Abstract
Objective
Tissue stem cells are central regulators of organ homoeostasis. We looked for a protein that is exclusively expressed and functionally involved in stem cell activity in rapidly proliferating isthmus stem cells in the stomach corpus.
Design
We uncovered the specific expression of Iqgap3 in proliferating isthmus stem cells through immunofluorescence and in situ hybridisation. We performed lineage tracing and transcriptomic analysis of Iqgap3 +isthmus stem cells with the Iqgap3-2A-tdTomato mouse model. Depletion of Iqgap3 revealed its functional importance in maintenance and proliferation of stem cells. We further studied Iqgap3 expression and the associated gene expression changes during tissue repair after tamoxifen-induced damage. Immunohistochemistry revealed elevated expression of Iqgap3 in proliferating regions of gastric tumours from patient samples.
Results
Iqgap3 is a highly specific marker of proliferating isthmus stem cells during homoeostasis. Iqgap3+isthmus stem cells give rise to major cell types of the corpus unit. Iqgap3 expression is essential for the maintenance of stem potential. The Ras pathway is a critical partner of Iqgap3 in promoting strong proliferation in isthmus stem cells. The robust induction of Iqgap3 expression following tissue damage indicates an active role for Iqgap3 in tissue regeneration.
Conclusion
IQGAP3 is a major regulator of stomach epithelial tissue homoeostasis and repair. The upregulation of IQGAP3 in gastric cancer suggests that IQGAP3 plays an important role in cancer cell proliferation.
Keywords: cell proliferation, stem cells, gastric cancer, gastric pre-cancer, gastric wound repair
Significance of this study.
What is already known on this subject?
Highly proliferative isthmus stem cells in the stomach corpus are multipotent.
Iqgap3 is necessary and sufficient for cell proliferation.
What are the new findings?
Identification of cytoskeletal scaffold protein Iqgap3 as a specific marker for actively cycling isthmus stem cells.
Iqgap3 is required for promoting Ras-driven proliferation of isthmus stem cells.
Iqgap3 is required for the maintenance of the stem cell transcriptional programme.
Robust induction of Iqgap3 during tissue repair is associated with expansion of isthmus stem cells and dedifferentiation of chief cells.
Tissue repair is accompanied by gene expression signatures associated with carcinogenesis. The commonality of increased Iqgap3 expression in tissue repair and cancer potentially indicates a driver role for Iqgap3 in ‘a wound that does not heal’.
How might it impact on clinical practice in the foreseeable future?
Identification of Iqgap3 as a specific marker for rapidly proliferating isthmus stem cells and cancer cells may provide molecular insights to proliferation of gastric cancer stem cells.
Robust induction of Iqgap3 during tissue repair indicates a potential mechanism for gastric cancer initiation.
Introduction
Characterisation of the mechanisms underlying the proliferation, plasticity and lineage commitment of adult stem cells is necessary for understanding the preneoplastic events leading to gastric cancer. In the stomach corpus, the gastric gland can be subdivided into four zones: the pit, the isthmus, the neck and the base. Classical radioactive tracer experiments indicated rapidly proliferating cells in the isthmus, which were revealed by electron microscopy to be granule-free stem cells.1 The isthmus region is therefore accepted to be the active stem cell zone of the corpus. Zymogenic chief cells located at the base of the stomach corpus are fully differentiated, postmitotic cells. Interestingly, in 2010, various studies reported that chief cells reacquire proliferative capacity, suggesting that chief cells may be progenitors for the preneoplastic spasmolytic polypeptide-expressing metaplasia.2–4 In 2013, Troy+chief cells were reported to serve as quiescent stem cells, which can be induced to proliferate during tissue damage to regenerate the entire gastric unit.5
A different view emerged in 2015, when Mist1+quiescent stem cells were identified in the isthmus and proposed to be the origin of gastric cancer.6 Notwithstanding this finding, Choi et al used targeted expression of oncogenic KrasG12D in Mist1+chief cells to contend that metaplasia arises from transdifferentiation or dedifferentiation of chief cells.7 Leushacke et al identified the expression of established stem cell factor Lgr5 in a subset of chief cells and observed that Kras(G12D) induction in Lgr5 +chief cells, in the presence of injury, led to metaplastic lesions. Together with the analysis of Lgr5-expressing human gastric cancer tissues, they proposed that gastric cancer originated from Lgr5-expressing chief cells.8
In 2017, we uncovered strong activity of an enhancer element of Runx1 (eR1), a haematopoietic and human hair follicle stem cell factor, in proliferating isthmus stem cells, as well as a small number of chief cells.9 KrasG12D expression in eR1+isthmus stem cells induced foveolar hyperplasia and elimination of parietal cells.9 However, Kinoshita et al detected induction of Lgr5 in isthmus stem cells after tissue injury and proposed that isthmus stem cells and cells from the neck lineage, instead of chief cells, contributed to metaplasia.10 Arguing that the contradictory data may have stemmed from the specificity of the chief cell markers, Hata et al identified Gpr30 as a specific marker of chief cells, with no expression in isthmus stem cells.11 Expression of KrasG12D in Gpr30+chief cells did not result in metaplasia but, instead, led to reduced chief cell population and a compensatory expansion of neck lineage, derived from Kitl +isthmus stem cells.11 It was further argued that since most chief cells are lost during metaplasia, they are unlikely to be involved in gastric carcinogenesis.11 Certainly, after acute oxyntic injury, the increased activity of Lrig1-expressing isthmus stem cells has been shown to contribute to tissue regeneration.12 During Helicobacter pylori-induced chronic injury, it was reported that increased proliferation and accelerated differentiation of Lrig1-expressing cells gave rise to surface mucous cells and chief cells, which subsequently produced spasmolytic polypeptide-expressing metaplasia.13 The origin of metaplasia continues to be highly debated, in part due to the expression of some markers in both isthmus and chief cell populations, progenitor cell plasticity, effects from stem cell niche as well as injury.
Two models have been suggested for the homoeostasis of the corpus: (1) long-lived stem cells supported by the stem cell niche at the isthmus zone is proposed to maintain the gastric epithelia11 and (2) two independent stem cell populations potentially maintain the corpus—the rapidly cycling isthmus stem cells support the pit, isthmus and neck regions, while the mostly quiescent chief cells support the base region.14 15
The study of isthmus stem cell behaviour will provide insights to the molecular mechanism underlying cancer initiation. Sox2, Lrig1, TFF2, Mist1, eR1, Bmi1 and Kitl have all been reported to mark stem/progenitor cells in the isthmus; however, many of these markers were expressed in other cell types and, thus not specific for proliferating isthmus stem cells.4 6 9 11 12 16–18 Actively cycling isthmus stem cells were also studied via their expression of proliferation markers Stathmin1 (Stmn1) and Mki67.14 Several signalling pathways, such as Notch, Sonic hedgehog and AMP-dependent protein kinase, were reported to govern proliferation of gastric epithelial cells.19–21 In particular, isthmus stem cell proliferation was found to be driven by the Notch pathway.22 Yet, these studies did not reveal the presence of an exclusive and definitive stem cell marker in proliferative isthmus stem cells.
In this study, we show that eR1+isthmus stem cell proliferation is driven by Iqgap3, a member of the Iqgap (IQ motif containing GTPase activating protein) cytoskeletal scaffold family that was reported to be necessary and sufficient for cell proliferation.23 Knockdown of Iqgap3 in undifferentiated cell lines, such as embryonal carcinoma NTERA-2 and gastric carcinoma HGC-27, resulted in reduction of stem cell-associated NANOG and OCT4 gene expression, as well as induction of differentiation. Therefore, Iqgap3 represents a functionally indispensable stem cell specific factor, which regulates stem cell function in homoeostasis and tissue damage repair. Our work further revealed that Iqgap3 induction during tissue repair is associated the acquisition of oncogenic traits. Finally, we found a strong correlation between Iqgap3 expression and proliferating cancer cells in tumours isolated from gastric cancer patients. Together, these data reveal a mechanism by which Ras is hyperactivated in preneoplasia and identify a role for Iqgap3 in lineage plasticity and proliferation in cancer initiation.
Material and methods
Mice and treatment
Wild-type (WT) C57BL/6JInv mice were obtained from InVivos Pte Ltd. The eR1-enhanced green fluorescent protein mice and the eR1-CreERT2;Rosa-Lox-Stop-Lox (LSL)-tdTomato (eR1-CreERT2;Rosa-tdTomato) mice were described previously.9 24 To induce tdTomato expression in eR1-CreERT2;Rosa-tdTomato mice for short lineage tracing experiments, 6–8 weeks old mice were given a single intraperitoneal injection of 2 mg of tamoxifen (Merck) diluted in corn oil (Merck). The mice were analysed at 16 or 24 hours post-tamoxifen injection. Iqgap3-2A-tdTomato mice were generated to monitor Iqgap3 expression in the stomach (Cyagen). The 2A-tdTomato sequence was inserted before the stop codon in exon 38 (online supplemental figure S1A). Iqgap3-2A-CreERT2 mice were generated for inducing or repressing expression of the gene of interest in Iqgap3 +cells (Cyagen). The 2A-CreERT2 sequence was inserted before the stop codon in exon 38 (online supplemental figure S2A). Iqgap3-2A-CreERT2 mice were bred to Rosa-tdTomato mice to generate Iqgap3-2A-CreERT2;Rosa-tdTomato mice. To induce tdTomato expression, 6–8 weeks old mice were given a single intraperitoneal injection of 2 mg of tamoxifen. The mice were analysed 1-day, 3-month, 6-month and 1-year post-tamoxifen injections. Iqgap3-2A-CreERT2 mice were also bred to LSL-KrasG12D (KrasG12D/+ ) mice to generate Iqgap3-2A-CreERT2;KrasG12D/+ mice. To induce KrasG12D expression, 6–8 weeks old mice were given a single intraperitoneal injection of 2 mg of tamoxifen. The mice were analysed 3 months post-tamoxifen injection. To induce tissue damage in the murine stomach, 6–8 weeks mice were given a single intraperitoneal injection of a high dose of tamoxifen (HDT) (5 mg per 20 g body weight). The mice were analysed at indicated time points post-tamoxifen injection.
gutjnl-2020-322779supp001.pdf (3.5MB, pdf)
gutjnl-2020-322779supp002.pdf (199.3KB, pdf)
Detailed materials and methods are described in online supplemental methods.
Results
Iqgap3 is specifically expressed in rapidly proliferating isthmus stem cells
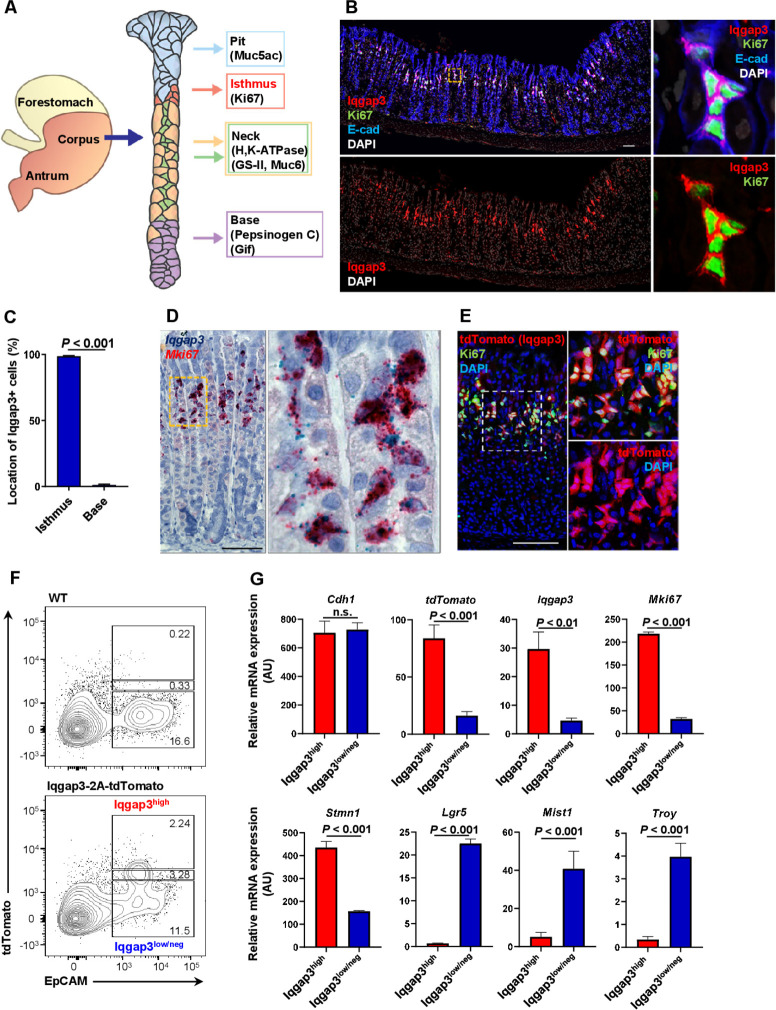
Using immunofluorescence staining and RNA in situ hybridisation (ISH), we found that Iqgap3 is strongly coexpressed with the proliferation marker Ki67 (figure 1A–D), thereby identifying Iqgap3 as a marker of rapidly proliferating cells at the isthmus zone. Using an Iqgap3-2A-tdTomato mouse, we found robust tdTomato expression in Ki67+cells at the isthmus (figure 1E; online supplemental figure S1A, B). We next isolated Iqgap3-tdTomatohigh EpCAMhigh epithelial cells by flow cytometry (figure 1F). Quantitative PCR (qPCR) showed that tdTomato, Iqgap3 and Mki67 mRNA were strongly enriched in the Iqgap3-tdTomatohighEpCAMhigh epithelial cells, relative to the Iqgap3-tdTomatolow/neg population (figure 1G, upper panel). Strong enrichment of Stmn1 mRNA—previously shown to mark cycling isthmus stem cells14—further confirmed the proliferative nature of Iqgap3-tdTomatohigh cells (figure 1G, lower panel). Lgr5 and Troy expression were low in Iqgap3-tdTomatohigh cells, suggesting that Iqgap3+cells are stem cells distinct from Lgr5+cells (figure 1G, lower panel). Similarly, Mist1 expression—earlier reported to mark chief cells and quiescent isthmus stem cells in the corpus3 6 7—was relatively low in Iqgap3-tdTomatohigh cells (figure 1G, lower panel). We had previously shown that the activity of the Runx1 enhancer element, eR1, could be used to identify adult stem cells in stomach tissues.9 eR1-CreERT2;Rosa-tdTomato mice were treated with tamoxifen to examine eR1+and Iqgap3 +cells in vivo. After 16 hours of short lineage tracing, eR1+cells partially coincided with Iqgap3-expressing cells at the isthmus, likely because of the variegated expression intrinsic to the mouse model (online supplemental figure S1C). This indicated that Iqgap3+cells harbour isthmus stem cells.
Figure 1.
Expression of Iqgap3 in the isthmus of corpus epithelium. (A) Schematic diagram of a gastric unit in the corpus of the mouse stomach. (B) Immunofluorescence (IF) staining for Iqgap3, Ki67 and E-cadherin (E-cad) on the corpus of wild-type (WT) mouse stomach (n=3). (C) Quantification of Iqgap3+cells in isthmus or base (n=3). Error bar represents SD from 2025 Iqgap3+cells of 3 mice. The Ki67+region was defined as the isthmus zone. (D) In situ hybridisation (ISH) for Iqgap3 (green) and Mki67 (red) on the corpus of WT mice (n=2). (E) IF staining for tdTomato and Ki67 on the corpus of Iqgap3-2A-tdTomato mice (n=3). (F) Flow cytometry to isolate tdTomato/Iqgap3 high expression epithelial cell fraction (Iqgap3high) and tdTomato/Iqgap3 low or negative expression epithelial cell fraction (Iqgap3low/neg) from stomach of Iqgap3-2A-tdTomato reporter mice (n=5). (G) qPCR for Cdh1, tdTomato, Iqgap3, Mki67, Stathmin1 (Stmn1), Lgr5, Bhlha15 (Mist1) and Tnfrsf19 (Troy) mRNA from isolated tdTomatohigh (Iqgap3high) and tdTomatolow/neg (Iqgap3low/neg) gastric epithelial cells. mRNA expression was normalised by Gapdh expression (n=3). Error bars represent SD scale bar=100 µm. qPCR, quantitative PCR.
The Iqgap family comprises three genes, namely Iqgap1, 2 and 3.25 Unlike Iqgap3, the expression of Iqgap1 and 2 were not confined to the isthmus, but observed throughout the gastric unit (online supplemental figure S1D–G). Therefore, Iqgap3—alone in the Iqgap family—plays a unique role in driving proliferation of isthmus stem cells.
Iqgap3+ isthmus stem cells are multipotent and responsible for core homoeostasis of corpus glands
We next used Iqgap3-2A-CreERT2;Rosa-tdTomato mice to perform lineage tracing of Iqgap3+cells (figure 2A, B; online supplemental figure S2A, B). One-day post-tamoxifen injection revealed strong coexpression of Ki67 and tdTomato at the isthmus (figure 2C; online supplemental figure S2B); at 3-month, 6-month and 1-year post-tamoxifen injection, we observed progressive expansion of tdTomato+cells to span almost the entire gastric gland (figure 2C). tdTomato expression overlapped with markers of various corpus lineages, including mucous pit, neck, parietal and chief cells (figures 1A and 2D, E). Analysis of the lineage tracing events at 6-month post-tamoxifen injection revealed that ~50% of the glands showed tdTomato positivity spanning from the pit to neck region, while ~40% showed tdTomato positivity from the pit to the transition regions (see #1 and #2 in figure 2F). About 10% of the glands showed tdTomato+cells spanning from the pit to base region (see #3 in figure 2F). At 1-year post-tamoxifen injection, tdTomato positivity spanning the pit to base region had increased to ~55% of the glands (figure 2F). XZ-plane imaging further confirmed tdTomato +cells in the neck (GS-II+/Gif–), transition (GS-II+/Gif+) and chief cell populations (GS-II–/Gif+) (online supplemental figure S2C). The self-replicating nature of chief cells15 may be the reason why not all chief cells were labelled with tdTomato, even after 1 year of tracing. Nevertheless, our time course data indicated that lineage tracing initiated at the isthmus and that tdTomato +cells are multipotent stem cells.
Figure 2.
The Iqgap3-expressing cells in the isthmus are multipotent stem cells. (A) Iqgap3-2A-CreERT2;Rosa-tdTomato mouse model. (B) Experimental strategy for lineage tracing time course. (C) IF staining for Ki67 and tdTomato on the corpus of Iqgap3-2A-CreERT2;Rosa-tdTomato mice at 1 day (1 d), 3 months (3 m), 6 months (6 m) and 1 year (1 y) post-tamoxifen induction (p.i.) (n=3). Lineage tracing, LT. (D, E) IF staining for tdTomato and markers of major stomach differentiated cells (Muc5ac, H, K-ATPase, GS-II and Gif) on the corpus of Iqgap3-2A-CreERT2;Rosa-tdTomato mice at 1 year post-tamoxifen induction (n=3). (F) Quantification of lineage tracing on the corpus of Iqgap3-2A-CreERT2;Rosa-tdTomato mice at 6 months and 1 year post-tamoxifen induction. (n=3). The tdTomato+lineage tracing glands were categorised into pit to neck (LT #1: Pit - Neck), pit to mucus-neck/chief cell transition (LT #2: Pit-Transition) and pit to base (LT #3: Pit – Base). A total of 167 tdTomato+glands from three mice (6 months) or 172 tdTomato+glands from three mice (1 year) were counted. Error bars represent SD data sets were analysed by one-way ANOVA. ***P<0.001. (G) tdTomato expression in the isolated corpus gastric units from Iqgap3-2A-CreERT2;Rosa-tdTomato mice at 20–24 hours post-tamoxifen administration (top). Corpus organoids were generated from tdTomato +cells (n=2). (H) qPCR for IQGAP3, NANOG, OCT4, KLF4, cMYC, SOX2, CD44v9 and GFAP mRNA from IQGAP3 knockdown embryonic stem cell line NTERA-2. mRNA expression was normalised by GAPDH expression (n=3). Error bars represent SD data sets were analysed by Student’s t-test. *P<0.05, **p<0.01. (I) Immunoblot for IQGAP3, Nanog, Oct4, KLF4, CD44v9, GFAP and GAPDH from knockdown embryonic stem cell line NTERA-2 (n=3). Scale bar=100 µm. ANOVA, analysis of variance; GFAP, glial fibrillar acidic protein; IF, immunofluorescence; qPCR, quantitative PCR.
tdTomato+isthmus cells from corpus units isolated from Iqgap3-2A-CreERT2;Rosa-tdTomato mice readily formed organoids, further confirming the stem potential of Iqgap3+isthmus cells (figure 2G). Induction of differentiation via removal of Wnt3a and R-spondin1 from the organoid culture media (termed ENFG as opposed to WENFRG)26 led to decreased Iqgap3 mRNA, accompanied by a sharp increase in differentiation marker Muc5ac (online supplemental figure S2D, E). Iqgap3 expression was therefore specific to stem cells and shows rapid reduction on differentiation in ENFG media. Iqgap3+/tdTomato +organoids showed upregulation of Muc5ac protein on differentiation (online supplemental figure S2F). The organoids derived from Iqgap3 +cells possessed PgC+/GS-II+, PgC+/GS-II– and PgC–/GS-II +cells, as well as Gif+/GS-II +and Gif+/GS-II– cells, demonstrating that Iqgap3 +isthmus cells differentiated to the chief cell, mucus-neck cell and transition cell lineages in our organoid culture system (online supplemental figure S2G, H). In addition, the Iqgap3 +cell-derived organoids also showed expression of Ki67, but not H, K-ATPase (online supplemental figure S2I, J). Iqgap3+/tdTomato +cells can therefore generate pit, neck and chief cells in vitro.
To understand the function of IQGAP3 in maintaining stemness, we performed siRNA-mediated knockdown of IQGAP3 in a well-established model for pluripotency and differentiation, the human embryonal carcinoma NTERA-2 cell line. Depletion of IQGAP3 led to significant reductions in mRNA and protein expression levels of stem cell factors such as NANOG, OCT4 and KLF4 (figure 2H, I). We also observed an increase of the glial fibrillar acidic protein, which is expressed on differentiation to astrocytes (figure 2H, I). This proof-of-concept experiment indicates that IQGAP3 is necessary for the maintenance of stem cell gene expression signature and not just a marker for proliferation.
Iqgap3 drives stem cell proliferation by promoting the Ras‐extracellular signal-regulated kinase signalling pathway
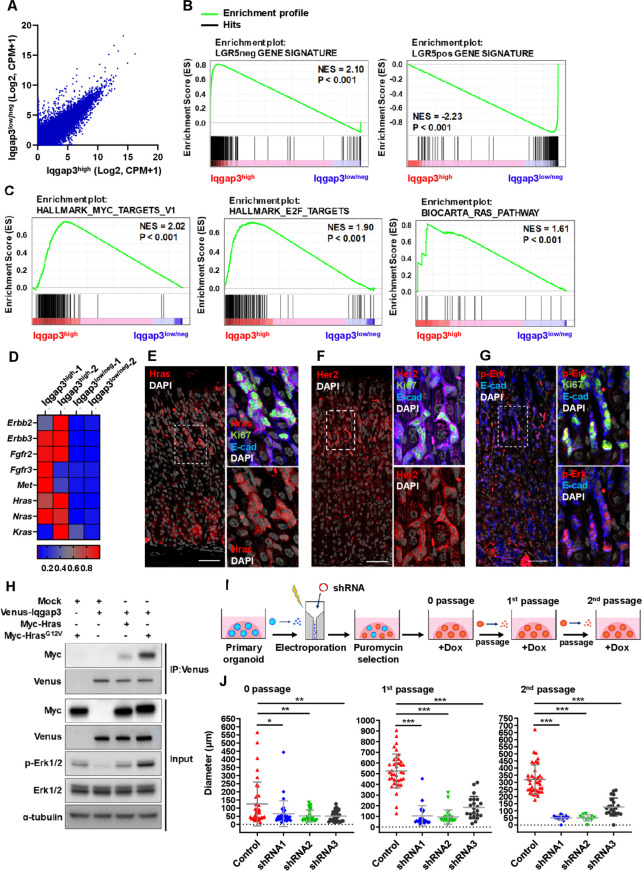
To identify the transcriptional programmes integral to the isthmus stem cell state, we next isolated Iqgap3-tdTomatohigh and Iqgap3-tdTomatolow/neg expressing cells by flow cytometry (figure 3A). RNA-sequencing revealed that Iqgap3-tdTomatohigh cells were highly enriched in Iqgap3, Mki67, Stmn1, Kitl mRNA, while showing low expression levels of chief cell markers PgC and Gpr30 (online supplemental figure S3A). Gene set enrichment analysis (GSEA) confirmed that Iqgap3-tdTomatohigh stem cells are distinct from Lgr5 +cells (figure 3B; online supplemental figure S3B). The transcriptional profile of Iqgap3-tdTomatohigh cells mapped closely with the short-term haematopoietic stem cell gene signature ST-HSC (online supplemental figure S3C, D). The enriched expression of HSC self-renewal—and asymmetric cell division-associated genes in the Iqgap3-tdTomatohigh fraction further reinforced the notion that Iqgap3 +cells possess stem cell properties (online supplemental figure S3E). Notch, Hedgehog and Wnt pathways were not significantly upregulated (online supplemental figure S3F). Conversely, the strong upregulation of Myc-target, E2F-target and Ras signalling genes in Iqgap3-tdTomatohigh cells identified these pathways as core programmes driving isthmus stem cell proliferation (figure 3C). Notably, key components of the Ras signalling pathway, namely Erbb2, Erbb3, Fgfr2, Fgfr3, Met and Ras, were enriched in Iqgap3-tdTomatohigh stem cells (figure 3D).
Figure 3.
Iqgap3 regulates stem cell proliferation via Ras-ERK pathway. (A) Average of RNA expressions in sorted Iqgap3high and Iqgap3low/neg cell fractions are shown. (B) Gene set enrichment analysis (GSEA) showing enrichment of Lgr5-negative (LGR5neg) and—high (LGR5pos) corpus epithelial cell gene signature from public datasets (GSE86603) in Iqgap3high and Iqgap3low/neg cell fractions. P values determined by a weighted Kolmogorov–Smirnov-like statistic and adjusted for multiple hypothesis testing. (C) GSEA showing enrichment of Myc target gene signature, E2F targets gene signature, Ras pathway gene signature in Iqgap3high and Iqgap3low/neg cell fractions. (D) Heat MAP showing expression of representative Ras-ERK pathway genes in Iqgap3high and Iqgap3low/neg cell fractions. Gene expression levels are shown in Z-score of CPM of RNA-sequencing. (E–G) IF staining for HRAS, Ki67, E-cad, HER2 and phosphorylated ERK (p-Erk) on the corpus of wild-type (WT) mice (n=3). (H) Immunoprecipitation for the interaction of Venus-tagged Iqgap3 (Venus-Iqgap3) with Myc-tagged HRAS (Myc-Hras) or Myc-tagged HrasG12V (Myc-HrasG12V) and immunoblot for cell lysate from Venus-Iqgap3/Myc-Hras/Myc-HrasG12V expressed 293 T cells. (I) Experimental strategy to suppress Iqgap3 expression by shRNA in organoids from WT mice. (J) Quantification of size of Iqgap3 knockdown organoids from 0 passage (6 day post doxycycline (Dox) treatment, before passage), first passage (8 days postpassage) and second passage (7 days postpassage) (n=2). Error bars represent SD from each population. Data sets were analysed by one-way ANOVA. Scale bar=100 µm. ANOVA, analysis of variance; CPM, counts per million; ERK, extracellular signal-regulated kinase; NES, normalised enrichment score.
We, therefore, investigated the activity of the Ras pathway in isthmus stem cells in vivo. Immunostaining showed that Hras, Nras and Kras were all expressed in the corpus, partially overlapping with Ki67 expression (figure 3E; online supplemental figure S4A, B). Her2 is known to promote Ras pathway activation.27 28 Immunostaining indicated the expression of Her2 and Ras downstream effector phosphorylated Erk (p-Erk) in the Ki67 +proliferating cells at the isthmus (figure 3F, G). Her2high and Her2low-expressing cells isolated by flow cytometry revealed enrichment of Erbb2, Iqgap3 and Mki67 mRNA in Her2high cells, and elevated Lgr5 and Mist1 mRNA in Her2low cells (online supplemental figure S4C, D). Kras, Hras and Nras mRNA were detected in both Her2high and Her2low cells, with Kras and Hras showing significantly higher expression in Her2high cells (online supplemental figure S4E). Iqgap3 expression, therefore, positively correlates with the activity of Erk signalling cascade. Indeed, immunoprecipitation revealed that the interaction of Iqgap3 with the constitutively active HrasG12V mutant was associated with increased p-Erk in the total cell lysate (figure 3H). To ascertain if the Iqgap3-Ras-Erk axis contributes to organoid growth, we subjected organoids to shRNA mediated depletion of Iqgap3 (figure 3I). Depletion of Iqgap3 using three different shRNAs (online supplemental figure S5A) resulted in drastically smaller organoids with reduced ability to be serially passaged (figure 3J; online supplemental figure S5B–D). Corpus glands were treated with two different inhibitors of the Erk pathway, U0126 and MK-8353. Immunostaining of the organoids revealed that both inhibitors resulted in severe depletion of p-Erk (online supplemental figure S5E). After 96 and 72 hours, organoid growth was strongly inhibited in a concentration-dependent manner by U0126 and MK-8353, respectively (online supplemental figure S5F, G).
Iqgap3 is strongly induced following tissue injury
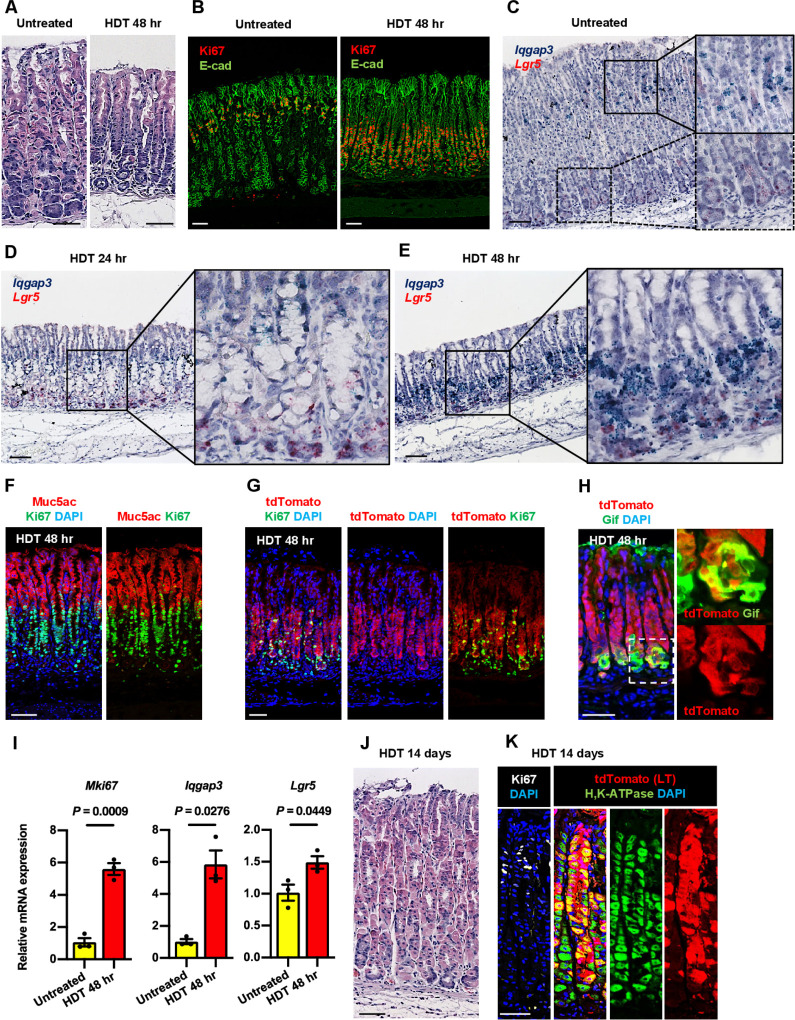
We next examined the expression of Iqgap3 during tissue injury. Parietal cell protonophores such as tamoxifen, DMP-777 and L635 have been used to study the development of metaplasia.29 Tamoxifen was reported to cause the back wash of acid into parietal cells, resulting in parietal cell death.30 In addition to induction of reversible atrophy, tamoxifen treatment has been associated with progenitor cell proliferation and metaplasia in the mouse stomach.31 32 Accordingly, an intraperitoneal injection of a HDT resulted in drastic changes in gland morphology and loss of the H, K-ATPase-producing parietal cells after 48 hours (figure 4A; online supplemental figure S6A). At 24 hours and 48 hours post-HDT, we observed dramatic increases in proliferating Ki67+epithelial cells, although at different positions, which subsided to the normal staining pattern 14 days after HDT (figure 4B; online supplemental figure S6B). At 48 hours post-HDT, Ki67 expression can be seen at the lower part of the gland (figure 4B). In untreated WT mice, co-ISH showed Iqgap3 mRNA at the isthmus zone and Lgr5 mRNA at the base of the gland (figure 4C). At 24 hours post-HDT, Iqgap3 mRNA was predominantly detected above the damaged parietal cells, suggesting enhanced proliferation of Iqgap3+isthmus stem cells (figure 4D). At 48 hours post-HDT, we observed a robust induction of Iqgap3 mRNA expression near the base of the gland (figure 4E). Coexpression of Iqgap3 and Lgr5 mRNA in some chief cells was observed by XZ-plane imaging (online supplemental figure S6C). The apparent downward shift in Ki67 and Iqgap3 positioning at 48 hours is due to HDT-induced massive expansion of foveolar cell population, as indicated by strong Muc5ac immunostaining covering the upper half of the gland (figure 4F). Our results indicated the coexpression of Iqgap3 and Ki67 at the region spanning from the isthmus to the base. Iqgap3-2A-tdTomato mice at 48 hours post-HDT also showed a dramatic expansion of tdTomato +cells, with strong tdTomato staining at the lower half of the gland where proliferative Ki67 +cells are enriched (figure 4G; online supplemental figure S6D). Moreover, a subpopulation of Gif +cells at the gland base was labelled by tdTomato, indicating expression of Iqgap3 in chief cells (figure 4H). qPCR showed that the induction of Mki67 mRNA reflected that of Iqgap3 during tissue injury, whereas Lgr5 showed moderate increase in expression (figure 4I). There was a population of Ki67 +cells near the gland bottom, in the area outside of the epithelium (figure 4G). These CD45 +Ki67+E-cad-cells (online supplemental figure S6E, F) are CD45 +leucocytes, most likely macrophages as described previously.33 To investigate whether cells with robust expression of Iqgap3 are involved in the repair of HDT-damaged tissue, Iqgap3-2A-CeERT2;Rosa-tdTomato mice were treated with HDT. At 14 days post HDT, the tissue morphology, confined Ki67 staining at the isthmus, and regeneration of H, K-ATPase-producing parietal cells indicated near complete repair (figure 4J, K; online supplemental figure S6G). The regenerated gastric glands showed patches of tdTomato-labelling from the bottom to the top, suggesting that Iqgap3 +cells, including Lgr5+chief cells, gave rise to the regenerated parietal cells as well as multiple lineages during repair (online supplemental figure S6G).
Figure 4.
Iqgap3-expressing cells drive corpus epithelial regeneration post-tissue damage. (A) H&E staining on untreated and high dose tamoxifen (HDT) treated WT corpus at 48 hours post-tamoxifen administration. (B) IF staining for Ki67 and E-cad on untreated and 48 hours post-HDT treated WT corpus. (C–E) ISH for Iqgap3(green) and Lgr5(red) on untreated, 24 hours and 48 hours post-HDT treated WT corpus. Boxes indicate enlarged regions. (F) IF staining for Muc5ac and Ki67 on 48 hours post-HDT treated WT corpus. (G, H) IF staining for tdTomato, Ki67 and GIF on 48 hours post-HDT treated corpus from Iqgap3-2A-tdTomato mice. (I) qPCR for Mki67, Iqgap3 and Lgr5 from isolated WT corpus tissue of untreated and 48 hours post-HDT treated mice (n=3). Error bars represent SE of mean. Data were analysed by Student’s t-test. (J) H&E staining on HDT treated corpus at 14 days post-tamoxifen administration. (K) IF staining of Ki67, tdTomato and H, K-ATPase on 14 days post-HDT treated corpus of Iqgap3-2A-CreERT2;Rosa-tdTomato mice. Scale bars=50 µm. IF, immunofluorescence; ISH, in situ hybridisation; LT, lineage tracing; qPCR, quantitative PCR; WT, wild-type.
HDT treatment promotes stem cell activity and neoplastic features
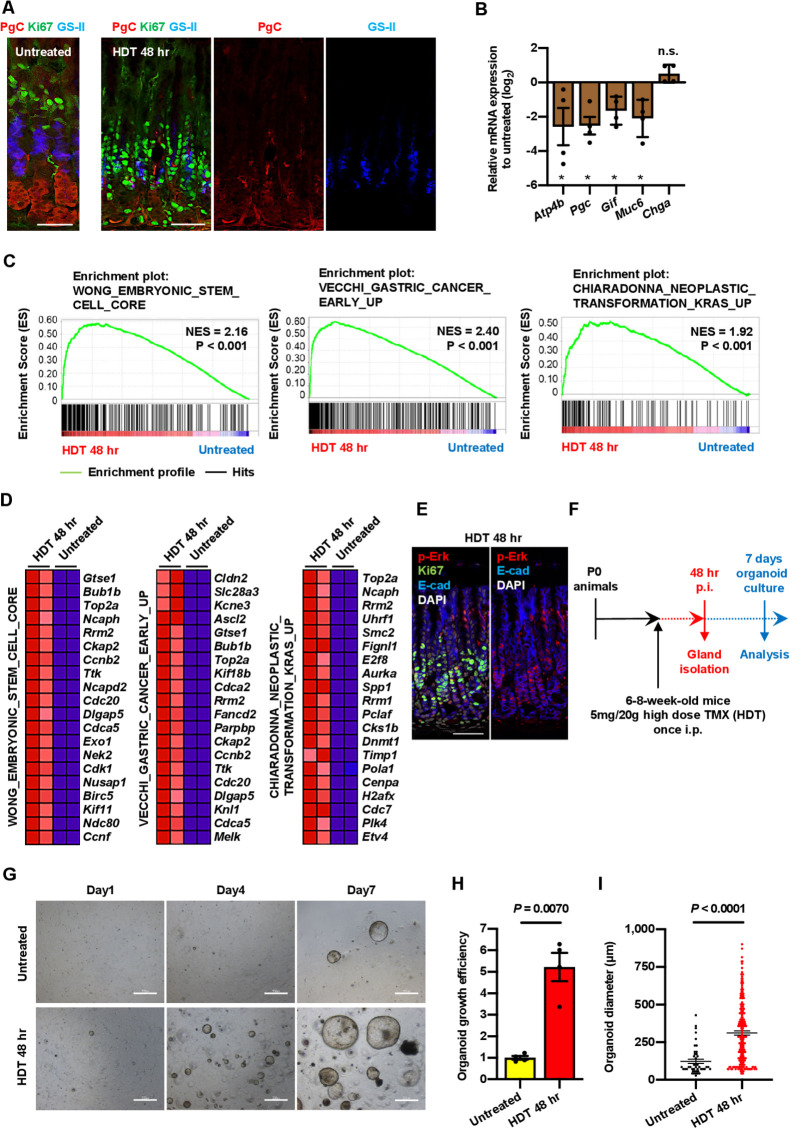
Immunostaining of the injured tissue showed reduction of neck and chief cell markers, such as GS-II and PgC (figure 5A); mRNA of differentiated cell markers Atp4b, PgC, Gif, and Muc6 were reduced as well (figure 5B). HDT-associated depletion of chief cells has been reported previously.11 34 Taken together with the observations by Radyk et al,34 the decreased expression of chief cell markers in our work suggests that the injury-induced Iqgap3+Ki67+cells at the base were stem-like cells possibly derived from reprogramming of chief cells. Conversely, Hata et al had reported that chief cells do not dedifferentiate after HDT and that the compensatory response from neck progenitors contributed to the replacement of chief cells.11 Our work indicated that two complementary events, namely the expansion of Iqgap3-expressing cells from the isthmus zone and the Iqgap3-associated dedifferentiation of chief cells to stem-like cells, accelerated tissue regeneration.
Figure 5.
HDT-induced tissue damage promotes stem cell activity and neoplastic characteristics. (A) IF staining for PGC, Ki67 and GS-II on untreated and 48 hours post-HDT treated WT corpus. (B) qPCR for Atp4b, Pgc, Gif, Muc6 and Chga from isolated corpus tissue of untreated or 48 hours post-HDT treated WT mice (n=4, expressed as Log2 scale). Data were analysed by Student’s t-test. ∗P<0.05. NS, not significant. (C) GSEA showing enrichment of embryonic stem cell gene signature, early gastric cancer gene signature and neoplastic transformation KRAS gene signature in 48 hours post-HDT treated WT corpus (n=2). P values determined by a weighted Kolmogorov-Smirnov-like statistic and adjusted for multiple hypothesis testing. (D) Heat MAP showing top 20 genes upregulated in (C) based on RNA-sequencing data from untreated and 48 hours post-HDT corpus tissue. (E) IF staining for p-Erk, Ki67 and E-cad on 48 hours post-HDT treated WT corpus. (F) Experimental strategy to generate corpus organoids from HDT treated mice. (G) Microscopic image of corpus organoids derived from untreated and 48 hours post-HDT-treated WT mice. (H, I) Organoid growth efficiency and diameter of corpus organoids derived from untreated or 48 hours post-HDT treated WT mice at 7 days of organoid culture (n=3). Data were analysed by Student’s t-test. Scale bars=50 µm (A), 100 µm (E), 500 µm (G). Error bars represent SEM. GSEA, gene set enrichment analysis; HDT, high dose of tamoxifen; IF, immunofluorescence; p-Erk, phosphorylated extracellular signal-regulated kinase; qPCR, quantitative PCR; SEM, SE of mean; WT, wild-type.
RNA-sequencing and GSEA of epithelial and non-epithelial cells from tissues isolated from HDT-treated mice revealed that mitotic genes, E2F- and Myc-target genes were upregulated, reflecting increased proliferation state of the HDT-treated cells (online supplemental figure S7A, B). Myc expression itself was strongly upregulated (2.5-fold increase) after tissue damage (online supplemental figure S7B). Myc induction was reported to activate an embryonic stem cell (ESC)-like transcriptional signature associated with diverse epithelial cancers.35 We observed that the ESC-like transcriptional programme,35 together with transcriptional programmes associated with Kras transformation,36 early gastric cancer37 and inflammation were significantly upregulated after HDT-induced injury (figure 5C, D; online supplemental figure S7C, E). An earlier study by Leushacke et al reported upregulation of matrix metalloproteinase-7 and downregulation of sclerostin domain containing 1 after injury, which they suggested might amplify Wnt signalling to drive tissue regeneration.8 While we detected upregulation of various Wnt-related genes (eg, Axin2, Notch1 and Myc), there was no statistically significant trend of Wnt activation (online supplemental figure S7D, E). Instead, our data suggest that a combination of dysregulated Myc and Kras signalling—driven by injury-induced Iqgap3 expression—contributed to a less differentiated state, and a more proliferative, stem-like and neoplastic phenotype. Importantly, HDT-treated tissues showed elevated p-Erk staining at Ki67 enriched regions, thereby confirming that Ras activation is associated with proliferation during tissue repair (figure 5E). The increased efficiency of organoid formation following injury further confirmed the increased stemness and proliferative potential of the regenerating cells (figure 5F–I).
Elevated Iqgap3 expression is associated with proliferation in gastric cancer
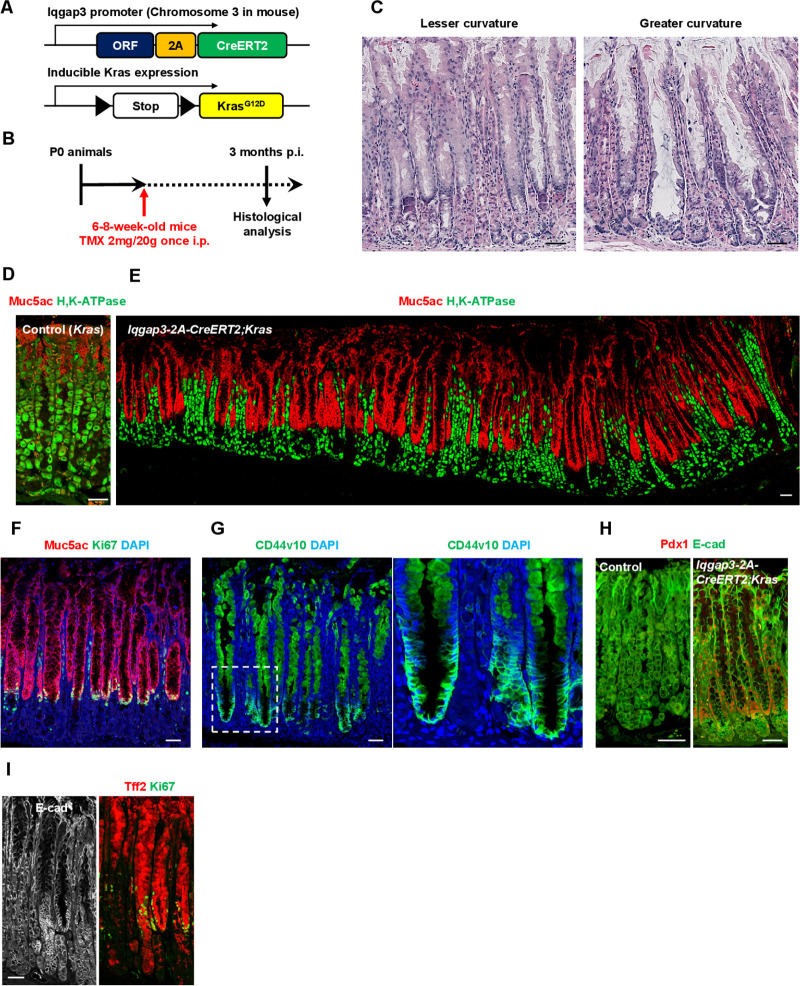
The Iqgap3-2A-CreERT2;KrasG12D/+ mouse model was used to investigate the effects of oncogenic Ras activation in Iqgap3 +cells (figure 6A, B). Three months after tamoxifen injection to induce KrasG12D expression, we observed the development of pseudopyloric metaplasia, which was characterised by a massive induction of Muc5ac+surface mucous cells and a reduction of parietal cells (figure 6C–E). The Ki67+proliferative cell zone was restricted to the lower neck zone (figure 6F). We observed cancer stem cell marker CD44v10 expression at the base of the metaplastic gland (figure 6G). Pdx1, frequently expressed in pseudopyloric glands and intestinal metaplasia,38 was also detected in corpus units with oncogenic Ras signalling (figure 6H). We also note the induction of Tff2 throughout the aberrant gland (compare figure 6I with the U-shaped Muc5ac staining pattern in figure 6E). The induced characteristics of the Iqgap3-2A-CreERT2;KrasG12D/+ mouse model resembled to an extent, Menetrier’s disease. Menetrier’s disease has been attributed to TGFα and receptor tyrosine kinase RTK EGFR.39 It could be that the specific expression of hyperactivated Ras in isthmus stem cells elicited features that mimicked Menetrier’s disease, which has been tenuously linked to gastric cancer.40 Together, our findings suggest that induction of oncogenic signalling in Iqgap3 +cells can give rise to hyperproliferative disorders and perhaps, preneoplastic lesions.
Figure 6.
Iqgap3-2A-CreERT2;KrasG12D/+ mice present pseudopyloric metaplasia. (A) Schematic representation of the genetic construct used to establish the Iqgap3-2A-CreERT2;KrasG12D/+ mouse model. (B) Experimental strategy for inducing Iqgap3-driven active KrasG12D expression. (C) H&E staining of the lesser and greater curvature on the corpus of Iqgap3-2A-CreERT2;KrasG12D/+ mouse. (D, E) IF staining for Muc5ac and H, K-ATPase on the corpus of control and Iqgap3-2A-CreERT2;KrasG12D/+ mouse. (F–I) IF staining for Muc5ac/Ki67 (F), CD44v10 (G), Pdx1/E-cad (H) and Tff2/Ki67/E-cad (I) on the corpus of Iqgap3-2A-CreERT2;KrasG12D/+ mice (n=3). Box indicates enlarged region. Scale bars=50 µm. IF, immunofluorescence; p.i, post-tamoxifen induction.
We, therefore, investigated the expression of IQGAP3 in human cancer. Similar to the mouse, we detected coexpression of IQGAP3 with Ki67 at the isthmus of the normal human stomach corpus (figure 7A). Microarray and RNA-sequencing showed significantly higher IQGAP3 expression levels in gastric cancer tissues obtained from Singapore patients (figure 7B). Tissue microarray analysis of gastric tumour tissues showed that although the tumour and adjacent normal tissues showed IQGAP3 expression, IQGAP3 levels were specifically elevated in neoplastic regions, relative to the adjacent normal tissue (figure 7C; online supplemental figure S8A, B). IQGAP3 expression was elevated in intestinal and mixed gastric cancers (figure 7D). We examined the effects of depleting IQGAP3 in the undifferentiated human gastric carcinoma cell line HGC-27. siRNA-mediated knockdown of IQGAP3 in HGC-27 resulted in drastic reduction of stem cell factors such as NANOG, OCT4, SOX2 and an increase of chief cell marker PGC (see mRNA and protein levels in online supplemental figure S8C, D). Expectedly, KLF4 was significantly increased following IQGAP3 knockdown. Despite its designation as a Yamanaka factor, KLF4 is associated with differentiation in the stomach—the work by Miao et al indicated that KLF4 repressed isthmus stem cell proliferation and induced differentiation to pit and parietal cells.19 Interestingly, cancer stem cell marker CD44v9 was also reduced after IQGAP3 depletion (online supplemental figure S8C, D). Our data, therefore, suggested that IQGAP3 expression contributed to a stem-like state and that its depletion promoted a more differentiated state in HGC-27.
Figure 7.
IQGAP3 is coexpressed with Ki67 in human gastric cancer. (A) IF staining for IQGAP3, Ki67 and E-cad on normal corpus in the human stomach (n=3). (B) Microarray and RNA-sequencing analysis for IQGAP3 expression in gastric tumour and normal stomach tissue (Singapore cohort). Error bars for microarray (tumour=185, normal=89) and RNA-sequencing (tumour=27, normal=18) represent SEM from each population. Data were analysed by two-tailed Wilcoxon RANK sum test. (C) IQGAP3 tissue microarray (TMA) of gastric cancer and paired-adjacent normal stomach tissue (n=237). (D) Box plot for comparing IQGAP3 TMA score in adjacent normal stomach, intestinal type gastric cancer, diffuse type gastric cancer and mixed type gastric cancer. Error bars represent SEM from each population. Data sets were analysed by one-way ANOVA. (E) H&E staining on human gastric tumour (n=3). (F) IF staining for IQGAP3, Ki67 and E-cad on the human gastric tumour (n=5). (G) IF staining for IQGAP3, Ki67 and CD44v9 on the human gastric tumour (n=7). Scale bar=100 µm. ANOVA, analysis of variance; IF, immunofluorescence; SEM, SE of mean.
The Cancer Genome Atlas database showed that IQGAP3 expression positively correlated with proliferation markers MKI67 and PCNA in gastric cancer (online supplemental figure S8E). We examined whether IQGAP3 also regulates proliferation of cancer stem cells. We found that the relationship between IQGAP3, Ki67 and CD44v9 expression varied across human cancer tissues. Whereas IQGAP3 invariably coexpressed with Ki67 in all examined human gastric cancer tissues, there was partial overlap of IQGAP3 and CD44v9 expression (figure 7E–G; online supplemental figure S9A–C). Importantly, IQGAP3 was expressed in all CD44v9+Ki67+double positive cells in the tumour samples examined. Our observations suggested that IQGAP3 was expressed in activated, but not quiescent, cancer stem cells.
Discussion
Here, we established Iqgap3 as a novel stem cell factor that is required not only for rapid proliferation of isthmus stem cells, but also for the maintenance of stem cell properties. Although Iqgap3 expression pattern reflected closely that of Ki67, the fact that IQGAP3 depletion in NTERA-2 cells resulted in reduced transcription of several pluripotency-related genes (ie, KLF4, NANOG and OCT4) and the onset of differentiation indicate a complex role for Iqgap3 in regulating stem cell behaviour. Likewise, we found that IQGAP3 expression contributes to a stem-like state in the gastric cancer cell line HGC-27. It is tempting to speculate that IQGAP3 may be therapeutically targeted to induce differentiation, or an irreversible exit from cell cycle, and thereby improve gastric cancer treatment.
Iqgap3 belongs to a three-membered cytoskeletal, scaffold protein family known to regulate diverse cellular processes such as signal transduction, cell-cell adhesion, cell motility and cytokinesis. The multiple protein binding domains in the Iqgap protein afford the tethering of signalling pathway components, which increases efficiency of complex formation and signalling intensity.25 41 Many key components of cell signalling, such E-cadherin, β-catenin, calmodulin and ERK, have been identified as Iqgap binding partners.25 Of note, Iqgap3 specifically interacts with the active, GTP-bound form of Ras, and through Ras-dependent Erk activation, constitutes an essential aspect of cell proliferation.23 Moreover, IQGAP3, alone in the IQGAP family, is indispensable for cell proliferation and motility during zebrafish embryonic development.42 IQGAP3 functions downstream of FGFR1-Ras signalling to regulate proliferation during zebrafish embryogenesis.42
Here, we show that the promotion of the Ras-Erk signalling pathway by Iqgap3 is a key underlying mechanistic basis for stem cell proliferation in the corpus. Khurana et al had previously showed that ERK signalling regulates isthmus stem cell proliferation through CD44 signalling.43 Our work, therefore, suggests a cascade involving IQGAP3, ERK and CD44 as main players in the regulation of proliferation at the isthmus zone. Aside from proliferation, the FGF-ERK cascade has also been shown to promote self-renewal in human ESC.44 Erk1/2 signalling can also induce chromatin remodelling in mouse ESCs, underscoring the importance of Erk1/2 signalling in stem cells.45 It is, therefore, tempting to hypothesise that Iqgap3-Erk signalling influences chromatin architecture to direct stem cell-specific transcription programme.
The roles of isthmus stem cells and chief cells during tissue repair have long been mired in controversy. Two disparate models, both supported by extensive evidence, have been proposed. One model holds that chief cells acquire plasticity and contribute to repair. The other model contends that quiescent Mist1-expressing isthmus stem cells are solely responsible for repair of the entire pit. Our work not only shows that rapidly proliferating isthmus stem cells contribute to the repair process, but also clearly supports the self-replicating and plasticity properties of chief cells. However, we cannot rule out the contribution of Mist1-expressing isthmus quiescent stem cells. We posit that expression of Iqgap3, be it in isthmus or chief cells, bestows stem-like behaviour to drive tissue repair. It is interesting that pERK was also detected in the Iqgap3-expressing cells after HDT. In view of the findings by Khurana et al,43 it is tempting to propose that an Iqgap3-ERK-CD44 axis drives stem cell proliferation during homoeostasis and tissue repair.
Interestingly, Iqgap3 induction during tissue repair was associated with the upregulation of c-Myc and genes associated with ESC. The abilities of c-Myc to increase cancer stem cell population and induce dedifferentiation suggest its involvement in tumour initiation. Moreover, our transcriptomic analysis reveals the enrichment of gene expression signatures associated with early gastric cancer, Kras-linked neoplastic transformation in HDT treated cells. Our results suggest that Iqgap3 is not simply a proliferation factor; rather, the injury-related induction of Iqgap3 and associated gene signatures represent the initial step of cancer development. Accordingly, we found that IQGAP3 is expressed in rapidly proliferating regions of gastric cancer. Since multiple cell types respond to HDT by expressing Iqgap3, we were not able to specify the cell of origin of cancer following injury. Nevertheless, by using the mouse strain harbouring Iqgap3-2A-CreERT2 to express KrasG12D in Iqgap3-expressing cells, we observed rapid generation of pseudopyloric metaplasia. Aberrant activation of Ras in Iqgap3-expressing cells are thus likely to be responsible for induction of early stage of cancer development.
The expression of Iqgap3 in isthmus or chief cells may be necessary to trigger Ras-driven cell proliferation and subsequent metaplastic transformation. Moreover, IQGAP3 ablation has been shown to reduce proliferation of breast and gastric cancer cell lines.46 47 We propose that an IQGAP3-Ras-associated mechanism might be conserved for proliferation of cancer cells.
Acknowledgments
The authors thank Dr. David Hershel Alpers for generously providing the GIF antibody.
Footnotes
Contributors: YI, JM and ST conceived the study. YI supervised the study. YI, JM and DD designed the experiments. JM, DD, KM, AY, NNM, DLH, SC, NAM, NN and KK performed experiments. YL, SWTH, NL, HKL, JMC and HY performed bioinformatics analysis. PT, SS, TM, JBYS, WPY, K-GY provided and analysed human clinical samples. ST, AT and MA provided reagents and conceptual advice. All authors commented on the results and discussed implications. YI, LSHC and JM analysed the data and wrote the manuscript.
Funding: This research is supported by the National Research Foundation Singapore and the Singapore Ministry of Education under its Research Centres of Excellence initiative, by the National Research Foundation under its Translational and Clinical Research Flagship Programme (Grant Number NMRC/TCR/009-NUHS/2013), Singapore Ministry of Health’s National Medical Research Council under its Clinician-Scientist Individual Research Grant (NMRC/CIRG/1452/2016) as well as National Medical Research Council’s Open Fund Large Collaborative Grant (OFLCG18 May-0023).
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
All data relevant to the study are included in the article or uploaded as online supplemental information. RNA-sequencing datasets generated in this study are deposited in Gene Expression Omnibus (GEO) database under accession codes GSE161443 for Iqgap3-2A-tdTomato reporter mice and GSE161442 for HDT treatment respectively. All supporting data are available from the corresponding authors upon reasonable request.
Ethics statements
Patient consent for publication
Not required.
Ethics approval
Institution of Animal Care and Use Committee, and the work was approved by the Institution of Animal Care and Use Committee and the Office of Safety, Health, and Environment at the National University of Singapore.
References
- 1.Karam SM, Leblond CP. Dynamics of epithelial cells in the corpus of the mouse stomach. I. Identification of proliferative cell types and Pinpointing of the stem cell. Anat Rec 1993;236:259–79. 10.1002/ar.1092360202 [DOI] [PubMed] [Google Scholar]
- 2.Lennerz JKM, Kim S-H, Oates EL, et al. The transcription factor MIST1 is a novel human gastric chief cell marker whose expression is lost in metaplasia, dysplasia, and carcinoma. Am J Pathol 2010;177:1514–33. 10.2353/ajpath.2010.100328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nam KT, Lee H-J, Sousa JF, et al. Mature chief cells are cryptic progenitors for metaplasia in the stomach. Gastroenterology 2010;139:2028–37. 10.1053/j.gastro.2010.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quante M, Marrache F, Goldenring JR, et al. TFF2 mRNA transcript expression marks a gland progenitor cell of the gastric oxyntic mucosa. Gastroenterology 2010;139:2018–27. 10.1053/j.gastro.2010.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stange DE, Koo B-K, Huch M, et al. Differentiated Troy+ chief cells act as reserve stem cells to generate all lineages of the stomach epithelium. Cell 2013;155:357–68. 10.1016/j.cell.2013.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hayakawa Y, Ariyama H, Stancikova J, et al. Mist1 expressing gastric stem cells maintain the normal and neoplastic gastric epithelium and are supported by a perivascular stem cell niche. Cancer Cell 2015;28:800–14. 10.1016/j.ccell.2015.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi E, Hendley AM, Bailey JM, et al. Expression of activated Ras in gastric chief cells of mice leads to the full spectrum of metaplastic lineage transitions. Gastroenterology 2016;150:918–30. 10.1053/j.gastro.2015.11.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leushacke M, Tan SH, Wong A, et al. Lgr5-expressing chief cells drive epithelial regeneration and cancer in the oxyntic stomach. Nat Cell Biol 2017;19:774–86. 10.1038/ncb3541 [DOI] [PubMed] [Google Scholar]
- 9.Matsuo J, Kimura S, Yamamura A, et al. Identification of stem cells in the epithelium of the stomach corpus and antrum of mice. Gastroenterology 2017;152:218–31. 10.1053/j.gastro.2016.09.018 [DOI] [PubMed] [Google Scholar]
- 10.Kinoshita H, Hayakawa Y, Niu Z, et al. Mature gastric chief cells are not required for the development of metaplasia. Am J Physiol Gastrointest Liver Physiol 2018;314:G583–96. 10.1152/ajpgi.00351.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hata M, Kinoshita H, Hayakawa Y, et al. GPR30-Expressing gastric chief cells do not Dedifferentiate but are eliminated via PDK-Dependent cell competition during development of metaplasia. Gastroenterology 2020;158:1650–66. 10.1053/j.gastro.2020.01.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi E, Lantz TL, Vlacich G, et al. Lrig1+ gastric isthmal progenitor cells restore normal gastric lineage cells during damage recovery in adult mouse stomach. Gut 2018;67:1595–605. 10.1136/gutjnl-2017-313874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wroblewski LE, Choi E, Petersen C, et al. Targeted mobilization of Lrig1+ gastric epithelial stem cell populations by a carcinogenic Helicobacter pylori type IV secretion system. Proc Natl Acad Sci U S A 2019;116:19652–8. 10.1073/pnas.1903798116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han S, Fink J, Jörg DJ, et al. Defining the identity and dynamics of adult gastric isthmus stem cells. Cell Stem Cell 2019;25:342–56. 10.1016/j.stem.2019.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burclaff J, Willet SG, Sáenz JB, et al. Proliferation and differentiation of gastric mucous neck and chief cells during homeostasis and injury-induced metaplasia. Gastroenterology 2020;158:598–609. 10.1053/j.gastro.2019.09.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arnold K, Sarkar A, Yram MA, et al. Sox2(+) adult stem and progenitor cells are important for tissue regeneration and survival of mice. Cell Stem Cell 2011;9:317–29. 10.1016/j.stem.2011.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schweiger PJ, Clement DL, Page ME, et al. Lrig1 marks a population of gastric epithelial cells capable of long-term tissue maintenance and growth in vitro. Sci Rep 2018;8:15255. 10.1038/s41598-018-33578-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoshioka T, Fukuda A, Araki O, et al. Bmi1 marks gastric stem cells located in the isthmus in mice. J Pathol 2019;248:179–90. 10.1002/path.5244 [DOI] [PubMed] [Google Scholar]
- 19.Miao Z-F, Adkins-Threats M, Burclaff JR, et al. A Metformin-Responsive metabolic pathway controls distinct steps in gastric progenitor fate decisions and maturation. Cell Stem Cell 2020;26:910–25. 10.1016/j.stem.2020.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Konstantinou D, Bertaux-Skeirik N, Zavros Y. Hedgehog signaling in the stomach. Curr Opin Pharmacol 2016;31:76–82. 10.1016/j.coph.2016.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Demitrack ES, Samuelson LC. Notch as a driver of gastric epithelial cell proliferation. Cell Mol Gastroenterol Hepatol 2017;3:323–30. 10.1016/j.jcmgh.2017.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Demitrack ES, Gifford GB, Keeley TM, et al. Notch1 and Notch2 regulate epithelial cell proliferation in mouse and human gastric corpus. Am J Physiol Gastrointest Liver Physiol 2017;312:G133–44. 10.1152/ajpgi.00325.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nojima H, Adachi M, Matsui T, et al. IQGAP3 regulates cell proliferation through the Ras/ERK signalling cascade. Nat Cell Biol 2008;10:971–8. 10.1038/ncb1757 [DOI] [PubMed] [Google Scholar]
- 24.Ng CEL, Yokomizo T, Yamashita N, et al. A Runx1 intronic enhancer marks hemogenic endothelial cells and hematopoietic stem cells. Stem Cells 2010;28:1869–81. 10.1002/stem.507 [DOI] [PubMed] [Google Scholar]
- 25.Hedman AC, Smith JM, Sacks DB. The biology of IQGAP proteins: beyond the cytoskeleton. EMBO Rep 2015;16:427–46. 10.15252/embr.201439834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barker N, Huch M, Kujala P, et al. Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell 2010;6:25–36. 10.1016/j.stem.2009.11.013 [DOI] [PubMed] [Google Scholar]
- 27.Moasser MM. The oncogene HER2: its signaling and transforming functions and its role in human cancer pathogenesis. Oncogene 2007;26:6469–87. 10.1038/sj.onc.1210477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karunagaran D, Tzahar E, Beerli RR, et al. ErbB-2 is a common auxiliary subunit of NDF and EGF receptors: implications for breast cancer. Embo J 1996;15:254–64. 10.1002/j.1460-2075.1996.tb00356.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goldenring JR, Nam KT. Oxyntic atrophy, metaplasia, and gastric cancer. Prog Mol Biol Transl Sci 2010;96:117–31. 10.1016/B978-0-12-381280-3.00005-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manning EH, Lapierre LA, Mills JC, et al. Tamoxifen acts as a parietal cell protonophore. Cell Mol Gastroenterol Hepatol 2020;10:655–7. 10.1016/j.jcmgh.2020.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huh WJ, Khurana SS, Geahlen JH, et al. Tamoxifen induces rapid, reversible atrophy, and metaplasia in mouse stomach. Gastroenterology 2012;142:21–4. 10.1053/j.gastro.2011.09.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keeley TM, Horita N, Samuelson LC. Tamoxifen-Induced gastric injury: effects of dose and method of administration. Cell Mol Gastroenterol Hepatol 2019;8:365–7. 10.1016/j.jcmgh.2019.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petersen CP, Weis VG, Nam KT, et al. Macrophages promote progression of spasmolytic polypeptide-expressing metaplasia after acute loss of parietal cells. Gastroenterology 2014;146:1727–38. 10.1053/j.gastro.2014.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Radyk MD, Burclaff J, Willet SG, et al. Metaplastic cells in the stomach arise, independently of stem cells, via dedifferentiation or transdifferentiation of chief cells. Gastroenterology 2018;154:839–43. 10.1053/j.gastro.2017.11.278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wong DJ, Liu H, Ridky TW, et al. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell 2008;2:333–44. 10.1016/j.stem.2008.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chiaradonna F, Sacco E, Manzoni R, et al. Ras-Dependent carbon metabolism and transformation in mouse fibroblasts. Oncogene 2006;25:5391–404. 10.1038/sj.onc.1209528 [DOI] [PubMed] [Google Scholar]
- 37.Vecchi M, Nuciforo P, Romagnoli S, et al. Gene expression analysis of early and advanced gastric cancers. Oncogene 2007;26:4284–94. 10.1038/sj.onc.1210208 [DOI] [PubMed] [Google Scholar]
- 38.Sakai H, Eishi Y, Li X-L, et al. PDX1 homeobox protein expression in pseudopyloric glands and gastric carcinomas. Gut 2004;53:323–30. 10.1136/gut.2003.026609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nomura S, Settle SH, Leys CM, et al. Evidence for repatterning of the gastric fundic epithelium associated with Ménétrier's disease and TGFalpha overexpression. Gastroenterology 2005;128:1292–305. 10.1053/j.gastro.2005.03.019 [DOI] [PubMed] [Google Scholar]
- 40.Coffey RJ, Washington MK, Corless CL, et al. Ménétrier disease and gastrointestinal stromal tumors: hyperproliferative disorders of the stomach. J Clin Invest 2007;117:70–80. 10.1172/JCI30491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanchez-Laorden B, Viros A, Marais R. Mind the IQGAP. Cancer Cell 2013;23:715–7. 10.1016/j.ccr.2013.05.017 [DOI] [PubMed] [Google Scholar]
- 42.Fang X, Zhang B, Thisse B, et al. IQGAP3 is essential for cell proliferation and motility during zebrafish embryonic development. Cytoskeleton 2015;72:422–33. 10.1002/cm.21237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khurana SS, Riehl TE, Moore BD, et al. The hyaluronic acid receptor CD44 coordinates normal and metaplastic gastric epithelial progenitor cell proliferation. J Biol Chem 2013;288:16085–97. 10.1074/jbc.M112.445551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Greber B, Coulon P, Zhang M, et al. FGF signalling inhibits neural induction in human embryonic stem cells. Embo J 2011;30:4874–84. 10.1038/emboj.2011.407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tee W-W, Shen SS, Oksuz O, et al. Erk1/2 activity promotes chromatin features and RNAPII phosphorylation at developmental promoters in mouse ESCs. Cell 2014;156:678–90. 10.1016/j.cell.2014.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oue N, Yamamoto Y, Oshima T, et al. Overexpression of the transmembrane protein IQGAP3 is associated with poor survival of patients with gastric cancer. Pathobiology 2018;85:192–200. 10.1159/000481890 [DOI] [PubMed] [Google Scholar]
- 47.Hu G, Xu Y, Chen W, et al. Rna interference of IQ motif containing GTPase-activating protein 3 (IQGAP3) inhibits cell proliferation and invasion in breast carcinoma cells. Oncol Res 2016;24:455–61. 10.3727/096504016X14685034103635 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2020-322779supp001.pdf (3.5MB, pdf)
gutjnl-2020-322779supp002.pdf (199.3KB, pdf)
Data Availability Statement
All data relevant to the study are included in the article or uploaded as online supplemental information. RNA-sequencing datasets generated in this study are deposited in Gene Expression Omnibus (GEO) database under accession codes GSE161443 for Iqgap3-2A-tdTomato reporter mice and GSE161442 for HDT treatment respectively. All supporting data are available from the corresponding authors upon reasonable request.