Abstract
Restriction fragment analysis and sequencing of a serotype-specific region were used to type 12 and 2 clinical isolates, respectively. Both molecular methods produced clear-cut results that completely correlated with that of the neutralization test.
Some human adenovirus serotypes (Ads), such as Ad1 to Ad8, Ad40, and Ad41, are frequently associated with epidemic outbreaks of acute respiratory tract, gastroenteric tract, and conjunctiva infections (10, 17, 20). Identification of the serotype is important for the epidemiological survey of the adenoviruses.
A serotype is defined mainly on the basis of a neutralization test which either exhibits no cross-reaction with others or shows a homologous/heterologous titer ratio greater than 16 in both directions. In the case of homologous/heterologous titer ratios of 8 or 16, the hemagglutination inhibition test must be applied and manifest a lack of crossreaction (19). Repeated experiments are often required, especially for the uncommon and unexpected serotypes. Sometimes, ambiguous results appear because of cross-reactions between different serotypes and the so-called intermediates. Moreover, the number of serotypes is still increasing (11), making serotyping even more complicated. Therefore, an alternative efficient and rapid typing method is desired.
Restriction fragment analysis (RFA) is one of the powerful methods that can be used to study the molecular epidemiology of adenoviruses (1–6, 11–16, 21). A comparison of all the precise restriction patterns revealed that all the genome types of a serotype usually present similar patterns. We therefore reasoned that RFA could be used for the identification of serotypes.
Our previous work has demonstrated that the adenovirus hexon possesses seven hypervariable regions, of which three regions (A1, A3, and B1) are obviously serotype specific (9, 18). Therefore, it seemed feasible to try to identify serotypes by sequencing of a serotype-specific region (SSR), especially the variable B1 region of the hexon loop 2.
In Northern Sweden, 20 to 60 adenovirus isolates have been obtained each winter since the late 1980s at the Department of Clinical Virology, University Hospital, Umeå, Sweden. However, during winter 1994–1995, 122 adenovirus strains were isolated. The significantly increased number of cases over the seasonal norm indicated that an epidemic outbreak of adenovirus infections had appeared. All the adenovirus strains were isolated in A549 cells and identified by a direct immunofluorescence test or a latex agglutination test.
Twelve isolates were selected from the 122 strains, especially those from patients with severe diseases (Table 1). Four standard prototypes of Ad2, Ad3, Ad5, and Ad7 were originally obtained from the American Type Culture Collection.
TABLE 1.
Origins of the 12 adenovirus isolates
| Isolate name | Specimen type | Collection date (mo/yr) | Main symptom | Age of patient (yr) | Genome type |
|---|---|---|---|---|---|
| 942000 | Conjunctiva swab | Nov. 1994 | Conjunctivitis | 39 | Ad7b |
| 942053 | Conjunctiva swab | Dec. 1994 | Conjunctivitis | 29 | Ad7b |
| 942055 | Pharynx swab | Dec. 1994 | Rhinopharyngitis | 2 | Ad7b |
| 942058 | Feces | Dec. 1994 | Rhinopharyngitis | 2 | Ad7b |
| 942154 | Feces | Dec. 1994 | Diarrhea | 82 | Ad5b |
| 942196 | Feces | Dec. 1994 | Diarrhea | 19 | Ad5b |
| 9544 | Conjunctiva swab | Jan. 1995 | Conjunctivitis | 73 | Ad3a9 |
| 9558 | Nasopharynx swab | Jan. 1995 | Pneumonia | 1 | Ad2p1 |
| 9576 | Feces | Jan. 1995 | Diarrhea | 1 | Ad5b |
| 95355 | Cerebrospinal fluid | Feb. 1995 | Meningoencephalitis | 26 | Ad5b |
| 95524 | Conjunctiva swab | Mar. 1995 | Conjunctivitis | 34 | Ad7b |
| 95553 | Feces | Mar. 1995 | Pneumonia | 1 | Ad7b |
All adenovirus isolates were propagated in A549 cells. Viral DNA preparation was performed by the method described previously (16, 22). DNA restriction enzyme cleavages were carried out according to the manufacturer’s instructions. The DNA restriction fragments were separated and analyzed by the methods described previously (16). The adenovirus genome types were denominated according to the nomenclature system described previously (13, 17).
The primer pair for PCR was designed according to the alignment result of 14 different hexons described previously (18). They were located at the conserved region of loop 2 and were QG65 (GGAACTGCCTAATTACTGTTTTCC) and QG66 (TTGGAGTACAGAAAACTTCTCCA). The PCR fragment covered the hypervariable and SSR B1 of the hexon. The templates were obtained from culture medium by chloroform extraction. The PCR fragments from strains 95355 and 95553 were ligated into the pT7-Blue plasmid and then were transformed into NovaBlue cells. The nucleotide sequences for both strands were determined by the dideoxynucleotide chain determination method.
The alignment of the predicted amino acid sequences of two strains, 95553 and 95355, and of the 18 serotypes was determined with the program MegAlign (DNAStar Inc., Madison, Wis.).
Results and discussion.
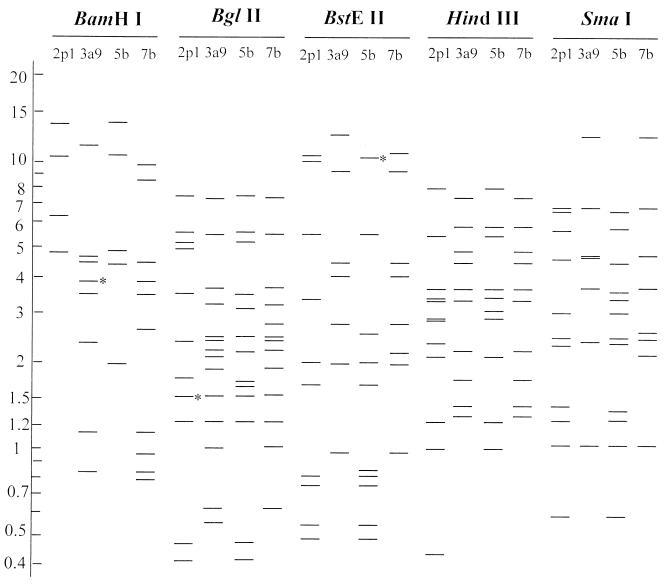
According to the highest pairwise comigrating restriction fragments (13) between a new strain and a known serotype, the four representative strains (95553, 95355, 9544, and 9558) (Table 1) were predicted to belong to the serotypes Ad7, Ad5, Ad3, and Ad2, respectively (Fig. 1). Strain 95553 was identical to genome type Ad7b (17). Strain 95355 shared the same patterns as Ad5 genome types with the exception of the BamHI pattern (4, 8). It was consequently recognized as a new genome type, which was named Ad5b. Strain 9544 shared the same patterns as Ad3a and Ad3a1 through Ad3a8 with the exception of the BglII pattern (14). It therefore represented a new genome type of Ad3, which was named Ad3a9. Strain 9558 shared the same restriction patterns as Ad2 genome types with the exception of HindIII pattern (4, 7). It represented a new genome type of Ad2, which was named Ad2p1.
FIG. 1.
Diagrammatic representation of DNA restriction patterns of four genome types digested with BamHI, BglII, BstEII, HindIII, and SmaI. The restriction patterns of 2p1, 3a9, 5b, and 7b were from representative strains 9558, 9544, 95355, and 95553, respectively. Numbers on the left indicate fragment sizes (in kilobase pairs). Double bands (indicated with asterisks) were unable to be separated by agarose gel electrophoresis as used in this study.
Of the 12 selected adenovirus isolates, six were determined to be genome type Ad7b and four were Ad5b. They were the dominating genome types among the 12 isolates studied.
The alignment of the amino acid sequences of the hexon B1 region from two new isolates and 18 prototypes demonstrated serotype-specific features. Strain 95553 and Ad7 and strain 95355 and Ad2 had distinctly higher percentages of similarity than the mean homology between members of each subgenus, 72.9 and 90.4%, respectively (Table 2). Therefore, according to the highest percentage of similarity to a known serotype, strain 95553 was determined to be serotype Ad7 and strain 95355 was determined to be Ad2.
TABLE 2.
Amino acid sequence similarities of the B1 regions between clinical isolates and 18 different adenovirus serotypesa
| Strain | Sequence similarity (%) of subgenus:
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A
|
B
|
C
|
D
|
E
|
F
|
|||||||||||||
| Ad12 | Ad31 | Ad3 | Ad7 | Ad16 | Ad1 | Ad2 | Ad5 | Ad6 | Ad8 | Ad9 | Ad15 | Ad19 | Ad37 | Ad48 | Ad4 | Ad40 | Ad41 | |
| 95355 | 39.2 | 36.7 | 40.0 | 41.7 | 44.2 | 48.1 | 46.2 | 90.4 | 46.2 | 34.6 | 32.7 | 36.5 | 36.5 | 36.5 | 39.2 | 48.1 | 38.0 | 40.8 |
| 95553 | 45.8 | 45.8 | 58.3 | 72.9 | 52.1 | 43.8 | 52.1 | 35.4 | 43.8 | 47.9 | 41.7 | 47.9 | 52.1 | 41.7 | 52.1 | 50.0 | 43.8 | 35.4 |
Eighteen amino acid sequences of the B1 region of hexon loop 2 with the following accession numbers were obtained from GenBank: Ad1, X67709; Ad2, J01917; Ad3, X76549; Ad4, X84646; Ad5, M73260; Ad6, X67710; Ad7, Z48571; Ad8, X74663; Ad9, X74664; Ad12, X73487; Ad15, X74666; Ad16, X74662; Ad19, X78359; Ad31, X74661; Ad37, X78360; Ad40, L19443; Ad41, X51783; Ad48, U20821.
To evaluate the two molecular methods, conventional neutralization tests were performed. The representative strains 9558, 9544, 95355, and 95553 were identified as Ad2, Ad3, Ad5, and Ad7, respectively. This result completely corroborated both RFA and SSR sequencing. The processing of the neutralization test takes at least 3 weeks, whereas each of the other two methods takes only 1 week.
The two molecular methods applied herein for typing are based on accumulated data obtained over more than 2 decades. The two methods, at present, can be efficiently used only for the identification of common Ads, i.e., those serotypes of which sequences of the hexon B1 region and restriction patterns have been studied. To type all of the known serotypes of any clinical isolate, the restriction patterns of as many presently known genome types as possible are required. Also, the sequences of the hexon B1 region from uncommon serotypes have to be determined.
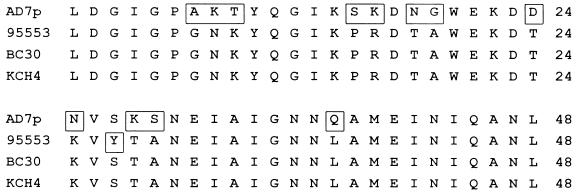
When the amino acid sequence of the B1 region of strain 95553 was aligned with the sequences of two earlier characterized Ad7b strains (17), an amino acid substitution at the tip of loop 2 (2) was discovered: 27Ser in BC30 and KCH4 was replaced by Tyr in 95553 (Fig. 2). This indicates that genetic drift had occurred within a certain genome type.
FIG. 2.
Comparison of the hexon B1 region of prototype Ad7p and those of three strains of genome type Ad7b (95553, BC30, and KCH4). Unconserved amino acids are shown in boxes.
Nucleotide sequence accession numbers.
Accession numbers for the two sequences determined in this study were obtained from GenBank, AF51950 for strain 95553 and AF51949 for strain 95355.
Acknowledgments
This work was supported by grants from the Swedish Foundation for Strategical Research and the Swedish Medical Research Council (grant K97-06X-05688-18A).
REFERENCES
- 1.Adrian T, Best B, Wigand R. A proposal for naming adenovirus genome types, exemplified by adenovirus type 6. J Gen Virol. 1985;66:2685–2691. doi: 10.1099/0022-1317-66-12-2685. [DOI] [PubMed] [Google Scholar]
- 2.Adrian T, Wadell G, Hierholzer J C, Wigand R. DNA restriction analysis of adenovirus prototype 1 to 41. Arch Virol. 1986;91:277–290. doi: 10.1007/BF01314287. [DOI] [PubMed] [Google Scholar]
- 3.Adrian T, de Jong J C, Wermenbol A G, van der Avoort H G A M, Wigand R. Genome type analysis of adenovirus 37 isolates. J Med Virol. 1988;25:77–83. doi: 10.1002/jmv.1890250111. [DOI] [PubMed] [Google Scholar]
- 4.Adrian T, Sassinek J, Wigand R. Genome type analysis of 480 isolates of adenovirus types 1, 2, and 5. Arch Virol. 1990;112:235–248. doi: 10.1007/BF01323168. [DOI] [PubMed] [Google Scholar]
- 5.Adrian T, Wolf U, Lauer H J, Wigand R. Restriction site mapping of adenovirus type 8 genome types. Res Virol. 1990;141:611–624. doi: 10.1016/0923-2516(90)90034-g. [DOI] [PubMed] [Google Scholar]
- 6.Adrian T, Wigand R. Restriction site mapping of subgenus D adenoviruses, prototypes 42-47, intermediate and atypical strains. Int J Med Microbiol. 1991;276:107–119. doi: 10.1016/s0934-8840(11)80224-0. [DOI] [PubMed] [Google Scholar]
- 7.Aird F, King J J, Younghusband H B. Identification of a new strain of adenovirus type 2 by restriction endonuclease analysis. Gene. 1983;22:133–134. doi: 10.1016/0378-1119(83)90072-0. [DOI] [PubMed] [Google Scholar]
- 8.Bruckova M, Wadell G, Sundell G, Syrucek L, Kunzova L. An outbreak of respiratory disease due to a type 5 adenovirus identified as genome type 5a. Acta Virol. 1980;24:161–165. [PubMed] [Google Scholar]
- 9.Crawford-Miksza L, Schnurr D P. Analysis of 15 adenovirus hexon proteins reveals the location and structure of seven hypervariable regions containing serotype-specific residues. J Virol. 1996;70:1836–1844. doi: 10.1128/jvi.70.3.1836-1844.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Jong J C, Wigand R, Kidd A H, Wadell G, Kapsenberg J G, Muzerie C J, Wermenbol A G, Firtzlaff R G. Candidate adenoviruses 40 and 41: fastidious adenoviruses from human infant stool. J Med Virol. 1983;11:215–231. doi: 10.1002/jmv.1890110305. [DOI] [PubMed] [Google Scholar]
- 11.De Jong J C, Wermenbol A G, Verweij-Uijterwaal M W, Slaterus K W, Wertheim-van Dillen P, van Doornum G J J, Khoo S H, Hierholzer J C. Xth International Congress of Virology. Jerusalem, Israel. 1996. Adenoviruses from AIDS patients, including two new candidate serotypes Ad50 and Ad51 of subgenus D and B1, respectively; p. 230. [Google Scholar]
- 12.Johansson M E, Brown M, Hierhozer J C, Thörner Å, Ushijima H, Wadell G. Genome analysis of adenovirus type 31 strains from immunocompromised and immunocompetent patients. J Infect Dis. 1991;163:293–299. doi: 10.1093/infdis/163.2.293. [DOI] [PubMed] [Google Scholar]
- 13.Li Q-G, Wadell G. Analysis of 15 different genome types of adenovirus type 7 isolated on five continents. J Virol. 1986;60:331–335. doi: 10.1128/jvi.60.1.331-335.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Q-G, Wadell G. Comparison of 17 genome types of adenovirus type 3 identified among strains recovered from six continents. J Clin Microbiol. 1988;26:1009–1015. doi: 10.1128/jcm.26.5.1009-1015.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Q-G, Wadell G. The degree of genetic variability among adenovirus type 4 strains isolated from man and chimpanzee. Arch Virol. 1988;101:65–77. doi: 10.1007/BF01314652. [DOI] [PubMed] [Google Scholar]
- 16.Li Q-G, Hambraeus J, Wadell G. Genetic relationship between thirteen genome types of adenovirus 11, 34, and 35 with different tropisms. Intervirology. 1991;32:338–350. doi: 10.1159/000150218. [DOI] [PubMed] [Google Scholar]
- 17.Li Q-G, Zheng Q-J, Liu Y-H, Wadell G. Molecular epidemiology of adenovirus types 3 and 7 isolated from children with pneumonia in Beijing. J Med Virol. 1996;49:170–177. doi: 10.1002/(SICI)1096-9071(199607)49:3<170::AID-JMV3>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 18.Li Q-G, Lindman K, Wadell G. Hydropathic characteristics of adenovirus hexons. Arch Virol. 1997;142:1307–1322. doi: 10.1007/s007050050162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Russell W C, Adrian T, Bartha A, Fujinaga K, Ginsberg H S, Hierholzer J C, Li Q-G, Mautner V, Nasz I, Wadell G. Adenoviridae. In: Murphy F A, Fauguet C M, Bishop D H L, Ghabrial S A, Jarvis A W, Martelli G P, Mayo M D, editors. Virus taxonomy: sixth report of the International Committee on Taxonomy of Viruses. New York, N.Y: Springer-Verlag Wien; 1995. pp. 128–133. [Google Scholar]
- 20.Schmitz H, Wigand R, Heinrich W. Worldwide epidemiology of human adenovirus infections. Am J Epidemiol. 1983;117:455–466. doi: 10.1093/oxfordjournals.aje.a113563. [DOI] [PubMed] [Google Scholar]
- 21.Schnurr D, Dondero M E. Two new candidate adenovirus serotypes. Intervirology. 1993;36:79–83. doi: 10.1159/000150325. [DOI] [PubMed] [Google Scholar]
- 22.Shinagawa M, Matsuda A, Ishiyama T, Goto H, Sato G. A rapid and simple method for preparation of adenovirus DNA from infected cells. Microbiol Immunol. 1983;27:817–822. doi: 10.1111/j.1348-0421.1983.tb00638.x. [DOI] [PubMed] [Google Scholar]