Abstract
Red palm weevil (RPW) is the most aggressive date palm parasite in the Middle East, and especially in the Gulf region. Originated in Southeast Asia, this pest has been detected in the entire Arabian Peninsula, North Africa, Italy, Latin America, and other territories. It is important to local from obtrusive species, which help augmenting the pest control strategies. In the present study we collected 21 RPW samples from 21 different locations in the Eastern Province, Saudi Arabia to genetically characterize them using RAPD- and ISSR-based clustering. Unweighted pair group method with arithmetic mean (UPGMA) for RAPD data categorized the 21 accessions into seven distinct groups, with Al-Oyonn and Juaymah each categorized in solitary group, meanwhile, UPGMA for ISSR indicated six different groups, with Battaliyah, Al-Oyoon, and Juaymah each assigned to a separate group. Combining RAPD and ISSR data revealed two accession; Al-Oyoon and Juaymah that might be considered obtrusive species. Based on distance calculations, we proposed that the potential origins of RPW collected from these locations are Iran and the United Arab Emirates. However, this assumption needs further studies for confirmation.
Keywords: RPW, RAPD, ISSR, Fingerprinting, Saudi Arabia
1. Introduction
The Red Palm Weevil (RPW) Rhynchophorus ferrugineus is a disturbing wide-spread pest that infest and kill date palm trees after unadorned infestation (Faleiro et al., 2014). It belongs to Order Coleoptera and Family Curculionidae (Faghih, 1996, Fiaboe et al., 2012). It has been thought that this pest is native to Southeast Asia and Melanesia (Musmeci et al., 2018, Rugman-Jones et al., 2013). Due to shifting of the cultivation of the ornamental palms, RPW has widely dispersed across the Arabian Peninsula during the last four decades (Mukhtar et al., 2011) and has expanded over the last three decades to invade nearly all countries of the Mediterranean Basin, sub-Saharan Africa (Naji Mordi et al., 2016), Caribbean, Central and Northern America (Fiaboe et al., 2012). RPW can affect more 40 date palm species belonging to 23 genera and 3 families causing enormous economic losses (Wang et al., 2017, Sun et al., 2016). Date palm trees are widespread because of its nutritional value for many people over the globe (Mulley et al., 2019, Rasool et al., 2018). These trees can grow between 10° and 39° in the northern hemisphere of the Earth, however, for the time being it covers areas from 0 to 1500 m above the sea level (Huda et al., 2019, Shabani et al., 2014). Accordingly, RPW could be also detected in these environments, makes it difficult to correctly classify this pest. Categorizing RPW to its right taxonomy is crucial to clearly identify and hence to control this pest that has different names i.e., red strip weevil, sago palm weevil, coconut weevil, and Asiatic weevil.
Nonetheless, in the Middle East still there is a controversy that RPW is a Pakistani weevil (Duguma et al., 2015), and this necessitate a clear genetic characterization to reduce the misclassification of this parasite. Furthermore, it is important to distinguish local species of RPW from those obtrusive ones, and this could be achieved by using molecular genetic-based identification tools such as Inter Simple Sequence Repeats (ISSR) (Hashem, 2016) and Random Amplified Polymorphic DNA (RAPD) (Rugman-Jones et al., 2013). However, it is recommended, for realistic DNA fingerprinting and phylogeny, to use minimum two tools to obtain reliable results.
These molecular genetic tools have proven powerful for identification of genotypes (Ishizaki et al., 2016), as it is relatively simple and accurate for molecular characterization of insect species. We used ISSR and RAPD for identification and the evolutionary divergence of palm weevils collected from 21 different locations in the Eastern Province, Saudi Arabia. The present report might be the first to shed light on the RPW genotypes in the Eastern Region and to identify a proposed obtrusive species.
2. Material and methods
2.1. Sample collection
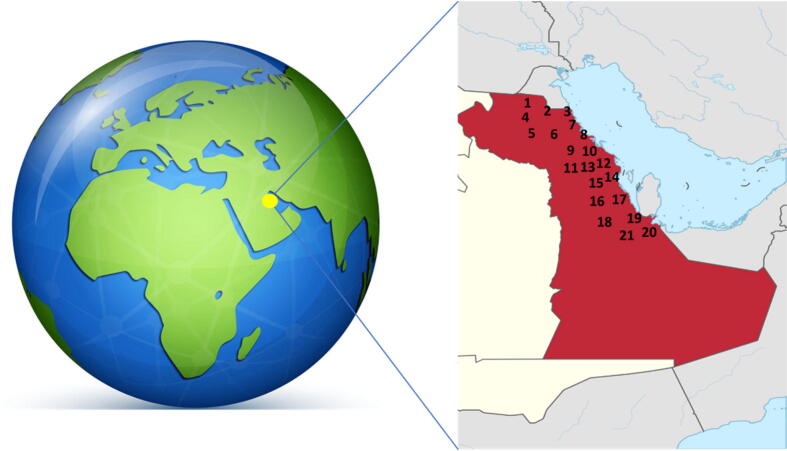
In the present study, 21 red palm weevil samples were collected between April 21 and June 6, 2019 from different geographical regions in the Eastern Province (daytime temperature: 20–45.5 °C, nighttime temperature: 9–30 °C, average relative humidity: 65%, hours of sunshine: 5.9–8 h, latitudes: between 17.10 m, and 29.10 m, and coordinates: 22°30′N 51°00′E), Saudi Arabia (Fig. 1, Fig. 2). From each location, two insects were collected, and the samples were preserved in ethanol and stored at − 20 °C until being subjected to analysis.
Fig. 1.
The Eastern Province map showing the sampling locations. 1: Al-Taraf, 2:Al-Jabil, 3:Al-Jabil-West 4: AL-Jaroudiya, 5: Umm Al Hamam, 6: Al-Sahimia, 7: Safwa, 8: Tarout, 9: Al-Qarah, 10: Sifah, 11: Haql, 12: Battaliyah-2, 13: Battaliyah, 14: Al-Oyoon, 15: Al-Asfar, 16: Juaymah, 17: Sahimia, 18: Bo-Sahabl, 19: AL-Kuwailidiya, 20: Al-Ghowaybah and 21: Safira.
Fig. 2.
Representative samples showing the morphological difference between different RPW from different locations.
2.2. DNA extraction
Total DNA was extracted from each sample by excising a small piece of the soft tissues located in the insect’s abdomen. DNA was extracted using the DNeasy Blood & Tissue Kit (Qiagen, Germany). We followed the kit’s instructions with some modifications, where we grinded the soft tissue in liquid nitrogen before applying the samples to the columns.
2.3. ISSR analysis
About 50 ng of the extracted DNA was subjected to amplification using thermal cycler (BIOMETRA UNO-Thermoblock Thermal Cycler) against 9 Inter Simple Sequence Repeats (ISSR) primers (Table 1). Briefly, to each tube, 1 µL (50 ng) of DNA was added, followed by 12.5 µL master mix, 0.5 µL of the ISSR primer and the total volume was brought to 25 µL using deionized water. The thermal profile was 95 °C for 5 min as a pre-PCR step followed by 35 cycles of 95 °C for 45 sec, 33 °C for 45 sec, and 72 °C for 1 min. A final extension at 72 °C for 5 min was performed as a post-PCR step. The protocol was conducted according to (Ibrahim et al., 2011).
Table 1.
Primer sequences of ISSR and RAPD used in this study.
| Primer code | Sequence | GC content | Ref. | |
|---|---|---|---|---|
| RAPD | OPA16 | AGCCAGGCAA | 60 | (Piątczak et al., 2015) |
| OPA18 | AGGTGACCGT | 60 | (Zhang et al., 1997) | |
| OPA19 | CAAACGTCGG | 60 | (Ikegami et al., 2009) | |
| OPA20 | GTTGCGATCC | 60 | (Ikegami et al., 2009) | |
| OPC18 | TGAGTGGGTG | 60 | (Kawai and Mitsuhashi, 1997) | |
| OPZ10 | CCGACAAACC | 60 | (Saxena et al., 2014) | |
| OPO14 | AGCATGGCTC | 60 | (Thilaga et al., 2017) | |
| OPD19 | CTGGGGACTT | 60 | (Sharma et al., 2018) | |
| OPC20 | ACCCGGTCAC | 70 | (Sharma et al., 2018) | |
| ISSR | HB8 | (GA)6GG | 57.1 | (Goyal et al., 2014) |
| HB9 | (GT)6GG | 57.1 | (Goyal et al., 2014) | |
| HB10 | (GA)6CC | 57.1 | (Goyal et al., 2014) | |
| HB11 | (GT)6CC | 57.1 | (Goyal et al., 2014) | |
| HB13 | (GAG)3GC | 72.7 | (Goyal et al., 2014) | |
| HB15 | (GTG)3GC | 72.7 | (Goyal et al., 2014) | |
| 17898A | (CA)6AC | 50 | (Goyal et al., 2014) | |
| 17898B | (CA)6GT | 50 | (Goyal et al., 2014) | |
| 874 | CCTCCTCCTCCT | 66.6 | (Chen et al., 2011) |
2.4. RAPD analysis
The extracted DNA was also subjected to amplification against 9 RAPD primers (Table 1). The same reaction volumes were used, while the thermal profile used was as follows: 95 °C for 5 min as a pre-PCR step followed by 35 cycles of 95 °C for 45 sec, 35 °C for 1 min, and 72 °C for 1 min. a final extension at 72 °C for 5 min was performed as a post-PCR step. The protocol was conducted according to (Rugman-Jones et al., 2013).
2.5. Gel electrophoresis
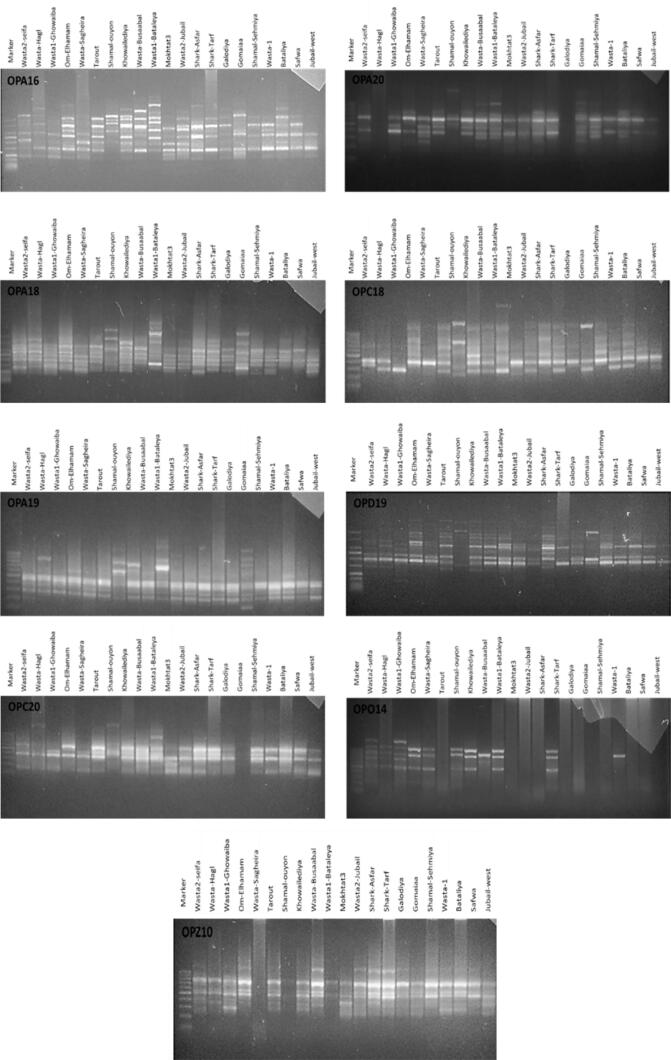
To visualize the ISSR and RAPD amplified fragments, PCR products were separated on 1.6% agarose gel and photographed after being stained with ethidium bromide. Ladder with 100 bp (Qiagen, Germany) was used to determine the lengths of different DNA fragments. ISSR's and RAPD's banding patterns were scored as 1 for the present band and 0 for the absent one.
2.6. Gel documentation
All photos were analyzed using GelPro Analyzer (Media Cybernetics, USA) to identify the common and unique bands. Relative front and molecular weights were also measured (data not shown).
2.7. Similarity scoring
Cluster analysis was performed by Un-weighted pair group method of arithmetic means (UPGMA). The similarity matrix and phylogenetic relationship were determined using Diversity Database Version 2.1 Windows (Bio Rad).
3. Results
3.1. RAPD analysis
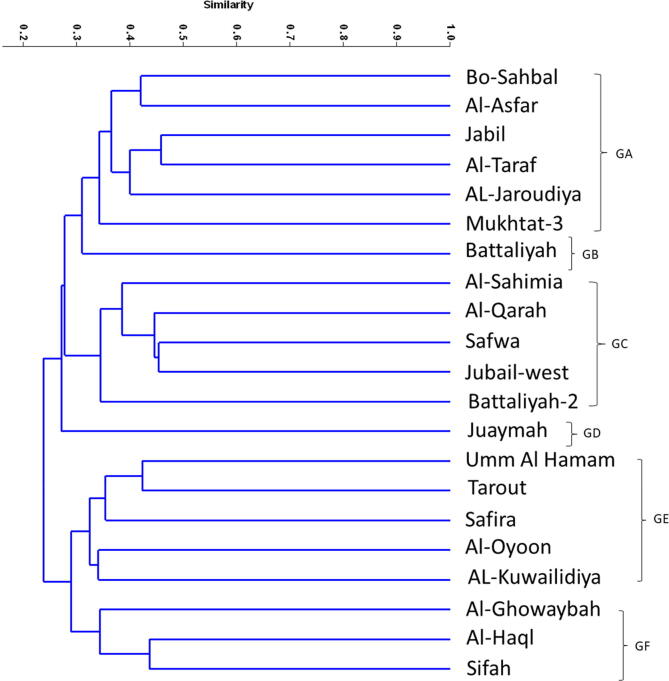
The similarity coefficient between the 21 accessions ranged from 0.11 to 0.52 with a mean of 0.32 based on RAPD markers (Table 2). The highest similarity coefficient is between Sifah and Haql (0.52), Sahimia and Battaliyah (0.50), Battaliyah and Safwa (0.49) and between Sahimia and Al-Qarah (0.48). While the lowest similarity was between Tarout and Al-Qarah (0.11). According to these data, the clustering analysis classified the 21 accessions into seven groups (Fig. 3, Fig. 4). Group A (GA) contains only one accession (Al-Oyoon), while GB contains three accessions (Al-Ghowaybah and Sifah). GC contains two accessions (Umm Al Hamam and Safira), while GD contains only accession (Juaymah). GE contains the eight accessions (Tarout, AL-Kuwailidiya, Bo-Sahabl, AL-Jaroudiya, Al-Jabil, Al-Taraf, Mukhatat-3, Al-Asfar and Battaliyah-2), while GF contains five accessions (Al-Sahimia, Jubail-west, Al-Qarah, Battaliyah and Safwa). Some samples did not generate banding profile because either failure of PCR amplifications or any unexpected errors.
Table 2.
Similarity coefficient of 21 accessions based on RAPD fingerprinting.
| Sifa | Haql | Al-Ghowaybah | Umm AlHamam | Safira | Tarout | Al-Oyoon | Al-kuwailidiya | Bo-Sahabl | Battaliyah | Mukatat-3 | Jabil | Al-Asfar | Al-Taraf | Al-Jaroudiya | Juaymah | Al-Sahimia | Al-Qarah | Battaliyah-2 | Safwa | Jubail-west | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sifa | 1 | ||||||||||||||||||||
| Haql | 0.52 | 1 | |||||||||||||||||||
| Al-Ghowaybah | 0.33 | 0.32 | 1 | ||||||||||||||||||
| Umm AlHamam | 0.24 | 0.28 | 0.23 | 1 | |||||||||||||||||
| Safira | 0.19 | 0.2 | 0.21 | 0.33 | 1 | ||||||||||||||||
| Tarout | 0.18 | 0.19 | 0.11 | 0.34 | 0.26 | 1 | |||||||||||||||
| Al-Oyoon | 0.25 | 0.24 | 0.13 | 0.2 | 0.24 | 0.16 | 1 | ||||||||||||||
| Al-kuwailidiya | 0.17 | 0.24 | 0.16 | 0.22 | 0.27 | 0.44 | 0.17 | 1 | |||||||||||||
| Bo-Sahabl | 0.16 | 0.12 | 0.14 | 0.28 | 0.23 | 0.41 | 0.17 | 0.39 | 1 | ||||||||||||
| Battaliyah | 0.16 | 0.15 | 0.19 | 0.25 | 0.17 | 0.27 | 0.23 | 0.27 | 0.33 | 1 | |||||||||||
| Mukatat-3 | 0.14 | 0.18 | 0.13 | 0.25 | 0.14 | 0.31 | 0.13 | 0.38 | 0.37 | 0.34 | 1 | ||||||||||
| Jabil | 0.2 | 0.17 | 0.13 | 0.33 | 0.18 | 0.32 | 0.21 | 0.24 | 0.41 | 0.32 | 0.26 | 1 | |||||||||
| Al-Asfar | 0.19 | 0.2 | 0.14 | 0.2 | 0.13 | 0.35 | 0.14 | 0.24 | 0.34 | 0.28 | 0.37 | 0.38 | 1 | ||||||||
| Al-Taraf | 0.25 | 0.25 | 0.16 | 0.29 | 0.15 | 0.33 | 0.17 | 0.24 | 0.31 | 0.25 | 0.23 | 0.42 | 0.27 | 1 | |||||||
| Al-Jaroudiya | 0.2 | 0.23 | 0.15 | 0.37 | 0.3 | 0.38 | 0.15 | 0.29 | 0.54 | 0.23 | 0.35 | 0.41 | 0.29 | 0.4 | 1 | ||||||
| Juaymah | 0.23 | 0.25 | 0.19 | 0.29 | 0.25 | 0.26 | 0.2 | 0.28 | 0.21 | 0.18 | 0.27 | 0.28 | 0.21 | 0.37 | 0.28 | 1 | |||||
| Al-Sahimia | 0.22 | 0.19 | 0.2 | 0.21 | 0.24 | 0.14 | 0.2 | 0.27 | 0.27 | 0.19 | 0.29 | 0.27 | 0.26 | 0.24 | 0.28 | 0.21 | 1 | ||||
| Al-Qarah | 0.18 | 0.16 | 0.22 | 0.15 | 0.17 | 0.11 | 0.21 | 0.22 | 0.25 | 0.22 | 0.18 | 0.27 | 0.24 | 0.28 | 0.26 | 0.18 | 0.48 | 1 | |||
| Battaliyah-2 | 0.18 | 0.18 | 0.22 | 0.2 | 0.22 | 0.17 | 0.18 | 0.27 | 0.27 | 0.19 | 0.26 | 0.25 | 0.23 | 0.26 | 0.28 | 0.27 | 0.36 | 0.5 | 1 | ||
| Safwa | 0.13 | 0.15 | 0.24 | 0.14 | 0.16 | 0.15 | 0.19 | 0.27 | 0.24 | 0.2 | 0.26 | 0.24 | 0.17 | 0.19 | 0.26 | 0.27 | 0.35 | 0.44 | 0.49 | 1 | |
| Jubail-west | 0.26 | 0.19 | 0.26 | 0.14 | 0.21 | 0.09 | 0.17 | 0.2 | 0.17 | 0.15 | 0.26 | 0.18 | 0.16 | 0.17 | 0.19 | 0.16 | 0.42 | 0.43 | 0.33 | 0.37 | 1 |
Fig. 3.
Unweighted pair group method with arithmetic mean (UPGMA) dendrogram illustrating the genetic relationships between 21 accessions based on RAPD fingerprinting.
Fig. 4.
Electrophoretic separation of the amplified RAPD fragments of 21 RPW samples collected from Eastern Province, Saudi Arabia.
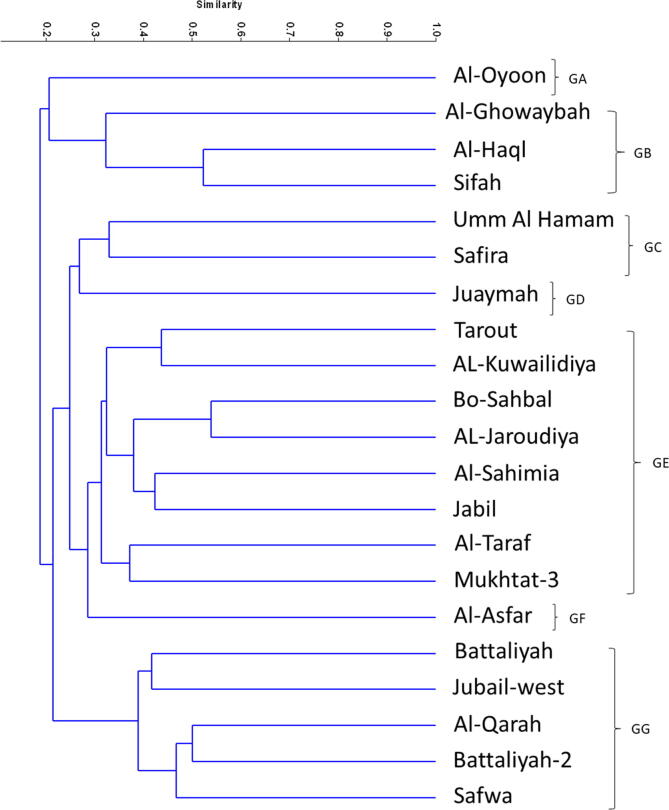
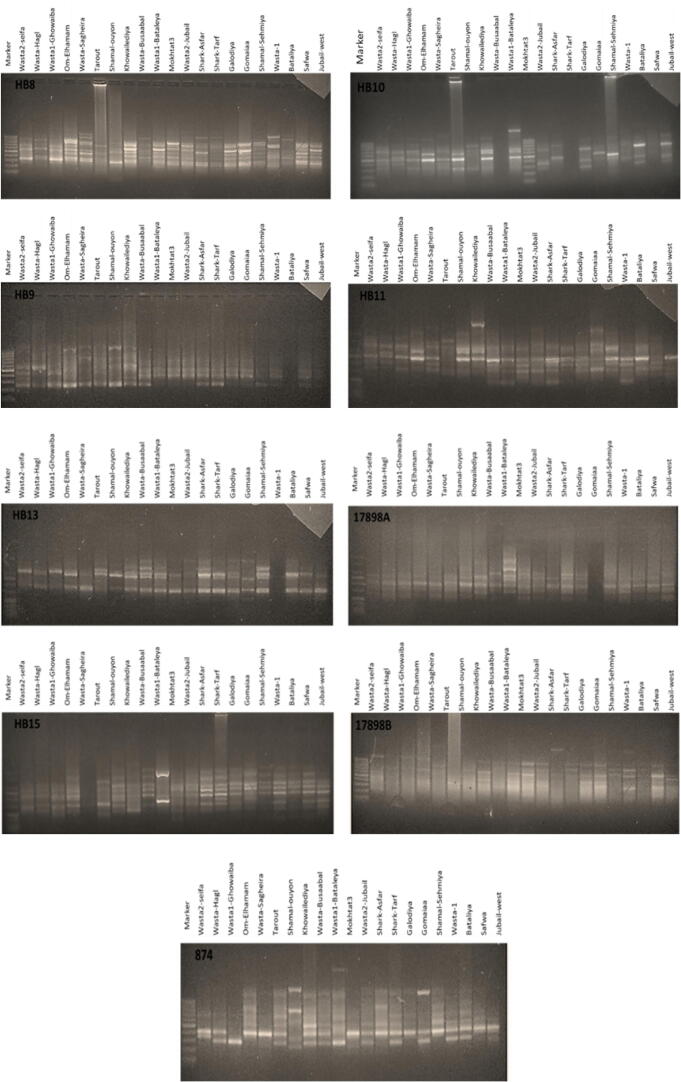
3.2. ISSR analysis
Based on ISSR banding, the similarity coefficient between the 21 accessions ranged from 0.16 to 0.45 with mean of 0.31 (Table 3). The highest similarity coefficient was between Mokhtat3 and Jabil (0.46), Jubail-west and Al-Qarah (44) and between Sifah and Haql (0.44). UPGMA analysis clustered the 21 accessions in 6 groups (Fig. 5, Fig. 6). The first group (GA) contains six accessions. Battaliyah and Juaymah accessions each was separated in solitary groups (GB and GD, respectively). Each of the 3rd and 5th group (GC and GE) contains five accessions, but the 6th one (GF) contains three accessions. Here also, we have some samples without banding profile or very faint to be visualized using the available tools.
Table 3.
Similarity coefficient of 21 accessions. based on ISSR banding.
| Sifa | Haql | Al-Ghowaybah | Umm AlHamam | Safira | Tarout | Al-Oyoon | Al-kuwailidiya | Bo-Sahabl | Battaliyah | Mukatat-3 | Jabil | Al-Asfar | Al-Taraf | Al-Jaroudiya | Juaymah | Al-Sahimia | Al-Qarah | Battaliyah-2 | Safwa | Jubail-west | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sifa | 1 | ||||||||||||||||||||
| Haql | 0.44 | 1 | |||||||||||||||||||
| Al-Ghowaybah | 0.36 | 0.33 | 1 | ||||||||||||||||||
| Umm AlHamam | 0.28 | 0.35 | 0.26 | 1 | |||||||||||||||||
| Safira | 0.3 | 0.35 | 0.26 | 0.38 | 1 | ||||||||||||||||
| Tarout | 0.26 | 0.29 | 0.29 | 0.42 | 0.33 | 1 | |||||||||||||||
| Al-Oyoon | 0.31 | 0.31 | 0.29 | 0.33 | 0.29 | 0.32 | 1 | ||||||||||||||
| Al-kuwailidiya | 0.25 | 0.29 | 0.26 | 0.28 | 0.34 | 0.39 | 0.34 | 1 | |||||||||||||
| Bo-Sahabl | 0.22 | 0.27 | 0.21 | 0.36 | 0.29 | 0.37 | 0.24 | 0.33 | 1 | ||||||||||||
| Battaliyah | 0.18 | 0.22 | 0.2 | 0.31 | 0.24 | 0.28 | 0.26 | 0.28 | 0.39 | 1 | |||||||||||
| Mukatat-3 | 0.19 | 0.22 | 0.17 | 0.26 | 0.26 | 0.27 | 0.17 | 0.29 | 0.29 | 0.31 | 1 | ||||||||||
| Jabil | 0.21 | 0.22 | 0.17 | 0.29 | 0.21 | 0.26 | 0.2 | 0.27 | 0.36 | 0.35 | 0.46 | 1 | |||||||||
| Al-Asfar | 0.2 | 0.28 | 0.24 | 0.32 | 0.23 | 0.33 | 0.27 | 0.29 | 0.42 | 0.25 | 0.4 | 0.38 | 1 | ||||||||
| Al-Taraf | 0.23 | 0.27 | 0.21 | 0.38 | 0.2 | 0.29 | 0.27 | 0.3 | 0.39 | 0.27 | 0.28 | 0.46 | 0.36 | 1 | |||||||
| Al-Jaroudiya | 0.22 | 0.3]23 | 0.2 | 0.3 | 0.2 | 0.29 | 0.16 | 0.22 | 0.39 | 0.29 | 0.29 | 0.43 | 0.32 | 0.37 | 1 | ||||||
| Juaymah | 0.24 | 0.22 | 0.22 | 0.34 | 0.21 | 0.25 | 0.21 | 0.26 | 0.27 | 0.28 | 0.25 | 0.26 | 0.25 | 0.3 | 0.33 | 1 | |||||
| Al-Sahimia | 0.18 | 0.19 | 0.2 | 0.25 | 0.23 | 0.17 | 0.25 | 0.26 | 0.25 | 0.28 | 0.27 | 0.33 | 0.3 | 0.36 | 0.39 | 0.24 | 1 | ||||
| Al-Qarah | 0.24 | 0.17 | 0.23 | 0.25 | 0.24 | 0.2 | 0.23 | 0.22 | 0.24 | 0.2 | 0.29 | 0.32 | 0.33 | 0.29 | 0.34 | 0.29 | 0.43 | 1 | |||
| Battaliyah-2 | 0.16 | 0.17 | 0.2 | 0.2 | 0.21 | 0.2 | 0.24 | 0.29 | 0.23 | 0.21 | 0.22 | 0.28 | 0.18 | 0.31 | 0.27 | 0.24 | 0.32 | 0.36 | 1 | ||
| Safwa | 0.23 | 0.19 | 0.26 | 0.16 | 0.23 | 0.17 | 0.21 | 0.23 | 0.26 | 0.29 | 0,27 | 0.33 | 0.26 | 0.24 | 0.35 | 0.31 | 0.33 | 0.45 | 0.4 | 1 | |
| Jubail-west | 0.27 | 0.23 | 0.26 | 0.2 | 0.25 | 0.21 | 0.23 | 0.27 | 0.25 | 0.18 | 0.25 | 0.25 | 0.3 | 0.28 | 0.34 | 0.25 | 0.39 | 0.44 | 0.3 | 0.45 |
Fig. 5.
Unweighted pair group method with arithmetic mean (UPGMA) dendrogram illustrating the genetic relationships between 21 accessions based on ISSR fingerprinting.
Fig. 6.
Electrophoretic separation of the amplified ISSR fragments of 21 RPW samples collected from Eastern Province, Saudi Arabia.
By combining RAPD and ISSR banding in one analysis, the similarity coefficient between the 21 accessions ranged from 0.15 to 0.48 with average 0.32 (Table 4). The highest similarity coefficient was between Sifah and Haql. The UPGMA analysis resulted in clustering the 21 accessions into 7 groups (Fig. 7). Each of Battaliyah, Juaymah and Al-Oyoon separated in single groups (GB, GD and GF, respectively). The first group (GA) contains six accessions, the 3rd group (GC) contains four accessions, the 5th group (GE) clustered five accessions and the 7th group consist of three accessions.
Table 4.
Similarity coefficient of 21 accessions based on RAPD and ISSR banding.
| Sifa | Haql | Al-Ghowaybah | Umm AlHamam | Safira | Tarout | Al-Oyoon | Al-kuwailidiya | Bo-Sahabl | Battaliyah | Mukatat-3 | Jabil | Al-Asfar | Al-Taraf | Al-Jaroudiya | Juaymah | Al-Sahimia | Al-Qarah | Battaliyah-2 | Safwa | Jubail-west | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sifa | 1 | ||||||||||||||||||||
| Haql | 0.48 | 1 | |||||||||||||||||||
| Al-Ghowaybah | 0.34 | 0.32 | 1 | ||||||||||||||||||
| Umm AlHamam | 0.26 | 0.31 | 0.24 | 1 | |||||||||||||||||
| Safira | 0.24 | 0.27 | 0.23 | 0.35 | 1 | ||||||||||||||||
| Tarout | 0.21 | 0.24 | 0.19 | 0.38 | 0.3 | 1 | |||||||||||||||
| Al-Oyoon | 0.28 | 0.27 | 0.2 | 0.26 | 0.26 | 0.23 | 1 | ||||||||||||||
| Al-kuwailidiya | 0.21 | 0.27 | 0.2 | 0.25 | 0.31 | 0.41 | 0.25 | 1 | |||||||||||||
| Bo-Sahabl | 0.19 | 0.19 | 0.17 | 0.32 | 0.26 | 0.39 | 0.2 | 0.36 | 1 | ||||||||||||
| Battaliyah | 0.17 | 0.19 | 0.2 | 0.28 | 0.2 | 0.27 | 0.24 | 0.27 | 0.36 | 1 | |||||||||||
| Mukatat-3 | 0.16 | 0.2 | 0.15 | 0.25 | 0.2 | 0.29 | 0.15 | 0.33 | 0.33 | 0.32 | 1 | ||||||||||
| Jabil | 0.2 | 0.19 | 0.15 | 0.31 | 0.2 | 0.29 | 0.21 | 0.26 | 0.38 | 0.34 | 0.36 | 1 | |||||||||
| Al-Asfar | 0.2 | 0.24 | 0.19 | 0.26 | 0.18 | 0.34 | 0.21 | 0.26 | 0.38 | 0.26 | 0.39 | 0.38 | 1 | ||||||||
| Al-Taraf | 0.24 | 0.26 | 0.19 | 0.33 | 0.18 | 0.31 | 0.22 | 0.27 | 0.34 | 0.26 | 0.25 | 0.44 | 0.31 | 1 | |||||||
| Al-Jaroudiya | 0.21 | 0.23 | 0.17 | 0.34 | 0.25 | 0.33 | 0.16 | 0.25 | 0.46 | 0.26 | 0.32 | 0.42 | 0.3 | 0.38 | 1 | ||||||
| Juaymah | 0.24 | 0.23 | 0.2 | 0.31 | 0.23 | 0.25 | 0.2 | 0.27 | 0.24 | 0.23 | 0.26 | 0.27 | 0.23 | 0.34 | 0.3 | 1 | |||||
| Al-Sahimia | 0.2 | 0.19 | 0.2 | 0.23 | 0.23 | 0.15 | 0.22 | 0.26 | 0.26 | 0.23 | 0.28 | 0.3 | 0.28 | 0.3 | 0.33 | 0.22 | 1 | ||||
| Al-Qarah | 0.21 | 0.17 | 0.22 | 0.2 | 0.21 | 0.15 | 0.22 | 0.22 | 0.24 | 0.21 | 0.24 | 0.3 | 0.29 | 0.28 | 0.3 | 0.23 | 0.45 | 1 | |||
| Battaliyah-2 | 0.17 | 0.18 | 0.21 | 0.2 | 0.22 | 0.18 | 0.21 | 0.27 | 0.25 | 0.2 | 0.24 | 0.27 | 0.2 | 0.28 | 0.28 | 0.26 | 0.34 | 43 | 1 | ||
| Safwa | 0.18 | 0.17 | 0.25 | 0.15 | 0.2 | 0.16 | 0.2 | 0.25 | 0.25 | 0.24 | 0.27 | 0.28 | 0.22 | 0.21 | 0.3 | 0.29 | 0.34 | 0.44 | 44 | 1 | |
| Jubail-west | 0.27 | 0.21 | 0.26 | 0.17 | 0.23 | 0.15 | 0.2 | 0.23 | 0.21 | 0.16 | 0.25 | 0.21 | 0.23 | 0.23 | 0.26 | 0.2 | 0.41 | 0.44 | 32 | 0.42 | 1 |
Fig. 7.
Unweighted pair group method with arithmetic mean (UPGMA) dendrogram illustrating the genetic relationships between 21 accessions based on RAPD and ISSR banding.
Table Generally, RAPD and the combination between RAPD and ISSR separated three accessions; Battaliyah, Juaymah and Al-Oyoon to solitary groups as they showed low similarity coefficient with other accessions. While ISSR separated Battaliyah and Juaymah to solitary groups (Fig. 8).
Fig. 8.
The two proposed obtrusive RPWs.
4. Discussion
Red Palm Weevil is an invasive, polyphagous palm pest that causes huge economic loses in the date palm industry, especially in the Arabian Peninsula. Genetic classification is controversial in the species status of R. vulneratus and R. ferrugineus according to the similarity observed in their production and response to pheromones (Perez et al., 1996). This parasite entered the Arabian Gulf area in the mid-1980 (Bozbuga and Hazir, 2008), and reached the Eastern Province of the KSA by 1985 (Baker, 2002). During the last four decades foreign insects might be introduced to the local community by various methods. This calls for the urgency of genetically determine the invading species from the local ones, which might augment the integrated pest management (IPM) programs. The nearest source of invasion is Pakistan, Iran, Oman, UAE, and other GCC countries.
In this investigation, 21 RPW samples were collected from 21 different locations in the Eastern Province, Saudi Arabia to identify the genetic similarity/dissimilarity aiming to characterize the local species and its relationships with the invading species. RAPD analysis revealed that Al-Oyoon and Juaymah locations represent unique genetic fingerprints, which might indicate that RPWs collected from these locations might be considered as an obtrusive species to the local population in recent times. The same profile was obtained for which represented a unique genetic fingerprint, where it was in a separate group. Genetic classification using RADP is straightforward and powerful tool to characterize alien species, and has been employed by different research groups to identify RPW samples collected from the UAE, Egypt, and KSA (Salama and Saker, 2002, Al-Ayied et al., 2006, Enan, 2005).
Moreover, ISSR clustering data revealed a higher similarity coefficient between Mokhtat3 and Jabil (0.46), Jubail-west and Al-Qarah (44) and between Sifah and Haql (0.44). UPGMA analysis clustered the 21 accessions in 6 groups. Battaliyah and Juaymah accessions each was separated in solitary groups (GB and GD, respectively). This might indicate that Battaliyah and Juaymah represent different species with unique fingerprinting.
Generally, by combining ISSR and RAPD data, we can conclude that RPWs collected from Juaymah (located at the cost of Arabian Gulf; 25° 27.693′ N, 49° 36.411′ E) and Al-Oyoon (located 50 km to the west of the Arabian Gulf; 25° 35.341′ N, 49° 34.380′ E) represent obtrusive species. Although prevailed in several countries in the Middle East and North Africa (Mizzi et al., 2009), the proposed origins of this species are Iran (47 km to 370 km) and UAE (429 km). In this study, morphological analysis of the proposed obtrusive species revealed different thoracic spot pattern in Juaymah samples, although the thoracic spot pattern of insects collected from Al-Oyoon was similar to those collected from different locations.
Al-Ayied et al. (2006a) indicated that RPW samples collected from Al-Hassa governorate in Eastern Province, Saudi Arabia have yielded different genetic fingerprinting. This perhaps due to the presence of obtrusive species in Al-Hassa governorate (60 km to the west of the Arabian Gulf; 22.2954° N, 50.6794° E) as it involves Al-Oyoon location. Reducing the perplexity between weevils is crucial component in augmenting the management process, and this could be done via exploring the genetic fingerprint profiles of local and obtrusive species (El-Mergawy et al., 2011).
5. Conclusion
Red palm weevil is still the most aggressive date palm pest in the Arabian Peninsula, where it causes enormous losses in the date industry. In the present study we collected RPW from 21 different geographic locations in the Eastern Province, Saudi Arabia and characterized them using RAPD and ISSR. Data obtained indicated that based on RAPD analysis, the 21 accessions were categorized into seven groups with Juaymah and Al-Oyoon were categorized each in a solitary group. ISSR clustering analysis also categorized the 21 accessions into six groups with Bataliyah, Juaymah and Al-Oyoon were categorized each in a separate group. Combining ISSR and RAPD data indicated that the two RPW samples collected from Juaymah and Al-Oyoon might represent obtrusive species. However, to deeply identify the origin of these insects, further molecular studies are required.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This work has been carried out in the laboratories of the College of Biotechnology, Misr University for Science and Technology, Giza, Egypt. The authors thank M. H. Alkhazal, M. A. Altammar, and A. A. Alshawaf, Center of Date Palm and Dates, Al-Qatif Branch, Ministry of Environment, Water and Agriculture, Saudi Arabia for their efforts in collecting samples. The authors also thank Dr. Osama A. M. said for his help in conducting the practical work.
Footnotes
Peer review under responsibility of King Saud University.
References
- Al-Ayied H.Y., Alswailem A.M., Shair O., Al Jabr A.M. Evaluation of phylogenetic relationship between three phenotypically different forms of Red date palm weevil Rhynchophorus ferrugineus Oliv. using PCR-based RAPD technique. Archives of Phytopathology and Plant Protection. 2006;39:303–309. [Google Scholar]
- BAKER, W. 2002. Insects on Palms. F. W. Howard, D. Moore, R. M. Giblin-Davis & R. G. Abad, with contributions by J. W. Amrine, E. H. Erickson & M. R. Wilson. Wallingford: CABI Publishing. 2001. 400pp., 16pp. colour plates. ISBN 0 85199 326 5. £65.00 (hardback). Edinburgh Journal of Botany, 59, 459-466.
- Bozbuga R., Hazir Adalet. Pests of the palm (Palmae sp.) and date palm (Phoenix dactylifera) determined in Turkey and evaluation of red palm weevil (Rhynchophorus ferrugineus Olivier) (Coleoptera:Curculionidae) EPPO Bulletin. 2008;38(1):127–130. [Google Scholar]
- Chen C., Duan L.-N., Zhou X.-L., Chen B.-L., Fu C.-X. Molecular authentication of geo-authentic Scrophularia ningpoensis. Journal of Zhejiang University. Science. B. 2011;12:393–398. doi: 10.1631/jzus.B1000179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duguma D., Hall M.W., Rugman-Jones P., Stouthamer R., Terenius O., Neufeld J.D., Walton W.E., Sveriges L. Developmental succession of the microbiome of Culex mosquitoes. BMC MICROBIOLOGY. 2015;15:140. doi: 10.1186/s12866-015-0475-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Mergawy R.A.A.M., Ala Jlan A.M., Abdallah N.A., Nasr M.I., Silvain J.-F. Determination of different geographical populations of Rhynchophorus ferrugineus (Olivier) (Coleoptera: Curculionidae) using RAPD-PCR. International Journal of Agriculture and Biology. 2011;13:227–232. [Google Scholar]
- ENAN, G. G. G. A. M. R. 2005. Genetic Diversity Among Populations of Red Palm Weevil, Rhynchophorus ferrugineus Olivier (Coleoptera: Curculionidae), Determined by Random Amplified Polymorphic DNAPolymerase Chain Reaction (RAPD-PCR).
- Faghih A.A. The biology of red palm weevil, Rhynchophorus ferrugineus Oliv. (Coleoptera, Curculionidae) in Saravan region (Sistan and Balouchistan province, Iran) Applied Entomology and Phytopathology. 1996;63:61–86. [Google Scholar]
- Faleiro J.R., El-Shafie H.A.F., Ajlan A.M., Sallam A.A. Screening Date Palm Cultivars for Resistance to Red Palm Weevil, Rhynchophorus ferrugineus (Coleoptera: Curculionidae) Florida Entomologist. 2014;97:1529–1536. [Google Scholar]
- Fiaboe K.K.M., Peterson A.T., Kairo M.T.K., Roda A.L. Predicting the Potential Worldwide Distribution of the Red Palm Weevil Rhynchophorus ferrugineus (Olivier) (Coleoptera: Curculionidae) using Ecological Niche Modeling. Florida Entomologist. 2012;95:659–673. [Google Scholar]
- Goyal A.G., Manoharachary C.M., Aakash, Chakravarthula . NY, Springer; New York: 2014. Future Challenges in Crop Protection Against Fungal Pathogens. [Google Scholar]
- Hashem M. Genetic variations among the red palm weevil rhynchophorus ferrugineus populations collected from egypt. Egyptian Journal of Genetics and Cytology. 2016;45:33–45. [Google Scholar]
- Huda M.N., Hasan M., Abdullah H.M., Sarker U. Spatial distribution and genetic diversity of wild date palm (Phoenix sylvestris) growing in coastal Bangladesh. Tree Genetics & Genomes. 2019;15:1–11. [Google Scholar]
- Ibrahim A.I., Hemeida A.A., Abdelkader H.S., Girgis A.A., Abou-El-einin H. Genetic variance between some Egyptian Date Palm cultivars using PCR-based markers with emphasis on the prevalence of Al wijam disease. Archives Of Phytopathology And Plant Protection. 2011;44:732–742. [Google Scholar]
- Ikegami H., Ikegami H., Nogata H., Nogata H., Hirashima K., Hirashima K., Awamura M., Awamura M., Nakahara T., Nakahara T. Analysis of genetic diversity among European and Asian fig varieties (Ficus carica L.) using ISSR, RAPD, and SSR markers. Genetic Resources and Crop Evolution. 2009;56:201–209. [Google Scholar]
- Ishizaki K., Nishihama R., Yamato K.T., Kohchi T. Molecular Genetic Tools and Techniques forMarchantia polymorphaResearch. Plant and Cell Physiology. 2016;57:262–270. doi: 10.1093/pcp/pcv097. [DOI] [PubMed] [Google Scholar]
- Kawai Y., Mitsuhashi J. An insect cell line discrimination method by RAPD-PCR. Vitro Cell Dev Biol Anim. 1997;33:512–515. doi: 10.1007/s11626-997-0093-3. [DOI] [PubMed] [Google Scholar]
- Mizzi, S., D. D., D. M. & Longo, S. 2009. The Red Palm Weevil, Rhynchophorus ferrugineus (Olivier, 1790) in Malta (Coleoptera: Curculionoidea). Bulletin of the Entomological Society of Malta, 2.
- Mukhtar M., Rasool K.G., Parrella M.P., Sheikh Q.I., Pain A., Lopez-Llorca L.V., Aldryhim Y.N., Mankin R.W., Aldawood A.S. New Initiatives for Management of Red Palm Weevil Threats to Historical Arabian Date Palms. Florida Entomologist. 2011;94:733–736. [Google Scholar]
- Mulley, M., Kooistra, L. & Bierens, L. 2019. High-Resolution Multisensor Remote Sensing to Support Date Palm Farm Management. Agriculture-Basel, 9.
- Musmeci S., Belvedere S., Sasso R., Arnone S., Cristofaro M., Nobili P., la Marca A., de Biase A. Last-male sperm precedence in Rhynchophorus ferrugineus (Olivier): observations in laboratory mating experiments with irradiated males. Bulletin of Entomological Research. 2018;108:93–100. doi: 10.1017/S0007485317000840. [DOI] [PubMed] [Google Scholar]
- Naji Mordi N.A.-D., Al-Dobai S., Faleiro J.R. Review on the management of red palm weevil Rhynchophorus ferrugineus olivier in date palm Phoenix dactylifera L. Emirates Journal of Food and Agriculture. 2016;28:34. [Google Scholar]
- Perez A.L., Hallett R.H., Gries R., Gries G., Cameron Oehlschlager A., Borden J.H. Pheromone chirality of asian palm weevils, Rhynchophorus ferrugineus (Oliv.) andR. vulneratus (Panz.) (Coleoptera: Curculionidae) Journal of chemical ecology. 1996;22:357–368. doi: 10.1007/BF02055104. [DOI] [PubMed] [Google Scholar]
- Piątczak E., Kuźma Ł., Sitarek P., Wysokińska H. Shoot organogenesis, molecular analysis and secondary metabolite production of micropropagated Rehmannia glutinosa Libosch. Plant Cell. Tissue and Organ Culture (PCTOC) 2015;120:539–549. [Google Scholar]
- Rasool K.G., Khan M.A., Tufail M., Husain M., Mehmood K., Mukhtar M., Takeda M., Aldawood A.S. Differential Proteomic Analysis of Date Palm Leaves Infested with the Red Palm Weevil (Coleoptera: Curculionidae) Florida Entomologist. 2018;101:290–298. [Google Scholar]
- Rugman-Jones, P. F., Hoddle, C. D., Hoddle, M. S. & Stouthamer, R. 2013. The lesser of two weevils: molecular-genetics of pest palm weevil populations confirm Rhynchophorus vulneratus (Panzer 1798) as a valid species distinct from R. ferrugineus (Olivier 1790), and reveal the global extent of both. PloS one, 8, e78379-e78379. [DOI] [PMC free article] [PubMed]
- Salama H.S., Saker M.M. DNA Fingerprints of Three Different Forms of the Red Palm Weevil Collected from Egyptian Date Palm Orchards. Archives of Phytopathology and Plant Protection. 2002;35:299–306. [Google Scholar]
- Saxena S., Verma J., Shikha, Raj Modi D. RAPD-PCR and 16S rDNA phylogenetic analysis of alkaline protease producing bacteria isolated from soil of India: Identification and detection of genetic variability. Journal of Genetic Engineering and Biotechnology. 2014;12:27–35. [Google Scholar]
- Shabani F., Kumar L., Taylor S. Suitable regions for date palm cultivation in Iran are predicted to increase substantially under future climate change scenarios. Journal of Agricultural Science. 2014;152:543–557. [Google Scholar]
- Sharma R., Sharma S., Kumar S. Pair-wise combinations of RAPD primers for diversity analysis with reference to protein and single primer RAPD in soybean. Annals of Agrarian Science. 2018;16:243–249. [Google Scholar]
- Sun X.D., Yan W., Zhang J., Niu X.Q., Li F.H., Qin W.Q., Ma G.C. Frozen section and electron microscopy studies of the infection of the red palm weevil, Rhynchophorus ferrugineus (coleoptera: curculionidae) by the entomopathogenic fungus Metarhizium anisopliae. Springerplus. 2016;5 doi: 10.1186/s40064-016-3416-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thilaga S., Rahul Nair R., Rajesh Kannan M., Ganesh D. RAPD markers for screening shoot gall maker (Betousa stylophora Swinhoe) tolerant genotypes of amla (Phyllanthus emblica L.) Journal of Genetic Engineering and Biotechnology. 2017;15:323–330. doi: 10.1016/j.jgeb.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Hou Y., Zhang X., Zhang J., Li J., Chen Z. Strong population genetic structure of an invasive species, Rhynchophorus ferrugineus (Olivier), in southern China. Ecology and Evolution. 2017;7:10770–10781. doi: 10.1002/ece3.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Rajagopalan, M., Brown, B. A. & R. J. Wallace, J. 1997. Randomly amplified polymorphic DNA PCR for comparison of Mycobacterium abscessus strains from nosocomial outbreaks. Journal of Clinical Microbiology, 35, 3132-3139. [DOI] [PMC free article] [PubMed]