Abstract
Saccharomyces cerevisiae Ty elements are retrotransposons whose life cycles are strikingly similar to those of retroviruses. They transpose via an RNA intermediate that is converted to linear double-stranded cDNA and then inserted into the host genome. Although Ty integration is mediated by the element-encoded integrase, it has been proposed that host factors are involved in this process. Here, we show that the DNA end-binding protein Ku, which functions in DNA double-strand break repair, potentiates retrotransposition. Specifically, by using a galactose-inducible Ty1 system, we found that in vivo, Ty1 retrotransposition rates were substantially reduced in the absence of Ku. In contrast, this phenotype was not observed with yeast strains containing mutations in other genes that are involved in DNA repair. We present evidence that Ku associates with Ty1 viruslike particles both in vitro and in vivo. These results provide an additional role for Ku and suggest that it might function in the life cycles of retroelements in other systems.
Long terminal repeat-containing retrotransposons are closely related to retroviruses in structure and life cycle (6). Briefly, the retrotransposon RNA transcript is translated and the protein products assemble into viruslike particles (VLPs) in the cytosol. During this process, no double-strand (ds) breaks are created in the host genome because the original retrotransposon is not excised. The full-length RNA transcript contained within the VLP is then reverse transcribed by the retroelement-encoded reverse transcriptase (RT) into a linear dsDNA molecule, which is then inserted into the host genome by the retroelement-encoded integrase. In all of the cases characterized, integrase catalyzes direct nucleophilic attack by a 3′ hydroxyl of the transposable element onto a target DNA phosphodiester bond (12). Again, this does not result in ds breaks in the host genome. Instead, the product of the integration reaction is the retroelement flanked by single-stranded (ss) DNA gaps that are then thought to be repaired by host factors (6).
It has been demonstrated that a reaction mechanism very similar to that of transposable element integration is employed by the RAG1 and RAG2 proteins in the first steps of V(D)J recombination, a site-specific genomic rearrangement process that helps generate the diversity of antigen-binding sites of immunoglobulin and T-cell receptor proteins (46). V(D)J recombination is severely debilitated in cell lines or animals defective in components of the DNA-dependent protein kinase (DNA-PK; 33, 43). DNA-PK is made up of a catalytic subunit (DNA-PKcs) and a DNA-binding protein (Ku), which is a heterodimer of ∼70- and ∼80-kDa subunits (Ku70 and Ku80, respectively; 18, 24). Ku and DNA-PKcs also function in the repair of radiation- or restriction enzyme-induced DNA ds breaks in mammalian cells (33, 43). Given that V(D)J recombination is similar mechanistically to retrotransposition and retroviral integration, we speculated that Ku and DNA-PKcs might play a role in retroelement life cycles. A second reason to consider a role for Ku in this process is that during retroelement life cycles, ds ends and ss gaps are created as products of reverse transcription and integration into the genome, respectively, and Ku has been shown to bind with strong affinity to these structures in vitro (19, 43). While no direct homologue of DNA-PKcs has been identified in Saccharomyces cerevisiae, there are homologues of Ku70 and Ku80 (the genes for these are termed YKU70 or HDF1, and YKU80 or HDF2, respectively) and these yeast factors have been shown to be involved in the repair of DNA ds breaks (13, 27). We investigated the potential role of Ku in Ty1 retrotransposition in S. cerevisiae, and we discuss the results in terms of the mechanism of action of Ku in this and related processes.
MATERIALS AND METHODS
Disruption constructs.
A BamHI-EcoRI fragment from pFA6a-kanMX4 (47) was cloned into the BamHI-EcoRI sites of pGEM-Ku80 (9) to create the p80::KAN disruption construct. The yku80::URA3 disruption construct pJDG80U was created by cloning a URA3-containing EcoRI-KpnI fragment from pBSURA3 into the EcoRI and KpnI sites of pGEM-Ku80 (9). A BamHI-EcoRI fragment containing the TRP1 gene from pJA52 (2) was inserted into the BglII-EcoRI sites of pJDGS3F to create the SIR3 disruption construct pJDGS3KO. The RAD50 disruption construct was created by excising the XbaI fragment of pGEM-50 and replacing it with an XbaI fragment containing the LEU2 gene from pJA51 (2) to create prad50::LEU. The full-length YKU70 gene and its promoter were excised from p413-fl70 (7) with BamHI and XhoI and inserted into the same sites of pRS414 to create p414-fl70.
Yeast strains.
Yeast strains were maintained in accordance with standard protocols in synthetic complete (SC) medium with the appropriate selection (45). Unless otherwise stated, 2% glucose was used as the carbon source. Strains W303α (wild type) and hdf1α (yku70::LEU2) were gifts from H. Feldmann and E. L. Winnacker. All disruption strains were made in W303α-derived strains by transforming 1 to 2 μg of restriction-digested disruption construct plasmid DNA by the standard lithium acetate transformation method (5), and the transformation reactions were plated on the appropriate selective medium. YKU80-disrupted strains were created by transforming strain W303α or the yku70 mutant strain with p80::KAN digested with NotI and selected on G418 plates as previously described (47). RAD52-disrupted strains were created by transforming strain W303α and the yku70 mutant strain with SalI-digested pR52T (Gift of D. Weaver). SGS1-disrupted strains were created by transforming W303α with pPWSGS1 (Gift of I. Hickson) digested with NcoI and PstI. SIR3-disrupted strains were created by transformation of W303α with ApaI- and SacI-digested pJDGS3KO. Disruptions were confirmed by PCR using one gene-specific primer and one marker-specific primer. Strain Y661 (wild type) and mec1-21, tel1, and mec1-21/tel1 mutant strains were gifts from S. Elledge. Strain JCY297 was a gift of J. Curcio, and YKU80 was disrupted in this strain with NotI-digested pJDG80U to create strain JDY15.
Ty1 retrotransposition assays.
Yeast strains were transformed with pGTy1-H3mHIS3AI (14) (referred to as pGTy1 later in the text and in the figures) and plated on SC-ura (SC medium without uracil). These plasmids contain a Ty1 element under the control of the galactose-inducible GAL1 promoter and possess a HIS3 gene with an artificial intron. Expression of HIS3 is dependent on Ty retrotransposition (see Results for details). Colonies were picked and grown in SC-ura containing either glucose or galactose overnight at 23°C. Cells were counted by taking the optical density at 600 nm (OD600) and plating them on nonselective medium to ensure that the OD was measuring viable cells. Equal numbers of viable cells were plated on SC-his (without histidine) containing glucose, and transposition levels were calculated as His+ colonies/total cells. Complementation of the yku70 mutant phenotype was performed by transforming p414-fl70 into yku70 mutant cells already containing the pGTy1 plasmid. Retrotransposition assays were performed exactly as described above but with selection for the p414-fl70 plasmid maintained by eliminating tryptophan from the medium. All assays were conducted a minimum of three times.
β-Galactosidase activity measurements.
Wild-type or yku70 mutant yeast cells were transformed with pJK101, which contains the lacZ gene under the control of the GAL1 promoter. Transformed cells were grown overnight in SC-ura containing glucose at 23°C. The OD600 was measured, and cells were diluted to a density corresponding to an OD600 of 0.2. The cells were then grown in SC-ura containing galactose at 23°C. At the indicated times, 200-μl aliquots were taken and β-galactosidase activity was measured by o-nitrophenyl-β-d-galactopyranoside (ONPG) cleavage as previously described (41). Measurements were taken for three independent transformants of each strain.
VLP isolation and RT activity levels.
The protocol for isolation of VLPs was adapted from reference 20. Briefly, 50 ml of an overnight yeast culture containing pGTy1 (grown in SC-ura containing glucose) was pelleted, washed with water, and used to inoculate 500 ml of SC-ura containing either glucose or galactose. After growing for 24 h at 22°C, cells were harvested, washed with water, and lysed by glass bead disruption in the presence of 2 ml of buffer B/Mg (10 mM HEPES [pH 7.6], 15 mM KCl, 3 mM dithiothreitol, Boehringer Mannheim complete protease inhibitors, 5 mM MgCl2). Lysate was centrifuged at 12,000 × g, and the supernatant was loaded onto a sucrose gradient (consisting of 1 volume [1 or 5 ml] of 70% sucrose in buffer B without MgCl2 and with 10 mM EDTA, 1 volume [1 or 5 ml] of this solution containing 30% sucrose, and 4 volumes [4 or 20 ml] of this solution containing 20% sucrose). Gradients were centrifuged in a Beckman SW-40 rotor at 4°C for 4 h. Fractions (0.75 ml) were collected and tested for RT activity as previously described (20). Positive fractions were pelleted at 50,000 × g overnight at 4°C, resuspended in 10 μl of buffer B/Mg, and stored at 4°C.
Western blot analysis.
For analysis of sucrose gradient fractions, either 2 μl (for integrase analysis) or 5 μl (for Ku analysis) of each 10-μl pelleted sucrose gradient fraction was analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and transferred to a nylon membrane. The membrane was incubated with either monoclonal antibody 8B11 directed against a portion of integrase (gift of J. Boeke; 20) or rabbit polyclonal anti-Yku70p serum and visualized with horseradish peroxidase-coupled anti-mouse or anti-rabbit immunoglobulin G and enhanced chemiluminescence (Amersham).
Immunoprecipitations from VLP preparations.
Nine microliters of a VLP preparation from either strain W303α or the yku70 mutant strain was used for each immunoprecipitation. One microliter of the VLP preparation was diluted to 100 μl with buffer B/Mg and processed for either DNA or protein analysis as the input (I) fraction. Samples to be immunoprecipitated were incubated with 5 μl of anti-Yku70p antibody, rotating, for 1 h at 4°C. One hundred microliters of a 25% (vol/vol) solution of protein A-Sepharose beads was added, and the samples were incubated, rotating, for 1 h at 4°C. The samples were pelleted, washed 10 times with 1.5 ml of buffer B/Mg, and resuspended in 100 μl of buffer B/Mg. The immunoprecipitated pellet (P) fractions were processed for either DNA isolation or protein analysis. For DNA analysis, 10 μl of 10-mg/ml proteinase K in 0.25 M EDTA and 10 μl of 10% SDS were added to 50 μl of the I or P fraction. The samples were incubated for 3 h at room temperature. These were extracted once with phenol-chloroform and once with chloroform. The DNA was ethanol precipitated in the presence of 10 μg of glycogen, washed with 70% ethanol, and resuspended in 25 μl of 10 mM Tris · Cl (pH 8.0). One microliter was used for PCR analysis with primers Kai3 (5′-TCGTACAGTGAAAATGAGACTAATCATACA) and His3-3 (5′-GATTGTCTGCGAGGCAAGAATG). For protein analysis, 5 μl of 10× SDS loading buffer was added to 50 μl of the I or 50 μl of the P fraction and incubated for 5 min at 95°C, and 15 μl was analyzed by SDS-PAGE and Western blotting as described above.
Northern blot analysis.
RNA was isolated from 5-ml cultures of JC297 or JDY15 grown to mid-log phase at room temperature by using the Qiagen RNeasy reagents and protocol. The RNA was quantitated by measuring the OD260, and equal amounts of total RNA were electrophoresed on a 0.8% formaldehyde-agarose gel. The gel was transferred overnight in 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) transfer buffer onto Amersham Hybond-N nylon membrane. The RNA was UV cross-linked to the membrane and hybridized overnight at 68°C in 6× SSC–2× Denhardt’s reagent–0.1% SDS–100 μg of ss DNA per ml. The membrane was washed three times for 30 min each time at 65°C in 0.1× SSC–0.1% SDS and exposed to film. Probes against the Ty1 element and actin were generated by random prime labeling (Prime-a-Gene labeling system; Promega).
In vivo formaldehyde cross-linking.
One-hundred-milliliter cultures of the wild-type and yku70 mutant strains were grown under the indicated selective or inductive conditions and subjected to in vivo formaldehyde cross-linking (38, 44). Briefly, formaldehyde was added to the cultures to a final concentration of 1% and incubated with shaking at room temperature for 15 min. Glycine was added to a final concentration of 125 mM, and cultures were incubated at room temperature for 5 min. Cells were harvested by centrifugation, washed twice with Tris-buffered saline, and resuspended in 800 μl of lysis buffer (50 mM HEPES [pH 7.4], 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, Boehringer Mannheim complete protease inhibitors). Eight hundred microliters of glass beads was added, and the cells were lysed by vortexing 20 × 1 min with 1 min on ice between bursts. The lysate was moved to a microcentrifuge tube and clarified by centrifugation for 15 min at maximum speed in a microcentrifuge at 4°C.
Immunoprecipitation of whole-cell extracts prepared from formaldehyde-treated cultures.
Ten microliters of whole-cell extract was transferred to a new microcentrifuge tube and diluted to 100 μl with 10 mM Tris · Cl (pH 8.0) to be analyzed as I fractions. Five microliters of preimmune serum was added to the remaining whole-cell extract and incubated, rotating, for 1 h at 4°C. One hundred microliters of 50% protein A-Sepharose beads (vol/vol) was added, and the tubes were rotated for 1 h at 4°C. The samples were centrifuged, and the supernatants were moved to new tubes. To these, 5 μl of anti-Yku70p antibody was added and the tubes were incubated, rotating, for 3 h at 4°C. One hundred microliters of 50% (vol/vol) protein A-Sepharose beads was added, and the tubes were rotated for 1 h at 4°C. The immunoprecipitates were pelleted and washed vigorously by inverting and vortexing the tubes for 5 × 5 min with 1.4 ml of lysis buffer, 2 × 5 min with lysis buffer containing 500 mM NaCl, 2 × 5 min with lysis buffer containing 600 mM NaCl, and 5 min with 10 mM Tris · Cl (pH 8.0). The pellets were resuspended in 100 μl of 10 mM Tris · Cl (pH 8.0). To both the I and P fractions, 1.25 μl of 10% SDS and 1.25 μl of 10-mg/ml proteinase K in 0.25 M EDTA were added. Samples were incubated overnight at 37°C and for a further 6 h at 65°C. Samples were extracted once with phenol-chloroform, the organic phase was back extracted with 100 μl of 10 mM Tris · Cl (pH 8.0), and the combined products were extracted once with chloroform. The DNA was ethanol precipitated in the presence of 20 μg of glycogen and resuspended in 25 μl of 10 mM Tris · Cl (pH 8.0). One microliter was used in PCRs using primers His3-4 (5′-AACCAAGTTCGACAACTGCG) and His3-5 (5′-GCAGAAGCAGTAGCAGAACA). These primers amplify a region of the HIS3 marker present in the pGTy1 plasmid across the site of the artificial intron. The product from the plasmid is 422 bp, while the product from the cDNA in which the intron has been spliced out is 318 bp. PCR products were electrophoresed on a 5% nondenaturing polyacrylamide gel, and the region between the 396- and 220-bp molecular size markers was excised and transferred onto nylon membrane (the region of the gel was excised to avoid overwhelming signal levels from the PCR product of plasmid DNA that was present at significant levels in the whole-cell extract). The membrane was analyzed by Southern blotting with radiolabeled His3-5 as the probe.
RESULTS
Ku facilitates Ty1 retrotransposition.
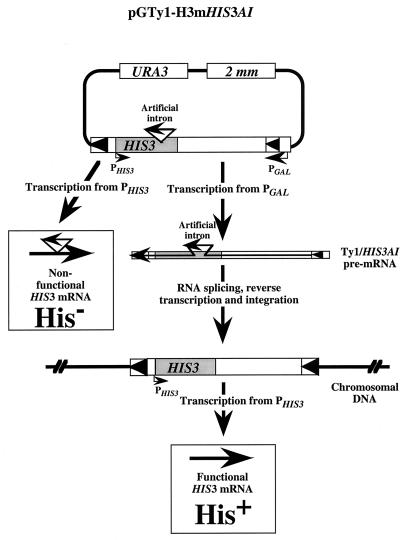
To test whether Ku affects Ty retrotransposition, we used a Ty1 retrotransposition assay developed by Curcio and Garfinkel (14; Fig. 1). In this assay, yeast strains that are mutated in HIS3 are transformed with a plasmid that directs the expression of Ty1 RNA from a galactose-inducible promoter. This Ty1 element contains a HIS3 gene that is interrupted by an artificially inserted intron in reverse (antisense) orientation so that it cannot be spliced, and yeast cells containing this plasmid are phenotypically His−. However, the intron is in sense orientation with respect to the galactose-inducible promoter, so it is removed by splicing of the Ty1 RNA. If this spliced product is reverse transcribed and integrated into the yeast genome, the resulting strain is His+ (Fig. 1). Therefore, the rate of His+ colony generation can be used as a measure of the retrotransposition rate in yeast (14).
FIG. 1.
Schematic of the Ty1 construct (pGTy1H3mHIS3AI or pGTy1 [14]) used to assay transposition frequencies. The artificial intron is in the sense direction relative to the transcription of full-length Ty1 but in the antisense direction relative to HIS3. Therefore, expression of functional HIS3 mRNA is primarily dependent on transcription, splicing, and integration of the reverse-transcribed Ty1 element into the yeast genome.
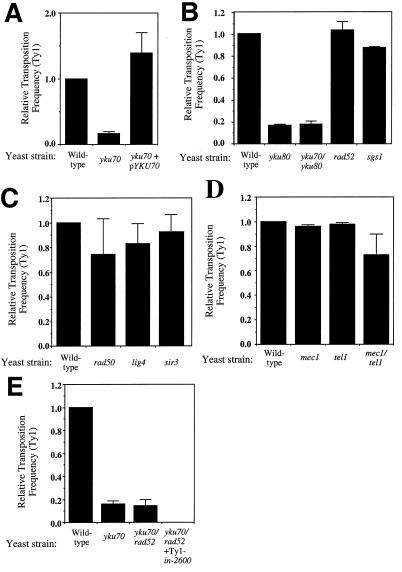
As shown in Fig. 2A, disruption of YKU70 leads to a >80% decrease in Ty1 retrotransposition. This is due to inactivation of YKU70, as transposition rates are restored when the yku70 mutant is complemented with an episomal plasmid containing the YKU70 gene under the control of its endogenous promoter (Fig. 2A). Similarly, yeast cells lacking functional YKU80 are debilitated for Ty1 retrotransposition (Fig. 2B). Furthermore, yeast cells lacking both Ku subunits are no more debilitated for Ty1 retrotransposition than either single mutant alone (Fig. 2B). In contrast, inactivation of several other genes involved in the maintenance of DNA integrity, such as RAD52 and SGS1, have little effect on retrotransposition rates (Fig. 2B). These results indicate that mutations in yeast Ku components reduce yeast Ty1 retrotransposition rates.
FIG. 2.
Inactivation of YKU70 or YKU80, but not other DNA maintenance genes, impairs Ty1 retrotransposition frequencies. (A) Transposition frequencies of Ty1 are decreased in a yku70 mutant background, and this is complemented by an episomal plasmid bearing YKU70. Frequencies are shown relative to that of the wild type, which had a mean Ty1 transposition frequency of 1.5 × 10−4. (B) Ty1 transposition frequencies in yku80, yku70/yku80, rad52, and sgs1 mutant strains relative to that of the wild type. (C) Ty1 transposition frequencies in lig4, rad50, and sir3 mutant strains relative to that of the wild type. (D) Ty1 transposition frequencies in tel1, mec1, and tel1/mec1 mutant strains relative to that of wild-type strain Y661. (E) Ty1 transposition frequencies in yku70, yku70/rad52, and yku70/rad52 strains with Ty1-in2600 relative to that of the wild type.
The role of Ku in Ty1 retrotransposition is specific.
The yeast homologues of mammalian ligase IV (termed Lig4p or Dnl4p), Rad50p, and Sir3p have been shown to work in the same pathway as Ku in repairing DNA ds breaks in yeast (13, 27). Additionally, Ku has been shown to play a role in telomeric silencing (8, 32, 37) and telomeric length maintenance (9, 35, 40) in a manner that is epistatic with Sir3p and Rad50p, respectively. In order to determine whether these other members of the Ku-associated DNA maintenance pathways are involved in facilitating retrotransposition, we examined strains that have disruptions in either RAD50, LIG4, or SIR3. Perhaps surprisingly, rad50, lig4, and sir3 mutant strains showed no significant decrease in retrotransposition rates relative to wild-type cells (Fig. 2C). Taken together, these results show that Ku facilitates retrotransposition in a manner that is distinct from its previously characterized roles and that the effect of Ku on Ty1 retrotransposition is not likely to be an indirect consequence of a defect in DNA ds break repair or telomeric maintenance.
While there is no direct DNA-PKcs homologue in yeast, there are two related nuclear kinases, Mec1p and Tel1p. Both Mec1p and Tel1p have been shown to be involved in DNA damage checkpoint responses (28), and strains with mutations in TEL1 have shorter telomeres than wild-type strains (25). Since Ku is involved in the repair of DNA damage and strains lacking Ku have shorter telomeres than wild-type strains, we addressed the possibility that Mec1p and/or Tel1p function in Ty1 retrotransposition. As shown in Fig. 2D, strains deficient in Mec1p or Tel1p are not impaired in Ty1 retrotransposition and mec1/tel1 double mutants are only impaired marginally. These results further indicate that the effect of Ku on Ty1 retrotransposition is highly specific.
Previous work has shown that, in addition to integrase-mediated Ty1 retrotransposition, Ty1 DNA is able to integrate at low levels into the yeast genome via homologous recombination (34, 42). We therefore tested the effect on Ty1 retrotransposition of mutating the key homologous recombination gene RAD52. As shown in Fig. 2E, residual retrotransposition in yku70 strains is not significantly reduced upon RAD52 inactivation, suggesting that the majority of residual integration in yku70 strains is not mediated via homologous recombination. It has also been shown that the His+ phenotype in the pGTy1 retrotransposition assay can be achieved by a mechanism that is independent of both integrase and homologous recombination and that many cells resulting from this alternative mechanism carry HIS3 on the plasmid (42). However, when residual His+ cells arising in yku70 mutant yeast were induced to lose the Ty1- and URA3-bearing plasmid by selection on 5-fluoro-orotic acid, all of the surviving colonies were still His+ (120 of 120 for the wild type and 118 of 118 for the yku70 mutant), indicating that the HIS3 gene was integrated into the genome. To confirm that the residual integration events seen in the absence of Ku are due to retrotransposition events, retrotransposition assays were performed with an integrase-deficient Ty1 element (pGTy1-in2600; 42). If a significant portion of the residual events that occur in the absence of Ku are a product of an alternative integrase-independent mechanism, there should still be detectable His+ colony formation in the absence of both RAD52 and a functional integrase. However, in yku70/rad52 background strains, the loss of a functional integrase results in retrotransposition rates of less than 10−7 (Fig. 2E). Taken together, these results demonstrate that the residual integration events that occur in the absence of Ku are still the products of integrase-mediated retrotransposition events. Therefore, it appears that Ku operates in the integrase-dependent pathway for Ty1 retrotransposition and that this pathway is facilitated by, but not totally dependent on, the presence of Ku.
Loss of Ku does not impair Ty1 transcription or VLP assembly.
In mammalian systems, there is evidence that Ku has a role in regulating transcription (23, 30, 31). This raised the possibility that the reduced rates of retrotransposition in the absence of Ku are due to a reduction in transcription of the Ty1 element. To address this possibility, we examined the levels of transcription in yku70 mutant cells by using a reporter plasmid containing the lacZ gene under the control of the same galactose-inducible promoter used in the pGTy1 construct. When this reporter construct was used, no reduction of β-galactosidase activity was observed in the absence of Ku (Fig. 3A), suggesting that the lower levels of retrotransposition are not the result of lower levels of Ty1 transcription.
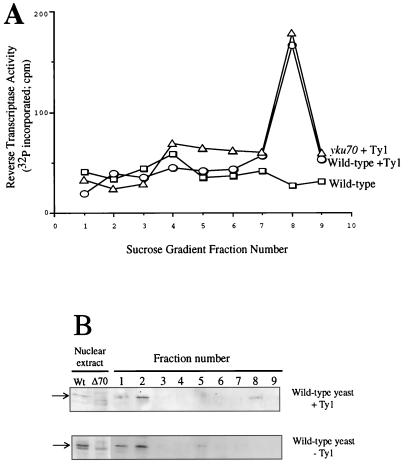
FIG. 3.
VLP formation is normal in yku70 mutant cells. (A) Transcription levels in wild-type (squares) and yku70 mutant (diamonds) cells assayed by using a reporter plasmid containing a lacZ construct under the control of the same galactose-inducible promoter that is on the Ty1 plasmid construct. Cells were grown in the presence of galactose and assayed at various time points after induction for β-galactosidase activity by measuring ONPG cleavage. (B) RT activity profile of sucrose gradient fractions. Whole-cell extracts of the wild-type and yku70 mutant strains containing pGTy1 and grown under inducing conditions were run over sucrose gradients and analyzed for RT activity. (C) Southern analysis of Ty1 cDNA prepared from VLPs from the wild-type and yku70 mutant strains. The DNA was hybridized to a radiolabeled fragment of the pGTy1 plasmid. One and 10 ng, respectively, of the BamHI-XhoI fragment of pGTy1 are shown in the last two lanes. (D) Ty1 cDNAs isolated from VLPs from the wild-type and yku70 mutant strains were digested with SpeI, radiolabeled, and analyzed by nondenaturing PAGE. (E) SpeI cleavage pattern of the Ty1 element. (F) Western blot analysis of VLPs prepared from the wild-type and yku70 mutant strains using monoclonal antibody 8B11 directed at a portion of the integrase (IN) coding sequence (20). It recognizes both the TYA/TYB precursor protein and the mature form of integrase (p90-TYB; IN).
Partial-purification protocols for Ty1 VLPs have been developed by using RT activity levels to monitor the VLPs (20). We used these procedures to determine whether the loss of Ku leads to any significant difference in either the amount or the integrity of Ty1 VLPs. We first analyzed whether the loss of Ku leads to defects either in the amount of RT activity present or in the RT activity profile over a sucrose gradient. To do this, whole-cell extracts from wild-type and yku70 mutant cells were generated from pGTy1-containing yeast that had been grown in the presence of either glucose or galactose. When equal concentrations of whole-cell extract were fractionated over a sucrose gradient, we found no significant difference in the amount of RT activity (Fig. 3B). In addition, we consistently found peak RT activity levels in the same sucrose gradient fractions from wild-type and yku70 mutant cell extracts (Fig. 3B), suggesting that, at least at a gross level, there is no defect in VLP assembly in the absence of Ku.
Ku has been shown to be able to bind to DNA ends in vitro, so one mechanism by which it could potentiate retrotransposition rates was protection of the Ty1 cDNA ends from nucleases. Therefore, we analyzed Ty1 cDNA isolated from VLP preparations generated from either a wild-type or a yku70 mutant yeast strain. Southern blot analysis revealed no detectable difference in the amount of Ty1 cDNA (Fig. 3C), suggesting that the reduction in retrotransposition rates seen in the absence of Ku is not due to lower amounts of cDNA. To look more specifically at the integrity of the ends of the Ty1 cDNA, DNA from VLPs prepared from a wild-type or yku70 mutant strain was digested with SpeI, which liberates a 287-bp fragment corresponding to the 3′ end of the Ty1 cDNA (Fig. 3D and E). The ends of the SpeI-digested DNA were radiolabeled by incubation with Klenow enzyme and analyzed by nondenaturing PAGE. Degradation of Ty1 cDNA ends in a yku70 mutant strain would result in either signal loss or increased mobility of the 287-bp fragment. However, as shown in Fig. 3D, no difference in either the amount or integrity of the DNA fragments was seen in the absence of Ku. These data suggest that there is no significant degradation of the Ty1 VLP cDNA in strains lacking Ku.
Finally, we used monoclonal antibody 8B11, directed against a portion of the integrase coding sequence (20), to determine whether loss of Ku leads to any significant differences in the amount of either the TYA/TYB precursor protein or the mature cleaved form of integrase, both of which are recognized by this antibody. Western blot analysis of VLPs prepared from the wild-type and yku70 mutant strains was performed, and no significant differences were apparent (Fig. 3F), suggesting that there is no loss of translation or proteolytic processing of these proteins in the absence of Ku. We conclude from these data that the reduction in Ty1 retrotransposition rates seen in the absence of Ku is due to a change in a step that occurs downstream of VLP formation.
Ku cofractionates with Ty1 VLPs.
Because Ku is able to bind specifically to DNA ds ends, and since the Ty1 RNA is reverse transcribed in the VLPs to produce a ds linear DNA element, we decided to investigate whether Ku is associated with the Ty1 VLPs. To do this, we fractionated VLPs by sucrose gradient sedimentation and then tested the resulting fractions for RT activity (Fig. 4A) and for the presence of Ku by Western blot analysis using a polyclonal antiserum directed against Yku70p (Fig. 4B; antibody specificity is demonstrated by detection of Yku70p in extracts of wild-type but not yku70 mutant cells). Yku70p was thus detected in sucrose gradient fractions 1 and 2 derived from wild-type cells either lacking or containing an induced Ty1 element (Fig. 4B). Moreover, Yku70p was also detected in fraction 8 of Ty-containing wild-type extracts. This is the same fraction that contains peak RT activity (Fig. 4B, top panel). In contrast, Yku70p was not detected in the equivalent fraction derived from cells lacking Ty1 (Fig. 4B, bottom panel). The presence of Yku70p was found consistently in fractions containing peak RT activity levels in independent VLP preparations, suggesting that Ku and the Ty1 VLPs can associate with one another.
FIG. 4.
Yku70p cofractionates with Ty1 VLPs. (A) Whole-cell extracts prepared from wild-type cells containing the induced Ty1 element (circles), wild-type cells without an induced Ty1 element (squares), and yku70 mutant cells containing the induced Ty1 element (triangles) were fractionated over a sucrose gradient and assayed for RT activity. (B) Western blot analysis of sucrose gradient fractions from panel A with anti-Yku70p antibody. Nuclear extracts from the wild-type (Wt) and yku70 (Δ70) strains were also analyzed. Yku70p is an ∼67-kDa protein recognized in wild-type extracts but not seen in yku70 extracts and is indicated by an arrow.
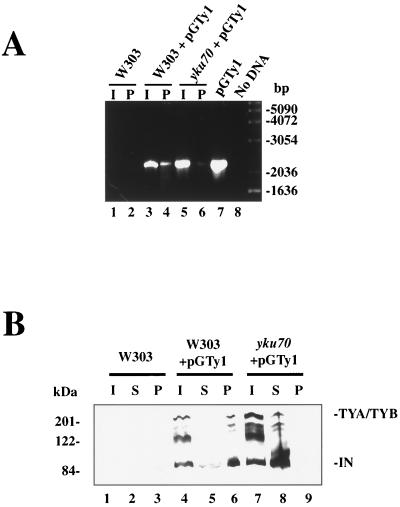
To verify that Ku is associated with the Ty1 VLPs, we used the anti-Yku70p polyclonal serum in immunoprecipitation assays from wild-type or yku70 mutant strains and then tested the immunoprecipitates for the presence of either Ty1 cDNA or the Ty1 integrase protein. To detect the Ty1 cDNA, the I and immunoprecipitated P fractions were used in PCR assays employing primers designed to amplify a portion of the pGTy1 element but not genomic sequences. As a control for primer specificity, we also performed immunoprecipitations from equivalent fractions from wild-type strains lacking the plasmid-borne Ty1 element. These studies yielded a significant level of Ty1 DNA in the immunoprecipitated fractions from wild-type, Ty1-containing strains (Fig. 5A, lane 4) but not in the immunoprecipitates from either a Ty1-containing yku70 mutant strain (lane 6) or a wild-type strain lacking pGTy1 (lane 2). We further analyzed these immunoprecipitates by Western blotting for the presence of integrase (Fig. 5B) and found both TYA/TYB and integrase present in the P fraction from the VLP-containing wild-type strain (lane 6) but not from the wild-type strain lacking VLPs (lane 3) or from the yku70 mutant strain containing the Ty1 VLPs (lane 9). These data provide additional evidence for a specific interaction between Ty1 VLPs and Ku.
FIG. 5.
Ku is associated with the Ty1 cDNA and integrase. (A) Immunoprecipitations were performed with a polyclonal antibody directed against Yku70p. These were done with VLPs prepared from the wild-type strain containing VLPs (W303 + pGTy1, lanes 3 and 4), the yku70 mutant strain (yku70 + pGTy1, lanes 5 and 6), or analogous fractions from the wild-type strain without pGTy1 (W303, lanes 1 and 2). DNA was isolated from the I and P fractions and analyzed by PCR using primers that amplify a portion of pGTy1. PCR products are shown from reactions containing pGTy1 (lane 7) or no DNA (lane 8). (B) Western blot analysis of the I, supernatant (S), and P fractions of the immunoprecipitations performed as described for panel A using monoclonal antibody 8B11 (20) directed against the integrase (IN) protein.
Ku associates with Ty1 VLPs in vivo.
To investigate the physiological relevance of the association detected between Ku and Ty1 VLPs, in vivo formaldehyde cross-linking experiments were performed. In these studies, wild-type and yku70 mutant strains containing the pGTy1 plasmid were grown in the presence of either glucose or galactose and then treated with formaldehyde. Whole-cell extracts were prepared, immunoprecipitations were performed by using the anti-Yku70p polyclonal antibody, and DNA was isolated from the I and immunoprecipitated P fractions. This DNA was analyzed for the presence of Ty1 cDNA by PCR, followed by Southern blotting. Because these experiments were performed with whole-cell extracts, we designed primers around the site of the artificial intron in the pGTy1 plasmid so that PCR products from the plasmid and the reverse-transcribed Ty1 cDNA could be distinguished from one another. With these primers, a product of 422 bp is generated with the pGTy1 plasmid DNA as a substrate (data not shown), whereas if the artificial intron has been spliced out and the RNA is subsequently reverse-transcribed, the expected PCR product is 318 bp. To avoid the overwhelming signal due to plasmid DNA present in the whole-cell extracts, only the region of the gel encompassing the anticipated product size for the spliced cDNA was analyzed by Southern blotting.
As shown in Fig. 6A, whereas PCR assays with DNA isolated from fractionated VLPs generate a product of the expected size (lane 9), when these assays are performed with pGTy1 plasmid DNA as the substrate, no such product is observed (lane 10). In addition, no such product is generated with anti-Yku70p immunoprecipitates from extracts of wild-type cells lacking pGTy1 (lane 11). Notably, however, immunoprecipitates from extracts of a pGTy1-containing wild-type strain generate a strong hybridization signal (lane 8), whereas much less product is detected when immunoprecipitates from extracts of Ty1-containing yku70 mutant cells are analyzed (lane 4). In all cases, and consistent with the galactose inducibility of the pGTy1 system, stronger signals are observed when extracts derived from cells grown in galactose are used than when those from cells grown in the presence of glucose are used.
FIG. 6.
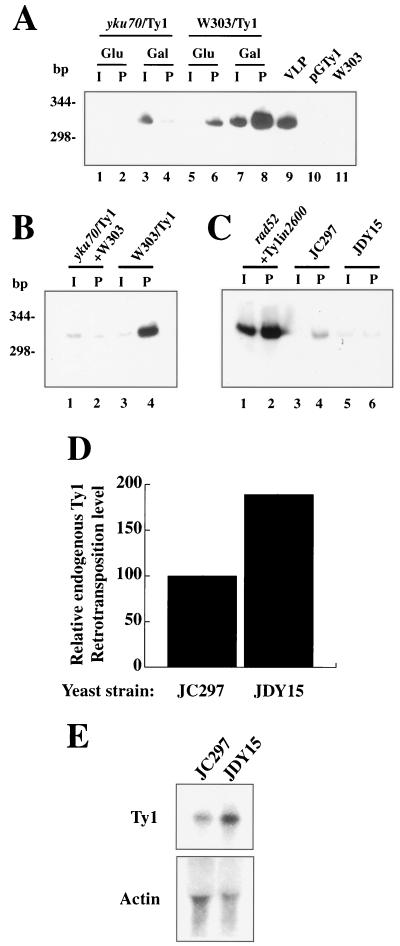
Ku is associated with the Ty1 cDNA in vivo. (A) In vivo formaldehyde cross-linking was performed on the wild-type and yku70 mutant strains, both carrying the pGTy1 plasmid. Whole-cell extracts were prepared and used in immunoprecipitation reactions with an antibody directed against Yku70p. DNA from the I or immunoprecipitated P fraction was isolated and analyzed by PCR and Southern blotting using primers that give a specific product for the Ty1 cDNA. Lanes: 1 and 2, I and P fractions from yku70 mutant cells grown in glucose; 3 and 4, I and P fractions from yku70 mutant cells grown in galactose; 5 and 6, I and P fractions from wild-type cells grown in glucose; 7 and 8, I and P fractions from wild-type cells grown in galactose. PCRs using DNA isolated from a partially purified VLP fraction (lane 9), plasmid DNA (pGTy1, lane 10), and the P fraction from wild-type yeast not containing the pGTy1 plasmid (W303, lane 11) as the substrate are shown, demonstrating the specificity of the PCR product. (B) Whole-cell extracts prepared from formaldehyde-treated wild-type cells not carrying the pGTy1 plasmid (W303) were mixed with whole-cell extracts from formaldehyde-treated yku70 mutant cells carrying the pGTy1 plasmid (yku70/Ty1). Immunoprecipitations and Ty1 DNA analysis using the mixed extracts were performed as described above, and I and P fractions are shown in lanes 1 and 2. In lanes 3 and 4, as a positive control, the immunoprecipitation and Ty1 DNA analysis were performed in parallel by using wild-type cells carrying the pGTy1 plasmid (W303/Ty1). (C) In vivo formaldehyde cross-linking and immunoprecipitations were performed as described above on strain JDY1 (rad52::TRP1) containing integration-deficient pGTy1-in2600 (lanes 1 and 2), JC297 (containing the genomic HIS3AI-marked Ty1 element under the control of its endogenous promoter [11]; lanes 3 and 4), or JDY15 (yku80::URA3 in JC297; lanes 5 and 6). (D) Transposition levels were measured for the genetically marked endogenous Ty1 element in wild-type (JC297) and yku80 mutant (JDY15) yeast strains. Measurements were done five times for each strain. (E) Northern blot analysis of 10 μg of total RNA using probes directed against either the Ty1 element or actin.
The above-described studies provided strong evidence for the binding of Ku to Ty1 VLPs in vivo. However, it remained a possibility that these experiments detected not in vivo interactions but, instead, interactions that might have taken place in vitro upon mixing of nuclear and cytosolic factors following cell lysis. To address this possibility, we performed mixing experiments. Wild-type cells lacking pGTy1 and yku70 mutant cells containing pGTy1 were grown overnight at 23°C in the presence of galactose and then treated with formaldehyde. Extracts from these cells were prepared and mixed prior to immunoprecipitation studies carried out as described above. To provide a positive control, wild-type cells containing pGTy1 were grown, treated with formaldehyde, immunoprecipitated, and analyzed in parallel. As shown in Fig. 6B, the spliced Ty1 product was detectable in the immunoprecipitated P fraction derived from the positive control Ty1-containing wild-type strain (lane 4). However, only background product levels were detected in the P fraction of the mixed samples (lane 2). This demonstrates that the assay system employed detects endogenous Ku-VLP complexes but not interactions that might take place between Ku and the VLPs after cell lysis and mixing.
It was possible that the association detected between Ku and Ty1 DNA was due to interaction of Ku with Ty1 elements that had already integrated into the genome. To determine whether the Ty1 DNA associated with Ku was derived from integrated Ty1 DNA, cross-linking experiments were performed with the integrase-deficient Ty1 element (42) in a rad52 mutant strain in which integration events are undetectable (Fig. 2E). Notably, Ty1 cDNA was still associated with Ku (Fig. 6C, lanes 1 and 2), indicating that Ku associates with the Ty1 cDNA element at a point after reverse transcription but prior to integration into the genome. It can therefore be concluded that a population of yeast Ku is associated with unintegrated Ty1 cDNA in vivo.
Although we had found Ku to be associated with Ty1 cDNA when pGTy1-containing cells were grown under noninduced conditions (in the presence of glucose), it was still possible that the association between Ku and Ty1 VLPs was a consequence of overexpression of the galactose-inducible Ty1 element. To address this issue, we examined the endogenous Ty1 element by using a strain carrying a genetically marked genomic copy of the Ty1 element under the control of its own promoter (11). The level of endogenous Ty1 retrotransposition was found to be almost twofold higher in the absence of Ku when assayed in the same manner employed with the pGTy1 system (Fig. 6D). In contrast to the pGTy1 system, however, we discovered that transcription of the endogenous element is consistently higher in the absence of Ku when analyzed by Northern blotting (Fig. 6E). This result is consistent with previous work showing that the endogenous Ty1 element forms part of the transcriptional response to DNA damage in yeast (10) and with work revealing that other DNA damage-responsive genes are upregulated in cells lacking Ku (3). These observations suggest that the effect of Ku on the transcription of the endogenous Ty1 element is a consequence of an increased level of unrepaired DNA damage in its absence. This transcriptional induction may, therefore, occlude a potential effect of Ku on the downstream events in the endogenous Ty1 life cycle. If Ku acted on downstream events in the endogenous Ty1 element life cycle, we reasoned that, as in the pGTy1 system, it would be associated with the Ty1 cDNA. To test this idea, a wild-type strain and a yku80 mutant strain harboring the endogenous Ty1 element were analyzed by formaldehyde cross-linking and anti-Yku70p immunoprecipitation as previously. Significantly, as shown in Fig. 6C, Ty1 cDNA was enriched in the P fraction from the wild-type strain (lanes 3 and 4) but not in that from the yku80 mutant (lanes 5 and 6). This provides evidence for an association between Ku and the endogenous Ty1 cDNA in vivo. Therefore, although we cannot rule out other explanations, the available evidence is consistent with the idea that Ku functions in a similar positive manner in downstream Ty1 retrotransposition events for both endogenous Ty1 and Ty1 generated by the pGTy1 system. However, such effects on Ty1 retrotransposition levels may be obscured as a consequence of changes in endogenous Ty1 transcription.
DISCUSSION
The results presented here indicate a role for yeast Ku in Ty1 retrotransposition. In contrast, the absence of other proteins that are involved in DNA repair and its maintenance, including other components of the Ku-mediated DNA ds break repair pathway, does not lead to markedly impaired retrotransposition in these assays. Furthermore, although Ku has been shown to affect telomere length and telomeric transcriptional silencing, other proteins examined in this assay that share these activities do not markedly affect Ty1 retrotransposition. Taken together, these observations suggest that the manner in which Ku facilitates retrotransposition is distinct from its other defined cellular roles and argue that the observed reduction of retrotransposition in Ku-deficient cells is not likely to be an indirect consequence of the perturbation of these processes. In addition, our data indicate that the reduction of retrotransposition in the absence of Ku does not result from impaired galactose-inducible transcription or defective VLP assembly. These results suggest that it is likely that Ku plays a role in the retrotransposon life cycle subsequent to VLP formation.
There are various ways in which Ku could potentiate integration of the retrotransposon. First, since Ty1 integrates preferentially into regions upstream of genes transcribed by RNA polymerase III (16), it is possible that Ku functions by targeting Ty1 to such locations. However, we found that Ty3 transposition, which is targeted to different sites in the genome from Ty1 (29), is also reduced significantly in the absence of Ku (48). This therefore argues that Ku is unlikely to facilitate Ty1 retrotransposition solely by targeting the retrotransposition machinery to specific sites in the genome and furthermore suggests that Ku might have a general role in retrotransposition processes.
Another possible role for Ku is suggested by our finding that it is associated with the Ty1 VLPs in vivo. Although this may reflect an association with VLP proteins, an attractive model is one in which Ku binds directly to the Ty1 cDNA. In this way, Ku might serve to prevent degradation of the Ty1 DNA by cellular nucleases, a possibility supported by the fact that Ku does protect dsDNA ends from nuclease-mediated degradation in vitro (17) and by the observation that Ty1 cDNA is susceptible to nuclease attack (15). Since a specific terminal structure of the retroelement DNA is necessary for efficient integration (21, 36), it is conceivable that even limited degradation of the Ty1 DNA in the absence of Ku might result in the loss of viable retroelements. We did not, however, find any significant or reproducible differences in either the amount or the apparent integrity of the Ty1 cDNA isolated from yeast lacking Ku. Nevertheless, it remains a possibility that degradation in the absence of Ku is limited to the few terminal bases of the linear retroelement DNA and/or that Ku protects against nuclease digestion only the subset of Ty1 cDNA that enters the nucleus on its way to integration into the host genome.
An additional potential role for Ku is during the Ty1 integration reaction itself. In this regard, in vitro studies have demonstrated that Ku can bind to DNA ss gaps with affinity comparable to that with which it binds to dsDNA ends (22, 39), raising the possibility that it is involved in the repair of the ss gaps that arise as retrotransposition intermediates. Through binding to such structures and displacement of integrase, Ku could also prevent the reversal of the transposition reaction, disintegration, which results in the removal of the newly integrated product from the substrate DNA. If Ku did function in any of these regards, it would make sense that, in order to be maximally effective, the retroelement would want to ensure that Ku is already present when it initiates its progression toward integration into the host genome. The binding of Ku to VLPs might reflect such a mechanism. Consistent with a role for Ku in facilitating integration of retroelement DNA, Ku and DNA-PKcs have been recently demonstrated to facilitate retroviral integration in mammalian cells (15a). Because transcription, translation, and viral-particle formation occur prior to infection of the new host cell, Ku must play a role in the retroviral life cycle subsequent to reverse transcription but prior to integration. These data are consistent with the role proposed here for Ku during Ty1 element retrotransposition and suggest that Ku may be involved in facilitating the integration of other retroelements in a diversity of eukaryotes.
Finally, previous work has established that Ku plays a role in Drosophila P-element DNA transposition (4). In DNA transposition, the transposable element is excised from one part of the genome before being integrated into a new site by the element-encoded transposase. Therefore, the decrease in P-element transposition rates in the absence of Ku might, at least in part, reflect a requirement for Ku to repair the DNA ds break in the host DNA after the transposable element has been excised. Similarly, it has been suggested that the mechanism by which Ku facilitates V(D)J recombination is repair of the DNA ds breaks created by the RAG1 and RAG2 proteins. Recent findings suggest that the V(D)J recombination machinery is evolutionarily related to that of DNA transposable elements (1, 26), implying that Ku could provide the same type of DNA ds break repair function in both of the above-described processes. Although ds break repair may be the only function of Ku in these processes, it is noteworthy that the mechanism by which retroelement integrases integrate the element into a new target site is essentially the same as that used by the RAG proteins and DNA transposon transposases, and we found a role for Ku in the Ty1 retrotransposon life cycle, where there is no obvious requirement for DNA ds break repair activity. This, therefore, raises the possibility that Ku has a common role in all of these events. It will be of great interest to investigate the association of Ku with V(D)J and DNA transposition intermediates and whether it plays a direct role in integrating these and other transposable elements into the host chromosome.
ACKNOWLEDGMENTS
We thank H. Feldmann and E. L. Winnacker for providing us with the W303α and YKU70/HDF1-disrupted yeast strains, D. Weaver for the RAD52 knockout construct, I. Hickson for the SGS1 knockout construct, S. Hwang-Teo for the LIG4 knockout construct, S. Buratowski for pJA51 and pJA52, S. Elledge for Y661-based strains, J. Boeke for monoclonal antibody 8B11, J. Curcio for strain JC297, and D. J. Garfinkel for pGTyH3mHIS3AI and pGTy1-in2600. Thanks also to L. Yieh, S. Sandmeyer, and members of the S.P.J. lab, particularly Steve Bell, for helpful discussions.
This work was funded by grants SP2143/0102 and SP2143/0401 from The Cancer Research Campaign (UK).
REFERENCES
- 1.Agrawal A, Eastman Q M, Schatz D G. Implications of transposition mediated by V(D)J-recombination proteins RAG1 and RAG2 for origins of antigen-specific immunity. Nature. 1998;394:744–751. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]
- 2.Allen J B, Elledge S J. A family of vectors that facilitate transposon and insertional mutagenesis of cloned genes in yeast. Yeast. 1994;10:1267–1272. doi: 10.1002/yea.320101003. [DOI] [PubMed] [Google Scholar]
- 3.Barnes G, Rio D. DNA double-strand-break sensitivity, DNA replication, and cell cycle arrest phenotypes of Ku-deficient Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1997;94:867–872. doi: 10.1073/pnas.94.3.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beall E L, Rio D C. Drosophila IRBP/Ku p70 corresponds to the mutagen sensitive mus309 gene and is involved in P-element excision in vivo. Genes Dev. 1996;10:921–933. doi: 10.1101/gad.10.8.921. [DOI] [PubMed] [Google Scholar]
- 5.Becker D M, Lundblad V. Transformation of yeast. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: Green Publishing Associates, Inc., and John Wiley & Sons, Inc.; 1994. pp. 13.7.1–13.7.10. [Google Scholar]
- 6.Boeke J D. Transposable elements in Saccharomyces cerevisiae. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C: American Society for Microbiology; 1989. pp. 335–374. [Google Scholar]
- 7.Boulton S J, Jackson S P. Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. 1996;15:5093–5103. [PMC free article] [PubMed] [Google Scholar]
- 8.Boulton S J, Jackson S P. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 1998;17:1819–1828. doi: 10.1093/emboj/17.6.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boulton S J, Jackson S P. Identification of a Saccharomyces cerevisiae Ku80 homologue: roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic Acids Res. 1996;24:4639–4648. doi: 10.1093/nar/24.23.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bradshaw V A, McEntee K. DNA damage activates transcription and transposition of yeast Ty retrotransposons. Mol Gen Genet. 1989;218:465–474. doi: 10.1007/BF00332411. [DOI] [PubMed] [Google Scholar]
- 11.Conte D, Barber E, Banerjee M, Garfinkel D J, Curcio M J. Posttranslational regulation of Ty1 retrotransposition by mitogen-activated protein kinase Fus3. Mol Cell Biol. 1998;18:2502–2513. doi: 10.1128/mcb.18.5.2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Craig N L. Unity in transposition reactions. Science. 1995;270:253–254. doi: 10.1126/science.270.5234.253. [DOI] [PubMed] [Google Scholar]
- 13.Critchlow S E, Jackson S P. DNA end joining: from yeast to man. Trends Biochem Sci. 1998;23:394–398. doi: 10.1016/s0968-0004(98)01284-5. [DOI] [PubMed] [Google Scholar]
- 14.Curcio M J, Garfinkel D J. Single-step selection for Ty1 element retrotransposition. Proc Natl Acad Sci USA. 1991;88:936–940. doi: 10.1073/pnas.88.3.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Curcio M J, Garfinkel D J. New lines of host defense: inhibition of Ty1 retrotransposition by Fus3p and NER/TFIIH. Trends Genet. 1999;15:43–45. doi: 10.1016/s0168-9525(98)01643-6. [DOI] [PubMed] [Google Scholar]
- 15a.Daniel R, Katz R A, Shalka A M. A role for DNA-PK in retroviral DNA integration. Science. 1999;284:644–647. doi: 10.1126/science.284.5414.644. [DOI] [PubMed] [Google Scholar]
- 16.Devine S E, Boeke J D. Integration of the yeast retrotransposon Ty1 is targeted to regions upstream of genes transcribed by RNA polymerase III. Genes Dev. 1996;10:620–633. doi: 10.1101/gad.10.5.620. [DOI] [PubMed] [Google Scholar]
- 17.de Vries E, van Driel W, Bergsma W G, Arnberg A C, van der Vliet P C. HeLa nuclear protein recognizing DNA termini and translocating on DNA forming a regular DNA-multimeric protein complex. J Mol Biol. 1989;208:65–78. doi: 10.1016/0022-2836(89)90088-0. [DOI] [PubMed] [Google Scholar]
- 18.Dvir A, Peterson S R, Knuth M W, Lu H, Dynan W S. Ku autoantigen is the regulatory component of a template associated protein kinase that phosphorylates RNA polymerase II. Proc Natl Acad Sci USA. 1992;89:11920–11924. doi: 10.1073/pnas.89.24.11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dynan W S, Yoo S. Interaction of Ku protein and DNA-dependent protein kinase catalytic subunit with nucleic acids. Nucleic Acids Res. 1998;26:1551–1559. doi: 10.1093/nar/26.7.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eichinger D J, Boeke J D. The DNA intermediate in yeast Ty1 element transposition copurifies with virus-like particles: cell-free Ty1 transposition. Cell. 1988;54:955–966. doi: 10.1016/0092-8674(88)90110-9. [DOI] [PubMed] [Google Scholar]
- 21.Eichinger D J, Boeke J D. A specific terminal structure is required for Ty1 transposition. Genes Dev. 1990;4:324–330. doi: 10.1101/gad.4.3.324. [DOI] [PubMed] [Google Scholar]
- 22.Falzon M, Fewell J W, Kuff E L. EBP-80, a transcription factor closely resembling the human autoantigen Ku, recognizes single- to double-strand transitions in DNA. J Biol Chem. 1993;268:10546–10552. [PubMed] [Google Scholar]
- 23.Giffen W, Torrance H, Rodda D J, Prefontaine G G, Pope L, Hache R. Sequence-specific DNA binding by Ku autoantigen and its effects on transcription. Nature. 1996;380:265–268. doi: 10.1038/380265a0. [DOI] [PubMed] [Google Scholar]
- 24.Gottlieb T M, Jackson S P. The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell. 1993;72:131–142. doi: 10.1016/0092-8674(93)90057-w. [DOI] [PubMed] [Google Scholar]
- 25.Greenwall P W, Kronmal S L, Porter S E, Gassenhuber J, Obermaier B, Petes T D. TEL1, a gene involved in controlling telomere length in S. cerevisiae, is homologous to the human ataxia telangiectasia gene. Cell. 1995;82:823–829. doi: 10.1016/0092-8674(95)90479-4. [DOI] [PubMed] [Google Scholar]
- 26.Hiom K, Melek M, Gellert M. DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell. 1998;94:463–470. doi: 10.1016/s0092-8674(00)81587-1. [DOI] [PubMed] [Google Scholar]
- 27.Kanaar R, Hoeijmakers J H J, van Gent D C. Molecular mechanisms of DNA double-strand break repair. Trends Cell Biol. 1998;8:483–489. doi: 10.1016/s0962-8924(98)01383-x. [DOI] [PubMed] [Google Scholar]
- 28.Keith C T, Schreiber S L. PIK-related kinases: DNA repair, recombination and cell cycle checkpoints. Science. 1995;270:50–51. doi: 10.1126/science.270.5233.50. [DOI] [PubMed] [Google Scholar]
- 29.Kirchner J, Connolly C M, Sandmeyer S B. Requirement of RNA polymerase III transcription factors for in vitro position-specific integration of a retroviruslike element. Science. 1995;267:1488–1491. doi: 10.1126/science.7878467. [DOI] [PubMed] [Google Scholar]
- 30.Kuhn A, Gottleib T M, Jackson S P, Grummt I. DNA-dependent protein kinase—a potent inhibitor of transcription by RNA polymerase I. Genes Dev. 1995;9:193–203. doi: 10.1101/gad.9.2.193. [DOI] [PubMed] [Google Scholar]
- 31.Labhart P. DNA-dependent protein kinase specifically represses promoter-directed transcription initiation by RNA polymerase I. Proc Natl Acad Sci USA. 1995;92:2934–2938. doi: 10.1073/pnas.92.7.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laroche T, Martin S G, Gotta M, Gorham H C, Pryde F E, Louis E J, Gasser S M. Mutation of yeast Ku genes disrupts the subnuclear organization of telomeres. Curr Biol. 1998;8:653–656. doi: 10.1016/s0960-9822(98)70252-0. [DOI] [PubMed] [Google Scholar]
- 33.Lieber M R, Grawunder U, Wu X T, Yaneva M. Tying loose ends: roles of Ku and DNA dependent protein kinase in the repair of double strand breaks. Curr Opin Genet Dev. 1997;7:99–104. doi: 10.1016/s0959-437x(97)80116-5. [DOI] [PubMed] [Google Scholar]
- 34.Melamed C, Nevo Y, Kupiec M. Involvement of cDNA in homologous recombination between Ty elements in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:1613–1620. doi: 10.1128/mcb.12.4.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Milne G T, Jin S, Shannon K B, Weaver D T. Mutations in two Ku homologs define a DNA end-joining repair pathway in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4189–4198. doi: 10.1128/mcb.16.8.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moore S P, Powers M, Garfinkel D J. Substrate specificity of Ty1 integrase. J Virol. 1995;69:4683–4692. doi: 10.1128/jvi.69.8.4683-4692.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nugent C I, Bosco G, Ross L O, Evans S K, Salinger A P, Moore J K, Haber J E, Lundblad V. Telomere maintenance is dependent on activities required for end repair of double strand breaks. Curr Biol. 1998;8:657–660. doi: 10.1016/s0960-9822(98)70253-2. [DOI] [PubMed] [Google Scholar]
- 38.Orlando V, Paro R. Mapping polycomb-repressed domains in the bithorax complex using in vivo formaldehyde cross-linked chromatin. Cell. 1993;75:1187–1198. doi: 10.1016/0092-8674(93)90328-n. [DOI] [PubMed] [Google Scholar]
- 39.Paillard S, Strauss F. Analysis of the mechanism of interaction of simian Ku protein with DNA. Nucleic Acids Res. 1991;19:5619–5624. doi: 10.1093/nar/19.20.5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Porter S E, Greenwell P W, Ritchie K B, Petes T D. The DNA-binding protein Hdf1p (a putative Ku homologue) is required for maintaining normal telomere length in Saccharomyces cerevisiae. Nucleic Acids Res. 1996;24:582–585. doi: 10.1093/nar/24.4.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reynolds A, Lundblad V. Assay for beta-galactosidase in liquid cultures. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: Greene Publishing Associates, Inc., and John Wiley & Sons, Inc.; 1994. pp. 13.6.1–13.6.4. [Google Scholar]
- 42.Sharon G, Burkett T J, Garfinkel D J. Efficient homologous recombination of Ty1 element cDNA when integration is blocked. Mol Cell Biol. 1994;14:6540–6551. doi: 10.1128/mcb.14.10.6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith G C M, Jackson S P. The DNA-dependent protein kinase. Genes Dev. 1999;13:916–934. doi: 10.1101/gad.13.8.916. [DOI] [PubMed] [Google Scholar]
- 44.Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- 45.Treco D A, Lundblad V. Yeast media. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: Greene Publishing Associates, Inc., and John Wiley & Sons, Inc.; 1994. pp. 13.1.1–13.1.7. [Google Scholar]
- 46.van Gent D C, Mizuuchi K, Gellert M. Similarities between initiation of V(D)J recombination and retroviral integration. Science. 1996;271:1592–1594. doi: 10.1126/science.271.5255.1592. [DOI] [PubMed] [Google Scholar]
- 47.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 48.Yieh, L., J. A. Downs, S. P. Jackson, and S. B. Sandmeyer. Unpublished data.