Abstract
Paramphistomosis is the most prevalent disease of domestic ruminants, causing heavy economic loss in many countries across the world. The morphological identification of these parasites is difficult, therefore molecular characterization is used to discriminate Paramphistomum species. The present study was conducted to identify Paramphistomum sp. at Mardan District, Khyber Pakhtunkhwa (KPK), Pakistan. All samples of these rumen flukes were collected from buffalo. The gDNA was isolated from the adult parasites and the ITS1 region was amplified for the sequence analysis. All flukes had 100% similarity and there was no intraspecific variation. The Blast results showed that all flukes were P. cervi as they form a single cluster with P. cervi reported from China. The results of the ITS1 sequences of the present study with reference sequencing from China showed eight specific SNPs. This was the first study in which P. cervi was genetically characterized through the ITS1 region of rDNA at District Mardan, Pakistan. It can also be used as a marker for the genetic identification of Paramphistomum species.
Keywords: Paramphistomosis, Genetic analysis, P. cervi, ITS1
1. Introduction
Paramphistomosis is the most important endo-parasitic diseases of ruminants that cause great economical loss to the livestock (Sivajothi et al., 2014). It has a wide geographical distribution in tropical and sub-tropical areas (Rolfe et al., 1991). This disease is caused by Paramphistomum species and also known as the ‘rumen flukes’ or amphistomes’ (O’Toole et al., 2014). Adult Paramphistome inhabit the rumen of host and immature flukes are present in the upper part of small intestine which can cause serious morbidity and even death. Death rates can be very high i.e., 80–90% due to immature Paramphistomes in domesticated cattles (Juyal et al., 2003, Ilha et al., 2005).
In spite of the morbidity and economic loss of Paramphistomiasis, there has been an important controversy about the taxonomy of Paramphistomum species (O’Toole et al., 2014). Most information does not identify the species of Paramphistomes which are responsible for the disease. Most of the studies were mainly focused on morphology and histology of Paramphistomum (Wang et al., 2006, Chaoudhary et al., 2015).
Genetic studies on this parasite is needed to characterize different species from endemic areas as genetic approaches provide more reliable results. There are some molecular-based studies which has revealed that the sequences of internal transcribed spacers (ITS1 and ITS2) region of ribosomal DNA (rDNA) provide valuable genetic markers for species identification of trematodes (Gasser and Chilton, 1995, Huang et al., 2004, Mufti et al., 2014).
This parasitic disease was also recorded from different areas of Pakistan (Khan et al., 2008; Kanwal et al., 2014, Kattak, 2017). But no attention has been given to genetically characterize Paramphistomum spp. in past. It is needed to be addressed as very few studies are reported recently and still controversial about the presence of P. epilictum or P. cervi or both (Ali et al., 2018; Khan et al., 2020; Rajput et al., 2020).
The present research aimed to study ITS1 region of rDNA of as genetic marker for the genetic characterization of Paramphistomum sp., reconstruction of its phylogenetic relationship and compare with already reported results from Pakistan and other regions.
2. Materials and methods
2.1. Study area
Study was conducted in the Mardan district of Khyber Pakhtunkhwa province of Pakistan. Coordinates of Mardan are: 34.05° to 34.32 N latitudes, 71.48 to 72.047° E longitudes.
2.2. Fluke collection and genomic DNA extraction
Adult flukes were collected from buffaloes slaughtered in the abattoirs of the Mardan district. The flukes were washed with normal saline solution and preserved in 70% alcohol at −20 °C. The Genomic DNA (gDNA) was extracted according to the standard Chloroform Phenol method (Sambrook et al., 1989) from 54 adult flukes randomly selected from 18 infected rumens. Extracted DNA was quantified by spectrophotometry at 260/280 nm. The DNA were stored at − 20 °C for genetic analysis.
2.3. PCR amplification and sequencing of ITS1 region of the rDNA
The primers set were designed by Primer 3 software for the first time for ITS1 region. Following BF (5′-GTGGTCTAAGCCTTTGGCTAGT-3′) as forward and BR (5′- CAGGCATGGCCCAAATAGGT-3′) as reverse primer were used to amplify the ITS1 region.
The 25 μl PCR reactions included 2 μl DNA template, 1 μl of each forward and reverse primer (10 pmol/μl), 12 μl master-mix containing Taq polymerase and MgCl2 (Advance Bioscience) and 9 μl of distilled H2O. The PCR reactions were performed under the following conditions: 95 °C for 5 min (initial denaturation), followed by 30 cycles of 94 °C for 30 sec, 60 °C for 1 min, 72 °C for 45 sec, and a final extension of 72 °C for 6 min.
2.4. Sequence alignments and phylogenetic comparison
All positive PCR products with very good bands were sequenced using the same primers set through AB 3730X1 DNA sequencing system. NCBI websites and MEGA-X software were utilized to identify species and pairwise alignments of sequences. The rDNA ITS-1 sequences of both strands from each individual fluke were assembled, aligned and edited to remove primers and poor-quality sequence on both ends using CLC sequence viewer 8.0 software. Edited sequence was submitted in GenBank for accession ID number. The phylogenetic tree was generated via the use of the maximum likelihood method with 100 Bootstrap value using MEGA-X software (Tamura et al., 2013).
3. Results
3.1. Phylogenetic characterization of Paramphistomum isolates
The analyses of ITS1 region produced 640 bp sequence, submitted in the NCBI blast search for the characterization of Paramphistomum isolates. All samples of current study showed 100% similarity in sequences indicating single haplotype and there was no intra-specific variation among isolates of current study. It was observed that all isolates showed 99% similarity with P. cervi reported from China. No separate sequence of ITS1 region was available in NCBI database; therefore, the only published complete sequence from China (Zheng et al., 2014) was used for the analysis of genetic characterization. All sequences in the alignment were trimmed to 555 bp, the length of the shortest sequence available that still contained all the informative sites. Eight variable sites showing single nucleotide polymorphism (SNPs) were detected in the current study isolates with reference sequence Accession no. KJ459934 and nine to KJ459935, KJ459936, KJ459937 and KJ459938 as shown in Table 1.
Table 1.
ITS1 region of rDNA of Paramphistomum sp. of current study (Par-pak*) showing position of SNPs, aligned with available reference sequences in GenBank.
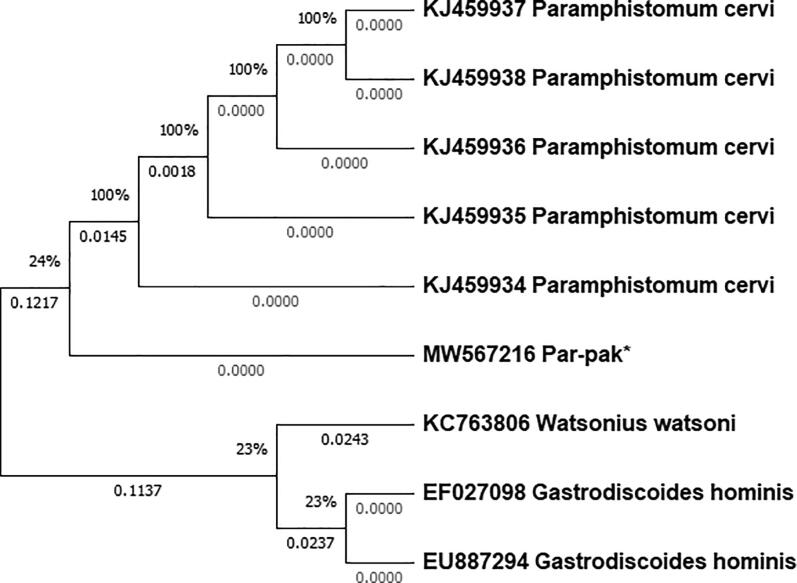
The phylogenetic analysis results showed two clades. One cluster of current study isolates with P. cervi reported from China and other with Watsonius watsoni of family Paramphistomidae and Gastrodiscoides hominis of family Gastrodiscidae (Fig. 1).
Fig. 1.
A Maximum likelihood phylogram of Paramphistoma sp. based on nucleotide sequences of the first internal transcribed spacer of ribosomal DNA (ITS-1). MW567216 Par-pak* = Paramphistomum cervi of current study published in GenBank.
4. Discussion
Morphological characteristics are generally used to identify adult flukes. However, sometimes morphological parameters are not sufficient for differentiation of species (Arbabi et al., 2012). Thus, molecular approaches are now being considered for working on the evolution and systematics of these species (Thompson et al., 2004). Use of different genetic markers through PCR-based techniques have made it convenient to differentiate between species. Generally, rDNA sequences show less intra-specific variation than inter-specific variation and are therefore, considered as reliable markers for species differentiation. The ITS1 and ITS2 are reliable genetic markers for genetic differentiation of species (Chilton et al., 2001). Internal repeats appear to be characteristic of the ITS1 evolution in different groups of organisms (Köhsler et al., 2006).
In the present study, ITS1 sequencing was used for identification of Paramphistomum sp. from buffaloes at District Mardan, KPK, Pakistan. The sequences of ITS-1 region from all collected isolates were similar, indicating no haplotypes. In a specific region same species is prevalent due to similar environmental conditions for continuation of life-cycle of the parasite in definitive vertebrate and intermediate snail host of that area. Other reason may be due to single type of host as all isolates were from buffaloes. These indicates buffaloes are more prone to this parasite. Isolates from different species of hosts may cause genetic variability. The BLAST results of our samples showed 98.6% similarity only with the P. cervi reference sequence reported from China (Zheng et al., 2014). No similarity was found with any other species of P. epiclitum or P. leydeni in blast results, which indicated that at Mardan District P. cervi is prominent species in buffaloes. These results are in contrast with Ali et al. (2018) who genetically identified presence of P. epiclitum through ITS-2 sequencing from Buffaloes at Punjab Province of Pakistan. This difference may be due to difference in sampling areas as District Mardan is in Khyber Pakhtunkhwa Province which is northern region of Pakistan and hilly area while Punjab is mainly plain area. Different environmental conditions of these regions may be the reason of this difference. Other reason may be presence of different snail host species in specific areas. Recent report from western Khyber Pakhtunkhwa indicated the presence of P. epiclitum on the basis of morphology (Khan et al., 2020). While phylogenetic analysis of ITS1 and 5.8S sequence of the same study clustering with P. cervi. We believe that it is P. cervi because the morphological parameters used for identification in this study were compared with that of P. epilictum of Thailand. Environmental conditions of Thailand are different from KPK province of Pakistan and may facilitate progression of other species. Further genetic analysis result is more important and reliable for specie identification.
In the current study single nucleotide polymorphism (SNP) transversions were noted at 8 sites when compared with the reference sequence KJ459934 and 9 with KJ459935, KJ459936, KJ459937 and KJ459938. These SNPs indicating evolutionary process. The phylogenetic tree analysis was carried out by maximum likelihood method. The ITS1 sequence analysis developed a close clade with P. cervi of China. This similarity may be due to the reason that these areas are adjacent to KPK province of Pakistan and share more or less same geographical and environmental conditions. The clade formed with other flukes showed phylogenetic relation with Watsonius watsoni and Gastrodiscoides hominis of family Paramphistomidae, respectively. These results are in agreement with the data reported by Khan et al., 2020, Firdausy et al., 2019. They also reported phylogenetic relation of their isolates of Paramphistomum with these two flukes. The similarities indicated sharing of same evolutionary history.
The present study determined the partial sequence of ITS-1 region of Paramphistomum for the first time at district Mardan, KPK, Pakistan. In our opinion, P. cervi is dominant species in this region and ITS1 marker is a useful genetic marker to study genetic variation with mitochondrial and ITS-2 markers. It can be used compare other isolates of Paramphistoma species not only from other areas of Pakistan but also from other countries.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Peer review under responsibility of King Saud University.
References
- Ali Q., Rashid I., Shabbir M.Z., Akbar H., Shahzad K., Ashraf K., Chaudhry U. First genetic evidence for the presence of the rumen fluke Paramphistomum epiclitum in Pakistan. Parasitol. Int. 2018;67:533–537. doi: 10.1016/j.parint.2018.05.005. [DOI] [PubMed] [Google Scholar]
- Arbabi M., Dalimi-Asl A., Ghafarifar F., Foorozandeh-Moghadam M. Morphological and molecular characterization of Dicrocoelium isolated from sheep in the north and center of Iran. Feyz. J. Kashan. Univ. Med. Sci. 2012;16:135–145. [Google Scholar]
- Chaoudhary V., Hasnani J.J., Khyalia M.K., Pandey S., Chauhan V.D., Pandya S.S., Patel P.V. Morphological and histological identification of Paramphistomum cervi (Trematoda: Paramiphistoma) in the rumen of infected sheep. Vet. world. 2015;8(1):125–129. doi: 10.14202/vetworld.2015.125-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilton N.B., Newton L.A., Beveridge I., Gasser R.B. Evolutionary relationships of trichostrongyloid nematodes (Strongylida) inferred from ribosomal DNA sequence data. Mol. Phylogenet. Evol. 2001;19:367–386. doi: 10.1006/mpev.2001.0938. [DOI] [PubMed] [Google Scholar]
- Gasser R.B., Chilton N.B. Characterisation of taeniid cestode species by PCR-RFLP of ITS2 ribosomal DNA.Acta. Trop. 1995;59:1–40. doi: 10.1016/0001-706x(94)00085-f. [DOI] [PubMed] [Google Scholar]
- Huang W.Y., He B., Wang C.R., Zhu X.Q. Characterization of Fasciola species from Mainland China by ITS-2 ribosomal DNA sequence. Vet. Parasitol. 2004;120:75–83. doi: 10.1016/j.vetpar.2003.12.006. [DOI] [PubMed] [Google Scholar]
- Ilha M.R., Loretti A.P., Reis A.C. Wasting and mortality in beef cattle parasitized by Eurytrema coelamaticum in the state of Parana, southern Brazil. Vet. Parasitol. 2005;133:49–60. doi: 10.1016/j.vetpar.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Juyal P.D., Kaur K., Hassan S.S., Paramjit K. Epidemiological status of paramphistomosis in domestic ruminants in Punjab. Parasites Dis. 2003:231–235. [Google Scholar]
- Kanwal S., Wadood S., Naiz S., Jamil M., Irshad M., Ilyas M., Malook I. Prevalence of Gastrointestinal Parasite (Paramphistoma cervi) In Domestic Animals of District Buner, KPK. Pakistan. Rev. Prog. J. 2014;1:1–12. [Google Scholar]
- Khan I., Afshan K., Shah S., Akhtar S., Komal M., Firasat S. Morphological and Molecular Identifcation of Paramphistomum epiclitum from Bufaloes in Pakistan. Acta. Parasitol. 2020;65:225–236. doi: 10.2478/s11686-019-00155-4. [DOI] [PubMed] [Google Scholar]
- Khan U.J., Tanveer A., Maqbool A., Masood S. Epidemiological studies of paramphistomosis in cattle. Vet. Arhiv. 2008;78:243–251. [Google Scholar]
- Kattak A. Prevalence of gastrointestinal parasite, Paramphistomum in domestic animals (Cows and Buffaloes) of district Swat and Charsadda, KP. Pakistan. J. Entomol. 2017;5(3):907–911. [Google Scholar]
- Köhsler M., Leitner B., Blaschitz M., Michel R., Aspöck H., Walochnik J. ITS1 sequence variabilities correlate with 18S rDNA sequence types in the genus Acanthamoeba (Protozoa: Amoebozoa) Parasitol. Res. 2006;98:86–93. doi: 10.1007/s00436-005-0022-x. [DOI] [PubMed] [Google Scholar]
- Mufti S., Afshan K., Khan I.A., Zafar Y., Raza Rizvi S.S., Nazir F., Qayyum M. Genetic Characterization of Fasciola Samples from Bovine Hosts in Pakistan by Sequences of Ribosomal Internal Transcribed Spacer Regions. Pak. Vet. J. 2014;34(3):361–366. [Google Scholar]
- O’Toole A., Browne J.A., Hogan S., Bassière T., Dewaal T., Mulcahy G., Zintl A. Identity of rumen fluke in deer. Parasitol. Res. 2014;113:4097–4103. doi: 10.1007/s00436-014-4078-3. [DOI] [PubMed] [Google Scholar]
- Rajput, M., Arijo, A.G., Bhutto, M.B., Buriro, R.S., Javaid Ali Gadahi, J.A., Naeem, M., Laghari, Z.A (2020). Morphological and Molecular Characterization of Rumen Fluke Species from Sheep in Southeastern Pakistan. Pakistan J. Zool., 52(5) 1921–1930.
- Rolfe P.F., Boray J.C., Nichols P., Collins G.H. Epidemiology of paramphistomosis in cattle. Int. J. Parasitol. 1991;21:813–819. doi: 10.1016/0020-7519(91)90150-6. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritschi E.F., Maniatis T. Cold Spring Harbor Laboratory Press; New York: 1989. Molecular cloning: a laboratory manual. [Google Scholar]
- Sivajothi S., Reddy B.S. Immature paramphistomosis in a sheep herd. Int. J. Biol. Res. 2014;2:140–142. [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson R.C.A., Zarlenga D.S., La Rosa G., Pozio E., Rosenthal B., Bandi C., Hu M. Advances in the diagnosis and systematics of parasites of veterinary importance: new and exciting prospects. Mol. Syst. Diag. Vet. Parasitol. 2004;125:69–92. [PubMed] [Google Scholar]
- Wang C.R., Qiu J.H., Zhu X.Q. Survey of helminthes in adult sheep in Heilongjiang Province, People’s Republic of China. Vet. Parasitol. 2006;140:378–382. doi: 10.1016/j.vetpar.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Zheng X., Chang Q.C., Zhang Y., Tian S.Q., Lou Y., Duan H., Zhu X.Q. Characterization of the complete nuclear ribosomal DNA sequences of Paramphistomum cervi. Sci. World J. 2014:1–12. doi: 10.1155/2014/751907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firdausy L.W., Prahardani R., Wusahaningtyas L.S., Indarjulianto S., Wahyu M., Nursalim M.T., Nurcahyo W. Morphological and molecular identification of Pfenderius heterocaeca (Trematode: Paramphistomoidea) from Sumatran elephant (Elephas maximus sumatranus) Vet. World. 2019;12(8):1341–1345. doi: 10.14202/vetworld.2019.1341-1345. [DOI] [PMC free article] [PubMed] [Google Scholar]