Abstract
Cyclic nucleotide gated ion channels (CNGCs) in plants have very important role in signaling and development. The study reports role of CNGC19 and CNGC20 in salt stress in A. thaliana. In-silico, genome wide analysis showed that CNGC19 and CNGC20 are related to salt stress with maximum expression after 6 h in A. thaliana. The position of inserted T-DNA was determined (in-vivo) through TAIL-PCR for activation tagged mutants of CNGC19 and CNGC20 under salt stress. The expression of AtCNGC19 and AtCNGC20 after cloning under 35S promoter of expression vectors pBCH1 and pEarleyGate100 was determined in A. thaliana by real-time PCR analysis. Genome wide analysis showed that AtCNGC11 had lowest and AtCNGC20 highest molecular weight as well as number of amino acid residues. In-vivo expression of AtCNGC19 and AtCNGC20 was enhanced through T-DNA insertion and 35S promoter in over-expressed plants under high salt concentration. AtCNGC19 was activated twice in control and about five times under 150 mM NaCl stress level, and expression value was also higher than AtCNGC20. Phenotypically, over-expressed plants and calli were healthier while knock-out plants and calli showed retarded growth under salinity stress. The study provides new insight for the role of AtCNGC19 and AtCNGC20 under salt stress regulation in A. thaliana and will be helpful for improvement of crop plants for salt stress to combat food shortage and security.
Keywords: In-silico expression, AtCNGCs, TAIL-PCR, Salinity, Over-expression, Knock out, Food crops
1. Introduction
Stresses to crop plants are main causes for loss in productivity, and a threat to food security worldwide. Abiotic stresses like salinity and drought affect plant growth, induce modifications in plant homeostasis and ultimately decrease crops production (Marsch-Martinez et al., 2002, Gobert et al., 2006, Peters et al., 2013). The survival of plants under different biotic and abiotic stresses depends upon the complex mechanisms to perceive external signals and optimal adaptive response (Thoen et al., 2017). Moreover, improvement of plant's stress resistance is significantly important for environmental sustainability and agricultural productivity because crops consume more fertilizer and water with poor stress resistance (Nieves-Cordones et al., 2019).
Abiotic stress tolerance is mediated by the molecular control mechanism and regulation of the stress related genes (Jha et al., 2016). Different kinds of calcium channels in plants contribute to the accumulation of calcium in cytosol and cyclic nucleotide gated ion channels are one of them (Chin et al., 2009). Cyclic nucleotide gated ion channels (CNGCs) are non-selective cation channels found both in plants and animals. CNGCs are considered as significant calcium transporters in plants (Talke et al., 2003).
Many types of researches have been started to explore the functions and underlying regulatory mechanisms, evolution, and phylogenetic relationship with other CNGCs (Kakar et al., 2017). The yields are being improved by changing plant growing procedures and using advanced genome editing techniques (Chen and Gao, 2013). Genome analyses of an organism or species by using bioinformatics tools have significant importance in gene identification, prediction of its proteins and its role in plant physiology (Proost et al., 2009). Activation tagging by T-DNA insertion has emerged as strategic tools for the study of functional genomics in plants. Various T-DNA insertion lines and mutations have been created in A. thaliana, rice and barley. The recovery of genomic sequences flanking the insertions is important for the identification of tagged gene by T-DNA insertion (Jansen et al., 2002, Apse et al., 2003). Gene analysis and structural studies have predicted that group IVA CNGCs (AtCNGC19 and AtCNGC20) in A. thaliana may have a synergistic role in accelerating salinity tolerance. The role of these genes may be further applied to confer salt resistance genes in crops or food plants. This work will be helpful to combat salinity and food shortage by exploring the role of these salt-tolerant genes in food crops.
2. Materials and methods
2.1. Sequence retrieval and genome wide analysis
Complete genome assembly along with amino acid sequences of A. thaliana was extracted from The Arabidopsis Information Resource (TAIR) database. Physical location of each AtCNGC gene on chromosome was determined through database e-plant (http://bar.utoronto.ca/eplant/ActiveSpeciesArabidopsis20thaliana&Genes). Structures of introns and exons of AtCNGCs were analyzed by using database Dicots plaza 3.0 (https://bioinformatics.psb.ugent.be/plaza/versions/plaza_v3_dicots/genes/view). Complete overview of AtCNGC gene family of A. thaliana was obtained by conducting a genome wide analysis using various bioinformatics resources. Amino acid sequences of 20 AtCNGCs of A. thaliana were used to find out the complete description of proteins (http://suba3.plantenergy.uwa.edu.au). This study describes subcellular location in various parts of the plants, location on chromosomes, number of ESTs, nucleotide length, full length cDNAs, amino acid residues and molecular weight of 20 AtCNGCs of A. thaliana.
2.2. In-silico gene expression analysis under salt stress
Salt stress expression data of A. thaliana was obtained from Arabidopsis eFP Browser database (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgiprimaryGene). Salt stress was induced by treating 18 days old plants with 150 mM NaCl and samples were collected for total RNA extraction (Kilian et al., 2007). Expression data of AtCNGCs after salt stress of 0, 6 and 12 h was extracted from database and analyzed by plotting bar graphs.
2.3. Generation and screening of activation tagged lines
Activation tagged mutant lines (2–1-6) of A. thaliana, carrying one copy of pGA-cab-luc-rbcS-gus and pGA-cab-bar-rbcS-hph constructs on chromosome 4 and 5 were used in the study. Seeds were surface sterilized, kept at 4 °C for one week, and then transferred on solidified Murashige-Skoog (MS) medium containing 0.2% gellan gum and incubated in growth chamber (20 °C with fluorescent light) (Tsugane et al., 1999). Seedlings (15–20) were transferred into flask containing liquid MS medium and subsequently grown with shaking at 80 rpm for two weeks. Green tissues (stem and leaves) from cultured roots were cut into small pieces and transferred into callus induction medium (CIM) supplemented with 0.5 ug/mL 2, 4-D and 50 ng/mL kinetin and incubated in growth chamber up to one week.
The roots of these plants were infected with Agrobacterium tumefaciens GV3101 harboring binary vector pRi35ADEn4 for activation tagging (Ahmad et al., 2015). The roots were washed with liquid CIM (supplemented with 0.1 mg/mL of cefotaxime) (Sanofi Aventis, France) and incubated for three weeks on CIM supplemented with 0.2 mg/mL vancomycin and 0.1 mg/mL cefotaxime for transformant selection. Transgenic calli were transferred to CIM containing 0.2 mg/mL vancomycin, 0.1 mg/mL cefotaxime, 0.1 ug/mL chlorsulfuron and 150 mM NaCl for 3 weeks incubation. The mutants were primarily selected on CIM with 150 mM NaCl and secondarily on CIM with 200 mM to 250 mM NaCl.
2.4. Confirmation of activation tagged mutants by PCR
Genomic DNA was isolated from transgenic calli growing on CIM medium containing 0.1 μg/mL chlorsulfuron using Isoplant II DNA extraction kit (Nippon Gene, Japan) as previously described (Weigel et al., 2000). A fragment of P35S-ALS-SUr approximately 200-bp was amplified through PCR by using primers 35SminiL-fd and ALS22-rv (Table 1) and amplicons were resolved through 1.3% (w/v) agarose gel electrophoresis by using Agarose 21 (Nippon Gene, Japan) (Maser et al., 2001, Ahmad et al., 2015). Copy number of T-DNA in selected mutants was determined through Southern analysis. Genomic DNA was isolated from calli using Isoplant II (Nippon Gene, Japan) and then digested with HindIII, PstI and SphI. DNA was blotted onto a Hybond-N membrane (GE Healthcare) after electrophoresis through a 0.8% agarose gel, cross-linked by UV irradiation and then hybridized with a 32P-labeled 2.6-kbp pUC18 backbone fragment at 65 °C according to standard procedures (Niwa et al., 2006). T-DNA position on chromosome was determined by TAIL-PCR. Genomic DNA was extracted from mutants and TAIL-PCR was performed by using AD and TDNA end primers (Table 1) (Ahmad et al., 2015). Purified fragments following tertiary PCR were sequenced directly and the flanking sequences obtained were subjected to BLAST search using the Arabidopsis Information Resource (TAIR, http//:www.arabidopsis.org). Specific primers were used in combination with T-DNA specific primers for amplification of specific fragments which were subsequently sequenced to confirm T-DNA insertion sites and distance from CNGCs.
Table 1.
PCR primers used for activation tagging, TAIL PCR, Real-time PCR and over-expression of AtCNGC19 and AtCNGC20.
| Name | Nucleotide sequence from 5 to 3 direction | Reference |
|---|---|---|
| 35SminiL-fd | GCAAGACCCTTCCTCTATATAAGG | Niwa et al., 1999 |
| ALS22-rv | TCTAGGGAGGAGGGTGGAGGATTT | Niwa et al., 2006 |
| LB1 | CCTTATATAGAGGAAGGGTCTTGC | Niwa et al., 2006 |
| LB2 | TGGGATTGTGCGTCATCCCTTACG | Niwa et al., 2006 |
| LB3 | TTGGAGTAGCCCCGATTGCCCTCA | Niwa et al., 2006 |
| AD2 | NGTCGA(G/C)(A/T)GANA(A/T)GAA | (Liu et al., 1995) |
| AD3 | (A/T)GTGNAG(A/T)ANCANAGA | (Liu et al., 1995) |
| AD2-2′ | GTNCGA(G/C)(A/T)CANA(A/T)GTT | (Liu et al., 2005) |
| RB3 | GGATTGATGTGATATCTAGATCCG | Niwa et al., 2006 |
| RBCS-lc-fd | CCGCAACAAGTGGATTCCTTGTG | Niwa et al., 2006 |
| RBCS-3B-s-rv | AATGAGCAGAGATAATTCATAAGAATG | Niwa et al., 2006 |
| ACT2-fd | GAAGATTAAGGTCGTTGCACCACCTG | Niwa et al., 2006 |
| ACT2-rv2 | ATTCCTGGACCTGCCTCATCATACTC | Niwa et al., 2006 |
| RLK-fd | AATCTCATAGTCCATGTGTGGAATC | Niwa et al., 2006 |
| RLK-rv | CTTCAAATTCAGTAGCTCATCTTGC | Niwa et al., 2006 |
| CNGC19-fd | GGTGTGTGACAAACGTGGAG | This study |
| CNGC19-rv: | ACGGTTGGAATTGGAGTGAG | This study |
| CNGC20-fd | GACAAATGTGGAGGCGTTTT | This study |
| CNGC20-rv | GCCACTTGAATCTGCCTAGC | This study |
| attB1-fd | GGGGACAAGTTTGTACAAAAAAGCAGGCTTCGAAGGAGATAGAACCATG- | This study |
| attB2-rv | GGGGACCACTTTGTACAAGAAAGCTGGGTCTCCACCTCCGGATC | This study |
| GW-CNGC20-fd | GGAGATAGAACCATGGCTTCCCACAACGAAAACGAT | This study |
| GW-CNGC20-rv | TCCACCTCCGGATCACTAAAGGCTATAACTAGACTG | This study |
2.5. Over-expression of AtCNGC19 and AtCNGC20
Open reading frame (ORF) of AtCNGC19 was cloned through PCR using specific primers (Table 1) and KOD polymerase to produce blunt ends and amplification of gene as well. This PCR product was cloned under cauliflower mosaic virus (CaMV) 35S promoter into pBluescript (Agilent, USA), previously digested with EcoRV. AtCNGC19 was cloned into pBCHI through XhoI and SacI under the control of CaMV 35S promoter to generate pBCH1-35S-ORF and transformed into Agrobacterium (Ito et al., 2001). AtCNGC20 was cloned with specific primers (Table 1) with addition of recombination sites to the 5′ and 3′ends as PCR product: attB1-CNGC20-attB2 and ligated into pDONR221 (Thermo Fisher Scientific, USA). The entry clone was sequenced and transformed into E. coli DH5α. The propagated entry clone was isolated from DH5α cells and subsequently cloned into destination vector pEarleyGate100 to form the expression clone. The attR sites of destination vector recombined with the attL sites of the entry clone with LR Clonase II (Thermo Fisher Scientific, USA) (containing excisionase, integrase and integration host factor) and form the expression clone containing attB sites flanking the gene of interest (Hartley, 2000). The expression clone containing the gene controlled by a CaMV 35S promoter was transformed into E. coli DH5α cells. The constructs pBCH1 and pEarleyGate100 containing genes of AtCNGC19 and AtCNGC20 were transformed into A. tumefaciens GV3101 by heat shock method. It was transformed through floral dip method into four to five weeks old wild type (col-0) A. thaliana seedlings with A. tumefaciens GV3101 containing plant expression vector (Clough et al., 2000).
2.6. Real-time PCR analysis of over-expressed and knock-out AtCNGC19 and AtCNGC20
Real-time PCR analysis was carried out to determine gene expression in wild type (Col-0), knock-out AtCNGC19 and AtCNGC20, and over-expressed AtCNGC19 and AtCNGC20. cDNA was synthesized from mRNA first strand cDNA synthesis kit (Roche, Indianapolis). Real time PCR was carried out with 200 pg cDNA mixed with 10 µl of SYBR green PCR master mix (Bio-Rad) and 10 pmol of each forward and reverse primer (Table 1) to final volume of 20 µl. The PCR conditions comprised of 35 cycles at 95 °C for 5 s and 60 °C for 20 s.
3. Results
3.1. Genome wide analysis of CNGCs in A. thaliana
The sub-cellular analysis of 20 AtCNGCs showed that AtCNGC 1, 5, 6, 7, 9 and 13 were located in cytosol; AtCNGC 2, 13 and 16 in mitochondria; AtCNGC3, 4, 8, 10, 11, 12, 14, 15, 17, 18 in plasma membrane, while AtCNGC19 and AtCNGC20 were located in plastids (Table 2). All 20 AtCNGCs of A. thaliana were unevenly distributed on five chromosomes. It was found that chromosome 1 carried 3 AtCNGCs (7, 8 and 10), while chromosome 2 carried 6 AtCNGCs (3, 6, 11, 12, 14 and 15). Chromosome 3 carried 3 AtCNGCs (17, 19 and 20) and both AtCNGC19 and AtCNGC20 are present at close proximity with difference of only 285 base pairs. AtCNGC 9, 13 and 17 were found on chromosome 4 while AtCNGC 2, 4, 5 and 18 were present on chromosome 5. The maximum number of AtCNGCs was located on chromosome 2. AtCNGC2 had the longest gene with 5897 bp and AtCNGC8 was the shortest with 2965 bp. The expressed sequence tags (ESTs) were highest (84) for AtCNGC2 while no EST was found for AtCNGC16. AtCNGC6 had maximum number of cDNAs (38), while AtCNGCs7, 8, 9, 13, 15, 18 and 19 had no cDNAs. AtCNGC20 had highest number of amino acid residues (7 6 4) with MW 86.95 kDa, while lowest for AtCNGC11 (6 2 1) MW 71.47 kDa. The number of exons in all CNGCs ranged from 5 to 11 with AtCNGC20 having maximum (11) while AtCNGC7 with minimum (5) exons. The number of introns ranged from 5 to 10. AtCNGC19 had maximum isoelectric point while AtCNGC4 had minimum 7.99 (Table 2).
Table 2.
Genome-wide analysis of 20 CNGCs of A. thalian.
| Genes | Gene name | Location SUBAcon | Chr locus | EST | cDNAs | Nucleotide length (bp) | Genomic position | Number of |
AAR | M.W (kDa) | PI | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Introns | Exons | |||||||||||
| AT5G53130 | CNGC1 | Cytosol | 5 | 75 | 4 | 3309 | 21537721–21541029 | 7 | 8 | 716 | 83096.3 | 9.39 |
| AT5G15410 | CNGC2 | Mitochondrion | 5 | 87 | 2 | 5897 | 5003226–5006882 | 7 | 8 | 726 | 83240.3 | 9.98 |
| AT2G46430 | CNGC3 | Plasma membrane | 2 | 16 | 2 | 3198 | 19058248–19061436 | 7 | 8 | 706 | 81668.1 | 10.0 |
| AT5G54250 | CNGC4 | Plasma membrane | 5 | 40 | 5 | 4545 | 22025536–22030080 | 7 | 8 | 694 | 80080.7 | 7.99 |
| AT5G57940 | CNGC5 | Cytosol | 5 | 25 | 8 | 3947 | 23456667–23460613 | 7 | 8 | 717 | 81967.8 | 9.27 |
| AT2G23980 | CNGC6 | Cytosol | 2 | 44 | 3 | 4319 | 10201104–10205422 | 7 | 8 | 747 | 85439.9 | 9.65 |
| AT1G15990 | CNGC7 | Cytosol | 1 | 5 | 0 | 3060 | 5491133–5494192 | 4 | 5 | 738 | 84633.6 | 9.53 |
| AT1G19780 | CNGC8 | Plasma membrane | 1 | 4 | 0 | 2965 | 6833669–6836633 | 5 | 6 | 753 | 85981.9 | 9.311 |
| AT4G30560 | CNGC9 | Cytosol | 4 | 9 | 0 | 3522 | 14926834–1493035 | 6 | 7 | 733 | 83623.0 | 9.85 |
| AT1G01340 | CNGC10 | Plasma membrane | 1 | 4 | 1 | 3655 | 132270–135924 | 8 | 9 | 711 | 81894.3 | 9.1 |
| AT2G46440 | CNGC11 | Plasma membrane | 2 | 16 | 2 | 3250 | 19061688–19064937 | 7 | 8 | 621 | 71473.7 | 9.75 |
| AT2G46450 | CNGC12 | Plasma membrane | 2 | 21 | 1 | 4034 | 19065536–19069569 | 8 | 9 | 649 | 74805.5 | 9.79 |
| AT4G01010 | CNGC13 | Cytosol | 4 | 5 | 0 | 3375 | 434217–437591 | 6 | 7 | 696 | 80472.8 | 9.68 |
| AT2G24610 | CNGC14 | Plasma membrane | 2 | 3 | 0 | 3693 | 10456879–10460571 | 6 | 7 | 726 | 83453.6 | 9.46 |
| AT2G28260 | CNGC15 | Plasma membrane | 2 | 3 | 0 | 3142 | 12049443–12052584 | 5 | 6 | 678 | 78722.1 | 10.01 |
| AT3G48010 | CNGC16 | Mitochondrion | 3 | 0 | 1 | 3004 | 17721137–17724140 | 6 | 7 | 705 | 81954.1 | 8.65 |
| AT4G30360 | CNGC17 | Plasma membrane | 4 | 12 | 3 | 3326 | 14854694–14858019 | 5 | 6 | 720 | 83360.4 | 8.66 |
| AT5G14870 | CNGC18 | Plasma membrane | 5 | 2 | 0 | 3003 | 4808091–4811093 | 5 | 6 | 706 | 80363.1 | 8.26 |
| AT3G17690 | CNGC19 | Plastids | 3 | 3 | 0 | 3528 | 6045002–6048529 | 9 | 10 | 729 | 84048.2 | 10.37 |
| AT3G17700 | CNGC20 | Plastids | 3 | 13 | 1 | 3862 | 6048814–6052675 | 10 | 11 | 764 | 86949.5 | 9.82 |
3.2. In-silico analysis of expression of AtCNGCs under salt stress
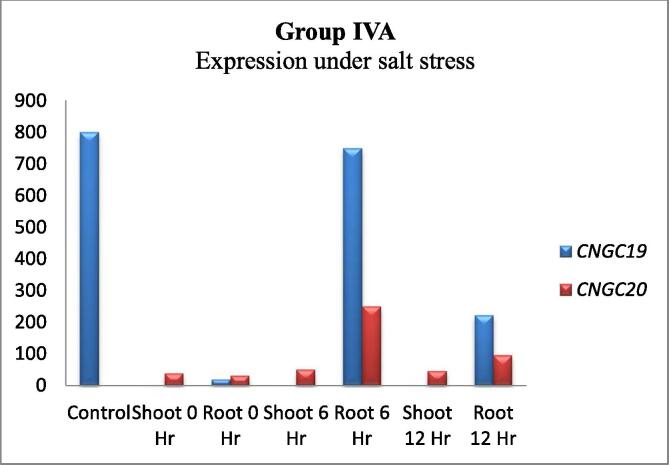
The In-silico expression analysis combined all AtCNGCs into five groups. Group I AtCNGCs (1, 3, 10, 11, 12 and 13) expressed differentially under salt stress in shoots and roots of A. thaliana after different time intervals except AtCNGC3 and AtCNGC11 which expressed similarly in both tissues. The expression of AtCNGC13 was slightly lower as compared to other AtCNGCs in this group (Fig. 1). Group II AtCNGCs (5, 6, 7, 8 and 9) analysis showed that the expression of AtCNGC5 was higher in shoots while AtCNGC6 was higher in roots. AtCNGC7 and AtCNGC8 showed very slight or no expression as well as AtCNGC9 with smaller expression (Fig. 2). Group III AtCNGCs (14, 15, 16, 17 and 18) showed higher expression of AtCNGC14 in roots as compared to other AtCNGCs of this group (Fig. 3). The expression of group IV AtCNGCs is subdivided into IVA (19, 20) and IVB (2, 4). It was observed that AtCNGC19 and AtCNGC20 were highly expressed under salt stress after 6 h and decreased after 12 h (Fig. 4). The expression of AtCNGC19 was higher as compared to all members of AtCNGC family. The group IVB AtCNGC2 was highly expressed under salt stress in shoots and its similar trend was observed in shoots after 12 h duration. The expression of AtCNGC4 was lower as compared to AtCNGC2 (Fig. 5).
Fig. 1.
Expression of CNGCs of group I under salt stress. The Gene Chip Operating Software (GCOS) was used for calculation of the expression values with the specified parameters: TGT = 100 and Bkg = 20. X-axis showed time intervals of treatment and tissue types whereas gene expression signals are shown along Y-axis.
Fig. 2.
Expression of CNGCs of group II under salt stress. The Gene Chip Operating Software (GCOS) was used for calculation of the expression values with the specified parameters: TGT = 100 and Bkg = 20. X-axis showed time intervals of treatment and tissue types whereas gene expression signals are shown along Y-axis.
Fig. 3.
Expression of CNGCs of group III under salt stress. The Gene Chip Operating Software (GCOS) was used for calculation of the expression values with the specified parameters: TGT = 100 and Bkg = 20. X-axis showed time intervals of treatment and tissue types whereas gene expression signals are shown along Y-axis.
Fig. 4.
Expression of CNGCs of group IVA under salt stress. The Gene Chip Operating Software (GCOS) was used for calculation of the expression values with the specified parameters: TGT = 100 and Bkg = 20. X-axis showed time intervals of treatment and tissue types whereas gene expression signals are shown along Y-axis.
Fig. 5.
Expression of CNGCs of group IVB under salt stress. The Gene Chip Operating Software (GCOS) was used for calculation of the expression values with the specified parameters: TGT = 100 and Bkg = 20. X-axis showed time intervals of treatment and tissue types whereas gene expression signals are shown along Y-axis.
3.3. Expression of salt tolerance through activation tagging
The activation tagging analysis through TAIL-PCR for 18 mutant calli showed that T-DNA insertion increased expression of salt tolerance genes in stc13 calli to maximum. The sequencing of TAIL-PCR product showed that T-DNA was inserted on chromosome number 3, and distance between activated gene and T-DNA was 1509 bp. AtCNGC19 and AtCNGC20 are present on the same chromosome number 3 at much closed vicinity. Real PCR analysis of salt tolerance genes for this stc13 mutant calli showed higher expression as compared to its parental line (2–1-6). The expression of salt tolerance genes also increased at higher concentration of salt (150 mM) (Fig. 6).
Fig. 6.
The expression of stc13 and parental line (2–1-6) of A. thaliana. Total RNA was extracted from calli grown on normal CIM and CIM supplemented with 150 mM NaCl and expression was determined by real time PCR.
3.4. Over-expression of AtCNGC19 and AtCNGC20
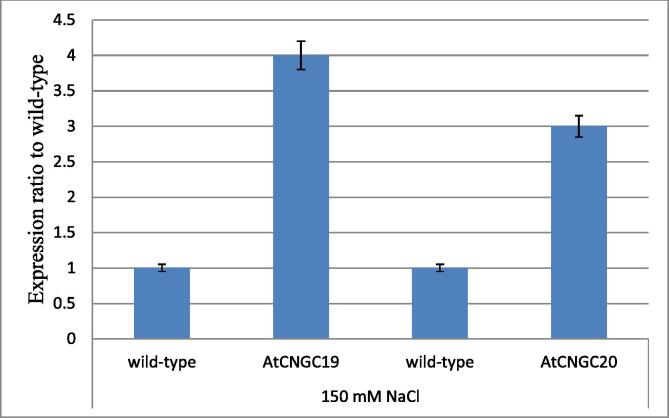
The AtCNGC19 and AtCNGC20 were further over-expressed in A. thaliana for the confirmation of their role in salt stress. The expression of both genes was enhanced by 35S promoter of pBCH1 and pEarleyGate100. The expression of AtCNGC19 and AtCNGC20 in over-expressed A. thaliana lines was higher as compared to wild-type up to 150 mM salt stress. The expression of AtCNGC19 was higher as compared to AtCNGC20 which implies that both genes play important role to confer salt tolerance mechanism with major contribution of AtCNGC19 (Fig. 7).
Fig. 7.
Expression of AtCNGC19 and AtCNGC20 in wild-type and over-expression lines grown on callus induction media with 150 mM NaCl. Total RNA was extracted from wild type (col-0), over-expressed AtCNGC19 and AtCNGC20 mutants and subjected to real time PCR.
3.5. CNGC19 and CNGC20 expression analysis in knock-out lines of A. thaliana
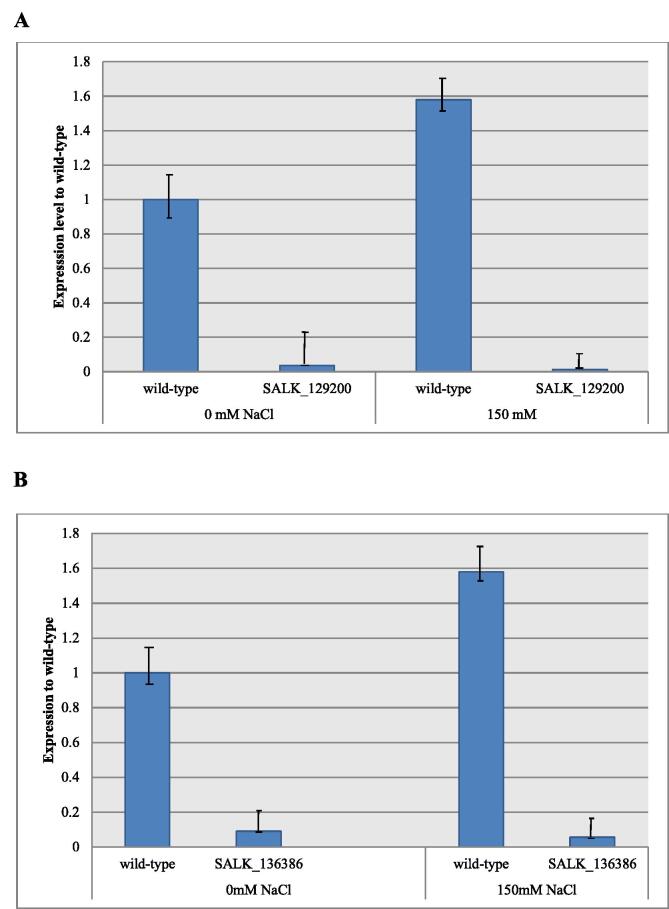
Real time PCR analysis of knock out A. thaliana lines SALK_129200 and SALK_136386 showed that T-DNA insertion suppressed expression of AtCNGC19 and AtCNGC20. It was observed that the wild type plants having all AtCNGCs intact, showed expression of these genes while in case of knock-out plants, the expression was significantly suppressed as the concentration of salt was increased from 0 mM to 150 mM NaCl in the MS media (Fig. 8).
Fig. 8.
Expression of AtCNGC19 and AtCNGC20 in wild-type and knock-out lines (A) SALK_129200, knock out line for AtCNGC19 (B) SALK_136386, knock-out line for AtCNGC20 Expression AtCNGC19 and AtCNGC20 in wild type (col-0) plants was higher as compared to the knock-out lines SALK_129200 and SALK_136386 respectively.
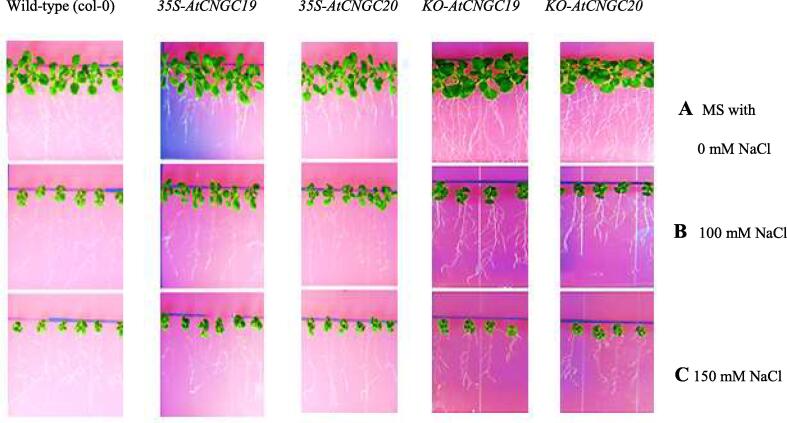
3.6. Phenotypes of over-expressed and knock-out plants
The phenotypes of over-expressed and knock-out AtCNGC19 and AtCNGC20 plants showed that the over-expressed AtCNGC19 and AtCNGC20 plants were healthy and grew well in high salt concentration up to 150 mM, compared to wild-type plants and knock-out plants which were unhealthy with retarded growth (Fig. 9). This observation depicted that both of these AtCNGCs might have synergistic effect and one of AtCNGC complementing the effect of other in its absence that is in knock-out plants, which could be further confirmed by making double mutants of both AtCNGC19 and AtCNGC20.
Fig. 9.
Phenotypes of over-expression lines and knock-out lines of AtCNGC19 and AtCNGC20 compared with wild-type (A) Only MS (B) MS with 100 mM NaCl (C) MS with 150 mM NaCl. Plants were transferred to NaCl supplemented MS plates after 4 days of germination on normal MS. Over-expressed plants showed better growth in 100 mM of NaCl as compared to knock-out and wild type plants. The phenotypic differences of over-expressed and knock-out AtCNGC19 and AtCNGC20 plants depicted that both of these CNGCs are complementing the effect of each other.
4. Discussion
Cyclic nucleotide gated ion channels play vital role in plant response to salt tolerance. A. thaliana contains twenty AtCNGCs distributed throughout different parts of the plant and are divided into four groups (I-IV) (Yuen and Christopher, 2016). The online databases showed that AtCNGCs are located in plasma membrane, cytosol as well as mitochondria and plastids (Nawaz et al., 2014). All CNGCs are involved in regulation of membrane potential. The available information on these genes showed that biological processes of AtCNGC 9, 13, 14, 15 and 17 have not yet been defined except regulation of membrane potential. AtCNGCs are involved in cell signaling, calcium uptake potassium ion transport, regulation of membrane potential defense response, nitric oxide mediated signal transduction and disease resistance (Ma et al., 2006, Kaplan et al., 2007; (Crawford et al., 2018; Zhang et al., 2018). AtCNGC19 and AtCNGC20 are involved in abiotic stress tolerance (Guo, 2008); Kugler et al., 2009).
Enhancer sequences within the activation tagging vectors may enhance either constitutive or ectopic expression, and the majority of enhanced genes have been reported as being located adjacent to the T-DNA (Weigel et al., 2000). T-DNA insertion is involved in regulation of salt tolerance genes in A. thaliana (Chalfun et al., 2003). A transgenic parent line 2–1-6 was transformed with activation tagging binary vector pRi35ADEn4 having four copies of a 339-bp CaMV 35S enhancer at its right border (Hayashi et al., 1992; Niwa et al., 2006). T-DNA insertion in A. thanliana close to AtCNGC19 and AtCNGC20 has enhanced the expression of these genes under salt stress which shows that these two genes are related to salinity tolerance in A. thaliana.
The expression of AtCNGC19 and AtCNGC20 through pBCH1 and pEarlygate100 under CaMV35S promoter has increased the expression of these genes in stc13 under high salt concentration. CaMV35S promoter effectively controls the regulation of the host genome and expresses the objective genes at approximately 2 to 3 orders of magnitude higher (Yoo et al., 2005).
The comparison of mutant lines with its wild-type plants showed that the expression of AtCNGC19 and AtCNGC20 was increased about 3–4 fold in high salt concentration. AtCNGC19 showed higher expression than AtCNGC20 at high salt concentration up to 150 mM. The higher expression of AtCNGC19 in roots at early stage of growth also confirms that this gene is involved in high root uptakability in salt stress. AtCNGC20 expressed predominant promoter activity in mesophyll cells surrounding the vessels, and its expression increases gradually during development and get saturated in mature leaves (Jha et al., 2016). The high concentration of sodium ions disturbs the pH and effects the ion channels and transporters under salinity stress (Bose, 2017). It was depicted that AtCNGC19 and AtCNGC20 both might be synergistically responsible for salinity tolerance in A. thaliana.
The phenotypic analysis expressed that AtCNGC19 and AtCNGC20 was greatly up regulated in over-expression line as compared to wild type. It was observed that the over-expressed plants were grown properly even at high concentration of salt. The plants grown in higher salt concentration showed retarded growth. Salinity stress increased the thickness of epidermal cells and decreased the intracellular spaces in epidermal cells of leaves. The decrease of surface area of the leaf and opening of stomata are also result of salt stress (Li and Li, 2017).
5. Conclusion
A. thaliana has five groups of CNGCs, and AtCNGC19 and AtCNGC20 belong to group IVA. In-silico expression analysis of AtCNGCs under salt stress in A. thaliana showed that these AtCNGCs express differentially in roots and shoots. It was observed that AtCNGC19 and AtCNGC20 expressed highly in roots under salt stress. The activation tagging analysis confirmed that AtCNGC19 and AtCNGC20 were expressed along with other genes of this family. The over-expression of AtCNGC19 and AtCNGC20 under 35S promoter showed high salt tolerance in recombinant A. thaliana. The analysis of gene expression in SALK knock-out lines of A. thaliana showed that AtCNGC19 and AtCNGC20 are involved in tolerance of high salt concentration in this plant. The phenotypic analysis indicated that both AtCNGC19 and AtCNGC20 are responsible for effective growth of recombinant plants. This can be further confirmed by making double knock-out mutants of AtCNGC19 and AtCNGC20. The findings of this study can be employed for genetic engineering for these genes in conventional food crops salt resistance for abiotic stresses and overall increase in yield of crops.
Funding
No funding was available. All the research work was done with lab resources.
7. Note
Sadaf Oranab and Bushra Munir have equal contribution in this manuscript and both should be considered as first authors.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Peer review under responsibility of King Saud University.
Contributor Information
Abdul Ghaffar, Email: abdulghaffar@gcuf.edu.pk.
Aftab Ahmad, Email: aftab.ahmad@uaf.edu.pk.
References
- Ahmad A., Niwa Y., Goto S., Kobayashi K., Shimizu M., Ito S., Usui Y., Nakayama T., Kobayashi H. Genome-wide screening of salt tolerant genes by activation-tagging using dedifferentiated calli of Arabidopsis and its application to finding gene for myo-inositol-1-P-synthase. PloS one. 2015;10(5) doi: 10.1371/journal.pone.0115502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apse M.P., Sottosanto J.B., Blumwald E. Vacuolar cation/H+exchange, ion homeostasis, and leaf development are altered in a T-DNA insertional mutant ofAtNHX1, the Arabidopsis vacuolar Na+/H+antiporter. Plant J. 2003;36:229–239. doi: 10.1046/j.1365-313x.2003.01871.x. [DOI] [PubMed] [Google Scholar]
- Chen K., Gao C. TALENs: customizable molecular DNA scissors for genome engineering of plants. J. Genet. Genom. 2013;40(6):271–279. doi: 10.1016/j.jgg.2013.03.009. [DOI] [PubMed] [Google Scholar]
- Chin K., Moeder W., Yoshioka K. Biological roles of cyclic nucleotide-gated ion channels in plants: what we know and don’t know about this 20 member ion channel family. Bot. 2009;87(7):668–677. [Google Scholar]
- Clough S.J., Fengler K.A., Yu I.C., Lippok B., Smith R.K., Bent A.F. The Arabidopsis dnd1 “defense, no death” gene encodes a mutated cyclic nucleotide-gated ion channel. Proc Natl. Acad. Sci. 2000;97(16):9323–9328. doi: 10.1073/pnas.150005697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose, Munns, R., Shabala, S., Gilliham, M., Pogson, B., Tyerman, S.D. Chloroplast function and ion regulation in plants growing on saline soils: lessons from halophytes. J. Exp. Bot. 2017;68:3129–3143. doi: 10.1093/jxb/erx142. [DOI] [PubMed] [Google Scholar]
- Chalfun-Junior A., Mes J.J., Mlynarova L., Aarts M.G., Angenent G.C. Low frequency of T-DNA based activation tagging in Arabidopsis is correlated with methylation of CaMV 35S enhancer sequences. FEBS Lett. 2003;555:459–463. doi: 10.1016/s0014-5793(03)01300-0. [DOI] [PubMed] [Google Scholar]
- Crawford, Lehotai, N., Strand. A. The role of retrograde signals during plant stress responses. J. Exp. Bot. 2018;69:2783–2795. doi: 10.1093/jxb/erx481. [DOI] [PubMed] [Google Scholar]
- Gobert A., Park G., Amtmann A., Sanders D., Maathuis F.J. Arabidopsis thaliana cyclic nucleotide gated channel 3 forms a non-selective ion transporter involved in germination and cation transport. J. Exp. Bot. 2006;57(4):791–800. doi: 10.1093/jxb/erj064. [DOI] [PubMed] [Google Scholar]
- Guo, Babourina, O., Christopher, D. A., Borsics, T., Rengel, Z. The cyclic nucleotide gated channel, AtCNGC10, influences salt tolerance in Arabidopsis. Physiol. Plant. 2008;134:499–507. doi: 10.1111/j.1399-3054.2008.01157.x. [DOI] [PubMed] [Google Scholar]
- Hartley J, l, Temple, G. F., Brasch, M. A. DNA cloning using in vitro site-specific recombination. Genome. Res. 2000;10:1788–1795. doi: 10.1101/gr.143000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi, Czaja, I., Lubenow, H., Schell, J., Walden, R. Activation of a plant gene by T-DNA tagging: auxin-independent growth in vitro. Sci. 1992;258:1350–1353. doi: 10.1126/science.1455228. [DOI] [PubMed] [Google Scholar]
- Ito, Chiba, T., Ozawa, R., Yoshida, M., Hattori, M., Sakaki, Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R., Embden J.D., Gaastra W., Schouls L.M. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Micro. 2002;43:1565–1575. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- Zhang X, R., Xu, Y. P., Cai, X. Z. SlCNGC1 and SlCNGC14 Suppress Xanthomonas oryzae pv. oryzicola-Induced Hypersensitive Response and Non-host Resistance in Tomato 285. Front. Plant Sci. 2018;9:285. doi: 10.3389/fpls.2018.00285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha, S., Sharma, M., K Pandey, G., 2016. Role of cyclic nucleotide gated channels in stress management in plants. Curr. Genom. 17(4), 315-329. [DOI] [PMC free article] [PubMed]
- Kakar K.U., Nawaz Z., Kakar K., Ali E., Almoneafy A.A., Ullah R., Ren X., Shu Q. Comprehensive genomic analysis of the CNGC gene family in brassica oleracea: Novel insights into synteny, structures, and transcript profiles. BMC Genom. 2017;18(1) doi: 10.1186/s12864-017-4244-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan B., Sherman T., Fromm H. Cyclic nucleotide-gated channels in plants. FEBS Lett. 2007;581:2237–2246. doi: 10.1016/j.febslet.2007.02.017. [DOI] [PubMed] [Google Scholar]
- Kilian J., Whitehead D., Horak J., Wanke D., Weinl S., Batistic O., D’Angelo C., Bornberg-Bauer E., Kudla J., Harter K. The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 2007;50(2):347–363. doi: 10.1111/j.1365-313X.2007.03052.x. [DOI] [PubMed] [Google Scholar]
- Kugler A., Kohler B., Palme K., Wolff P., Dietrich P. Salt-dependent regulation of a CNG channel subfamily in Arabidopsis. BMC Plant Biol. 2009;9:140. doi: 10.1186/1471-2229-9-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W., Li, Q. 2017. Effect of environmental salt stress on plants and the molecular mechanism of salt stress tolerance. Int. J. Environ. Sci. Nat. Res. 7, 1-6.
- Ma, W., Yoshioka, K., Berkowitz, G.A., 2006. Cyclic nucleotide gated channels and Ca2+-mediated signal transduction during plant innate immune response to pathogens. Plant Signal. Behav. 2, 548–550. [DOI] [PMC free article] [PubMed]
- Liu, Whittier, R. F., Thermal asymmetric interlaced PCR: Automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genom. 1995;25:674–681. doi: 10.1016/0888-7543(95)80010-j. [DOI] [PubMed] [Google Scholar]
- Liu, Chen, Y.,Zhang, Q. Amplification of genomics sequences flanking T-DNA insertion by thermal asymmetric interlaced polymerase chain reaction. Trans. Plant. 2005:341–348. doi: 10.1385/1-59259-827-7:341. [DOI] [PubMed] [Google Scholar]
- Marsch-Martinez N., Greco R., Van-Arkel G., Herrera-Estrella L., Pereira A. Activation tagging using the En-I maize transposon system in Arabidopsis. Plant Physiol. 2002;129:1544–1556. doi: 10.1104/pp.003327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maser P., Thomine S., Schroeder J.I., Ward J.M., Hirschi K., Sze H., Harper J.F., Tchieu J., Gribskoy M., Persans M.W., Salt D.E., Kim S.A., Guerinot M.L. Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol. 2001;126:1646–1667. doi: 10.1104/pp.126.4.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawaz F., Ashraf M.Y., Ahmad R., Waraich E.A., Shabbir R.N. Selenium (Se) Regulates Seedling Growth in Wheat under Drought Stress. Adv. Chem. 2014:1–7. [Google Scholar]
- Nieves-Cordones, Lopez-Delacalle, M., Ródenas, R., Martínez, V., Rubio, F., Rivero, R. M., Critical responses to nutrient deprivation: A comprehensive review on the role of ROS and RNS. Environ. Exp. Bot. 2019;161:74–85. [Google Scholar]
- Niwa Y., Goto S., Nakano T., Sakaiya M., Hirano T., Tsukaya H., Yoshibumi K., Kobayashi H. Arabidopsis mutants by activation tagging in which photosynthesis genes are expressed in dedifferentiated calli. Plant. Cell Physiol. 2006;47:319–331. doi: 10.1093/pcp/pci242. [DOI] [PubMed] [Google Scholar]
- Niwa Y., Hirano T., Yoshimoto K., Shimizu M., Kobayashi H. Non-invasive quantitative detection and applications of non-toxic, S65T-type green fluorescent protein in living plants. Plant J. 1999;18:455–463. doi: 10.1046/j.1365-313x.1999.00464.x. [DOI] [PubMed] [Google Scholar]
- Peters M., Herrero M., Fisher M., Erb K.H., Rao I., Subbarao G.V., Van der Hoek R. Challenges and opportunities for improving eco-efficiency of tropical forage-based systems to mitigate greenhouse gas emissions. Trop. Grassl.-Forrajes Trop. 2013;1:156–167. [Google Scholar]
- Proost S., Van Bel M., Sterck L., Billiau K., Van Parys T., Van de Peer Y., Vandepoele K. PLAZA: a comparative genomics resource to study gene and genome evolution in plants. Plant Cell. 2009;21:3718–3731. doi: 10.1105/tpc.109.071506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talke I.N., Blaudez D., Maathuis F.J., Sanders D. CNGCs: prime targets of plant cyclic nucleotide signalling. Trend. Plant Sci. 2003;8:286–293. doi: 10.1016/S1360-1385(03)00099-2. [DOI] [PubMed] [Google Scholar]
- Thoen M, P, Davila Olivas, N. H., Kloth, K. J ., Coolen, S., Huang, P., Aarts, M. G., Bac-Molenaar, J. A., Bakker, J., Bouwmeester, H. J., Broekgaarden, C., Bucher, J., Busscher-Lange, J., Cheng, X., Fradin,E. F., Jongsma, M. A., Julkowska, M. M., Keurentjes, J. J., Ligterink, W., Pieterse, C. M., Ruyter-Spira, C., Smant, G., Testerink, C., Usadel, B., Loon, J. J., Pelt, J. A., Schaik, C. C., Wees, S. C., Visser, R., Voorrips, R., Vosman, B., Vreugdenhil, D., Warmerdam, S., Wiegers, G. L., Heerwaarden, J. V., Kruijer, W., Eeuwijk, F. A., Dicke, M., Genetic architecture of plant stress resistance: multi‐trait genome‐wide association mapping. New Phyto. 2017;213:1346–1362. doi: 10.1111/nph.14220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugane K., Kobayashi K., Niwa Y., Ohba Y., Wada K., Kobayashi H. A recessive Arabidopsis mutant that grows photoautotrophically under salt stress shows enhanced active oxygen detoxification. Plant Cell. 1999;11:1195–1206. doi: 10.1105/tpc.11.7.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D., Ahn J.H., Blázquez M.A., Borevitz J.O., Christensen S.K., Fankhauser C., Chory J. Activation tagging in Arabidopsis. Plant Physiol. 2000;122(4):1003–1014. doi: 10.1104/pp.122.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.Y., Bomblies K., Yoo S.K., Yang J.W., Choi M.S., Lee J.S., Ahn J.H. The 35S promoter used in a selectable marker gene of a plant transformation vector affects the expression of the transgene. Plant. 2005;221:523–530. doi: 10.1007/s00425-004-1466-4. [DOI] [PubMed] [Google Scholar]
- Yuen C.C., Christopher D.D. The group IV-A cyclic nucleotide-gated channels, CNGC19 and CNGC20, localize to the vacuole membrane in Arabidopsis thaliana. AoB Plant. 2016;5:12. [Google Scholar]