Abstract
All of the mitochondrial tRNAs of Trypanosoma brucei have been shown to be encoded in the nucleus and must be imported into the mitochondrion. The import of nuclearly encoded tRNAs into the mitochondrion has been demonstrated in a variety of organisms and is essential for proper function in the mitochondrion. An in vitro import assay has been developed to study the pathway of tRNA import in T. brucei. The in vitro system utilizes crude isolated trypanosome mitochondria and synthetic RNAs transcribed from a cloned nucleus-encoded tRNA gene cluster. The substrate, composed of tRNASer and tRNALeu, is transcribed in tandem with a 59-nucleotide intergenic region. The tandem tRNA substrate is imported rapidly, while the mature-size tRNALeu fails to be imported in this system. These results suggest that the preferred substrate for tRNA import into trypanosome mitochondria is a precursor molecule composed of tandemly linked tRNAs. Import of the tandem tRNA substrate requires (i) a protein component that is associated with the surface of the mitochondrion, (ii) ATP pools both outside and within the mitochondrion, and (iii) a membrane potential. Dissipation of the proton gradient across the inner mitochondrial membrane by treatment with an uncoupling agent inhibits import of the tandem tRNA substrate. Characterization of the import requirements indicates that mitochondrial RNA import proceeds by a pathway including a protein component associated with the outer mitochondrial membrane, ATP-dependent steps, and a mitochondrial membrane potential.
The mitochondrial genome of eukaryotic cells typically encodes the mitochondrial rRNAs, tRNAs, and mRNAs. However, it has been shown in a number of organisms that some tRNAs necessary for protein translation within the mitochondrion are encoded in the nucleus and imported into the mitochondrion. The number and the identity of the nuclearly encoded mitochondrial tRNAs vary from a few in higher plants (15), at least 9 in Acanthamoeba (3), and 26 in Tetrahymena (27) to the entire set of mitochondrial tRNAs in the kinetoplastid protozoans Trypanosoma brucei (6) and Leishmania tarentolae (26). Although the mitochondrial genome for Saccharomyces cerevisiae encodes all the tRNAs necessary for protein synthesis, a single nuclearly encoded tRNALys (CUU) has been shown to be imported into mitochondria (16, 30). The purpose for importing this tRNALys has yet to be determined.
The pathways of protein import into mitochondria have been extensively studied. Protein import is posttranslational, and translocation to the mitochondrial matrix typically begins with a precursor protein that must be at least partially unfolded by cytoplasmic chaperones (19, 22). Mitochondrial import of precursor proteins usually requires internal and external ATP hydrolysis, an electrical potential across the inner mitochondrial membrane, and protein involvement in maintaining the transport-competent conformation of the precursor proteins (19, 22). However, several unfolded precursor proteins can be imported in the absence of cytosolic factors (20), suggesting that cytosolic factors are not required for the targeting of all precursor proteins to the mitochondrion.
In contrast, little is known about the mechanism of RNA import into the mitochondrion (23). Much of the work defining the import pathway for nuclearly encoded tRNAs into mitochondria has focused on determining the substrate requirements necessary for import. Some evidence exists for the involvement of precursor tRNAs as substrates for mitochondrial import in T. brucei (7, 10). However, in S. cerevisiae the import substrate is the mature tRNALys (16, 30). Precursor tRNAs with either 5′ or 3′ extensions have not been detected in yeast mitochondria. The import of tRNALys in S. cerevisiae has been defined in vivo and in vitro as requiring ATP, a membrane potential, and a cytosolic fraction containing the cytosolic tRNA synthetases (29, 30). The in vitro system also requires an intact protein translocating system (28).
For Leishmania donovani, an in vitro assay was employed to study the import of an antisense transcript containing the 5′ untranslated region and 25 nucleotides (nt) of the β-tubulin mRNA (14). Import of this transcript was ATP dependent but did not require addition of a soluble cytosolic fraction. When the same in vitro system was used, import of tRNATyr from Leishmania also demonstrated ATP-dependent import into isolated mitochondria (13). The tRNATyr was shown to bind specifically to two mitochondrial surface proteins of 15 and 22 kDa (1, 13).
In this paper, we describe the development of an in vitro import assay for T. brucei nuclearly encoded tRNAs. Efficient import of tandemly linked tRNASer and tRNALeu but not mature-size tRNAs was detected. Trypanosome tRNA genes are typically organized as clusters (17, 18). These genes appear to be transcribed as precursor transcripts (10). One cluster containing tandemly arranged tRNASer and tRNALeu separated by a 59-nt intergenic sequence has been recently shown to be transcribed as a precursor molecule that is found in both the cytosol and mitochondria of T. brucei (10). Import of the tandem tRNA occurs rapidly, as early as 1 min after incubation with isolated mitochondria, and continues for 15 min. Import of the tandem tRNASer-tRNALeu requires a mitochondrial fraction, a protein component on the surface of the mitochondrion, ATP hydrolysis, and a membrane potential.
MATERIALS AND METHODS
Construction of plasmids.
The plasmid pBSD1 was constructed from pBluescript digested with BssHII to remove the multiple cloning site containing the T7 promoter. A portion of the T. brucei genomic clone (D1) containing tRNASer, a 59-nt intergenic region, and tRNALeu (10) was PCR amplified with primer 002 (sense) (5′-GCA TGG CGC GCC TAA TAC GAC TCA CTA TAG GGT GGC GGG GCG GTG TCA CC-3′) and primer 003 (antisense) (5′-GGT TAA GGC GCG cct cgt cct cgt gca aaA CCT GAC AAG AGT GGG GTT-3′. The primers contain template sequence (underlined), the T7 promoter (boldface), AscI restriction sites (italics), and an MnlI restriction site (primer 003 only; lowercase). The AscI restriction sites were digested and used as the cloning sites into Bluescript (BssHII and AscI have the same overhangs). The MnlI site was used to create a blunt-end digestion for T7 transcription. The tandem tRNA and mature tRNALeu clones were digested with MnlI (New England Biolabs) to produce 241- and 87-nt templates for runoff transcription, respectively. pTE was derived from the vector pTF-EMC (NcoI) where the encephalomyocarditis virus (EMC) leader was added to the BamHI site of pTF7.5 (4). When pTF-EMC (NcoI) is digested with HindIII, a 241-nt fragment containing a T7 promoter is produced. The rRNA control was from the T7-MEGAshortscript kit (Ambion) containing a 116-bp cDNA fragment from the 18S rRNA, inserted downstream from the T7 promoter sequence of pUC18 in the antisense orientation.
Transcription and purification of RNA.
All three substrates were incubated with T7 RNA polymerase and [α-32P]UTP according to protocols from Promega to produce uniformly labeled transcripts. Reactions were carried out for 30 min to 2 h and stopped by the addition of formamide loading buffer, and the products were loaded onto a 6% acrylamide–8 M urea gel. The wet gel was exposed for 10 s to X-Omat AR film (Kodak), and the appropriate bands were excised and incubated overnight at 37°C in 0.5 M ammonium acetate, 0.1% sodium dodecyl sulfate (SDS), and 1.0 mM EDTA (pH 8.0). The substrates were precipitated with 95% ethanol and washed with 70% ethanol. The pellet was dried in a Speed Vac (Savant) and resuspended in 30 μl of distilled water. For each import assay, 5 × 104 cpm of substrate was used.
Trypanosome growth and isolation of mitochondria.
T. brucei procyclic cells (TREU 667) were grown at 26°C in semidefined medium (5) containing 10% heat-inactivated fetal bovine serum (Summit Biotechnology) and 20 μg of gentamicin sulfate (Life Technologies, Inc.) per ml. Mitochondria were isolated according to the method of Priest and Hajduk (21). Briefly, cells were grown to a density of 1 × 107 to 1.5 × 107 cells/ml. Two liters was collected by centrifugation at 3,000 × g for 15 min at 4°C. The remainder of the procedure was performed at 4°C. The cells were washed in a buffer containing 0.15 M NaCl, 20 mM glucose, and 20 mM NaH2PO4 (pH 7.9) and centrifuged at 3,000 × g for 10 min. The cells were resuspended at a density of 1010 cells/ml in buffer containing 0.25 M sucrose, 20 mM MOPS (morpholinepropanesulfonic acid)-KOH (pH 7.2), and 1 mM EDTA (SME-20 buffer). The cells were equilibrated with N2 at 1,000 lb/in2 for 15 min in a nitrogen cavitation bomb (minibomb cell disruption chamber; Kontes, Vineland, N.J.). Release from the bomb resulted in the disruption of >95% of the cells. The cells were diluted with 1 volume of SME-20 buffer and centrifuged in an SW-41 rotor at 1,500 × g for 10 min to remove intact cells, cell ghosts, and large cellular debris. The supernatant was then centrifuged at 16,000 × g for 15 min. The pellet was resuspended in the original volume of SME-20 buffer and centrifuged again at 1,500 × g for 10 min. The final pellet was resuspended in a storage buffer containing SME-20 buffer and 50% glycerol at a final concentration of 3 × 109 cells/ml. After homogenization with a Dounce homogenizer and B pestle (10 strokes), 500-μl aliquots were frozen at −70°C. The mitochondria maintain import activity for up to 2 weeks under these conditions. The mitochondrial preparation is able to import and process the mitochondrial Rieske iron-sulfur protein (ISP) (21), cytochrome c1 protein (21a), and RNA.
Standard in vitro import assay protocol.
Crude mitochondria in SME-20 and 50% glycerol were thawed on ice at 4°C. The mitochondria were aliquoted and diluted with 9 volumes of SME-20 buffer before centrifugation at 14,000 × g for 10 min. Each 100 μl of assay mixture (final volume) contained the mitochondria from 3 × 108 cells, in vitro-transcribed T7 substrate (5 × 104 cpm/assay), 0.25 M sucrose, 80 mM KCl, 5 mM MgCl2, 1 mg of bovine serum albumin per ml, 2 mM ATP, 10 mM creatine phosphate, 0.1 mg of creatine kinase (Boehringer Mannheim) per ml, and 20 mM MOPS-KOH (pH 7.2). The ATP, creatine phosphate, and creatine kinase were added as an ATP regeneration system, except when specified as import buffer (−ATP). The tRNA import assay mixtures were incubated at 26°C for 30 min, unless otherwise stated. Assays were terminated by centrifugation at 14,000 × g for 5 min at 4°C. The pellet was resuspended in 25 μl of SME-20 buffer with final concentrations of 5 mM CaCl2 and 100 mM KCl.
Postimport treatment of mitochondria and RNAs.
To remove labeled tRNAs that may be nonspecifically associated with the outer mitochondrial membrane or bound through protein interactions, proteinase K (Boehringer Mannheim) was added to a final concentration of 30 μg/ml (21). Digestion was carried out for 10 min at room temperature. One volume of cold SME-20 buffer containing 4 mM phenylmethylsulfonyl fluoride (PMSF) was added to terminate the reaction. The mitochondria were then collected by centrifugation at 14,000 × g for 5 min at 4°C. To remove RNAs not imported, micrococcal nuclease (Pharmacia) was added to a final amount of 10 U. Digestion was carried out for 10 min at room temperature. The reaction was terminated with 10 mM EGTA, and the mitochondria and tRNAs were collected by centrifugation as described above. As a control for nuclease digestion, following import CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate} was added to a final concentration of 2% before proteinase K treatment and the mixture was incubated at room temperature for 10 min. After the addition of 25 μl of SME-20, CaCl2, and KCl, the reaction mixtures were then treated as before with proteinase K and micrococcal nuclease. For all samples, the RNAs were collected by resuspending the pellet in 0.3 M sodium acetate and 0.5% SDS, extraction with phenol-chloroform, precipitation with 95% ethanol, and a 70% ethanol wash. For the CHAPS control, the reaction product was phenol-chloroform extracted and ethanol precipitated. The pellets were dried and resuspended in 5 μl of formamide loading buffer.
Analysis of import reactions.
The entire RNA sample was resolved by gel electrophoresis on a 6% polyacrylamide–8 M urea gel. The gels were dried and exposed to X-Omat AR film (Kodak) with an intensifying screen (Lightning Plus; NEN-DuPont) at −70°C. Quantitation was performed with a Molecular Dynamics PhosphorImager (model STORM-860).
Additional procedures.
Stocks of proteinase K (1 mg/ml or 100 μg/ml) were added prior to import to a final concentration as specified above and incubated for 15 min at room temperature. Concentrated stocks of potato apyrase (grade VIII, 1,000 U/ml) and carboxyatractyloside (CAT) were made in 10 mM MOPS-KOH at pH 7.2 and distilled H2O, respectively. The apyrase and CAT were incubated with the mitochondria at room temperature for 10 min. Concentrated oligomycin and carbonylcyanide m-chlorophenylhydrazone (CCCP) stocks were resuspended in 100% ethanol. The reagents were incubated with the mitochondria at 4°C for 10 min prior to addition of labeled substrate. All reagents were from Sigma unless otherwise specified.
RESULTS
Isolated mitochondria from procyclic T. brucei cells import a tandemly transcribed tRNA.
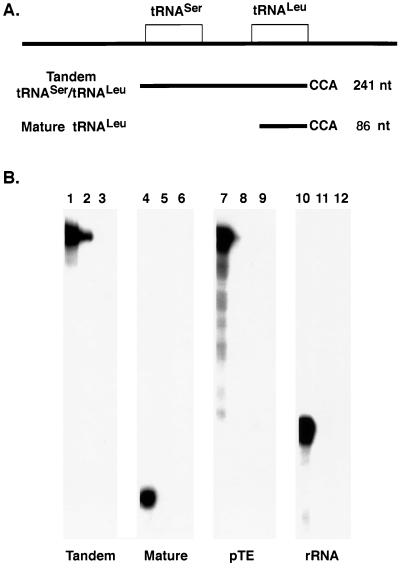
Mitochondria isolated for in vitro import were prepared by nitrogen cavitation lysis followed by differential centrifugation (21). This procedure minimizes disruption of the mitochondria while efficiently lysing the T. brucei cells. The RNAs used as substrates for import were prepared by in vitro transcription and are diagramed in Fig. 1A. The full-length tandem substrate consists of the 82-nt tRNASer with 14 nt immediately upstream, a 59-nt intergenic region, and the 86-nt tRNALeu with a mature 3′ end containing a CCA. pTE, a plasmid that contains sequence from the EMC virus leader and produces a transcript of 241 nt (the same size as the tandem tRNA substrate), was used as a negative control. The mature tRNA substrate consists of the 86-nt tRNALeu with a fully processed 5′ end and a CCA at the 3′ end. The 116-nt antisense rRNA transcript serves as a negative control in the same size range as the mature tRNALeu.
FIG. 1.
The tandem tRNASer-tRNALeu is imported into isolated T. brucei mitochondria. (A) Schematic of the organization of the T. brucei tRNASer and tRNALeu genes and the tRNA substrates transcribed in vitro with T7 RNA polymerase in the presence of [α-32P]UTP for uniform labeling. (B) A 100-μl standard import assay mixture contained 5 × 104 cpm of labeled substrate and isolated mitochondria from 3 × 108 cells. Following import, the mitochondria were collected by centrifugation and resuspended in 25 μl of SME-20 containing 5 mM CaCl2 and 100 mM KCl. Proteinase K was added to a final concentration of 30 μg/ml and the mixture was incubated for 10 min; then 1 volume of SME-20 and 4 mM PMSF were added. The mitochondria were centrifuged, and the pellet was resuspended in 0.3 M sodium acetate–0.5% SDS and phenol-chloroform extracted. The RNAs were collected by precipitation in the presence of 95% ethanol (lanes 1, 4, 7, and 10). In the remaining reactions, proteinase K digestion preceded the addition of 10 U of micrococcal nuclease followed by 10 mM EGTA, lysis of the mitochondria, phenol-chloroform extraction, and ethanol precipitation of the RNAs (lanes 2, 5, 8, and 11). Other reaction mixtures were first treated with 2% CHAPS before proteinase K and micrococcal nuclease treatment (lanes 3, 6, 9, and 12). The isolated RNAs were run on a 6% polyacrylamide–8 M urea gel and visualized by autoradiography.
The RNAs were tested for their ability to bind to, as well as be imported into, the mitochondria by incubating [α-32P]UTP-labeled RNAs with isolated mitochondria and import buffer. To test for binding, RNA substrates were incubated with the mitochondria and the mitochondria were then centrifuged, resuspended in SME-20 with CaCl2 and KCl, and treated with proteinase K. This treatment removes RNAs associated with the outer mitochondrial membrane by RNA-protein interactions that are not actively engaged in import. All four of the RNAs were able to bind the mitochondria (Fig. 1B, lanes 1, 4, 7, and 10). Apparent binding of the tandem tRNA substrate was 15 to 20% higher than that of the control RNAs, antisense rRNA, and pTE. Binding of the mature tRNALeu was 60% less than that of the tandem tRNA, suggesting that sequences outside the mature tRNA contribute to the binding of the tandem tRNA.
In order to evaluate whether the isolated mitochondria were able to import RNA, the mitochondria were treated with micrococcal nuclease following incubation with the radiolabeled substrate RNAs (Fig. 1B, lanes 2, 5, 8, and 11). In these samples, only those RNAs that were imported and therefore protected by the mitochondrial membrane would be able to escape digestion by micrococcal nuclease. Only the tandem tRNASer-tRNALeu was protected from micrococcal nuclease digestion (Fig. 1B, lane 2). In contrast, complete digestion of the mature tRNALeu, pTE, and the antisense rRNA (Fig. 1B, lanes 5, 8, and 11) shows that these RNAs are not import substrates. We have also examined the import of fully modified mature tRNA isolated from T. brucei mitochondria. These tRNAs also fail to be imported (6). Therefore, while the mature tRNALeu, pTE, and the antisense rRNA each have the ability to associate with the outer mitochondrial membrane, binding alone does not facilitate import. As a control, when import reaction mixtures were treated with CHAPS prior to micrococcal nuclease treatment to disrupt the mitochondrial membranes, all RNAs were completely digested (Fig. 1B, lanes 3, 6, 9, and 12). These results are consistent with the tandem tRNA being imported into a membrane-bound organelle.
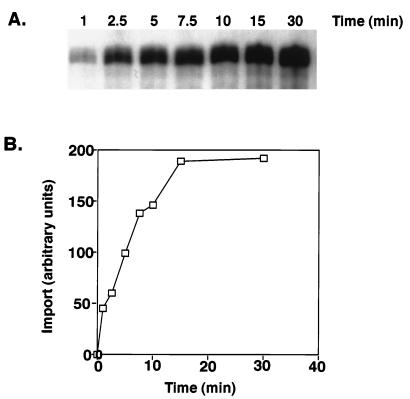
Time course of import for the tandem tRNA into isolated mitochondria.
Import of the labeled tandem tRNASer-tRNALeu is time dependent (Fig. 2A). The half-life for import of the tandem tRNASer-tRNALeu was 7 min, and import reached a maximum after 15 min, a time frame similar to that reported for protein import with this in vitro system (21). Longer incubation (60 min) resulted in a decrease in the amount of imported full-length tandem substrate. This is likely due to either degradation of imported tandem tRNASer-tRNALeu or processing within the mitochondria. The results of the time course were quantitated by PhosphorImager analysis (Fig. 2B).
FIG. 2.
Time course of import of the tandem tRNASer-tRNALeu into mitochondria. (A) Standard import reaction mixtures (100 μl) containing 5 × 104 cpm of labeled tandem substrate and isolated mitochondria from 3 × 108 cells were incubated at 26°C for the indicated time points. Samples were centrifuged at 4°C, resuspended in 25 μl of SME-20 containing 5 mM CaCl2 and 100 mM KCl, and digested with 30 μg of proteinase K per ml for 10 min at room temperature. The proteinase K was inhibited with addition of 1 volume of SME-20 and 4 mM PMSF. The reaction mixtures were incubated for 10 min at room temperature with 10 U of micrococcal nuclease, followed by the addition of 10 mM EGTA. The RNAs were precipitated and resolved on a 6% polyacrylamide–8 M urea gel and visualized by autoradiography. (B) The import level of the labeled tRNA was determined with a PhosphorImager.
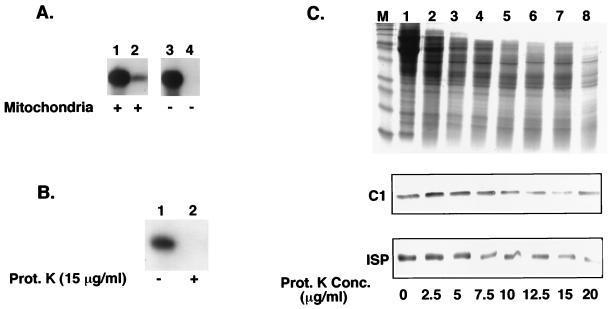
Import of the tandem tRNA requires a mitochondrial fraction and a protein component located on the exterior surface of the mitochondrion.
To confirm that the protection of the tandem tRNASer-tRNALeu from micrococcal nuclease digestion was dependent upon mitochondrial import, assays were performed in the absence of a mitochondrial fraction. A fraction of the added labeled tandem tRNASer-tRNALeu was protected from micrococcal nuclease when incubated with mitochondria (Fig. 3A, lane 2). When the mitochondrial fraction was omitted from the import reaction, the tandem tRNASer-tRNALeu was still present after incubation with proteinase K only (Fig. 3A, lane 3). However, in the absence of the mitochondrial fraction the tandem complex was completely digested by micrococcal nuclease (Fig. 3A, lane 4). This indicates that protection from micrococcal nuclease digestion results from import into mitochondria in the assay and not a conformational change of the tandem tRNA which renders it resistant to micrococcal nuclease.
FIG. 3.
Protection from micrococcal nuclease degradation is dependent on the addition of intact mitochondria. (A) Standard import reaction mixtures (100 μl) contained 5 × 104 cpm of labeled tandem tRNA substrate with or without mitochondria from 3 × 108 cells. The reaction mixtures were treated with 30 μg of proteinase K per ml and washed with 1 volume of SME-20 and 4 mM PMSF (lanes 1 and 3), followed by 10 U of micrococcal nuclease and 10 mM EGTA (lanes 2 and 4). The RNAs were collected by ethanol precipitation and run on a 6% polyacrylamide–8 M urea gel. (B) Standard import reaction mixtures (100 μl) containing isolated mitochondria from 3 × 108 cells were incubated with 15 μg of proteinase K per ml prior to incubation with substrate. The mitochondria were washed with SME-20 containing 4 mM PMSF and resuspended in import buffer, and [α-32P]UTP-labeled tandem tRNA substrate was added (lane 2). To the reaction mixtures, 30 μg of proteinase K per ml was added, followed by 1 volume of SME-20 and 4 mM PMSF, 10 U of micrococcal nuclease, and finally 10 mM EGTA. RNAs were collected by ethanol precipitation and run on a 6% polyacrylamide–8 M urea gel. (C) The mitochondria from 3 × 108 cells were pretreated as described for panel B with increasing concentrations of proteinase K. The mitochondria were washed in SME-20 and resuspended in SDS loading buffer. The Coomassie blue-stained protein profiles of the pretreated mitochondria are shown at the top. Below are the corresponding Western blots that have been probed with polyclonal antibodies to cytochrome c1 (C1) and ISP.
To determine whether a protein component associated with the outer mitochondrial membrane was essential for import, isolated mitochondria were treated with proteinase K (15 μg/ml) prior to incubation with the labeled tandem tRNASer-tRNALeu (Fig. 3B). Pretreatment of the mitochondria with proteinase K completely abolished import of the tandem tRNASer-tRNALeu (Fig. 3B, lane 2), whereas untreated mitochondria maintained import competency (Fig. 3B, lane 1). The labeled tandem tRNASer-tRNALeu from the import reactions with the proteinase K-pretreated mitochondria was recovered in the supernatant (data not shown). This result suggests the necessity for a protein factor or factors on the outer mitochondrial membrane. The same result has been previously obtained for import of the ISP by using a mitochondrial fraction prepared by the same method (21).
To characterize the extent of mitochondrial digestion by the pretreatment with proteinase K, isolated mitochondria were incubated with increasing concentrations of proteinase K. The protein profile shows a range of proteins that are hypersensitive to the protease digestion (Fig. 3C). To ensure that the inner mitochondrial membrane remained primarily intact throughout the protease treatment, the pretreated mitochondrial proteins were probed with polyclonal antibodies to two components of the cytochrome c reductase complex, cytochrome c1 and the ISP. The results show that at proteinase K concentrations as high as 20 μg/ml, the ISP and cytochrome c1 are not digested (Fig. 3C, lane 8). Since cytochrome c1 is largely exposed to the inner membrane space, these results indicate that both the outer and inner mitochondrial membranes remain intact. After treatment with CHAPS detergent, followed by proteinase K treatment at 15 μg/ml, the overall protein profile was significantly reduced and the inner membrane proteins, c1 and ISP, were no longer present (data not shown). An outer membrane protein component may play a role in maintaining the conformation of the tandem tRNASer-tRNALeu necessary for import to occur, act as a specific recognition site to present the tandem tRNASer-tRNALeu to the import machinery, or both.
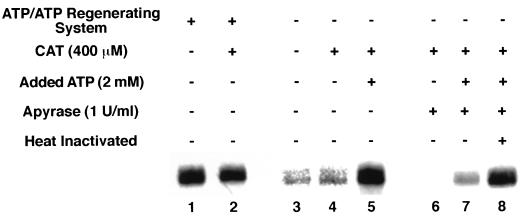
Import of the tandem tRNA requires an external pool of ATP.
To define the ATP requirements for import, mitochondria were incubated with labeled tandem tRNASer-tRNALeu and import buffer that contained (Fig. 4, lanes 1 and 2) or lacked (Fig. 4, lanes 3 to 8) ATP and an ATP regenerating system. Comparison of import levels in the presence (Fig. 4, lane 1) and absence (Fig. 4, lane 3) of ATP and a regenerating system shows a dramatic decrease in the amount of tRNASer-tRNALeu imported with the omission of ATP and the ATP regenerating system. A low level of import in lanes 3 and 4 may be due to an external pool of ATP that existed in the mitochondrial preparations.
FIG. 4.
Import of tandem tRNASer-tRNALeu into mitochondria requires an external pool of ATP. Standard import reaction mixtures (100 μl) containing isolated mitochondria from 3 × 108 cells and 5 × 104 cpm of labeled tandem substrate were incubated in the presence or absence of ATP and an ATP regenerating system. In some cases, mitochondria were preincubated with CAT and apyrase. The apyrase in lane 8 was heat inactivated at 95°C for 10 min. Some reaction mixtures received added ATP before the addition of substrates. The reaction mixtures were then treated sequentially with 30 μg of proteinase K per ml, 1 volume of SME-20, 4 mM PMSF, 10 U of micrococcal nuclease, and 10 mM EGTA. The RNAs were collected by ethanol precipitation and run on a 6% polyacrylamide–8 M urea gel.
To distinguish between an external and an internal ATP requirement, CAT was added to prevent movement of ATP into or out of the matrix. The addition of CAT (400 μM) in the presence of an ATP regenerating system does not inhibit import of tRNASer-tRNALeu (Fig. 4, lane 2) and has no effect on import levels in the absence of an ATP regenerating system (Fig. 4, lane 4). When 2 mM ATP was added to the import reaction mixtures lacking an ATP regenerating system and containing CAT, import was restored to a level comparable to that seen in the presence of a regenerating system (Fig. 4, lane 5). These results demonstrate that the addition of exogenous ATP or an ATP regenerating system enhances tRNA import. Import in the presence of CAT shows that if there is an external ATP requirement for import that it is not supplied by movement of ATP from the mitochondrial matrix by the ADP-ATP translocase.
Since a low level of import was still able to proceed in the absence of exogenously added ATP or ATP regenerating system, import was carried out in import buffer (−ATP), CAT, and potato apyrase (1 U/ml). Apyrase-catalyzed hydrolysis of ATP completely abolished import (Fig. 4, lane 6). Addition of ATP in the presence of apyrase was able to restore low levels of import (Fig. 4, lane 7). However, import was completely restored in the presence of apyrase that was heat inactivated at 95°C for 10 min and exogenously added ATP (Fig. 4, lane 8). These results clearly demonstrate an ATP requirement for import of the tandem tRNA substrate, which exists as an external pool.
Import of the tandem tRNA requires both an internal ATP pool and a membrane potential.
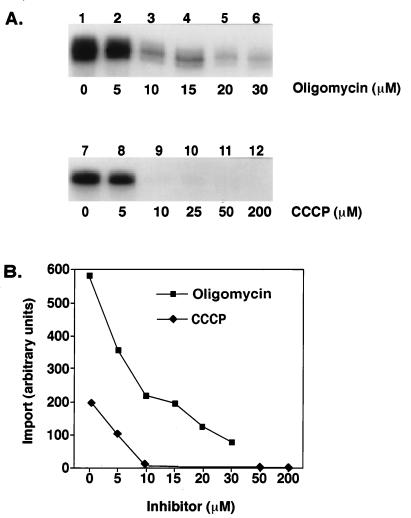
Different inhibitors were used to determine if an internal ATP pool and a membrane potential were required for RNA import. Oligomycin was used to test whether import of the tandem tRNASer-tRNALeu could be driven by an energized membrane in the absence of ATP by inhibiting the H+-translocating F1F0-ATPase. CCCP is a protonophore that uncouples oxidative phosphorylation, thereby dissipating the membrane potential by eliminating the pH gradient. Each of the inhibitors was incubated with the mitochondria and import buffer prior to addition of the labeled tandem tRNASer-tRNALeu.
At increasing concentrations of oligomycin, there was a linear decrease in the amount of import for the tandem tRNASer-tRNALeu (Fig. 5B). At 10 μM oligomycin, import was inhibited by more than 50% (Fig. 5A, lane 3). However, a low level of import was retained at oligomycin concentrations as high as 30 μM (Fig. 5A, lane 6). A 48-h exposure of the import reaction products in the presence of oligomycin was required in order to clearly visualize and quantitate (Fig. 5B) the small amount of import occurring in lanes 5 and 6. In the presence of CAT and 30 μM oligomycin, import was completely inhibited (data not shown). This suggests that import requires an internal pool of ATP.
FIG. 5.
Import of tandem tRNASer-tRNALeu into mitochondria requires an internal pool of ATP and a membrane potential. (A) Standard import reaction mixtures (100 μl) contained mitochondria from 3 × 108 cells. The mitochondria were first incubated with increasing concentrations of oligomycin and CCCP at 4°C for 10 min. Ethanol alone was added to lanes 1 and 7. Then the labeled tandem substrate at 5 × 104 cpm was added. (B) The import level of the labeled tandem substrate was determined with a PhosphorImager.
For import reactions incubated with CCCP, a concentration of 10 μM was sufficient to abolish import of the tandem tRNASer-tRNALeu (Fig. 5A, lane 9). A 16-h exposure of the import reaction products in the presence of CCCP was quantitated (Fig. 5B). A longer exposure did not show any import at the higher concentrations. The inhibitory effect of oligomycin and CCCP was not due to the chemicals being dissolved in ethanol, since the solvent by itself had only a slight effect on import (Fig. 5A, lanes 1 and 7) compared with import reactions containing no ethanol (Fig. 2A). These results are consistent with a membrane potential being required for import of the tandem tRNASer-tRNALeu into the mitochondrial matrix and with an internal ATP requirement. Import of mitochondrial precursor proteins show similar inhibitions with both CCCP and oligomycin (21).
DISCUSSION
In this paper we report the development of the first in vitro system to examine the import pathway for mitochondrial tRNAs in T. brucei. Using this system, we have shown that import occurs rapidly and is dependent upon a mitochondrial protein component. In addition, import is ATP dependent and requires a membrane potential. The results presented here also demonstrate in vitro import of a nuclearly encoded tandem tRNA into the mitochondrion of T. brucei.
Import for the tandem tRNA occurs within 5 min of incubation with isolated mitochondria (Fig. 2), which is consistent with the import time course seen for proteins (21). This is much more rapid than the import time reported for the antisense transcript containing the 5′ untranslated region and 25 nt of the β-tubulin mRNA in L. donovani, which was shown to appear after about 30 min and reach a maximum import level at 45 min (14). This difference in import rate may be due to the different mitochondrial preparation techniques used. Mitochondria isolated for the in vitro import assay in T. brucei were prepared by the nitrogen cavitation lysis protocol. The mitochondria isolated by this procedure contain an increased amount of impurities compared to mitochondria isolated by the hypotonic lysis and Percoll gradient technique (8) used for Leishmania in vitro import. However, mitochondria prepared by the nitrogen cavitation protocol have been shown to be at least fivefold more efficient at specific import of mitochondrial precursor proteins (21), which suggests that specific factors involved in import remain associated with the mitochondria isolated by nitrogen cavitation.
When isolated mitochondria were preincubated with proteinase K to digest any proteins associated with the outer mitochondrial membrane and then incubated with the labeled tandem tRNA, no import was detected (Fig. 3B). These results correspond to what has also been observed in S. cerevisiae (29) and L. donovani (14), demonstrating that there is a protein requirement at some level for import. In the yeast in vitro system, incubation of isolated mitochondria with labeled tRNAs under the conditions described for protein import did not promote introduction of the tRNAs into the mitochondrion (30). The yeast import required a 30-min preincubation of the tRNA substrate with an S-100 cytosolic fraction. We carried out import reactions in the presence of concentrated cytosol from T. brucei. The addition of the cytosol had no apparent effect on the substrate specificity or level of import (data not shown). It has also been shown that cytosolic factors are not required for in vitro mitochondrial RNA import in Leishmania (14) and mitochondrial protein import when the protein is already in an unfolded state (20, 23).
Import of a yeast cytoplasmic tRNA, in an in vitro system, requires intact protein translocating machinery (28). Partial ATP depletion of the yeast in vitro import system leads to the stabilization of a translocation intermediate containing the tRNA. Import of the tRNA could then be completed by the addition of ATP. In L. donovani, import of antisense 5′ untranslated region and 25 nt of the β-tubulin mRNA was dependent upon the addition of ATP (14). In the in vitro import assay described here, we showed that low levels of import of the tandem tRNASer-tRNALeu occur in the absence of an ATP regenerating system and additional ATP (Fig. 4). However, addition of either the regenerating system or additional ATP restores import. The import that did occur in the absence of exogenously added ATP may be from ATP already associated with the isolated mitochondria or movement of internal ATP. Import still occurs in the presence of CAT, supporting the presence of an external supply of ATP. To demonstrate this, the isolated mitochondria and import mix (−ATP) were preincubated with CAT and then apyrase to hydrolyze any pre-existing ATP, and then the labeled tandem tRNA was added. In the presence of apyrase, import was completely blocked (Fig. 4). Low and normal levels of import were seen with the addition of ATP to reaction mixtures containing apyrase and heat-inactivated apyrase, respectively.
Based on these results, it is clear that an external supply of ATP is a necessary component in the import of this tandem tRNA. It is still uncertain whether the protein translocating machinery is involved in RNA import or whether import of the tandem tRNASer-tRNALeu utilizes a separate pathway with similar requirements. It is unlikely that an RNA could be imported as a naked molecule. The net negative membrane potential across the mitochondrial membrane would have a repulsive effect on the negatively charged tRNA. However, it has been suggested that tRNAs are coimported with a carrier protein, perhaps as part of the protein import pathway (28). If this is the case, then import of RNA should also require an internal pool of ATP and a membrane potential.
In the in vitro import assay described here, import of the tandem tRNASer-tRNALeu requires both an internal supply of ATP and a membrane potential (Fig. 5). It is evident that depletion of matrix ATP in the presence of oligomycin reduces import significantly (Fig. 5A) and completely abolishes import with the addition of CAT. It is also quite clear that dissipating the membrane potential by eliminating the pH gradient across the inner mitochondrial membrane by including CCCP in the import reactions has dramatic effects upon import levels, with complete inhibition of import in the presence of 10 μM CCCP (Fig. 5A). However, results in the presence of valinomycin, a potassium ionophore that dissipates the membrane potential while leaving the pH gradient intact, appear to show a decrease in import but were inconsistent. These results were inconclusive in reference to a pH gradient requirement across the inner mitochondrial membrane, yet the matrix ATP pool and the membrane potential are necessary components of the mitochondrial tRNA import pathway in T. brucei.
The specific substrate requirements necessary for recognition of the tRNA by the import machinery remain undetermined. Results presented in this paper demonstrate the preference for a tandem tRNA as an import substrate over the mature tRNALeu and heterologous RNAs. Much of the research thus far has supported mature tRNAs as the mitochondrial import substrates. The yeast in vitro import system appears to be specific for only one nuclearly encoded mature tRNALys (30). In vitro RNA import systems developed in Leishmania imported a wide variety of RNAs, including the 5′ untranslated region and 25 nt of the β-tubulin mRNA, small rRNAs, 5S RNA, and a yeast tRNA (14, 28). The functional significance of importing these heterologous RNAs has yet to be determined.
In vivo import studies with Leishmania have compared import of a tRNATyr (GUA), tRNAIle (UAU), and tRNAGln (CUG) (12, 13, 25). The tRNAIle and tRNAGln were shown to be localized solely within the mitochondrion (25) and cytosol (12) in vivo, respectively. Exchange or deletion of the tRNAIle and tRNAGln 5′ flanking sequences had no effect on the expression or localization of the tRNAs (11). Hybridization with genomic probes was unsuccessful in detecting 5′ extension products for the tRNAIle and tRNAGln in L. tarentolae (2). This suggested that the signals for import were located within the mature tRNA. Replacing the D loop-stem from the cytoplasmic tRNAGln with that of the mitochondrial tRNAIle led to partial mitochondrial localization of the tRNAGln (11). However, D-loop replacement in the mitochondrial tRNAIle with the cytoplasmic tRNAGln still resulted in mitochondrial localization (11), suggesting the involvement of additional sequences or structural elements in the import signal.
Import of a T. brucei tRNATyr has been demonstrated in transfection studies with T. brucei (24). An episomal expression system was used to show that the 5′ and 3′ flanking sequences were not necessary for import of the tRNATyr (9). However, the system was able to import not only the tRNA from T. brucei but also cytosolic tRNAs from yeast and humans, tRNAHis and tRNALys, respectively. These heterologous tRNAs were able to be imported in the presence of T. brucei 5′ and 3′ flanking sequence, as well as in the presence of their own genomic flanking sequences. The conclusion that import of tRNAs into the mitochondria of T. brucei required no specific targeting signals was drawn.
With the in vitro import system described here for T. brucei, we have seen that in the presence of high concentrations of RNA the assay was able to detect import of nonspecific RNAs at low levels (data not shown). This may have been a result of either an overload of the import system, causing it to lose specificity, or the inability of the micrococcal nuclease to digest all of the RNA not protected by the mitochondrion. Overexpression of substrate RNAs would also be a concern in the episomal expression system, where numerous copies are being introduced within the trypanosome (24).
Our import assay is selective, in that only the tandem tRNASer-tRNALeu substrate would be imported (Fig. 1B). The tandem tRNA substrate used in determining the necessary requirements for import was previously identified in the mitochondrion of T. brucei (10). The substrate contains a tRNASer, a 59-nt intergenic region, and a tRNALeu (Fig. 1A). The RNA, pTE, contains sequence from the EMC leader and was chosen as an import substrate because when transcribed by T7 RNA polymerase, it is the same size (241 nt) as the tandem tRNASer-tRNALeu. The 116-nt antisense rRNA was chosen as an import substrate because it is approximately the same size as the mature tRNALeu. The mature tRNALeu, pTE, and the antisense rRNA substrates all failed to be imported. The efficiency of import for the tandem tRNASer-tRNALeu was typically between 5 and 10% of the amount bound to the mitochondrion.
T. brucei has an unusual genomic organization containing clusters of tRNA genes and other small RNAs that are tightly linked by intergenic sequences ranging from 58 to 210 bp (17, 18). It is possible that multiple tRNAs in T. brucei are transcribed in tandem and that their import pathways would be similar to that of the tandem tRNASer-tRNALeu substrate. While our results argue for an import pathway dependent on tandemly transcribed tRNAs, multiple import pathways could exist for other tRNAs, as has been seen in mitochondrial protein import (19). Further experiments are under way to determine the minimum sequence requirements for import and the structural characteristics of the imported RNAs.
ACKNOWLEDGMENTS
We thank Brian Adler, Karen Bertrand, Jayleen Grams, Allen J. LeBlanc, Susan Madison-Antenucci, Jeff Priest, Bob Sabatini, Lynn Sherrer, and other members of the Hajduk lab for their insight and helpful discussions.
This work was supported by National Institutes of Health grant AI21401 (to S.L.H.).
REFERENCES
- 1.Adhya S, Ghosh T, Das A, Bera S K, Mahapatra S. Role of an RNA-binding protein in import of tRNA into Leishmania mitochondria. J Biol Chem. 1997;272:21396–21402. doi: 10.1074/jbc.272.34.21396. [DOI] [PubMed] [Google Scholar]
- 2.Aphasizhev R, Karmarkar U, Simpson L. Are tRNAs imported into the mitochondria of kinetoplastid protozoa as 5′-extended precursors? Mol Biochem Parasitol. 1998;93:73–80. doi: 10.1016/s0166-6851(98)00022-x. [DOI] [PubMed] [Google Scholar]
- 3.Burger G, Plante I, Lonergan K M, Gray M W. The mitochondrial DNA of the amoeboid protozoon, Acanthamoeba castellanii: complete sequence, gene content and genome organization. J Mol Biol. 1995;245:522–537. doi: 10.1006/jmbi.1994.0043. [DOI] [PubMed] [Google Scholar]
- 4.Caterina N C, Windsor L J, Yermovsky A E, Bodden M K, Taylor K B, Birkedal-Hansen H, Engler J A. Replacement of conserved cysteines in human tissue inhibitor of metalloproteinases-1. J Biol Chem. 1997;272:32141–32149. doi: 10.1074/jbc.272.51.32141. [DOI] [PubMed] [Google Scholar]
- 5.Cunningham I. New culture medium for maintenance of tsetse tissues and growth of trypanosomatids. J Protozool. 1977;24:325–329. doi: 10.1111/j.1550-7408.1977.tb00987.x. [DOI] [PubMed] [Google Scholar]
- 6.Hancock K, Hajduk S L. The mitochondrial tRNAs of Trypanosoma brucei are nuclear encoded. J Biol Chem. 1990;265:19208–19215. [PubMed] [Google Scholar]
- 7.Hancock K, LeBlanc A J, Donze D, Hajduk S L. Identification of nuclear encoded precursor tRNAs within the mitochondrion of Trypanosoma brucei. J Biol Chem. 1992;267:23963–23971. [PubMed] [Google Scholar]
- 8.Harris M E, Moore D R, Hajduk S L. Addition of uridines to edited RNAs in trypanosome mitochondria occurs independently of transcription. J Biol Chem. 1990;265:11368–11376. [PubMed] [Google Scholar]
- 9.Hauser R, Schneider A. tRNAs are imported into mitochondria of Trypanosoma brucei independently of their genomic context and genetic origin. EMBO J. 1995;14:4212–4220. doi: 10.1002/j.1460-2075.1995.tb00095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.LeBlanc, A. J., A. E. Yermovsky-Kammerer, and S. L. Hajduk. Identification of a nuclear encoded and mitochondrial imported precursor tRNAleu in Trypanosoma brucei. J. Biol. Chem., in press. [DOI] [PubMed]
- 11.Lima B D, Simpson L. Sequence-dependent in vivo importation of tRNAs into the mitochondrion of Leishmania tarentolae. RNA. 1996;2:429–440. [PMC free article] [PubMed] [Google Scholar]
- 12.Lye L F, Chen D H, Suyama Y. Selective import of nuclear-encoded tRNAs into mitochondria of the protozoan Leishmania tarentolae. Mol Biochem Parasitol. 1993;58:233–245. doi: 10.1016/0166-6851(93)90045-y. [DOI] [PubMed] [Google Scholar]
- 13.Mahapatra S, Adhya S. Import of RNA into Leishmania mitochondria occurs through direct interaction with membrane-bound receptors. J Biol Chem. 1996;271:20432–20437. doi: 10.1074/jbc.271.34.20432. [DOI] [PubMed] [Google Scholar]
- 14.Mahapatra S, Ghosh T, Adhya S. Import of small RNAs into Leishmania mitochondria in vitro. Nucleic Acids Res. 1994;22:3381–3386. doi: 10.1093/nar/22.16.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marechal-Drouard L, Weil J H, Guillemaut P. Import of several tRNAs from the cytoplasm into the mitochondria in bean Phaseolus vulgaris. Nucleic Acids Res. 1988;16:4777–4788. doi: 10.1093/nar/16.11.4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin R P, Schneller J M, Stahl A J, Dirheimer G. Import of nuclear deoxyribonucleic acid coded lysine-accepting transfer ribonucleic acid (anticodon C-U-U) into yeast mitochondria. Biochemistry. 1979;18:4600–4605. doi: 10.1021/bi00588a021. [DOI] [PubMed] [Google Scholar]
- 17.Mottram J C, Bell S D, Nelson R G, Barry J D. tRNAs of Trypanosoma brucei. Unusual gene organization and mitochondrial importation. J Biol Chem. 1991;266:18313–18317. [PubMed] [Google Scholar]
- 18.Mottram J C, Shafi Y, Barry J D. Sequence of a tRNA gene cluster in Trypanosoma brucei. Nucleic Acids Res. 1991;19:3995. doi: 10.1093/nar/19.14.3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neupert W. Protein import into mitochondria. Annu Rev Biochem. 1997;66:863–917. doi: 10.1146/annurev.biochem.66.1.863. [DOI] [PubMed] [Google Scholar]
- 20.Pfaller R, Neupert W. High-affinity binding sites involved in the import of porin into mitochondria. EMBO J. 1987;6:2635–2642. doi: 10.1002/j.1460-2075.1987.tb02554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Priest J W, Hajduk S L. In vitro import of the Rieske iron-sulfur protein by trypanosome mitochondria. J Biol Chem. 1996;271:20060–20069. doi: 10.1074/jbc.271.33.20060. [DOI] [PubMed] [Google Scholar]
- 21a.Priest, J. W., and S. L. Hajduk. Unpublished results.
- 22.Schatz G, Dobberstein B. Common principles of protein translocation across membranes. Science. 1996;271:1519–1526. doi: 10.1126/science.271.5255.1519. [DOI] [PubMed] [Google Scholar]
- 23.Schneider A. Import of RNA into mitochondria. Trends Cell Biol. 1994;4:282–286. doi: 10.1016/0962-8924(94)90218-6. [DOI] [PubMed] [Google Scholar]
- 24.Schneider A, Martin J, Agabian N. A nuclear encoded tRNA of Trypanosoma brucei is imported into mitochondria. Mol Cell Biol. 1994;14:2317–2322. doi: 10.1128/mcb.14.4.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi X, Chen D H, Suyama Y. A nuclear tRNA gene cluster in the protozoan Leishmania tarentolae and differential distribution of nuclear-encoded tRNAs between the cytosol and mitochondria. Mol Biochem Parasitol. 1994;65:23–37. doi: 10.1016/0166-6851(94)90112-0. [DOI] [PubMed] [Google Scholar]
- 26.Simpson A M, Suyama Y, Dewes H, Campbell D A, Simpson L. Kinetoplastid mitochondria contain functional tRNAs which are encoded in nuclear DNA and also contain small minicircle and maxicircle transcripts of unknown function. Nucleic Acids Res. 1989;17:5427–5445. doi: 10.1093/nar/17.14.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suyama Y. Two dimensional polyacrylamide gel electrophoresis analysis of Tetrahymena mitochondrial tRNA. Curr Genet. 1986;10:411–420. doi: 10.1007/BF00418415. [DOI] [PubMed] [Google Scholar]
- 28.Tarassov I, Entelis N, Martin R P. An intact protein translocating machinery is required for mitochondrial import of a yeast cytoplasmic tRNA. J Mol Biol. 1995;245:315–323. doi: 10.1006/jmbi.1994.0026. [DOI] [PubMed] [Google Scholar]
- 29.Tarassov I, Entelis N, Martin R P. Mitochondrial import of a cytoplasmic lysine-tRNA in yeast is mediated by cooperation of cytoplasmic and mitochondrial lysyl-tRNA synthetases. EMBO J. 1995;14:3461–3471. doi: 10.1002/j.1460-2075.1995.tb07352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tarassov I A, Entelis N S. Mitochondrially-imported cytoplasmic tRNA(Lys)(CUU) of Saccharomyces cerevisiae: in vivo and in vitro targetting systems. Nucleic Acids Res. 1992;20:1277–1281. doi: 10.1093/nar/20.6.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]