Abstract
Our study aimed to identify single nucleotide polymorphisms (SNPs) with a significant impact on the innate immunity represented by antibody response against lipopolysaccharide (LPS) and lipoteichoid acid (LTA) and the adaptive immune response represented toward keyhole limpet hemocyanin (KLH) using the SNP prioritization method. Data set consisted of 288 F2 experimental individuals, created by crossing Green-legged Partridgelike and White Leghorn. The analyzed SNPs were located within 24 short genomic regions of GGA1, GGA2, GGA3, GGA4, GGA9, GGA10, GGA14, GGA18, and GGZ, pre-targeted based on literature references and database information. For the specific antibody response toward KLH at d 0 the most highly prioritized SNP for additive and dominance effects were located on GGA2 in the 3’UTR of MYD88. For the response at d 7, the most highly prioritized SNP pointed at the 3’UTR of MYD88, but potential causal additive variants were located within ADIPOQ and one in PROCR. The highest priority for additive and dominance effects in the antibody response toward lipoteichoic acid at d 0 was attributed to the same SNP, located on GGA2 in the 3’UTR region of MYD88. Two SNPs among the top-10 for additive effect were located in the exon of NOCT. SNPs selected for their additive effect on antibody response toward lipopolysaccharide at d 0 marked 3 genes – NOCT, MYD88, and SNX8, while SNPs selected for their dominance effect marked – NOCT, ADIPOQ, and MYD88. The top-10 variants identified in our study were located in different functional parts of the genome. In the context of causality three groups can be distinguished: variants located in exons of protein coding genes (ADIPOQ, NOCT, PROCR, SNX8), variants within exons of non-coding transcripts, and variants located in genes’ UTR regions. Variants from the first group influence protein structure and variants from both latter groups’ exhibit regulatory roles on DNA (UTR) or RNA (lncRNA).
Key words: adaptive immunity, amplicon, innate immunity, mixed model
INTRODUCTION
Immune response is a complex quantitative trait and the knowledge on its genetic background in chickens is still limited (Biscarini et al., 2010; Berghof et al., 2018). The overall immunity is composed of innate and acquired parts. The innate immunity is the first line of the host organism defence against pathogens. The humoral part of the innate immunity is composed of several components, among them are the Natural Antibodies (NAbs) (Shishido et al., 2012). The NAbs are defined as immunoglobulins, which are constitutively generated in the host in the absence of immunization or pathological condition (Baumgarth et al., 2005; Maddur et al., 2020). The major features of NAbs are low antibody affinity, high avidity, and polyreactivity against antigens (Ochsenbein and Zinkernagel, 2000). Therefore, they are very effective as a first line of defense against pathogen invasion before the acquired antibodies are generated (Siwek and Knol, 2005). Some NAbs have the ability to recognize the evolutionary conserved epitopes, which was used in this study to differentiate between the NAbs populations. In the current study, the innate immunity was represented by NAbs against lipopolysaccharide (LPS) and lipoteichoid acid (LTA). LPS is a molecule present on the outer membrane of the Gram-negative bacteria whereas LTA is an ingredient of the cell wall of Gram-positive bacteria (Siwek et al., 2006). The action of the innate immunity alone is in some cases insufficient to defend the host against pathogen invasion. Therefore, the adaptive immunity is required for successful elimination of the pathogen. The acquired immunity involves humoral and cellular immune responses (Erf, 2004). The adaptive immune response was represented by a specific antibody response towards keyhole limpet hemocyanin (KLH), which was determined in individuals specifically immunized with KLH antigen. This is a specific antigen obtained from the hemolymph of a certain sea mollusc species. The important characteristic of the KLH antigen is that birds never encountered this antigen before (Siwek et al., 2010).
The genetic background of the immune responses toward: LPS, LTA and KLH has been extensively analyzed in a unique experimental population composed of a cross between White Leghorn and Green-legged Partidgelike (WL × GP) (Siwek et al., 2010). These analyses covered all the steps of the classical quantitative trait loci (QTL) approach. The final step of each QTL analysis is a candidate gene selection. Identification of the candidate genes was performed in silico and the selected genes were verified in an association study based on a custom Illumina SNP genotyping panel (Siwek et al., 2015). One of the conclusions from this study was that the most significant single-nucleotide polymorphisms (SNPs) for immune responses toward KLH and LTA were located outside of the QTL regions originally proposed by the linkage analysis. Therefore, in the current study a different approach was undertaken. A selection of the candidate genes was based not only on the location near the QTL region, but also outside of the QTL region if the biological function matched the phenotypic trait of interest.
However, from the statistical perspective, performing an association study on tightly linked SNPs located within targeted regions poses problems in selecting specific polymorphisms (and genes) that most potently influence the variation of immune response measures. In such scenario, one can no longer rely on P-values resulting from testing SNP effects estimated in an association model. It is due to the location of the SNPs in genomic regions already known to be associated with trait variation, and because there is a very high intercorrelation between SNPs attributed to very high linkage disequilibrium. Causal mutations, due to their significant impact on the phenotypes, typically do not exhibit high genotype variation, and therefore they have low frequency of the negative allele in a population. As a consequence, it is more difficult to estimate a significant association for those SNPs as compared to non-causal SNPs which typically have higher frequency of the minor allele (Auer and Lettre, 2015). Testing of the SNP significance in the targeted regions sequenced with the high throughput technologies is also biased by the sequencing quality. In a typical NGS application, the quality of the sequencing is very nonuniform across regions. It impacts the number of successfully detected genotypes and their accuracy, which may further be a confounding factor in significance testing. These drawbacks have been described in the literature, especially in the context of the human genomic data (Cantor et al., 2010). In this study, we propose a SNP prioritization approach based on the utilization of the variant sequencing information available in the variant calling format (VCF) file.
The goal of the current study was to investigate a new method of SNP prioritization and to identify genes, and possibly SNPs, located within those genes, that are potential candidates of being causal for the innate and specific immune response in chicken. In the context of the quantitative phenotypes, the causality is defined as a significant impact on a trait variation (Auer and Lettre, 2015).
MATERIALS AND METHODS
Animals and Phenotypes
Experimental dataset consisted of 288 F2 individuals, created by crossing Green-legged Partridgelike and White Leghorn. The population was described in details by Siwek et al. (2010). Birds were kept on a floor system on a farm at the University of Life Sciences in Lublin. The entire population was vaccinated according to the routine vaccination schedule, which incorporated a vaccine against Salmonella, Gumboro disease, bronchitis, bursa of Fabricius disease, and encephalomyelitis. Total antibody responses to KLH were measured in individual plasma samples obtained at 7 d after s.c. immunization with 1 mg of KLH (Cal Biochem-Novabiochem Co., La Jolla, CA) in 1 mL of PBS (pH 7.2) at 12 wk of age. Antibody titers to KLH of all birds were measured by an indirect ELISA as described by Sijben et al. (2000). The assay for 2 homotopes LPS and LTA was performed according to the methodology reported by Siwek et al. (2006). The titration plates were coated with 4 μg/mL of LPS from Escherichia coli (Sigma-Aldrich GmbH) and 10 μg/ mL of LTA from Staphylococcus aureus (Sigma-Aldrich GmbH, St. Louis, MO). Serum samples were applied on the coated plates. Binding of NAb to LPS and LTA antigens was detected by 1:20,000 diluted rabbit anti-chicken antibody (IgGH+L; Nordic, Tilburg, the Netherlands) conjugated to peroxidase (PO; RAch/IgG H+L/PO). Substrate for enzymatic color reaction was 0.05% H2O2 with addition of tetramethylbenzidine. Absorbances were measured with a multiscan (Labsystems, Helsinki, Finland) at 450 nm. All of the phenotypes were expressed by titers as the log2 values of the highest dilution giving a positive reaction as described by Siwek et al. (2003) for KLH and by Siwek et al. (2006) for LTA and LPS. KLH, LTA and LPS phenotypes were measured at d 0 without immunization (KLHd0, LTAd0, and LPSd0, respectively), to estimate the level of natural antibodies (innate humoral immune response). Additionally, the individuals were immunized with KLH and the second measurement of antibodies against KLH was taken at d 7 (KLHd7) to estimate the level of specific antibodies (adaptive humoral immune response). Descriptive statistics of the phenotypic data were summarized in Table 1.
Table 1.
Descriptive statistics of phenotypes.
| Trait name | Mean | Standard deviation | Minimum | Maximum |
|---|---|---|---|---|
| KLHd0 | 3.81 | 1.55 | 1.3 | 9.6 |
| KLHd7 | 11.55 | 1.90 | 5.5 | 16.7 |
| LTAd0 | 5.67 | 1.79 | 1.0 | 12.0 |
| LPSd0 | 2.96 | 1.28 | 1.1 | 8.5 |
SNP Genotyping
Selection of the candidate genes was performed based on literature references and data base information: the InnateDB (www.innatedb.com), the KEGG (www.kegg.jp), and the Reactome (reactome.org). All gene functions were assessed based on the NCBI Gene (www.ncbi.nlm.nih.gov) and the AmiGO (amigo.geneontology.org) databases to classify genes as immune-related. The primers for Long-Range PCR were designed according to the NEB (New England Biolabs, Ipswich, MA) guidelines for reactions using Q5 High-Fidelity DNA Polymerase. The list of the selected genes and their primers is presented in Table 2. Primer3 was used for the primer design (bioinfo.ut.ee/primer3-0.4.0/). Amplicons’ quality was verified based on band integrity (1% agarose gel) and the concentration (QUBIT4, ThermoFisher Scientific, measurement). The bands were assessed based on the size marker. Only PCR products with similar gel intensity and concentration were used in further analysis. The amplicons were cleaned up with magnetic beads and checked again on an agarose gel.
Table 2.
The list of selected genes.
| Gene name | Gene ID | Chromosome | F primer | R primer |
|---|---|---|---|---|
| PIK3R1 | 427171 | Z | GCATGAAAGGACAGCACTGA | AAGGTACCATCGGCAGTGTC |
| AP3B1 | 427646 | Z | GGCTCAGCAGGCAGTAAATC | TCGCAGAGACACCAGAATTG |
| CD36 | 417730 | 1 | GGCCTCTTCTCTCGTGTGAC | ACATGTTTCAGTGGGCAACA |
| MYD88 | 420420 | 2 | GGCCCTGACTGCTACTTGAG | TCTGACCTTGCAGATGTTGC |
| TLR5 | 554217 | 3 | ACATCAACGGCTAATAATTGTCTT | TATAGTTTGCAGCTCTATACCACT |
| TAB2 | 421622 | 3 | TCTGTTTTCTCCAGCCTCGT | GCAGAAGTGGCTTCCTGAAC |
| CCRN4L (NOCT) | 404779 | 4 | TTCCATGGGAAACAGCACCAG | TTATAACAGTCTGTCAGGGTCTTG |
| MAP3K13 | 424876 | 9 | TGTTCTGTGAGCGAGAATGG | GAGGCAAGGTTTGCAGAAAG |
| ADIPOQ | 404536 | 9 | CAGCTCTCCAGCTTGCTTCT | TCCATCTTTTCCATCCTTGC |
| PROCR | 424867 | 9 | CACGCATCACCTACAGCACT | TGACTTTCACCCCTCTCCAC |
| MAP2K1 | 415549 | 10 | TGTTGTGGTGAGAGCAGGAG | ACAAGGTTCCAAGGATGCAC |
| SNX8 | 416468 | 14 | TTGAATTGCCAGTTGCTGAG | CCTGACCCCACAAAGACTGT |
| E4F1 | 416560 | 14 | AAAGAAGGGCGGTATGTGTG | GCTCAGTGGAAGACGGAAAG |
| PDGFA | 374196 | 14 | TCAGGCTCTTTTCGTGAGGT | CATTACGGAGCACATGGTTG |
| SOCS1 | 416630 | 14 | TGGTGCAGCACTGCTAATTC | CTGCAAAGCAAGGAGGTTTC |
| IL21R | 416586 | 14 | ATGGGGCAGTTAGGTGAGTG | TGAGACTCGCTCTCAAAGCA |
| IL20RB | 768437 | 14 | ATTTCCATGGGTTTGGATGA | GCTAAGATGGCAGCAACACA |
| PECAM1 | 771243 | 18 | CCAGGCTGCACAGATGAGTA | TGCATGTCCTCCTGTCTCTG |
Amplicons’ libraries were prepared and indexed with the use of Nextera XT DNA library preparation kit and Nextera XT Index Kit v2 (Illumina, San Diego, CA). Size of final library constructs was assessed by on-chip electrophoresis Agilent Bioanalyzer and Agilent High Sensitivity DNA Kit. Concentrations of libraries were measured with QUBIT 4 fluorometer and QUBIT dsDNA HS assay (Thermo Fisher Scientific, Waltham, MA). All libraries were diluted to 4 nM concentration, pooled and denatured. Final concentration of the libraries used for clustering was 10 pM, with 10% of PhiX control (Illumina) added in each run. For each individual, a panel of 18 amplicons spanning 18 genes on eight autosomes and the Z chromosome were sequenced in four paired-end runs (2 × 250 bp) with the use of Illumina MiSeq System and MiSeq Reagent Kits v2 (500 cycles). Over 70M of high quality, passing filter pair-end reads were generated from all runs (average 250K reads/per sample). Quality control of each FASTQ file was performed using FastQC software (Andrews, 2010). Data was then filtered to remove adapter sequences and the threshold of 30 was set in order to remove low-quality bases from the 3’ and 5’ ends using CutAdapt software (Martin, 2011). After quality control, reads were aligned to the GRCg6a reference genome (https://www.ncbi.nlm.nih.gov/assembly/GCF_000002315.5/) using BWA-MEM software (Li et al., 2009). Resulting BAM files were sorted and indexed using SAMtools (Li et al., 2009). The processed files were calibrated using BaseRecalibrator package in GATK (McKenna et al., 2010) in order to adjust base quality scores. Variant calling for each individual was performed using HyplotypeCaller and GenotypeGVCFs packages also from GATK software. The resulting multisample VCF file describing identified variants was further downstream processed. InDels and variants that did not overlap with amplicon locations were removed by VCFtools (Danecek et al., 2011), leaving the total number of 262 SNPs. SNP preselection criteria for downstream analyses were based on a minor allele frequency of at least 0.01 and technical quality of genotyping expressed by a minimum call rate of 80%. Variants that did not match flanked regions by PCR primers designed for this study were excluded. Occurrence of these SNPs was caused by alignment of a sequence to a domain that is shared by 2 genes of high homology in amplicon sequences. Genomic annotation of SNPs was assigned by the Variant Effect Predictor software (McLaren et al., 2016).
Estimation of Additive and Dominance Effects of SNPs
For the estimation of cumulative additive and dominance effects of animals the following mixed model was applied (Vitezica et al., 2018):
| (1) |
where represents a vector of phenotypes (i.e., KLHd0, KLHd7, LTAd0, or LPSd0), is a general mean, is a vector of fixed effects representing sex and batch with a corresponding design matrix , is a vector of random additive animal effects, is a vector of random dominance animal effects with corresponding design matrices and , is a vector of residuals with , where is an identity matrix and represents the residual variance. The covariance structure of and was assumed as and respectively with
| (2) |
and
| (3) |
where and were respectively calculated for homozygous, heterozygous, and alternative homozygous genotypes for ith SNP of jth individual, represent frequency of the reference/alternative allele for ith SNP and is a total number of SNPs. and denote additive, dominance, and residual variances, respectively, which were assumed to be known as , and .
In the next step additive (vector ) and dominance (vector ) SNP effects were calculated based on animal effects (a, d) using the back-solve method proposed by Liu et al. (2014, 2016) for additive effects:
| (4) |
and for dominance effects
| (5) |
where k is tuning parameter set in our analysis to 0.2 and represents the numerator relationship matrix among animals calculated based on their pedigree records. Other elements of Equations (4) and (5) are the same as in Equations (2) and (3).
For testing the significance of additive and dominance SNP effects we used the Wald test, which under H0 follows the standard normal distribution.
SNP Prioritization
Based on SNP descriptive information available in the VCF file the prioritization of SNPs was carried out separately for additive and dominance effects, based on the following mixed linear model:
| (6) |
where is the nominal P-value of the Wald test for the additive or dominance effect of each SNP and each trait from, is the frequency of an alternative allele, is the Z-score from Wilcoxon rank sum test of alternative vs. reference base qualities, is the root mean square of the mapping quality of reads across all animals, is the symmetric odds ratio to detect strand bias, is a random vector of SNP priorities with the covariance matrix with R2 LD measure between pairs of SNPs as the off-diagonal elements and ones on the diagonal and representing the variance of SNP associated P-values, is the residual term with , where is an identity matrix and represents the residual variance.
In model (6) SNP priorities are expressed by their P-values corrected for the SNP allele frequency (), sequencing quality ( and the LD between SNPs. These estimated priorities () were used to rank SNPs according to their importance on the variation of immune response traits.
RESULTS
Genomic Annotation of SNPs From Targeted Sequencing
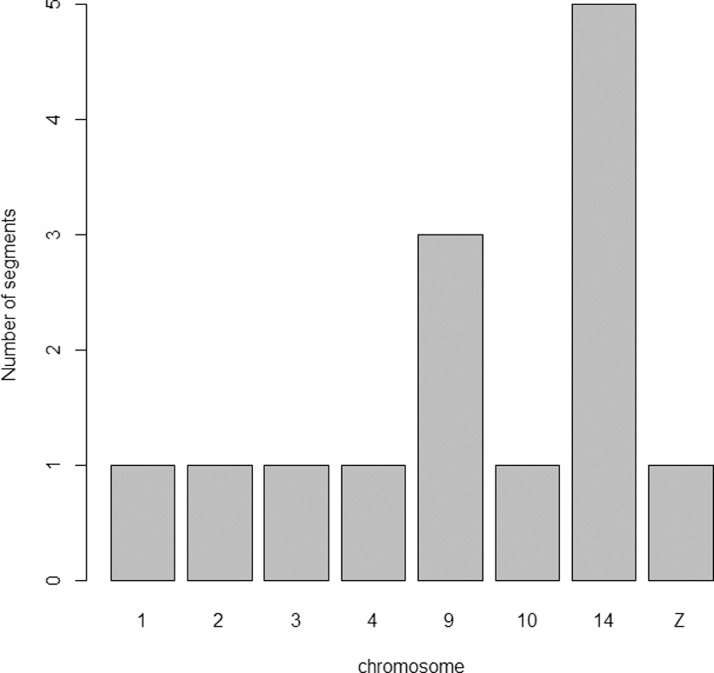
The 114 SNPs located within 24 short genomic fragments (Figure 1) assigned to eight autosomes (GGA1, GGA2, GGA3, GGA4, GGA9, GGA10, GGA14, and GGA18) and the sex chromosome (GGZ) was used in the downstream analysis. The average segment length was 435 ± 492 bp ranging between 89 bp and 2,205 bp. Genomic annotation of SNPs showed that nearly 57% of SNPs (n = 65) were localized in the intronic regions while 20% (n = 23) of SNPs overlapped exons. The remaining SNPs were located in the UTR and downstream/upstream regions of genes. SNPs located in exons are represented by 30% (n = 7) of missense variants and 70% (n = 16) of synonymous variants. Of all SNPs, 79% SNPs (n = 90), 6% (n = 7), and 15% (n = 17) had potential modifier, moderate, and low impact on the effectiveness of translated proteins respectively. The distribution of SNPs and segments on chromosomes with descriptive statistics was presented in Table S1.
Figure 1.
Number of segments with genetic markers per chromosome.
SNP Priority
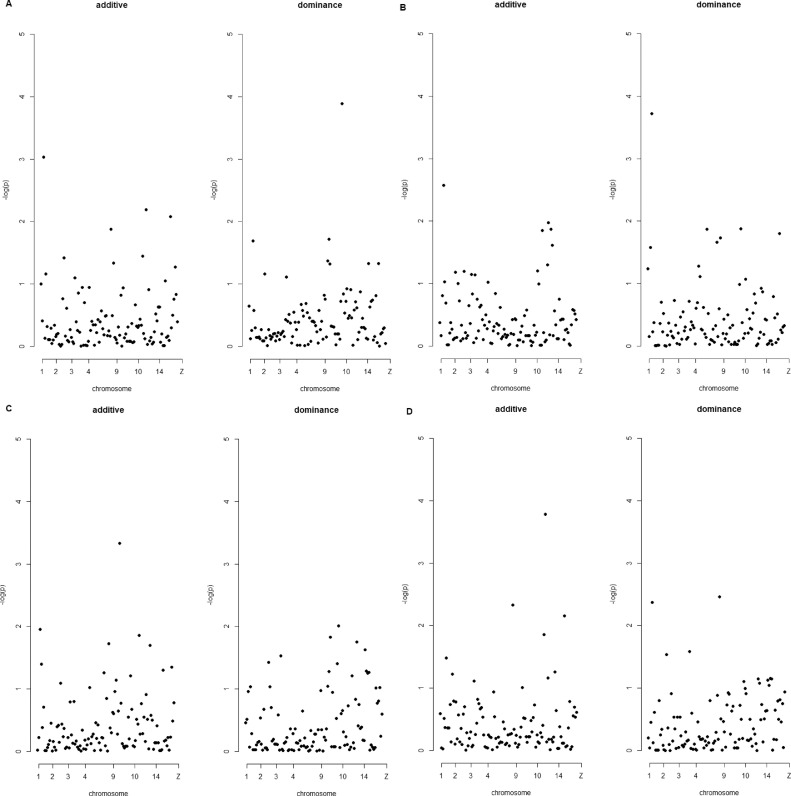
The results of a mixed model analysis of all genotyped SNPs Figure 2) in the form of nominal P-values from the Wald test on SNP effects estimated by Equations (4) and ((5) enhanced by the sequencing quality and polymorphism information from the VCF files after SNP calling were utilized to construct a SNP priority list separately for additive and dominance effects on each trait. For the specific antibody response toward KLH at d 0 (KLHd0), the most highly prioritized SNP for additive and dominance effects was located on GGA2 in the 3’UTR of MYD88. For the additive effect, all the remaining nine polymorphisms from the top-10 list marked the ADIPOQ gene on GGA9, SNX8 on GGA14, MYD88 on GGA2, and NOCT on GGA4. One SNP was exonic and thus candidate for being a causal mutation. For the dominance effect, all the remaining nine polymorphisms from the top-10 list marked the same genes. Moreover, 2 SNPs were exonic and indicated potential for being a causal mutation. For the specific antibody response toward KLH at d 7 (KLHd7), there were 4 potential causal additive variants – 3 in ADIPOQ and one in PROCR on GGA9. The most highly prioritized SNP pointed at the 3’UTR of MYD88. The top-10 SNPs composition for dominance effects comprised polymorphisms located in 3’UTR regions of MYD88 (1st SNP), within ADIPOQ (including 2 exonic variants and 3 intronic variants), NOCT (one exonic and 2 intronic variants), and SNX8 (one intronic variant). The gene set related to the antibody response toward LTA at d 0 (LTAd0) comprised SNPs located in MYD88, NOCT, and ADIPOQ (additive), and MYD88, and NOCT (dominance). The highest priority for additive and dominance effects was attributed to the same SNP located on GGA2 in the 3’UTR region of MYD88. Two SNPs among the top-10 for additive effect were located in the exon of NOCT (2 of them were also in the top 10 list for KLHd0), 4 in the intron of NOCT, and one more SNP in the 3’UTR region of MYD88. Eight SNPs among the top-10 for dominance effect were located in the NOCT (3 exonic and 5 intronic), and one more SNP in the 3’UTR region of MYD88. SNPs selected for their additive effect on antibody response toward LPS at d 0 (LPSd0) marked three genes – NOCT (with SNP ranked 4th located in an exon), MYD88 (one SNP located in the 3’UTR region), and SNX8 (with SNP ranked 7th located in an exon). SNPs selected for their dominance effect marked – NOCT (including four exonic SNPs ranked 4th, 5th, 7th, and 9th), ADIPOQ, and MYD88. The lists of top-10 ranked SNPs were presented in Tables S2-S5.
Figure 2.
Manhattan plots of the amplicon association study.
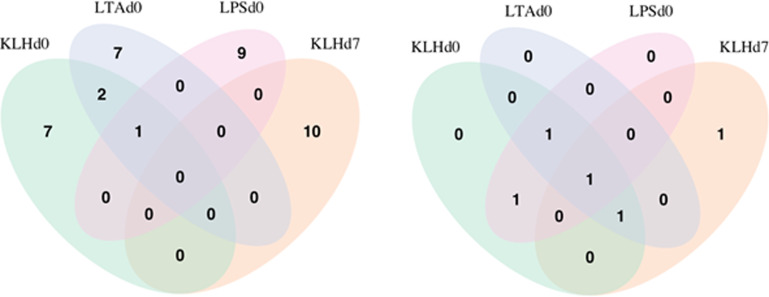
Considering the SNPs with top-10 additive effects, there was no polymorphism common to all 4 traits. Three SNPs were common between KLHd0 and LTAd0 (rs14131328, rs316037040, rs14131330). The SNP rs14131328 was in the top 10 for KLHd0, LTAd0 and LPSd0 (Figure 3A). KLHd7 was represented by the unique set of SNPs (no intersection with the other traits). However, shifting to gene level, MYD88 was represented among top 10 additive SNPs in all 4 traits. The other genes represented by selected SNPs were NOCT and ADIPOQ present for three traits (NOCT in KLHd0, LTAd0, and LPSd0; ADIPOQ in KLHd0, KLHd7, and LTAd0), as well as SNX8 represented in the top 10 for KLHd0 and LPSd0 (Figure 3B for additive effects).
Figure 3.
SNPs (A) and genes (B) represented among the top-10 additive effects.
DISCUSSION
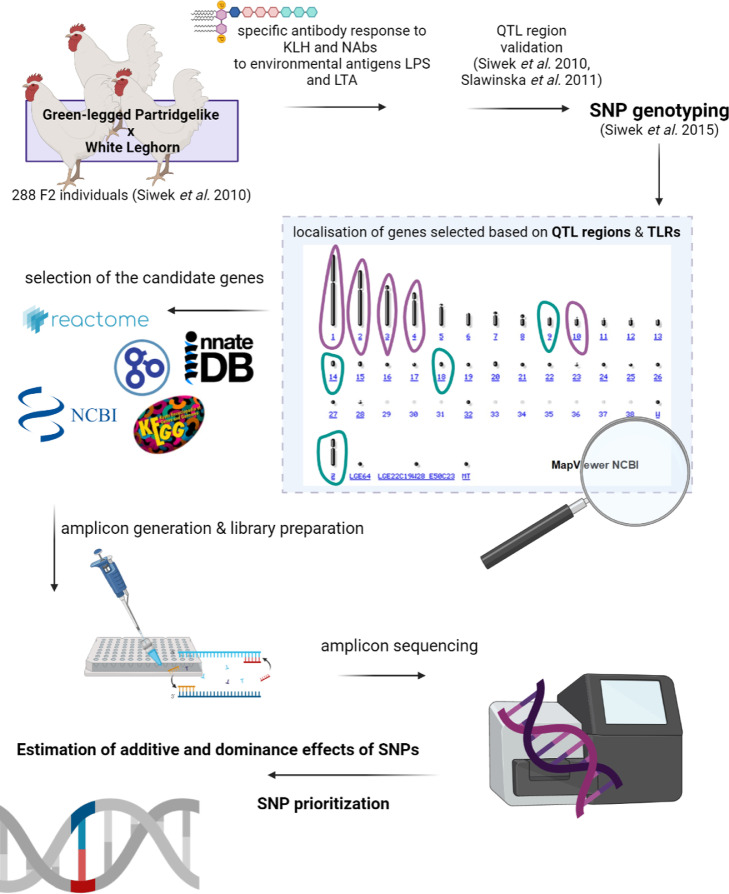
The analysis of the genetic background of the immune responses in the experimental population created by crossing White Leghorn layer with Green-legged Partridgelike dual-purpose native chicken goes back to 2010 (Siwek et al., 2010; Sławińska et al., 2011) (Figure 4). However, the QTL for innate immune responses toward LPS and LTA were initially detected in 2 independent populations of laying hens: a cross between 2 chicken lines expressing different feather pecking behavior and a cross between 2 lines divergently selected toward SRBC (Sheep Red Blood Cells) response (Siwek et al., 2006). Three QTL located on chicken chromosome (Gallus gallus domesticus [GGA]): GGA9, GGA18, and GGAZ were confirmed to be linked with NAbs for LPS, and 2 QTL located on: GGA5 and GGA14 were validated as linked to NAbs for LTA. Similar approach was undertaken for a primary antibody response toward KLH. The initial QTL for this antigen was detected in 2 independent populations of laying hens (Siwek et al., 2003). The validation of this QTL was performed also in WL × GP population (Siwek et al., 2010). A QTL for primary antibody response toward KLH located on GGA14 was further fine-mapped with a 2-QTL model (Siwek et al., 2012). The QTL detected for the innate immune responses toward LTA and LPS, and adaptive immune responses toward KLH in different experiments were narrowed down with 2 bioinformatics approaches: a meta-analysis and a combined analysis (Slawinska and Siwek, 2013). The information content used at each step of the analysis varied following the increasing genomic resolution. At first, the microsatellite markers pointed out to different chromosomal regions linked to each of the immune parameters (Siwek et al., 2003). An exception was GGA14 that harbored 2 QTL for LTA and KLH. The current experimental population was foreseen as a cross for a QTL validation study. Therefore, the selection of the microsatellite markers was focused on the previously identified QTL regions (Siwek et al., 2003, 2006). The availability of the SNP custom assays gave the opportunity to run an association analysis (Siwek et al., 2015). The major conclusion of the association study was that the most significant SNPs for the immune responses toward KLH and LTA are located outside of the QTL regions originally proposed by the linkage analysis. Therefore, in the search for causal mutations in candidate genes, a much higher marker resolution is necessary. Second, innate and adaptive immunity have some genes in common. The results of the current analysis confirmed this finding. Furthermore, 3 traits representing innate immunity (KLHd0, LTAd0, LPSd0) share a single variant located in MYD88 gene.
Figure 4.
The overview of the different steps of the analysis. Created with BioRender.com.
MYD88 participates in several KEGG pathways related to the immune responses, such as MAPK signaling pathway (gga04010), Toll-like receptor signaling pathway (gga04620), NOD-like receptor signaling pathway (gga04621), Salmonella infection (gga05132), Influenza A (gga05164), Herpes simplex virus 1 infection (gga05168). MYD88 encodes a protein which plays a central role in the innate and adaptive immune responses (Stelzer et al., 2016). The essential functions of this protein are related to signal transduction in the interleukin-1 and Toll like receptor signaling pathways, which regulate activation of proinflammatory genes (Stelzer et al., 2016).
Remaining genes pinpointed by the analysis in the current study are ADIPOQ, NOCT, SNX8, and PROCR. ADIPOQ encodes the adiponectin protein, whose role in the immune response and inflammation processes has been known for humans and mice (Fantuzzi, 2013) albeit not for poultry. Adiponectin is known for its anti-inflammatory activities such as inhibition of proinflammatory cytokines, induction of anti-inflammatory cytokines, downregulation of adhesion molecule expression, antagonism of toll like receptors (TLR) including their ligands for example, LPS (Fantuzzi, 2013). NOCT also known as Nocturnin is identified as a circadian clock regulated gene in Xenopus laevis (Stelzer et al., 2016). In vertebrates, this gene is known as Noct, Noc, CCR4L, CCRN4L or Ccr4c [22]. Niu et al. (2011) proved that Nocturnin is induced by LPS treatment in mouse embryonic fibroblasts. SNX8, sorting nexin 8 belongs to the family of proteins involved in endocytosis, endosomal sorting, and signaling (Stelzer et al., 2016). The study performed by Wei et al. (2017) showed that SNX8 is a part of the IFN gamma noncanonical signaling pathway and participates in the host defense to microbial infection. PROCR, also known as Activated Protein C Receptor, is involved in direct cellular signaling function through inhibition of activation of NFkB expression of proinflammatory cytokines (IL-1, IL-6, TNF-α) (Cai et al., 2019). PROCR together with CD1 were identified as a distinct subfamily of MHC class-I-like genes. The function of the class-I-like genes ranges from specialized antigen presenting to the transport of IgG and lipid mobilization and catabolism (Maruoka et al., 2005).
SNP Effect
Three of the selected SNP variants showed pleiotropic effects. The most pronounced pleiotropy was attributed to rs14131328 located in the 3’UTR of MYD88, which was associated with KLHd0, LTAd0, and LPSd0. This single SNP has been identified as nonsynonymous mutation in the study on susceptibility to Salmonella pullorum in chickens (Liu et al., 2015). The 3 traits KLHd0, LTAd0, and LPSd0, represent innate immunity. This part of the immune response is involved in the first line of defense against microbial pathogens and plays a key role in bacterial infections. As it has been already described in the introduction, both LTA and LPS are present in the bacterial membranes. They belong to the pathogen-associated molecular patterns (PAMPs) which activate pattern recognition receptors (PRR) of the host organisms. One of the major classes of PRR is TLR.
There are 2 additional SNPs common to KLHd0 and LTAd0. The variant rs14131330 is located within 3’UTR of MYD88 gene. This SNP has been also identified as a nonsynonymous mutation in the study on susceptibility to Salmonella pullorum in chickens (Liu et al., 2015). The variant rs316037040 is located in ADIPOQ gene. The literature does not report previous phenotype association with this variant.
Considering inheritance modes, most of the selected SNPs exhibit either a strong additive or a strong dominance effect. Interestingly, most of the variants located within MYD88 (rs14131328, rs14131330, rs731606804, and rs14131330) and ADIPOQ (rs16662812, rs317362477, rs313075798, rs736460279, and rs316037040) revealed both, additive and dominance effects. This observation is important in view of the practical application of the results in breeding programs, since additive effects are independent on the commercial-cross design, while the dominance are.
Since all of the considered SNPs are located inside the genes representing genomic regions it is impossible to rely on P-values resulting from an association analysis anymore. This is because of 2 main factors: 1) huge intercorrelation between SNPs due to huge linkage disequilibrium (Biernacka and Cordell, 2007); 2) not high genotype variation due to low frequency of the negative allele of causal mutations (Auer and Lettre, 2015). Because of this we used the novel SNP prioritization method using 2 sources of information: 1) P-values from an association analysis and 2) information coming out from variant calling file. Such statistical solution could improve the accuracy of detection association between phenotype and causal variant. Also, other authors suggested that prioritization methods are effective in such approaches (Hou and Zhao, 2013; Suravajhala and Benso, 2017).
From within the protein coding regions, 2 of the SNPs are good candidates for being causal mutations: for the adaptive response – a deleterious (SIFT = 0.02) variant rs741071044 located within PROCR, which changes valine to methionine and for the innate response – a tolerated (SIFT = 0.33) variant rs315673797 located within NOCT, which changes serine to asparagine. However, an interesting observation is that many of the top 10 SNPs were identified within regulatory sequences. It indicates the genetic determination of immune response traits seems to be shaped by the regulation of protein expression. Long non-coding RNAs are defined as non-protein coding transcripts longer than 200 nucleotides. Their role in immune response has been demonstrated by numerous studies (Heward and Lindsay, 2014). lncRNAs are expressed in tissue-specific or developmental stage-specific manner (Kwon et al., 2019). It was also reported that some lncRNAs regulate post-transcriptional gene regulation through biding to specific RBP (Kwon et al., 2019). Through the interaction with various molecules (protein, mRNA, miRNA) lncRNAs regulate transcription, splicing, RNA degradation and translation (Fitzgerald and Caffrey, 2014). There is also a demonstrated link between lncRNAs and 3’UTRs in the regulation of immune response (Schwerk and Savan, 2015). The 3’UTRs located downstream of the coding sequence are related to regulatory process of RNA stability, mRNA translation and localization. Any variation in 3’UTR both the length and the sequence may change the binding for miRNA and RNA binding proteins (RBP) leading to changes in the gene expression (Steri et al., 2018).
CONCLUSIONS
The major goal underlying our study was to identify causal mutations. Of course, in the case of complex traits, measured on a quantitative scale, no single causal mutation is expected to cause the response. Quantitative traits typically undergo a complex mode of inheritance determined by many genes with varying effect sizes. The top-10 variants identified in our study were located in different functional parts of the genome. In the context of causality, we want to highlight 3 groups: 1) variants located in exons of protein coding genes (ADIPOQ, NOCT, PROCR, SNX8) 2) variants within exons of non-coding transcripts, and 3) variants located in genes’ UTR regions. Variants from the first group influence protein structure. While variants from both latter groups’ exhibit regulatory roles on DNA (UTR) or RNA (lncRNA).
Summarizing, our study has demonstrated the important role of non-coding DNA variation in the determination of both innate and adaptive immune response in chicken, which complements coding genes with know impact on immune response.
Acknowledgments
ACKNOWLEDGMENTS
The research is co-financed by the: National Research Centre Grant number UMO/2014/13/B/NZ9/02123; UTP University of Science and Technology Grant number BN-8/2021 and The publication is co-financed under the Leading Research Groups support project from the subsidy increased for the period 2020–2025 in the amount of 2% of the subsidy referred to Art. 387 (3) of the Law of 20 July 2018 on Higher Education and Science, obtained in 2019.
DISCLOSURES
All authors declare no potential conflicts of interest.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.psj.2021.101433.
Appendix. Supplementary materials
S1 Table. Top-10 SNPs for specific antibody response toward keyhole limpet hemocyanin at day 0 (KLHd0) after prioritization. Genomic annotation is based on GRCg6a and on the RefSeq (in italics). SNPs located in exons are marked with gray.
S2 Table. Top-10 SNPs for specific antibody response toward keyhole limpet hemocyanin at day 7 (KLHd7) after prioritization. Genomic annotation is based on GRCg6a and on the RefSeq (in italics). SNPs located in exons are marked with gray.
S3 Table. Top-10 SNPs for innate antibody response toward lipoteichoic acid at day 0 (LTAd0) after prioritization. Genomic annotation is based on GRCg6a and on the RefSeq (in italics). SNPs located in exons are marked with gray.
S4 Table. Top-10 SNPs for innate antibody response toward lipopolysaccharide at day 0 (LPSd0) after prioritization. Genomic annotation is based on GRCg6a and on the RefSeq (in italics). SNPs located in exons are marked with gray.
S5 Table. Distribution of SNPs and segments on chromosomes with descriptive statistics
REFERENCES
- Andrews, S. 2010. FastQC: a quality control tool for high throughput sequence data. Accessed November 2, 2020. http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- Auer P.L., Lettre G. Rare variant association studies: considerations, challenges and opportunities. Genome Med. 2015;7:16. doi: 10.1186/s13073-015-0138-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgarth N., Tung J.W., Herzenberg L.A. Inherent specificities in natural antibodies: a key to immune defense against pathogen invasion. Spring. Semin. Immuno. 2005;26:347–362. doi: 10.1007/s00281-004-0182-2. [DOI] [PubMed] [Google Scholar]
- Berghof T.V.L., Visker M.H.P.W., Arts J.A.J., Parmentier H.K., van der Poel J.J., Vereijken A.L.J., Bovenhuis H. Genomic region containing toll-like receptor genes has a major impact on total IgM antibodies including KLH-binding IgM natural antibodies in chickens. Front. Immunol. 2018;8:1879. doi: 10.3389/fimmu.2017.01879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biernacka J.M., Cordell H.J. Exploring causality via identification of SNPs or haplotypes responsible for a linkage signal. Gen. Epidem. 2007;31:727–740. doi: 10.1002/gepi.20236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biscarini F., Bovenhuis H., van Arendonk J.A.M., Parmentier H.K., Jungerius A.P., van der Poel J.J. Across-line SNP association study of innate and adaptive immune response in laying hens. Ani. Genet. 2010;41:26–38. doi: 10.1111/j.1365-2052.2009.01960.x. [DOI] [PubMed] [Google Scholar]
- Cai X., Biswas I., Panicker S.R., Giri H., Rezaie A.R. Activated protein C inhibits lipopolysaccharide-mediated acetylation and secretion of high-mobility group box 1 in endothelial cells. J. Thromb. Haemost. 2019;17:803–817. doi: 10.1111/jth.14425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor R.M., Lange K., Sinsheimer J.S. Prioritizing GWAS Results: a review of statistical methods and recommendations for their application. Am. J. Hum. Gen. 2010;86:6–22. doi: 10.1016/j.ajhg.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T., McVean G., Durbin R., 1000 Genomes Project Analysis Group The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erf G.F. Cell-mediated immunity in poultry. Poult. Sci. 2004;3:580–590. doi: 10.1093/ps/83.4.580. [DOI] [PubMed] [Google Scholar]
- Fantuzzi G. Adiponectin in inflammatory and immune-mediated diseases. Cytokine. 2013;64:1–10. doi: 10.1016/j.cyto.2013.06.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald K.A., Caffrey D.R. Long noncoding RNAs in innate and adaptive immunity. Curr. Opin. Immunol. 2014;26:140–146. doi: 10.1016/j.coi.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heward J.A., Lindsay M.A. Long non-coding RNAs in the regulation of the immune response. Trends Immunol. 2014;35:408–419. doi: 10.1016/j.it.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou L., Zhao H. A review of post-GWAS prioritization approaches. Front. Genet. 2013;4:280. doi: 10.3389/fgene.2013.00280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon G.J., Henao-Mejia J., Williams A. Editorial: long non-coding RNAs and immunity. Front. Immunol. 2019;10:2378. doi: 10.3389/fimmu.2019.02378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Goddard M.E., Hayes B.J., Reinhardt F., Reents R. Technical note: equivalent genomic models with a residual polygenic effect. J. Dairy Sci. 2016;99:2016–2025. doi: 10.3168/jds.2015-10394. [DOI] [PubMed] [Google Scholar]
- Liu Z., Goddard M.E., Reinhardt F., Reents R. A single-step genomic model with direct estimation of marker effects. J. Dairy Sci. 2014;97:5833–5850. doi: 10.3168/jds.2014-7924. [DOI] [PubMed] [Google Scholar]
- Liu X.-Q., Wang F., Jin J., Zhou Y.-G., Ran J.-S., Feng Z.-Q., Wang Y., Liu Y.-P. MyD88 polymorphisms and association with susceptibility to salmonella pullorum. BioMed Res. Internat. 2015;2015:1–7. doi: 10.1155/2015/692973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddur M.S., Lacroix-Desmazes S., Dimitrov J.D., Kazatchkine M.D., Bayry J., Kaveri S.V. Natural antibodies: from first-line defense against pathogens to perpetual immune homeostasis. Clin. Rev. Aller. Immuno. 2020;58:213–228. doi: 10.1007/s12016-019-08746-9. [DOI] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10. [Google Scholar]
- Maruoka T., Tanabe H., Chiba M., Kasahara M. Chicken CD1 genes are located in the MHC: CD1 and endothelial protein C receptor genes constitute a distinct subfamily of class-I-like genes that predates the emergence of mammals. Immunogenetics. 2005;57:590–600. doi: 10.1007/s00251-005-0016-y. [DOI] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome. Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R., Thormann A., Flicek P., Cunningham F. The ensembl variant effect predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu S., Shingle D.L., Garbarino-Pico E., Kojima S., Gilbert M., Green C.B. The circadian deadenylase nocturnin is necessary for stabilization of the iNOS mRNA in mice. PLoS One. 2011;6:e26954. doi: 10.1371/journal.pone.0026954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsenbein A.F., Zinkernagel R.M. Natural antibodies and complement link innate and acquired immunity. Immuno. Today. 2000;21:624–630. doi: 10.1016/s0167-5699(00)01754-0. [DOI] [PubMed] [Google Scholar]
- Schwerk J., Savan R. Translating the untranslated region. J. Immunol. 2015;195:2963–2971. doi: 10.4049/jimmunol.1500756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shishido S., Varahan S., Yuan K, Li X., Fleming S. Humoral innate immune response and disease. Clin Immunol. 2012;144:142–158. doi: 10.1016/j.clim.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijben J.W., de Groot H., Nieuwland M.G.B., Schrama J.W., Parmentier H.K. Dietary linoleic acid divergently affects immune responsiveness of growing layer hens. Poult. Sci. 2000;79:1106–1115. doi: 10.1093/ps/79.8.1106. [DOI] [PubMed] [Google Scholar]
- Siwek M., Buitenhuis A.J., Cornelissen S.J.B., Nieuwland M.G.B., Bovenhuis H., Crooijmans R.P.M.A., Groenen M.a.M., de Vries-Reilingh G., Parmentier H.K., van der Poel J.J. Detection of different quantitative trait loci for antibody responses to keyhole lympet hemocyanin and Mycobacterium butyricum in two unrelated populations of laying hens. Poult. Sci. 2003;82:1845–1852. doi: 10.1093/ps/82.12.1845. [DOI] [PubMed] [Google Scholar]
- Siwek M., Buitenhuis B., Cornelissen S., Nieuwland M., Knol E.F., Crooijmans R., Groenen M., Parmentier H., van der Poel J. Detection of QTL for innate: non-specific antibody levels binding LPS and LTA in two independent populations of laying hens. Dev. Comp. Immunol. 2006;30:659–666. doi: 10.1016/j.dci.2005.09.004. [DOI] [PubMed] [Google Scholar]
- Siwek M., Knol Egbert.F. Genetic aspects of biological processes underlying the defense system in the neonate. Folia Biol. (krakow). 2005;53:39–43. [Google Scholar]
- Siwek M., Sławińska A., Nieuwland M., Witkowski A., Zięba G., Minozzi G., Knol E.F., Bednarczyk M. A quantitative trait locus for a primary antibody response to keyhole limpet hemocyanin on chicken chromosome 14—confirmation and candidate gene approach. Poult. Sci. 2010;89:1850–1857. doi: 10.3382/ps.2010-00755. [DOI] [PubMed] [Google Scholar]
- Siwek M., Slawinska A., Rydzanicz M., Wesoly J., Fraszczak M., Suchocki T., Skiba J., Skiba K., Szyda J. Identification of candidate genes and mutations in QTL regions for immune responses in chicken. Anim. Genet. 2015;46:247–254. doi: 10.1111/age.12280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siwek M., Szyda J., Sławińska A., Bednarczyk M. Detection of two QTL on chicken chromosome 14 for keyhole lymphet heamocyanin. J Appl. Genet. 2012;53:115–119. doi: 10.1007/s13353-011-0074-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slawinska A., Siwek M. Meta - and combined - QTL analysis of different experiments on immune traits in chickens. J Appl. Genet. 2013;54:483–487. doi: 10.1007/s13353-013-0177-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sławińska A., Witkowski A., Nieuwland M., Minozzi G., Bednarczyk M., Siwek M. Quantitative trait loci associated with the humoral innate immune response in chickens were confirmed in a cross between green-legged Partridgelike and White Leghorn. Poult. Sci. 2011;90:1909–1915. doi: 10.3382/ps.2011-01465. [DOI] [PubMed] [Google Scholar]
- Stelzer G., Rosen N., Plaschkes I., Zimmerman S., Twik M., Fishilevich S., Stein T.I., Nudel R., Lieder I., Mazor Y., Kaplan S., Dahary D., Warshawsky D., Guan-Golan Y., Kohn A., Rappaport N., Safran M., Lancet D. The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr. Prot. Bioinf. 2016;54:1.30.1–1.30.33. doi: 10.1002/cpbi.5. [DOI] [PubMed] [Google Scholar]
- Steri M., Idda M.L., Whalen M.B., Orrù V. Genetic variants in mRNA untranslated regions. WIREs RNA. 2018;9:e1474. doi: 10.1002/wrna.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suravajhala P., Benso A. Prioritizing single-nucleotide polymorphisms and variants associated with clinical mastitis. AABC. 2017;10:57–64. doi: 10.2147/AABC.S123604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitezica Z.G., Reverter A., Herring W., Legarra A. Dominance and epistatic genetic variances for litter size in pigs using genomic models. Genet. Sel. Evol. 2018;50:71. doi: 10.1186/s12711-018-0437-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J., Guo W., Lian H., Yang Q., Lin H., Li S., Shu H.-B. SNX8 mediates IFNγ-triggered noncanonical signaling pathway and host defense against Listeria monocytogenes. Proc. Natl. Acad. Sci. U. S. A. 2017;114:13000–13005. doi: 10.1073/pnas.1713462114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S1 Table. Top-10 SNPs for specific antibody response toward keyhole limpet hemocyanin at day 0 (KLHd0) after prioritization. Genomic annotation is based on GRCg6a and on the RefSeq (in italics). SNPs located in exons are marked with gray.
S2 Table. Top-10 SNPs for specific antibody response toward keyhole limpet hemocyanin at day 7 (KLHd7) after prioritization. Genomic annotation is based on GRCg6a and on the RefSeq (in italics). SNPs located in exons are marked with gray.
S3 Table. Top-10 SNPs for innate antibody response toward lipoteichoic acid at day 0 (LTAd0) after prioritization. Genomic annotation is based on GRCg6a and on the RefSeq (in italics). SNPs located in exons are marked with gray.
S4 Table. Top-10 SNPs for innate antibody response toward lipopolysaccharide at day 0 (LPSd0) after prioritization. Genomic annotation is based on GRCg6a and on the RefSeq (in italics). SNPs located in exons are marked with gray.
S5 Table. Distribution of SNPs and segments on chromosomes with descriptive statistics