Abstract
Objective
O-linked N-acetylglucosaminylation (O-GlcNAcylation), controlled by O-GlcNAcase (OGA) and O-GlcNAc transferase (OGT), is an important post-translational modification of eukaryotic proteins and plays an essential role in regulating gut inflammation. Gut microbiota encode various enzymes involved in O-GlcNAcylation. However, the characteristics, abundance and function of these enzymes are unknown.
Design
We first investigated the structure and taxonomic distribution of bacterial OGAs and OGTs. Then, we performed metagenomic analysis to explore the OGA genes abundance in health samples and different diseases. Finally, we employed in vitro and in vivo experiments to determine the effects and mechanisms of bacterial OGAs to hydrolyse O-GlcNAcylated proteins in host cells and suppress inflammatory response in the gut.
Results
We found OGAs, instead of OGTs, are enriched in Bacteroidetes and Firmicutes, the major bacterial divisions in the human gut. Most bacterial OGAs are secreted enzymes with the same conserved catalytic domain as human OGAs. A pooled analysis on 1999 metagenomic samples encompassed six diseases revealed that bacterial OGA genes were conserved in healthy human gut with high abundance, and reduced exclusively in ulcerative colitis. In vitro studies showed that bacterial OGAs could hydrolyse O-GlcNAcylated proteins in host cells, including O-GlcNAcylated NF-κB-p65 subunit, which is important for activating NF-κB signalling. In vivo studies demonstrated that gut bacteria-derived OGAs could protect mice from chemically induced colonic inflammation through hydrolysing O-GlcNAcylated proteins.
Conclusion
Our results reveal a previously unrecognised enzymatic activity by which gut microbiota influence intestinal physiology and highlight bacterial OGAs as a promising therapeutic strategy in colonic inflammation.
Keywords: intestinal bacteria, inflammatory bowel disease, gut inflammation, intestinal enzymes, probiotics
Significance of this study.
What is already known on this subject?
Protein O-linked N-acetylglucosaminylation (O-GlcNAcylation), controlled by O-GlcNAcase (OGA) and O-GlcNAc transferase, plays an essential role in regulating gut inflammation.
In addition to human beings, several bacteria also encode OGAs, some of which have similar catalytic activities as human OGAs.
Little is known about the role of gut microbiota derived OGAs on human physiology and pathology.
What are the new findings?
Most of the gut bacterial OGAs are secreted proteins with similar conserved catalytic domain as human OGAs.
Bacterial OGA genes are conserved in healthy human gut with high abundance, and reduce exclusively in ulcerative colitis (UC) patients.
OGAs from Akkermansia muciniphila and Bacteroides thetaiotaomicron can hydrolyze O-GlcNAcylated proteins in both epithelial cells and immune cells, including O-GlcNAcylated NF-κB-p65 subunit, which is important for activating NF-κB signalling.
Gut bacteria-derived OGAs can protect mice from colonic inflammation in multiple models through reducing colonic protein O-GlcNAcylation.
How might it impact on clinical practice in the foreseeable future?
Deficiency of bacterial OGAs may contribute to the pathogenesis of UC, which needs to be further addressed in future work.
Bacteria-derived OGAs might evolve as new and promising therapeutic methods for treating UC.
Introduction
Post-translational modification of proteins enables cells to respond promptly to internal and external cues through direct and dynamic control of protein function. O-linked N-acetylglucosaminylation (O-GlcNAcylation) is an important post-translational modification of serine and threonine residues with O-GlcNAc on cytoplasmic, nuclear and mitochondrial proteins.1 Protein O-GlcNAcylation plays an essential role in regulating inflammation and metabolism.2–6 Given the wide range of physical functions associated with O-GlcNAcylation, it is not surprising that disruption of O-GlcNAc homoeostasis has been implicated in the pathogenesis of a plethora of human diseases, including gut inflammatory disease, diabetes and neurodegeneration.3 4 7
O-GlcNAcylation is controlled by a single pair of enzymes: O-GlcNAcase (OGA), which catalyses the hydrolysis of the O-GlcNAc modification and O-GlcNAc transferase (OGT), which catalyses the transfer of a GlcNAc moiety from the substrate uridine diphosphate-GlcNAc to the hydroxyl groups of target serine and threonine residues. In humans, this pair of enzymes is distributed in many organs including brain tissues, liver and intestine.1
In addition to human beings, human symbionts and pathogens also encode this pair of enzymes with similar or distinct structure.8 9 In recent years, evidences have shown that bacteria-derived OGTs could modify host protein O-GlcNAcylation. For example, OGTs from Clostridium perfringens, Legionella and Photorhabdus can mono-O-GlcNAcylate Rho GTPase or Ras proteins in host to induce apoptosis in target cells.8 10 11 Furthermore, in vitro experiments revealed Clostridium perfringens OGA can remove O-GlcNAc from transforming growth factor β activated kinase-1 binding protein-1 in mammalian cells.12 These studies indicated that in addition to being regulated by human enzymes, the host protein O-GlcNAcylation may also be manipulated by bacterial OGAs and OGTs. However, the distribution of this pair of enzymes in human gut microbiota and its function on host proteins O-GlcNAcylation and subsequent physiology remain unclear.
In this study, we found that bacterial OGA-encoding genes are enriched in healthy human gut samples and reduced significantly in ulcerative colitis (UC) patients. Furthermore, we demonstrated that supplement of specific bacterial OGAs could ameliorate colitis in multiple animal models.
Results
Taxonomic profiling and structure of bacterial OGAs and OGTs
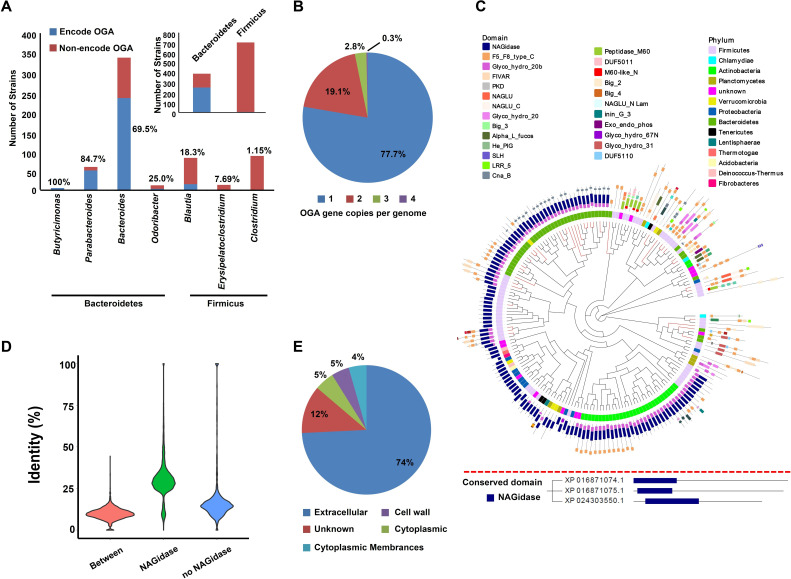
At first, we obtained all available (total 225) OGAs from UniRef90, which were found to be distributed in 50 genera from 13 phyla (online supplemental table 1). Then, we tried to identify the taxonomy distribution of these 225 OGAs in human gut microbiota. 1520 reference genomes of human gut bacteria were downloaded,13 and OGA sequences in these genomes were searched. Overall, 314 (20.6%) strains encode at least one copy of OGA gene (online supplemental table 2). In detail, more than half of the strains from phylum Bacteroidetes encode OGAs, while the proportion of OGA-encoding strains from Firmicus were relatively lower (figure 1A). Specially, the proportion of OGA-encoding strains from Butyricimonas and Parabacteroides are extremely high (>80%, figure 1A). Furthermore, we found most of the strains (77.7%) only encoding one copy of OGA gene (figure 1B).
Figure 1.
Taxonomic distribution, phylogenetic analysis and sequence characteristics of bacterial OGAs. (A) Proportion of OGA-encoding strains at phylum or genus level. (B) The proportion of different OGA gene copies in different strains. Detail is shown in online supplemental table 1. (C) Upper panel: a phylogenetic tree of OGAs with phylum (inner circle) and conserved domain (outside circle) annotation. Tips of the phylogenetic tree in red represent OGA sequences have homologues in human gut microbiota (>80% identity and >80% of coverage). Lower panel: the human OGAs annotated with conserved domain. (D) Density distribution of identity of OGA homologues according to conserved domain NAGidase. (E) The predicted sub-cellular location of OGAs. OGA, O-GlcNAcase.
gutjnl-2020-322468supp001.xlsx (1.9MB, xlsx)
The OGA sequences from UniRef90 were clustered at 70% and the representative sequences were used to build a phylogenetic tree (figure 1C). An OGA sequence from Akkermansia (A0A139TME1) interrupted OGAs sequences from Bacteroidetes, indicating it might be gained by Akkermansia through horizontal transferring (figure 1C). However, the phylogenetic tree of A0A139TME1 homologues (online supplemental figure 1A) and genomic analysis of Akkermansia showed no evidence for this gene to be gained through horizontal transferring (online supplemental figure 1B–I). Furthermore, DarkHorse analysis of other 82 genomes harbouring different OGA sequences showed that all the OGA genes had high lineage probability indexes14 (online supplemental table 3), indicating low possibility of them to be gained through horizontal transferring.
gutjnl-2020-322468supp002.pdf (272.1KB, pdf)
Conserved domain analysis showed that most of the bacterial OGAs have a catalytic β-N-acetyl-D-glucosarninidase (NAGidase) domain, which is also conserved in human OGAs (figure 1C). Sequence similarity analysis revealed OGAs differed much from each other (figure 1D). Protein location prediction indicated that the most of these enzymes are secreted proteins (figure 1E). These results indicated that bacterial OGAs may have a role in manipulating the host proteins O-GlcNAcylation.
Afterward, we obtained all available (total 121) OGTs from UniRef90, which were found to be distributed in 20 genera from 15 phyla. Most of them are from phylum Planctomycetes, Verrucomicrobia and Proteobacteria (online supplemental figure 2A), which are not the common symbionts in the human gut.15 Notably, some OGT encoding genus are even pathogens to human beings such as Burkholderia and Acetobacter (online supplemental table 4).16 17
gutjnl-2020-322468supp003.pdf (240.3KB, pdf)
Most of the bacterial OGTs were found to have one or more protein–protein interaction domain consisting of over 11 tetratricopeptide repeats (abbreviate to TPRs in online supplemental figure 2B), which were also found in human OGTs. However, the human OGTs also had a catalytic domain glycosyltransferase family 41 (Glyco_transf_41), which occurred in only two bacterial OGT sequences (online supplemental figure 2B). Sequence similarity analysis revealed that bacterial OGT homologues differed much with each other (online supplemental figure 2C). Protein location prediction indicated that most of the bacterial OGTs were intracellular proteins (online supplemental figure 2D). These results indicate that in contrast to bacterial OGAs, bacterial OGTs are less likely to modify host protein O-GlcNAcylation, considering their distinct catalytic domain from human OGT and intracellular location.
Abundance of bacterial OGA-encoding genes in healthy human gut
To investigate the abundance of bacterial OGA-encoding and OGT-encoding genes in human gut ecology, the above identified enzymes were searched in the integrated gene catalogue (IGC) of the human gut microbiome.18 The results showed that 110 OGA homologues, but no OGTs were identified from IGC (online supplemental table 5). Thus, the following analyses were focused on OGAs from gut microbiota.
The above identified OGA-encoding genes were quantified in 1130 metagenomic samples derived from health controls from 11 publicly available datasets that originated from six countries (online supplemental table 6).18–28 OGA-encoding genes were detected in most samples (more than 95%) at mean relative abundance of 5.61×10−5, which were higher than 80% of other gene families annotated by eggNOG database. The abundance of OGA-encoding genes was shown in online supplemental figure 3A and online supplemental table 7. OGA-encoding genes from Firmicutes or Verrucomicrobia had relatively low abundances in most samples, compared with those from Bacteroidetes (online supplemental figure 3B). The abundance of genes encoding OGAs with NAGidase domain exceeded those without this domain (online supplemental figure 3C). Considering the influence of several factors on microbiota among individuals, we compared differences in the abundance of OGA-encoding genes based on country, gender, age and body mass index (BMI) using single factor analysis. No significant differences were found between males and females (online supplemental figure 3D, P) =0.267). The OGA-encoding genes differed in people from different country: China has the highest mean relative abundance while those from Austria have the lowest (7.01×10−5 vs 1.26×10−5, p<0.01) (online supplemental figure 3E). Correlation analysis revealed a negative correlation between the abundance of OGA-encoding genes and BMI or age (online supplemental figure 3F) of individuals. However, further linear regression of multivariable analysis showed only countries contribute to the differences in the abundance of OGA-encoding genes (data not shown).
gutjnl-2020-322468supp004.pdf (195.2KB, pdf)
The abundance of OGA-encoding genes decreased in UC patients
To further investigate the functional implications of OGAs in gut microbiota, we analysed the abundance of OGA-encoding genes in different disease status, including rheumatoid arthritis, type 2 diabetes mellitus, obesity, liver cirrhosis, Crohn’s disease (CD) and UC (online supplemental table 6).
No significant alterations of OGA-encoding genes in extra-intestinal disease were found (online supplemental figure 4A–F). Although the abundance of OGA-encoding genes elevated in Spain-CD patients, the results needed to be further validated since the CD sample size is too small compared with controls (CD: 17 vs Control: 73). In addition, this tendency was not found in USA-CD cohort (figure 2A, B). Notably, UC patients from two independent studies exhibited decreased abundance of OGA-encoding genes (figure 2A, B). To confirm these changes in UC patients, we employed Shortbred to target analysed OGA-encoding genes in these two datasets, and the same results were found (figure 2C, D). Logistic analysis indicated a significantly negative association between the abundance of OGA-encoding genes and UC, instead of extra-intestinal diseases (figure 2E).
Figure 2.
Association of the abundance of OGA-encoding genes with inflammatory bowel diseases (IBD). Alteration of the abundance of OGAs-encoding genes in Spain-IBD cohort (A, C) or US-IBD cohort (B, D). (E) Forest plot reporting effect size (OR) of the abundance of OGA-encoding genes on different diseases. The gene relative abundances were quantified by method 1 (Bowtie2) and RPKM was quantified by method 2 (Shortbred), as shown in online supplemental materials. P values were computed by Manne-Whitney U tests and corrected by false discovery rate, *P<0.05, **P<0.01. OR was calculated using logistic regression analysis of different datasets. IBD, inflammatory bowel diseases. OGA, O-GlcNAcase; RPKM, reads per kilobase per million mapped reads.
gutjnl-2020-322468supp005.pdf (252.4KB, pdf)
gutjnl-2020-322468supp014.pdf (126.3KB, pdf)
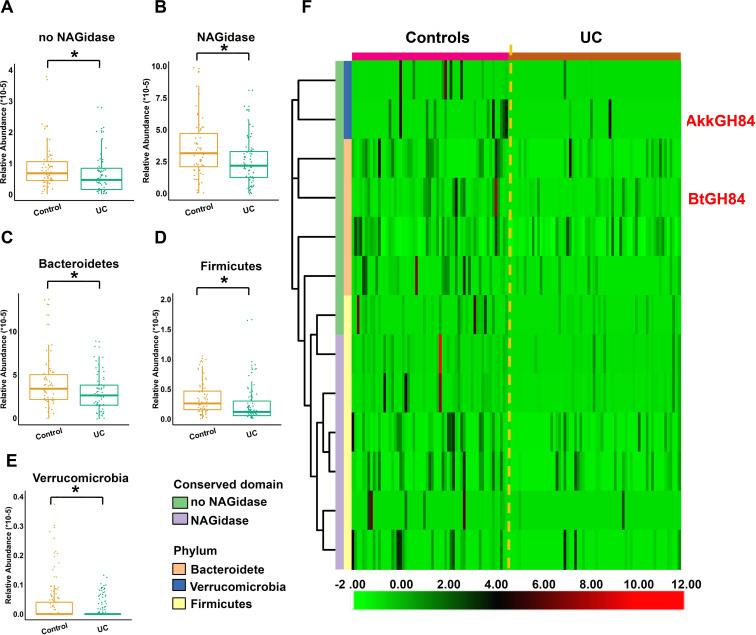
To gain more detailed information, we analysed the gene abundance in Spain-IBD cohort according to conserved domains and phylum of OGA sequences, and found that the gene abundances of both the OGAs with or without NAGidase domain were decreased in UC (figure 3A, B). The abundances of OGA-encoding genes from all three phyla reduced significantly in UC (figure 3C–E). Further analysis of each significantly different genes revealed they cover all the three phyla and both the protein with or without NAGidase domains (figure 3F, online supplemental table 8).
Figure 3.
Detail of the abundance of OGA-encoding genes in UC. (A, B) Gene abundance of OGAs with or without NAGidase domain in controls and UC. (C–E) Abundance of OGA-encoding genes from different phylum in controls and UC. (F) Heatmap of OGA-encoding genes with significantly different abundance between controls and UC. P values were computed by Manne-Whitney U test and corrected by false discovery rate, *P<0.05. OGA, O-GlcNAcase; UC, ulcerative colitis.
Further, we try to figure out if the reduced abundance of these OGA-encoding genes in UC patients were caused by the absence of certain species or strains. We found most of the species that positively correlated with the abundance of OGA-encoding genes were species from Bacteroides (online supplemental figure 5), indicating these species contributed most to the OGAs in the human gut. However, only a few OGA-encoding species reduced significantly in UC patients, which could only explain part of the gene abundance reduction, indicating a community level shift of this function in UC patients (online supplemental figures 6 and 7, online supplemental table 8).
gutjnl-2020-322468supp006.pdf (141.5KB, pdf)
gutjnl-2020-322468supp007.pdf (437.7KB, pdf)
gutjnl-2020-322468supp008.pdf (360KB, pdf)
Then we investigated if there were more truncated or deleterious variants of OGAs in UC or CD patients. OGA homologues were retrieved from metagenomic datasets and we found that OGA homologues from different samples groups had similar lengths (online supplemental figure 8A). Then the sequences of NAGIdase domains were extracted and analysed with Oligotype.29 Results showed that the 11 amino acids around the ligand binding sites were conserved among all the OGA sequences from UC, CD or control (online supplemental figures 8B9, online supplemental table 9), indicating low possibility for more truncated or deleterious proteins in patients.
gutjnl-2020-322468supp009.pdf (194.8KB, pdf)
gutjnl-2020-322468supp010.pdf (874.8KB, pdf)
Gut bacterial secreted OGAs can hydrolyse O-GlcNAcylated proteins in host cells
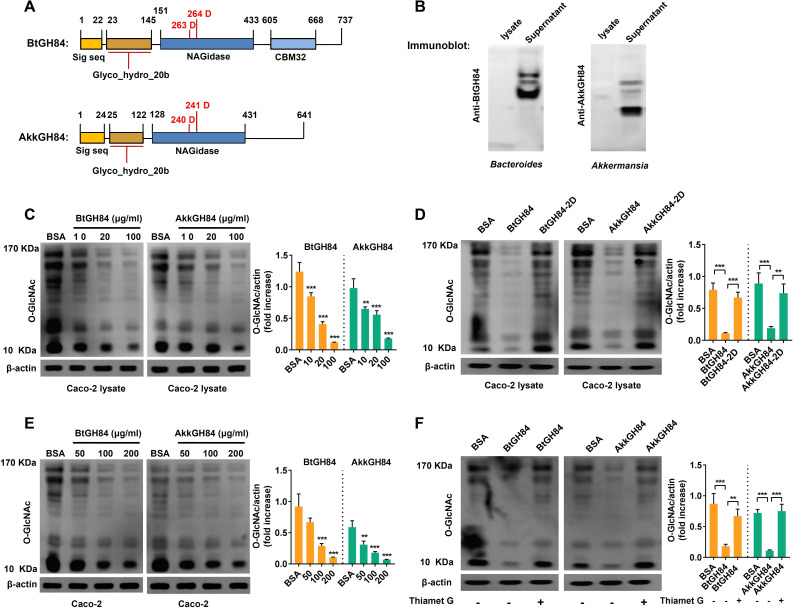
Since most of the bacterial OGAs were predicted to be secreted enzymes and have similar conserved domains with human OGAs, we speculated they might hydrolyse O-GlcNAcylated proteins in host cells. We focused on two bacterial OGAs whose gene abundance reduced significantly in UC patients (figure 3F): one is an experimentally validated OGA named BtGH84 from Bacteroides thetaiotaomicron (KXT29508.1),9 the other is a predicted OGA from Akkermansia muciniphila (AYR30073.1), named AkkGH84 here (figure 4A). First, we obtained purified His-tag BtGH84 and AkkGH84, and using them to generate anti-BtGH84-antibody and anti-AkkGH84-antibody. As shown in immunoblot assay in figure 4B, the supernatants of B.thetaiotaomicron and A. muciniphila, but not their bacterial lysates, showed a positive signal, indicating that both BtGH84 and AkkGH84 are secreted proteins. Furthermore, we generated the active site mutants of BtGH84 (D263A and D264A, named BtGH84-2D) and AkkGH84 (D240A and D241A, named AkkGH84-2D), and found that both BtGH84 and AkkGH84 can hydrolyse O-GlcNAcylated proteins in Caco-2 cell lysates (figure 4C), while the activate sites mutants loss this ability (figure 4D). Considering that protein O-GlcNAcylation or de-O-GlcNAcylation is an intracellular process, we next investigated if BtGH84 and AkkGH84 could hydrolyse O-GlcNAcylated proteins in cells. As shown in figure 4E, when given in the cell culture medium of Caco-2, both BtGH84 and AkkGH84 could also hydrolyse the O-GlcNAcylated proteins in a dose dependent manner. In agreement with the competitive inhibition of Thiamet G, this compound effectively inhibited the activity of AkkGH84 and BtGH84 on O-GlcNAcylated proteins in Caco-2 cells (figure 4F). These results prove that bacteria secreted OGAs can hydrolyse O-GlcNAcylated proteins in host cells and their functions can be inhibited by Thiamet G, a potent and selective OGA inhibitor.
Figure 4.
Bacterial secreted OGAs can hydrolyse O-GlcNAcylated proteins in human enterocyte-like Caco-2 cells. (A) Schematic of BtGH84 and AkkGH84 domain organisation, the active catalytic sites were indicated in red. (B) Polyclonal antibody against BtGH84 or AkkGH84 was generated and used to detect OGAs in bacterial lysates and supernatant by immunoblot analysis. (C, D) Immunoblot analysis of O-GlcNAc levels in Caco-2 lysates, after incubating the cell lysates with different amounts of BtGH84 or AkkGH84 (C) or their active sites mutants BtGH84-2D or AkkGH84-2D (D). (E, F) Immunoblot analysis of O-GlcNAc levels in Caco-2, after adding into the cell culture supernatant with different amounts of BtGH84 or AkkGH84 (E) or the Thiament G, an OGA inhibitor (F). Data represent the mean of three repeats per group with the SD; **P<0.01, ***P<0.001. OGA, O-GlcNAcase.
Bacterial OGAs can protect mice from colitis in multiple models
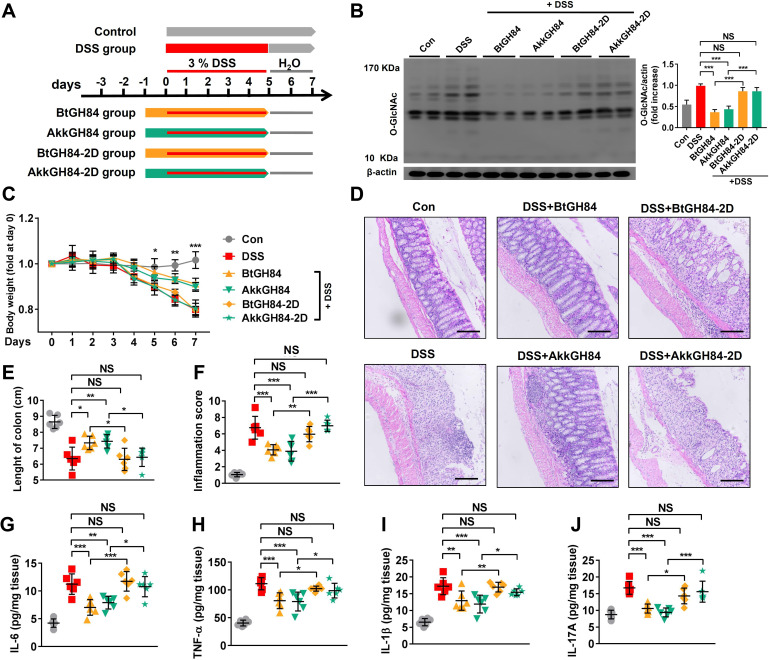
Considering that elevated protein O-GlcNAcylation promotes colonic inflammation4 and the bacterial OGA genes abundance decreases significantly in UC patients, we next investigate if supply with bacterial OGAs could protect mice from dextran sulfate sodium (DSS)-induced colitis, a well established IBD model. Mice were administrated with OGAs three or 1 day before inducing colitis by DSS (figure 5A and online supplemental figure 10A, F). To avoid being destroyed in the gastrointestinal tract, the pectin/zein beads delivery system was employed to deliver OGAs,30 and immunofluorescence staining showed OGAs could deliver into the colon cells (online supplemental figure 11). Seven days following colitis induction, loss of body weight, shorten of colon length, histopathological damage and inflammatory response were observed in the DSS group (figure 5B–J, online supplemental figure 10). Pretreatment with BtGH84 or AkkGH84 significantly ameliorated these parameters (figure 5B–J, (online supplemental figure 10), indicating bacterial OGAs can prevent mice from DSS-induced colitis.
Figure 5.
OGA enzymatic functions are required for bacterial OGAs’ protective effects on DSS-induced colitis. (A) Schematic outline of the DSS-colitis model and the treatment approach. (B) Immunoblot analysis of O-GlcNAc levels in colonic tissue lysates of mice. (C–F) Disease severity of DSS-colitis mice treated with wild type enzymes or their active sites mutants: body weight change (C), histology manifestation on H&E staining (D), scale bar: 200 µm, colon length (E) and inflammation score (F). (G–J) Proinflammatory cytokines in colonic tissue lysates. Data represent the mean of three repeats per group with the SD; *P<0.05, **P<0.01, ***P<0.001. DSS, dextran sulfate sodium; IL, interleukin; OGA, O-GlcNAcase; TNF, tumour necrosis factor.
gutjnl-2020-322468supp011.pdf (697.9KB, pdf)
gutjnl-2020-322468supp012.pdf (404.5KB, pdf)
To further evaluate the ability of bacterial OGAs to ameliorate established colitis (treatment effects), mice were administrated with bacterial OGAs at varies time points after DSS colitis induction (day 0–3, (online supplemental figure 10A, F). The results showed that bacterial OGAs can efficiently ameliorate DSS-induced colitis when given from day 0 and day 1, partially lose its protective function when given from day 2, and completely lose these effects when given from day 3 (online supplemental figure 10). Taken together, these data suggest that bacterial OGAs exert both preventive and therapeutic effects on DSS-induced colitis and their therapeutic effects are better when administrated at the initial stage of inflammation.
Furthermore, we showed that bacterial OGAs-treated mice were also resistant to colitis induced by 2,4,6-trinitrobenzenesulfonic acid or oxazolone (figure 6), suggesting the protective effects of bacterial OGAs were active against diverse chemically induced colitis.
Figure 6.
The protective effects of bacterial-derived OGAs BtGH84 and AkkGH84 against TNBS-induced or OXA-induced colitis. (A, F) Schematic outline of the TNBS-colitis model (A), OXA-colitis model (F) and their treatment approach. (B–E, G–J) Disease severity of colitis mice: body weight change (B, G), colon length (C, H); histology manifestation on H&E staining (D, I), scale bar: 200 µm and inflammation score (E, J). Data represent the mean of three repeats per group with the SD; *P<0.05, **P<0.01, ***P<0.001. OGA, O-GlcNAcase’ OXA, oxazolone; TNBS, 2,4,6-trinitrobenzenesulfonic acid.
Bacterial OGAs can ameliorate colitis through its OGA enzymatic activity
To test if the enzymatic activity was required for bacterial OGA’s protective effects, mice were pretreated with the wild-type enzymes or their active sites mutants (BtGH84-2D, AkkGH84-2D) and then induced colitis by DSS. Immunoblot showed that global O-GlcNAcylation level was elevated in colonic tissues of DSS-colitis mice. BtGH84 and AkkGH84, but not their active sites mutants, normalised the O-GlcNAcylation level (figure 5B). In accordance, BtGH84 and AkkGH84 ameliorated DSS-induced colitis, including reducing weight loss (figure 5C), improving colonic tissue damage (figure 5D, E) and suppressing gut inflammation (figure 5F–J), while their active sites mutants lost these abilities. These results indicate that the OGA enzymatic activities are required for the colitis protective effects of BtGH84 and AkkGH84.
Finally, we tested if BtGH84 and AkkGH84 protect mice from colitis through hydrolyzing O-GlcNAcylated proteins (online supplemental figure 12A). As shown in online supplemental figure 12B–J, Thiamet G, a selective OGA inhibitor, significantly inhibited the bacterial OGAs’ enzymatic functions and also abolished their protective effect on colitis. These results prove that bacterial OGAs can alleviate colitis through decreasing protein O-GlcNAcylation in colonic tissues.
gutjnl-2020-322468supp013.pdf (245.5KB, pdf)
Bacterial OGAs can hydrolase O-GlcNAcylated NF-κB-p65 and IKKβ to inhibit NF-κB signalling
T cell is an important cell type involved in gut inflammation pathogenesis.31 32 We have shown bacterial OGAs can hydrolase O-GlcNAcylated proteins in Caco-2 cells (figure 4C). To comprehensively investigate the cell type that responsible for bacterial OGAs’ protective effects, we further test their effects on human T cells. As expected, we found that both BtGH84 and AkkGH84 could also hydrolyse O-GlcNAcylated proteins in T cells (figure 7A). Next, to decipher the target molecules that might mediate bacterial OGAs’ protective effects, we performed mass spectrum analysis of O-GlcNAcylated peptides enriched from T cells’ lysates. The results showed that 916 O-GlcNAcylated peptides (belong to 848 proteins) elevated in lipopolysaccharide (LPS)-treated T cell group compared with the control. Among them, 573 O-GlcNAcylated peptides (belong to 538 proteins) decreased both in BtGH84 + LPS and AkkGH84 + LPS group (online supplemental table 10). Specificity, NF-κB-p65 subunit and IKKβ, whose O-GlcNAcylation have been reported to activate NF-κB signalling and subsequent inflammation,4 33 34 were only detected in the LPS-treated T cell group. Further, succinylated wheat germ agglutinin agarose pull down and immunoprecipitation experiments validated that LPS could induce NF-κB-p65 and IKKβ O-GlcNAcylation in both T cells and Caco-2 cells, which could be re-normalised by bacterial OGAs (figure 7B, C, online supplemental figure 13B, C).
Figure 7.
LPS-induced NF-κB-p65 subunit and IKKβ O-GlcNAcylation and subsequent NF-κB signalling activation in Jurkat cells were blocked by BtGH84 and AkkGH84. Jurkat cells were pretreated with BtGH84, AkkGH84, BtGH84-2D or AkkGH84-2D, followed by LPS incubation for. (A) Whole cell extracts were analysed with immunoblots for O-GlcNAc, NF-κB-p65, IKKβ and IκBα. β-actin serves as a loading control. (B) O-GlcNAcylated proteins in Jurkat were pulled down using sWGA beads. O-GlcNAc, NF-κB-p65, IKKβ and IκBα in the pull-down complexes were detected using immunoblots. (C) Whole cell extracts of Jurkat were immunoprecipiated with anti-NF-κB-p65 or anti-IKKβ antibody. The O-GlcNAcylated NF-κB-p65 and IKKβ were detected using anti-O-GlcNAc antibody. (D–F) Cytosolic and nuclear sections were analysed by immunoblots for IκBα and NF-κB-p65, respectively. β-actin serves as a loading control of cytosolic section; histone-H3 serves as a loading control of nuclear. (G) Jurkat were transfected with pGL3/NF-κB and PRL, followed by treated with BtGH84, AkkGH84, BtGH84-2D or AkkGH84-2D as described above. Afterward, cells were harvested for luciferase activity assay. (H, I) Proinflammatory cytokines levels in treated Jurkat. Data represent the mean of three repeats per group with the SD; *P<0.05, **P<0.01, ***P<0.001. LPS, lipopolysaccharide; NS, not significant; sWGA, succinylated wheat germ agglutinin agarose.
gutjnl-2020-322468supp015.pdf (340.9KB, pdf)
To investigate if the NF-κB signalling was inhibited following NF-κB-p65/IKKβ de-O-GlcNAcylation by bacterial OGAs, we tested the nuclear transduction of NF-κB-p65 and inflammatory cytokines release in different groups. The results showed that all these events were elevated after LPS stimulation and suppressed by bacterial OGAs (figure 7D–I, online supplemental material). Finally, we validated the NF-κB-p65/IKKβ de-O-GlcNAcylation functions and NF-κB signalling-suppressive effects of bacterial OGAs on colon tissues of DSS colitis models (figures 8 and 5G, H). Taken together, these results prove that bacterial OGAs can hydrolase O-GlcNAcylated NF-κB-p65/IKKβ to inhibit NF-κB signalling, which may be responsible for their colitis protective effects.
Figure 8.
DSS-mediated NF-κB-p65 subunit/IKKβ O-GlcNAcylation and subsequent NF-κB signalling activation in mouse were alleviated by BtGH84 and AkkGH84. Mice were treated with control, DSS, DSS+BtGH84, DSS+AkkGH84, DSS+BtGH84-2D and DSS+AkkGH84-2D as described in methods. (A, C, D) Colonic tissue lysates were immunoprecipitated with anti-NF-κB-p65 or anti-IKKβ antibody. The O-GlcNAcylated NF-κB-p65 and IKKβ were detected using anti-O-GlcNAc antibody. (B, E, F) Cytosolic and nuclear sections from colonic tissue lysates were analysed by immunoblots for IκBα and NF-κB-p65, respectively. β-actin serves as a loading control of cytosolic section; histone-H3 serves as a loading control of nuclear. Data represent the mean of 3 repeats per group with the SD; *P<0.05, **P<0.01, ***P<0.001. DSS, dextran sulfate sodium; NS, not significant.
Discussion
Gut microbiome derived enzymes have long been recognised to have a great implication on intestinal physiology and pathology.35 36 Most of them act through generating small molecules such as indoleproprionic acid, bile acids, trimethylamine and short-chain fatty acids.36–39 Here, we first link intestinal microbial derived OGAs to host protein O-GlcNAc modification and intestinal inflammation.
The most profound result of this study is that bacteria-derived OGAs can modify host protein O-GlcNAcylation, an essential intracellular process involved in inflammation, ageing and tumourigenesis. Human protein O-GlcNAcylation have long been thought to control by a pair of human encoding enzymes: OGT and OGA.1 Though bacterial OGAs have the same catalytic domain as human OGAs, their roles in modifying host protein O-GlcNAcylations are never investigated. The main concern is that protein O-GlcNAcylation is an intracellular process. In fact, bacterial proteins can entry into host cells through two main ways: secretion system or endocytosis. Although most of the bacterial OGAs are predicted to be secreted proteins, only seldom of them are predicted to be secretion system effectors. Here, we proved that two secreted bacterial OGAs, when used independent of the bacteria, can entry the host cells and hydrolyse O-GlcNAcylated proteins in host cells both in vitro and in vivo, indicating that endocytosis may act as the main pathway through which bacterial OGAs enter into host cells. Considering that OGA genes are widely distributed in human gut microbiota and have a relatively high abundance in healthy human gut samples (online supplemental figure 3A), we speculated that bacteria-derived OGAs may play a more fundamental role in regulating host physiology than we have ever thought.
The role of O-GlcNAc modifications in gut inflammation is controversial. One important question to discuss is how O-GlcNAcylation level changes during gut inflammation. Although multiple studies showed that O-GlcNAcylation level elevated in colonic tissue of colitis mice.4 34 40 Zhao et al showed in a small sample size (Con: 14, UC: 13, CD: 10) that epithelial O-GlcNAcylation level decreased in patients with gut inflammation.41 This results need to be further validated since a recent study found a contrary change in a larger sample size (Con 45, CD 58).34 The other question is what are the effects of protein O-GlcNAcylation on gut inflammation. Zhao et al showed that deletion of O-GlcNAcylation in paneth cell led to gut inflammation.41 This contrary effects of O-GlcNAcylation to other studies4 34 may be explained by the fact that Zhao et al use a paneth cell-specific OGT knock out model, which can’t comprehensively evaluate the overall effects of O-GlcNAcylation in gut tissues. Yang et al and Sun et al have shown that reduced the overall O-GlcNAcylation levels in gut tissues could lead to gut inflammation, in which NF-κB signalling plays a critical role.4 34 Accordantly, here, we show bacterial OGAs can hydrolase O-GlcNAcylation on NF-κB p65 subunit, suppress subsequent inflammation in both immune cells and epithelial cells, and protect mice from colitis in multiple models. Considering the pivotal effect of NF-κB signalling mediated inflammation on UC pathogenesis,42 43 we speculated that de-O-GlcNAcylation on NF-κB-p65 subunit may serve as the main mechanism of bacterial OGA’s protective effects. However, other proteins responsible for the protective effects of bacterial OGAs would be necessary to study, and our mass spectrum analysis profiling results may provide a useful resource for these future studies.
Our study first links intestinal microbial OGAs to intestinal physiology and pathology, by combining bioinformatics methods, metagenomics analysis and bioexperiments. Given the myriad function of O-GlcNAcylation in gut physiology and the wide distribution of OGAs in gut microbe, further explore the mechanism of bacterial OGAs to control host protein O-GlcNAcylation homeostasis may provide new microbial therapeutic strategy for UC.
Methods
Bioinformatic analysis
The detailed methods of bioinformatic analysis and metagenomic quantifying are provided in online supplemental materials. All the bioinformatic tools used in this study are summarised in online supplemental table 11.
Protein expression, purification and generation of polyclonal antibodies
The gene sequence encoding BtGH84 or AkkGH84 was amplified with synthetic oligonucleotides, and D263/D264 (BtGH84) and D240/D241 (AkkGH84) were mutated to alanine by PCR mutagenesis. These genes sequences were subcloned into the expression vector pET-28a (Addgene). The DNA sequence was verified by sequencing and transformed into Escherichia coli BL21(DE3). After induction of protein expression by isopropyl-β-d-thiogalactoside (Sigma-Aldrich), bacteria were harvested and lysed by sonication. The protein in the soluble fraction was purified using His-Bind purification kit (Novagen). Recombinant BtGH84, AkkGH84 and their mutant protein were passed through an EndoTrap Red endotoxin removal column (Hyglos) to remove potential endotoxin according to the manufacturer’s instructions.
Female New Zealand rabbits were immunised via repeated intradermal injections of full-length BtGH84 or AkkGH84 with an equal volume of Freund’s adjuvant (Sigma-Aldrich). The rabbits were sacrificed and their blood collected after the final boosts. Anti-sera were collected from blood and used to perform the immunoblot analysis as described below.
Cell culture and treatment
Human colon carcinoma cell line Caco-2 and human acute T-cell leukaemia cell line Jurkat were purchased from Shanghai Institute of Cell Biology (Shanghai, China). Cells were routinely cultured in Eagle’s Minimum Essential Medium with 10% heat inactivated fetal bovine serum (PAN Biotech, Aidenbach, Germany), 1% penicillin-streptomycin (HyClone, USA) in a humidified atmosphere of 5% CO2. To determine the hydrolysis effect of OGAs, Caco-2 or Jurkat was seeded into 6-well plates for immunoblot analysis. The next day, the cells were stimulated with nonserum medium containing LPS (100 ng/mL), BtGH84, BtGH84-2D, AkkGH84 or AkkGH84-2D (10–200 µg/mL) for 4 hours. For the experiments studying the effect of inhibitor, the cells were prestimulated with Thiamet G (Sigma-Aldrich) for 2 hours before stimulation with OGAs. After treatment, cells were lysed and assayed for the O-GlcNAcylation using immunoblot as described below.
Animals and DSS-induced colitis
All research involving animals has been approved by the ethics committee and performed strictly according to the guidelines for animal care in Southern Medical University. C57BL/6J mice and New Zealand rabbits were purchased from Animal Experimental Center of Southern Medical University. Animals were housed under a 12-hour light-12-hour dark cycle, and they had free access to autoclave chow and water. Animal care and experimental procedures were performed under anaesthesia with ketamine and lidocaine, and utmost efforts were taken to minimise suffering.
DSS colitis was induced in age-matched and sex-matched C57BL/6J mice by oral administration of 3% (w/v) DSS (Sigma-Aldrich, 40 KDa) in drinking water for 5 days, followed by a tap water for 2 days. Mice were orally administered protein beads daily until termination of experiments. Body weight was monitored daily. Mice were sacrificed on day 8, the colon length was measured. Colon tissues were fixed in 4% paraformaldehyde and embedded in paraffin, followed by H&E staining. Severity of inflammation was scored based on body weight loss, occult blood and stool consistency in a blind manner.
Additional protocols, material and methods are described in online supplemental material.
Acknowledgments
We thank Dr Yang Wu for the professional advices on statistical analysis of this study.
Footnotes
XH, JG and LP contributed equally.
Contributors: HC, PZ, MZ and XH conceived and designed the study. HC, JG, PZ, LP, MZ and XH contributed data analysis and drafting the manuscript. XH, JG, LP, TH, PZ, MZ and YW performed bioinformatic and bioexperiments analysis. All authors read and approved the final manuscript.
Funding: This work was supported by the National Natural Science Foundation of China (NSFC-31900101, 81801985).
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
All data relevant to the study are included in the article or uploaded as online supplemental information.
Ethics statements
Patient consent for publication
Not required.
Ethics approval
All animal procedures were approved by the Medical Ethics Committee of Southern Medical University.
References
- 1.Yang X, Qian K. Protein O-GlcNAcylation: emerging mechanisms and functions. Nat Rev Mol Cell Biol 2017;18:452–65. 10.1038/nrm.2017.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hart GW. Three Decades of Research on O-GlcNAcylation – A Major Nutrient Sensor That Regulates Signaling, Transcription and Cellular Metabolism. Front Endocrinol 2014;5:183. 10.3389/fendo.2014.00183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanover JA, Krause MW, Love DC. Linking metabolism to epigenetics through O-GlcNAcylation. Nat Rev Mol Cell Biol 2012;13:312–21. 10.1038/nrm3334 [DOI] [PubMed] [Google Scholar]
- 4.Yang YR, Kim DH, Seo Y-K, et al. Elevated O-GlcNAcylation promotes colonic inflammation and tumorigenesis by modulating NF-κB signaling. Oncotarget 2015;6:12529–42. 10.18632/oncotarget.3725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Y, Xie M, Men L, et al. O-Glcnacylation in immunity and inflammation: an intricate system (review). Int J Mol Med 2019;44:363–74. 10.3892/ijmm.2019.4238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baudoin L, Issad T. O-Glcnacylation and inflammation: a vast Territory to explore. Front Endocrinol 2014;5:235. 10.3389/fendo.2014.00235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu F, Iqbal K, Grundke-Iqbal I, et al. O-GlcNAcylation regulates phosphorylation of tau: A mechanism involved in Alzheimer’s disease. Proc Natl Acad Sci U S A 2004;101:10804–9. 10.1073/pnas.0400348101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu Q, Li S, Shao F. Sweet talk: protein glycosylation in bacterial interaction with the host. Trends Microbiol 2015;23:630–41. 10.1016/j.tim.2015.07.003 [DOI] [PubMed] [Google Scholar]
- 9.Dennis RJ, Taylor EJ, Macauley MS, et al. Structure and mechanism of a bacterial β-glucosaminidase having O-GlcNAcase activity. Nat Struct Mol Biol 2006;13:365–71. 10.1038/nsmb1079 [DOI] [PubMed] [Google Scholar]
- 10.Shi W-W, Jiang Y-L, Zhu F, et al. Structure of a novel O-linked N-acetyl-D-glucosamine (O-GlcNAc) transferase, GtfA, reveals insights into the glycosylation of pneumococcal serine-rich repeat adhesins. J Biol Chem 2014;289:20898–907. 10.1074/jbc.M114.581934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guttenberg G, Hornei S, Jank T, et al. Molecular Characteristics of Clostridium perfringens TpeL Toxin and Consequences of Mono- O -GlcNAcylation of Ras in Living Cells. J. Biol. Chem. 2012;287:24929–40. 10.1074/jbc.M112.347773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pathak S, Borodkin VS, Albarbarawi O, et al. O -GlcNAcylation of TAB1 modulates TAK1-mediated cytokine release. Embo J 2012;31:1394–404. 10.1038/emboj.2012.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zou Y, Xue W, Luo G, et al. 1,520 reference genomes from cultivated human gut bacteria enable functional microbiome analyses. Nat Biotechnol 2019;37:179–85. 10.1038/s41587-018-0008-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Podell S, Gaasterland T. DarkHorse: a method for genome-wide prediction of horizontal gene transfer. Genome Biol 2007;8:R16. 10.1186/gb-2007-8-2-r16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huttenhower C, Gevers D, Knight R, et al. Structure, function and diversity of the healthy human microbiome. Nature 2012;486:207–14. 10.1038/nature11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benanti EL, Nguyen CM, Welch MD. Virulent Burkholderia species mimic host actin polymerases to drive actin-based motility. Cell 2015;161:348–60. 10.1016/j.cell.2015.02.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bittar F, Reynaud-Gaubert M, Thomas P, et al. Acetobacter indonesiensis pneumonia after lung transplant. Emerg Infect Dis 2008;14:997–8. 10.3201/eid1406.071236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J, Jia H, Cai X, et al. An integrated catalog of reference genes in the human gut microbiome. Nat Biotechnol 2014;32:834–41. 10.1038/nbt.2942 [DOI] [PubMed] [Google Scholar]
- 19.Human Microbiome Project Consortium . A framework for human microbiome research. Nature 2012;486:215–21. 10.1038/nature11209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qin N, Yang F, Li A, et al. Alterations of the human gut microbiome in liver cirrhosis. Nature 2014;513:59–64. 10.1038/nature13568 [DOI] [PubMed] [Google Scholar]
- 21.Le Chatelier E, Nielsen T, Qin J, et al. Richness of human gut microbiome correlates with metabolic markers. Nature 2013;500:541–6. 10.1038/nature12506 [DOI] [PubMed] [Google Scholar]
- 22.Zhang X, Zhang D, Jia H, et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med 2015;21:895–905. 10.1038/nm.3914 [DOI] [PubMed] [Google Scholar]
- 23.Qin J, Li Y, Cai Z, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012;490:55–60. 10.1038/nature11450 [DOI] [PubMed] [Google Scholar]
- 24.Karlsson FH, Tremaroli V, Nookaew I, et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 2013;498:99–103. 10.1038/nature12198 [DOI] [PubMed] [Google Scholar]
- 25.Nielsen HB, Almeida M, Juncker AS, et al. Identification and assembly of genomes and genetic elements in complex metagenomic samples without using reference genomes. Nat Biotechnol 2014;32:822–8. 10.1038/nbt.2939 [DOI] [PubMed] [Google Scholar]
- 26.Lloyd-Price J, Arze C, Ananthakrishnan AN, et al. Multi-Omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 2019;569:655–62. 10.1038/s41586-019-1237-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng Q, Liang S, Jia H, et al. Gut microbiome development along the colorectal adenoma–carcinoma sequence. Nat Commun 2015;6:6528. 10.1038/ncomms7528 [DOI] [PubMed] [Google Scholar]
- 28.Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010;464:59–65. 10.1038/nature08821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eren AM, Maignien L, Sul WJ, et al. Oligotyping: differentiating between closely related microbial taxa using 16S rRNA gene data. Methods Ecol Evol 2013;4:1111–9. 10.1111/2041-210X.12114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao J, Li Y, Wan Y, et al. A novel Postbiotic from Lactobacillus rhamnosus GG with a beneficial effect on intestinal barrier function. Front Microbiol 2019;10:477. 10.3389/fmicb.2019.00477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pelczar P, Witkowski M, Perez LG, et al. A pathogenic role for T cell–derived IL-22BP in inflammatory bowel disease. Science 2016;354:358–62. 10.1126/science.aah5903 [DOI] [PubMed] [Google Scholar]
- 32.Zundler S, Schillinger D, Fischer A, et al. Blockade of αEβ7 integrin suppresses accumulation of CD8+ and Th9 lymphocytes from patients with IBD in the inflamed gut in vivo. Gut 2017;66:1936–48. 10.1136/gutjnl-2016-312439 [DOI] [PubMed] [Google Scholar]
- 33.Yang WH, Park SY, Nam HW, et al. Nfkappab activation is associated with its O-GlcNAcylation state under hyperglycemic conditions. Proc Natl Acad Sci U S A 2008;105:17345–50. 10.1073/pnas.0806198105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun Q-H, Wang Y-S, Liu G, et al. Enhanced O-linked Glcnacylation in Crohn’s disease promotes intestinal inflammation. E Bio Medicine 2020;53:102693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown JM, Hazen SL. Targeting of microbe-derived metabolites to improve human health: the next frontier for drug discovery. J Biol Chem 2017;292:8560–8. 10.1074/jbc.R116.765388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rath S, Rud T, Karch A, et al. Pathogenic functions of host microbiota. Microbiome 2018;6:174. 10.1186/s40168-018-0542-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song Z, Cai Y, Lao X, et al. Taxonomic profiling and populational patterns of bacterial bile salt hydrolase (BSH) genes based on worldwide human gut microbiome. Microbiome 2019;7:9. 10.1186/s40168-019-0628-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones BV, Begley M, Hill C, et al. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc Natl Acad Sci U S A 2008;105:13580–5. 10.1073/pnas.0804437105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roager HM, Licht TR. Microbial tryptophan catabolites in health and disease. Nat Commun 2018;9:3294. 10.1038/s41467-018-05470-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li X, Zhang Z, Li L, et al. Myeloid-Derived Cullin 3 promotes STAT3 phosphorylation by inhibiting OGT expression and protects against intestinal inflammation. J Exp Med 2017;214:1093–109. 10.1084/jem.20161105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao M, Xiong X, Ren K, et al. Deficiency in intestinal epithelial O-GlcNAcylation predisposes to gut inflammation. EMBO Mol Med 2018;10:e8736. 10.15252/emmm.201708736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang G, Xu B, Shi F, et al. Protective Effect of Methane-Rich Saline on Acetic Acid-Induced Ulcerative Colitis via Blocking the TLR4/NF-κB/MAPK Pathway and Promoting IL-10/JAK1/STAT3-Mediated Anti-inflammatory Response. Oxid Med Cell Longev 2019;2019:7850324. 10.1155/2019/7850324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andresen L, Jørgensen VL, Perner A, et al. Activation of nuclear factor kappaB in colonic mucosa from patients with collagenous and ulcerative colitis. Gut 2005;54:503–9. 10.1136/gut.2003.034165 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2020-322468supp001.xlsx (1.9MB, xlsx)
gutjnl-2020-322468supp002.pdf (272.1KB, pdf)
gutjnl-2020-322468supp003.pdf (240.3KB, pdf)
gutjnl-2020-322468supp004.pdf (195.2KB, pdf)
gutjnl-2020-322468supp005.pdf (252.4KB, pdf)
gutjnl-2020-322468supp014.pdf (126.3KB, pdf)
gutjnl-2020-322468supp006.pdf (141.5KB, pdf)
gutjnl-2020-322468supp007.pdf (437.7KB, pdf)
gutjnl-2020-322468supp008.pdf (360KB, pdf)
gutjnl-2020-322468supp009.pdf (194.8KB, pdf)
gutjnl-2020-322468supp010.pdf (874.8KB, pdf)
gutjnl-2020-322468supp011.pdf (697.9KB, pdf)
gutjnl-2020-322468supp012.pdf (404.5KB, pdf)
gutjnl-2020-322468supp013.pdf (245.5KB, pdf)
gutjnl-2020-322468supp015.pdf (340.9KB, pdf)
Data Availability Statement
All data relevant to the study are included in the article or uploaded as online supplemental information.