Abstract
Serological tests play an important role in the fight against Coronavirus Disease 2019 (COVID-19), including monitoring the dynamic immune response after vaccination, identifying past infection and determining community infection rate. Conventional methods for serological testing, such as enzyme-linked immunosorbent assays and chemiluminescence immunoassays, provide reliable and sensitive antibody detection but require sophisticated laboratory infrastructure and/or lengthy assay time. Conversely, lateral flow immunoassays are suitable for rapid point-of-care tests but have limited sensitivity. Here, we describe the development of a rapid and sensitive magnetofluidic immuno-PCR platform that can address the current gap in point-of-care serological testing for COVID-19. Our magnetofluidic immuno-PCR platform automates a magnetic bead-based, single-binding, and one-wash immuno-PCR assay in a palm-sized magnetofluidic device and delivers results in ∼30 min. In the device, a programmable magnetic arm attracts and transports magnetically-captured antibodies through assay reagents pre-loaded in a companion plastic cartridge, and a miniaturized thermocycler and a fluorescence detector perform immuno-PCR to detect the antibodies. We evaluated our magnetofluidic immuno-PCR with 108 clinical serum/plasma samples and achieved 93.8% (45/48) sensitivity and 98.3% (59/60) specificity, demonstrating its potential as a rapid and sensitive point-of-care serological test for COVID-19.
Keywords: COVID-19, Serological testing, Point-of-care, Immuno-PCR, Magnetofluidic device
1. Introduction
Coronavirus Disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has spread throughout the world at an alarming rate and caused more than 170 million infections with 3.5 million lives claimed as of May of 2021 according to the Johns Hopkins University Coronavirus Resource Center (“COVID-19 Dashboard by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins Univeristy (JHU)”). Serological tests, which measure the antibodies against specific proteins of SARS-CoV-2, play an important role in detecting past infection (Yong et al., 2020), determining community infection rate (Klumpp-Thomas et al., 2021), characterizing the disease progression (Amanat et al., 2020; Long et al., 2020a), and guiding return-to-work decision-making (Cheng et al., 2020; Hansen et al., 2021; Krammer and Simon, 2020; Mina and Andersen, 2021; Peeling et al., 2020; Winter and Hegde, 2020).
Conventional COVID-19 serological tests include enzyme-linked immunosorbent assays (ELISA), chemiluminescence immunoassays (CLIA), and lateral flow immunoassays (LFIA). Each method has pros and cons (Duong et al., 2020; Espejo et al., 2020). ELISA is the gold standard for serological testing and provides quantitative and highly sensitive measurement (Amanat et al., 2020). However, ELISA requires sophisticated laboratory infrastructure and a protracted workflow that includes multiple steps of incubation and washing, thus leading to a turnaround time of 2–5 h. CLIA can provide high sensitivity as well and is amenable to automation with high-throughput capability (Long et al, 2020a, 2020b). However, it is also limited by the requirement of bulky equipment and/or long turnaround times. The need for sample delivery to centralized laboratories can result in further delay of clinical reporting. LFIA, on the other hand, enables rapid, point-of-care (POC) serological testing in 15–30 min but is less sensitive than ELISA and CLIA and thus prone to yielding false-negative results and potentially consequential underestimates of infection rates (Duong et al., 2020; Lin et al., 2020; Lisboa Bastos et al., 2020; Montesinos et al., 2020; Whitman et al., 2020). For example, LFIA showed a pooled sensitivity of 66.0% (95% confidence interval: [49.3%, 79.3%]) while ELISA and CLIA demonstrated pooled sensitivities of 84.3% (95% confidence interval: [75.6%, 90.9%]) and 97.8% (95% confidence interval: [46.2%, 100%]), respectively (Lisboa Bastos et al., 2020). An ideal POC serological test would combine the best of each of these approaches, providing the diagnostic performance of ELISA and CLIA with the short turnaround time afforded by LFIA.
Immuno-PCR is a particularly promising approach for highly sensitive antibody detection but translating it to POC settings remains challenging. By combining antigen-antibody recognition with the unparalleled sensitivity of PCR, immuno-PCR has demonstrated ultrasensitive antibody-based detection in benchtop assays (Hendrickson et al., 1995; Malou and Raoult, 2011; Niemeyer et al, 2005, 2007). However, immuno-PCR involves a complicated workflow that has hindered its use in POC settings. Immuno-PCR follows a protocol similar to ELISA but uses oligo-tag-initiated PCR as a readout instead of enzyme-based color development as in ELISA. Generally, immuno-PCR is lengthier than ELISA because the PCR detection, which requires 1–2 h, is much longer than the colorimetric detection in conventional ELISA (Niemeyer et al., 2007). Moreover, in conventional immuno-PCR on benchtop, the PCR step ordinarily necessitates a bulky thermocycler, which, together with the lengthy workflow, complicates the translation of immuno-PCR into a POC test. Consequently, even though in theory immuno-PCR can improve the sensitivity of serological testing, its translation to the point of care is a technical challenge.
In this work, we report successful achievement of a POC immuno-PCR assay for anti-SARS-CoV-2 Immunoglobulin G (IgG) detection using a palm-sized magnetofluidic instrument. This platform, termed magnetofluidic immuno-PCR, provides reliable, high-sensitivity detection of targeted epitopes in approximately 30 min. To develop this approach, we first streamline the lengthy immunobinding workflow, including IgG capture and oligo-tagged goat anti-human IgG antibody (i.e., oligo-anti-IgG) binding, into a 15 min single-step, one-pot reaction. We then implement the streamlined assay into the palm-sized magnetofluidic device, which uses magnetic particles to transport bimolecular targets through discrete reagent droplets, enabling the integration of biomolecular assays without the need for complex fluidic cartridges and instrumentation (Chen et al., 2019; Shin et al, 2017, 2018; Trick et al., 2021; Zhang and Nguyen, 2017; Zhang and Wang, 2013). Our magnetofluidic device automates magnetic bead transfer across different wells in a USB drive-sized disposable cartridge, performs an ultrafast 15 min PCR using a miniature thermocycler and a fluorescence detector, and finally reports the testing results to a smartphone app via Bluetooth. With a streamlined and automated workflow, our magnetofluidic immuno-PCR significantly shortened the immuno-PCR assay time to ∼30 min. To demonstrate the clinical performance of our immuno-PCR platform, we test 108 clinical samples, including 48 convalescent serum/plasma samples from SARS-CoV-2 patients and 60 negative serum/plasma samples. Overall, our platform demonstrates strong potential as a POC solution for serological testing for COVID-19 or other immunoassay-based applications.
2. Experimental
2.1. Biological samples
Cohort 1 of thirty-four convalescent plasma samples from SARS-CoV-2 patients confirmed with Roche ECLIA test (Elecsys® Anti-SARS-CoV-2) were provided by the Advanced Clinical Chemistry Diagnostics Laboratory at Johns Hopkins Hospital. Cohort 2 of fourteen convalescent serum samples confirmed by a custom-developed magnetic bead-based serological test was provided by the Environmental Health Microbiology and Immunology Laboratory at Johns Hopkins Bloomberg School of Public Health. The two series of progression samples were also provided by the Advanced Clinical Chemistry Diagnostics Lab at Johns Hopkins Hospital. The negative control samples, including 40 serum samples and 20 plasma samples, were collected before pandemic. All the plasma/serum samples were de-identified and stored at −80 °C before the test. The study protocol was approved by the respective Institutional Review Board (IRB00247886).
2.2. Materials
The magnetic beads (Dynabeads® MyOne™ Carboxylic Acid, 65011) were bought from Life Technologies Corporation (Carlsbad, CA). 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC, E1769-5G), N-hydroxy succinimide (NHS, 130672-5G), negative control human serum (H4522-20 ML), and single-stranded salmon sperm DNA (ssDNA, D7656-1 ML) were bought from Sigma (St. Louis, MO). Human whole blood was bought from BioIVT (Westbury, NY). Recombinant SARS-CoV-2 spike subunit 1 (S1) protein (40591-V08H, 40591-V08B1) and chimeric anti-SARS-CoV-2 IgG (40150-D006) were bought from Sino Biological (Chesterbrook, PA). Recombinant SARS-CoV-2 S1 protein (RP01261) was from ABclonal Technology (Woburn, MA). Recombinant SARS-CoV-2 S1 protein (PNA002) and receptor-binding domain (RBD) protein (PNA004) were bought from Sanyou Biopharmaceuticals Co., Ltd (Shanghai, China). Another recombinant RBD protein (NR-52306) was provided by BEI Resources. The goat anti-human IgG (AffiniPure Goat Anti-Human IgG, 109-005-170) was bought from Jackson ImmunoResearch Laboratories, Inc (Pennsylvania, USA).
2.3. Protein coating
We use the EDC/NHS chemistry to conjugate recombinant SARS-CoV-2 protein on magnetic beads. Briefly, 20 μL well-mixed 10 mg/mL Dynabeads were recovered back to room temperature (r.t.) and then washed in 40 μL activation buffer (100 mM MES, 0.01% Tween-20, pH 4.5) four times before finally resuspended in 180 μL activation buffer. Next, 20 μL 80 mg/ml EDC & 80 mg/ml NHS were prepared and immediately mixed with the magnetic particles solution to activate the surface for protein conjugation for 15 min. The activated magnetic beads were washed twice in activation buffer to remove remaining EDC/NHS and immediately resuspended in 15 μL activation buffer before adding 5 μL 1 mg/mL protein solution for overnight conjugation at 4 °C. The magnetic beads after protein conjugation were quenched in 40 μL quenching buffer (100 mM boric acid, pH 8.5, 30 mM 2-(2-aminoethoxy) ethanol, 0.01% Tween-20) for 30 min followed by six washings in 120 μL washing buffer (10 mM PBS/0.05% Tween 20). The coated beads were finally resuspended in 200 μL storage buffer (10 mM PBS, 0.1% BSA, 0.02% Tween-20, 0.05% sodium azide, pH 7.4) for immuno-PCR test. We note that we have tuned the EDC/NHS concentration for optimal conjugation condition (see Supplementary Information). Additionally, for the initial ELISA tests we sought to screen optimal recombinant SARS-CoV-2 antigens, we used 250 μg/mL protein instead for magnetic bead conjugation and the conjugated beads were resuspended in 40 μL storage buffer at a bead concentration of 5 mg/mL.
2.4. Oligo-anti-human IgG antibody conjugation
We chose a short luciferase sequence as the amplification target for our immuno-PCR assay (Johnson et al., 2005). To conjugate the oligo and the goat anti-human IgG, the oligonucleotides synthesized with a 5′-thiol modifier (C6 S–S) (Integrated DNA Technologies, see sequence in the Supplementary Table 1) was first reduced using dithiothreitol (DTT) to restore thiol groups (Assarsson et al., 2014). To do that, the oligo was resuspended at 0.5 mM in 100 mM phosphate buffer with 20 mM EDTA and 2.6 μL oligo solution was distributed in a tube. Then, 4.4 μL 40 mM DTT (1x PBS buffer) was added to the 2.6 μL oligonucleotide, mixed, and incubated at 95°, 2 min, followed by a 1 h incubation step at 37°. The excess DTT was removed using two consecutive 7 kDa Zeba columns (Thermo Scientific, Rockford, IL) equilibrated with 100 mM phosphate buffer after mixing the oligo solution with 17 μL PBS buffer (100 mM PBS) with 20 mM EDTA. The reduced oligo concentration was measured using Nanodrop and immediately stored in a freezer until usage.
At the same time, the goat anti-human IgG antibody (anti-IgG) was activated using maleimide before conjugating with the reduced oligo. First, the anti-IgG was resuspended at 1 mg/mL in PBS and 14 μL anti-IgG was taken out and purified using a 40 kDa Zeba column equilibrated four times with 100 mM phosphate buffer, pH 7.3. Then, 2 μL 3.33 mM sulfosuccinimidyl 4-(N-maleimidomethyl) cyclohexane-1-carboxylate (sulfo-SMCC, Thermo Scientific) in 100 mM phosphate buffer was added to the anti-IgG solution, mixed, and incubated at 4° for 2 h with three times intermittent mixing. The excess sulfo-SMCC was then removed using a new 40 kDa Zeba column equilibrated four times with 100 mM phosphate buffer with 20 mM EDTA. The purified anti-IgG concentration was measured using Nanodrop.
Finally, the sulfo-SMCC-treated anti-IgG was mixed with the DTT treated oligonucleotides at 10× molar excess of oligo to anti-IgG and incubated overnight at 4 °C for conjugation. To purify the oligo-anti-IgG conjugate, a 0.5 mL 40 kDa Zeba spin desalting column was equilibrated four times with 100 mM phosphate buffer, pH 7.3. The oligo-anti-IgG conjugation solution was added to the desalting column, spun at 1000×g for 3 min, and collected in a new tube. The conjugation was confirmed with polyacrylamide gel electrophoresis (PAGE) and the concentration of the purified oligo-anti-IgG conjugate was measured using Pierce™ BCA Protein Assay Kit (Thermo Scientific). The oligo-anti-IgG conjugate was stored at −20 °C until usage.
2.5. ELISA tests
To start ELISA tests, the S1/RBD-coated beads were first blocked in 1% fish gelatin for 1 h to minimize nonspecific adsorption. The magnetic beads were then washed three times in washing buffer (1× PBS, 0.05% Tween-20, 150 mM NaCl) before being resuspended in 0.1% BSA solution in PBST (1× PBS, 0.05% Tween-20) at 5 mg/mL. At the same time, a series of 10-fold dilutions of chimeric anti-S1 IgG (40150-D006) in 0.1% BSA PBST solution (0, 2, 20, 200, 2000 ng/mL) was prepared and kept on ice. Before mixing the beads and anti-SARS-CoV-2 IgG, the reaction tubes were washed to minimize any nonspecific adsorption. Then, 10 μL of S1/RBD-coated beads and 10 μL of IgG spike-in sample were mixed and incubated for 1 h at r.t. with continuous mixing on an end-over-end mixer. Following incubation, the beads were separated using magnetics for 2 min and washed three times with 60 μL washing buffer for 20 s. The washed beads were resuspended in 20 μL of HRP-conjugated goat-anti human IgG antibody (1:2000 dilution in 0.5% BSA PBST solution), followed by another 60 min incubation at r.t. with continuous mixing. Then, the magnetic beads were washed three times with 60 μL of washing buffer. At the third wash, the beads were transferred to a new tube to avoid nonspecific adsorption on the tube wall. The magnetic beads were then incubated with 20 μL 3,3′,5,5′-Tetramethylbenzidine (TMB) for 30 min before taking out the supernatant for optical density (OD) measurement at 655 nm using a microplate reader.
2.6. Benchtop 2-step immuno-PCR
For the 2-step immuno-PCR, the RBD-coated beads were first incubated in 0.25 mg/mL RBD solution for 2 h at 4 °C followed by 1 h blocking in 10% goat serum in 1xTBS buffer (20 mM Tris-HCl, 150 mM NaCl, 5 mM EDTA, 100 μg/mL ssDNA, pH 7.5) at r.t. The magnetic beads were then washed three times in washing buffer before being resuspended in goat serum at 1 mg/mL. Similarly, a series of 10-fold dilutions of chimeric anti-S1 IgG (40150-D006) in 1/100 (for Fig. 2C) or 1/500 (for Fig. 2B) diluted human serum solution (0, 2, 20, 200, 2000 ng/mL) was prepared and kept on ice. Then, 10 μL of RBD-coated beads and 10 μL of IgG spike-in sample were mixed and incubated for 1 h at r.t. with continuous end-over-end mixing. Instead of adding HRP-conjugated goat-anti human IgG antibody for measurement, 20 μL 1 fM oligo-anti-IgG conjugate was used to incubate with IgG-captured beads for 60 min at r.t. with continuous mixing. Again, the magnetic beads were washed three times with 60 μL of washing buffer and at the third wash, the beads were transferred to a new tube to avoid nonspecific adsorption. For thorough cleaning of remaining oligo-anti-IgG conjugate, we added another two washes before resuspending the magnetic beads in 10 μL PBS buffer. To perform ultrafast PCR, 1 μL bead aliquot was mixed with 1 μL 10 × FAST buffer, 0.13 μL primer/probe mix (40 μM forward/reverse primers, 20 μM TaqMan probe, see primer and TaqMan probe sequences in Supplementary Tables 1) and 1 μL 2 mM dNTP, 0.2 μL 5000 U/mL SpeedSTAR polymerase, 2 μL 10% Tween-20, and 5.67 μL water. The PCR was performed using BioRad CFX96 real-time detector. The ΔCt for each sample was calculated by subtracting Ct values from the average Ct of negative control for quantification measurement. The limit of detection (LOD) was calculated at three standard deviations above the background.
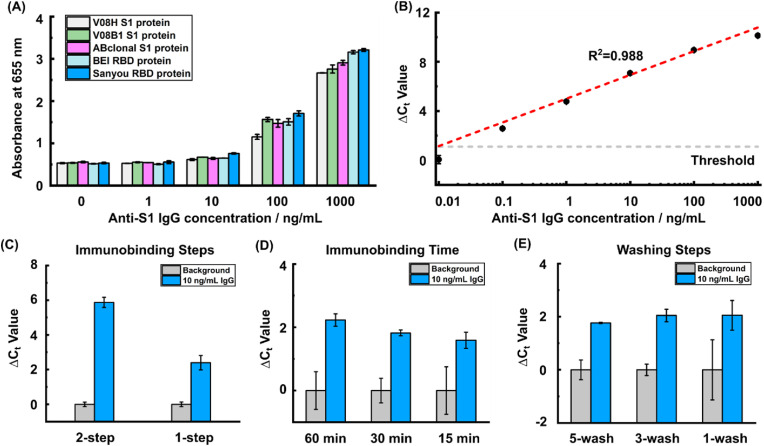
Fig. 2.
Development of a streamlined immuno-PCR assay. (A) Five different recombinant SARS-CoV-2 antigens were tested using ELISA for assessing anti-SARS-CoV-2 IgG capture efficiency. (B) A dilution series of anti-SARS-CoV-2 IgG spike-in in diluted human serum were tested using benchtop immuno-PCR to evaluate the assay sensitivity. (C) The 1-step, one-pot immunobinding showed differentiable Ct values between background and 10 ng/mL spike-in sample even though the ΔCt decreased compared with the 2-step workflow. (D) The immunobinding time was reduced from 60 min to 15 min to shorten the assay time. (E) The immuno-PCR was further simplified by reducing the number of washing steps from 5 times to 1 time. The error bars are standard deviations from duplicated experiments.
2.7. Benchtop 1-step immuno-PCR
The initial beads preparation was similar to 2-step immuno-PCR. However, for the 1-step immuno-PCR workflow, we directly mixed 10 μL 1 mg/mL RBD-coated beads, 20 μL of IgG spike-in sample and 10 μL 0.4 pM oligo-anti-IgG conjugate and incubated for different amount of time at r.t. with continuous mixing on an end-over-end mixer (60 min for Fig. 2C and Supplementary Fig. 5, 30 min for Fig. 2E, and 15 min for Supplementary Fig. 6). Then, the beads were transferred to a new tube and washed with the different number of washing steps (5 times for Fig. 2C and D, and Supplementary Figs. 5 and 1 time for Supplementary Fig. 6). All other remaining steps were same with 2-step immuno-PCR. For the carrier bead blocking, we incubated 2 mg Dynabeads in 200 μL whole goat serum at 4 °C for 2 h followed by an additional 1 h blocking using 200 μL 10/50 μg/mL goat anti-human IgG in 1xTBS buffer at r.t. The carrier beads were then washed three times before using. We note that 1/100 diluted human serum was used for results in Fig. 2C, 2D, 2E and Supplementary Fig. 5 while 1/500 diluted human serum was used for Supplementary Fig. 6 to improve ΔCt for better differentiation.
2.8. Magnetofluidic immuno-PCR for point-of-care serological testing
A detailed description of our POC magnetofluidic device can be found in our previous publications (Shin et al, 2017, 2018). The disposable cartridges used for the immuno-PCR test were fabricated as described before. Briefly, the bottom polypropylene layer was thermoformed with three extruded wells, including sample well, washing well and PCR well. Next, the middle acrylic spacer was laser cut and bonded with the bottom layer by a pressure-sensitive adhesive. Then, the washing well and the PCR well were preloaded with 50 μL washing buffer containing 100 μg fully-blocked blank magnetic beads and 10 μL PCR buffer, respectively, followed by filling the remaining space with 50 cSt silicone oil after sealing the cartridge with a top lid layer. Assembled cartridges were kept on ice before loading samples and being processed in our portable device.
For the POC test, the 40 μL solution after 15 min immunobinding (including 10 μL 1 mg/mL RBD-coated beads, 20 μL of IgG spike-in sample (1:500 serum dilution) and 10 μL 0.4 pM oligo-anti-IgG conjugate) was added into the sample well of the cartridge, which was then loaded into our portable magnetofluidic device to start bead transfer. Our device relies on a pre-programmed dual-axis (Z-θ) manipulator to move beads from one well to another using two opposing permanent magnets. The carrier beads in the washing well were first moved to the sample well to aid the enrichment and transfer of immunomagnetic (RBD-coated with captured IgG and oligo-anti-IgG) beads to washing well. After washing and removing carryover of the nonspecifically-adsorbed oligo-anti-IgG conjugate in the washing well, the magnetic beads were further transferred into PCR well to initiate PCR. The ultrafast PCR recipe was the same as the bulk test. The denature time and annealing time was set to 2 s and 5 s, respectively, slightly longer than the bulk test (1 s for both denature and annealing) to ensure stable amplification. Importantly, the entire process was completed in an automated manner and controlled by a smartphone app via Bluetooth communication or a computer user interface via serial communication.
Each experiment was replicated twice to ensure the reproducibility of the assays. To quantify the result, the real-time amplification curve was fitted using a logistic regression to calculate a Ct value for each amplification, which was then used to calculate ΔCt values by being subtracted from the average Ct of the negative control. The ΔCt values were then plotted against spike-in IgG concentrations and a linear fitting between the ΔCt values and IgG concentrations was used to calculate the limit of detection by setting a threshold as three standard deviations above the negative control.
2.9. Testing of clinical samples
For all the clinical sample tests, the same workflow was followed as just described for spike-in samples. To counterpart the batch-to-batch variation of RBD coating, duplicated negative control serum tests were included for each batch of beads to set a Ct threshold (mean Ct value of control serum minus three times standard deviation). The ΔCt for each clinical sample was then calculated by subtracting Ct values from the threshold. Any ΔCt values below and equal to zero (meaning sample Ct values are higher than the threshold) would be count as IgG-negative and vice versa.
3. Results and discussion
3.1. Overview of magnetofluidic immuno-PCR platform for SARS-CoV-2 serological testing
Our magnetofluidic immuno-PCR was realized through a streamlined immuno-PCR assay implemented within a portable magnetofluidic device (Fig. 1 A). First, magnetic beads functionalized with the receptor-binding domain (RBD) of SARS-CoV-2 spike protein capture SARS-CoV-2 IgG from the sample and then bind with the oligo-anti-IgG to form bead-IgG-(oligo-anti-IgG) complexes (Fig. 1B). The bead-IgG-(oligo-anti-IgG) complexes were loaded into the sample well of a disposable thermoplastic cartridge, which was subsequently mounted onto a heat block within the faceplate of our palm-sized magnetofluidic device to start the POC test. The magnetic beads were transferred into the washing well to remove unbound oligo-anti-IgG before being moved into PCR well for PCR detection. The readout in immuno-PCR – threshold cycle (Ct) values – quantifies the oligos on the bead-IgG-(oligo-anti-IgG) complexes, of which concentration corresponds to the SARS-CoV-2 IgG titer from the sample. Importantly, our magnetofluidic device automates the magnetic bead transfer across different cartridge wells, controls the PCR thermal cycling and real-time fluorescence measurement, and reports the results to a smartphone app via Bluetooth, thus enabling a completely self-contained POC test (Supplementary Fig. 1). Moreover, by optimizing the PCR chemistry and minimizing the thermal mass of the heat block, we were able to achieve PCR amplification in as little as ∼15 min, which significantly shortens the turnaround time of immuno-PCR from 4 to 7 h (Niemeyer et al., 2007) to ∼0.5 h.
Fig. 1.
Overview of the magnetofluidic immuno-PCR platform. (A) The magnetofluidic immuno-PCR features a streamlined immuno-PCR assay performed in a disposable 3-well cartridge using a portable magnetofluidic device, which automates magnetic bead transfer and PCR thermal cycling and communicates results via Bluetooth to a smartphone app. (B) Receptor-binding domain (RBD)-coated magnetic beads capture anti-SARS-CoV-2 IgG together with oligo-goat anti-human IgG antibody (oligo-anti-IgG) conjugates before being loaded into the sample well of the cartridge. The IgG-captured magnetic beads are then transferred to the washing well to remove nonspecifically-adsorbed oligo-anti-IgG conjugates followed by another bead transfer to the PCR well to initiate PCR. The entire process is completed in ∼0.5 h, including 15 min immunobinding, 2 min washing, and 15 min for rapid PCR.
3.2. Development of a streamlined immuno-PCR assay for SARS-CoV-2
We first developed a protocol for conjugating recombinant SARS-CoV-2 antigen with magnetic beads for capture of anti-SARS-CoV-2 IgG from serum samples. We leveraged the carboxylic group on Dynabeads' surface to react with the primary amine group from recombinant antigens via 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC)/N-hydroxy succinimide (NHS) activation (Supplementary Note 1, Supplementary Fig. 2A and Supplementary Fig. 2B). We then developed a protocol for conjugation between an oligo and goat anti-human IgG antibody (i.e., anti-IgG) that would be used as the target for PCR. We chose sulfosuccinimidyl 4-(N-maleimidomethyl) cyclohexane-1-carboxylate (Sulfo-SMCC) to activate the anti-IgG, which subsequently mediated the conjugation with freshly reduced 5′-thiol modified oligos (Supplementary Fig. 2C, see Methods). The oligo-anti-IgG conjugate was then purified via spin desalting column and confirmed via polyacrylamide gel electrophoresis (PAGE) (Supplementary Note 2 and Supplementary Fig. 3).
Using the SARS-CoV-2 antigen conjugated magnetic beads and the oligo conjugated anti-IgG, we then established the immuno-PCR assay for anti-SARS-CoV-2 IgG detection on benchtop. We first assessed the efficiency of recombinant SARS-CoV-2 antigens for capture of anti-SARS-CoV-2 IgG and optimized the blocking buffer to minimize nonspecific interactions. We first tested five different recombinant antigens, including two RBD proteins and three spike subunit 1 (S1) proteins of the SARS-CoV-2, by creating a 10-fold dilution series of chimeric anti-S1 IgG spike-in using a magnetic bead-based ELISA assay. We note that we focused on the S1 and RBD protein here because the antibodies against these epitopes correlate well with virus neutralization (Ju et al., 2020; Zost et al., 2020) and we tested different sources for the same antigen because the difference in recombinant protein expression can also affect the result (Jiang et al., 2020). The results showed that the RBD protein from Sanyou Biopharmaceuticals produced highest signal and provided the best sensitivity among the five recombinant antigens (Fig. 2A). Moreover, our results indicated that 10% goat serum performed best among the blocking buffers for minimizing nonspecific binding from human serum (Supplementary Note 3 and Supplementary Fig. 4). Using the identified capture antigen and optimized blocking buffer, we established the immuno-PCR assay on benchtop with the dilution series of chimeric IgG spike-in samples in diluted human serum (see Methods). We observed a strong linear relationship between the spike-in IgG concentrations and the ΔCt values (defined as the Ct difference between each spike-in IgG concentration and control serum without IgG spike-in (i.e., background)) with a dynamic range across 5 orders of magnitude. The calculated limit of detection (LOD) of our immuno-PCR assay was determined to be 21 pg/mL (Fig. 2B).
To transition benchtop immuno-PCR to POC serological testing, we streamlined, simplified, and accelerated the binding and washing steps. To simplify the characterization and optimization, we spiked in 10 ng/mL anti-S1 IgG in diluted human serum as a positive control and used diluted serum without spiked-in IgG as background. The conventional immuno-PCR workflow consists of a 2-step immunobinding, including a 1-h anti-SARS-CoV-2 IgG capture followed by another 1-h oligo-anti-IgG conjugate binding with multiple washing cycles after each immunobinding. We first sought to simplify this lengthy 2-step immunobinding into a 1-step, one-pot reaction to reduce overall assay time. To test the feasibility of the 1-step immunobinding, we mixed RBD-coated magnetic beads with both the chimeric anti-S1 IgG and oligo-anti-IgG conjugate for simultaneous anti-S1 IgG capture and oligo-anti-IgG binding. We observed that with 1-step immunobinding, the ΔCt between the positive control and background increased proportionally when the oligo-anti-IgG conjugate concentration was raised from 1 fM to 100 fM (Supplementary Note 4 and Supplementary Fig. 5), though it was ultimately lower than the ΔCt achieved with the conventional 2-step workflow (Fig. 2C). To accelerate the 1-step immunobinding, we tested 60 min, 30 min, and 15 min incubation time. We found that even 15 min provided sufficient time for IgG and oligo-anti-IgG to bind and achieved IgG detection (Fig. 2D). We then further simplified the workflow by reducing the washing steps after immunobinding. We tested 5 washes, 3 washes, and 1 wash, and found that IgG remained detectable even after only 1 wash (Fig. 2E).
The remaining challenge for implementation of the streamlined immuno-PCR assay in our magnetofluidic device was achieving adequate bead concentration to overcome surface tension (Zhang and Wang, 2013) and ensure successful bead transfer across the water-oil interface between wells. The 10 μg RBD-coated beads are not enough to overcome the high surface tension and therefore resulted in less efficient transport of beads between the reagent droplets. To address this problem, we decided to include unconjugated magnetic beads as “carrier beads” to enhance the bead transfer and determined that consecutive goat serum and anti-IgG blocking steps minimized reagent carryover from the carrier beads, maintaining a consistent ΔCt between the positive control and background (Supplementary Note 5 and Supplementary Fig. 6). We noted that adding carrier beads also suppressed the background variation which further improved the sensitivity (Supplementary Fig. 6).
3.3. Implementation of SARS-CoV-2 immuno-PCR in the magnetofluidic device
We next adapted the magnetic-based immuno-PCR assay for POC use by employing a palm-sized magnetofluidic device (Shin et al., 2018; Trick et al., 2021), previously developed by our group, that utilizes a disposable thermoplastic cartridge to automate its sample processing and PCR detection. The magnetofluidic device relies on a pre-programmed biaxial manipulator to move magnetic beads from one well to another in the cartridge using two opposing permanent magnets that are attached to a servo motor assembly (Supplementary Fig. 7). It provides automated thermal cycling using a thermoelectric element-driven miniature heat block while real-time fluorescence signal is acquired and recorded using a fluorescence detector and software control interface, respectively. The disposable thermoplastic cartridge is assembled from a combination of low-cost thermoformed and laser-cut components that can be easily laminated together, and is thus well suited for use in POC testing (Supplementary Fig. 8).
To perform the immuno-PCR assay using the portable magnetofluidic instrument, the sample together with RBD-coated magnetic beads and oligo-anti-IgG is first loaded into the sample well immediately after a 15 min 1-step immunobinding, followed by automated bead transfer to the washing well that is initiated via smartphone app connected through Bluetooth communication (Supplementary Fig. 1). After the beads are washed to remove nonspecifically-adsorbed oligo-anti-IgG conjugate, they are then transferred to the PCR reaction well and PCR is initiated. To speed up PCR for POC testing, we improved the reaction kinetics with enhanced assay chemistry and accelerated the temperature ramping by optimizing heat block geometry (Trick et al., 2021). As a result, we achieved a maximal thermal ramping rate of 10.4 °C/s and a maximal cooling rate of 18.5 °C/s between 60 and 100 °C. The ultra-fast ramping and cooling enable an ultrafast PCR with 40 thermal cycles in only ∼15 min (Supplementary Fig. 9) for a total assay turnaround time of ∼0.5 h.
To evaluate our magnetofluidic immuno-PCR, we tested a 10-fold dilution series of anti-S1 IgG samples in diluted human serum. The resulted real-time fluorescence curves showed clear amplification in which the higher IgG concentration amplified with earlier Ct values (Fig. 3 A). The ΔCt values between samples and background were plotted against the input IgG concentrations in which a strong linear relationship was observed, suggesting the potential quantification capability of our magnetofluidic immuno-PCR (Fig. 3B). Additionally, the LOD of 2 ng/mL is comparable to recently developed platforms for anti-SARS-CoV-2 antibody detection (Fig. 3B) (Liu et al., 2020). To demonstrate the feasibility of using whole blood as sample input for our magnetofluidic immuno-PCR, we tested a 10-fold dilution series of anti-S1 IgG samples spiked in diluted human whole blood. We also observed a linear increase of ΔCt values against the input IgG concentrations. Though we observed higher background from whole blood, the LOD of our magnetofluidic immuno-PCR can still maintain at 22 ng/mL, which shows the potential of our magnetofluidic immuno-PCR to directly use whole blood for POC serological testing (Fig. 3C).
Fig. 3.
Implementation of SARS-CoV-2 immuno-PCR assay in the magnetofluidic device. (A) The real-time amplification curves of the magnetofluidic immuno-PCR tests showed earlier amplification with higher spike-in anti-S1 IgG concentrations. (B)As a result, the ΔCt values of IgG spike-in series in diluted human serum increased linearly with the IgG concentrations across three orders of magnitude with an estimated LOD of 2 ng/mL. (C) Our magnetofluidic immuno-PCR was also compatible with whole blood sample input though the LOD (∼22 ng/mL) was inferior to that of serum.
3.4. Clinical evaluation of magnetofluidic immuno-PCR
After analytical validation using serum/whole blood spike-in samples, we proceeded to test clinical samples with our magnetofluidic immuno-PCR platform. We performed the magnetofluidic immuno-PCR in two independent cohorts of convalescent serum/plasma samples from SARS-CoV-2 patients, including a cohort of 34 plasma samples that were confirmed IgG-positive using an electrochemiluminescence immunoassay (ECLIA) from Roche and a cohort of 14 serum samples confirmed IgG-positive using a previously-reported Luminex bead-based platform (Pisanic et al., 2020). Our magnetofluidic immuno-PCR test was able to detect 32/34 of ECLIA cohort samples and 13/14 of Luminex cohort samples, equivalent to 94.1% and 92.9% sensitivity, respectively, and overall sensitivity of 93.8% (45/48) (Fig. 4 A). To validate the specificity of the portable serological test, we tested 60 archived serum/plasma samples that were collected before the pandemic. Of these 60 pre-pandemic samples, only one showed a positive ΔCt when compared to our control serum, indicating 98.3% specificity. In total, we achieved an area under the curve (AUC) of 0.968 (95% CI: [0.933, 1.000]) via receiver operating characteristic curve analysis, demonstrating the high sensitivity and specificity of our magnetofluidic immuno-PCR platform (Fig. 4B).
Fig. 4.
Clinical evaluation of the magnetofluidic immuno-PCR using convalescent serum/plasma samples. We tested two independent cohorts, 48 in total, of convalescent serum/plasma samples from SARS-CoV-2 patients and 60 pre-pandemic plasma/serum samples to characterize the sensitivity and specificity of our magnetofluidic immuno-PCR. (A) ΔCt values from magnetofluidic immuno-PCR analysis of clinical specimens. Negative specimens (black) are from archived samples collected prior to the pandemic. Red and blue dots indicate test-positive samples from ECLIA and custom Luminex analyses, respectively. (B) Receiver operating curve (ROC) analysis was performed on the ΔCt values of 60 pre-pandemic samples and 48 convalescent samples, resulting in an area under the curve (AUC) of 0.968 (95% CI: [0.933, 1.000]). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Of note, our magnetofluidic immuno-PCR workflow is designed with a balance between sensitivity and speed. By slightly increasing the immunobinding time, we can further improve the sensitivity of the POC serological testing (Fig. 2D). To confirm this, we chose the three convalescent samples that resulted in false-negative calls and increased the immunobinding time from 15 min to 30 min. Indeed, we were able to detect the anti-SARS-CoV-2 IgG in these samples and increased the sensitivity to 100% and the AUC to 0.999 (95% CI: [0.997, 1.000]) with approximately 45 min turnaround time (Supplementary Fig. 10).
We also tested two series of progression samples from recovered COVID-19 patients. We observed generally increased anti-SARS-CoV-2 IgG levels over time after symptom onset and this increasing trend in concordance with benchtop Roche ECLIA results (Pearson's Correlation Coefficient = 0.5). Further investigations may be warranted to further demonstrate the quantification capability of our POC serological test (Supplementary Fig. 11).
4. Conclusion
Serological tests play an important role in the management and surveillance of the COVID-19 pandemic. The goal of our study was to develop an improved serological test that combines the best attributes of ELISA/CLIA and LFIA as a potentially useful tool in the fight against COVID-19. Toward this end, we developed a streamlined immuno-PCR assay and applied it to a portable magnetofluidic device for POC SARS-CoV-2 serological testing. With an only slightly longer turnaround time than LFIA, our magnetofluidic immuno-PCR demonstrated higher sensitivity (93.8%) and specificity (98.3%) than most LFIA tests (Supplementary Table 2 and Supplementary Table 3). Moreover, our magnetofluidic immuno-PCR is designed with a balance between speed and sensitivity, and 100% sensitivity was achieved by just increasing the total assay time from 30 min to 45 min. The assay time of our magnetofluidic immuno-PCR assay can be set to 45 min when sensitivity is more desirable such as confirm past infection for individuals, or to 30 min when speed is more important such as entrance screening.
Our magnetofluidic immuno-PCR is made possible by three enabling features. First, the highly sensitive immuno-PCR assay, which was established for COVID-19 serological testing for the first time, takes advantage of the exponential amplification power of PCR for protein analysis and lays the foundation of the improved sensitivity of the magnetofluidic immuno-PCR. Second, the simplified and accelerated immuno-PCR protocol significantly shortens the assay time and simplifies the conventional complicated workflow for more POC amendable testing. Last, our portable magnetofluidic device provides a self-contained platform for automated and ultrafast sample preparation and PCR, which significantly minimizes hands-on operation time compared with benchtop immuno-PCR. Importantly, the magnetofluidic device enables implementation of ultrafast (∼15 min) PCR by the enhanced PCR assay chemistry as well as the optimized heat block. Taken together, our magnetofluidic immuno-PCR significantly shortens the lengthy workflow of conventional immuno-PCR and facilitates a rapid and sensitive POC serological test for COVID-19. To the best of our knowledge, our magnetofluidic immuno-PCR is the first to successfully demonstrate immuno-PCR for sensitive POC test by overcoming the lengthy workflow and obviating the need for heavy instrumentation.
It is worth noting the limitations in the current study and opportunities for future improvements. We validated our platform using positive convalescent samples that were confirmed with benchtop serological tests, but due to limited access to appropriate negative controls, we were not able to test the cross-reactivity of our platform with other virus infections, including influenza viruses and other coronaviruses (Liu et al., 2020). More extensive tests are warranted to further validate the clinical translatability of our magnetofluidic immuno-PCR in real clinical settings in the future. Nevertheless, with further development, our magnetofluidic immuno-PCR offer a useful diagnostic tool for rapid and sensitive serological testing for COVID-19 and other infectious agents.
CRediT authorship contribution statement
Pengfei Zhang: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Resources, Data curation, Writing – original draft, Writing – review & editing, Visualization. Liben Chen: Conceptualization, Methodology, Investigation, Writing – review & editing. Jiumei Hu: Methodology, Validation, Data curation, Writing – review & editing. Alexander Y. Trick: Methodology, Investigation, Resources, Writing – review & editing. Fan-En Chen: Methodology, Investigation, Resources, Writing – review & editing. Kuangwen Hsieh: Methodology, Investigation, Writing – review & editing. Yang Zhao: Resources. Branch Coleman: Resources. Kate Kruczynski: Resources. Thomas R. Pisanic: Methodology, Resources, Writing – review & editing. Christopher D. Heaney: Resources, Supervision. William A. Clarke: Investigation, Resources, Writing – review & editing, Supervision. Tza-Huei Wang: Conceptualization, Investigation, Resources, Writing – review & editing, Supervision, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Research reported in this publication is financially supported by the National Institutes of Health (R01AI138978 and R61AI154628). The following reagent was produced under HHSN272201400008C and obtained through BEI Resources, NIAID, NIH: Spike Glycoprotein Receptor Binding Domain (RBD) from SARS-Related Coronavirus 2, Wuhan-Hu-1 with C-Terminal Histidine Tag, Recombinant from HEK293 Cells, NR-52306. We also thank Mr. Joon Soo Park for his help with experiments for running PAGE gel.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2021.113656.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H.O., Chromikova V., McMahon M., Jiang K., Arunkumar G.A., Jurczyszak D., Polanco J., Bermudez-Gonzalez M., Kleiner G., Aydillo T., Miorin L., Fierer D.S., Lugo L.A., Kojic E.M., Stoever J., Liu S.T.H., Cunningham-Rundles C., Felgner P.L., Moran T., García-Sastre A., Caplivski D., Cheng A.C., Kedzierska K., Vapalahti O., Hepojoki J.M., Simon V., Krammer F. Nat. Med. 2020;26:1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assarsson E., Lundberg M., Holmquist G., Björkesten J., Thorsen S.B., Ekman D., Eriksson A., Dickens E.R., Ohlsson S., Edfeldt G., Andersson A.C., Lindstedt P., Stenvang J., Gullberg M., Fredriksson S. PloS One. 2014;9 doi: 10.1371/journal.pone.0095192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F.E., Trick A.Y., Hsieh W., Shin D.J., Chen L., Chang E., Kaushik A., Wang T.H. Int. Conf. Solid-State Sensors, Actuators Microsystems Eurosensors XXXIII. 2019. TRANSDUCERS 2019 EUROSENSORS XXXIII 1116–1119, 2019 20th. [DOI] [Google Scholar]
- Cheng M.P., Yansouni C.P., Basta N.E., Desjardins M., Kanjilal S., Paquette K., Caya C., Semret M., Quach C., Libman M., Mazzola L., Sacks J.A., Dittrich S., Papenburg J. Ann. Intern. Med. 2020;173:450–460. doi: 10.7326/M20-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CSSE COVID-19 dashboard by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins Univeristy (JHU) https://coronavirus.jhu.edu/map.html URL. 12.30.20.
- Duong Y.T., Wright C.G., Justman J. BMJ. 2020;370:1–2. doi: 10.1136/bmj.m2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espejo, A.P., Akgun, Y., Al Mana, A.F., Tjendra, Y., Millan, N.C., Gomez-Fernandez, C., Cray, C., 2020. 154, 293–304. 10.1093/ajcp/aqaa112. [DOI] [PMC free article] [PubMed]
- Hansen C.H., Michlmayr D., Gubbels S.M., Mølbak K., Ethelberg S. Lancet. 2021;6736:1–9. doi: 10.1016/S0140-6736(21)00575-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson E.R., Truby T.M.H., Joerger R.D., Majarian W.R., Ebersole R.C. Nucleic Acids Res. 1995;23:522–529. doi: 10.1093/nar/23.3.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H. wei, Li Y., Zhang H. nan, Wang W., Yang X., Qi H., Li H., Men D., Zhou J., Tao S. ce. Nat. Commun. 2020;11:1–11. doi: 10.1038/s41467-020-17488-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D.R., Lee P.K.H., Holmes V.F., Alvarez-Cohen L. Appl. Environ. Microbiol. 2005;71:3866–3871. doi: 10.1128/AEM.71.7.3866-3871.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu Jiazhen, Shan S., Zhou B., Song S., Tang X., Yu Jinfang, Lan J., Yuan J., Wang H., Zhao Juanjuan, Zhang S., Wang Y., Shi X., Liu L., Zhao Jincun, Wang X., Zhang Z., Zhang L. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- Klumpp-Thomas C., Kalish H., Drew M., Hunsberger S., Snead K., Fay M.P., Mehalko J., Shunmugavel A., Wall V., Frank P., Denson J.P., Hong M., Gulten G., Messing S., Hicks J., Michael S., Gillette W., Hall M.D., Memoli M.J., Esposito D., Sadtler K. Nat. Commun. 2021;12:1–13. doi: 10.1038/s41467-020-20383-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F., Simon V. Science. 2020;368(80-):1060–1061. doi: 10.1126/science.abc1227. [DOI] [PubMed] [Google Scholar]
- Lin D., Liu L., Zhang M., Hu Y., Yang Q., Guo J., Dai Y., Xu Y., Cai Y., Chen X., Huang K., Zhang Z. Eur. J. Clin. Microbiol. Infect. Dis. 2020;39:2271–2277. doi: 10.1007/s10096-020-03978-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisboa Bastos M., Tavaziva G., Abidi S.K., Campbell J.R., Haraoui L.P., Johnston J.C., Lan Z., Law S., MacLean E., Trajman A., Menzies D., Benedetti A., Khan F.A. BMJ. 2020;370 doi: 10.1136/bmj.m2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Hsiung J., Zhao S., Kost J., Sreedhar D., Hanson C.V., Olson K., Keare D., Chang S.T., Bliden K.P., Gurbel P.A., Tantry U.S., Roche J., Press C., Boggs J., Rodriguez-Soto J.P., Montoya J.G., Tang M., Dai H. Nat. Biomed. Eng. 2020;4:1188–1196. doi: 10.1038/s41551-020-00642-4. [DOI] [PubMed] [Google Scholar]
- Long Q.X., Liu B.Z., Deng H.J., Wu G.C., Deng K., Chen Y.K., Liao P., Qiu J.F., Lin Y., Cai X.F., Wang D.Q., Hu Y., Ren J.H., Tang N., Xu Y.Y., Yu L.H., Mo Z., Gong F., Zhang X.L., Tian W.G., Hu L., Zhang X.X., Xiang J.L., Du H.X., Liu H.W., Lang C.H., Luo X.H., Wu S.B., Cui X.P., Zhou Z., Zhu M.M., Wang J., Xue C.J., Li X.F., Wang L., Li Z.J., Wang K., Niu C.C., Yang Q.J., Tang X.J., Zhang Y., Liu X.M., Li J.J., Zhang D.C., Zhang F., Liu P., Yuan J., Li Q., Hu J.L., Chen J., Huang A.L. Nat. Med. 2020;26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- Long Q.X., Tang X.J., Shi Q.L., Li Q., Deng H.J., Yuan J., Hu J.L., Xu W., Zhang Y., Lv F.J., Su K., Zhang F., Gong J., Wu B., Liu X.M., Li J.J., Qiu J.F., Chen J., Huang A.L. Nat. Med. 2020;26:1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- Malou N., Raoult D. Trends Microbiol. 2011;19:295–302. doi: 10.1016/j.tim.2011.03.004. [DOI] [PubMed] [Google Scholar]
- Mina B.M.J., Andersen K.G. Science. 2021;37(80-):126–128. [Google Scholar]
- Niemeyer C.M., Adler M., Wacker R. Nat. Protoc. 2007;2:1918–1930. doi: 10.1038/nprot.2007.267. [DOI] [PubMed] [Google Scholar]
- Montesinos I., Gruson D., Kabamba B., Dahma H., Wijngaert S. Van Den. J. Clin. Virol. 2020;128:104413. doi: 10.1016/j.jcv.2020.104413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemeyer C.M., Adler M., Wacker R. Trends Biotechnol. 2005;23:208–216. doi: 10.1016/j.tibtech.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Peeling R.W., Wedderburn C.J., Garcia P.J., Boeras D., Fongwen N., Nkengasong J., Sall A., Tanuri A., Heymann D.L. Lancet Infect. Dis. 2020;20:e245–e249. doi: 10.1016/S1473-3099(20)30517-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisanic N., Randad P.R., Kruczynski K., Manabe Y.C., Thomas D.L., Pekosz A., Klein S.L., Betenbaugh M.J., Clarke W.A., Laeyendecker O., Caturegli P.P., Larman H.B., Detrick B., Fairley J.K., Sherman A.C., Rouphael N., Edupuganti S., Granger D.A., Granger S.W., Collins M.H., Heaney C.D. J. Clin. Microbiol. 2020;59:e02204–20. doi: 10.1128/jcm.02204-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D.J., Athamanolap P., Chen L., Hardick J., Lewis M., Hsieh Y.H., Rothman R.E., Gaydos C.A., Wang T.H. Sci. Rep. 2017;7:1–10. doi: 10.1038/s41598-017-04781-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D.J., Trick A.Y., Hsieh Y.H., Thomas D.L., Wang T.H. Sci. Rep. 2018;8:1–12. doi: 10.1038/s41598-018-28124-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trick A.Y., Melendez J.H., Chen F.-E., Chen L., Onzia A., Zawedde A., Nakku-Joloba E., Kyambadde P., Mande E., Matovu J., Atuheirwe M., Kwizera R., Gilliams E.A., Hsieh Y.-H., Gaydos C.A., Manabe Y.C., Hamill M.M., Wang T.-H. Sci. Transl. Med. 2021;13:eabf6356. doi: 10.1126/scitranslmed.abf6356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman J.D., Hiatt J., Mowery C.T., Shy B.R., Yu R., Yamamoto T.N., Rathore U., Goldgof G.M., Whitty C., Woo J.M., Gallman A.E., Miller T.E., Levine A.G., Nguyen D.N., Bapat S.P., Balcerek J., Bylsma S.A., Lyons A.M., Li S., Wong A.W. yi, Gillis-Buck E.M., Steinhart Z.B., Lee Y., Apathy R., Lipke M.J., Smith J.A., Zheng T., Boothby I.C., Isaza E., Chan J., Acenas D.D., Lee J., Macrae T.A., Kyaw T.S., Wu D., Ng D.L., Gu W., York V.A., Eskandarian H.A., Callaway P.C., Warrier L., Moreno M.E., Levan J., Torres L., Farrington L.A., Loudermilk R.P., Koshal K., Zorn K.C., Garcia-Beltran W.F., Yang D., Astudillo M.G., Bernstein B.E., Gelfand J.A., Ryan E.T., Charles R.C., Iafrate A.J., Lennerz J.K., Miller S., Chiu C.Y., Stramer S.L., Wilson M.R., Manglik A., Ye C.J., Krogan N.J., Anderson M.S., Cyster J.G., Ernst J.D., Wu A.H.B., Lynch K.L., Bern C., Hsu P.D., Marson A. Nat. Biotechnol. 2020;38:1174–1183. doi: 10.1038/s41587-020-0659-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter A.K., Hegde S.T. Lancet. 2020;20:758–759. doi: 10.1093/nsr/nwaa036.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong S.E.F., Anderson D.E., Wei W.E., Pang J., Chia W.N., Tan C.W., Teoh Y.L., Rajendram P., Toh M.P.H.S., Poh C., Koh V.T.J., Lum J., Suhaimi N.A.M., Chia P.Y., Chen M.I.C., Vasoo S., Ong B., Leo Y.S., Wang L., Lee V.J.M. Lancet Infect. Dis. 2020;20:809–815. doi: 10.1016/S1473-3099(20)30273-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Nguyen N.T. Lab Chip. 2017;17:994–1008. doi: 10.1039/c7lc00025a. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Wang T.H. Adv. Mater. 2013;25:2903–2908. doi: 10.1002/adma.201300383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost S.J., Gilchuk P., Case J.B., Binshtein E., Chen R.E., Nkolola J.P., Schäfer A., Reidy J.X., Trivette A., Nargi R.S., Sutton R.E., Suryadevara N., Martinez D.R., Williamson L.E., Chen E.C., Jones T., Day S., Myers L., Hassan A.O., Kafai N.M., Winkler E.S., Fox J.M., Shrihari S., Mueller B.K., Meiler J., Chandrashekar A., Mercado N.B., Steinhardt J.J., Ren K., Loo Y.M., Kallewaard N.L., McCune B.T., Keeler S.P., Holtzman M.J., Barouch D.H., Gralinski L.E., Baric R.S., Thackray L.B., Diamond M.S., Carnahan R.H., Crowe J.E. Nature. 2020;584:443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.