Abstract
Bitter taste perception in sheep can lead to avoidance of specific types of forage, such as sagebrush, which is present on many rangeland grazing systems in the Intermountain West. In humans, bitter taste perception is influenced by variation in several TAS2R genes, including more extensively studied TAS2R38 and TAS2R16. We hypothesize that variation in taste receptor genes in sheep is associated with bitter taste. Therefore, the objective of this study was to examine variation in TAS2R genes in relation to consumption of a bitter tasting compound phenylthiocarbamide (PTC) which determines bitter “taster” and “non-taster” status in humans. Rambouillet and Targhee rams (n = 26) were offered various concentrations of PTC solution (0.2–12.29 mM) and water in a side-by-side presentation during two experiments. Blood was collected for DNA isolation and sequencing. Nineteen TAS2R genes were amplified and sequenced with long read Oxford Nanopore MinION technology. A total of 1,049 single nucleotide polymorphisms (SNPs) and 26 haplotypes were identified in these genes. Of these, 24 SNPs and 11 haplotypes were significantly (P < 0.05) associated with PTC consumption in TAS2R3, TAS2R5, TAS2R8, TAS2R9, TAS2R16, TAS2R31-like, TAS2R38, TAS2R39, and TAS2R42-like. Over 50% of the SNPs resulted in a change in amino acid sequence and several resided in potential regulatory regions, which could have downstream functional consequences and influence bitter taste perception in sheep. Further research is needed to validate these associations and elucidate the mechanisms that link variation in TAS2R genes to bitter taste perception in sheep. This may enable producers to select sheep more likely to consume bitter forage such as sagebrush as a flock and rangeland management strategy.
Keywords: bitter taste, genetics, grazing, rangeland, sagebrush, sheep
INTRODUCTION
Dietary preference in humans is genetically associated with the 7-transmembrane G-protein coupled receptors known as type-two taste receptors (TAS2R) (Drewnowski and Rock, 1995). These are the only known taste receptors to perceive bitterness (Adler et al., 2000). Bitterness is the most sensitive of the five taste senses in sheep, often resulting in the avoidance of some forages (Goatcher and Church, 1970). Therefore, selection of sheep that could be genetically inclined to graze on less palatable and more bitter tasting vegetation could have economic and production applications in the areas of grazing management, although further research is needed for practical application.
TAS2R38 and TAS2R16 are among the variety of these genes responsible for bitter taste perception (Bufe et al., 2002; Kim et al., 2003). For instance, activation of TAS2R16 receptors is initiated by bitter tasting β-glucopyranosides, such as salicylic acid (Bufe et al., 2002). The TAS2R38 receptors detect chemicals such as phenylthiocarbamide (PTC), which has been used extensively to test sensitivity to bitterness in humans and mice (Kim et al., 2003). While PTC is not a naturally occurring substance, the ability to perceive bitter tasting chemical compounds, such as PTC, has been correlated with the ability of animals to taste other bitter-tasting foods (Glanville and Kaplan 1965). In humans, PTC avoidance has specifically been linked to amino acid haplotype variations (PAV for “tasters” and AVI for “non-tasters”) on the TAS2R38 gene (Kim et al., 2003).
Much like humans, rams have varying levels of sensitivity to PTC (Henslee et al., 2019). To date, there have been no studies conducted in sheep aimed at investigating the relationships between TAS2R genes and phenotypic traits, such as dietary preferences. Since TAS2R38 and similar TAS2R genes are also found in sheep (Ferreira et al., 2013; Henslee et al., 2020), we hypothesized that there may exist similar genetic and phenotypic expression of bitterness avoidance. Thus, the purpose of this study was to evaluate the relationship between the consumption level of PTC and haplotype of various TAS2R genes within the ram.
MATERIALS AND METHODS
Animals and Phenotype Collection
All animal procedures were approved by an Institutional Animal Care and Use Committee (USDA, ARS, Dubois, ID) in accordance with the USDA, APHIS Animal Welfare Regulations (2013; 9 C.F.R. § 2.30−2.38 2013) and the Guide for the Care and Use of Agricultural Animals in Research and Teaching (FASS, 2010). Two trials were conducted at the USDA, ARS U.S. Sheep Experiment Station, Dubois, ID, in the spring of 2018. Phenylthiocarbamide (PTC) intake data was collected from yearling Rambouillet and Targhee rams (n = 26; n = 11 Rambouillet and n = 15 Targhee) where various concentrations of PTC solution (0.20–12.29 mM) and water were offered in a side-by-side presentation (Henslee et al., 2019). The experimental design, animal treatment, and animals were previously described by Henslee et al., 2019. Briefly, rams were housed in individual pens indoors and the environment was controlled at 10 °C with a 12 h light:dark cycle (Henslee et al., 2019). Feed, water and the treatment PTC solution were offered for ad libidum intake except between 1700 and 0700 h each day (Henslee et al., 2019). Rams were randomly assigned to pens within a breed and five treatments were assigned in a cross-over design (Henslee et al., 2019). The buckets with water and PTC solution were randomly assigned a location each time they were delivered (Henslee et al., 2019). Individual variation in the amount of PTC consumed was detected and rams clustered into low, medium, and high PTC consumption groups (Henslee et al., 2019). Blood was collected using EDTA tubes (BD Vacutainer EDTA tube, BD, USA) for DNA isolation post-treatment.
Classification of PTC consumption into high, medium, and low categories were calculated from total fluid intake (TFI) data (Henslee et al., 2019). In the absence of external influences, theoretically 50% of TFI should be consumed from each bucket (Goatcher and Church, 1970). Thus, classifications were formulated using a mean of 50% PTC consumption of TFI and divided into thirds, which resulted in three categories of equal range (low ≤ 16.6%; 16.6% < intermediate < 33.4%; high ≥ 33.4%). The number of rams that fell into the low, intermediate, and high categories was 8 (n = 5 Rambouillet and n = 3 Targhee), 10 (n = 4 Rambouillet and n = 6 Targhee), and 8 (n = 2 Rambouillet and n = 6 Targhee), respectively.
DNA Extraction and Quantification
DNA was extracted from 500 µL of blood from each ram (n = 26) following the phenol chloroform method previously described and eluted in nuclease free water (Sambrook et al., 1989). Concentration and quality of DNA were determined with the Nanodrop Spectrophotometer (Thermo Fisher Scientific, Waltham, MA). All DNA extracted was above 20ng/ul in concentration and had a 260/280 ratio of >1.8 and 260/230 ratio of >1.9. Each sample was then diluted to 5 ng/µL with nuclease free water to perform PCR amplification.
PCR Amplification of Target Genes
Amplification of target taste receptor genes in sheep was performed with FastStart High Fidelity PCR System, dNTPack (cat. 4738292001, Millipore Sigma, Burlington, MA) according to manufacturer’s instructions. Thermocycling conditions included an initial denature at 95 °C for 2 min followed by 35 cycles of denature at 95 °C for 30 s, annealing at 60 °C for 20 s, and extension at 72 °C for 40 s, followed by a final extension of 72 °C for 5 min. Amplicons were stored at 4 °C until sequencing. A list of genes, forward and reverse primers, and amplicon length which span the entire gene amplified are displayed in Table 1.
Table 1.
Primers designed to amplify the entire family of known TAS2R genes in sheep
| Gene | Gene Length (bp) | Forward primer* | Reverse primer* | Amplicon length (bp)** |
|---|---|---|---|---|
| TAS2R3 | 2841 | CAGCTAACGGTCTGGAGGTC | CAGTAACAGCTTCACCGCCT | 3111 |
| TAS2R4 | 1710 | CCCAGGTTCACTTTGGTGGT | CCACAGTCCTGCTGTTCCAA | 2119 |
| TAS2R5 | 2645 | AGATTGCAGAAGGGTAAGACCA | TATCTCAAAACAGTCTCCTGACCAC | 2807 |
| TAS2R7 | 939 | GGGACCGACAACTGCATTAC | TCCTCTGGCAGTTACTGTTAAGAT | 1456 |
| TAS2R8 | 929 | GAGCTTGGAACTTTCGGAGGA | GTGCACTTTAGTAGGGGCCA | 1399 |
| TAS2R9 | 1164 | TTTGAAGTCCCTGGCCAACA | TGGTGTGAAGTGTGAACGTGA | 1500 |
| TAS2R10 (LOC101115110) | 930 | AGGCATTCAGTCTGGGTGTG | GGGAGAAACCACTGGCAAGA | 1398 |
| TAS2R10-like (LOC101122269) | 900 | TGGAGGCATCTCTGTCAAGC | GGGAGAAACCACTCCAAGGG | 1349 |
| TAS2R12 (LOC101114857) | 912 | AGCAGTGGCGACACATACAT | TGAGAGGTCATCATCACTTCAGG | 1180 |
| TAS2R16 | 928 | AGATGGCTGTGGGCAAAGAG | GGAACCTGGTCCCAAACTGG | 1126 |
| TAS2R31-like (LOC101121003) | 914 | TCCATCCCATAGTAGGGCAC | AGACACTTTTTGTTATTAGCTCAGG | 1318 |
| TAS2R38 | 1002 | GTGGAAGGGCCCATTGATGTA | AGCTTCTGCATCACCCAAGG | 1448 |
| TAS2R39 | 1092 | CACACCAGCGCATCCAAAAA | CAGCCCCGGAAATCTTGACT | 1536 |
| TAS2R40 | 1364 | TAAACCGGGACTCTTGCCCT | TGACTCTGGGTTAGTGGGGT | 1880 |
| TAS2R41 | 936 | GAGCTCAGTCACAGACACCC | TCCCAAAGGAGAAAGCCCAC | 1391 |
| TAS2R42 | 930 | TGCCGATGATGAATGCACAC | GCCTCTTCTCCCAAATACGAGT | 1519 |
| TAS2R42-like (LOC101120742) | 936 | TGCCAGCACCAATGATGAGT | GGGCATGTCCAAATGATCGTG | 1522 |
| TAS2R60 | 954 | AATTCATGGACAGGCAGCGA | TCTTTGGCCACATCAGGTCC | 1438 |
| TAS2R67 (LOC101120486) | 939 | AGTGGGCACATTCACTGCTT | TGATGCCAGTGATGCTTGCT | 1416 |
* Gene specific primers do not include Oxford Nanopore forward and reverse sequence tags.
**Amplicon length was determined from sequence data.
Library Preparation and Sequencing
Target gene amplicons were prepared for sequencing following the PCR Barcoding (96) Amplicons (SQK-LSK109) protocol available through the Oxford Nanopore community (www.nanoporetech.com). Barcoded amplicons were purified using MagBio HighPrep PCR magnetic beads (cat. AC-60005, MagBio Genomics, Gaithersburg, MD) according to manufacturer’s instructions. Amplicons were quantified with a NanoDrop Spectrophotometer, pooled in equimolar amounts, and subjected to end repair and prep and adapter ligation. Amplicons had a 260/230 ratio of >1.8 and 260/230 ratio of >2.0. A final cleanup was performed with AMPure XP magnetic beads (cat. A63880, Beckman Coulter, Indianapolis, IN) according to manufacturer’s instructions and quantified with a Qubit fluorometer (Thermo Fisher Scientific, Waltham, MA). The library was sequenced with a MinION device using one flow cell (R9.4.1; www.nanoporetech.com) for 48 hours.
Bioinformatics and Data Processing
Base calling, demultiplexing by barcode, and trimming was performed with Guppy v3.2.2 (www.nanoporetech.com). The quality of FASTQ sequences were assessed with FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Sequences were then mapped to Oar_rambouillet_v1.0 with minimap2 v2.17 using default settings and indexed for visualization in the Integrative Genomics Viewer with SAMtools (Li et al, 2009; Li 2018; Salavati et al., 2020). Single nucleotide polymorphisms (SNPs) and haplotypes were identified in regions with greater than 100× coverage with marginPhase (Ebler et al., 2019). SNPs with a quality score <100 were discarded. Haplotypes were verified with visual inspection in the Integrative Genomics Viewer and manual examination of the sequence data (Robinson et al., 2011).
Genetic Associations Analyses with PTC Consumption
Individual SNPs identified in the target genes for each animal were imported into the SNP & Variation Suite software v8 (Golden Helix, Bozeman, MT). Basic, genotypic, and additive genetic association tests were performed with the PTC consumption categories to capture the implied additive nature of this phenotype described previously and also investigate dominant and recessive associations with individual alleles and genotypes (Henslee et al., 2019). Haplotype associations with PTC categories were examined with a trend regression. Significance for SNPs and haplotypes was declared at P < 0.05. Variation in each gene was tested individually and independently.
RESULTS
In total, 387,002 sequences were generated across all 19 TAS2R gene regions and 26 animals. A total of 1,049 SNPs were identified in these TAS2R gene target regions. A total of 24 SNPs were identified as significantly associated with PTC consumption category (P < 0.05) in seven TAS2R genes, specifically TAS2R42-like, TAS2R31-like, TAS2R9, TAS2R8, TAS2R16, TAS2R3, and TAS2R5. The precise genomic location of the SNP of each of the specific TAS2R genes are shown in Table 2. Significant SNPs were identified in the promoter 5’ and 3’ untranslated region of TAS2R42-like, TAS2R9 and TAS2R8, respectively. All other associated SNPs were identified in the coding regions of the TAS2R genes. Of these, 9 SNPs were non-amino acid changing, 11 SNPs were amino acid changing and one induced a stop codon. The most significant (P = 0.002) SNPs that were identified were in TAS2R16.
Table 2.
Significant SNPs in taste receptor genes associated with PTC intake in rams.
| Position* | Gene** | SNP ID number (dbSNP) | SNP model | SNP P-value |
Amino acid change |
|---|---|---|---|---|---|
| 3: 218872939 | upstream of TAS2R42-like | Novel | Genotypic | 0.045 | N/A |
| 3: 218873800 | TAS2R42-like | Novel | Genotypic | 0.009 | Ala/Ser |
| 3: 218909947 | TAS2R31-like | rs419195294 | Basic | 0.031 | Asp/Gly |
| 3: 219066474 | TAS2R9 | rs404321924 | Basic | 0.014 | Gly/Asp |
| 3: 219066528 | TAS2R9 | rs411528391 | Basic | 0.043 | Asn/Ser |
| 3: 219066529 | TAS2R9 | rs422591339 | Basic | 0.043 | Asn/Asn |
| 3: 219066621 | TAS2R9 | rs416718763 | Basic | 0.034 | Pro/Pro |
| 3: 219066682 | TAS2R9 | rs428638379 | Basic | 0.014 | Ile/Val |
| 3: 219066686 | TAS2R9 | rs403087052 | Basic | 0.007 | Arg/Lys |
| 3: 219068310 | TAS2R8 | rs412528176 | Additive | 0.034 | Tyr/Tyr |
| 3: 219068366 | TAS2R8 | rs423882545 | Additive | 0.034 | Leu/STOP |
| 3: 219068463 | TAS2R8 | rs404141749 | Additive | 0.034 | Lys/Lys |
| 3: 219068570 | TAS2R8 | rs426576409 | Additive | 0.010 | Lys/Asn |
| 3: 219068699 | TAS2R8 | rs416054395 | Additive | 0.010 | Thr/Thr |
| 3: 219068761 | TAS2R8 | rs410022040 | Additive | 0.010 | Thr/Lys |
| 3: 219069019 | Downstream of TAS2R8 | rs419056212 | Additive | 0.009 | N/A |
| 3: 219069021 | Downstream of TAS2R8 | rs401680429 | Additive | 0.009 | N/A |
| 4: 95494095 | TAS2R16 | rs420035590 | Basic | 0.002 | Lys/Lys |
| 4: 95494125 | TAS2R16 | rs400637145 | Basic | 0.002 | His/His |
| 4: 95494164 | TAS2R16 | rs412097853 | Basic | 0.015 | Asp/Asp |
| 4: 113926390 | TAS2R3 | rs425774520 | Basic | 0.023 | Ser/Ser |
| 4: 113927769 | TAS2R3 | rs413852493 | Basic | 0.013 | Asp/Gly |
| 4: 113928198 | TAS2R3 | rs415725026 | Basic | 0.004 | Arg/Lys |
| 4: 113956501 | TAS2R5 | rs422577624 | Basic | 0.048 | Tyr/Cys |
* Position is displayed as chromosome number: base pair location.
**The entire gene including upstream in the 5’ region and downstream in the 3’ region was amplified and sequenced to include coding and potential promoter and regulatory regions for each gene.
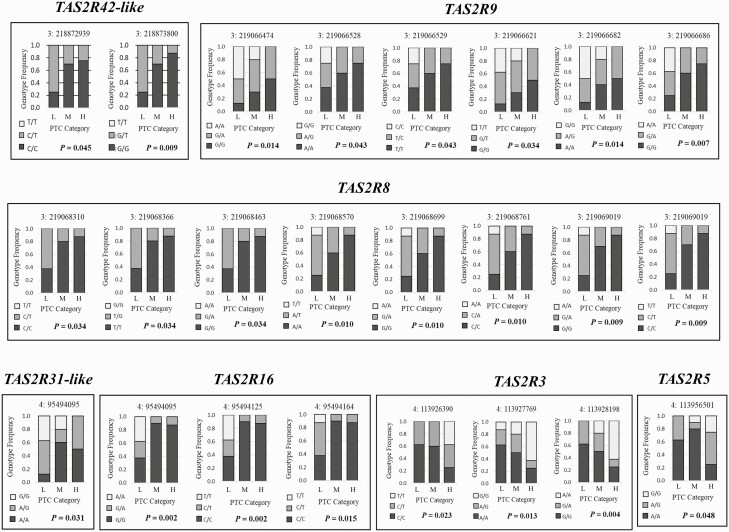
Next, we examined how each of the significant genotypes for each of the genes were associated with different amounts of PTC intake. Bar graphs for each significantly associated genotype were generated for low, medium and high PTC intake in order to gain a better understanding of the genetic relationship (Figure 1). In the TAS2R42-like gene, animals with C/C and G/G genotypes at 218,872,939 bp and 218,873,800 bp consumed more PTC. Whereas in the TAS2R9 gene animals with A/A, G/G, C/C, T/T, G/G, and A/A genotypes consumed less PTC. The TAS2R8 gene harbored the greatest number of significant SNPs, where animals with C/C, T/T, G/G, A/A, G/G, C/C, G/G, and C/C genotypes consumed more PTC. In contrast, TAS2R31-like only exhibited one significant SNP, where animals with a G/G consumed less PTC. Within the TAS2R16 gene, which encompasses the two most significant SNPs, animals with an A/A, T/T, and T/T genotypes at these three locations consumed less PTC. Lastly, animals with a T/T, G/G, and A/A in TAS2R3 significant SNP locations and a G/G in the single TAS2R5 significant SNP consumed a greater amount of PTC.
Figure 1.
Genotype frequency in each PTC consumption category of low (represented by L), medium (represented by M), and high (represented by H) across SNPs significantly associated with PTC consumption category in TAS2R42-like, TAS2R9, TAS2R8, TAS2R31-like, TAS2R16, TAS2R3, and TAS2R5.
Interestingly, animals with the alternative genotype at all of these loci exhibit increasing willingness to consume PTC in an additive manner. Although the significant SNPs in TAS2R9, TAS2R31-like, TAS2R16, TAS2R3, and TAS2R5 had the most significant p-value in the basic genetic test, the additive genetic model was also significant (P < 0.05) for all of these SNP locations. In other words, the amount of PTC intake increased with heterozygosity and further increased in animals that exhibited homozygosity for the other SNP at all of the significant loci.
Several p-values of significant SNPs within the same gene were identical. This prompted us to examine if haplotypes in TAS2R genes harboring several SNPs were significantly associated with PTC consumption. A total of 26 haplotype blocks were identified across all genes, with 11 reaching significance and 9 containing independently significant SNPs (Table 3). The TAS2R42-like gene had the most significantly associated (P = 5.07E−05) haplotype with PTC consumption in a large block of 22 SNPs. The TAS2R31-like gene contained 12 SNPs in a haplotype block, with 1 SNP individually associated. Just over half of the SNPs in the significant TAS2R9 haplotype block were independently associated with PTC consumption. Interestingly, the TAS2R8 gene had two different significant haplotype blocks, with the most significant of the two (P = 0.007) the end and past the gene, with all four SNPs in the block also individually associated with PTC consumption. Significant haplotype blocks in TAS2R16 and TAS2R3 also contained SNPs individually associated with PTC consumption, whereas within in TAS2R5, only one SNP was individually significant. Three additional genes had significant haplotype blocks without independently significant SNPs: TAS2R4, TAS2R38, and TAS2R39.
Table 3.
Significant haplotypes in taste receptor genes associated with PTC intake in rams.
| Gene | Locations and haplotype* | P-value |
|---|---|---|
| TAS2R42-like | 3:218872709, 3:218872798, 3:218872818, 3:218872939, 3:218872942, 3:218872977, 3:218873004, 3:218873008, 3:218873059, 3:218873295, 3:218873381, 3:218873404, 3:218873413, 3:218873518, 3:218873552, 3:218873678, 3:218873703, 3:218873722, 3:218873771, 3:218873800, 3:218873802, 3:218873869 | 5.067E−05 |
| G, T, A, T, C, G, A, G, G, C, T, T, T, T, G, C, C, G, G, T, G, A | ||
| TAS2R31-like | 3:218909211, 3:218909225, 3:218909242, 3:218909259, 3:218909471, 3:218909696, 3:218909813, 3:218909860, 3:218909873, 3:218909924, 3:218909947, 3:218910086 | 0.004 |
| T, T, G, A, T, T, G, C, T, G, A, C | ||
| TAS2R9 | 3:219066474, 3:219066528, 3:219066529, 3:219066621, 3:219066682, 3:219066686, 3:219067174, 3:219067187, 3:219067247, 3:219067271, 3:219067323 | 0.002 |
| G, A, T, G, A, G, A, A, G, G, G | ||
| TAS2R8 | 3:219068310, 3:219068366, 3:219068393, 3:219068463, 3:219068570, 3:219068572, 3:219068634 | 0.030 |
| T, G, A, A, T, A, C | ||
| TAS2R8 | 3:219068699, 3:219068761, 3:219069019, 3:219069021 | 0.007 |
| A, A, A, T | ||
| TAS2R16 | 4:95494095, 4:95494125, 4:95494164 | 0.009 |
| G, C, C | ||
| TAS2R3 | 4:113926390, 4:113927769 | 0.028 |
| T, G | ||
| TAS2R4 | 4:113940107, 4:113940522, 4:113941205, 4:113941293, 4:113941383, 4:113941740, 4:113941860, 4:113941862, 4:113941960 | 0.050 |
| C, G, G, C, C, T, C, G, T | ||
| TAS2R5 | 4:113955710, 4:113956096, 4:113956118, 4:113956150, 4:113956190, 4:113956216, 4:113956230, 4:113956501 | 0.043 |
| C, T, A, C, C, T, G, A | ||
| TAS2R38 | 4:114099430, 4:114099567, 4:114099648, 4:114099679, 4:114099708, 4:114099738, 4:114099818, 4:114099916, 4:114100153, 4:114100217 | 0.027 |
| A, A, T, G, T, A, G, G, C, T | ||
| TAS2R39 | 4:115407175, 4:115407221, 4:115407222, 4:115407601, 4:115407644, 4:115407656, 4:115407701, 4:115407842, 4:115407991, 4:115408072, 4:115408190, 4:115408410, 4:115408492 | 0.018 |
| T, T, G, A, C, G, G, A, T, T, T, C, T, C |
* Position is displayed as chromosome number: base pair location. Locations and nucleotides in bold are SNPs that are significant individually as well as within the haplotype.
DISCUSSION
A total of 24 SNPs and 11 haplotype blocks in TAS2R genes were associated with PTC consumption in rams enrolled in this study. All SNPs significantly associated with PTC consumption except three were in coding regions of TAS2R genes, which is not surprising given that many of these genes are comprised of only a single exon. Of the significant SNPs, over half resulted in a change in an amino acid and one resulted in a stop codon, which may have downstream consequences related to the function of TAS2Rs and bitter taste perception. Upstream of TAS2R42-like also displayed a significant SNP, which is within the 5′ untranslated region (UTR) of the gene and likely contains a promoter. Two significant SNPs reside in the 3′ UTR region of TAS2R8 which may also be a regulatory region. These changes in exon and regulatory regions may have significant influence on TAS2R function, however additional studies are needed to directly link these genetic changes to functional consequences.
The two most significant SNPs reside in TAS2R16, which has been found in humans to detect bitter tasting β-glucopyranosides, such as salicylic acid in willow bark, and may be important for sheep encountering similar substances in their grazing regime (Bufe et al., 2002). In addition, polymorphisms in TAS2R3, TAS2R5, TAS2R9, and TAS2R31 have been linked to human intake of brassica vegetables, grapefruits and even the bitter undertones of artificial sweeteners (Mikołajczyk-Stecyna et al. 2020; Allen et al., 2013). This study suggests that more than one taste receptor gene potentially influences bitter taste perception as assessed by PTC but will need validation in a larger population of animals. Further, it is more difficult to characterize bitter taste perception in animals than it is in humans.
In addition to individual SNPs, haplotypes from 10 genes were associated with PTC consumption, including TAS2R38 which also displays significant haplotypes (PAV and AVI) related to PTC taster status in humans (Kim et al., 2003). This gene exhibits 76.45% homology between sheep and humans (Henslee et al., 2020). Taken together with significant SNPs, these results suggest conservation of genetic influence on bitter taste perception across mammalian species. Interestingly, differences between the two breeds, Targhee and Rambouillet, were not detected however a larger dataset is needed to investigate any potential breed differences. Further research is needed to examine breed differences in the genetic relationship to bitter taste perceptions in sheep. In summary, this study identified several SNPs and haplotypes significantly associated with consumption of the bitter tasting PTC compound in sheep. This is important as genomic information could be informative in selecting sheep to graze specific forages available in the production environment. Further, sheep can serve as rangeland management tool by controlling overgrowth and weed-like behavior of shrubs which can lead to a reduction of rangeland plant diversity, carrying capacity, and wildlife abundance (Johnson et al., 1996; Launchbaugh, 2003). Sheep grazing can be utilized as a natural and practical alternative to other brush control strategies such as plowing, burning, and spraying when selected for specific management goals using genetic tools (Wambolt and Payne, 1986).
IMPLICATIONS
This study identified variation in TAS2R genes significantly associated with consumption of the bitter tasting compound PTC. While PTC is not a naturally occurring substance, sensitivity to these compounds have been correlated with the perceived intensity of other bitter foods in mammals (Glanville and Kaplan, 1965). Understanding genetic relationships to bitter taste perception could have implications in grazing systems by enabling producers to select sheep that are more likely to consume bitter forages, such as sagebrush, and implement management strategies that compliment both sheep production and rangeland habitat.
ACKNOWLEDGMENTS
This project was supported by Hatch grant no. IDA01566 from the USDA National Institute of Food and Agriculture.
LITERATURE CITED
- Adler, E., Hoon M. A., Mueller K. L., Chandrashekar J., Ryba N. J., and Zuker C. S.. . 2000. A novel family of mammalian taste receptors. Cell 100:693–702. doi: 10.1016/s0092-8674(00)80705-9. [DOI] [PubMed] [Google Scholar]
- Allen, Alissa L., Mcgeary John E., Knopik Valerie S., and Hayes John E.. . 2013. Bitterness of the non-nutritive sweetener acesulfame potassium varies with polymorphisms in TAS2R9 and TAS2R31. Chem. Senses 38(5):379–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bufe, B., Hofmann T., Krautwurst D., Raguse J.-D., and Meyerhof W.. . 2002. The human TAS2R16 receptor mediates bitter taste in response to β-glucopyranosides. Nat. Genet. 32:397. [DOI] [PubMed] [Google Scholar]
- Drewnowski, A., and Rock C. L.. . 1995. The influence of genetic taste markers on food acceptance. Am. J. Clin. Nutr. 62:506–511. doi: 10.1093/ajcn/62.3.506. [DOI] [PubMed] [Google Scholar]
- Ebler, J., Haukness M., Pesout T., Marschall T., and Paten B.. . 2019. Haplotype-aware diplotyping from noisy long reads. Genome Biol. 20:116. doi: 10.1186/s13059-019-1709-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira, A. M., Araújo S. S., Sales-Baptista E., and Almeida A. M.. . 2013. Identification of novel genes for bitter taste receptors in sheep (Ovis aries). Animal 7:547–554. doi: 10.1017/S1751731112002030. [DOI] [PubMed] [Google Scholar]
- Glanville, E. V., and Kaplan A. R.. . 1965. Food preference and sensitivity of taste for bitter compounds. Nature. 205:851–853. [Google Scholar]
- Goatcher, W. D., and Church D. C.. . 1970. Taste responses in ruminants. II. Reactions of sheep to acids, quinine, urea and sodium hydroxide. J. Anim. Sci. 30:784–790. [DOI] [PubMed] [Google Scholar]
- Henslee, D., Murdoch B., Yelich J., Taylor J. B., and Ellison M.. . 2020. Comparative genomics of the sheep Tas2r repertoire to cattle, goat, human, dog, and mice. Animal Gene. 17-18 Volumes 17–18. [Google Scholar]
- Henslee, D., Yelich J., Taylor J. B., and Ellison M.. . 2019. Avoidance of phenylthiocarbamide in mature Targhee and Rambouillet rams. Transl. Anim. Sci. 3:1194–1204. doi: 10.1093/tas/txz125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, K. H., Olson R. A., and Whitson T. D.. . 1996. Composition and diversity of plant and small mammal communities in tebuthiuron-treated big sagebrush (Artesmisa tridentata). Weed Technol. 10:404–416. [Google Scholar]
- Kim, U. K., Jorgenson E.Coon H.Leppert H.Risch N., Drayna D., 2003. Positional cloning of the human quantitative trait locus underlying taste sensitivity to phenylthiocarbamide. Science. 299:1221–1225. doi: 10.1126/science.1080190. [DOI] [PubMed] [Google Scholar]
- Launchbaugh, K. L. 2003. Prescription grazing for rangeland weed management: a new look at an old tool. Rangelands. 25:43–47. [Google Scholar]
- Li, H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34(18):3094–3100. 10.1093/bioinformatics/bty191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., and Durbin R.; 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikołajczyk-Stecyna, J., Malinowska A. M., and Chmurzynska A.. . 2020. Polymorphism of TAS2R3, TAS2R5, TAS2R19, and TAS2R50 genes and bitter food intake frequency in elderly woman. Acta Scient. Polon. Technol. Aliment., 19(1):109–122. doi: 10.17306/J.AFS.0729 [DOI] [PubMed] [Google Scholar]
- Robinson, J.Thorvaldsdóttir T., H., Winckler, W.Guttman, M.Lander, E. S.Getz, and G.Mesirov J. P.. 2011. Integrative genomics viewer. Nat. Biotech. 29:24–26. doi: 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salavati, M., Caulton A., Clark R., Gazova I., Smith T. P. L., Worley K. C., Cockett N. E., Archibald A. L., Clarke S. M., Murdoch B. M., . et al. 2020. Global analysis of transcription start sites in the new ovine reference genome (Oar rambouillet v1.0). Front. Genet. 11:580580. doi: 10.3389/fgene.2020.580580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch E. F., and Maniatis T.. . 1989. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor Laboratory Press. [Google Scholar]
- Wambolt, C. L., and Payne G. F.. . 1986. An 18-year comparison of control methods for Wyoming big sagebrush in southwestern Montana. J. Range Manag. 39: 314–319. [Google Scholar]