between actual and fake image pairs.
Fig. 3.
Generator G is provided with input chest X-ray image y and generates the synthesized segmented mask image . The discriminator D aspires to classify the fake image pair (includes y and ) and real image pair (includes y and ground truth mask image y).
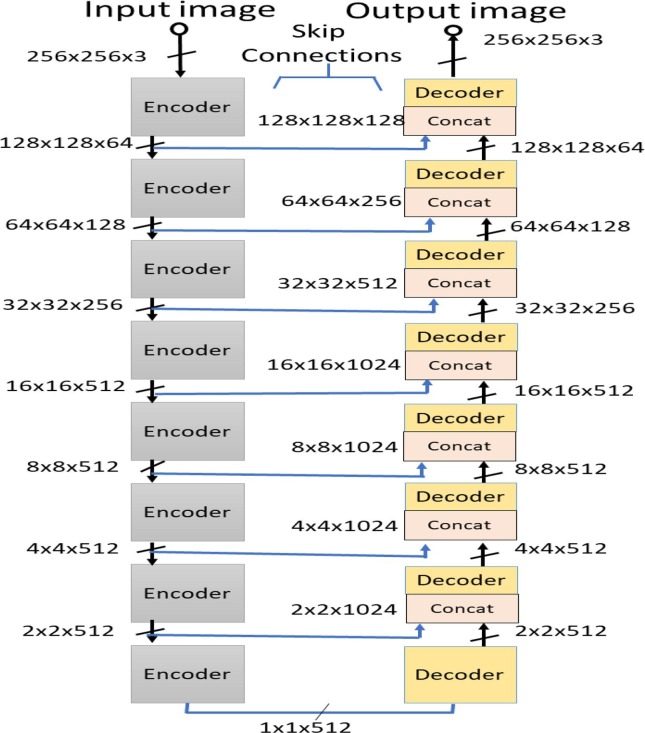
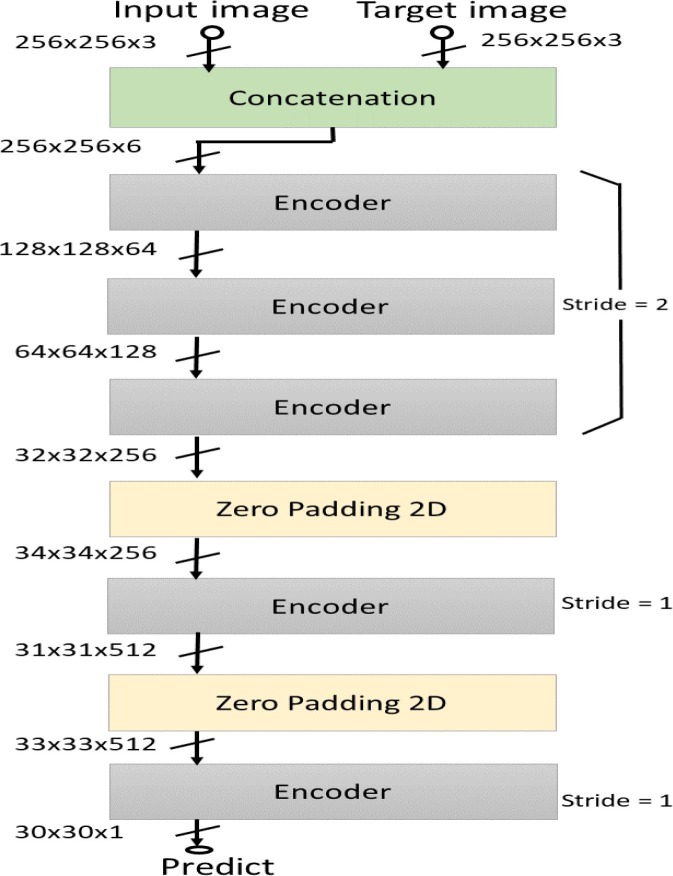
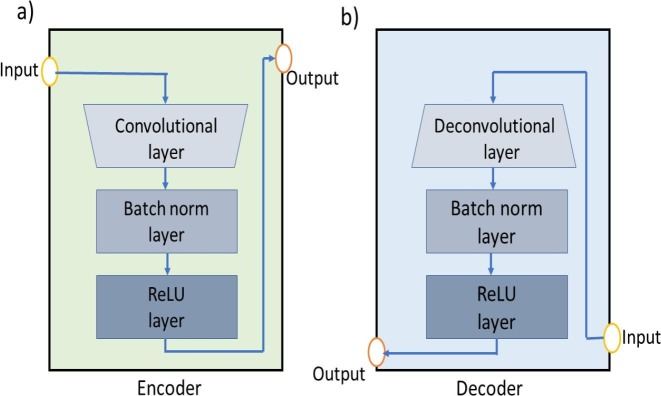
For carrying out chest X-ray image segmentation, we specifically employ the Pix2Pix C-GAN model proposed by Isola et al. [21]. In this model, generator G is a conventional encoder-decoder network, whereas discriminator D is a Patch-GAN encoder network. The architectural diagram of generator and discriminators are presented in Fig. 4 and Fig. 5 , respectively. Also, the encoder and decoder blocks used in these models are also shown in Fig. 6 . Irrespective of generator or discriminator, each encoder block comprises convolutional, batch-norm, and rectified linear unit (ReLU) layers, whereas each decoder block includes deconvolutional, batch-norm, and ReLU layers. The encoder block outputs a compact data representation contrary to the decoder block. The random noise is provided to the generator in the form of dropout in the first three decoder blocks during both training and testing time. The generator network also has skip connections from the encoder to the decoder for pattern reinforcement at different levels. The last block in the discriminator outputs a shape/patch of size 3030, where each pixel of this patch classifies a portion of the input image.
Fig. 4.
Architectural diagram of the generator network used in C-GAN. The architecture is inspired from the U-Net model [32]. The generator is a cascade of encoder blocks followed by a cascade of decoder blocks, with skip connections for reinforcing patterns in di.fferent levels.
Fig. 5.
Architectural diagram of discriminator network used in C-GAN. The discriminator follows the Patch-GAN network architecture, which is composed of a cascade of encoder blocks that outputs a compact data representation.
Fig. 6.
The representation of encoder and decoder blocks used in the generator and discriminator networks: (a) encoder block that considers a input image and performs convolution, batch normalisation, and evaluates the ReLu activation for generating output image. (b) decoder block, that performs same operations of the encoder except deconvolution instead of convolution.
3.1.1. Training of segmentation network
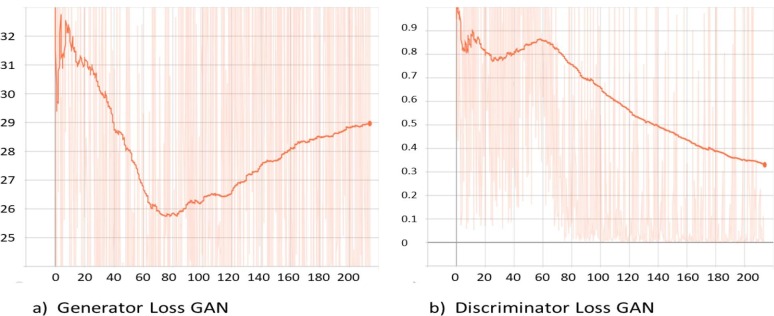
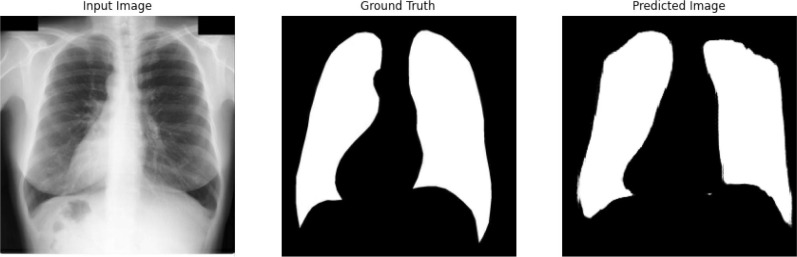
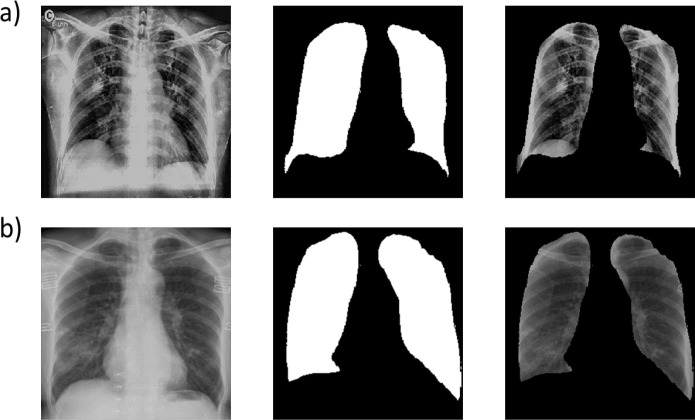
As mentioned earlier, image segmentation is performed by training a C-GAN based chest X-ray segmentation model, basically a U-net architecture [32] in a broader level. More specifically, the pix-to-pix algorithm was used to train the generator network, and the patch GAN like classifier was utilized for the discriminator network. The training loss functions for generator and discriminator networks are shown in Fig. 7 (a) and Fig. 7 (b), respectively. Both the networks compete against each other to optimise themselves and try to become better than the other network. As it can be observed in Fig. 7 (a) and Fig. 7 (b) that the generator loss starts decreasing when then the discriminator loss increases and vice versa. The whole graph, in general, converges for both models when the trends are seen on the macro level. After training, the generator is capable of generating the lung masks for the input chest X-ray images. Fig. 8 presents the input chest X-ray, ground truth mask, and lung mask produced by the generator. It is clear that the predicted lung mask possesses almost similar morphological structure as the ground truth mask. After applying the generated lung masks to the input chest X-ray images, we obtain the lung images. Fig. 9 shows the segmented lung images for AP and PA view chest X-ray images with the generated masks. In the next stage, these segmented lung images are used for training of the proposed classification pipeline.
Fig. 7.
Generator and discriminator loss function graphs for C-GAN during the training process.
Fig. 8.
A sample chest X-ray image (left), it’s original lung mask (center) and it’s predicted lung mask (right).
Fig. 9.
Segmentation of AP and PA view chest X-rays using the proposed C-GAN based model: (a) Top row shows AP view chest X-ray, generated lung mask, and segmented lungs. (b) Bottom row shows PA view chest X-ray, generated lung mask, and segmented lungs.
3.2. Hidden pattern extraction and classification of lung images
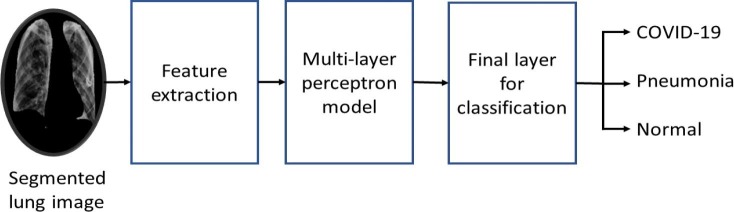
This section focuses on the automatic extraction of salient features from the segmented lung images, enabling the efficient detection of COVID-19 and pneumonia cases. Extraction of features is considered one of the essential steps in the image classification process. Traditional hand-engineered features have been found helpful in the literature for image classification tasks. In recent times, DL methods are widely used to automatically find hidden patterns associated with the analyzed data, providing significant improvement in the classification performance compared to traditional feature extraction methods. This study introduces a pipeline that employs DL architectures and traditional feature extraction techniques for the in–hand image classification task. In addition, a rigorous analysis of the classification performance is also carried out when deep architectures are applied in stand-alone mode. The pipeline for the classification process is represented by the flow diagram shown in Fig. 10 . The flow diagram consists of three major blocks/components: feature extraction layer, multi-layer perceptron (MLP) layer, and final classification layer.
Fig. 10.
The flow diagram of proposed method for discrimination of COVID-19, pneumonia, and normal chest X-ray images.
3.2.1. Feature extraction layer
This layer of the pipeline includes two significant parts, deep CNN models and the other part employing traditional hand-crafted features. The simplest form of this layer is presented in Fig. 11 , without adding any hyper-parameters, loss-functions, optimizer, etc. The parameters that were used during the training phase are mentioned in Table 1 .
Fig. 11.
Flow diagram explaining the elements in the feature extraction block (part of main pipeline (see Fig. 10)).
Table 1.
The various parameters used during the feature extraction model training process.
| Variables | Values |
|---|---|
| Optimizer | Adam |
| Learning rate | 0.01 |
| Loss function | Categorical cross entropy |
| Evaluation matrix | Accuracy |
| Image pre-processing | Predefined pre-process input function of related transfer learning model |
| Callbacks | Model checkpoint (save best only), Reduce LR on Plateau |
| Epochs | 100 |
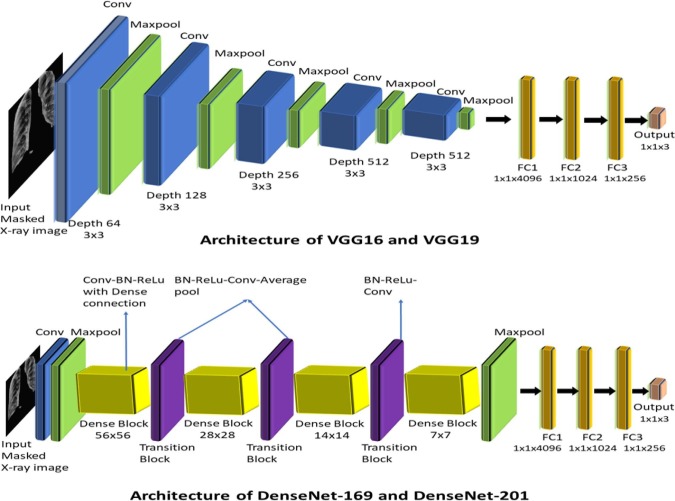
The DL models with conventional transfer learning methods are used in this work. The selected transfer learning models are DenseNet169, DenseNet201, VGG16, and VGG19. The DenseNet architectures have been chosen in this work because of their following inherent advantages [24]: direct connectivity exists between any two layers with feature-map size unaltered, identity mapping properties are naturally integrated, provides diversified depth and deep supervision, and feature reuse capability. On the other hand, the VGG models with depth of 16–19 weight layers and very small size (33) convolutional kernel provided significant improvement over state-of-the-art models in terms of classification accuracy and validation error [25].
The flatten layers of these selected models are customized as presented in Table 2 and Fig. 12 represents the architectural diagram of the transfer learning models that were created. Along with these models, we have also built a sCNN model with five two-dimensional (2D) convolutional layers followed by five 2D max-pooling layers and three flatten layers attached at the end, with other specified parameters mentioned in Table 2. The intention behind using sCNN model is to investigate the potential of a basic CNN model in extracting the discriminatory features from segmented lung images and compare its performance with the transfer learning models. The sCNN model was trained from scratch, unlike the transfer learning models. Once a particular DL model gets trained, the features from the FC3 layer of the trained model are computed. It is worth mentioning that the feature vector’s dimension computed from the FC3 layer becomes 256 1 after considering each of the 256 units/perceptrons in this layer.
Table 2.
Additional dense layers for the transfer learning models at the end.
| FC 1 | FC 2 | FC 3 | Output | |
|---|---|---|---|---|
| No. of perceptrons | 4096 | 1024 | 256 | 3 |
| Activation function | ReLU | ReLU | ReLU | Softmax |
Fig. 12.
Architectural diagram of the selected transfer learning models in the feature extraction block. (Here Conv, FC and BN stands for convolution, fully connected and batch normalization, respectively.)
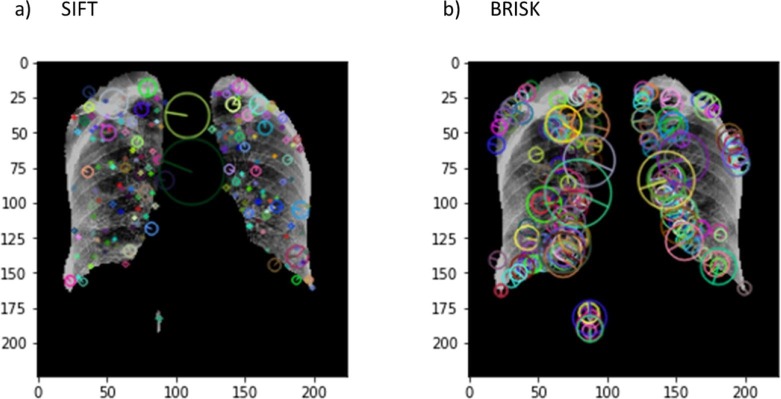
The second part of the feature extraction block (see Fig. 11) uses computer vision algorithms to extract the key points from the image. These key points are generally the local features that are in the form of blobs in an image. For detecting these features in the segmented lung images, we individually used two algorithms in the pipeline namely SIFT [22] and BRISK [23]. SIFT is one of the most popular algorithms in computer vision used to extract the key-points and build the corresponding features descriptor which are invariant to scaling or rotation. The SIFT algorithm can be explained using four steps [22]. In the first step, over all image locations and scales, the difference-of-Gaussian function is used to find the interest points (scale and orientation invariant). The second step is the keypoint localization that finds the location and scale of each candidate using a detailed model. The keypoints are identified based on their stability measures. In the third step, each keypoint location is assigned with one or more orientations using local image gradient directions. In the fourth step, local image gradients are computed at the chosen scale around every keypoint. The key points identified by the SIFT algorithm on the masked lung X-ray image as an input can be seen in Fig. 13 (a).
Fig. 13.
Images visually represent the detected features by SIFT and BRISK algorithms on sample lung images.
BRISK is a low computational feature detector scheme with an appreciable performance and accuracy [23]. In BRISK, keypoints are identified in both image and scale dimensions. The scale and location of the keypoints in the continuous domain are obtained using quadratic function fitting. For the keypoint description, a sampling pattern (points lying on scaled concentric circles) is employed at each keypoint neighborhood. A sample of how the BRISK algorithm performed on the masked lung X-ray image as an input can be seen in Fig. 13(b).
Once identification of the key points in the lung images using SIFT/BRISK algorithm gets completed, they are clustered into a total of 128 groups using the k-means clustering algorithm. Then, the mean values of the descriptors belonging from each of the groups are computed, resulted in a feature vector of dimension 128 1.
Afterward, the aforementioned feature vector’s dimension is increased by augmenting the feature vector (256 1) obtained from the FC3 layer of DL models. Thus, the resultant feature vector obtained from the masked lung images becomes of dimension 384 1. The block diagram in Fig. 11 represents the aforementioned process. In the subsequent step, the augmented feature vector is fed to a dense MLP model, a part of our main pipeline (see Fig. 10).
3.2.2. Multi-layer perceptron model
The next part of this pipeline is a DNN (MLP model), accepts concatenated features as input from the previous block. Then, based on the labels assigned to these input features, the MLP model is trained for predicting the unseen correct labels. The MLP model input is a vector of length 384 with 256 features coming from the transfer learning model’s second last or FC3 layer and the remaining 128 features coming from the 128 clusters mean points of the extracted features using SIFT or BRISK algorithm. The MLP model consists of seven dense layers without any new hyper-parameters. The activation functions used were ReLU in the first six layers and then softmax in the last layer. Table 3 provides the basic structural knowledge of the MLP network. Other related details to the network can be seen by referring to Table 1.
Table 3.
The layer description of the MLP model.
| Namely layer | Output shape | Activation function |
|---|---|---|
| Dense_1 | 1024 | ReLU |
| Dense_2 | 800 | ReLU |
| Dense_3 | 512 | ReLU |
| Dense_4 | 300 | ReLU |
| Dense_5 | 256 | ReLU |
| Dense_6 | 128 | ReLU |
| Output_layer | 3 | Softmax |
3.2.3. Final layer for prediction
In this study, apart from softmax, we have also employed three other ML techniques, namely SVM with radial basis function (RBF) kernel [33], XG-boost [34], and random forest (RF) [35] in the final layer. These ML classifiers were used in numerous studies because of their simplicity, effectiveness, and ease of implementation [36], [37]. Hence, they are considered to classify the lung images into the three labeled classes based on the features obtained from the Dense_6 layer of the MLP network.
3.2.4. Training of deep classification networks
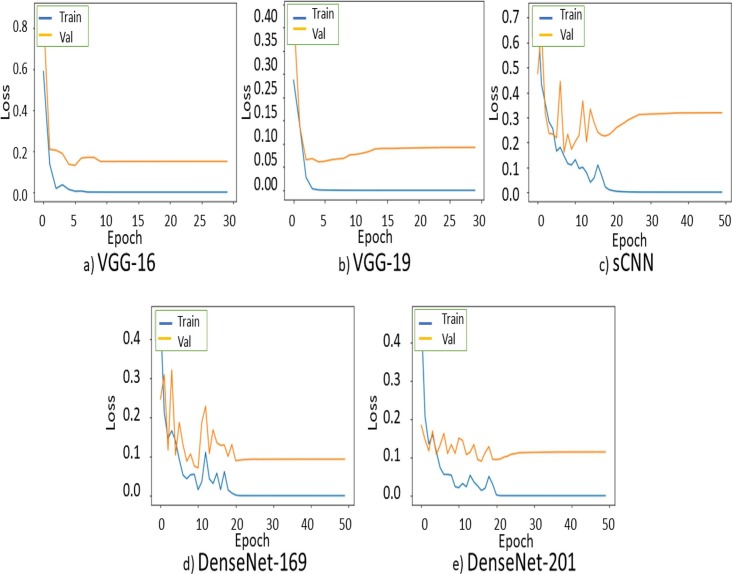
After the segmentation network training, the generator model was used for segmenting the new chest X-ray images, which were further used for the training of deep classification networks. For proper training of the deep classification architectures with an extensive training dataset, image augmentation was performed, including affine transformations such as rotations and shearing of the segmented images. Four transfer learning models (VGG-16, VGG19, DenseNet-169, and DenseNet-201) and one simple customized model (sCNN) were studied as deep feature extraction techniques. These models were also tested in combination with SIFT and BRISK-based feature extraction algorithms. Several ML techniques were employed as final classification layers. Fig. 14 shows the convergence of the loss functions of the specified DL models. From the graphs, one can be observe that the transfer learning models converge faster as compared to the sCNN model, which was trained from scratch.
Fig. 14.
Loss function convergence graphs for different transfer learning models. Here “train” and “val” represent the training and validation loss convergence curves, respectively.
To assess the computational complexity of the deep feature extraction models, this study reports and compares the execution time of those models in their training part, as exhibited in Table 4 . The proposed method was implemented on the TensorFlow platform in a system with a 7th Gen Intel® Core™ i7 Quad-Core processor, NVIDIA® GeForce® GTX 1050 Ti 4 GB graphics card, and 8 GB RAM. The VGG-16 model took 1046 s for training, with an average training time per step is 174.3 ms. The VGG-19 model training was completed in 1289 s with 214.8 ms as the average training time per step. For DenseNet-169, the training time was 811 s, and 135.1 ms was the average training time per step. DenseNet-201 model needed the highest training time of 2298 s with 383.0 ms as the average training time per step. The sCNN model required the lowest training time of 200 s with 33.3 ms as the average training time per step.
Table 4.
Training time for different considered CNN architectures.
| Training time | Average time per step | |
|---|---|---|
| VGG-16 | 1046s | 174.3 ms |
| VGG-19 | 1289s | 214.8 ms |
| DenseNet-169 | 811s | 135.1 ms |
| DenseNet-201 | 2298s | 383.0 ms |
| sCNN | 200s | 33.3 ms |
4. Experimental results
This section presents the effectiveness of the proposed deep features extraction methods in classifying COVID-19, pneumonia, and normal segmented chest X-ray images. Once all the models are trained, the confusion matrix based performance parameters are computed from every stage of the proposed methodology, which effectively combines the handcrafted and deep feature extraction methods. The performance parameters, namely accuracy (Acc), sensitivity (Sens), specificity (Spec), precision (Prec), false positive rate (FPR), false negative rate (FNR), and F1 Score are expressed as,
| (5) |
| (6) |
| (7) |
| (8) |
| (9) |
| (10) |
| (11) |
TP, TN, FP, and FN are the confusion matrix elements (see Table 5 ) and represent true positive, true negative, false positive, and false negative rates, respectively. The obtained confusion matrix is denoted as , representing the predicted and actual classes (i.e, COVID-19, normal, and pneumonia) of the segmented X-ray images. The matrix C is further resized to matrices (see Table 5) respective to each of the three classes for evaluating the binary classification parameters. Finally, each of the performance parameters is averaged (Avg) over the three considered classes. Table 6 presents the performance of the deep features extracted from customized sCNN and several other transfer learning models (VGG-16, VGG-19, DenseNet-169, and DenseNet-201). The generated feature matrices were classified using several ML techniques (softmax, SVM, RF, and XG-Boost) employed in the final layer.
Table 5.
Confusion matrix for mathematical analysis.
| Predicted negative (PN)-0 | Predicted positive (PP)-1 | |
|---|---|---|
| Actual negative (AN)-0 | True negative (TN) | False positive (FP) |
| Actual positive (AP)- 1 | False negative (FN) | True positive (TP) |
Table 6.
Experimental results obtained with deep features and different ML methods.
| Model | Final layer | Avg Acc (%) | Avg Sens (%) | Avg Spec (%) | Avg Prec (%) | Avg F1-score (%) |
|---|---|---|---|---|---|---|
| VGG-16 | Softmax | 95.9 | 93.9 | 96.9 | 93.9 | 93.9 |
| SVM | 95.9 | 93.9 | 96.9 | 94.0 | 93.9 | |
| RF | 96.4 | 94.6 | 97.3 | 94.3 | 94.6 | |
| XG-Boost | 94.9 | 92.4 | 96.2 | 92.5 | 92.5 | |
| VGG-19 | Softmax | 96.4 | 94.7 | 97.2 | 94.6 | 94.6 |
| SVM | 95.9 | 93.9 | 96.9 | 94.0 | 93.9 | |
| RF | 95.4 | 93.3 | 96.6 | 93.3 | 93.1 | |
| XG-Boost | 95.4 | 93.5 | 96.7 | 93.4 | 93.1 | |
| sCNN | Softmax | 94.0 | 91.0 | 95.5 | 91.2 | 91.0 |
| SVM | 93.5 | 90.3 | 95.1 | 90.4 | 90.3 | |
| RF | 93.7 | 90.7 | 95.3 | 90.7 | 90.7 | |
| XG-Boost | 93.3 | 90.0 | 94.9 | 90.0 | 90.0 | |
| DenseNet169 | Softmax | 96.1 | 94.3 | 97.1 | 94.4 | 94.2 |
| SVM | 96.1 | 94.3 | 97.1 | 94.4 | 94.2 | |
| RF | 96.1 | 94.3 | 97.1 | 94.4 | 94.2 | |
| XG-Boost | 95.2 | 92.8 | 96.4 | 92.9 | 92.8 | |
| DenseNet201 | Softmax | 94.2 | 91.6 | 95.6 | 91.4 | 91.5 |
| SVM | 94.5 | 91.9 | 95.8 | 91.8 | 91.8 | |
| RF | 94.2 | 91.6 | 95.6 | 91.4 | 91.5 | |
| XG-Boost | 94.9 | 92.5 | 96.2 | 92.5 | 92.5 |
It can be observed from Table 6 that transfer learning models performed better than sCNN model in terms of average Acc, Sens, Spec, Prec, and F1-score. It can also be noticed that the RF classifier has outperformed other ML classifiers with regard to obtained classification performance parameters. The best classification performance is reported when deep features obtained from the VGG-16 model are fed to the RF classifier, average Acc, Sens, Spec, Prec, F1-score are obtained as 96.4%, 94.6%, 97.3%, 94.3%, and 94.6%, respectively. The sCNN model with XG-Boost as the final layer provided average Acc, Sens, Spec, Prec, F1-score of 93.3%, 90.0%, 94.9%, 90.0%, 90.0%, respectively. The performance of other transfer learning models has been found comparable to the best performing VGG-16 model.
Further, Table 7, Table 8 present the obtained performance parameters when deep features were fused with the hand crafted features, extracted using SIFT and BRISK algorithms, respectively. It can be observed from Table 7 that the obtained classification performance parameters did not change significantly after the inclusion of SIFT algorithm-based features. It can be noticed that deep features extracted from DenseNet 169 model, when fused with SIFT-based features, provided average Acc, Sens, Spec, Prec, F1-score as 96.1%, 94.2%, 97.1%, 94.3%, and 94.2%, respectively with SVM as the classification layer. However, there is a significant improvement over classification performance when deep features were fused with BRISK-based key-point features and fed to different ML algorithms, as it can be observed in Table 8. The sCNN model-based deep features and BRISK-based features provided an average increased Acc, Sens, Spec, Prec, F1-score of 95.4%, 93.3%, 96.5%, 93.1%, 93.2%, respectively, with RF as the final layer. These values are approximately 2% higher as compared to when only sCNN based deep features were classified using RF classifier. The VGG-19 model-based deep features along with BRISK features and RF classifier in the last layer provided the best classification performance among all the models; the average Acc, Sens, Spec, Prec, F1-score are reported as 96.6%, 95%, 97.4%, 95.0%, 95.0%, respectively.
Table 7.
Experimental results obtained with deep and SIFT based handcrafted features and different ML methods.
| Model & Feature extraction | Final layer | Avg Acc (%) | Avg Sens (%) | Avg Spec (%) | Avg Prec (%) | Avg F1-score (%) |
|---|---|---|---|---|---|---|
| SIFT VGG-16 | Softmax | 96.1 | 94.2 | 97.1 | 94.3 | 94.2 |
| SVM | 96.1 | 94.2 | 97.1 | 94.3 | 94.2 | |
| RF | 96.1 | 94.2 | 97.1 | 94.3 | 94.2 | |
| XG-Boost | 96.1 | 94.2 | 97.1 | 94.3 | 92.2 | |
| SIFT VGG-19 | Softmax | 95.9 | 94.0 | 96.9 | 93.9 | 93.9 |
| SVM | 95.9 | 94.0 | 96.9 | 93.9 | 93.9 | |
| RF | 95.9 | 94.0 | 96.9 | 93.9 | 93.9 | |
| XG-Boost | 94.7 | 92.5 | 96.0 | 92.0 | 92.2 | |
| SIFT sCNN | Softmax | 93.3 | 90.0 | 94.9 | 90.0 | 90.0 |
| SVM | 93.3 | 90.0 | 94.9 | 90.0 | 90.0 | |
| RF | 93.5 | 90.3 | 95.1 | 90.4 | 90.3 | |
| XG-Boost | 93.5 | 90.3 | 95.1 | 90.4 | 90.3 | |
| SIFT DenseNet 169 | Softmax | 95.6 | 93.6 | 96.7 | 93.6 | 93.6 |
| SVM | 96.1 | 94.2 | 97.1 | 94.3 | 94.2 | |
| RF | 96.1 | 94.2 | 97.1 | 94.3 | 94.2 | |
| XG-Boost | 96.1 | 94.2 | 97.1 | 94.3 | 94.2 | |
| SIFT DenseNet 201 | Softmax | 94.7 | 92.2 | 96.0 | 92.1 | 92.1 |
| SVM | 94.0 | 91.2 | 95.5 | 91.0 | 91.1 | |
| RF | 94.7 | 92.2 | 96.0 | 92.1 | 92.1 | |
| XG-Boost | 94.2 | 91.5 | 95.6 | 91.5 | 91.4 |
Table 8.
Experimental results obtained with deep and BRISK based handcrafted features and different ML methods.
| Model & Feature extraction | Final layer | Avg Acc (%) | Avg Sens (%) | Avg Spec (%) | Avg Prec (%) | Avg F1-score (%) |
|---|---|---|---|---|---|---|
| BRISK VGG-16 | Softmax | 95.9 | 93.9 | 96.9 | 93.9 | 93.9 |
| SVM | 95.9 | 93.9 | 96.9 | 93.9 | 93.9 | |
| RF | 95.9 | 93.8 | 96.9 | 93.9 | 93.9 | |
| XG-Boost | 95.6 | 93.6 | 96.8 | 93.7 | 93.5 | |
| BRISK VGG-19 | Softmax | 96.5 | 94.6 | 97.2 | 94.6 | 94.6 |
| SVM | 96.4 | 94.6 | 97.2 | 94.6 | 94.6 | |
| RF | 96.6 | 95.0 | 97.4 | 95.0 | 95.0 | |
| XG-Boost | 96.1 | 94.4 | 97.1 | 94.2 | 94.3 | |
| BRISK sCNN | Softmax | 94.7 | 92.3 | 96.0 | 92.1 | 92.1 |
| SVM | 95.2 | 93.0 | 96.4 | 92.8 | 92.8 | |
| RF | 95.4 | 93.3 | 96.5 | 93.1 | 93.2 | |
| XG-Boost | 95.4 | 93.3 | 96.5 | 93.1 | 93.2 | |
| BRISK DenseNet 169 | Softmax | 95.2 | 92.9 | 96.5 | 93.0 | 92.7 |
| SVM | 94.9 | 92.5 | 96.2 | 92.5 | 92.5 | |
| RF | 95.2 | 92.8 | 96.4 | 93.0 | 92.8 | |
| XG-Boost | 95.2 | 92.9 | 96.5 | 93.0 | 92.7 | |
| BRISK DenseNet 201 | Softmax | 94.9 | 92.4 | 96.2 | 92.5 | 92.4 |
| SVM | 94.7 | 92.2 | 96.0 | 92.1 | 92.1 | |
| RF | 94.5 | 91.8 | 95.8 | 91.8 | 91.8 | |
| XG-Boost | 94.5 | 91.8 | 95.8 | 91.8 | 91.8 |
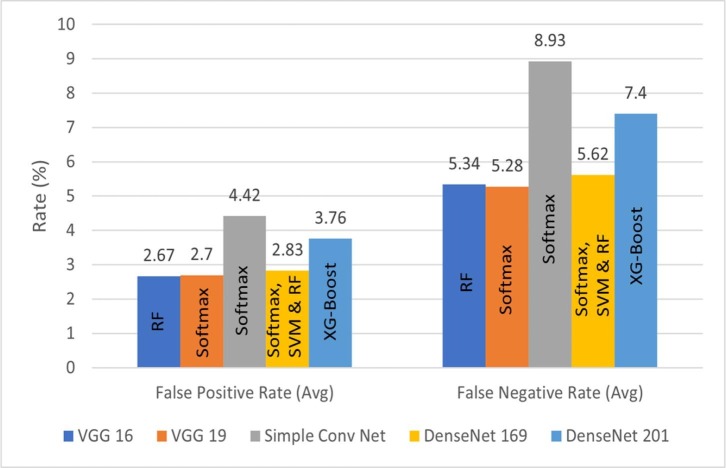
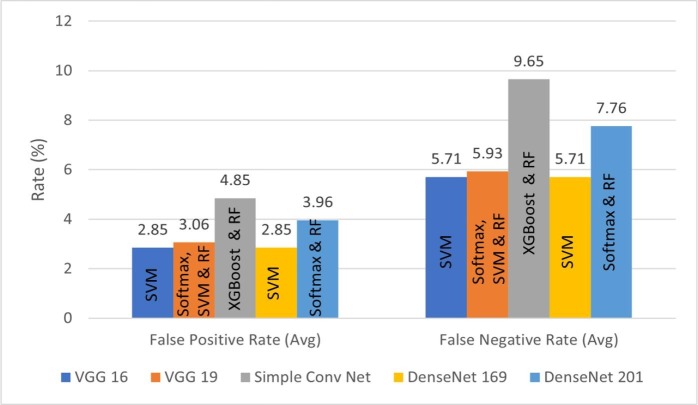
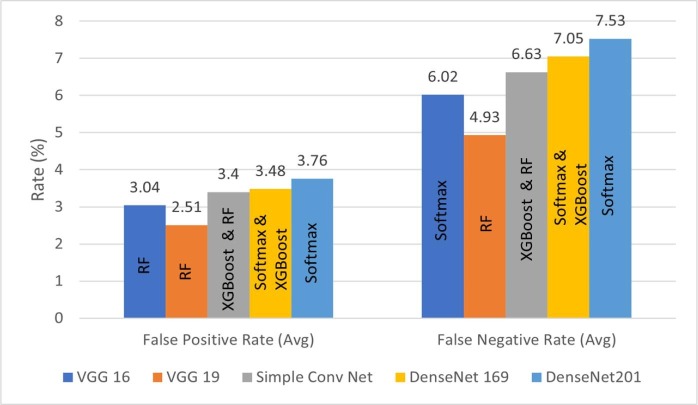
The classification performance of the proposed method is further analyzed in terms of average FPR and FNR bar plots, as illustrated by Fig. 15, Fig. 16, Fig. 17 . For better quality of classification performance, the FPR and FNR values should be significantly lower. It can be seen in Fig. 15 that the FPR of VGG-16 with RF as the last layer is the lowest, which is 2.67%. The lowest FNR of 5.28% is obtained using VGG-19 with softmax function as the last layer. In Fig. 16, it can be observed that the combination of VGG-16/DenseNet-169 and SIFT model achieved the lowest FPR and FNR of 2.85% and 5.71% with SVM in the final layer. From Fig. 17 it can be seen that VGG-19 with BRISK achieved the lowest FPR and FNR of 2.15% and 4.93% with RF as the last layer, making it the best combination among all the feature extraction models proposed.
Fig. 15.
FPR and FNR bar graphs for deep features with different ML techniques.
Fig. 16.
FPR and FNR bar graphs for deep and SIFT features with different ML techniques.
Fig. 17.
FPR and FNR bar graphs for deep and BRISK features with different ML techniques.
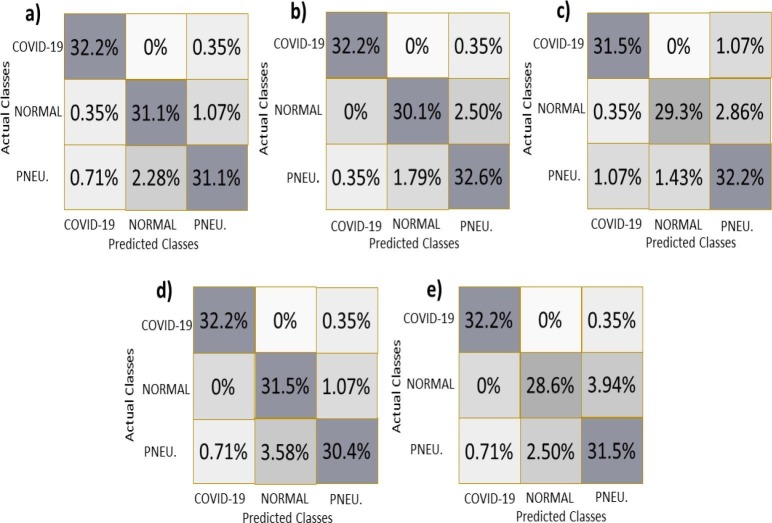
In order to illustrate more class-specific performance evidence of our proposed feature extraction models, Fig. 18 presents the confusion matrices corresponding to a total of 279 test images (92 “COVID-19” images, 91 “normal” images, and 97 “pneumonia” images). These matrices represent the data in the form of a percentage, where each cell of the confusion matrix indicates the percentage of total test images included in it. A specific model’s accuracy can be computed by summing the diagonal values (percentages) in the matrix with a diagonal starting from the matrix’s leftmost corner. It can be seen in Fig. 18 (a) that the VGG-16 model with RF in the final layer correctly classified 32.2%, 31.1%, and 31.1% of total test images as COVID-19, normal, and pneumonia images, respectively, achieving the overall classification Acc to 94.4%. Similar class-specific performance can be observed for other models namely VGG-19-BRISK and RF, sCNN-BRISK and RF/XG-Boost, DenseNet 169 and softmax/SVM/RF, Densenet 201 and XG-Boost in Fig. 18 (b), Fig. 18 (c), Fig. 18 (d), Fig. 18 (e), respectively.
Fig. 18.
Confusion matrices for the different proposed feature extraction networks: (a) VGG-16 (Final layer is RF) (b) VGG-19 and BRISK (Final layer is RF) (c) sCNN and BRISK (Final layer is RF/XG-Boost) (d) DenseNet 169 (Final layer is softmax/SVM/RF) (e) DenseNet 201 (Final layer is XG-Boost). A cell of each matrix represents the percentage share of total data in it.
Finally, it is worth mentioning that the VGG-19 model combined with the BRISK algorithm with RF as the last layer has performed the best among all the models proposed in this work.
4.1. Comparison with existing methods
We have compared our best performing model with other existing state-of-the-art DL approaches for detecting COVID-19 from chest X-ray images, illustrated in Table 9 . All the existing methods in the table have been re-implemented for classification of COVID-19, pneumonia, and normal X-ray images present in our classification dataset (see Section 2.0.2). The fusion [38] of local binary patterns (LBP) features with the deep features (via Inception V3 architecture) computed from Gaussian filtered images has provided average Acc, Sens, and Spec of 95.11%, 93.15%, and 96.5% respectively. The MobileNet v2 (with transfer learning) based features [39] has achieved average classification Acc, Sens, and Spec of 93.33%, 90.66%, and 95.23%, respectively. Using the transfer learning model ResNet-34 based features [20] the average Acc, Sens, and Spec of 95.29%, 92.97%, and 96.46% have been obtained. The ResNet-50 model with SVM (proposed in [19]) has provided average classification Acc, Sens, and Spec of 93.33%, 90.41%, 95.07%, respectively. A fine tuned AlexNet model (proposed in [40]) has achieved average classification Acc, Sens, Spec of 95.72%, 93.59%, and 96.78%, respectively. On the other hand, our proposed framework combining the VGG-19 model, BRISK feature extraction method, and RF classifier outperformed the existing methods for COVID-19 and pneumonia detection reported in the Table. Further, it is worth to mention that the proposed feature extraction framework considers segmented lung images as input. This is contrary to the existing methods reported in the table which compute features directly from the chest X-ray images.
Table 9.
Comparison with the other existing state-of-the-art methods for classification of COVID-19, pneumonia, and normal chest X-ray images.
| Authors | Method | Avg | Avg | Avg |
|---|---|---|---|---|
| Acc (%) | Sens (%) | Spec (%) | ||
| Shankar and | LBP and | 95.11 | 93.15 | 96.5 |
| Perumal [38] | Inception V3 | |||
| Apostolopoulos | MobileNet v2 | 93.33 | 90.66 | 95.23 |
| and Mpesiana [39] | ||||
| Nayak et al. [20] | ResNet-34 | 95.29 | 92.97 | 96.46 |
| Sethy and | ResNet-50 | 93.33 | 90.41 | 95.07 |
| Behera [19] | and SVM | |||
| Pham [40] | AlexNet | 95.72 | 93.59 | 96.78 |
| Proposed | VGG-19, BRISK, | 96.60 | 95.0 | 97.4 |
| method | and RF |
5. Discussion
We explored all three crucial steps for image classification in this work, namely, image segmentation, feature extraction, and classification. For segmentation, we initially employed simple supervised learning models where we tested various U-net architectures. However, the C-GAN model provided the best outcomes among all the tested supervised learning algorithms. The feature extraction pipeline has been built with the combination of keypoint extraction algorithms and deep CNN models. It was demonstrated that the keypoint descriptor algorithms are well suited for obtaining intensity information of the objects present in the image [41]. Thus, we employ the keypoint descriptors (SIFT and BRISK) to extract potential key intensity points that contribute significantly to the classification of segmented lung images. As mentioned, the framework also extracts the deep features using CNN models. All these models have provided convincing accuracy with convergence nature in significantly increased depth. The computed features in the final layer are classified using DL classifier (i.e. softmax) and also using various ML methods. The proposed segmentation and classification methods have been trained on publicly available datasets. We have presented a comprehensive study of the performance of proposed framework with and without considering the intensity keypoint features. The experimental results indicate that all the transfer learning models performed better than sCNN model in all the occasions. This study’s obtained results strongly support the usefulness of the proposed method in discriminating COVID-19, pneumonia, and normal chest X-ray images and can be employed as an assistive tool for the radiologists in their diagnosis. The primary objective of this work is to develop a cost-effective diagnostic method which can rapidly identify COVID-19 patients based on their chest radiographs. Because of nonavailability of larger dataset till date, this study has been carried out in a database containing a low number of COVID-19 chest X-ray images, which can be considered as a limitation of our work. In future, we plan to validate the proposed framework on a large dataset incorporating more COVID-19 chest X-ray images. In addition, we also intend to train our framework on a dataset containing CT images of COVID-19 patients and compare the performance when the method is trained with X-ray images.
6. Conclusion
In this paper, a method is proposed combining DL-based image segmentation and classification architectures to efficiently detect COVID-19, pneumonia, and normal chest X-ray images, improving the performances. The lung images were segmented using conditional GAN with the pix-to-pix algorithm, which was trained effectively using chest X-ray images with available ground truth masks. The trained segmentation network, which is basically the generator part of the C-GAN, is used for the segmentation of the preprocessed images. Afterward, segmented lung images were put into the feature extraction network, which is a composition of different DNNs (VGG-16, VGG-19, DenseNet 160, DenseNet 200, sCNN) and also key point detection methods such as SIFT and BRISK. Finally, the extracted features were classified using various ML techniques, namely softmax, RF, SVM, XG Boost, to detect different class images. In this study, VGG 19 model combined with the BRISK keypoints extraction method with RF as the final layer achieved the highest average classification accuracy of 96.6%. The model also provided average FPR and FNR of 2.51% and 4.93%, respectively, which are the lowest among all the other different architectures proposed in this work. The proposed method outperformed the other existing methods for the diagnosis of COVID-19 and pneumonia using chest X-ray images. In the future, the proposed method can be trained and tested using a larger dataset, and several other transfer learning methods also be included for performance evaluation.
CRediT authorship contribution statement
Abhijit Bhattacharyya: Conceptualization; Methodology; Data curation; Formal analysis; Investigation; Visualization; Validation; Writing - original draft; Supervision. Divyanshu Bhaik: Data curation; Software; Methodology; Formal analysis; Validation; Visualization; Writing - review & editing. Sunil Kumar: Data curation; Validation; Visualization; Writing - review & editing. Prayas Thakur: Data curation; Validation; Visualization; Writing - review & editing. Rahul Sharma: Data curation; Validation; Visualization; Writing - review & editing. Ram Bilas Pachori: Writing - review & editing; Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.C. Huang, Y. Wang, X. Li, L. Ren, J. Zhao, Y. Hu, L. Zhang, G. Fan, J. Xu, X. Gu, Z. Cheng, T. Yu, J. Xia, Y. Wei, W. Wu, X. Xie, W. Yin, H. Li, M. Liu, B. Cao, Clinical features of patients infected with 2019 novel coronavirus in wuhan, china, The Lancet 395. doi:10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed]
- 2.WHO, Weekly epidemiological update on covid-19 - 20 July 2021. url:https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports.
- 3.Chung M., Bernheim A., Mei X., Zhang N., Huang M., Zeng X., Cui J., Xu W., Yang Y., Fayad Z.A., Jacobi A., Li K., Li S., Shan H. Ct imaging features of 2019 novel coronavirus (2019-ncov) Radiology. 2020;295(1):202–207. doi: 10.1148/radiol.2020200230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.J. Zhang, Y. Xie, Z. Liao, G. Pang, J. Verjans, W. Li, Z. Sun, J. He, Y. Li, C. Shen, et al., Viral pneumonia screening on chest x-ray images using confidence-aware anomaly detection (2020), arXiv preprint arXiv:2003.12338. [DOI] [PMC free article] [PubMed]
- 5.Oh Y., Park S., Ye J.C. Deep learning covid-19 features on cxr using limited training data sets. IEEE Trans. Med. Imaging. 2020;39(8):2688–2700. doi: 10.1109/TMI.2020.2993291. [DOI] [PubMed] [Google Scholar]
- 6.H.-C. Shin, H.R. Roth, M. Gao, L. Lu, Z. Xu, I. Nogues, J. Yao, D. Mollura, R.M. Summers, Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning, IEEE Transactions on Medical Imaging 35 (5) 1285. (5). [DOI] [PMC free article] [PubMed]
- 7.Litjens G., Kooi T., Bejnordi B.E., Setio A.A.A., Ciompi F., Ghafoorian M., van der Laak J.A.W.M., van Ginneken B., Sánchez C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017;42 doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 8.Y. LeCun, Y. Bengio, G. Hinton, Deep learning, Nature 521 (7553) 436. (7553). [DOI] [PubMed]
- 9.Nayak D.R., Dash R., Majhi B., Pachori R.B., Zhang Y. A deep stacked random vector functional link network autoencoder for diagnosis of brain abnormalities and breast cancer. Biomed. Signal Process. Control. 2020;58 [Google Scholar]
- 10.A. Esteva, B. Kuprel, R.A. Novoa, J. Ko, S.M. Swetter, H.M. Blau, S. Thrun, Dermatologist-level classification of skin cancer with deep neural networks, no. 7639, Nature 542 (7639), 2017. [DOI] [PMC free article] [PubMed]
- 11.Chaudhary P.K., Pachori R.B. Fbsed based automatic diagnosis of covid-19 using x-ray and ct images. Comput. Biol. Med. 2021;104454 doi: 10.1016/j.compbiomed.2021.104454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.N. Ghassemi, A. Shoeibi, M. Khodatars, J. Heras, A. Rahimi, A. Zare, R.B. Pachori, J.M. Gorriz, Automatic diagnosis of covid-19 from ct images using cyclegan and transfer learning, arXiv preprint arXiv:2104.11949. [DOI] [PMC free article] [PubMed]
- 13.V. Chouhan, S.K. Singh, A. Khamparia, D. Gupta, P. Tiwari, C. Moreira, R. Damaaˇevicius, V.H.C. de Albuquerque, A novel transfer learning based approach for pneumonia detection in chest X-ray images, Applied Sciences 10 (2) 559. (2).
- 14.Gu X., Pan L., Liang H., Yang R. Proceedings of the 3rd International Conference on Multimedia and Image Processing. 2018. Classification of bacterial and viral childhood pneumonia using deep learning in chest radiography; pp. 88–93. [Google Scholar]
- 15.P. Lakhani, B. Sundaram, Deep learning at chest radiography: automated classification of pulmonary tuberculosis by using convolutional neural networks, no. 2, Radiology 284 (2), 2017. [DOI] [PubMed]
- 16.Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Rajendra Acharya U. Automated detection of covid-19 cases using deep neural networks with x-ray images. Computers Biology Med. 2020;121 doi: 10.1016/j.compbiomed.2020.103792. url:https://www.sciencedirect.com/science/article/pii/S0010482520301621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.E.E.-D. Hemdan, M.A. Shouman, M.E. Karar, Covidx-net: A framework of deep learning classifiers to diagnose covid-19 in x-ray images, arXiv preprint arXiv:2003.11055.
- 18.Wang L., Lin Z.Q., Wong A. Covid-net: A tailored deep convolutional neural network design for detection of covid-19 cases from chest x-ray images. Scientific Reports. 2020;10(1):1–12. doi: 10.1038/s41598-020-76550-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.P. Sethy, S. Behera, Detection of coronavirus disease (covid-19) based on deep features. preprints, Preprint posted online March 19.
- 20.Nayak S.R., Nayak D., Sinha U., Arora V., Pachori R. Application of deep learning techniques for detection of covid-19 cases using chest x-ray images: A comprehensive study. Biomed. Signal Process. Control. 2021;64:1–12. doi: 10.1016/j.bspc.2020.102365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Isola P., Zhu J.-Y., Zhou T., Efros A.A. Proceedings of the IEEE conference on computer vision and pattern recognition. 2017. Image-to-image translation with conditional adversarial networks; pp. 1125–1134. [Google Scholar]
- 22.Lowe D.G. Distinctive image features from scale-invariant keypoints. Int. J. Computer Vision. 2004;60(2):91–110. [Google Scholar]
- 23.Leutenegger S., Chli M., Siegwart R.Y. 2011 International Conference on Computer Vision. 2011. Brisk: Binary robust invariant scalable keypoints; pp. 2548–2555. [DOI] [Google Scholar]
- 24.Huang G., Liu Z., Van Der Maaten L., Weinberger K.Q. Proceedings of the IEEE conference on computer vision and pattern recognition. 2017. Densely connected convolutional networks; pp. 4700–4708. [Google Scholar]
- 25.K. Simonyan, A. Zisserman, Very deep convolutional networks for large-scale image recognition, arXiv preprint arXiv:1409.1556.
- 26.Shiraishi J., Katsuragawa S., Ikezoe J., Matsumoto T., Kobayashi T., Komatsu K.-I., Matsui M., Fujita H., Kodera Y., Doi K. Development of a digital image database for chest radiographs with and without a lung nodule: receiver operating characteristic analysis of radiologists’ detection of pulmonary nodules. Am. J. Roentgenol. 2000;174(1):71–74. doi: 10.2214/ajr.174.1.1740071. [DOI] [PubMed] [Google Scholar]
- 27.B. van Ginneken, M. Stegmann, M. Loog, Segmentation of anatomical structures in chest radiographs using supervised methods: a comparative study on a public database, Medical Image Analysis 10 (1) (2006) 19–40. url:http://www.isi.uu.nl/Research/Databases/SCR/. [DOI] [PubMed]
- 28.J.P. Cohen, P. Morrison, L. Dao, Covid-19 image data collection, arXiv 2003.11597. url:https://github.com/ieee8023/covid-chestxray-dataset.
- 29.P. Mooney, Chest x-ray images (pneumonia). url:https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia/metadata.
- 30.Heidari M., Mirniaharikandehei S., Khuzani A.Z., Danala G., Qiu Y., Zheng B. Improving the performance of cnn to predict the likelihood of covid-19 using chest x-ray images with preprocessing algorithms. Int. J. Med. Inform. 2020;144 doi: 10.1016/j.ijmedinf.2020.104284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goodfellow I., Pouget-Abadie J., Mirza M., Xu B., Warde-Farley D., Ozair S., Courville A., Bengio Y. Generative adversarial nets. Adva. Neural Information Processing Syst. 2014;27:2672–2680. [Google Scholar]
- 32.O. Ronneberger, P. Fischer, T. Brox, U-net: Convolutional networks for biomedical image segmentation, in: International Conference on Medical image computing and computer-assisted intervention, Springer, 2015, pp. 234–241.
- 33.Hearst M.A., Dumais S.T., Osuna E., Platt J., Scholkopf B. Support vector machines. IEEE Intell. Syst. Appl. 1998;13(4):18–28. doi: 10.1109/5254.708428. [DOI] [Google Scholar]
- 34.T. Chen, C. Guestrin, Xgboost, Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Miningdoi:10.1145/2939672.2939785.
- 35.Tin Kam Ho, Random decision forests, in: Proceedings of 3rd International Conference on Document Analysis and Recognition, Vol. 1, 1995, pp. 278–282 vol 1. doi:10.1109/ICDAR.1995.598994.
- 36.Bhattacharyya A., Pachori R.B. A multivariate approach for patient-specific EEG seizure detection using empirical wavelet transform. IEEE Trans. Biomed. Eng. 2017;64(9):2003–2015. doi: 10.1109/TBME.2017.2650259. [DOI] [PubMed] [Google Scholar]
- 37.A. Bhattacharyya, R.K. Tripathy, L. Garg, R.B. Pachori, A Novel Multivariate-Multiscale Approach for Computing EEG Spectral and Temporal Complexity for Human Emotion Recognition, IEEE Sensors Journal.
- 38.Shankar K., Perumal E. A novel hand-crafted with deep learning features based fusion model for covid-19 diagnosis and classification using chest x-ray images. Complex Intell. Syst. 2021;7(3):1277–1293. doi: 10.1007/s40747-020-00216-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Apostolopoulos I.D., Mpesiana T.A. Covid-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Phys. Eng. Sci. Med. 2020;43(2):635–640. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pham T.D. Classification of covid-19 chest x-rays with deep learning: new models or fine tuning? Health Inform. Sci. Syst. 2021;9(1):1–11. doi: 10.1007/s13755-020-00135-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xie J., Zhang L., You J., Shiu S. Effective texture classification by texton encoding induced statistical features. Pattern Recogn. 2015;48(2):447–457. [Google Scholar]