Abstract
Rapid and accurate testing tools for SARS-CoV-2 detection are urgently needed to prevent the spreading of the virus and to take timely governmental actions. Internet of things (IoT)-based diagnostic devices would be an ideal platform for point-of-care (POC) screening of COVID-19 and ubiquitous healthcare monitoring for patients. Herein, we present an advanced IoT-based POC device for real-time direct reverse-transcription-loop mediated isothermal amplification assay to detect SARS-CoV-2. The diagnostic system is miniaturized (10 cm [height] × 9 cm [width] × 5.5 cm [length]) and lightweight (320 g), which can be operated with a portable battery and a smartphone. Once a liquid sample was loaded into an integrated microfluidic chip, a series of sample lysis, nucleic amplification, and real-time monitoring of the fluorescent signals of amplicons were automatically performed. Four reaction chambers were patterned on the chip, targeting As1e, N, E genes and a negative control, so multiple genes of SARS-CoV-2 could be simultaneously analyzed. The fluorescence intensities in each chamber were measured by a CMOS camera upon excitation with a 488 nm LED light source. The recorded data were processed by a microprocessor inside the IoT-based POC device and transferred and displayed on the wirelessly connected smartphone in real-time. The positive results could be obtained using three primer sets of SARS-CoV-2 with a limit of detection of 2 × 101 genome copies/μL, and the clinical sample of SARS-CoV-2 was successfully analyzed with high sensitivity and accuracy. Our platform could provide an advanced molecular diagnostic tool to test SARS-CoV-2 anytime and anywhere.
Keywords: Internet of things (IoT), Loop-mediated isothermal amplification (LAMP), SARS-CoV-2, Smartphone, Molecular diagnostic
1. Introduction
In the last two decades, world health organizations and many countries around the globe have been fighting tirelessly to control the spread of coronavirus diseases. The coronaviruses, the positive-sense single-strand RNA viruses with crown-like spikes on their surface, are able to transmit among humans and usually cause mild respiratory diseases (WHO, 2020). The first severe acute respiratory syndrome-Coronavirus (SARS-CoV), emerged in Guangdong, China in 2002, leads to more than 8000 confirmed cases worldwide with an approximately 10% mortality rate (WHO, 2003). In 2012, the Middle East respiratory syndrome (MERS) was first reported in Jeddah, Saudi Arabia. A beta-coronavirus (MERS-CoV) was spread over 27 countries and caused around 2500 confirmed cases and a higher mortality rate of approximately 34% (WHO, 2018; Zaki et al., 2012). In December 2019, the outbreak of another novel coronavirus (SARS-CoV-2) was reported in Wuhan, China. As of July 6, 2021, after one and a half years since the outbreak, there are over 183 million cases and almost 4.0 million deaths (WHO, 2021). While the epidemics in some of the countries have started to show signs of slowing down, many countries continue to struggle with the spread of new SARS-CoV-2 variants. Health organizations and countries are overburdened to control the pandemics by treating the increased number of SARS-CoV-2 testing, finding effective treatment therapies, developing and distributing vaccines (Taleghani and Taghipour, 2021). The development of the vaccines shows positive results with known variants, but there is no guaranty that they retain the same immune effect against upcoming variants of SARS-CoV-2. One of the keys to holding this pandemic under control may lie in developing a portable and rapid diagnostic system and isolate the infected individuals timely and properly.
The conventional detection methods for SARS-CoV-2, similar to those used for other forms of viral infectious pneumonia, are generally based on computed tomography, serology tests, and molecular diagnostics. Among these methods, the molecular diagnosis was reported as the most specific and sensitive method to detect the SARS-CoV-2 (Kubina and Dziedzic, 2020). However, the molecular test such as PCR typically requires well-resourced facilities, multiple reagents and steps, and skilled personnel (Lei et al., 2021). To overcome the drawbacks of the conventional genetic processes, the development of the POC nucleic acid testing systems has been recently focused. The World Health Organization recommends the ASSURED (Affordable, Sensitive, Specific, User-friendly, Rapid and robust, Equipment-free, and Deliverable to end-users) criteria for defining the most appropriate POC diagnostic tests for resource-limited settings (Kosack et al., 2017; Mauk et al., 2018). These ASSURED criteria have driven the recent trends in the development of the POC diagnostics, which include minimally instrumented formats, like battery-powered or electricity-free diagnostics devices, and the utilization of smartphones for remote mobile communication, and GPS locations. Such a portable device normally employs a plastic microfluidic chip or a cartridge, which is disposable, single-use, and pre-loaded with reagents. Owing to the benefits of the microfluidic technology, the integrated POC diagnostic system is capable of short testing times, reduced reagent consumption, and minimal human interferences. In addition, the combination of the IoT technology with the POCT system would enable ubiquitous healthcare monitoring between the doctors and the patients by sharing the diagnostic data stored in a cloud service. Such an IoT-based platform can analyze and store the data and be automatically synchronized with a healthcare center so that the government can catch a prompt and clear picture of pandemic situations to make better decisions accordingly. Therefore, the development of the future POC diagnostic device integrated with an IoT technology will be essential to facilitate a number of immediate tests in limited-resource environments, while maintaining the accuracy and sensitivity comparable to the conventional diagnostic methods.
On-chip molecular diagnostics are typically composed of the following sequential steps: (1) Lysis of cells or virus particles; (2) extraction of DNA or RNA from the lysate, (3) amplification of specific nucleic acid target sequences, (4) endpoint or real-time detection of the resultant amplicons by optical or electrochemical instruments (Nguyen et al., 2019b; Taleghani and Taghipour, 2021). Molecular diagnostics mostly rely on the polymerase chain reaction (PCR) for enzymatic amplification of target sequences (Park et al., 2011; Saiki et al., 1988). PCR requires relatively expensive, sophisticated, and bulky instrumentation to achieve precisely (±0.5 °C) and rapid (>10 °C/s) thermal cycling. To simplify the thermal cycling process as well as the heater system, a variety of isothermal amplification methods have been proposed. These methods not only amplify the target gene at a constant temperature but also show better tolerance for the impurities than PCR, making them more suitable for POC diagnostics (Taleghani and Taghipour, 2021). Among the isothermal amplification technologies including LAMP, recombinase polymerase amplification, helicase dependent amplification, nucleic acid sequence-based amplification, rolling circle amplification, and strand displacement amplification (Oh et al., 2016), the LAMP is one of the promising methods in terms of sensitivity and specificity. It utilizes four primers for stringent binding with a template and two sets of loop primers to boost up the amplification efficiency (Inaba et al., 2021; Khan et al., 2018; Singh et al., 2019). The formation of the air bubble and the evaporation issue is avoided due to mild reaction temperatures (63–65 °C) (Dan Van et al., 2019; Lee et al., 2016; Mauk et al., 2018; Nguyen et al., 2020), and Bst polymerase in the LAMP reaction has a higher tolerance for inhibitors that are commonly found in clinical specimens (Francois et al., 2011; Kaneko et al., 2007; Nkouawa et al., 2010). In addition, the “direct” LAMP reaction was reported in which the cell lysis and the genetic amplification occurred simultaneously without DNA/RNA purification (Dudley et al., 2020; Gadkar et al., 2018; Lee et al., 2016). Thus, the combination of the direct lysis buffer and the LAMP reaction allows us to set up a simplified automated POC molecular diagnostic system. Recent reports for the COVID-19 diagnostics using a POCT platform were commonly based on the isothermal amplification methods, because the integration of qPCR into a POCT system is somewhat challenging due to the need for high-purity of nucleic acids and precise thermal control (Song et al., 2021). For example, Broughton et al. and Pang et al. presented a POC testing method with RT-LAMP reaction to rapidly detect the virus in an extracted RNA samples with the assist of a CRISPR-Cas system (Broughton et al., 2020; Pang et al., 2020). Ramachandrana et al. reported an electric field-driven microfluidic chip for rapid LAMP-and CRISPR-based SARS-COV-2 diagnostics (Ramachandran et al., 2020). However, they mainly are endpoint detection and depended on the prior RNA extraction by a conventional method or needed a separate module for the RNA extraction, which causes prolonged and complicates the process for the POC COVID-19 testing.
In this study, we developed a rapid SARS-CoV-2 diagnostic platform to confront the COVID-19 pandemic. We proposed an advanced IoT-based POC device for direct real-time RT-LAMP reaction to detect SARS-CoV-2, which holds several advantages of portability, connectivity, user-friendliness, and low cost compared with previous reports (G Soares et al., 2021; Nguyen et al., 2019a; Shen et al., 2019). Powered by a portable battery, our device can perform the viral lysis and the RT-LAMP reaction on an integrated microfluidic chip. During the reaction, the fluorescent signals of each reaction chamber were simultaneously and automatically measured, processed, and transmitted to a smartphone via a Wi-Fi network in real-time. Three target genes (As1e, N, and E genes) of SARS-CoV-2 were analyzed to impose a stringent standard for decision by preventing false-positive results. Clinical samples of SARS-CoV-2 were also analyzed with other respiratory viruses (Influenza A, RSV A, and RSV B) to demonstrate the practicability of our proposed molecular diagnostic platform in the fields.
2. Materials and methods
2.1. Chemicals and reagents
An EzWay Direct PCR lysis buffer (5×) was purchased from Komabiotech (Cat: KBT-K0568001, Korea), and a LAMP Reaction Mix (RM) and an Enzyme Mix (EM) which contains Bst DNA Polymerase with reverse transcriptase enzymes were purchased from Eiken Genome (Japan). A 10 × SYBR dye was ordered from Invitrogen (USA), and the LAMP primers were synthesized by Macrogen (Korea). Heat-inactivated SARS-CoV-2 was obtained from ATCC (VR-1986HK™, Lot # 70036071, USA). Three mm-thick PMMA sheets were ordered from Acrytal (Korea), and a pressure-sensitive adhesive (PSA) film was purchased from HJ-Bioanalytik GmbH (Cat. 900360, Germany). Paraffin wax with a melting point of 58–62 °C was obtained from Sigma (ASTM D 87, USA). The super-hydrophobic reagent was delivered from Ultratech (USA).
2.2. A portable POC genetic analyzer
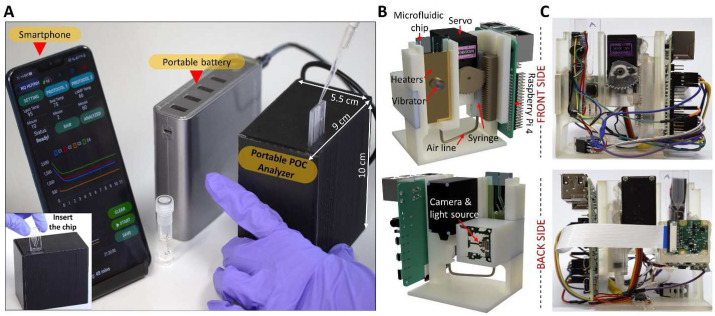
A POC genetic analyzer (w × h × l: 9 cm × 10 cm × 5.5 cm, weight: 320 g) powered by a portable battery was constructed together with a microfluidic chip to detect COVID-19 (a black box in Fig. 1 A). The scaffold of the portable genetic analyzer was designed with 3D CAD design software (Autodesk® Fusion 360, USA) (Fig. 1B) and manufactured using an SLA 3D printer (Form 3, Form lab, USA) with a precision of 0.1 mm (Fig. 1C). This platform contains four functional elements for (1) fluid control, (2) temperature control, (3) vibration, and (4) real-time fluorescence detection. These four subsystems are managed by a Raspberry Pi 4, and the electrical connection between the subsystems is shown in Fig. S1A. The fluid control subsystem includes a high torque servo (TowerPro, MG996R) that operates a set of gears to pull or push a 3 mL syringe. For the aspiration of the syringe, the servo turns counterclockwise, while the servo turns clockwise for exhaustion. When an integrated microfluidic chip is inserted into the portable genetic analyzer (Inset of Fig. 1A), the bottom of the chip is automatically clicked with the adapter which is connected with a syringe via a silicon tube (2 mm in diameter). Thus, the transfer of the solution inside the microfluidic chip can be manipulated by pushing or pulling the syringe. A subsystem for temperature control consists of three independent heaters, each of which is designed to tune the temperature of three zones of the chip (a lysis chamber, wax valves, and reaction chambers) (Fig. S1B). These customized heaters were fabricated in a 1.6 mm-thick copper clad laminate sheet (the thickness of copper foil: 18 μm) by a CNC machine with a 0.3 mm end mill. The fabricated heater for the lysis, the wax sealing, and the LAMP reaction has a resistance of 2.4, 1.6, 2.4 Ω, respectively. A subsystem for the viral lysis includes a small vibration motor (dia. 2.7 mm, 3.5 V, 10000 RPM) on the back of the lysis heater, which is vibrating the sample solution during the lysis step to accelerate the lysis efficiency of virus particles (Fig. S1C). A subsystem for real-time fluorescence detection consists of a high-intensity (60 mW) light-emitting diode (LED) with an excitation wavelength at 488 nm (488T-60 DS5, China) and a Sony IMX219 8-megapixel CMOS sensor. The LED is placed at 3 cm away from the microfluidic chip surface with a tilted angle of 45° to illuminate the reaction chambers. A CMOS sensor covered by a bandpass filter (525 nm CWL, Dia. 25 mm, 70 nm bandwidth, Edmund Optics, USA) is placed at 3.5 cm away from the front side of the microfluidic chip to capture the emitted light from the reaction chambers (Fig. S1C).
Fig. 1.
A POC genetic analyzer for on-site molecular diagnostics. (A) The portable genetic analyzer equipped with an integrated microchip is powered by a portable battery and controlled by a smartphone via Wi-Fi connection. (B) The 3D model of the inner structure of the POC genetic analyzer, and (C) the digital images of the front and back sides of the manufactured POC genetic analyzer.
2.3. Fabrication of the microfluidic chip
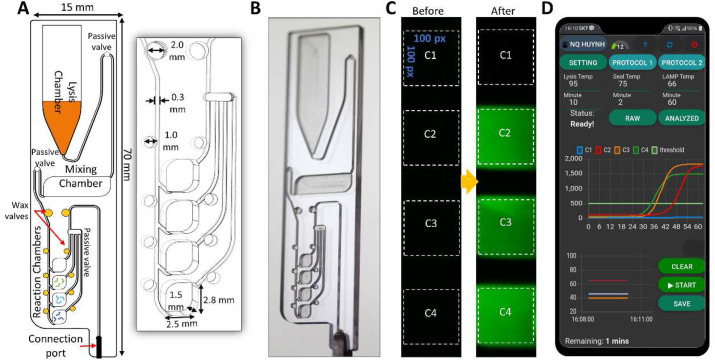
As illustrated in Fig. 2 A, the disposable microfluidic chip (w × l × h: 15 mm × 70 mm × 3 mm, weight: 3 g) consists of a lysis chamber, a mixing chamber, four reaction chambers, and ten wax valves. It was designed using Cut2D software (Vectric, USA) and etched in a 3.0 mm thick PMMA sheet by a CNC machine (Tinyrobo30, Korea). The dimension of one reaction chamber was 2.5 mm [width] × 2.8 mm [length] × 1.5 mm [height] and had a volume of 10 μL. Microfluidic circuits such as microchannels, chambers, and manifolds were patterned on one side and sealed by a PSA film. The fabricated chips were cleaned with a sonicator in a detergent solution, following by 70% ethanol and DNase-free water. Fig. 2B shows a real digital image of the integrated chip. Three passive valves in Fig. 2A were coated with a super-hydrophobic reagent by dropping 3 μL on each valve. Paraffin wax with a melting point of 58–62 °C was placed in the wax valves (Fig. 2A, yellow dots). 1.2 μL of Bst DNA polymerase and primers were freeze-dried in the mixing chamber and the reaction chambers using a Concentrator plus (Eppendorf, Germany), respectively.
Fig. 2.
The integrated microfluidic chip. (A) (Left) Schematic of the microfluidic chip layout, which consists of a lysis chamber, a mixing chamber, reaction chambers, passive valves, and wax valves. (Right) A enlarged view of the reaction chambers and the wax valves with the information of dimensions. (B) A digital image of the fabricated microfluidic chip. (C) The fluorescence images of the reaction chambers before and after the RT-LAMP reaction. Chamber C2, C3, C4 were pre-dried with the SARS-CoV-2 specific primers, while Chamber C1 is a negative control. (D) The real-time fluorescence signal is transmitted and displayed on a smartphone together with the temperature profiles of each heater.
2.4. Structure of an IoT unit and the fluorescent measurement on a chip
A Raspberry Pi 4, a small single-board computer developed by the Raspberry Pi Foundation, was used as a central processing unit of the POC device. The software was written in JavaScript and Node-Red programming tool to control the input and output pins of the Raspberry Pi 4. Node-RED is a free, open-source logic engine for controlling a single IoT device as well as the network of multiple IoT devices that allows us to build a cloud-based system. The operation of the POC genetic analyzer is wirelessly manipulated by a smartphone via a Wi-Fi connection and was powered with a 5 V- 4 A (20 W) charger or battery (Fig. 1A). The temperatures of the heaters were separately monitored using three 1-wire digital temperature sensors (DS18S20) and controlled with proportional integral derivative algorithms and three MOSFETs (IRLB3813) (Fig. S1A).
Fluorescence images of the reaction chambers were captured every minute during the RT-LAMP reaction by the CMOS sensor with an excitation of 488 nm-LED (Fig. 2C). The fluorescent signal was then calculated as the sum of green intensities in the dotted region of each reaction chamber (100 pixels × 100 pixels). The RT-LAMP amplification curve was fitted with a 5-parameter log-logistic curve and was displayed on a smartphone in real-time (Fig. 2D).
2.5. Preparation of the RT-LAMP reaction
The LAMP primer sets (F3, B3, FIP, BIP, LF, and LB) for targeting three genes of SARS-CoV-2 were selected from the previous reports (Dudley et al., 2020; Zhang et al., 2020). Information of the primers and target sequences is shown in Table S1. For the on-chip experiment, 58 μL of the RT-LAMP solution including 12 μL of an EzWay Direct PCR lysis buffer (5 × ), 26.4 μL of the LAMP RM, 2.4 μL of 10 × SYBR green and 17.2 μL of nuclease-free water were prepared and injected into the lysis chamber of the chip. The microfluidic chip was stored at −20 °C prior to use in order to preserve the quality of the dried enzymes. When the genetic analysis of a sample started on the POC system, 2 μL of a virus sample was loaded directly into the lysis chamber, and then the viral lysis, the enzyme mixing, the RT-LAMP reaction, and the optical detection were sequentially executed.
The RT-LAMP reaction in a 200 μL tube was also carried out as a reference using a commercial qPCR machine. The volume of the RT-LAMP mixture was 10 μL containing 2 μL of EzWay Direct PCR lysis buffer, 4.4 μL of the LAMP RM, 0.4 μL of 10 × SYBR green, 2.4 μL of the primer mixture (Table S1), 0.26 μL of nuclease-free water, and 0.34 μL of a virus sample, and was incubated at 95 °C for 15 min to lyse the viral particles. Then, 0.2 μL of an EM was added to the RT-LAMP mixture. One-step RT-LAMP reaction proceeded at 65 °C for 1 h. The fluorescence intensity of the RT-LAMP mixture was measured at an interval of 1 min.
2.6. Clinical sample test
The clinical samples were obtained from Kyung Hee University Hospital (Gangdong, Seoul). The nasopharyngeal swab samples from the patients infected by SARS-CoV-2, Influenza A, RSV A, and RSV B were suspended in 3 mL Viral Smart Transport Medium (MedSchenker™) and stored at −80 °C. We treated all the clinical samples in accordance with the Institutional Review Board requirements of Kyung Hee University.
3. Results and discussion
3.1. Microfluidic chip operation
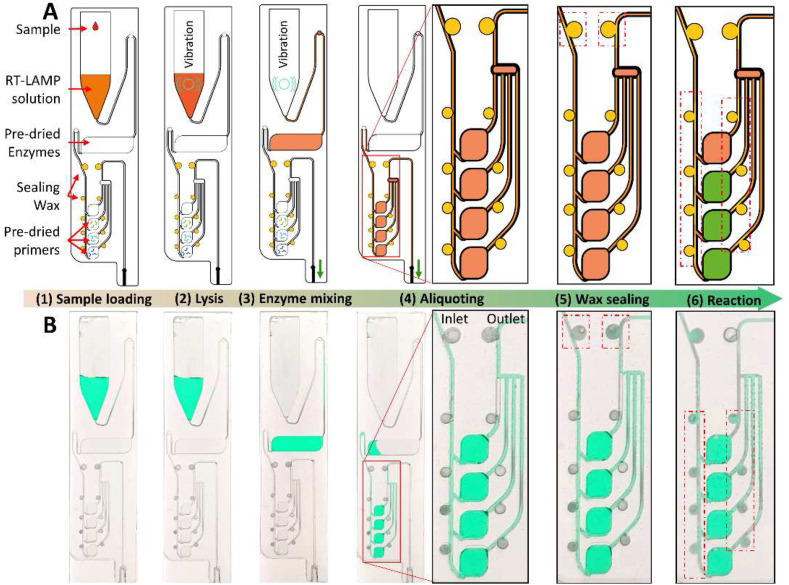
The combination of the integrated microfluidic chip with the POC device is capable of SARS-CoV-2 diagnostics in a sample-to-answer-out manner. When the microfluidic chip is inserted on the top of the POC device, the syringe is linked to the chip via the connection port (Fig. 2A). The microfluidic chip contains a pre-loaded RT-LAMP solution in the lysis chamber, freeze-dried enzymes in the mixing chamber, sealing wax in the wax valves, and freeze-dried primers in the designated reaction chamber. A typical workflow and the control program of the chip are shown in Fig. 3 A, and it consists of 6 steps. (1) A sample loading step: 2.0 μL of the viral sample is loaded into the lysis chamber of the microfluidic chip, in which 58 μL of the RT-LAMP solution was preloaded. (2) A viral lysis step: the lysis heater is heating up to 95 °C for 10 min, and the vibrator shakes the solution for 30 s every minute. (3) An enzyme mixing step: the syringe is aspirated (70 μL) to transfer the lysed sample (60 μL) into the mixing chamber, where the EM (Bst DNA polymerase and reverse transcriptase) is pre-dried, and the vibrator turns on for 30 s for the efficient mixing between the EM and the RT-LAMP solution. (4) An aliquoting step: After the enzyme mixing step, the RT-LAMP mixture (60 μL) is slowly aspirated into the aliquoting structure. The last passive valve at the end of the aliquoting structure was coated with a hydrophobic reagent, so the RT-LAMP mixture could fill in all the reaction chambers gently. (5) A wax sealing step: After the reaction chambers are filled with the RT-LAMP mixture, the temperature of the wax sealing heater is ramping up to 75 °C for 2 min. The wax in the two upper wax valves is melted and flows into the connected microchannel to block the inlet and the outlet of the aliquoting structure. Step (5) in Fig. 3B (red dotted boxes) shows the release of wax into the adjacent microchannel and the disconnection of the green solution in the microchannels due to the wax. The wax sealing process is important to prevent the RT-LAMP mixture from being evaporated during the RT-LAMP reaction. (6) An RT-LAMP reaction step: The temperature of the reaction heater increases, and the mixture is then incubated at 65 °C for 60 min for the one-step RT-LAMP reaction. At this temperature, the wax inside the eight wax valves beside the reaction chambers is melted and flowed in the microchannels, resulting in the isolation of the reaction chambers. These wax valves not only help to block the evaporation but also prevent cross-contamination between the reaction chambers. During the incubation time, the fluorescence images of each reaction chamber are recorded every minute. The heating profiles of the lysis heater, the wax sealing heater, and the RT-LAMP reaction heater are shown in Fig. S2.
Fig. 3.
Illustration of the microfluidic chip operation. (A) The sketch and (B) the digital images of the microfluidic chip illustrate the operation of a molecular diagnosis on a chip, starting from (1) the sample loading into the preloaded RT-LAMP solution, (2) the viral lysis, (3) the mixing with enzymes, (4) the aliquoting of the RT-LAMP mixture, (5) the wax sealing, and (6) the RT-LAMP reaction.
3.2. Efficient thermal lysis of virus particles with vibration
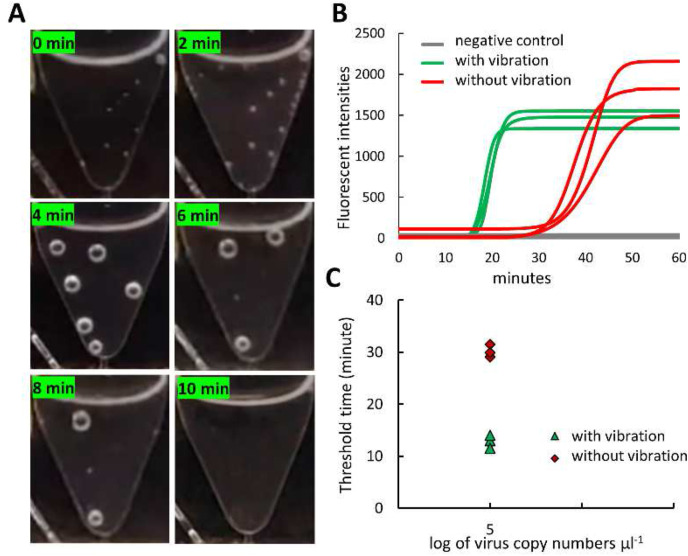
According to the manufacture protocol, an EzWay Direct PCR lysis buffer requires a high temperature (95 °C) for 15 min to lyse the virus particles. For rapid on-chip viral detection, we reduced the lysis time from 15 min to 10 min and enhanced the lysis efficiency with a high-frequency vibrator (10000 RPM) for 30 s every 1 min. A sample of SARS-CoV-2 with a concentration of 2 × 105 copies/μL was used to validate our modified lysis protocol with the As1e primer set. As shown in Fig. 4 A, the vibration was useful to remove the air bubbles that were generated inside the lysis chamber during the heating. The vibration of the lysis chamber in addition to the heating also significantly increased the on-chip lysis efficiency. The sample lysed with the vibration was amplified faster than that without the vibration (Fig. 4B). The threshold time was dramatically reduced from 30.2 min to 12.8 min, demonstrating the excellent on-chip lysis performance by combining the heating and the vibration function (Fig. 4C).
Fig. 4.
On-chip lysis process. (A) The digital images of the lysis chamber throughout the lysis step with the assistance of a vibrator, showing the removal of the generated air bubbles. (B) The amplification curve of the lysate sample with and without the vibration. (C) Threshold time of the lysis sample with and without the vibration, demonstrating the high efficiency of lysis owing to the vibration mode.
3.3. Cross-contamination test
Since our chip design contains the four reaction chambers for multiplex analysis of target genes, it is necessary to confirm no cross-contamination in the RT-LAMP reaction. To do this, we employed the wax valves surrounding the reaction chambers, and the melted wax was filled in the microchannel to isolate the reaction chambers at the temperature of 65 °C. To ascertain whether cross-contamination occurs between the adjacent reaction chambers, the primer sets of the As1e gene for SARS-CoV-2 were pre-dried in the reaction chamber C1 and C3, or C2 and C4 (Fig. 2C). Then, a sample of SARS-CoV-2 with the concentration of 2 × 102 copies/μL was added and the whole process was performed on the microfluidic chip. During the LAMP reaction, the fluorescence intensities of all the reaction chambers were monitored by the CMOS sensor. As shown in Fig. S3, the fluorescence intensity gradually increased only in the reaction chamber C1 and C3, where the As1e primers were pre-dried (Fig. S3A). Similarly, only the reaction chamber C2 and C4 show the amplification profiles, in which the As1e primers were pre-dried only in the chamber C2 and C4 (Fig. S3B). Thus, the incorporation of the wax valves on a chip eliminates the cross-contamination issue, resulting in reliable data for multiplex target gene detection. In addition, we often put our miniaturized POCT system in a clean bench equipped with a UV light for sterilization to eliminate a cross-contamination issue between the microfluidic chip and the POC device.
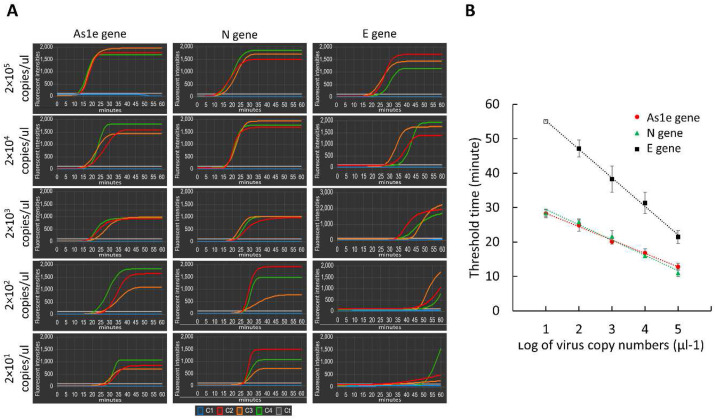
3.4. Limit of detection test
The low limit of detection (LOD) is important in the diagnostic methods, considering that a few virus particles of SARS-CoV-2 could infect the respiratory tract of a person. Thus, the evaluation of the detection sensitivity of our proposed platform is necessary. For this purpose, the heat-inactivated SARS-CoV-2 sample was serially diluted to prepare a variety of concentrations: 2 × 105, 2 × 104, 2 × 103, 2 × 102, and 2 × 101 genomic copy numbers/μL, and the LOD test was carried out. The amplification curves of the three genes of SARS-CoV-2 were plotted in Fig. 5 A depending on the concentrations. Most of the genes of SARS-CoV-2 samples were successfully detected between 13 min and 51 min of the reaction time. While the amplification efficiency of the As1e and N genes looks similar, that of the E gene is slower. At the lowest concentration of 2 × 101 copies/μL, the amplification of the E gene was not reproducible. We set the threshold value of fluorescent intensity as 100 (white lines), and accordingly, determined the threshold time. The threshold times of the three genes are plotted in Fig. 5B. All experiments were repeated at least three times, and the calculated standard deviation was expressed by error bars. The threshold time of the E gene was much larger than that of As1e and N genes, while the threshold times of As1e and N genes were quite equivalent. As shown in the blue line in Fig. 5A, the negative control experiments show no fluorescence signals all the time. Thus, if we want to identify SARS-CoV-2 using three genes (As1e, N, and E genes) with high confidence, the LOD was 2 × 102 copies/μL. However, if we employ two genes (As1e and N gene), the LOD is lower by 10-fold, namely 2 × 101 copies/μL, which is equivalent to 10 copies/reaction chamber. Recent reports show that the detectable viral load of clinical SARS-CoV-2 obtained from nasopharyngeal swabs ranged from 101.8 – 107.8 copies/mL (Fajnzylber et al., 2020). In this case, RNAs were extracted using ultra-high-speed centrifugation (21,000 g) and concentrated by a small volume of elution solution, and then a standard qPCR was performed. Another report presented the detectable range of 104–1.14 × 1010 copies/mL (Dudley et al., 2020). These data were also obtained by an extraction-based method and a qPCR on a benchtop thermocycler. Although our LOD value was relatively higher than that of the conventional method, it was quite comparable to those produced by other POC systems with a direct RT-LAMP reaction (Table S2). Considering that exponential growth of virus occurs within 1–2 days after viral infection during which the viral load rise to 1000-fold higher than the LOD of qPCR (105–107 copies/mL) (Flynn et al., 2020; Larremore et al., 2021), our proposed system can detect virus spikes by performing the COVID-19 surveillance testing.
Fig. 5.
On-chip LOD test for SARS-CoV-2. (A) Amplification profiles of three genes (As1e, N, and E genes) of SARS-CoV-2 depending on the viral concentration. The blue, red, orange, and green color, representing the amplification curve in the chamber C1 (negative control), C2, C3, and C4, respectively. (B) The correspondent threshold times for three genes of SARS-CoV-2. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
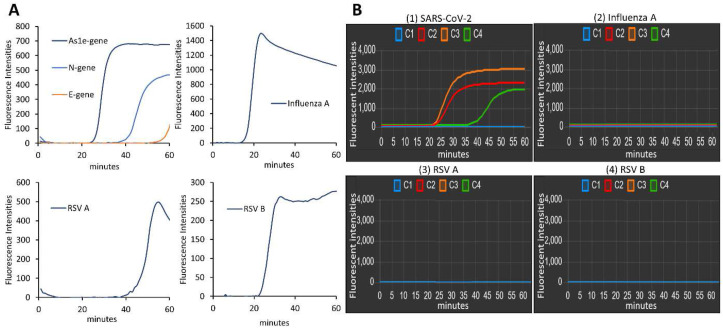
3.5. Clinical sample testing
To demonstrate the real applicability of our POC molecular diagnostic platform for analyzing clinical samples, we obtained the four clinical respiratory virus samples (SARS-CoV-2, Influenza A, RSV A, and RSV B) from four patients and tested them. First, the clinical samples were analyzed using a commercial real-time PCR machine, and target genes of each virus were amplified with their specific primer sets: As1e, N, and E genes for SARS-CoV-2, Neuraminidase gene for Influenza A, fusion glycoprotein gene for RSV A and RSV B (Fig. 6 A) (Table S1). The results show the positivity of all four clinical samples (Fig. 6A). Then, we coated the reaction chamber C2, C3, and C4 with the primer sets targeting As1e, N, and E gene, respectively, while the chamber C1 contained no primers as a negative control to ensure that there was no cross-contamination between the reaction chambers, and between the runs. The amplification curves of the three reaction chambers (C2, C3, and C4) were only detectable when the clinical sample of SARS-CoV-2 was loaded (Fig. 6B (1)). Other respiratory viruses (Fig. 6B (2) for Influenza A, Fig. 6B (3) for RSV A, Fig. 6B (4) for RSV B) produced no fluorescence signal. The entire process was completed within 60 min. These results indicate that the RT-LAMP primers for SARS-CoV-2 were designed with high specificity, and the portable genetic analyzer could accurately and simultaneously detect clinical samples of SARS-CoV-2 with high fidelity. We furthermore analyzed the clinical samples of Influenza A (2 samples), RSV A (2 samples), and RSV B (2 samples) from six patients. As shown in Fig. S4, each virus was identified reproducibly from the triplicated reactions. Thus, our POCT system can be expanded for a variety of respiratory virus testing.
Fig. 6.
Diagnosis of clinical respiratory virus samples. (A) The amplification profiles show the positivity of each virus that have been tested using a commercial qPCR machine. (B) Clinical sample analysis on our platform. The amplification profiles were shown when the viral sample of (1) SARS-CoV-2, (2) Influenza A, (3) RSV A, and (4) RSV B was injected.
4. Conclusion
In summary, we developed a palm-sized and IoT-based genetic analyzer for POC molecular diagnostics of COVID-19, while fulfilling the ASSURED criteria (Table S3). Our platform needs low power consumption, so it can be operated with a portable battery. The fluidic control, temperature control, vibrator function, and data display for RT-LAMP can be manipulated by a smartphone. The whole processes such as the viral lysis, the direct RT-LAMP reaction, and the real-time fluorescence detection could be automatically performed on a microfluidic chip. We could simultaneously analyze multiplex genes of SARS-CoV-2 to improve the accuracy of the judgment with low LOD. Respiratory clinical samples were successfully evaluated to demonstrate the practicality of our platform in the fields. Thus, we believe that our advanced diagnostic system can be applied for on-site COVID-19 screening anytime, anywhere, and by anyone.
CRediT authorship contribution statement
Huynh Quoc Nguyen: Conceptualization, Methodology, Software, Validation, Formal analysis, Writing – original draft, Visualization. Hoang Khang Bui: Conceptualization, Methodology. Vu Minh Phan: Investigation, Formal analysis. Tae Seok Seo: Conceptualization, Methodology, Validation, Resources, Writing – review & editing, Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the Engineering Research Center of Excellence Program of Korea Ministry of Science, ICT & Future Planning (MSIP)/National Research Foundation of Korea (NRF) (2021R1A5A6002853) and Korea Health Technology R&D Project/Korea Health Industry Development Institute (KHIDI), Ministry of Health & Welfare of South Korea (HI20C0644). We were grateful to Kyung Hee University Hospital for the donation of the clinical samples.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2021.113655.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C.-Y., Guevara H., Wadford D.A., Chen J.S., Chiu C.Y. CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020;38(387):870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan Van N., Van Hau N., Seok Seo T. Quantification of colorimetric loop-mediated isothermal amplification process. BioChip J. 2019;13:158–164. [Google Scholar]
- Dudley D.M., Newman C.M., Weiler A.M., Ramuta M.D., Shortreed C.G., Heffron A.S., Accola M.A., Rehrauer W.M., Friedrich T.C., O'Connor D.H. Optimizing direct RT-LAMP to detect transmissible SARS-CoV-2 from primary nasopharyngeal swab samples. PLoS One. 2020;15 doi: 10.1371/journal.pone.0244882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajnzylber J., Regan J., Coxen K., Corry H., Wong C., Rosenthal A., Worrall D., Giguel F., Piechocka-Trocha A., Atyeo C., Fischinger S., Chan A., Flaherty K.T., Hall K., Dougan M., Ryan E.T., Gillespie E., Chishti R., Li Y., Jilg N., Hanidziar D., Baron R.M., Baden L., Tsibris A.M., Armstrong K.A., Kuritzkes D.R., Alter G., Walker B.D., Yu X., Li J.Z. Vol. 11. Nat Commun; 2020. SARS-CoV-2 Viral Load is Associated with Increased Disease Severity and Mortality. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn M.J., Snitser O., Flynn J., Green S., Yelin I., Szwarcwort-Cohen M., Kishony R., Elowitz M.B. medRxiv; 2020. A Simple Direct RT-LAMP SARS-CoV-2 Saliva Diagnostic. [Google Scholar]
- Francois P., Tangomo M., Hibbs J., Bonetti E.J., Boehme C.C., Notomi T., Perkins M.D., Schrenzel J. Robustness of a loop-mediated isothermal amplification reaction for diagnostic applications. FEMS Immunol. Med. Microbiol. 2011;62:41–48. doi: 10.1111/j.1574-695X.2011.00785.x. [DOI] [PubMed] [Google Scholar]
- G Soares R.R., Akhtar A.S., Pinto I.F., Lapins N., Barrett D., Sandh G., Yin X., Vicent Pelechano bde. Vol. 21. Lab Chip; 2021. pp. 2932–2944. (Lab on a Chip Sample-to-answer COVID-19 Nucleic Acid Testing using a Low-cost Centrifugal Microfluidic Platform with Bead-based Signal Enhancement and Smartphone Read-out †). [DOI] [PubMed] [Google Scholar]
- Gadkar V.J., Goldfarb D.M., Gantt S., Tilley P.A.G. Real-time detection and monitoring of loop mediated amplification (LAMP) reaction using self-quenching and de-quenching fluorogenic probes. Sci. Rep. 2018;8:5548. doi: 10.1038/s41598-018-23930-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba M., Higashimoto Y., Toyama Y., Horiguchi T., Hibino M., Iwata M., Imaizumi K., Doi Y. Diagnostic accuracy of LAMP versus PCR over the course of SARS-CoV-2 infection. Int. J. Infect. Dis. 2021;107:195–200. doi: 10.1016/j.ijid.2021.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko H., Kawana T., Fukushima E., Suzutani T. Tolerance of loop-mediated isothermal amplification to a culture medium and biological substances. J. Biochem. Biophys. Methods. 2007;70:499–501. doi: 10.1016/j.jbbm.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Khan M., Wang R., Li B., Liu P., Weng Q., Chen Q. Comparative evaluation of the LAMP assay and PCR-based assays for the rapid detection of Alternaria solani. Front. Microbiol. 2018;9:2089. doi: 10.3389/fmicb.2018.02089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosack C.S., Page A.-L., Klatser P.R. A guide to aid the selection of diagnostic tests. Bull. World Health Organ. 2017;95:639–645. doi: 10.2471/BLT.16.187468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubina R., Dziedzic A. Molecular and serological tests for COVID-19. A comparative review of SARS-CoV-2 coronavirus laboratory and point-of-care diagnostics. Diagnostics. 2020;10(6) doi: 10.3390/diagnostics10060434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larremore D.B., Wilder B., Lester E., Shehata S., Burke J.M., Hay J.A., Tambe M., Mina M.J., Parker R. Vol. 7. Science Advances; 2021. Test Sensitivity is Secondary to Frequency and Turnaround Time for COVID-19 Surveillance. eabd5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D., Kim Y.T., Lee J.W., Kim D.H., Seo T.S. An integrated direct loop-mediated isothermal amplification microdevice incorporated with an immunochromatographic strip for bacteria detection in human whole blood and milk without a sample preparation step. Biosens. Bioelectron. 2016;79:273–279. doi: 10.1016/j.bios.2015.12.044. [DOI] [PubMed] [Google Scholar]
- Lei Z., Haixia L., Junli Z., Kang L. Different methods of COVID-19 detection. Health Sci. J. 2021;(3):001. [Google Scholar]
- Mauk M.G., Song J., Liu C., Bau H.H. Simple approaches to minimally-instrumented, microfluidic-based point-of-care nucleic acid amplification tests. Biosensors. 2018;8(1):17. doi: 10.3390/bios8010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H.Q., Nguyen V.D., Van Nguyen H., Seo T.S. Quantification of colorimetric isothermal amplification on the smartphone and its open-source app for point-of-care pathogen detection. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-72095-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H. Van, Nguyen V.D., Lee E.Y., Seo T.S. Point-of-care genetic analysis for multiplex pathogenic bacteria on a fully integrated centrifugal microdevice with a large-volume sample. Biosens. Bioelectron. 2019;136:132–139. doi: 10.1016/j.bios.2019.04.035. [DOI] [PubMed] [Google Scholar]
- Nguyen H. Van, Nguyen V.D., Nguyen H.Q., Chau T.H.T., Lee E.Y., Seo T.S. Nucleic acid diagnostics on the total integrated lab-on-a-disc for point-of-care testing. Biosens. Bioelectron. 2019;141:111466. doi: 10.1016/j.bios.2019.111466. [DOI] [PubMed] [Google Scholar]
- Nkouawa A., Sako Y., Li T., Chen X., Wandra T., Swastika I.K., Nakao M., Yanagida T., Nakaya K., Qiu D., Ito A. Evaluation of a loop-mediated isothermal amplification method using fecal specimens for differential detection of Taenia species from humans. J. Clin. Microbiol. 2010;48:3350–3352. doi: 10.1128/JCM.00697-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S.J., Park B.H., Choi G., Seo J.H., Jung J.H., Choi J.S., Kim D.H., Seo T.S. Fully automated and colorimetric foodborne pathogen detection on an integrated centrifugal microfluidic device. Lab Chip. 2016;16:1917–1926. doi: 10.1039/c6lc00326e. [DOI] [PubMed] [Google Scholar]
- Pang B., Xu J., Liu Y., Peng H., Feng W., Cao Y., Wu J., Xiao H., Pabbaraju K., Tipples G., Joyce M.A., Saffran H.A., Tyrrell D.L., Zhang H., Le X.C. Isothermal amplification and ambient visualization in a single tube for the detection of SARS-CoV-2 using loop-mediated amplification and CRISPR technology. Anal. Chem. 2020;92:16204–16212. doi: 10.1021/acs.analchem.0c04047. [DOI] [PubMed] [Google Scholar]
- Park S., Zhang Y., Lin S., Wang T.H., Yang S. Advances in microfluidic PCR for point-of-care infectious disease diagnostics. Biotechnol. Adv. 2011;29(6):830–839. doi: 10.1016/j.biotechadv.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran A., Huyke D.A., Sharma E., Sahoo M.K., Huang C., Banaei N., Pinsky B.A., Santiago J.G. Electric field-driven microfluidics for rapid CRISPR-based diagnostics and its application to detection of SARS-CoV-2. Proc. Natl. Acad. Sci. Unit. States Am. 2020;117:29518–29525. doi: 10.1073/pnas.2010254117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R., Gelfand D., Stoffel S., Scharf S., Higuchi R., Horn G., Mullis K., Erlich H. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science (80-. ) 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Shen K.M., Sabbavarapu N.M., Fu C.Y., Jan J.T., Wang J.R., Hung S.C., Lee G. Bin. An integrated microfluidic system for rapid detection and multiple subtyping of influenza A viruses by using glycan-coated magnetic beads and RT-PCR. Lab Chip. 2019;19:1277–1286. doi: 10.1039/c8lc01369a. [DOI] [PubMed] [Google Scholar]
- Singh P., Kanade S., Nataraj G. European Respiratory Journal. European Respiratory Society (ERS); 2019. Sensitivity and specificity of loop-mediated isothermal amplification assay for diagnosis of extra-pulmonary tuberculosis: a cross-sectional study; p. PA554. [Google Scholar]
- Song Q., Sun X., Dai Z., Gao Y., Gong X., Zhou B., Wu J., Wen W. Point-of-care testing detection methods for COVID-19. Lab Chip. 2021;21:1634–1660. doi: 10.1039/d0lc01156h. [DOI] [PubMed] [Google Scholar]
- Taleghani N., Taghipour F. Diagnosis of COVID-19 for controlling the pandemic: a review of the state-of-the-art. Biosens. Bioelectron. 2021;174:112830. doi: 10.1016/j.bios.2020.112830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . 2021. Weekly Epidemiological Update on COVID-19 - 6 July 2021. [WWW Document] [Google Scholar]
- WHO . 2020. WHO Director-General’s Opening Remarks at the Media Briefing on COVID-19 - 11 March 2020. [WWW Document] [Google Scholar]
- WHO . 2018. Middle East Respiratory Syndrome Coronavirus (MERS-CoV) WHO MERS Global Summary and Assessment of Risk Global Summary. [Google Scholar]
- WHO . Meeting, World Health Organization, World Health Organization. Expert Committee on Biological Standardization - Google Books; 2003. WHO Expert Committee on biological standardization: fifty-fourth report - WHO Expert Committee on biological standardization. [WWW Document] [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D.M.E., Fouchier R.A.M. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Ren G., Buss J., Barry A.J., Patton G.C., Tanner N.A. Enhancing colorimetric loop-mediated isothermal amplification speed and sensitivity with guanidine chloride. Biotechniques. 2020;69:179–185. doi: 10.2144/btn-2020-0078. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.