Abstract
Cumin (Cuminum cyminum L.) is an important spice crop worldwide and its production is hampered by the infection of Alternaria blight. Cultivation of cumin in Bangladesh is very limited due to the lack of appropriate germplasm and adequate scientific information regarding the prevalence of Alternaria blight. Field trials were conducted with four advanced lines of cumin viz. CN026, CN028, CN031 and CN038 in five agro-ecological zones (AEZ) to know the adaptation possibility of these lines against the incidence and severity of Alternaria blight of cumin in Bangladesh. Among all lines, CN026 was found as the best in germination capacity and other yield parameters in all locations. The incidence and severity of the disease was observed as high as 98% and 88%, respectively, however, out of the five locations, the incidence and severity of the disease was the lowest in Bogura for the line CN026. In an attempt to identify the causal organism of the Alternaria blight of cumin by using molecular tools, a total of twenty three isolates were collected from the plants showing Alternaria blight symptoms from different AEZ in Bangladesh. Based on the molecular analysis, the isolates were identified as Alternaria alternata, A. burnsii, A. gaisen and A. tenuissima. A. alternata was the most prevalent species followed by A. tenuissima. The isolates of the identified species were found to have genetic, morphological and pathogenic variation. An isolate of A. alternata was observed as the most virulent among the isolates. This is the first report of A. alternata causing Alternaria blight disease of cumin in Bangladesh. The findings of this experiment will help in selecting suitable cumin germplasm and designing proper management strategies against Alternaria blight of cumin in Bangladesh.
Keywords: Cumin, Alternaria blight, Alternaria spp., Bangladesh
1. Introduction
Cumin (Cuminum cyminum L.) belongs to the family Apiaceae (Umbelliferae) and is believed to be the native of the mediterranean and near eastern regions of the globe (Nabhan, 2014). It is widely cultivated in Uzbekistan, Tajikistan, Turkey, Morocco, Egypt, India, Syria, Mexico, Pakistan and Chile (Azeez et al., 2008). It grows best on well drained sandy loam to loamy soil with a pH range of 6.8 to 8.3 (Weiss, 2002). Cumin seeds are used as spice in culinary for flavoring soups, sauces, pickles and for seasoning breads and cakes. Cumin is an important ingredient in Bangladeshi cuisine. To meet the internal demand, a huge amount of cumin is imported every year in Bangladesh in expense of costly foreign exchange. In 2019–20, about 27,000 to 28,000 tons of cumin was imported in Bangladesh (The Financial Express, 2020). Therefore, cultivation of cumin in Bangladesh might reduce its dependency on other countries. Despite having congenial climatic and edaphic conditions, commercial cultivation of cumin is not well adopted in Bangladesh due to many socio-economic reasons including the lack of appropriate germplasm and adequate scientific information regarding the infestation of different pathogens.
Production of cumin is seriously affected by the infection of Alternaria blight caused by Alternaria spp. The disease appears in devastating form every year in the most cumin-growing areas in the world and can cause up to 80% of yield loss (Gemawat and Prasad, 1972). Initially, blight affected plants show minute whitish to black isolated necrotic areas on the aerial parts, especially on tips of young leaves leading to the death of the whole plant or the affected parts (Uppal et al., 1938, Sharma, 2010). Diseased seeds are small, de-shaped, shriveled, light weight and black in colour (Gemawat and Prasad, 1972). High humidity during flowering and fruit setting, the most vulnerable stage of the growth of the crop, is conducive for the development of Alternaria blight in cumin (Uppal et al., 1938, Patel et al., 1957, Gemawat and Prasad, 1971). Correct species identification is an imperative for designing successful management approaches against any plant pathogen. Different species and isolates of Alternaria vary in radial mycelial growth, conidia structure, colony character, sporulation and pathogenic capability (Ansari et al., 1989, Patni et al., 2005, Kaur et al., 2007). For delimiting the species of fungal pathogens and to study their diversity, molecular tools are being used increasingly including other morphological characters (Benali et al., 2011). To the best of our knowledge, there is no extensive study conducted so far in Bangladesh about the prevalence of the Alternaria blight of cumin and its causal organisms.
The present study was undertaken 1) to know the prevalence of the Alternaria blight of cumin in Bangladesh 2) to study the feasibility of introducing some advanced lines of cumin, better adapted against the Alternaria blight, in Bangladesh 3) to identify and characterize species of fungus associated with the Alternaria blight of cumin in Bangladesh using morphological and molecular features 4) to discriminate the pathogenicity of the isolates of fungus associated with the Alternaria blight of cumin collected from different locations of Bangladesh.
2. Materials and methods
Assessment of germination and yield parameters
The experiment was conducted at five sub-centers under Spices Research Centre (SRC) of Bangladesh Agricultural Research Institute situated in different Agro-ecological Zones (AEZ) in Bangladesh (Table 1, Table 2). The experiment was laid out in a randomized complete block design (RCBD) with four replications during November 2019 to March 2020. Standard procedures of cultivating cumin were followed for the land preparation and subsequent intercultural operations (Verma et al., 2018). Four advanced cumin lines viz. CN026, CN028, CN031 and CN038 were used in the experiment. One hundred seeds of each cumin line were sown in each location and percentage of germination was determined by calculating the number of plants grown. For assessing the field performance of four cumin lines, mean of yield parameters of five locations were used (Table 6).
Table 1.
The list of isolates of Alternaria spp. collected from different locations of Bangladesh with their corresponding GenBank Accession number.
| Serial No. | Location | *AEZ | Isolate Code (GenBank Accession number) |
|---|---|---|---|
| 1 | Bogura | 3 | BoCA1 (MN989187) |
| 2 | Bogura | 3 | BoCA4 (MN989188) |
| 3 | Bogura | 3 | BoCA5 (MN989189) |
| 4 | Bogura | 3 | BoCA8 (MN989190) |
| 5 | Bogura | 3 | BoCA11 (MN989191) |
| 6 | Bogura | 3 | BoCA12 (MN989192) |
| 7 | Magura | 11 | MaCA2 (MN989193) |
| 8 | Magura | 11 | MaCA3 (MN989194) |
| 9 | Magura | 11 | MaCA4 (MN989195) |
| 10 | Faridpur | 12 | FaCA3 (MN989196) |
| 11 | Lalmonirhat | 2 | LaCA1 (MN989197) |
| 12 | Lalmonirhat | 2 | LaCA3 (MN989198) |
| 13 | Bogura | 3 | BoCA6 (MN989199) |
| 14 | Bogura | 3 | BoCA3 (MN989200) |
| 15 | Bogura | 3 | BoCA2 (MN989213) |
| 16 | Bogura | 3 | BoCA9 (MN989214) |
| 17 | Bogura | 3 | BoCA10 (MN989215) |
| 18 | Bogura | 3 | BoCA13 (MN989216) |
| 19 | Gazipur | 8 | GaCA1 (MN989217) |
| 20 | Gazipur | 8 | GaCA5 (MN989218) |
| 21 | Magura | 11 | MaCA1 (MN989219) |
| 22 | Faridpur | 12 | FaCA4 (MN989220) |
| 23 | Lalmonirhat | 2 | LaCA2 (MN989221) |
Agro-ecological Zones (AEZ).
Table 2.
Weather parameters of experiment sites during November 2019 to March 2020.
| Location |
* AEZ |
Weather parameter | November, 2019 |
December, 2019 |
January, 2020 |
February, 2020 |
March, 2020 |
Average weather parameter |
|---|---|---|---|---|---|---|---|---|
| Bogura | 3 | Temperature (0c) | 25.32 | 21.34 | 19.18 | 22.11 | 25.18 | 22.63 |
| Rainfall (mm) | 0.033 | 0.00 | 0.00 | 0.138 | 0.90 | 0.214 | ||
| Relative Humidity (%) | 87.08 | 91.44 | 93.58 | 86.0 | 75.97 | 86.81 | ||
| Gazipur | 8 | Temperature (0c) | 24.6 | 19.23 | 18.24 | 22.71 | 24.92 | 21.94 |
| Rainfall (mm) | 0.70 | 0.00 | 0.94 | 0.00 | 0.06 | 0.34 | ||
| Relative Humidity (%) | 89.55 | 86.48 | 80.53 | 79.31 | 75.97 | 82.37 | ||
| Lalmonirhat | 2 | Temperature (0c) | 18.56 | 15.68 | 13.15 | 17.58 | 22.45 | 17.48 |
| Rainfall (mm) | 0.81 | 0.00 | 0.11 | 0.116 | 0.65 | 0.337 | ||
| Relative Humidity (%) | 87.60 | 88.60 | 90.12 | 82.50 | 74.60 | 84.68 | ||
| Faridpur | 12 | Temperature (0c) | 23.14 | 17.56 | 16.41 | 19.26 | 22.84 | 19.84 |
| Rainfall (mm) | 32.4 | 6.2 | 16.6 | 0.6 | 3.8 | 11.92 | ||
| Relative Humidity (%) | 86.84 | 87.18 | 88.03 | 79.62 | 77.16 | 83.77 | ||
| Magura | 11 | Temperature (0c) | 23.8 | 19.70 | 20.20 | 24.6 | 27.85 | 23.23 |
| Rainfall (mm) | 1.6 | 1.6 | 0.00 | 2.0 | 0.4 | 1.12 | ||
| Relative Humidity (%) | 85.50 | 86.20 | 87.03 | 75.32 | 74.11 | 81.63 |
Agro-ecological Zones (AEZ).
Table 6.
Yield parameters of different cumin lines.
| Cumin line | Plant height (cm) | Primary Branch/plant (no.) | Umbel/plant (No.) | Umbel let/umbel (No.) | Seeds/umbel let (No.) | 1000 seed wt. (g) | Yield (kg/ha) |
|---|---|---|---|---|---|---|---|
| CN026 | 51.30 a | 5.17 a | 95.46 a | 6.19 a | 5.75 a | 5.82 a | 592.85 a |
| CN028 | 44.53 ab | 4.83 ab | 83.88 ab | 5.05 abc | 5.03 ab | 5.10 ab | 450.60 abc |
| CN031 | 42.75 bc | 68 abc | 79.97 a-d | 5.23b | 4.67 ab | 4.83 abc | 279.34 cde |
| CN038 | 38.98b-e | 4.50 a-d | 81.44 abc | 4.50 bc | 4.59 ab | 4.33 bc | 485.31 ab |
| L.S. | ** | ** | ** | ** | ** | ** | ** |
| CV (%) | 6.48 | 7.42 | 13.22 | 9.41 | 8.45 | 9.67 | 24.42 |
1% level of probability; Means followed by the same letter in a column did not differ significantly.
Incidence and Severity of the disease
The incidence and severity of Alternaria blight were recorded for three times starting from the fifty five days after emergence (DAE) of cumin seedlings at the interval of 10 days. For determining disease severity, ten plants were randomly selected from each plot of all locations and percent disease index (PDI) was calculated.
No. of infected plants per unit area × 100
% Disease incidence (Chester, 1959, Wheeler, 1969) = ----------------------------------------------
Total no. of plants per unit area
Sum of all disease rating × 100
PDI (Chester, 1959, Wheeler, 1969) = ----------------------------------------------------------------
Total no. of plants assessed × Maximum possible rating
The disease intensity was calculated with the help of disease rating scale (0–5) where, (0 = Free from disease, 1 = 1–10% area of leaf & umbel blighted, 2 = 11–20% area of leaf, stem & umbel blighted, 3 = 21–35% area of leaf, stem & umbel blighted, 4 = 36–60% area of leaf, stem & umbel blighted and 5 = More than 60% area of leaf, stem & umbel blighted (Jat, 2015).
Isolation, purification and identification of the pathogen
A total of twenty three samples of Alternaria blight of cumin were collected from five locations (Table 1). The diseased parts were cut into small pieces of 3 mm diameter with some healthy part. The cut pieces were washed in tap water and surface sterilized with 10% NaOCl for 30 sec and blotted to dry on sterilized blotting paper. After drying, three pieces were aseptically placed into petri dishes containing Potato Dextrose Agar (PDA) medium. The PDA with small pieces of infected plant parts were incubated at a temperature of 26 ± 1 °C for 7 days (ISTA, 1976). Fungal pathogens were identified on the basis of their morphological, cultural and pathogenic characteristics (Rifai, 1969, Aneja, 2004).
Cultural variability
Purified culture of each isolate was prepared following the hyphae tip culture method (Tutte, 1969). The growth patterns of twenty three isolates of the pathogen were recorded on PDA medium by inoculating with 5 mm discs cut from PDA culture of the isolates. The discs were placed at the center of the Petri plates containing PDA. Three plates of each isolate were incubated at 26 ± 1 °C and inoculated. The growth rate, colony characters, growth habit and sporulation were recorded every 4 days after incubation and continued to 11 days after incubation. Observations on variation in conidial dimension were recorded with the help of a compound microscope.
Measurement of fungal sporulation
To determine the conidial concentration of each isolate, 5 ml of double distilled water (DDW) and 2.5 ml of ethanol was added in each petri plate containing 14 days old pure culture. The surface of the fungal growth on PDA was brushed gently with a toothbrush to disperse the spores. Three ml of suspension and 22 ml of DDW were taken in a test tube and mixed well by shaking. The final suspension (30 µl) was placed in a haemocytometer and the numbers of conidia/ml were counted under a compound microscope and number of conidia/ml was estimated (Nur-E-Nasreen et al., 2017). The procedure was repeated for thrice and a mean number of conidia/ml was estimated.
Extraction of genomic DNA
Fungal DNA was extracted following the modified CTAB method (Doyle and Doyle, 1987). The dried mycelia were ground into powder in liquid nitrogen. The homogenized mycelia were transferred to an eppendorf and CTAB extraction buffer was added. The sample was incubated at 65 °C for 30 min in a hot water bath and centrifuged at 10000 rpm for 10 min. Equal volume of Phenol:Chloroform:Iso-amylalcohol (25:24:1) were mixed and centrifuged at 10000 rpm for 10 min. The supernatant and equal volume of ice-cold isopropanol was added. The sample was incubated at −20 °C for 1.30 h and followed by centrifugation at 13000 rpm for 15 min. The DNA pellet left at the bottom was washed with 70% ethanol, air-dried and dissolved in TE buffer. The DNA stock solution was stored at −20 °C.
PCR amplification
The internal transcribed spacer (ITS) region of the ribosomal DNA (rDNA) of 23 isolates were amplified using primer set ITS1 (5ˊ-TCCGTAGGTGAACCTGCGG-3ˊ) and ITS4 (5ˊ-TCCTCCGCTTATTGATATGC-3ˊ) (White et al., 1990). The amplification reaction for the simplex PCR consisted of a volume of 25 μl PCR mix that was made up by adding 12.5 μl ready to use master mix (Promega, Madison, WI, USA), 9.5 μl nuclease-free water, 1 μl of each 10 μM forward and reverse primer, and 1 μl of the respective isolate’s DNA. PCR cycle for ITS sequence amplification consisted of initial denaturation (94 °C for 5 min) followed by 35 cycles of denaturation (94 °C for 30 sec), annealing (56 °C for 30 sec), extension (72 °C for 1.0 min 30 sec) and a final extension of 10 min at 72 °C.
Gel electrophoresis
After DNA amplification, a 10 μl of PCR product from each sample was loaded into 1% agarose gel and stained with Ethidium Bromide (0.5 mg/ml). Electrophoresis was conducted in the Tris-Borate-EDTA (TBE) buffer at 80 V for 45 min. DNA bands were visualized and photographed under UV light by GelView Master (Dynamica Scientific Ltd.). The length of each amplified DNA fragment was compared with a 1 kb DNA ladder (Promega, Madison, WI, USA).
Sequencing
The amplified fragments were sent to the National Institute of Biotechnology (NIB), Dhaka, Bangladesh and were sequenced bi-directionally. The nucleotide sequences were submitted to the National Center for Biotechnology Information (NCBI) GenBank and their accession were obtained (Table1). Primers used for the sequencing reactions were the same as those used in the amplification step.
Phylogenetic analyses
The sequences of Alternaria spp. used in the phylogenetic analyses were downloaded from the NCBI Genbank based on the highest match of Nucleotide Basic Local Alignment Search Tool (BLASTN) results against the isolates of Alternaria spp. identified in this study. Evolutionary analyses were conducted in MEGA7 (Kumar et al., 2016). The evolutionary history was inferred using the Neighbor-Joining method (Saitou and Nei, 1987). The evolutionary distances were computed using the Maximum Composite Likelihood method (Tamura et al., 2004) and are in the units of the number of base substitutions per site. All positions containing gaps and missing data were eliminated. A bootstrap analysis using 1000 replications of the sequence data was carried out (Felsenstein, 1985).
Pathogenic variability
Pathogenicity of all the isolates of Alternaria spp. was tested on the cumin line CN026 grown on sterilized soil in pots by the spray inoculation technique (Dhingra and Sinclair, 1995). This line was selected for the experiment because it showed better adaptability in the climatic conditions of Bangladesh in terms of germination capacity, yield, incidence and severity of Alternaria blight of cumin. Each pot received 10–15 seeds of the cumin line (Shekhawat et al., 2013). The concentration of the spores was maintained at 1 × 103 ml−1 during inoculation. Forty to fifty days old plants were inoculated by spraying the conidial suspension with a hand-held atomizer. The inoculated plants were kept in a humid chamber for 24 h before transferring to a cage house. High humidity was maintained throughout the disease development period by frequent irrigations. Experiments were conducted in Completely Randomized Design (CRD). Each treatment was replicated three times. The disease intensity was calculated with the help of a disease rating scale (***Jat, 2013). Re-isolation was done by collecting the infected plant parts after 10 days of inoculation. The isolated cultures were compared with the original one to confirm the pathogenicity.
2.1. Statistical analysis
Statistical analyses were done by R statistical software (http://www.R-project.org). ANOVA was performed on germination, yield and yield contributing characters of cumin lines, disease incidence and disease severity, radial mycelia growth, length and breadth of conidia, beak length, and number of vertical and horizontal septa by the Agricolae R package and AOV function (R Core Team, 2020).
3. Results
Germination percentage and yield parameters of cumin lines: One of the objectives of this study was to introduce some advanced lines of cumin in Bangladesh. To achieve this objective, germination capacity and different yield parameters of four cumin lines were assessed. The overall germination percentage of four lines varied significantly at different locations (Table 3). The highest (80.12%) germination was recorded at SRC, Bogura and the lowest (64.06%) was at SRC, Lalmanirhat. Significant variation in the germination percentage was observed in the four cumin lines (Table 4). The cumin lines CN026 and CN038 had significantly higher germination than the other two lines. The interaction effect of the cumin lines and locations on the germination percentage was significant (Table 5). All of the four cumin lines had higher germination at SRC, Bogura. The highest germination was recorded in the CN026 (85.00%) and CN038 (82.75%) at SRC, Bogura. There was a significant difference among the yield parameters of the cumin lines in all of the locations (Table 6). All of the yields contributing characters were higher for the line CN026. The highest seed (592.85 kg/ha) yield was recorded from the line CN026 and the lowest (279.34 kg/ha) was obtained in the CN031.
Table 3.
The germination (%) of cumin seeds in different locations.
| Location | Germination (%) |
|---|---|
| SRC, Bogura | 80.12 a |
| SRC, Gazipur | 75.06b |
| SRC, Lalmanirhat | 64.06 e |
| SRC, Magura | 72.81c |
| SRC, Faridpur | 69.87 d |
| Level of significance | ** |
| CV (%) | 4.088 |
Spice Research Centre (SRC); **1% level of probability; Means followed by the same letter in a column did not differ significantly.
Table 4.
The germination (%) of different cumin lines.
| Cumin line | Germination (%) |
|---|---|
| CN026 | 75.20 a |
| CN028 | 71.10b |
| CN031 | 69.30b |
| CN038 | 73.95 a |
| L.S. | ** |
| CV (%) | 4.088 |
1% level of probability; Means followed by the same letter in a column did not differ significantly.
Table 5.
The germination (%) of different cumin lines in different locations.
| Location | Cumin line | Germination (%) |
|---|---|---|
| SRC, Bogura | CN026 | 85.00 a |
| CN028 | 77.50 bc | |
| CN031 | 75.25b-e | |
| CN038 | 82.75 a | |
| SRC, Gazipur | CN026 | 78.00b |
| CN028 | 74.75b-e | |
| CN031 | 73.25 d-f | |
| CN038 | 74.25b-e | |
| SRC, Lalmanirhat | CN026 | 65.25 h-j |
| CN028 | 64.75 ij | |
| CN031 | 63.25 j | |
| CN038 | 63 j | |
| SRC, Magura | CN026 | 76.25b-d |
| CN028 | 70 fg | |
| CN031 | 69 gh | |
| CN038 | 76.00b-d | |
| SRC, Faridpur | CN026 | 71.50 e-g |
| CN028 | 68.50 g-i | |
| CN031 | 65.75 h-j | |
| CN038 | 73.75c-f | |
| L.S. | ** | |
| CV (%) | 4.089 | |
Spice Research Centre (SRC); **1% level of probability. Means followed by the same letter in a column did not differ significantly.
Incidence and severity of Alternaria blight disease: The incidence and severity of the Alternaria blight was observed at five locations on four cumin lines at different DAE of seedlings. Significant variation in the incidence and severity of the disease was observed at five locations at different DAE (Table 7). Both incidence and severity of the disease increased with the progress of time during the experiment period at all locations for all cumin lines and reached the maximum of 98% and 88%, respectively, at 75 DAE. At all intervals of time the highest incidence of the disease was found at SRC, Faridpur whereas the lowest was recorded at SRC, Bogura. However, at all locations except SRC, Bogura, the incidence of the disease was statistically similar at 75 DAE. At 55 DAE, the highest severity of the disease was seen at Faridpur (55%), but at 65 and 75 DAE, Lalmonirhat had the highest severity of 79% and 88%, respectively. The lowest severity of the disease was observed at Bogura at all intervals of time. The incidence and severity of Alternaria blight varied significantly among the four cumin lines at 55 and 65 DAE (Table 8). The incidence and severity of the disease was the highest for the line CN031 and the lowest for the CN026 at all DAE, however, at 75 DAE the difference was non-significant. The interaction effect of locations and cumin lines on the incidence of the disease was found to be significant at 55 DAE and 65 DAE, while it was non-significant at 75 DAE (Table 9). On the other hand, interaction effect on the severity of the disease was significant at all DAE. The cumin line CN026 had the lowest incidence and severity of the disease at Bogura at all intervals of time. The data obtained in this experiment suggested that the cumin line CN026 was a better candidate to be adapted in Bangladesh as it had shown higher germination capacity and better yield parameters with lower incidence and severity of being affected by the Alternaria blight.
Table 7.
The incidence and severity of Alternaria blight of cumin in different locations.
| Location | Disease incidence (%) |
Disease severity (%) |
||||
|---|---|---|---|---|---|---|
| 55 DAE | 65 DAE | 75 DAE | 55 DAE | 65 DAE | 75 DAE | |
| SRC, Bogura | 30.14 e | 75.73c | 91.69b | 48.96c | 69.71c | 79.90b |
| SRC, Gazipur | 44.56b | 77.93 bc | 94.20 ab | 52.81b | 77.12 a | 85.80 a |
| SRC, Lalmanirhat | 43.22c | 77.82 bc | 97.73 a | 55.31 a | 78.74 a | 87.97 a |
| SRC, Magura | 38.05 d | 79.34b | 97.71 a | 53.33 ab | 73.27b | 85.41 a |
| SRC, Faridpur | 46.03 a | 83.89 a | 97.77 a | 55.01 ab | 69.86c | 82.28b |
| L.S. | ** | ** | * | ** | ** | ** |
| CV (%) | 4.025 | 4.155 | 6.297 | 6.653 | 4.675 | 5.039 |
Spice Research Centre (SRC); Days after Emergence (DAE) **1% level of probability; *5% level of probability; Means followed by the same letter in a column did not differ significantly.
Table 8.
The incidence and severity of Alternaria blight of cumin in different lines.
| Cumin line | Disease incidence (%) |
Disease severity (%) |
||||
|---|---|---|---|---|---|---|
| 55 DAE | 65 DAE | 75 DAE | 55 DAE | 65 DAE | 75 DAE | |
| CN026 | 39.52b | 76.09c | 95.01 | 47.96c | 70.39b | 82.93 |
| CN028 | 40.18 ab | 80.04 ab | 95.96 | 55.04 ab | 74.99 a | 85.33 |
| CN031 | 41.05 a | 81.56 a | 96.69 | 56.01 a | 75.47 a | 84.82 |
| CN038 | 40.85 a | 78.09 bc | 95.62 | 53.36b | 74.11 a | 84.02 |
| L.S. | * | ** | NS | ** | ** | NS |
| CV (%) | 4.0246 | 4.155 | 6.297 | 6.653 | 4.675 | 5.039 |
Days after Emergence (DAE); **1% level of probability; *5% level of probability; Non Significant (NS); Means followed by the same letter in a column did not differ significantly.
Table 9.
The incidence and severity of Alternaria blight of cumin in different lines in different locations.
| Location Cumin line | Disease severity (%) |
Disease severity (%) |
|||||
|---|---|---|---|---|---|---|---|
| 55 DAE | 65 DAE | 75 DAE | 55 DAE | 65 DAE | 75 DAE | ||
| SRC, Bogura | CN026 | 25.58 i | 71.21 h | 91.22 | 40.37 h | 62.47 h | 74.37e |
| CN028 | 31.32 h | 79.26 d-f | 92.12 | 51.47 d-g | 71.37 d-g | 84.37 ab | |
| CN031 | 32.16 h | 83.61 a-d | 92.60 | 53.82b-f | 76.85 a-c | 77.75 de | |
| CN038 | 31.47 h | 77.66 e-g | 90.85 | 50.17 e-g | 68.75 fg | 83.10 a-d | |
| SRC, Gazipur | CN026 | 44.75 cd | 70.98 h | 94.07 | 48.12 g | 73.87b-e | 82.72 a-d |
| CN028 | 44.98 cd | 78.26 ef | 94.40 | 52.62c-g | 76.62 a-c | 85.85 ab | |
| CN031 | 44.34 cd | 80.08b-f | 97.10 | 53.72b-f | 78.25 ab | 88.25 a | |
| CN038 | 44.16 cd | 73.61 gh | 91.22 | 56.75 a-c | 79.75 a | 86.38 ab | |
| SRC, Lalmonirhat | CN026 | 43.15 d | 76.64 fg | 94.07 | 50.00 fg | 76.85 a-c | 88.10 ab |
| CN028 | 45.63 bc | 77.29 fg | 94.40 | 55.10 a-e | 81.27 a | 87.17 ab | |
| CN031 | 43.33 cd | 80.22b-f | 97.10 | 59.52 a | 80.00 a | 88.25 a | |
| CN038 | 40.77 e | 77.12 fg | 98.12 | 56.62 a-c | 76.85 a-c | 88.37 a | |
| SRC, Magura | CN026 | 40.02 ef | 82.28 a-e | 97.35 | 51.25 d-g | 71.12 d-g | 83.87 bc |
| CN028 | 47.60 ab | 84.42 ab | 98.00 | 58.00 ab | 74.74b-d | 86.50 ab | |
| CN031 | 47.91 ab | 83.95 a-c | 97.60 | 57.25 a-c | 72.87c-f | 87.25 ab | |
| CN038 | 40.24 ef | 77.13 fg | 98.12 | 53.75b-f | 74.35b-d | 84.00 a-c | |
| SRC, Faridpur | CN026 | 38.34 fg | 79.32c-f | 96.86 | 50.07 fg | 67.62 g | 82.20b-d |
| CN028 | 37.10 g | 80.98 a-f | 97.64 | 58.00 ab | 71.35 d-g | 82.75 a-d | |
| CN031 | 36.49 g | 79.92b-f | 98.23 | 55.75 a-d | 69.37 e-g | 85.96 ab | |
| CN038 | 48.61 ab | 84.94 a | 98.12 | 49.50 fg | 70.47 d-g | 78.22c-e | |
| L.S. | ** | * | NS | * | * | * | |
| CV (%) | 4.022 | 4.155 | 6.297 | 6.653 | 4.675 | 5.039 | |
Spice Research Centre (SRC); Days after Emergence (DAE) **1% level of probability; * 5% level of probability; NS = Non significant; Means followed by the same letter in a column did not differ significantly.
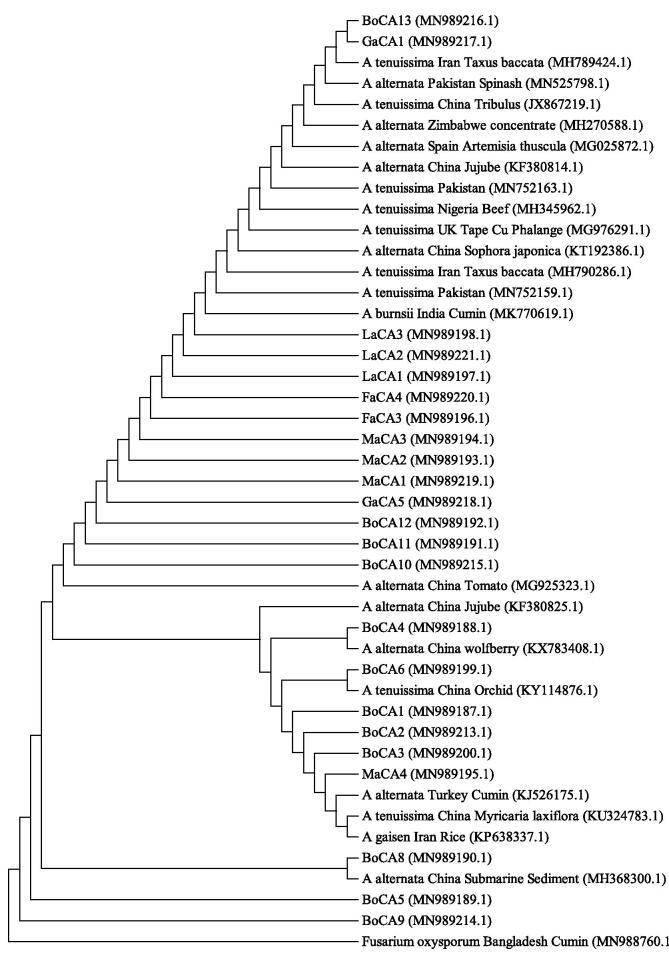
Phylogeny of Alternaria spp.: A total of twenty three fungal isolates were collected from the infected leaf, stem and twig of cumin having the symptom of the Alternaria blight (Table 1). DNA was successfully extracted from the fungal isolates using CTAB method. The preliminary fingerprinting of the twenty three isolates using the primers ITS1 and ITS4 confirmed these isolates as fungi (Fig. 1). The PCR amplification yielded DNA fragments of approximately 570 bp for all isolates that is the characteristic band size of Alternaria spp. The PCR products were sequenced bi-directionally and BLASTN search was conducted. The fungal isolates were found in match with the GenBank accessions of A. alternata, A. tenuissima, A. burnsii and A. gaisen. The phylogenetic tree constructed with the sequence data of twenty three isolates and the corresponding accessions of GenBank revealed the isolates as different species of Alternaria (Table 1 and Fig. 2). Twelve isolates out of twenty three (isolate no. 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12) had been found in homology with the GenBank accessions of A. alternata, nine (isolate no. 15, 16, 17, 18, 19, 20, 21, 22, 23) with A. tenuissima, one (isolate no. 13) with A. burnsii, one (isolate no. 14) with A. gaisen (Table 1). The findings suggested that A. alternata and A. tenuissima were the most prevalent species in causing the Alternaria blight in cumin among the sampled locations. A. alternata and A. tenuissima were detected in all locations whereas A. gaisen and A. burnsii were detected in Bogura only. The colony characters of the isolates revealed that the isolates molecularly identified as A. alternata, A. tenuissima, A. burnsii and A. gaisen had diverse up-side and reverse side color with distinct appearance, texture and margin of the colony (Table 10).
Fig. 1.
(a, b) Electrophoresis in agarose gel of polymerase chain reaction products obtained by using the primer ITS1 and ITS4. L: 1 kb ladder (Promega). Lane 1 – 25 are the PCR products of the DNA samples obtained from the different locations of Bangladesh (Table 1). A product length of 570 bp is the characteristic of fungus obtained by ITS1 and ITS4 primers.
Fig. 2.
The evolutionary relationship among the populations was inferred using the Neighbor-Joining method. The evolutionary distances were computed using the Maximum Composite Likelihood method and are in the units of the number of base substitutions per site. The sequences used in the phylogenetic analysis were chosen from the best matches in GenBank based on BLASTN against the sequences of four Alternaria species recovered from this study. Fusarium oxysporum was used as out-group. For isolate codes, please see Table 1.
Table 10.
Colony characters of different isolates of Alternaria spp. of cumin collected from different locations of Bangladesh.
| SL No. | Isolate code | Location | Colony characters |
Pathogen isolated | ||
|---|---|---|---|---|---|---|
| Appearance, texture and margin | Up side of colony | Reverse side colony | ||||
| 1 | BoCA1 | Bogura | Umbronate, velveti, White, irregular margin | Olive green | Dark black | Alternaria alternata |
| 2 | BoCA4 | Bogura | Umbronate, velveti, Whitish, irregular margin | Green | Light green | |
| 3 | BoCA5 | Bogura | Umbronate, velveti, White, regular margin | Whitish black | Light brown | |
| 4 | BoCA8 | Bogura | Umbronate, velveti, Olive white, regular margin | Olive white | Brownish white or light brown | |
| 5 | BoCA11 | Bogura | Whitish, regular margin | Olive green | Dark black | |
| 6 | BoCA12 | Bogura | Umbronate, cottony and whitish regular margin | Green | Light black | |
| 7 | MaCA2 | Magura | Umbronate, velveti, Blackish white, irregular margin | Whitish olive green | Brown | |
| 8 | MaCA3 | Magura | Umbronate, velveti,White, regular margin | Greenish white | Black | |
| 9 | MaCA4 | Magura | Umbronate, velveti, White, regular margin | Greenish white | Dark black | |
| 10 | FaCA3 | Faridpur | Rugase, velveti,Whitish olive, irregular margin | Greenish white | Blackish white | |
| 11 | LaCA1 | Lalmonirhat | Umbronate, velveti, Whitish, regular margin | Blackish white | Light black | |
| 12 | LaCA3 | Lalmonirhat | Greenish white, irregular margin | Greenish ash | Blackish white | |
| 13 | BoCA6 | Bogura | Umbronate, cottony, Olive white, regular margin | Olive green | Dark black | A. burnsii |
| 14 | BoCA3 | Bogura | Umbronate, velveti, White, irregular margin | Green | Black | A. gaisen |
| 15 | BoCA2 | Bogura | Brown, irregular margin | Blackish ash | Dark black | A. tenuissima |
| 16 | BoCA9 | Bogura | Velveti, White, irregular margin | Greenish white | Dark black | |
| 17 | BoCA10 | Bogura | Rugose, velveti, White, regular margin | Greenish white | Light brown | |
| 18 | BoCA13 | Bogura | Whitish, regular margin | Blackish white | Black | |
| 19 | GaCA1 | Gazipur | Umbronate, cottony, Greenish white, regular margin | Green | Dark black | |
| 20 | GaCA5 | Gazipur | Cottony, White, regular margin | Olive green | Black | |
| 21 | MaCA1 | Magura | Ash, regular margin | Greenish white | Dark black | |
| 22 | FaCA4 | Faridpur | Rugose, velveti, Greenish white, irregular margin | Dark green | Black | |
| 23 | LaCA2 | Lalmonirhat | Umbronate, velveti, Blackish white, regular margin | Dark black | Dark black | |
Morphological variation of Alternaria spp.: The isolates significantly varied in radial mycelial growth (Table 11). The radial mycelial growths of the isolates were measured from four to eleven days after inoculation (DAI) on PDA media. Maximum growth (39.03 mm) was recorded in the MaCA3 which was followed by the BoCA11, MaCA2 and LaCA3 at eleven DAI and the minimum (33.02 mm) was observed in the FaCA3. In addition to these, color, size and shape of the conidia with their spore production capacity were found varied significantly among the stains (Table 12). When cumin plants were inoculated with the spore suspensions of the Alternaria isolates, infection started as minute necrotic lesions on all of the above ground plant parts which turned purple at advanced stages and later turned brown to black in 36 to 72 h resulting into blighting of the affected plant. These typical blight symptoms were found at 6 to 7 DAI. The symptoms were not observed in control plants treated with water. The pathogens were re-isolated from the infected plant parts at 10 days after inoculation. The isolated cultures were compared with the original one to confirm the pathogenicity. The pathogenic variation among the isolates was examined by inoculating the line CN026. There were significant differences in the incidence and severity of the disease among the isolates of the four Alternaria species (Table 13). The MaCA2 of A. alternata was observed as the most pathogenic isolate.
Table 11.
Radial mycelial growth of different isolates of Alternaria spp. on culture medium collected from different locations of Bangladesh.
| Isolate code | Radial mycelial growth (mm) at different days after inoculation (DAI) |
|||||||
|---|---|---|---|---|---|---|---|---|
| 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | |
| BoCA1 | 19.33 hi | 22.12f-i | 25.33 e-i | 29.25 d-h | 30.58f-i | 32.67 hi | 34.83 e-h | 37.17 d-i |
| BoCA4 | 20.92 a-e | 24.25 ab | 25.45 d-h | 28.56f-i | 29.80 i | 31.42 jk | 35.92 bcd | 37.83b-f |
| BoCA5 | 20.80b-e | 22.50 e-h | 25.67c-g | 29.42 d-g | 31.67 de | 34.08c-f | 35.77b-e | 37.08 d-i |
| BoCA8 | 19.47 ghi | 22.67 d-g | 25.25 e-i | 29.50c-f | 31.58 def | 32.75 hi | 34.67f-i | 36.42 hij |
| BoCA11 | 20.60c-f | 23.25b-e | 27.05b | 30.79 ab | 32.92 abc | 34.17c-f | 35.56c-f | 38.80 abc |
| BoCA12 | 21.60 ab | 23.72 bcd | 25.21 e-j | 29.25 d-h | 32.44 bcd | 34.87 bc | 36.63b | 37.63c-g |
| MaCA2 | 21.42 abc | 23.25b-e | 26.08b-f | 29.92b-e | 32.33 bcd | 33.75 efg | 35.42c-g | 38.07 a-e |
| MaCA3 | 20.27 d-h | 23.42b-e | 26.58 bc | 29.55c-f | 33.17 ab | 36.00 a | 37.76 a | 39.03 a |
| MaCA4 | 20.75b-f | 23.42b-e | 26.33 bcd | 30.08 bcd | 32.08 cde | 33.50 fgh | 35.50c-f | 36.92 e-j |
| FaCA3 | 20.62c-f | 22.83 d-g | 26.13b-f | 27.00 L | 28.23 j | 29.20 L | 30.83 k | 33.02 k |
| LaCA1 | 21.33 abc | 23.50b-e | 26.63 bc | 30.47 bc | 32.13b-e | 34.67b-e | 36.33 bc | 37.50 d-h |
| LaCA3 | 21.83a | 23.33b-e | 25.17f-j | 28.47 g-j | 30.49 ghi | 33.90 def | 36.03 bc | 38.07 a-e |
| BoCA6 | 21.17 a-d | 25.00 a | 28.17 a | 31.67 a | 33.88 a | 35.60 ab | 37.67 a | 39.25 a |
| BoCA3 | 20. 42c-g | 21.97 ghi | 24.46 ijk | 29.02 e-i | 33.00 bc | 34.17c-f | 35.93 bcd | 37.25 d-i |
| BoCA2 | 19.50 ghi | 21.45 hi | 23.88 k | 27.13 kl | 31.25 efg | 32.92 ghi | 35.83 bcd | 38.92 ab |
| BoCA9 | 20.84 a-e | 23.25b-e | 26.08b-f | 29.67 cde | 31.17 e-h | 32.33 ij | 33.75 ij | 36.13 ij |
| BoCA10 | 20.08 e-h | 21.92 ghi | 24.67 h-k | 28.05 ijk | 30.52 ghi | 32.58 hi | 35.67b-e | 38.17 a-d |
| BoCA13 | 20.42c-g | 23.08c-f | 25.08 g-j | 29.08 d-h | 31.83 de | 34.08c-f | 35.01 d-h | 37.57 d-h |
| GaCA1 | 20.85 a-e | 24.08 abc | 26.92b | 30.48 bc | 33.67 a | 34.77 bcd | 36.13 bc | 36.92 e-j |
| GaCA5 | 19.92 e-h | 22.83 d-g | 25.17f-j | 27.48 jkl | 29.72 i | 30.57 k | 33.00 j | 35.87 j |
| MaCA1 | 19.75f-i | 22.58 efg | 25.00 g-j | 28.33 hij | 30.52 ghi | 32.17 ij | 34.33 hi | 36.08 ij |
| FaCA4 | 18.83 i | 21.17 i | 24.25 jk | 27.50 jkl | 30.18 hi | 33.35 fgh | 35.02 d-h | 36.58 g-j |
| LaCA2 | 20.25 d-h | 23.33b-e | 26.12b-e | 28.53f-i | 31.10 e-h | 32.77 hi | 34.51 ghi | 36.83f-j |
| L.S. | ** | ** | ** | ** | ** | ** | ** | ** |
| CV (%) | 3.05 | 2.79 | 2.30 | 2.13 | 2.02 | 1.74 | 1.67 | 1.94 |
**1% level of probability; Means followed by the same letter in a column did not differ significantly.
Table 12.
Morpho-physiological characters of conidia of different isolates of Alternaria spp. of cumin collected from different locations of Bangladesh.
| Isolate code | Characteristics of conidia |
Mean length of conidia (µm) | Mean breadth of conidia (µm) | Mean beak length of conidia (µm) | Number of vertical septa | Number of horizontal septa | Number of spores/ml (x106) | |
|---|---|---|---|---|---|---|---|---|
| Shape | Color | |||||||
| BoCA1 | Clavate to obclavate | Brown | 52.93 ab | 20.37 efg | 7.75 hijk | 0.94 e-i | 1.68 j | 2.38 e |
| BoCA4 | Obclavate | Light brown | 46.64 ef | 22.80 a-d | 8.24 ghi | 0.77 hi | 2.36 d-h | 3.62 a |
| BoCA5 | Obclavate to ovoid | Brown | 46.18 ef | 20.34 efg | 6.98 k | 0.78 hi | 2.44c-g | 2.26 ef |
| BoCA8 | Clavate | Dark brown | 56.09 a | 24.19 a | 8.39 fgh | 1.18c-h | 2.95 ab | 1.51 g |
| BoCA11 | Obclavate | Light brown | 44.90 fg | 17.13 ij | 9.86 bcd | 0.60 ij | 2.40c-h | 2.83 d |
| BoCA12 | Obclavate | Brown | 42.10 ghi | 19.45 fgh | 8.32 fghi | 0.88f-i | 2.48b-g | 1.39 gh |
| MaCA2 | Obclavate | Light brown | 50.87 bcd | 23.32 ab | 9.85 bcd | 1.20c-h | 2.30f-i | 3.36 ab |
| MaCA3 | Clavate to obclavate | Dark brown | 38.90 ijk | 19.97 efg | 10.12 abc | 1.03 d-i | 1.83 ij | 1.58 g |
| MaCA4 | Clavate | Dark brown | 44.51 fgh | 21.50 cde | 7.04 jk | 1.50 abc | 2.33 e-h | 0.953 ij |
| FaCA3 | Obclavate | Dark brown | 47.46 def | 19.32 fgh | 10.59 ab | 1.30c-f | 2.80 a-e | 1.33 gh |
| LaCA1 | Obclavate | Dark brown | 41.10 hij | 21.04 def | 9.61 cde | 1.93 a | 2.70 a-f | 2.46 e |
| LaCA3 | Clavate to Obclavate | Brown | 44.11 fgh | 19.86 e-h | 9.24 cdef | 1.60 abc | 3.10 a | 2.31 ef |
| BoCA6 | obovate to obclavate | Light brown | 48.45 cde | 16.66 ij | 10.19 abc | 0.67 i | 2.88 abc | 2.04f |
| BoCA3 | Ovoid | Brown | 37.67 jk | 20.38 efg | 7.41 ijk | 0.59 ij | 1.79 j | 2.24 ef |
| BoCA2 | Obclavate | Light brown | 37.20 k | 21.51 cde | 8.66 e-h | 0.82 ghi | 1.93 hij | 0.86 j |
| BoCA9 | |Obclavate | Light brown | 50.42 bcd | 22.40 a-d | 10.97 a | 1.26c-g | 2.86 abc | 2.89 cd |
| BoCA10 | Obclavate to ovoid | Brown | 54.75 a | 23.18 abc | 7.99 hij | 1.36 cde | 3.02 a | 2.83 d |
| BoCA13 | Obclavate | Brown | 47.48 def | 23.57 a | 8.70 e-h | 1.91 ab | 2.74 a-f | 3.12 bc |
| GaCA1 | Obclavate | Brown | 38.01 jk | 18.11 hi | 9.25 cdef | 0.60 ij | 2.13 g-j | 1.18 hi |
| GaCA5 | Obclavate | Dark brown | 50.82 bcd | 21.62b-e | 10.19 abc | 1.47 bcd | 3.00 a | 2.82 d |
| MaCA1 | Obclavate | Light brown | 48.57 cde | 23.16 abc | 6.92 k | 1.18c-h | 2.70 a-f | 3.21b |
| FaCA4 | Obclavate | Light brown | 42.41 gh | 15.86 j | 9.02 d-g | 0.17 j | 2.47c-g | 1.38 gh |
| LaCA2 | Oblavate | Dark brown | 51.23 bc | 19.24 gh | 10.11 abc | 0.93 e-i | 2.83 a-d | 2.38 e |
| L.S. | ** | ** | ** | ** | ** | ** | ||
| CV (%) | 4.59 | 5.29 | 6.62 | 25.73 | 11.58 | 7.34 | ||
**1% level of probability; Means followed by the same letter in a column did not differ significantly.
Table 13.
Pathogenic variation of the different isolates of Alternaria spp. on the cumin line CN026.
| Isolate code | Pathogen isolated | Disease incidence (%) |
Disease severity (%) |
||
|---|---|---|---|---|---|
| 50 DAS | 55 DAS | 50 DAS | 55 DAS | ||
| BoCA1 | Alternaria alternata | 33.16 ef (30.00) | 42.12 cd (45.00) | 25.57 ef (18.67) | 25.08 fg (18) |
| BoCA4 | 17.33 g (13.33) | 19.33 e-h (16.67) | 8.29 g (3.33) | 7.55 h (2.67) | |
| BoCA5 | 34.15 ef (31.67) | 29.92 def (25.00) | 26.04 ef (19.33) | 27.02f (20.67) | |
| BoCA8 | 12.40 gh (6.67) | 12.40 fgh (6.67) | 4.67 (10.31 g) | 11.06 gh (5.33) | |
| BoCA11 | 57.98b (71.67) | 72.67 ab (86.67) | 45.19 bc (50.33) | 62.57 bc (71.33) | |
| BoCA12 | 57.98b (71.67) | 70.66 ab (83.33) | 55.85b (68.33) | 72.67 ab (86.67) | |
| MaCA2 | 71.56 a (90.00) | 89.66 a (100.00) | 68.85 a (86.67) | 85.05 a (98.00) | |
| MaCA3 | 60.07 ab (75.00) | 73.44 ab (88.33) | 51.91b (61.33) | 59.85 bcd (74.67) | |
| MaCA4 | 52.77bcd (63.33) | 63.93b (76.67) | 48.28 bc (55.33) | 67.75b (80.00) | |
| FaCA3 | 41.16def (43.33) | 74.89 ab (90.00) | 29.92 def (25.00) | 39.21 ef (40.00) | |
| LaCA1 | 6.37 gh (3.33) | 6.37 gh (3.33) | 2.93 g (0.67) | 2.93 h (0.67) | |
| LaCA3 | 47.88 bcd (55.00) | 57.78 bc (70.00) | 38.44 cd (38.67) | 46.09 de (52.00) | |
| BoCA6 | A. burnsii | 9.08 gh (6.67) | 9.08 fgh (6.67) | 7.549 g (4.67) | 4.07 h (1.33) |
| BoCA3 | A. gaisen | 29.92f (25.00) | 22.59 d-g (15.00) | 22.98f (15.33) | 28.18f (22.33) |
| BoCA2 | A. tenuissima | 12.40 gh (6.67) | 12.40 fgh (6.67) | 11.28 g (4.00) | 8.29 h (3.33) |
| BoCA9 | 32.01f (28.33) | 42.12 cd (45.00) | 27.22 def (21.00) | 26.71f (20.33) | |
| BoCA10 | 30.99f (26.67) | 39.15 cde (40.00) | 22.98f (15.33) | 30.19f (25.33) | |
| BoCA13 | 56.99 bc (70.00) | 59.21 bc7 (3.33) | 52.24b (62.33) | 70.57b (87.33) | |
| GaCA1 | 48.84 bcd (56.67) | 72.67 ab (86.67) | 55.98b (68.33) | 53.14 de (64.00) | |
| GaCA5 | 45.00 cde (50.00) | 64.81b (73.33) | 48.16 bc (55.33) | 53.17 cde (64.00) | |
| MaCA1 | 51.14 bcd (60.00) | 63.93b (80.00) | 36.79cde (36.00) | 52.35 cde (62.67) | |
| FaCA4 | 42.12 def (45.00) | 59.89 bc (66.67) | 31.72 def (29.33) | 29.50f (24.33) | |
| LaCA2 | 35.01 ef (33.33) | 42.12 cd (45.00) | 25.63 ef (20.00) | 30.34f (25.67) | |
| Control | 0.3375 h (0.00) | 0.3375 h (0.00) | 0.3375 g (0.00) | 0.3375 h (0.00) | |
| L.S. | ** | ** | ** | ** | |
| C.V | 20.43 | 28.80 | 22.63 | 23.37 | |
1% level of probability; Means followed by the same letter in a column did not differ significantly; Arcsine transformed values are followed by raw data within parenthesis DAS = Days after sowing.
Discussions
Cumin is an important spice widely used in Bangladeshi cuisine. Its cultivation in Bangladesh is limited due to many socio-economic factors including the lack of high yielding and disease resistant variety. Alternaria blight is one of the major diseases of cumin causing huge economic loss in its production worldwide. In the present study, field trials were conducted with four advanced lines of cumin viz. CN026, CN028, CN031 and CN038 in five AEZ of Bangladesh to know their performances against the incidence and severity of the Alternaria blight. Further attempts were made to identify the causal organism of the Alternaria blight of cumin in Bangladesh and to study their diversity based on the morphological and molecular characteristics.
Among the five locations, all the cumin lines had the highest germination at Bogura and the lowest at Lalmonirhat. In this experiment, the incidence and severity of the Alternaria blight of four germplasms was observed ranging from 30% to 97% and 48% to 85%, respectively, at different locations. The incidence and severity of the disease was the least in Bogura and the highest in Faridpur. The findings of the present investigation are in agreement with Özer and Bayraktar (2015). They reported that the severity of the blight caused by A. burnsii and A. alternata was ranging from 90% to 98%. Singh et al. (2015) also observed the disease severity of 62% to 68% and 62% to 63%, by inoculating A. burnsii and A. alternata, respectively. Climatic and edaphic factors might have an impact on the production of cumin and the prevalence of the Alternaria blight. Gemawat and Prasad (1971) has reported that high humidity (90% and above) for about 3 days, temperature between 23 and 28 °C, rainfall, hours of sunlight and wind speed played important roles in the development of the cumin blight caused by A. burnsii. Sharma and Pandey (2013) has also noted that the maximum temperature between 29 and 35.5 °C and the minimum temperature between 9.6 and 19.7 °C, average relative humidity of more than 60% in the morning and 28% in the afternoon, wind speed of 2.1–4.8 km/hour and bright hours of sunshine were favorable for the development of the Alternaria blight of cumin. Singh and Shukla (1986) observed that the infection by A. alternata is favored by the temperature of 28.7 °C to 32.2 °C and the relative humidity of 74%. Higher incidences and severity of the Alternaria blight were reported from the higher elevations also (Lenne, 1991).
As Bangladesh has the climatic factors favorable for the development of Alternaria blight, the findings of this experiment outlined the importance of identifying the causal organism, use of the selective cultivars, careful selection of the location and timeliness of undertaking control measures for the safe cultivation of cumin to avoid the disease. In this experiment, four advanced lines of cumin were tested in five different locations to observe their germination capacity, yield performance and their propensity of being infected by Alternaria blight. In all locations, the line CN026 showed the highest germination capacity and yield with the lowest incidence and severity of the Alternaria blight that proved its better adaptability in the climatic conditions of Bangladesh. The primary means for identification of Alternaria species are morphological traits that include the appearance, texture, margin, color of the upsides & reverse sides of colonies, properties of conidia and pattern of sporulation (Simmons, 2007). However, molecular methods have been suggested by researchers to complement the morphology based approaches (Kang et al., 2001). On the basis of the band size of the PCR products, DNA sequence analysis and phylogenetic study, four species of Alternaria were identified in the present work. In the order of prevalence, these were A. alternata, A. tenuissima, A. burnsii and A. gaisen. Sharma et al., (2013) found that the band size of PCR products of Alternaria spp of cauliflower and mustard was ~550 to 600 bp in India. Zheng et al. (2015) also identified the Alternaria spp. of potato in China following the similar molecular methods. The analysis of the observed morphological traits and the molecular characters indicated the diversity of Alternaria isolates of cumin in Bangladesh. Uppal et al., 1938, Shakir et al., 1995 identified A. burnsii as the pathogen for causing Alternaria blight of cumin in India and Pakistan, respectively. On the other hand, A. alternata was found to be associated with Alternaria blight of cumin in Turkey (Özer and Bayraktar, 2016). However, this is the first report of A. alternata causing Alternaria blight of cumin in Bangladesh.
In this experiment, a considerable amount of morphological, cultural and pathogenic variability were recorded among the different isolates of A. alternata, A. burnsii, A. gaisen and A. tenuissima. All Alternaria strains obtained from five locations of Bangladesh were found to have separated in different phylogenetic groups. Likewise, pathogenic variability among the isolates was also observed. There are 30 AEZ in Bangladesh based on physiographic, soil, hydrological and agro-climatic characteristics (Bangladesh Agricultural Research Council, 2005). The locations from where samples were collected are situated in different AEZ. Navas et al. (2001) reported that ecological factors or the size of the area of origin of the populations might also be involved in the production of such variation. In this experiment, A. alternata was found as the most virulent species. In an experiment, Zheng et al. (2015) had also observed that among the three species of Alternaria, A. alternata was more virulent than A. tenuissima and A. solani to cause foliar diseases of potato in China.
To the best of our knowledge, this is the first extensive study regarding the prevalence of the Alternaria blight of cumin in Bangladesh. In this work, a promising line of cumin (CN026) was selected as having the better resilience against the incidence and severity of the Alternaria blight with higher germination and yield potential. The causal organisms of the Alternaria blight were also identified using molecular and morphological tools. All of these findings have formed the foundation of further research that might help in designing successful management options against the Alternaria blight of cumin leading to its increased cultivation and production in Bangladesh.
Declaration of Competing Interest
The author declare that there is no conflict of interest.
Acknowledgments
Acknowledgements
This work was a part of PhD research of the first author financially supported by the World Bank funded National Agricultural Technology Program, Phase – ΙΙ (NATP - 2) Project in Bangladesh.
Funding
This study was funded by World Bank sponsored National Agricultural Technology Program, Phase – ΙΙ (NATP - 2) Project in Bangladesh.
Footnotes
Peer review under responsibility of King Saud University.
References
- Ansari N.A., Khan M.W., Muheet A. Effect of some factors on growth and sporulation of Alternaria brassicae causing Alternaria blight of rapeseed and mustard. Acta Botanica Indica. 1989;17:49–53. [Google Scholar]
- Aneja K.R. 4th ed. New International (P) Limited Publisher; India: 2004. Experiment in microbiology, plant pathology and biotechnology; pp. 121–128. [Google Scholar]
- Azeez, S. V.A., Champakam, B., Zachariah, T.J., 2008. Cumin. In: Parthasarathy eds.: Chemistry of Spices. CABI International, Wallingford, UK. This review article deals with botany, distribution and medicinal properties of cumin, and with the chemistry of cumin oil.
- Bangladesh Agricultural Research Council. 2005. Dhaka.
- Benali S., Mohamed B., Eddine H.J., Neema C. Advances of molecular markers application in plant pathology research. Eur. J. Sci. Res. 2011;50:110–123. [Google Scholar]
- Chester, K.S., 1959. How sick the plant? In: Plant Pathology an advanced treatise (Eds. J.G. Horshfall and A.E. Diamond). Academic Press, New York. 1, 199-242.
- Dhingra, O.B., Sinclair, J.B., 1995. Basic Plant Pathology Methods. 2nd Edition, CRC Press, Boca Raton Florida, USA.
- Doyle J.J., Doyle J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987;19:11–15. [Google Scholar]
- Felsenstein, J., 1985. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 39,783-791. [DOI] [PubMed]
- Gemawat P.D., Prasad N. Epidemiological studies on Alternaria blight of Cuminum cyminum, Indian. J. Mycol. Pl. Pathol. 1971;2:65–75. [Google Scholar]
- Gemawat P.D., Prasad N. Epidemiological studies on Alternaria blight of cuminum cyminum, Indian. J. Mycol. Pl. Path. 1972;2(1):65–75. [Google Scholar]
- ISTA., International Seed Testing Association. 1976.
- Jat, V.D., 2015. Effect of Culture Filtrate of Blight Pathogen [Alternaria alternata (Fr.) Keissler] on Coriander and its Management. M. Sc. thesis, Sri Karan Narendra Agriculture University, Jobner, India.
- Kang, J.C., Crous, P.W., Schoch, C.L., 2001. Species concepts in the Cylindrocladium floridanum and Cy. spathiphylli complexes (Hypocreaceae) based on multi-allelic sequence data, sexual compatibility and morphology. Systematic and Applied Microbiology. 24, 206–17. [DOI] [PubMed]
- Kaur, S., G., Singh , Banga ,S.S., 2007. Documenting variation in Alternaria brassicae isolates based on conidial morphology, fungicidal sensitivity and molecular profile. Proceeding of the 12th International Rapeseed Congress, Wuhan, China. 4, 87-89.
- Kumar S., Stecher, G., Tamura, K., 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets.Molecular Biology and Evolution. 33, 1870-1874. [DOI] [PMC free article] [PubMed]
- Lenne J.M. Nature Research Institute Bulletin; No: 1991. Diseases and pests of sweetpotatoes: South East Asia, the pacific and East Africa; p. 46. [Google Scholar]
- Nabhan, G.P., 2014. My library My History Books on Google Play Cumin, Camels, and Caravans: A Spice Odyssey. Univ of California Press. ISBN 978-0-520-26720-6. p. 234.
- Nasreen, N.E., Meah, M.B., Tumpa, F.H., Hossain, M.A., 2017. Effect of Media Composition on Vegetative and Reproductive Growth of Alternaria brassicicola and Bipolaris sorokiniana. Curr. Agri. Res. 5(3), 266- 278.
- Navas A., Castagnone-Sereno P., Blazquez J. Genetic structure and diversity within local population of Meloidogyne (Nematoda: Meloidogynidae) Nematology. 2001;3:243–253. [Google Scholar]
- Özer, G., Bayraktar, H., 2015. Determination of fungal pathogens associated with Cuminum cyminum in Turkey. Plant Protect. Sci. 51, 74–79.
- Patel P.N., Prasad N., Mathur R.L., Mathur B.L. Fusarium wilt of cumin. Curr. Sci. 1957;26:181–182. [Google Scholar]
- Patni C.S., Kolte S.J., Awasthi R.P. Cultural variability of Alternaria brassicae, causing Alternaria blight of mustard. Ann. Plant Physiol. 2005;19:231–242. [Google Scholar]
- R Core Team. 2020. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL:http://www.r-project.org/index.html.
- Rifai A. A revision of the genus Trichoderma. Mycological Papers. 1969;116:1–56. [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Shakir A.S., Mirza J.H., Sahi S.T., Ansar M. First report of Alternaria burnsii, the causal organism of cumin blight in Pakistan. Pakistan J. Phytopathol. 1995;7(2):219. [Google Scholar]
- Sharma, S., 2010. Studies on cumin blight incited by Alternaria burnsii (Uppal Patel and Kamat) and its management. M. Sc. thesis, Anand Agricultural University, Anand, India.
- Sharma S., Pandey R.N. Survial, epiderminolgy and management of Alternata blight of cumin in Gujarat. Bioinfolet. 2013;10(2B):639–642. [Google Scholar]
- Shekhawat N., Trivedi A., Sharma S.K., Kumar A. Cultural, morphological and pathogenic variability in Alternaria burnsii causing blight of cumin. J. Mycol. Pl. Path. 2013;43(1):80–83. [Google Scholar]
- Simmons, E.G., 2007. Alternaria. An Identification Manual. CBS Biodiversity Series No. 6. CBS Fungal Biodiversity Centre, Utrecht, the Netherlands. pp.775.
- Singh M., Shukla T.N. Epidemiology of Alternaria leaf spot and fruit rot of brinjal. Indian Phytopathol. 1986;39:119–120. [Google Scholar]
- Singh S., Trivedi A., Mathur K., Padamini R. Assessment of yield loss of cumin (Cuminum cyminum) caused by Alternaria leaf blight and pathogen recovery from infected seeds. Indian Phytopath. 2015;68(3):350–352. [Google Scholar]
- Tamura K., Nei M., Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. (USA). 2004;101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Daily Financial Express. 5 May 2020. Dhaka, Bangladesh.
- Tutte J. Burgess Publishing Company; U.S.A.: 1969. Plant pathological methods: Fungi and bacteria; p. 229. [Google Scholar]
- Uppal B.N., Patel M.K., Kamat M.K. Alter naria blight of cumin. Indian J. Agric. Sci. 1938;8:9–62. [Google Scholar]
- Verma A.K., Singh R., Choudhary S., Lal G. Cultivation of dollar earning cumin crop for higher income. Acta Scientific Agriculture. 2018;2(3):46–48. [Google Scholar]
- Weiss, E.A., 2002. Spice Crops. CABI International, Wallingford,UK, [Family wise description of spice crops belonging to cruciferae, lauraceae, leguminosae, myristicaceae, myrtaceaeumbelliferae and zingiberaceae. All aspects of cultivation processing and industrial application are discussed]. P. 299.
- Wheeler, B.E.J., 1969. An introduction of plant diseases. John Willy & Sons Ltd., London. pp. 301.
- White, T.J., Bruns, T., Lee, S., Taylor, J., 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press, San Diego. pp. 315–322.
- Zheng, H.H., Zhao, J., Wang, T.Y., Wu, X.H., 2015. Characterization of Alternaria species associated with potato foliar diseases in China. Plant Pathology. 64, 425-433.