Abstract
Campylobacter jejuni and Salmonella typhimurium are the leading causes of bacterial food contamination in chicken carcasses. Contamination is particularly associated with the slaughtering process. The present study isolated C. jejuni and S. typhimurim from fifty chicken carcass samples, all of which were acquired from different companies in Riyadh, Saudi Arabia. The identification of C. jejuni was performed phenotypically by using a hippurate test and genetically using a polymerase chain reaction with primers for 16S rRNA and hippurate hydrolase (hipO gene). For the dentification of S. typhimurim, a serological Widal test was carried out using serum anti-S. typhimurium antibodies. Strains were genetically detected using invA gene primers. The positive isolates for C. jejuni showed a specific molecular size of 1448 bp for 16S rRNA and 1148 bp for hipO genes. However, the positive isolates of the invA gene exhibited a specific molecular size at 244 bp using polymerase chain reaction (PCR). Comparing sequencing was performed with respect to the invA gene and the BLAST nucleotide isolates that were identified as Salmonella enterica subsp. enterica serovar typhimurium strain ST45, thereby producing a similarity of 100%. The testing identified C. jejuni for hippuricase, GenBank: Z36940.1. While many isolates of Salmonella spp. that contained the invA gene were not necessarily identified as S. typhimurim, the limiting factor for the Widal test used antiS. typhimurum antibodies. The multidrug resistance (MDR) of C. jejuni isolates in chickens was compared with the standard C. jejuni strain ATCC 22931. Similarly, S. typhimurium isolates were compared with the standard S. typhimurium strain ATCC 14028.
Keywords: Campylobacter jejuni, chicken carcass, Hipo gene, invA gene, multidrug resistance, Salmonella typhimurim
Introduction
Campylobacter is a gram-negative, microaerophilic genus of bacteria that is responsible for multiple gastroenteric conditions in humans [1–3]. According to the European Food Safety Authority [4], the most common type of foodborne gastroenteritis is caused by Campylobacter jejuni [5]. Another common bacterial infection that manifests with severity comparable to gastroenteritis is campylobacteriosis, which is an infection also caused by the Campylobacter bacterium, most commonly C. jejuni. While this disease is seldom life-threatening in adults, complications can arise in young people and children, even when they are healthy. In addition, older people and immunocompromised individuals may also require antibiotic therapy [6].
The common routes for the transmission of C. jejuni from poultry waste to humans include exposure to bird faeces during the cleaning of the coops and poor bird handling practices (such as petting and kissing) [7]. Furthermore, the bacteria can be transmitted via the handling of contaminated eggs and meat [8], during the slaughtering process, and via the water used to clean the carcass. Placing ready-to-eat foods alongside raw chickens can result in cross-contamination, as poor practices can be related to food handling or the incorrect use of kitchen utensils, such as knives and cutting boards [9].
The hippuricase test (N-benzoylglycine amidohydrolase) is considered the moat important test whereby C. jejuni (hippuricase positive) can be differentiated from other campylobacters (hippuricase negative). Similarly, the hipO gene (hippuricase) is the main basis for the differentiation between C. jejuni and C. coli [10].
Microorganisms have the ability to produce the enzyme hippurate hydrolase, which can hydrolyse substrate hippurate into glycine and benzoic acid. The formation of this enzyme does not require microorganisms to grow. Rather, the enzyme detects the microorganisms by testing the presence of glycine, one of the end products of hydrolysis. If glycine is present, a blue or deep purple colour is manifest. Hippurate reactions have been proven to be effective in the identification of B. streptococci [11,12]. Recently, polymerase chain reaction (PCR)-based techniques have been found to be more specific, accurate, and sensitive than phenotypic methods as a means of differentiating between Campylobacter species [4,13–15]. However, as C. jejuni or C. coli are the dominant Campylobacter isolates in human cases and poultry, it is necessary for clinical and treatment purposes to utilize simple and economical tests that can distinguish between them.
The Salmonella genus comprises a group of rod-shaped, gram-negative bacteria with several flagella on their cell surfaces. These bacteria belong to the family Enterobacteriaceae and are facultative anaerobes. Salmonella has been proven to be urease negative, unlike Proteus sp. Moreover, Salmonella can ferment glucose, although it cannot ferment lactose. Furthermore, Salmonella infections cause acute gastroenteritis and are the most common cause of foodborne illness outbreaks in the world. In addition, the presence of Salmonella bacteria in the bloodstream can induce sepsis [16]. Gastrointestinal and typhoid fever S. typhimurium are also common consequences of Salmonella infections [17]. Salmonella also shows zoonosis since it can jump from nonhumans to humans [17].
The pathogenicity of Salmonella is related to numerous virulence genes, such as the invA, spv, fimA, and stn genes, which are associated with combined chromosomal and plasmid elements. The invA gene is located in the genome and contains coding for one of the most important proteins on the inner membrane, which is essential for the invasion of epithelial cells [18].
The rapid detection and identification of Salmonella can be achieved using PCR for the invA gene as per the diagnostic application [19,20]. Both S. typhimurium and S. enteritidis contain the invA gene virulence factor and are regarded as common and clinically significant genetic markers for the serovar that causes salmonellosis worldwide [21].
Antibacterial resistance (AMR) is recognized as one of the most significant public health challenges in the contemporary global environment. AMR impacts human, animal, and environmental health. AMR is exacerbated by multidrug resistance (MDR), which poses a major challenge for clinicians since it reduces potential therapeutic responses to bacteria, such as Campylobacter. The clinical treatment of campylobacteriosis is currently hampered by the ineffectiveness of many existing antibiotics, with inevitable increases in related mortality rates [22–24]. The recommended treatment for campylobacteriosis is erythromycin, which is often deemed the usual drug of choice when fighting bacterial infections, including severe intestinal infections caused by Campylobacter. Moreover, erythromycin is the drug of choice for use in immunocompromised patients [25,26]. In addition, Campylobacter is sometimes treated with other antibiotics, such as tetracycline, gentamycin, and fluoroquinolone [5]. One Canadian study found that Campylobacter spp. detected in chicken samples and slaughterhouses were highly resistant to several other classes of antibiotics, including tetracycline and fluoroquinolone [28–30]. In many other countries, including China and Poland, the detection of antibiotic resistance in Campylobacter spp. has also been reported [31–33].
The resistance of Salmonella and other pathogens to different classes of antibiotics can be ascertained from extrachromosomal genes. Moreover, resistance genes have been related to large transferable plasmids, which may be other DNA mobile elements, such as transposons and integrons [34]. Differences in resistance to antimicrobial determinants can amass within linked clusters because classes of antimicrobials, disinfectants, and heavy metals may be linked to MDR in bacteria [35,36]. The correlations between serotypes and drug resistance can be altered. Hence, there is a need for closer international monitoring of antimicrobial resistance phenotypes in Salmonella isolates that are human or animal in origin [34].
Many antimicrobial agents have been used to prevent infections in live poultry. Moreover, these agents have also been utilized as growth promotors in the poultry industry. Therefore, these antibiotics have been ineffective in human subjects when required to treat infections [27]. Excessive use of antibiotics in animal feed has also resulted in high resistance to antibiotics [31,32].
The current research has investigated the prevalence of Campylobacter jejuni and S. typhimurium in chicken carcasses, wherein PCR was employed to assess the presence of the hipO and invA genes, respectively. The presence of the invA gene was confirmed using serological tests with control serum antiS. typhimurium antibodies. Finally, the multidrug resistance (MDR) of C. jejuni and S. typhimurium isolates was determined using an array of antibiotics from different classes.
Sample collection
Fifty samples from chicken carcasses were collected from different companies (A, B, C, D and E) located in city markets in Al-Riyadh.
Methods used to isolate C. jejuni and S. typhimurium
To activate the standard strain of C. jejuni ATCC 33291 and isolate C. jejuni from chicken carcasses, this study utilized Bolton broth (Oxoid, CM0983, Solaar House, 19 Mercers Row, Cambridge, CB5 8BZ, U.K.). The medium was autoclaved and cooled to 50°C, after which horse blood (SR 0048) and an antibiotic supplement (SR0138) were added. The medium was inoculated with the C. jejuni standard strain, and the sample (cotton swab rubbing on carcasses) was incubated under microaerophilic conditions using gas generating kits (Oxoid BR38) at 42°C for 3–4 days. To study the morphological characterization of Campylobacter colonies using the streaking method on Campylobacter, blood-free selective medium agar (modified CCDA-Preston, Oxoid CM0739) was supplemented with CCDA Selective Supplement (SR0155) and incubated at 42°C for 48–72 h under microaerophilic conditions. The suspected colonies of chicken carcass isolates that were akin to the standard strain colonies were preserved in glycerol at −20°C for additional testing.
Fifty-five grams of chicken was added to 225 ml of lactose broth to isolate Salmonella from the chicken carcasses. The mixture was incubated at 37°C for 24 h as a pre-enrichment step. Lactose broth culture (1 ml) was transferred to selenite cysteine broth (used as selective enrichment medium) and incubated at 37°C for 24 h. One loop of selenite cysteine broth was streaked on xylose lysine deoxycholate agar (XLD) (Oxoid code, CM0469) and incubated at 37°C for 24 h. Suspected Salmonella spp. appeared as colonies with black centres surrounded by white halos. Such colonies were used for further analysis.
The hippurate test
The hippurate hydrolysis test was standardized with C. jejuni ATCC 33291 and C. coli ATCC 33559 reference strains. The manufacturer indicates that at least McFarland 4 turbidity should be used in the hippurate test. Moreover, it is important to use a high inoculum for Campylobacter. For standardization, the bacteria were grown on blood agar for 42 to 48 h at 42°C in a microaerobic atmosphere generated with Anoxomat (Mart®BV Microbiology Automation, Lichtenvoorde, Holland). The effect of cell suspension turbidity on the test results was determined. Thus, 14 suspensions of both strains with optical density (OD) values ranging from 0.2 to 2.8 were prepared in 0.9% NaCl. The OD of the cell suspensions was measured at 450 nm with a photometer (GENE-TRAK®, GENE-TRAK Systems, Hopkinton, MA, U.S.A.), and the turbidity was compared by eye and with a photometer in accordance with the McFarland turbidity standards. In addition, 0.5–12 was prepared from 1% barium chloride and 1% sulfuric acid. Diagnostic tablets were added, and the tubes were incubated for 4 h at +37°C. Five drops of 3.5% ninhydrin solution (Rosco Diagnostica A/S, Denmark) were added, and the results were read immediately after 10 min of reincubation. Based on the results, the optimal cell suspension turbidity for high hydrolysis testing was defined. The patient strains were retested for hippurate hydrolysis, and the bacterial suspensions were measured with a photometer and adjusted to between OD 450 0 0.8 (approximately McFarland 6) and 1.4 (approximately McFarland 10). Each strain was tested three times. All blue or purple colour reactions were read as positive, and colourless or yellow reactions were deemed to be negative.
DNA extraction from Campylobacter and Salmonella isolates
The DNA of Campylobacter and Salmonella isolates was extracted using a QIAamp DNA Mini Kit (50) Cat No./ID: 51304 (QIAGEN GmbH-Bezirksregierung Düsseldorf, Germany) in accordance with the manufacturer’s protocols.
Widal tests
Widal tests used anti-S. typhimurium and anti-S. entertidis control sera (SIFIN, Institüt fur Immunpräparate und Nährmedien GmbH, Berlin, Germany REF., TS 1624 and TS 1625, LOT., 880910 and 1090712, respectively) and assessed agglutination with antigens of Salmonella. A positive reaction is indicated by the degree of agglutination compared with the agglutination associated with the standard strains of S. typhimurium ATCC 14028 and S. entertidis ATCC 13076.
PCR amplification and electrophoresis
Campy 16S F TTGATCCTGGCTCAGAGT Campy 16S R TTCACCCCAGTCGCTGAT, hipO F ACTGCAAAATTAGTGGCG, hipO R GAGCTTTTAGCAAACCTTCC. PCR was used to detect the invasive encoding gene (invA gene) in Salmonella isolates. A final volume of 50 μl contained invA gene primers F: ACAGTGCTCGTTTACGACCTGAAT and R: AGACGACTGGTACTGATCGATAA [37]. The amplification process for C. jejuni genes involved incubation at 94°C for 5 min, followed by 35 cycles of 94°C for 1 min, annealing at 56°C for 30 s, and elongation at 72°C for 30 s. Subsequently, there was a final extension at 72°C for 10 min. The amplification process for S. typhimurium was performed using the same program with the exception of an annealing temperature of 52°C. Amplified PCR products were separated on 1% agarose gels with ethidium bromide at 100 v for approximately 1 h. DNA bands were then observed under ultraviolet light. The size of the DNA of C. jejuni for 16S rRNA and 1148 bp for hipO were compared with the DNA ladder run. In addition, the bands of the invA gene were used for the identification of S. typhimurium using the same DNA ladder run. Positive isolates were then sequenced, and the GenBank BLAST program was used to ensure that the proposed primers were consistent with the target species.
The susceptibility test for C. jejuni
A comparison of antimicrobial susceptibility tests for standard strains of C. jejuni ATCC 33291 and seven identified isolates was performed. The tested bacteria were obtained from overnight cultures inoculated from single colonies into Campylobacter blood-free selective agar (modified CCDA-Preston), (Oxoid CM0739), supplemented with CCDA selective supplement (SR 155E), which was applied to the surface of the same medium and used in the agar disk diffusion method.
Antimicrobial susceptibility testing was performed using the disk diffusion method, in accordance with the protocol of the Clinical and Laboratory Standards Institute [38]. A total of 25 different antibiotic discs (Oxoid, U.K.) were prepared and tested against C. jejuni strains. The diameters of the zones of inhibition (mm) were measured using the criteria recommended for C. jejuni [38].
Susceptibility test for S. typhimurium
Salmonella isolates were employed to compare the findings emerging from the susceptibility tests for S. typhimurium ATCC 14028 with the other results. Following the overnight incubation of single colonies in BHI media (Oxoid, U.K.), it was possible to extract the bacterial isolates. Cultures were spread on Mueller-Hinton agar (Oxoid, U.K.), and individual plates were used for agar disk diffusion assays.
Tests were conducted on 20 different antibiotic-impregnated disks (Oxoid, U.K.), with agents belonging to seven different classes, namely, β-lactam, aminoglycoside, cyclic peptide, sulfonamide, quinolone, fluoroquinolone, and Macrobid. The diameters of the zones of inhibition (mm) for each antibiotic and isolate were recorded using criteria recommended for Enterobacteriaceae [38]. Diameter measurements were used to classify isolates as sensitive (S) or resistant (R).
The preparation of cell extracts of Salmonella isolates for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE)
According to Yehia and Al-Dagal (2014) [39], electrophoresis was performed at room temperature in a vertical chamber (Biometra, Germany) using a constant voltage of 100 V until the bromophenol blue tracking dye reached the bottom of the gel. A comparison was made between the whole-cell protein profiles of presumptive Salmonella isolates and the standard Salmonella typhimurium strain ATCC 14028. A high degree of similarity with standard strains was further confirmed via positive anti-S. typhimurium agglutination tests.
Results and discussion
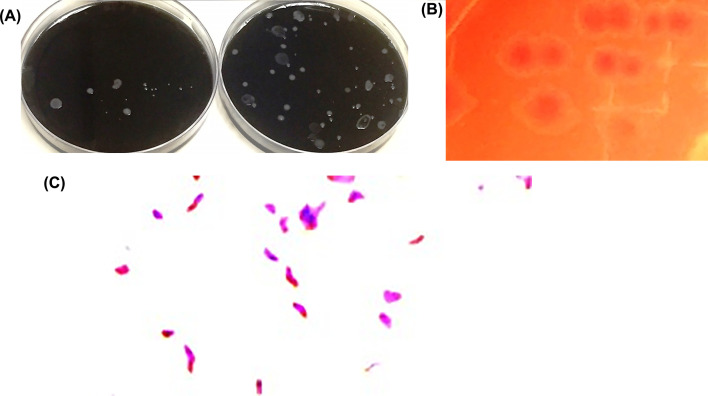
Fifty isolates comprising 10 poultry sourced from different companies were putatively identified as C. jejuni based on morphological characteristics and biochemical reactions. Colonies appeared on Campylobacter blood-free selective agar (modified CCDA-preston) and on Bolton selective enrichment agar supplemented with 25 ml of horse blood, as shown in Figure 1A and B. Some of the colonies that were greyish, low, flat, finely granular, and translucent may spread and swarm, whereas others were 1–2 mm in diameter, and others exhibited small raised convex, smooth, and glistening colonies with an entire edge [40]. Campylobacter spp. belongs to the family Campylobacteriaceae. It is spiral or S-shaped and sometimes curved in appearance when viewed under a compound microscope, as per Figure 1C. Moreover, it has Gram-negative rods when stained with Gram stain. These rods are motile, as in the motion of a corkscrew. The motility denotes the flagella, which may comprise a single form flagellum either at one end or at both ends of the cell. Nonmotile species or species with multiple flagella have also been described [41].
Figure 1. Colonies of C. jejuni.
Colonies on Campylobacter blood-free selective agar (modified CCDA-preston) (A) and on Bolton selective enrichment agar with agar supplements and with 25 ml of horse blood (B), creating a gram-negative spiral shape when stained with Gram stain (C).
To detect the ability of Campylobacter strains to hydrolyse hippurate, as mentioned by Hwang and Ederer (1975) [11], after a 24 h incubation period at 37°C, 50 μl of 3.5% (w/v) ninhydrin solution was added to each cupule containing a bacterial suspension and then thoroughly mixed. The samples were then viewed. All C. jejuni strains exhibited a dark purple color, indicative of hippurate hydrolysis. Figure 2 illustrates examples of the color changes identified in the high hydrolysis test.
Figure 2. Hippurate hydrolysis test.

For C. jejuni ATCC 33291 (+) and C. coli ATCC 33559 (-).
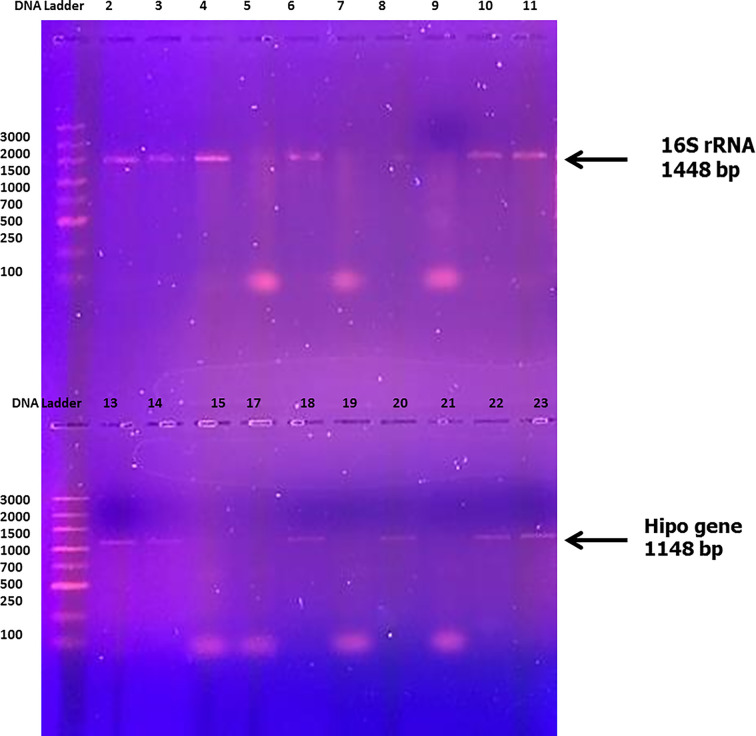
PCR amplification of the hippuricase (hipO) gene revealed that there were eight isolates that contained genes and that the other isolates failed as templates for amplification of the 1148 bp fragment (Figure 3). The isolates that were hipO-negative may be other Campylobacter species or represent hipO-negative C. jejuni [10,42]. Further analyses were required to determine their species identities. Linton et al. (1997) [42] used PCR amplification in relation to a portion of the hippuricase (hipO) gene isolated from humans and poultry and found that 56 of the 84 poultry isolates (67%) and 31 of the 32 human isolates (97%) contained the hipO gene. The outstanding 29 isolates failed as templates for the amplification of the 735 bp fragment. By using southern hybridization analyses, Linton et al. (1997) [42] found that 14 of 28 hipO-negative poultry isolates (50%) used a DIG-labelled 735 bp fragment of the hipO gene, thereby confirming that these isolates did not possess hipO. Thus far, the hipO gene has not been detected in any other species of Campylobacter; it appears to be exclusive to C. jejuni [43].
Figure 3. PCR amplification of 16S rRNA and hipO gene primers of C. jejuni isolates.
A number of PCR-based assays have been devised to identify specific Campylobacter species. However, a review of the literature failed to return any comparative analyses of the assay accuracy in classifying Campylobacter isolated from humans and poultry in different geographical areas. Six different published PCR assays were used to differentiate 116 Campylobacter isolates. Having the capacity to hydrolyse hippurate, the isolates had previously been determined to be C. jejuni. Our analysis verified that the majority (87 isolates) were C. jejuni, 28 were C. coli and the remaining isolate (ICPMR/6) was unidentifiable. Based upon its 16S rRNA gene sequence, this unique isolate was most homologous to the C. upsaliensis strain. Efforts to confirm this speculative identification of the species were unsuccessful using PCR-RFLP assays of Fermer and Engvall (1999) [44] and Jackson et al. (1996) [45]. Three PCR assays confirmed C. jejuni identification: PCR-RFLP [45], a putative oxidoreductase PCR assay [46], and hipO [42]. There was 100% agreement between the assays: 31 of the 32 human isolates and 56 of the 84 poultry were C. jejuni. Using the assay of Stucki et al. [42], the same number of human isolates was identified as C. jejuni; however, in contrast with the other assays, only 52 of the 84 poultry were identified as C. jejuni by this method. The poultry isolates that failed to amplify a PCR product using the assay of Stucki et al. were 98/E600/5, 98/E599/10, 99/3912/7 and 8. Possible reasons for these isolates failing to amplify a PCR product could be that there were PCR inhibitors present in the whole-cell DNA preparations or there were slight variations in the sequence of one or both primer sites. The least reliable assay was the PCR-RFLP assay devised by Fermer and Engvall (1999) [44]. In this assay, three isolates returned uncategorizable RFLP patterns, and no PCR amplicon was generated for 10 isolates. All 28 C. coli isolates were identified correctly by the C. coli-specific PCR assay [42]. However, this assay also identified one human and two poultry C. jejuni isolates as C. coli, indicating that more than one assay method should be used to verify the identification of C. coli species.
Totten et al. (1987) [10] recommended that the differentiation of thermophilic Campylobacter spp. not be centred on the single criterion of hippurate hydrolysis. They argue that while C. jejuni is the only Campylobacter species that has that capacity, some C. jejuni isolates are hippurate-negative. The findings from our study confirm this recommendation, as our high hydrolysis results were inconsistent in their reproducibility; additionally, weakly positive reactions were interpreted variably, and the strength of the reaction was influenced by the size of the inoculant. The uncertainty in identifying isolates is highlighted by a study conducted by Engvall et al. (2002) [47]. They recovered 174 Campylobacter isolates from various wild and domestic animals, of which 52 were identified incorrectly or unreliably as being either C. jejuni, C. coli, C. lari or C. upsaliensis. Together with our own, the results of Engvall et al. (2002) [47] indicate that phenotypic and biochemical assays are not reliable for determining Campylobacter spp., especially isolates obtained from animals.
Positive 16S rRNA and hipO gene amplification for C. jejuni ATCC 33291 (Lane 2) and isolates (Lanes 3, 4, 6, 7, 10 and 11). The DNA bands at 1448 and 1148 bp show that the marker is present.
The length of the 16S rRNA gene fragments for the eight sequenced Campylobacter isolates was 1448 nucleotides. Ambiguities were not detected in any of the sequences, indicating that the three 16S rRNA genes contained no sequence polymorphisms. The DNA sequence analysis of 16S rRNA genes for the eight isolates showed the highest identity to Campylobacter sp. strain RM12654 16S ribosomal RNA gene, GenBank: MW131451.1. While the DNA sequence of the hipO gene of the eight isolates shared identity with C. jejuni for hippuricase, GenBank: Z36940.1.
Table 1 presents data indicating that nine isolates of Campylobacter sp. were found in the 50 chicken carcass samples, equating to a ratio of 18%. Of the nine Campylobacter sp. isolates, five were C. jejuni (55.55%), as determined by PCR, which was conducted using primers for hipO and the 16S rRNA genes. In Indonesia, Campylobacter spp. in chicken meat was found to reach to 61.9%. Regarding the identification, 23 isolates (41.07%) were C. jejuni, 22 (39.29%) were C. coli, six (10.71%) were a mix between C. jejuni and C. coli, and five isolates (8.93%) were Campylobacter spp. The high prevalence of C. jejuni and C. coli in chicken meat in Indonesia indicates a high risk of campylobacteriosis in humans (Syarifah et al., 2020) [48].
Table 1. Results for Salmonella sp., invA gene, and serum antityphimurium and C. jejuni isolate detection in chicken samples.
| Microorganisms | Salmonella sp. | Sequence identification | Campylobacter sp. | Sequence identification | |||||
|---|---|---|---|---|---|---|---|---|---|
| Poultry companies | No. of samples | No. of isolates | invA gene | Widal test | No. of isolates | hipO gene | 16S rRNA | ||
| A | 10 | 7 | 5 | 2 | S. enterica subsp. enterica serovar typhimurium strain ST45 GenBank: CP050753.1 | 1 | 1 | 1 | C. jejuni for hippuricase, GenBank: Z36940.1. |
| B | 10 | 6 | 4 | 0 | 2 | 1 | 1 | ||
| C | 10 | 9 | 5 | 2 | 2 | 1 | 1 | ||
| D | 10 | 6 | 6 | 4 | 3 | 1 | 1 | ||
| E | 10 | 5 | 3 | 2 | 1 | 1 | 1 | ||
| Total | 50 | 33 | 23 | 10 | 10 | 9 | 5 | 5 | 5 |
| % | 100 | 66 | 69.69 | 43.47 | 43.47 | 18 | 55.55 | 55.55 | 55.55 |
Salmonella colonies on xylose lysine deoxylate agar (XLD) medium can be identified by their black centres and circular white halos. Microscopic examination of Salmonella colonies shows gram-negative rod-shaped cells. Out of 50 chicken carcasses, 33 samples (66%) were primarily identified by their growth on a specific medium, and the shape of the Salmonella sp. colony (see Table 1).
The results of Widal tests for visible agglutination between invA-positive Salmonella isolates and control sera are provided in Table 1. Anti-Salmonella typhimurium and anti-S. entertidis were applied to colonies grown on brain heart infusion agar for 24 h at 37°C. Of the 33 isolates, only 23 (69.69%) were positive for the invA gene, whereas 10 (43.47%) were positive in the Widal tests. The remaining 13 isolates (56.52%) were positive for the invA gene but negative in the Widal assays.
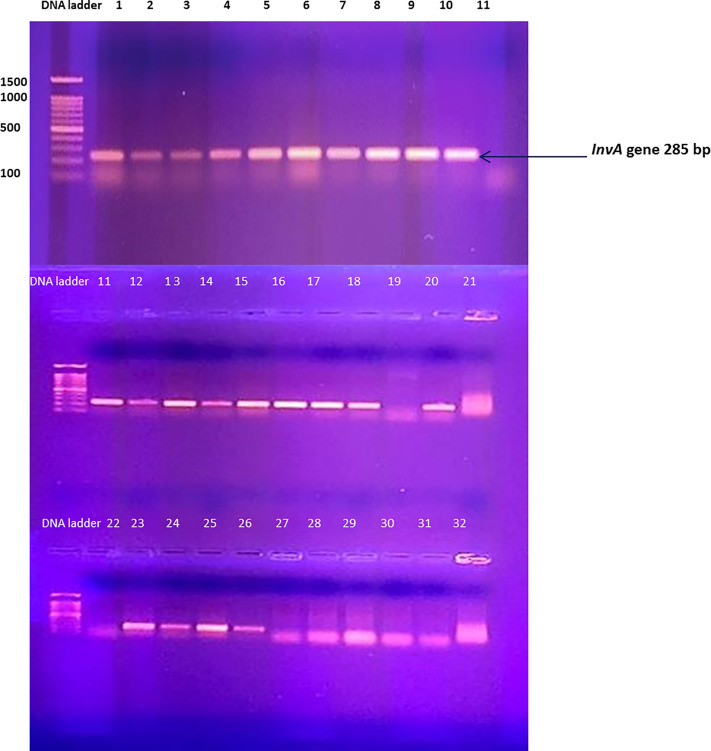
An examination of the incidence of Salmonella in poultry carcasses in different national contexts indicates that there is a huge variation in its presence. Specifically, proportions range from 3% to 66% [49,50]. The findings emerging from the current research indicate that there remains a need for substantial improvements to hygiene and safety standards in the poultry industry, in addition to the need for greater consumer awareness of this issue. Amplification results for all 33 Salmonella isolates revealed that only 23 isolates (69.69%) were positive for the invA gene, displaying a DNA marker of 244 bp (Figure 4). Submission of this 244 bp sequence to GenBank for BLAST alignment analysis indicated the presence of the genome of S. enterica subsp. In the enterica serovar typhimurium strain ST45 (Sequence ID: CP050753.1) for 10 isolates, the other 13 isolates were identified as Salmonella sp.
Figure 4. PCR amplification of S. typhimurium invA gene primers.
Positive invA gene amplification for S. typhimurium ATCC 14028 and S. enetritidis ATCC 13076 (Lane 1 and Lane 2) for 21 isolates (3-10), (11-18), (20) and (23-26). The DNA band at 244 bp shows marker presence, as visualized by gel electrophoresis using 1% agarose with an image analyser (SYNGENE) and DNA marker (1 kb ladder)
Amplification of the invA gene in the detection of the Salmonella gene is recognized as an international standard [51]. This gene encodes a protein in the inner membrane of bacteria that is responsible for invasion of host epithelial cells [52]. False-positives have been suggested to be be created by some bacterial isolates of Salmonella when an invA gene primer is utilized via PCR [53]. For this reason, attempts were made to assess performance in alternative published primers through the targeting of the invA gene.
Rahn et al. (1992) [53] used isolates from poultry such as Citrobacter spp., E. coli and Serratia sp., whereby it was possible to select invA gene primers with nonspecific signals. Comparable results have been reported in reactions containing genomic DNA from non-Salmonella isolates [51–55]. The DNA of the type of strain indicated a high specificity for invA PCR assays [56,57]. A study designed to evaluate the specificity of PCR assays based on an invA gene with a stain generated conflicting results and indicated that PCR assays based on invA gene amplification are not a reliable means of Salmonella detection.
The present study concluded that while many isolates of Salmonella spp. may contain the invA gene, not all of them may be identified as Salmonella typhimurium. Hence, additional tests, such as the Widal test, should be used to complete the identification of Salmonella strains.
The results in Table 2 reveal that all the identified C. jejuni isolated from the chicken carcass samples were resistant to different antibiotics in comparison with the C. jejuni ATCC 33291 standard strain. The highest level of antibiotic resistance was found in C. jejuni 2 A1, with a resistance ratio of 100%, followed by C. jejuni 2 E1, with a resistance ratio of 92.8%; C. jejuni 2 B, B2, with an 85.71% resistance rate; and C. jejuni 2 C5, C6 and D7-D9, which were 78.57% resistant. The lowest ratio was recorded for C. jejuni ATCC 33291, which demonstrated 57.15% resistance. Hence, there is a high change in the resistance ratio attributed to the fact that the microbes acquired resistance to multiple classes of antibiotics.
Table 2. The susceptibility test for C. jejuni isolates.
| Antibiotics classes | β- Lactam | Chloramphenicol | Quinolones | Oxazolidone | Aminoglycosides | Lipopeptides | Penicillin | Glycopeptide | Tetracyclines | Macrolides | % | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Isolates | AMC 30 ≥18 14-17 ≤13 | C 30 ≥ 1 8 13-17 ≤12 | CIP 5 ≥ 2116-20≤ 15 | NA 30 ≥ 1914-18 ≤13 | LZD 30 ≥ 21 – ≤ 20 | K 30 ≥ 18 14-17 ≤13 | N 30 ≥ 16 14-17 ≤12 | CT 25 ND | TIC 75 ≥20 15-19 ≤14 | AMP 25 ≥ 17-14-16 ≤ 13 | VA 5 ≥ 17 15-16 ≤ 14 | DO 30 ≥ 14 11-13 ≤ 10 | E 15 ≥ 23 14-22 ≤13 | F300 ≥ 1 7 15-16 ≤14 | Resistance | Intermediate | Sensitivity |
| C. jejuni ATCC 33291 | R | S | I | S | R | S | S | R | I | R | R | R | R | R | 57.15 | 14.28 | 28.57 |
| C. jejuni 2 A1 | R | R | R | R | R | R | R | R | R | R | R | R | R | R | 100 | 0 | 0 |
| C. jejuni 2 B1 | R | R | R | I | R | R | R | R | R | R | R | R | I | R | 85.71 | 14.28 | 0 |
| C. jejuni 2 B2 | R | R | R | I | R | R | R | R | R | R | R | R | R | S | 85.71 | 7.14 | 7.14 |
| C. jejuni 2 C5 | R | R | I | I | R | R | R | R | R | R | R | R | R | S | 78.57 | 14.28 | 7.14 |
| C. jejuni 2 C6 | R | R | I | I | R | R | R | R | R | R | R | R | R | S | 78.57 | 14.28 | 7.14 |
| C. jejuni 2 D7 | R | S | R | R | R | I | R | R | R | R | R | R | R | S | 78.57 | 7.14 | 14.28 |
| C. jejuni 2 D8 | R | S | R | R | R | I | R | R | R | R | R | R | R | S | 78.57 | 7.14 | 14.28 |
| C. jejuni 2 D9 | R | S | R | R | R | I | R | R | R | R | R | R | R | S | 78.57 | 7.14 | 14.28 |
| C. jejuni 2 E1 | R | R | R | R | R | R | R | R | R | R | R | R | R | S | 92.8 | 7.14 | 0 |
| Resistance % | 100 | 60 | 70 | 50 | 100 | 60 | 90 | 100 | 90 | 100 | 100 | 100 | 90 | 30 | - | - | - |
| Intermediate % | 0 | 0 | 30 | 40 | 0 | 30 | 0 | 0 | 10 | 0 | 0 | 0 | 10 | 0 | - | - | - |
| Sensitive % | 0 | 40 | 0 | 10 | 0 | 10 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 70 | |||
Mean zones of inhibition for common antibiotics: S = Sensitive, I = Intermediate, R = Resistant, except noted above and *ND = Not Detected: treated as a common antibiotic inhibition zone. AMC = Amoxy/clav.acid (30 μg), C 30 = Chloramphenicol (30 μg), CIP 5 = Ciprofloxacin (5 μg), LZD 30 = Linezolid (30 μg), K30 = Kanamycin (30 μg), N 30 = Neomycin (30 µg), CT 25 = Colistin sulfate (25 µg), TIC 75 = Ticarcillin (75 μg), AMP 25 = Ampicillin (25 μg), VA 30 = Vancomycin (30 µg), Do 30 = Doxycyclin (30 µg), E 15 = Erythromycin (15 μg), F 300 = Nitrofurantoin (300 μg).
Many antibiotic classes, such as the β-lactam group, were associated with resistance in all identified C. jejuni, including C. jejuni ATCC 33291, where the resistance ratio was 100%. A similar situation was found with the B-lactam groups oxazolidone, lipopeptides, penicillin, glycopeptide, glycopeptide and tetracyclines, which were all associated with resistance in C. jejuni.
From the chicken-carcass samples that were obtained from various companies (A-E), all of the Campylobacter strains were determined to be MDR. Numerous other studies have also found that C. jejuni isolated from chickens is resistant to antibiotics; this phenomenon is attributed to antibiotics being overused and/or improperly used in animal husbandry and in humans. Therefore, the incidence of antibiotic-resistant infections is rising, and more new and resistant strains are emerging. To limit AMR and devise novel treatments for human and animal populations, new strategies are needed to characterise the resistance mechanisms used by C. jejuni [58]. That C. jejuni is developing resistance mechanisms in response to the overuse of antibiotics presents a significant serious public health risk. Research conducted by Pollett et al. (2012) [59], Szczepanska et al. (2017) [60] and Tang et al. (2017) [61] has confirmed that C. jejuni strains bear antibiotic resistance mechanisms. There is consensus among these studies that most of the resistance mechanisms are the product of overusing antibiotics in chicken feed and animal production, as well as in human medicine. Carcasses retrieved from slaughterhouses had the highest load of C. jejuni, but equipment was also contaminated with antibiotic-resistant strains of C. coli and C. jejuni [62].
Mechanisms considered to be responsible for Campylobacter spp. Resistance to the β-lactam class of antibiotics includes intrinsic resistance and the production of β-lactamase, meaning that the efficacy of this class of drugs is limited [63].
C. jejuni strains are sensitive to the aminoglycoside class of antibiotics. Drug-modified proteins are responsible for C. jejuni resistance to antibiotics. According to Gaudreau and Gilbert (1997) [63], Campylobacter spp. have numerous enzymes that confer resistance against the aminoglycoside class of antibiotics. These enzymes include 3′,9-aminoglycoside adenyltransferase, 6-aminoglycoside adenyltransferase and 3′-aminoglycoside phosphotransferase types I, III, IV and VII. Campylobacter resistance to macrolides is facilitated by ribosomal targets; the resistance mechanism can arise from point mutations in the 23S rRNA and/or ribosomal proteins L4 and L22 or enzyme-mediated methylation [64,65]. Payot et al. (2006) [66] described C. rectus resistance to macrolides arising from rRNA methylation. However, in C. coli and C. jejuni, macrolide resistance is attributed to point mutations in domain V of the 23S rRNA [67].
C. jejuni has also demonstrated resistance to ciprofloxacin and nalidixic acid, which are quinolone antibiotics. Corcoran et al. (2006) [68] described the phenomenon by which ciprofloxacin-resistant mutant Campylobacter spp. are an inevitable outcome following exposure to fluoroquinolone (FQ). Multiple studies have explored the rapid expansion in the number of FQ-resistant mutants occurring in chickens. Previously, C. jejuni was susceptible to FQ, but resistance emerged following treatment with enrofloxacin [69–72].
The tet(O) gene, which encodes a ribosomal protective protein (Farnell et al., 2005) [73], has been found in Campylobacter isolates in diverse animal species; this gene makes the bacteria resistant to tetracycline [7]. Until recently, no other tet resistance genes had been identified in Campylobacter. In response to the gene binding to an open A site on the ribosome of Campylobacter spp., a conformational change takes place, which dislodges the tetracycline molecule that is bound to the ribosome [74]. The CmeABC multidrug efflux pump has also been implicated in conferring C. jejuni resistance to tetracycline [75,64]. Another Campylobacter spp. resistance mechanism to tetracycline that has been proposed is a plasmid-encoded tet(O) gene [76], described as being transferred by plasmids in a horizontal manner between C. jejuni and C. coli in the intestinal tracts of animals and humans [77,78].
Globally, there is an increase in the resistance exhibited by both C. jejuni and C. coli against various antibiotics [79,80]. Research has explored the high level of resistance that has emerged in human and animal isolates towards aminoglycosides, fluoroquinolones and macrolides [81,82].
Poultry diseases are frequently treated by broad-spectrum antibiotics, such as macrolides and tetracyclines. These antibiotics have also been used for more than three decades to promote the growth of poultry. They increase poultry production by inhibiting the pathogenic microflora and encourage the concentrations of cadaverine and putrescine. Broad-spectrum antibiotics are also used to treat humans and animals. For example, in humans, erythromycin is used to treat campylobacteriosis [83], and tetracycline is used to treat respiratory infections [84]. Meanwhile, due to their nephrotoxic activities, aminoglycosides are not used in general therapy.
Campylobacter spp. Resistance genes can be horizontally transferred between C. jejuni strains that are present in the intestinal tracts of food animals and humans. To limit the further development and spread of MDR in Campylobacter strains, the routine practice of adding antibiotics without a veterinary prescription to poultry feed or water to either kill pathogenic bacteria or to promote growth must cease.
Antibiotic resistance and virulence profiles exacerbate the risk of foodborne infection, which is further exacerbated by a reduction in antibiotic treatment options (Sithole et al., 2021) [85].
S. typhimurium strains no. 19 and no. 25 demonstrated the highest level of resistance (60%) to the antibiotics tested. In addition, S. typhimurium strains no. 15 and no. 26 revealed 55% resistance, and S. typhimurium 8 and 20 were 50% resistant. The standard strains S. typhimurium ATCC 14028 no. 1 and no. 3 were resistant to 45% of the antibiotics (see Table 3), whereas S. typhimurium 5 was resistant to fewer antibiotics (40%). There were fewer changes in the resistance to antibiotics when compared to the changes in the resistance in the standard strain.
Table 3. The susceptibility test for S. typhimurium isolates.
| Antibiotics classes | B-lactam | Aminoglycosides | Cyclicpeptides | Sulfonamide | Quinolone | Fluoroquinolone | Oxazolidone | Macrobid | Chloramphenicol | Glycopeptide | Lincosamide | % | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Isola- tes | R I S | FOX 30 ≥ 18 15-17 ≤14 | CAR 100 ND | TIC 75 ≥20 15-19 ≤14 | AMC 30 ≥18 14-17 ≤13 | CTX 30 ≥ 26 23-25 ≤22 | S 10 ≥ 15 12-14 ≤11 | CN 10 ≥ 15 13-14 ≤12 | K 30 ≥ 18 14-17 ≤13 | N 30 ≥ 16 14-17 ≤12 | TE 30 ≥ 15 13-15 ≤ 4 | RL 25 ND* | NA 30 ≥ 19 14-18 ≤13 | NOR 5 ≥ 17 13-16 ≤12 | OFX 5 ≥ 1 6 13-15 ≤12 | LZD 30 ≥ 21 — ≤10 | F 300 ≥ 1 7 15-16 ≤14 | E 15 5 ≥ 23 14-22 ≤13 | C 30 ≥ 1 8 13-17 ≤12 | VA 30 ≥ 17 15-16 ≤ 14 | MY 2 ND* | Resistance | Intermediate | Sensitive |
| S. typhimurium ATCC 14028 | R | R | S | S | S | R | S | I | R | S | R | S | S | S | R | I | R | S | R | R | 45 | 10 | 45 | |
| S. typhimurium 1 | R | R | I | S | S | R | S | I | I | R | R | S | S | S | R | I | R | S | R | R | 45 | 20 | 35 | |
| S. typhimurium 3 | R | R | I | S | S | R | S | I | I | S | R | R | S | S | R | I | R | S | R | R | 45 | 20 | 35 | |
| S. typhimurium 5 | R | R | I | S | S | R | S | I | I | S | R | R | S | S | S | I | R | S | R | R | 40 | 20 | 35 | |
| S. typhimurium 8 | R | R | R | S | S | R | S | I | I | R | R | S | S | S | R | I | R | S | R | R | 50 | 15 | 35 | |
| S. typhimurium 15 | R | R | I | S | S | R | S | I | R | R | R | S | S | S | S | R | R | R | R | R | 55 | 10 | 35 | |
| S. typhimurium 19 | R | R | R | S | S | R | S | I | I | R | R | R | S | S | R | R | R | S | R | R | 60 | 5 | 35 | |
| S. typhimurium 20 | R | R | I | S | S | R | S | I | R | S | R | R | S | S | R | I | R | S | R | R | 50 | 10 | 35 | |
| S. typhimurium 25 | R | R | R | S | S | R | S | I | R | R | R | R | S | S | R | I | R | S | R | R | 60 | 10 | 30 | |
| S. typhimurium 26 | R | R | R | S | S | R | S | I | R | S | R | R | S | S | R | I | R | S | R | R | 55 | 5 | 35 | |
| Resistance % | 100 | 100 | 40 | 0 | 0 | 100 | 0 | 0 | 60 | 50 | 100 | 40 | 0 | 0 | 100 | 20 | 100 | 0 | 100 | 100 | - | - | - | |
| Intermediate % | 0 | 0 | 50 | 0 | 0 | 0 | 0 | 100 | 0 | 0 | 0 | 60 | 100 | 0 | 0 | 80 | 0 | 0 | 0 | 0 | - | - | - | |
| Sensitive % | 0 | 0 | 10 | 100 | 100 | 0 | 100 | 0 | 40 | 50 | 0 | 0 | 0 | 100 | 0 | 0 | 00 | 100 | 0 | 0 | - | - | - | |
Mean zones of inhibition for common antibiotics: S = Sensitive, I = Intermediate, R = Resistant, except noted above and *ND = Not Detected: treated as a c100ommon antibiotic inhibition zone. FOX = Cefoxitin (30 μg), CAR = Carbenicillin 100 µg, TIC 75 = Ticarcillin (75 μg), AMC = Amoxy/clav.acid (30 μg), CTX = Cefotaxime 30 µg, S = Streptomycin (100 μg), CN = Gentamycin (10 µg), K30 = Kanamycin (30 μg), N = Neomycin (30 µg), TE = Tetracycline (30 μg), RL = Sulfamethoxazole (25 μg), NA = Naldioxic acid (30 μg), NOR = Norfloxacin (5 µg), OFX = Ofloxacin (5 µg), LZD 30 = Linezolid (30 μg), F 300 = Nitrofurantoin (300 μg), E 15 = Erythromycin (15 μg), C 30 = Chloramphenicol (30 μg), VA 30 = Vancomycin (30 µg). MY = Lincomycin (2 µg)
Resistance to antibiotics reached 100% for β-lactams (cefoxitin and carbencillin) and was high for sulfonamides (sulfamethoxazole trimethoprim), macrobids (erythromycin), oxazolidinone linezolid, glycopeptides (vancomycin) and lincosamides (lincomycin).
The data produced in the present study confirm that there is extensive resistance to some strains of S. typhimurium. The ACSSuT (AMP/CHL/STR/SMX/TET) phenotype may include resistance to amoxicillin-clavulanic acid (AUG), cefoxitin (FOX), and ceftiofur (TIO) and decreased susceptibility to AXO (MIC ≥ 4 μg/ml). TIO is a third-generation cephalosporin that was approved for use in animals in 1998 [84].
Resistance to antibiotics was 96.42% and 57.14% for aminoglycosides (streptomycin and neomycin, respectively), 78.57% for oxazolidone and linezolid and 64.28% for cyclopeptide and tetracycline. These data indicate that many Salmonella isolates are resistant to multiple antibiotics, which helps distribute these organisms and poses a serious problem to human public health. S. typhimurium, S. entertidis and all unidentified isolates remained sensitive to amoxycillin/clavulanic acid, cefotaxime, norfloxacin and ofloxacin with sensitivity ratios of 100%.
Salmonella strains were found to be multidrug-resistant (MDR) and have been detected in many serotypes, including S. enterica serotype typhimurium [83,85], in addition to S. enterica serotypesagona, anatum, choleraesuis, dublin, Heidelberg, Kentucky, Newport, Schwarzengrund, Senftenberg and Uganda, among others [86–88].
MDR S. typhimurium is no longer sensitive to ampicillin (AMP), chloramphenicol (CHL), streptomycin (STR), sulfonamides, and tetracycline (TET) and has been termed ACSSuT (AMP/CHL/STR/SMX/TET), referencing the strain carrying the blaCMY gene and others. Recently, many other strains belonging to the Enterobacteriaceae family of strains have exhibited the ACSSuT pattern and acquired MDR plasmids carrying the blaCMY gene [87].
Some strains may also display resistance to gentamicin (GEN), kanamycin (KAN), and trimethoprim-sulfamethoxazole ([SMX] COT), in addition to being resistant to disinfectants and heavy metals.
Resistance to third generation cephalosporins in Salmonella strains is of interest because these drugs are antibiotics of choice for treating salmonellosis in children, where fluoroquinolones are contraindicated [83].
Evidence of resistance to third generation cephalosporins in strains of Salmonella is significant because these antibiotics are the preferred therapeutic options in the treatment of children with salmonellosis in cases where medical reasons prohibit the use of fluoroquinolones [83]. The detection of pathogenic MDR Salmonella enterica serovars Typhimurium and Enteritidis from the caecal contents of healthy chickens in retail wet markets remains extremely alarming and has led to great public health concern [88].
Protein profiles for ten isolates (Lanes 2–9, 12, 13) on SDS-PAGE demonstrated similar major bands compared with S. typhimurium ATCC 14028 (see Figure 5). The presence of the invA gene with positive agglutination in control sera with anti-Salmonella typhimurium and protein similarity for the isolates confirms the accurate identification of S. typhimurium. Conversely, the isolates were positive for the invA gene but negative in the Widal assays, which did not demonstrate the same degree of similarity in major protein bands in SDS-PAGE with the standard strains S. typhimurium ATCC 14028 and S. eneteridis ATCC 13078 (see Figure 5).
Figure 5. Whole protein profiles of S. typhimurium isolates by SDS/PAGE.
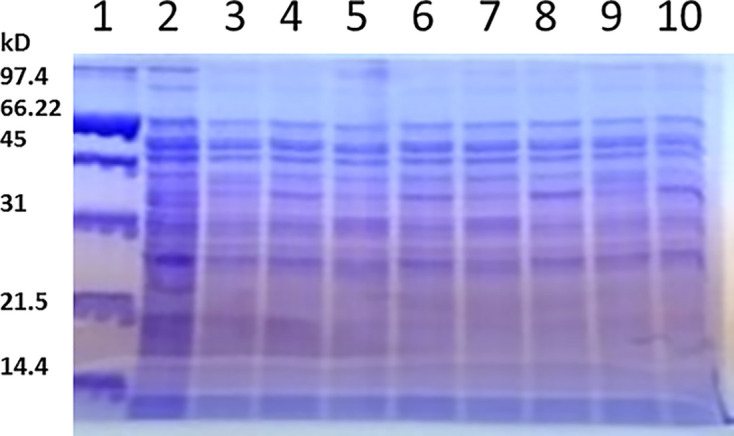
Lane M molecular weight standard. Lane 2 S. typhimurium ATCC 14028, Lanes 3–10 S. typhimurium total protein and a positive antisera typhimurium.
Conventional methods of Salmonella serovar identification and classification remain important for microbiological diagnosis [89]. Whole protein extracts of bacterial cells reflect the genomes of different strains and can support the identification and classification of bacteria. Comparative SDS-PAGE is also an important molecular technique used for identification at the species level [89].
Microbiological analysis related to an epidemiological investigation of outbreaks requires accurate identification and characterization of causative organisms. Many authors have used total protein extracts of Salmonella serovars on SDS-PAGE to evaluate whole cell lysates [90–94].
Our study is in agreement with Nakamura et al. (2002) [92], who reported that the total protein profiles of S. typhimurium ATCC 14028 and S. enteritidis 13076 showed major similarity in the pattern of bands on SDS-PAGE.
The finding emerging from the present study agrees with the conclusions reached by Nakamura et al. (2002) [92] that there were similarities in the SDS-PAGE band patterns in the total protein profiles for S. typhimurium ATCC 14028 and S. enteritidis 13076.
Major bands were noticed at 71.4, 67.7, 44.0, and 30.3 kDa (Nakamura et al., 2002) [92], while Ngwai et al. (2005) [94] noted that the total protein for S. typhimurium strains using SDS-PAGE detected 36.5 and 65 kDa proteins in all strains. Hassanain (2008) [96] observed that whole protein analysis of Salmonella by SDS-PAGE revealed that there were many bands between 11.4 and 77.5 kDa and that bands at 77.5, 55.2, 33.1, and 16.2 kDa were common. The total protein profiles of 54 Salmonella serovars, including S. typhimurium, S. enteritidis, S. agona, S. anatum, S. virchow, and S. corvallis, have also been compared using SDS-PAGE [95]. A protein band of 37.8 kDa was detected in all serovars. Furthermore, the protein profiles did not differ among the serovars. Acik et al. (2005) [93] argue that the electrophoretic banding patterns obtained using SDS-PAGE are insufficient to achieve reliable differentiation of Salmonella species.
Conclusions
Chicken carcasses are the main disseminators of C. jejuni and S. typhimurium. Testing revealed a high prevalence of alarming microbe rates in Saudi Arabia. In addition to testing for the presence of C. jejuni strains, virulence factors such as the hipO gene and 16S rRNA are considered useful tools to assess the potential risk of chicken meat as a pathogen disseminator. The invA gene virulence factor was associated with the detection of Salmonella typhimurium and must be verified with other tests, such as Widal. However, verification by other means, such as the Widal test, is required. The present study has confirmed that the C. jejuni and S. typhimurium strains detected in chicken carcasses have levels of antimicrobial resistance, which raises the risk to human and public health.
Abbreviations
- AMP
ampicillin
- CHL
chloramphenicol
- GEM
gentamicin
- MDR
multidrug resistance
- STR
streptomycin
- TET
tetracycline
- XLD
xylose lysine deoxylate agar
Data Availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This research was funded by the Deanship of Scientific Research at Princess Nourah bint Abdulrahman University through the Fast-Track Research Funding Program.
CRediT Author Contribution
Khaloud M. Alarjani: Conceptualization, Methodology. Manal F. Elkhadragy: Conceptualization, Data curation. Abdulrahman H. Al-Masoud: Software, Investigation, Methodology. Hany M. Yehia: Conceptualization, Software, Supervision, Methodology, Writing—original draft, Project administration.
References
- 1.Thomas M.K., Murray R., Flockhart L., Pintar K., Fazil A., Nesbitt A.et al. (2015) Estimates of foodborne illness-related hospitalizations and deaths in Canada for 30 specified pathogens and unspecified agents. Foodborne Pathog. Dis. 12, 820–827 10.1089/fpd.2015.1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scallan E., Hoekstra R.M., Mahon B.E., Jones T.F. and Griffin P.M. (2015) An assessment of the human health impact of seven leading foodborne pathogens in the United States using disability adjusted life years. Epidemiol. Infect. 143, 2795–2804 10.1017/S0950268814003185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kirk M.D., Pires S.M., Black R.E., Caipo M., Crump J.A., Devleesschauwer B.et al. (2015) World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: A data synthesis. PLoS Med. 12, e1001921 10.1371/journal.pmed.1001921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.EFSA (2005) Scientific report of the scientific panel on biological hazards on the request from the Commission related to Campylobacter in animals and foodstuffs. EFSA J. 173, 1–105, [Google Scholar] [Google Scholar]
- 5.Blaser M.J. and Engberg J. (2008) Clinical aspects of Campylobacter jejuni and Campylobacter coli infections. In Campylobactervol. 3, (Nachamkin I., Szymanski C.M. and Blaser M.J., eds), pp. 99–121, ASM Press, Washington DC, USA: 10.1128/9781555815554.ch6 [DOI] [Google Scholar]
- 6.Cox L.A. (2002) Re-examining the causes of campylobacteriosis. Int. J. Infect. Dis. 6, 26–36 10.1016/S1201-9712(02)90181-5 [DOI] [PubMed] [Google Scholar]
- 7.Blaser M.J., Berkowitz I.D., La Force F.M., Cravens J., Reller L.B.et al. (1979) Campylobacter enteritis: clinical and epidemiological features. Ann. Int. Med. 91, 179–185 10.7326/0003-4819-91-2-179 [DOI] [PubMed] [Google Scholar]
- 8.Sahin O., Kassem I.I., Shen Z., Lin J., Rajashekara G.et al. (2015) Campylobacter in poultry: Ecology and potential interventions. Avian Dis. 59:, 185–200 10.1637/11072-032315-Review [DOI] [PubMed] [Google Scholar]
- 9.Lorenz R.J. (1996) Grundbegriffe der Biometrie, 4th ed., G. Fischer Verlag, Stuttgart, Germany [Google Scholar]
- 10.Totten P.A., Patton C.M., Tenover F.C., Barrett T.J., Stamm W.E., Steigerwalt A.G.et al. (1987) Prevalence and characterisation of hippurate-negative Campylobacter jejuni in King County, Washington. J. Clin. Microbiol. 25, 1747–1752 10.1128/jcm.25.9.1747-1752.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hwang M.N. and Ederer G.M. (1975) Rapid hippurate hydrolysis method for presumptive identification of group B streptococci. JCM 1, 114–115, [PMC free article] [Google Scholar] 10.1128/jcm.1.1.114-115.1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mugg P.A. (1983) Rapid hippurate hydrolysis test for the presumptive identification of group B streptococci. Pathology 15, 251–252, [Google Scholar] 10.3109/00313028309083502 [DOI] [PubMed] [Google Scholar]
- 13.Didelot X. and Falush D. (2007) Inference of bacterial microevolution using multilocus sequence data. Genetics 175, 1251–1266, [PMC free article] [Google Scholar] 10.1534/genetics.106.063305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ridley A.M., Allen V.M., Sharma M., Harris J.A. and Newell D.G. (2008) Real-time PCR approach for detection of environmental sources of campylobacter strains colonizing broiler flocks. A E M. 74, 2492–2504, [PMC free article] [Google Scholar] 10.1128/AEM.01242-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adzitey F. and Nurul H. (2011) Campylobacter in poultry: incidences and possible control measures. Res. J. Microbiol. 6, 182–192, [Google Scholar] 10.3923/jm.2011.182.192 [DOI] [Google Scholar]
- 16.Pui C.F., Wong W.C., Chai L.C., Tunung R., Jeyaletchumi R., Noor H.M.S.et al. (2011) Salmonella: a foodborne pathogen. Int. Food Res. J. 18, 465–473 [Google Scholar]
- 17.Arsenault R.J., Scott N. and Kogut M.H. (2013) Salmonella enterica typhimurium infection 1′`causes metabolic changes in chicken muscle involving AMPK, fatty acid and insulin/mTOR signaling. Vet. Res. 44, 35 10.1186/1297-9716-44-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharma I. and Das K. (2016) Detection of invA gene in isolated Salmonella from marketed poultry meat by PCR assay. J. Food Process Technol. 7, 564 10.4172/2157-7110.1000564 [DOI] [Google Scholar]
- 19.Mohamed K. (2013) Detection of virulence gene (invA) in Salmonella isolated from meat and poultry products. Int. J. Genet. 3, 7–12 [Google Scholar]
- 20.Shanmugasamy M., Velayutham T. and Rajeswar J. (2011) InvA gene-specific PCR for detection of Salmonella from broilers. Vet World 4, 562–564 10.5455/vetworld.2011.562-564 [DOI] [Google Scholar]
- 21.Rodriguez J., Rondón I. and Verjan N. (2015) Serotypes of Salmonella in broiler carcasses marketed at Ibague, Colombia. Rev. Bras. Cienc. Avic. 17, 545–552 10.1590/1516-635X1704545-552 [DOI] [Google Scholar]
- 22.Moore J.E., Corcoran D., Dooley J.S., Fanning S., Lucey B., Matsuda M.et al. (2005) Campylobacter. Vet. Res. 36, 351–382, [Google Scholar] 10.1051/vetres:2005012 [DOI] [PubMed] [Google Scholar]
- 23.Thakur S., Zhao S., McDermott P.F., Harbottle H., Abbott J., English L.et al. (2010) Antimicrobial resistance, virulence, and genotypic profile comparison of Campylobacter jejuni and Campylobacter coli isolated from humans and retail meats. Foodborne Pathog. Dis. 7, 835–844 10.1089/fpd.2009.0487 [DOI] [PubMed] [Google Scholar]
- 24.Doyle M.E. (2015) Multidrug-resistant pathogens in the food supply. Foodborne Pathog. Dis. 12, 261–279 10.1089/fpd.2014.1865 [DOI] [PubMed] [Google Scholar]
- 25.Nachamkin I., Engberg J. and Aarestrup F.M. (2000) Diagnosis and antimicrobial susceptibility of Campylobacter spp. In Campylobacter 2nd edn(Nachamkin I. and Blaser M.J., eds), pp. 45–66, American Society for Microbiology, Washington [Google Scholar]
- 26.Petruccelli B.P., Murphy G.S., Sanchez J.L., Walz S., DeFraites R., Gelnett J.et al. (1992) Treatment of traveler’s diarrhea with ciprofloxacin and loperamide. J. Infect. Dis. 165, 557–560 10.1093/infdis/165.3.557 [DOI] [PubMed] [Google Scholar]
- 27.Agunos A., Le´ger D., ; Avery B.P., Parmley E.J., Deckert A., Carson C.A.et al. (2013) Ciprofloxacin-resistant Campylobacter spp. in retail chicken, western Canada. Emerg. Infect. Dis. 19, 1121–1124 10.3201/eid1907.111417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agunos A., Arsenault R.K., Avery B.P., Deckert A.E., Gow S.P., Janecko N.et al. (2018) Changes in antimicrobial resistance levels among Escherichia coli, Salmonella, and Campylobacter in Ontario broiler chickens between 2003 and 2015. Can. J. Vet. Res. 82, 163–177 [PMC free article] [PubMed] [Google Scholar]
- 29.Agunos A., Le´ger D., Avery B.P., Parmley E.J., Deckert A., Carson C.A.et al. (2013) Ciprofloxacin-resistant Campylobacter spp. in retail chicken, western Canada. Emerg. Infect. Dis. 19, 1121–1124 10.3201/eid1907.111417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Government of Canada (2018) Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS) 2016 Annual Report. Available from: http://publications.gc.ca/collections/collection_2018/aspc-phac/HP2-4-2016-eng.pdf [Google Scholar]
- 31.Woźniak-Biel A., Bugla-Płoskońska G., Kielsznia A., Korzekwa K., Tobiasz A., Korzeniowska-Kowal A.et al. (2018) High prevalence of resistance to fluoroquinolones and tetracycline Campylobacter spp. isolated from poultry in Poland. Microb. Drug Resist. 24, 314–322 10.1089/mdr.2016.0249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li B., Ma L., Li Y., Jia H., Wei J., Shao D.et al. (2017) Antimicrobial resistance of Campylobacter species isolated from broilers in live bird markets in Shanghai, China. Foodborne Pathog. Dis. 14:, 96–102 10.1089/fpd.2016.2186 [DOI] [PubMed] [Google Scholar]
- 33.Giacomelli M., Salata C., Martini M., Montesissa C. and Piccirillo A. (2014) Antimicrobial resistance of Campylobacter jejuni and Campylobacter coli from poultry in Italy. Microb. Drug Resist. 20:, 181–188 10.1089/mdr.2013.0110 [DOI] [PubMed] [Google Scholar]
- 34.Parveen S., Taabodi M., Schwarz J.S., Oscar T.P., Harter-Dennis J. and White D.G. (2007) Prevalence and Antimicrobial Resistance of Salmonella Recovered from Processed Poultry. J. Food Prot. 70, 2466–2472 10.4315/0362-028X-70.11.2466 [DOI] [PubMed] [Google Scholar]
- 35.Hall R.M., Collis C.M., Kim M.J., Partridge S.R., Recchia G.D. and Stokes H.W. (1999) Mobile gene cassettes and integrons in evolution. Ann. N. Y. Acad. Sci. 870, 68–80 10.1111/j.1749-6632.1999.tb08866.x [DOI] [PubMed] [Google Scholar]
- 36.Harbottle H., Thakur S., Zhao S. and White D.G. (2006) Genetics of antimicrobial resistance. Anim. Biotechnol. 17, 111–124 10.1080/10495390600957092 [DOI] [PubMed] [Google Scholar]
- 37.Chiu C.H. and Ou J.T. (1996) Rapid identification of Salmonella serovars in feces by specific detection of virulence genes, invA and spvC, by an enrichment broth culture-multiplex PCR combination assay. J. Clin. Microbiol. 34, 2619–2622 10.1128/jcm.34.10.2619-2622.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clinical and Laboratory Standards Institute. CLSI guidelines . (2018) Performance standards for antimicrobial susceptibility testing Vol. M-100, 28th ed., Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 39.Yehia H.M. and AL-Dagal M.M. (2014) Prevalence of Campylobacter jejuni in chicken produced by major poultry companies in Saudi Arabia. Int. J. Food Contam. 1, 2 10.1186/s40550-014-0002-y [DOI] [Google Scholar]
- 40.Bergey's Manual of Systemic Bacteriology . (1984) Aerobic/Microaerophilic, Motile, Helical/Vibroid Gram-Negative Bacteria, Vol. 1(Krieg N.R., ed.), p. 115, Williams and Wilkins, Baltimore, MD [Google Scholar]
- 41.Belkaid Y. and Hand T. (2014) Role of the Microbiota in Immunity and inflammation. Cell 157, 121–141 10.1016/j.cell.2014.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Linton D., Lawson A.J., Owen R.J. and Stanley J. (1997) PCR detection, identification to species level, and fingerprinting of Campylobacter jejuni and Campylobacter coli direct from diarrhoeic samples. J. Clin. Microbiol. 35, 2568–2572 10.1128/jcm.35.10.2568-2572.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rizal A., Kumar A. and Vidyarthi A. (2010) Prevalence of pathogenic genetics in Campylobacter jejuni. J. Food Saf. 12, 29–34 [Google Scholar]
- 44.Fermer C. and Engvall E.O. (1999) Specific PCR identification and differentiation of the thermophilic campylobacters, Campylobacter jejuni, C. coli, C. lari and C. upsaliensis. J. Clin. Microbiol. 37, 3370–3373 10.1128/JCM.37.10.3370-3373.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jackson C.J., Fox A.J. and Jones D.M. (1996) A novel polymerase chain reaction assay for the detection and speciation of thermophilic Campylobacter spp. J. Appl. Bacteriol. 81, 467–473 [DOI] [PubMed] [Google Scholar]
- 46.Harmon K.M., Ransom G.M. and Wesley I.V. (1997) Differentiation of Campylobacter jejuni and Campylobacter coli by polymerase chain reaction. Mol. Cell. Probes 11, 195–200 10.1006/mcpr.1997.0104 [DOI] [PubMed] [Google Scholar]
- 47.Engvall E.O., Bra«ndstro«m B., Gunnarsson A., Morner T., Wahlstro«m H. and Fermer C. (2002) Validation of a polymerase chain reaction/restriction enzyme analysis method for species identification of thermophilic campylobacters isolated from domestic and wild animals. J. Appl. Microbiol. 92, 47–54 10.1046/j.1365-2672.2002.01491.x [DOI] [PubMed] [Google Scholar]
- 48.Syarifah I.K., Latif H., Basri C. and Rahayu P. (2020) Identification and differentiation of Campylobacter isolated from chicken meat using real-time polymerase chain reaction and high-resolution melting analysis of hipO and glyA genes. Vet World 13, 1875–1883 10.14202/vetworld.2020.1875-1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao G., Ge B., De Villena J., Sudler R., Emily Yeh E., Zhao S.et al. (2001) Prevalence of Campylobacter spp.,Escherichia coli, and Salmonella serovars in retail chicken, turkey, pork, and beef from the Greater Washington, D. C., Area. Appl. Environ. Microbiol. 67, 5431–5436 10.1128/AEM.67.12.5431-5436.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Uyttendaele M.R., Debevere C.M., Lips R.M. and Neyts K.D. (1998) Prevalence of Salmonella in poultry carcasses and their products in Belgium. Int. J. Food Microbiol. 40, 1–8 10.1016/S0168-1605(98)00012-9 [DOI] [PubMed] [Google Scholar]
- 51.Malorny B., Hoorfar J., Bunge C. and Helmuth R. (2003) Multicenter validation of the analytical accuracy of Salmonella PCR: towards an international standard. Appl. Environ. Microbiol. 69, 290–296 10.1128/AEM.69.1.290-296.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Darwin K.H. and Miller V.L. (1999) Molecular basis of the interaction of Salmonella with the intestinal mucosa. Clin. Microbiol. Rev. 12, 405–428 10.1128/CMR.12.3.405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rahn K., De Grandis S.A., Clarke R.C., McEwen S.A., Galan J.E., Ginocchio C.et al. (1992) Amplification of an invA gene sequence of Salmonella typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Mol. Cell. Probes 6, 271–279 10.1016/0890-8508(92)90002-F [DOI] [PubMed] [Google Scholar]
- 54.Arnold T., Scholz H.C., Marg H., Rosler U. and Hensel A. (2004) Impact of invA-PCR and culture detection methods on occurrence and survival of salmonella in the flesh, internal organs and lymphoid tissues of experimentally infected pigs. J. Vet. Med. B Infect. Dis. Vet. Public Health 51, 459–463 10.1111/j.1439-0450.2004.00808.x [DOI] [PubMed] [Google Scholar]
- 55.Scholz H.C., Arnold T., Marg H., Rosler U. and Hensel A. (2001) Improvement of an invA-based PCR for the specific detection of Salmonella typhimurium in organs of pigs. Berl. Munch. Tierarztl. Wochenschr. 114, 401–403 10.31274/safepork-180809-1193 [DOI] [PubMed] [Google Scholar]
- 56.Heymans R., Vila A., van Heerwaarden C.A.M., Jansen C.C.C., Castelijn G.A.A., van der Voort M.et al. (2018) Rapid detection and differentiation of Salmonella species, Salmonella Typhimurium and Salmonella Enteritidis by multiplex quantitative PCR. PLoS ONE 13, e0206316, [PubMed Central: PMC6201931] 10.1371/journal.pone.0206316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bai J., Trinetta V., Shi X., Noll L.W., Magossi G., Zheng W.et al. (2018) A multiplex real-time PCR assay, based on invA and pagC genes, for the detection and quantification of Salmonella enterica from cattle lymph nodes. J. Microbiol. Methods 148, 110–116 10.1016/j.mimet.2018.03.019 [DOI] [PubMed] [Google Scholar]
- 58.Marotta F., Garofolo G., di Marcantonio L., Di Serafino G., Neri D., Romantini R.et al. (2019) Antimicrobial resistance genotypes and phenotypes of Campylobacter jejuni isolated in Italy from humans, birds from wild and urban habitats, and poultry. PLoS ONE 14, e0223804 10.1371/journal.pone.0223804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pollett S., Rocha C., Zerpa R., Patiño L., Valencia A., Camiña M.et al. (2012) Campylobacteria antimicrobial resistance in Peru: a ten-year observational study. BMC Infect. Dis. 16, 193 10.1186/1471-2334-12-193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Szczepanska B., Andrzejewska M., Spica D. and Klawe J.J. (2017) Prevalence and anti-microbial resistance of Campylobacter jejuni and Campylobacter coli isolated from children and environmental sources in urban and suburban areas. BMC Microbiol. 17, 80 10.1186/s12866-017-0991-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tang Y., Sahin O., Pavlovic N., LeJeune J., Carlson J., Wu Z.et al. (2017) Rising fluoroquinolone resistance in Campylobacteria isolated from feedlot cattle in the United States. Sci. Rep. 7, 494 10.1038/s41598-017-00584-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Torralbo A., Borge C., García-Bocanegra I., Méric G., Perea A. and Carbonero A. (2015) Higher resistance of Campylobacter coli compared to Campylobacter jejuni at chicken slaughterhouse. Comp. Immunol. Microbiol. Infect. Dis. 39, 47–52 10.1016/j.cimid.2015.02.003 [DOI] [PubMed] [Google Scholar]
- 63.Gaudreau C. and Gilbert H. (1997) Comparison of disc diffusion and agar dilution methods for antibiotic susceptibility testing of Campylobacter jejuni subsp. jejuni and Campylobacter coli. J. Antimicrob. Chemother. 39, 707–712 10.1093/jac/39.6.707 [DOI] [PubMed] [Google Scholar]
- 64.Gibreel A. and Taylor D.E. (2006) Macrolide resistance in Campylobacter jejuni and Campylobacter coli. J. Antimicrob. Chemother. 58, 243–255 10.1093/jac/dkl210 [DOI] [PubMed] [Google Scholar]
- 65.Cagliero C., Mouline C., Cloeckaert A. and Payot S. (2006) Synergy between efflux pump CmeABC and modifications in ribosomal proteins L4 and L22 in conferring macrolide resistance in Campylobacter jejuni and Campylobacter coli. Antimicrob. Agents Chemother. 50, 3893–3896 10.1128/AAC.00616-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Payot S., Bolla J.M., Corcoran D., Fanning S., Megraud F. and Zhang Q. (2006) Mechanisms of fluoroquinolone and macrolide resistance in Campylobacter spp. Microbes Infect. 8, 1967–1971 10.1016/j.micinf.2005.12.032 [DOI] [PubMed] [Google Scholar]
- 67.Roe D.E., Weinberg A. and Roberts M.C. (1995) Mobile rRNA methylase genes in Campylobacter (Wolinella) rectus. J. Antimicrob. Chemother. 36, 738–740 10.1093/jac/36.4.738 [DOI] [PubMed] [Google Scholar]
- 68.Corcoran D., Quinn T., Cotter L. and Fanning S. (2006) An investigation of the molecular mechanisms contributing to high-level erythromycin resistance in Campylobacter. Int. J. Antimicrob. Agents 27, 40–45, [Google Scholar] 10.1016/j.ijantimicag.2005.08.019 [DOI] [PubMed] [Google Scholar]
- 69.Han J., Sahin O., Barton Y.W. and Zhang Q. (2008) Key role of Mfd in the development of fluoroquinolone resistance in Campylobacter jejuni. PLoS Pathog. 4, e1000083 10.1371/journal.ppat.1000083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Luo N., Sahin O., Lin J., Michel L.O. and Zhang Q. (2003) In vivo selection of Campylobacter isolates with high levels of fluoroquinolone resistance associated with gyrA mutations and the function of the CmeABC efflux pump. Antimicrob. Agents Chemother. 47, 390–394 10.1128/AAC.47.1.390-394.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.van Boven M., Veldman K.T., de Jong M.C. and Mevius D.J. (2003) Rapid selection of quinolone resistance in Campylobacter jejuni but not in Escherichia coli in individually housed broilers. J. Antimicrob. Chemother. 52, 719–723, [Google Scholar] 10.1093/jac/dkg402 [DOI] [PubMed] [Google Scholar]
- 72.Griggs D.J., Johnson M.M., Frost J.A., Humphrey T., Jorgensen F. and Piddock L.J. (2005) Incidence and mechanism of ciprofloxacin resistance in Campylobacter spp isolated from commercial poultry flocks in the United Kingdom before, during, and after fluoroquinolone treatment. Antimicrob. Agents Chemother. 49, 699–707 10.1128/AAC.49.2.699-707.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Farnell M.B., Donoghue A.M., Cole K., Reyes-Herrera I., Blore P.J. and Donoghue D.J. (2005) Campylobacter susceptibility to ciprofloxacin and corresponding fluoroquinolone concentrations within the gastrointestinal tracts of chickens. J. Appl. Microbiol. 99, 1043–1050 10.1111/j.1365-2672.2005.02712.x [DOI] [PubMed] [Google Scholar]
- 74.Taylor D.E., Hiratsuka K., Ray H. and Manavathu E.K. (1987) Characterization and expression of a cloned tetracycline resistance determinant from Campylobacter jejuni plasmid pUA466. J. Bacteriol. 169, 2984–2989 10.1128/jb.169.7.2984-2989.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Alterkruse S.F., Stern N.J., Fields P.I. and Swerdlow D.L. (1999) Campylobacter jejuni an emerging food borne pathogen. Emerg. Infect. Dis. 5, 28–35 10.3201/eid0501.990104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Andersen S.R., Saadbye P., Shukri N.M., Rosenquist H., Nielsen N.L. and Boel J. (2006) Antimicrobial resistance among Campylobacter jejuni isolated from raw poultry meat at retail level in Denmark. Int. J. Food Microbiol. 107, 250–255 10.1016/j.ijfoodmicro.2005.04.029 [DOI] [PubMed] [Google Scholar]
- 77.Hakkinen M., Heiska H. and Hanninen M.L. (2007) Prevalence of Campylobacter spp in cattle in Finland and antimicrobial susceptibilities of bovine Campylobacter jejuni strains. Appl. Environ. Microbiol. 73, 3232–3238 10.1128/AEM.02579-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Samie A., Ramalivhana J., Igumbor E.O. and Obi C.L. (2007) Prevalence, haemolytic and haemagglutination activities and antibiotic susceptibility profiles of Campylobacter spp isolated from human diarrhoeal stools in Vhembe District, South Africa. J. Health Popul. Nutr. 25, 406–413 [PMC free article] [PubMed] [Google Scholar]
- 79.Gaudreau C. and Gilbert H. (2003) Antimicrobial resistance of Campylobacter jejuni subsp jejuni strains isolated from humans in 1998 to 2001 in Montreal, Canada. Antimicrob. Agents Chemother. 47, 2027–2029 10.1128/AAC.47.6.2027-2029.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Papavasileiou E., Voyatzi A., Papavasileiou K., Makri A., Andrianopoulou I. and Chatzipanagiotou S. (2007) Antimicrobial susceptibilities of Campylobacter jejuni isolates from hospitalized children in Athens, Greece, collected during 2004-2005. Eur. J. Epidemiol. 22, 77–78 10.1007/s10654-006-9080-3 [DOI] [PubMed] [Google Scholar]
- 81.Nachamkin I., Ung H. and Li M. (2002) Increasing fluoroquinolone resistance in Campylobacter jejuni, Pennsylvania, USA, 1982-2001. Emerg. Infect. Dis. 8, 1501–1503 10.3201/eid0812.020115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hakanen A.J., Lehtopolku M., Siitonen A., Huovinen P. and Kotilainen P. (2003) Multidrug resistance in Campylobacter jejuni strains collected from Finnish patients during 1995-2000. J. Antimicrob. Chemother. 52, 1035–1039 10.1093/jac/dkg489 [DOI] [PubMed] [Google Scholar]
- 83.Glynn M.K., Bopp C., Dewitt W., Dabney P., Mokhtar M. and Angulo F.J. (1998) Emergence of multidrug-resistant Salmonella enterica serotype typhimurium DT104 infections in the United States. N. Engl. J. Med. 338, 1333–1338 10.1056/NEJM199805073381901 [DOI] [PubMed] [Google Scholar]
- 84.Acheson D. and Hohmann E.L. (2001) Nontyphoidal salmonellosis. Clin. Infect. Dis. 32, 263–269 10.1086/318457 [DOI] [PubMed] [Google Scholar]
- 85.Sithole V., Amoako D.G., Abia A.L.K., Perrett K., Bester L.A. and Essack S.Y. (2021) Occurrence, antimicrobial resistance, and molecular characterization of Campylobacter spp. in Intensive Pig Production in South Africa. Pathogens 10, 439 10.3390/pathogens10040439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Threlfall E.J., Ward L.R., Frost J.A. and Willshaw G.A. (2000) The emergence and spread of antibiotic resistance in food-borne bacteria. Int. J. Food Microbiol. 62, 1–5 10.1016/S0168-1605(00)00351-2 [DOI] [PubMed] [Google Scholar]
- 87.Hsueh P.R., Teng L.J., Tseng S.P., Chang C.F., Wan J.H., Yan J.J.et al. (2004) Ciprofloxacin-resistant Salmonella enterica typhimurium and Choleraesuis from pigs to humans, Taiwan. Emerg. Infect. Dis. 10, 60–68 10.3201/eid1001.030171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Siddiky A., Sarker S., Khan S.R., Begum R., Kabir E., Karim R.et al. (2021) Virulence and antimicrobial resistance profiles of salmonella enterica serovars isolated from chicken at Wet Markets in Dhaka, Bangladesh Nure. Microorganisms 9, 952 10.3390/microorganisms9050952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhao S., McDermott P.F., White D.G., Qaiyumi S., Friedman S.L., Abbott J.W.et al. (2007) Characterization of multidrug resistant Salmonella recovered from diseased animals. Vet. Microbiol. 123, 122–132 10.1016/j.vetmic.2007.03.001 [DOI] [PubMed] [Google Scholar]
- 90.Zhao S., White D.G., Friedman S.L., Glenn A., Blickenstaff K., Ayers S.L.et al. (2008) Antimicrobial resistance in Salmonella enterica serovar Heidelberg isolates from retail meats, including poultry, from 2002 to 2006. Appl. Environ. Microbiol. 74, 6656–6662 10.1128/AEM.01249-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Durrani R., Abubakar M., Javed Arshed M., Saleha S., Ullah I. and Ali Q. (2008) Biological characterization and protein profiles of two model bacteria by SDS-PAGE and FT-IR. ARPN. J. Agric. Biol. Sci. 3, 6–16 [Google Scholar]
- 92.Nakamura A., Ota Y., Mizukami A., Ito T., Ngwai Y.B. and Adachi Y. (2002) Evaluation of aviguard, a commercial competitive exclusion product for efficacy and after-effect on the antibody response of chicks to Salmonella. Poult. Sci. 81, 1653–1660 10.1093/ps/81.11.1653 [DOI] [PubMed] [Google Scholar]
- 93.Acik L., Temiz A., Celebi A., Arslan S. and Yilmaz R. (2005) Protein patterns and plasmid profiles of the bacterial strains isolated from a poultry slaughterhouse in Ankara, Turkey. Food Technol. Biotechnol. 43, 255–262 [Google Scholar]
- 94.Ngwai Y.B., Ochi K., Ogawa Y. and Adachi Y. (2005) Analysis of the protein profiles of the antibiotic-resistant Salmonella typhimurium definitive phage type (dt) 104. Afr. J. Biotechnol. 4, 727–737 10.5897/AJB2005.000-3133 [DOI] [Google Scholar]
- 95.Begum F., Adachi Y. and Khan M.S.R. (2008) Characterization of Salmonella serovars in comparison with some Enterobacteria by SDS-PAGE analysis. Bangladesh J. Vet. Med. 6, 169–174 10.3329/bjvm.v6i2.2331 [DOI] [Google Scholar]
- 96.Hassanain N.A. (2008) Detection of antibodies against zoonotic food borne pathogens in sera of food handlers. Glob Vet. 2, 285–289 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.