Abstract
Antimicrobial resistance (AMR) is a major health crisis globally. Migratory birds could be a potential source for antibiotic resistant (ABR) bacteria. Not much is known about their role in the transmission of ABR in Bangladesh. In this study, a total of 66 freshly dropped fecal materials of migratory birds were analyzed. Bacterial isolation and identification were based on cultural properties, biochemical tests, and polymerase chain reaction (PCR). The disk diffusion method was employed to evaluate antibiogram profiles. By PCR, out of 66 samples, the detection rate of Enterococcus spp. (60.61%; 95% confidence interval: 48.55–71.50%) was found significantly higher than Salmonella spp. (21.21%; 95% CI: 13.08–32.51%) and Vibrio spp. (39.40%; 95% CI: 28.50–51.45%). Enterococcus isolates were frequently found resistant (100–40%) to ampicillin, streptomycin, meropenem, erythromycin, and gentamicin; Salmonella isolates were frequently resistant (72–43%) to chloramphenicol, tetracycline, ampicillin, streptomycin, and erythromycin; and Vibrio spp. isolates were frequently resistant (77–31%) to vancomycin, ampicillin, erythromycin, tetracycline, and streptomycin. In addition, 60% (95% CI: 44.60–73.65%) Enterococcus spp., 85.71% (95% CI: 60.06–97.46%) Salmonella spp., and 76.92% (95% CI: 57.95–88.97%) Vibrio spp. isolates were multi-drug resistant (MDR) in nature. Three isolates (one from each bacterium) were found resistant against six classes of antibiotics. The bivariate analysis revealed strong associations (both positive and negative) between several antibiotic pairs which were resistant to isolated organisms. To the best of our knowledge, this is the first study in detecting MDR Enterococcus spp., Salmonella spp., and Vibrio spp. from migratory birds travelling to Bangladesh. Frequent detection of MDR bacteria from migratory birds travelling to Bangladesh suggests that these birds have the potential to carry and spread ABR bacteria and could implicate potential risks to public health. We recommend that these birds should be kept under an AMR surveillance program to minimize the potential risk of contamination of the environment with ABR as well as to reduce their hazardous impacts on health.
Keywords: Migratory birds, Bacteria, Antibiotic resistance, MDR, Environment, Public health
1. Introduction
The usages of antibiotics have been gradually increased since their first utilization for the treatment of bacterial infections. Though the first treatment with antibiotics started in humans, antibiotics are widely used in veterinary medicine and livestock production. The randomized use of antibiotics has triggered to the development of antibiotic resistance or multi-drug resistance (MDR) among bacteria in different settings. Antibiotic resistant bacteria acquired in one setting can easily be transferred to other settings that disturbs the normal ecosystems globally. The misuse of antibiotics and defective healthcare systems are connected with the dissemination of resistant bacteria in diversified environmental components (Rashid et al., 2015). The level of use of antibiotic practices controls the abundance of resistant bacteria in environments (Ramey et al., 2018). Interestingly, pristine environments are being involved with bacterial antibiotic resistance, though they are not directly connected with human exposures (Rashid et al., 2015). These findings suggest that other factors are contributing to the dissemination of resistant bacteria to such pristine environments.
Migratory birds can be linked with the carriers and transmitters of antibiotic resistant bacteria having both human and veterinary importance (Islam et al., 2021). The ability of migratory birds in disseminating microorganisms came to the attention of biologists during the global avian influenza virus outbreak. Billions of birds migrate yearly and visit almost all continents globally (Guenther et al., 2012). For high mobility, migratory birds can easily acquire and disseminate antibiotic resistant bacteria from one place to another. During migration, they can interlace with the native birds, contact with feeds from different environments, and ultimately can pick up resistant bacteria from those environments (Hatha et al., 2013). Such way they can be reservoirs and spreaders of resistant bacteria to aquatic environments (haor, baor, lakes, ponds, rivers, etc.) via their fecal contaminations (Rashid et al., 2015, Islam et al., 2021). Previous studies recognized their role in transferring antibiotic resistant and/or MDR bacteria (Lin et al., 2020, Najdenski et al., 2018, Ramey et al., 2018, Atterby et al., 2016).
Antibiotic resistance in commensal pathogens e.g. Enterococcus spp., Escherichia coli; zoonotic enteropathogens e.g. Campylobacter spp., Salmonella spp., Vibrio spp. has been emerged in the last few decades (Díaz-Sánchez et al., 2012). Migratory birds can act as carriers and vectors of Enterococcus spp. (Klibi et al., 2015), Salmonella spp. (Wei et al., 2020), and Vibrio spp. (Fu et al., 2019). Enterococcus spp. - ubiquitous organisms in mammals and birds- are able to develop nosocomial pathogenicity and community-acquired infections in the human body. In addition, they also can develop infections in the urinary tract, intra-abdominal and pelvic regions of humans (Fisher and Phillips, 2009). Although enterococcal infections are rare in poultry, they can cause septicemia, endocarditis, encephalomalacia, tracheitis, and osteomyelitis (Chadfield et al., 2004). Salmonella spp. - major zoonotic pathogens- can cause salmonellosis in poultry and enteric fever, gastroenteritis, even life-threatening consequences in humans (Tawyabur et al., 2020). The food chain plays a significant role in the transmission of Salmonella spp. to humans. Another zoonotic pathogen Vibrio spp. can develop a severe foodborne illness to humans including severe gastroenteritis, vomiting, dehydration, and even fatal infections (Rahman et al., 2020). Vibrio spp. can be transmitted to humans via ingestion of contaminated foods and/or water.
Bangladesh is known as a riverine country due to having numerous transboundary rivers. Having suitable weather and water bodies, Bangladesh attracts thousands of migratory birds every year. Many people of Bangladesh are directly or indirectly connected with rivers and wetlands. Their settlements have been developed with such water bodies (Rashid et al., 2015). These settlements expose humans to migratory birds and can pollute the environments with antibiotic resistant bacteria. Resistant bacteria can be transmitted to humans from various water bodies contaminated with fecal materials originating from migratory birds. In addition, resistant bacteria can be transmitted to other avian species including poultry species by duck-like poultry species/ migratory birds (Islam et al., 2021). The role of migratory birds as a source for transmission of such resistant bacteria is still being neglected in many parts of the world. As per know, there is no published data on the detection of MDR Enterococcus spp., Salmonella spp., and Vibrio spp. from migratory birds travelling to Bangladesh. Therefore, in our present study, we explored MDR bacteria among migratory birds in a selected area of Bangladesh.
2. Materials and methods
2.1. Ethical approval
The methodologies and related protocols used in this study were approved by the Institutional Ethical Committee (AWEEC/BAU/2019(14)).
2.2. Sample collection and processing
Only freshly dropped wet fecal samples were collected from the ground and tree leaves where the migratory birds were available. The sterile conditions were maintained during sample collection. A total of 66 fecal materials of migratory birds were collected from Baojani Baor, Mohammadpur Upazila (23.4056° N, 89.5686° E) in Magura district of Bangladesh during November 2019 and November 2020. The samples were collected by following the previously described procedure (Akter et al., 2020). In brief, each sample, collected by whirling a sterilized cotton bud into migratory bird’s dropping, was shifted to a separate sterilized zip lock bag with a particular tag number and directly transferred to the laboratory maintaining a cool chain. Each transferred sample was divided into two parts: one for seeding into 5 ml nutrient broth (HiMedia, India) and the other for taking into 5 ml Brain heart infusion (BHI) broth (Becton, Dickinson and Company, USA) containing 2% NaCl. All the broth containing sterile test tubes were then incubated aerobically for 18–24 h at 37 °C for the enrichment of the bacteria.
2.3. Isolation of bacteria
After incubation, one loopful of the sample from the nutrient broth was streaked on Enterococcus agar media (EAM) and Xylose-lysine deoxycholate (XLD) agar media, whereas samples from BHI broth was transferred on Thiosulfate-citrate-bile salts-sucrose (TCBS) agar media (HiMedia, India). Agar media were then incubated aerobically overnight at 3°C to obtain specific colonies. Oval-shaped yellowish colonies on EAM, central black colonies on XLD, and yellowish or green colonies on TCBS agar media were primarily suspected as Enterococcus spp., Salmonella spp., and Vibrio spp., respectively. For further confirmation, suspected colonies were subjected to Gram’s staining and biochemical tests (Facklam et al., 2002, Tawyabur et al., 2020, Zereen et al., 2019).
2.4. Molecular detection of bacteria
The final confirmation of isolated bacteria were done by polymerase chain reaction (PCR) assays targeting specific genes as mentioned in Table 1.
Table 1.
Primers used for the detection of different bacteria from fecal materials of migratory birds.
| Bacteria | Targeted genes | Primer sequence (5′-3′) | Amplicon size (bp) |
Annealing temperature (◦C) |
References |
|---|---|---|---|---|---|
| Enterococcus spp. | ddl | F: GCAAGGCTTCTTAGAGA R: CATCGTGTAAGCTAACTTC |
550 | 50 | Dutka-Malen S et al.(1995) |
| Salmonella spp. | invA | F: ATCAGTACCAGTCGTCTTATCTTGAT R: TCTGTTTACCGGGCATACCAT |
211 | 58 | Fratamico, 2003 |
| Vibrio spp. | VG C2694352 | F: GTCARATTGAAAARCARTTYGGTAAAGG R: ACYTTRATRCGNGTTTCRTTRCC |
689 | 55 | Kim et al., 2015 |
For PCR, bacterial genomic DNA was extracted by boiling and freeze-thawing method as previously described (Ievy et al., 2020). In brief, 1 ml of overnight culture was centrifuged at 5000 rpm for 5 min, and the pellet after discarding supernatant was then suspended into deionized water. The suspension was then boiled and cooled for 10 min in each step and centrifuged at 10,000 rpm for 10 min. Finally, the supernatant was collected as a DNA template and stored at −20 °C for further use.
A total of 20 μl of a final volume containing 10 µl of master mixture 2X (Promega, USA), 1 μl of each primer (100 pmol), 4 μl genomic DNA, and 4 μl of nuclease-free water was used to amplify each gene. The amplified PCR products were then analyzed by gel electrophoresis using 1.5% agarose gel. After staining with ethidium bromide, amplicon products were visualized under ultraviolet trans-illuminator (Biometra, Germany). 1 kb DNA ladder (Promega, Madison, WI, USA) was used to check the targeted amplicon size.
2.5. Antibiotic susceptibility test
The disk-diffusion method (Bauer et al., 1966) was used to perform an antibiotic susceptibility test (AST) of the isolated bacteria. Initially, the concentration of freshly grown bacteria was adjusted with 0.5 McFarland standard and subsequently spread on Mueller Hinton agar (HiMedia, India) media. In our current study, frequently used ten antibiotics under nine classes were selected namely fluoroquinolones (ciprofloxacin- 5 µg), aminoglycosides (gentamicin- 10 µg; streptomycin- 10 µg), tetracycline (tetracycline- 30 µg), macrolides (erythromycin- 15 µg), cephalosporins (ceftriaxone- 30 µg), carbapenems (meropenem- 10 µg), penicillins (ampicillin- 25 µg), glycopeptides (vancomycin- 30 µg), and amphenicols (chloramphenicol- 10 µg). All the discs were purchased from HiMedia, India. The results were interpreted by the guidelines of Clinical and Laboratory Standard Institute (CLSI) (CLSI, 2018), and where not possible, following the European Committee on Antimicrobial Susceptibility Testing (EUCAST, 2019). Multi-drug resistant (MDR) isolates were defined with the resistance against three or more classes of antibiotics (Sweeney et al., 2018).
2.6. Statistical analysis
2.6.1. Descriptive analysis
Data obtained from the present study were inserted into the Microsoft Excel-2013 (Los Angeles, CA, USA) and analyzed by the Statistical Package for the Social Sciences (SPSS) software (IBM SPSS-25.0, USA) and GraphPad Prism 8.4.2 (GraphPad Software, Inc.). By SPSS (version 25), a Pearson chi-square test for goodness-of-fit was done to assess the existence of the possible variations among the occurrence of isolated bacteria. A probability (p) value<0.05 was considered as statistically significant. In addition, binomial 95% confidence intervals were calculated in Graphpad Prism following the Wilson/Brown Hybrid method as previously described (Brown et al., 2001).
2.6.2. Bivariate analysis
By SPSS (version 25), a Pearson correlation test was performed to evaluate the possible correlation in between any of two antibiotics resistance to isolated bacteria. The statistical significant p-value was<0.05.
3. Results
3.1. Prevalence of bacteria
Out of 66 samples, 40 (60.61%) were confirmed to be positive for Enterococcus spp., which was significantly higher (Chi-square test, 95% CI, p= <0.001) compared to Salmonella spp. (21.21%) and Vibrio spp. (39.40%). The overall results of occurrence of isolated bacteria are represented to Table 2.
Table 2.
Occurrence of Enterococcus spp., Salmonella spp., and Vibrio spp. in fecal materials of migratory birds.
| Name of organisms (n = 66) | Positive isolates (%) | 95% CI (%) | p-value |
|---|---|---|---|
| Enterococcus spp. | 40 (60.61) | 48.55–71.50 | <0.001 |
| Salmonella spp. | 14 (21.21) | 13.08–32.51 | |
| Vibrio spp. | 26 (39.40) | 28.50–51.45 |
Here, a p-value<0.05 was deemed as statistically significant; CI = Confidence interval
3.2. Antibiotic susceptibility test
3.2.1. Antibiogram of Enterococcus spp.
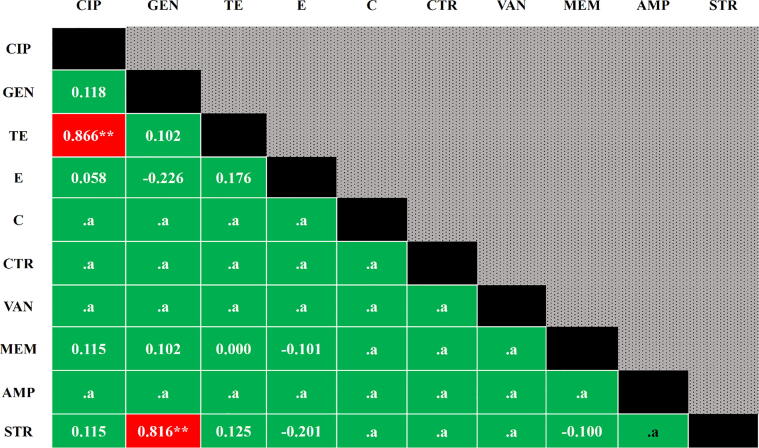
In antibiogram, all the Enterococcus isolates were found resistant to ampicillin, and frequent resistant to streptomycin (50%), meropenem (50%), erythromycin (45%), and gentamicin (40%) (Table 3). By bivariate analysis, high positive significant correlations were observed in between resistance profiles of ciprofloxacin and tetracycline (p=<0.001) and gentamicin and streptomycin (p=<0.001) (Fig. 1).
Table 3.
Overall antibiogram profile of isolated bacteria from fecal samples of migratory birds.
| Name of Antibiotics |
Enterococcus spp. (n = 40) |
Salmonella spp. (n = 14) |
Vibrio spp. (n = 26) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| R (%) | I (%) | S (%) | R (%) | I (%) | S (%) | R (%) | I (%) | S (%) | |
| CIP | 10 (25) | 5 (12.5) | 25 (62.5) | 2 (14.29) | 1 (7.14) | 11 (78.57) | 3 (11.54) | 5 (19.23) | 18 (69.23) |
| GEN | 16 (40) | 17 (42.5) | 7 (17.5) | 2 (14.29) | 0 (0) | 12 (85.71) | 3 (11.54) | 7 (26.92) | 16 (61.54) |
| TE | 8 (20) | 3 (7.5) | 29 (72.5) | 9 (64.28) | 3 (21.43) | 2 (14.29) | 9 (34.62) | 5 (19.23) | 12 (46.15) |
| E | 18 (45) | 22 (55) | 0 (0) | 6 (42.86) | 0 (0) | 8 (57.14) | 19 (73.08) | 5 (19.23) | 2 (7.69) |
| C | 0 (0) | 0 (0) | 40 (1 0 0) | 10 (71.42) | 2 (14.29) | 2 (14.29) | 3 (11.54) | 3 (11.54) | 20 (76.92) |
| CTR | 0 (0) | 5 (12.5) | 35 (87.5) | 0 (0) | 0 (0) | 14 (1 0 0) | 3 (11.54) | 0 (0) | 23 (88.46) |
| VAN | 0 (0) | 5 (12.5) | 35 (87.5) | 7 (50) | 3 (21.43) | 4 (28.57) | 20 (76.92) | 2 (7.69) | 4 (15.39) |
| MEM | 20 (50) | 0 (0) | 20 (50) | 3 (21.43) | 1 (7.14) | 10 (71.43) | 2 (7.69) | 4 (15.39) | 20 (76.92) |
| AMP | 40 (1 0 0) | 0 (0) | 0 (0) | 9 (64.28) | 3 (21.43) | 2 (14.29) | 20 (76.92) | 2 (7.69) | 4 (15.39) |
| STR | 20 (50) | 15 (37.5) | 5 (12.5) | 8 (57.14) | 3 (21.43) | 3 (21.43) | 8 (30.77) | 2 (7.69) | 16 (61.54) |
Here, CIP = Ciprofloxacin, GEN = Gentamicin, TE = Tetracycline, E = Erythromycin, C = Chloramphenicol, CTR = Ceftriaxone, VAN = Vancomycin, MEM = Meropenem, AMP = Ampicillin, STR = Streptomycin, R = Resistant, I = Intermediate, S = Sensitive, n = Number of isolates.
Fig. 1.
Pearson correlation coefficients in between any of two antibiotics resistance to Enterococcus isolates. A p-value <0.05 was deemed as statistically significant, ** Correlation is significant at the 0.01 level (2-tailed), .a cannot be computed because at least one of the variables is constant, Red = highly significant, Green = Non-significant, CIP = Ciprofloxacin, GEN = Gentamicin, TE = Tetracycline, E = Erythromycin, C = Chloramphenicol, CTR = Ceftriaxone, VAN = Vancomycin, MEM = Meropenem, AMP = Ampicillin, STR = Streptomycin.
3.2.2. Antibiogram of Salmonella spp.
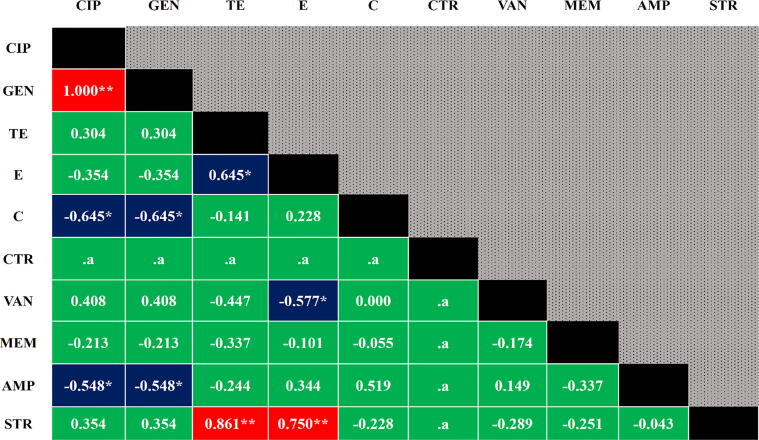
Our antibiogram study revealed that Salmonella isolates were frequently resistant to chloramphenicol (71.42%), tetracycline (64.28%), ampicillin (64.28%), streptomycin (57.14%), and erythromycin (42.86%) (Table 3). Based on bivariate analysis, ciprofloxacin and gentamicin (p=<0.001), tetracycline and streptomycin (p=<0.001), and erythromycin and streptomycin (p = 0.002) showed high positive significant correlation. Moderate significant correlations (both positive and negative) were also audited in between several antibiotic pairs which were resistant to Salmonella spp. (Fig. 2).
Fig. 2.
Pearson correlation coefficients in between any of two antibiotics resistance to Salmonella isolates. A p-value <0.05 was deemed as statistically significant, ** Correlation is significant at the 0.01 level (2-tailed), * Correlation is significant at the 0.05 level (2-tailed), .a cannot be computed because at least one of the variables is constant, Red = highly significant, Blue = moderately significant, Green = Non-significant, CIP = Ciprofloxacin, GEN = Gentamicin, TE = Tetracycline, E = Erythromycin, C = Chloramphenicol, CTR = Ceftriaxone, VAN = Vancomycin, MEM = Meropenem, AMP = Ampicillin, STR = Streptomycin.
3.2.3. Antibiogram of Vibrio spp.
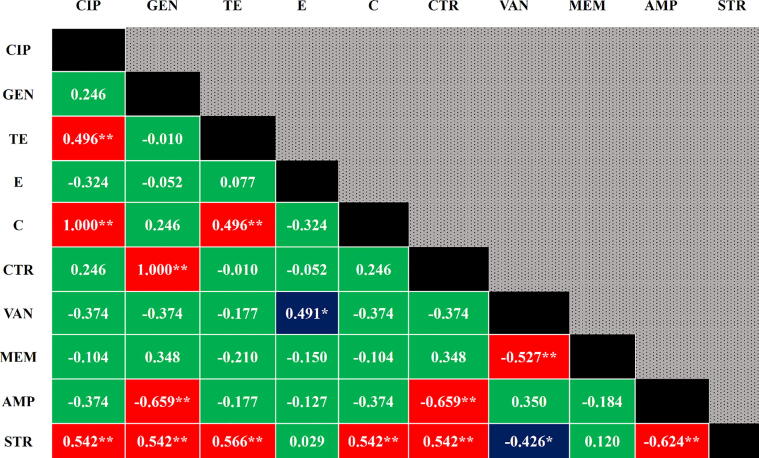
In the antibiogram study, 76.92% (20/26) Vibrio spp. isolates were found resistant to vancomycin and ampicillin, followed by 73.08% to erythromycin, 34.62% to tetracycline, and 30.77% to streptomycin (Table 3). High to moderate positive significant correlations were observed in between ciprofloxacin and chloramphenicol (p=<0.001), gentamicin and ceftriaxone (p=<0.001), tetracycline and streptomycin (p = 0.003), ciprofloxacin and streptomycin (p = 0.004), gentamicin and streptomycin (p = 0.004), chloramphenicol and streptomycin (p = 0.004), ceftriaxone and streptomycin (p = 0.004), ciprofloxacin and tetracycline (p = 0.01), chloramphenicol and tetracycline (p = 0.01), and erythromycin and vancomycin (p = 0.011). In addition, negative significant correlations were also observed (Fig. 3).
Fig. 3.
Pearson correlation coefficients in between any of two antibiotics resistance to Vibrio isolates. A p-valu e <0.05 was deemed as statistically significant, ** Correlation is significant at the 0.01 level (2-tailed), * Correlation is significant at the 0.05 level (2-tailed), Red = highly significant, Blue = moderately significant, Green = Non-significant, CIP = Ciprofloxacin, GEN = Gentamicin, TE = Tetracycline, E = Erythromycin, C = Chloramphenicol, CTR = Ceftriaxone, VAN = Vancomycin, MEM = Meropenem, AMP = Ampicillin, STR = Streptomycin.
3.3. MDR profiles of isolated bacteria
Of the isolated bacteria, 60% (24/40; 95% CI: 44.60–73.65%) Enterococcus spp., 85.71% (12/14; 95% CI: 60.06–97.46%) Salmonella spp., and 76.92% (20/26; 95% CI: 57.95–88.97%) Vibrio spp. isolates were MDR in nature. In addition, 13, six, and nine antibiotic resistance patterns were observed in Enterococcus spp., Salmonella spp., and Vibrio spp. isolates, respectively. Three isolates (one from each bacteria type) showed resistance against six (out of nine) classes of antibiotics (Table 4).
Table 4.
Multidrug resistance profile of the isolated bacteria from fecal materials of migratory birds.
| Pattern No. | Antibiotic resistance patterns | No. of antibiotics (Classes) | No. of MDR isolates (%) | Overall no. of MDR isolates (%) |
|---|---|---|---|---|
| Enterococcus spp. | ||||
| 1 | E, AMP, STR | 3 (3) | 2 (8.33) | 24/40 (60) |
| 2 | E, MEM, AMP | 3 (3) | 4 (16.67) | |
| 3 | CIP, MEM, AMP | 3 (3) | 1 (4.17) | |
| 4 | GEN, MEM, AMP, STR | 4 (3) | 5 (20.83) | |
| 5 | GEN, E, AMP, STR | 4 (3) | 2 (8.33) | |
| 6 | CIP, TE, AMP, STR | 4 (4) | 2 (8.33) | |
| 7 | CIP, TE, E, AMP | 4 (4) | 1 (4.17) | |
| 8 | GEN, E, MEM, AMP, STR | 5 (4) | 1 (4.17) | |
| 9 | CIP, GEN, MEM, AMP, STR | 5 (4) | 1 (4.17) | |
| 10 | CIP, GEN, TET, AMP, STR | 5 (4) | 1 (4.17) | |
| 11 | CIP, TE, E, MEM, AMP | 5 (5) | 2 (8.33) | |
| 12 | CIP, GEN, TET, MEM, AMP, STR | 6 (5) | 1 (4.17) | |
| 13 | CIP, GEN, TET, E, MEM, AMP, STR | 7 (6) | 1 (4.17) | |
| Salmonella spp. | ||||
| 1 | C, VAN, AMP | 3 (3) | 3 (25) | 12/14 (85.71) |
| 2 | TE, E, AMP, STR | 4 (4) | 1 (8.33) | |
| 3 | C, VAN, MEM, STR | 4 (4) | 1 (8.33) | |
| 4 | CIP, GEN, TE, VAN, STR | 5 (4) | 2 (16.67) | |
| 5 | TE, E, C, AMP, STR | 5 (5) | 4 (33.33) | |
| 6 | TE, E, C, VAN, AMP, STR | 6 (6) | 1 (8.33) | |
| Vibrio spp. | ||||
| 1 | E, VAN, AMP | 3 (3) | 9 (45) | 20/26 (76.92) |
| 2 | TE, E, STR | 3 (3) | 1 (5) | |
| 3 | TE, E, VAN, AMP | 4 (4) | 3 (15) | |
| 4 | GEN, E, CTR, VAN, STR | 5 (4) | 1 (5) | |
| 5 | GEN, E, CTR, MEM, STR | 5 (4) | 1 (5) | |
| 6 | TE, E, VAN, AMP, STR | 5 (5) | 2 (10) | |
| 7 | CIP, TE, C, AMP, STR | 5 (5) | 1 (5) | |
| 8 | CIP, GEN, TE, C, CTR, STR | 6 (5) | 1 (5) | |
| 9 | CIP, TE, E, C, VAN, STR | 6 (6) | 1 (5) | |
Here, CIP = Ciprofloxacin, GEN = Gentamicin, TE = Tetracycline, E = Erythromycin, C = Chloramphenicol, CTR = Ceftriaxone, VAN = Vancomycin, MEM = Meropenem, AMP = Ampicillin, STR = Streptomycin, MDR = Multi-drug resistant.
4. Discussion
Every year millions of migratory birds travel a long distance from one place to another across the globe. The migratory birds have a significant epidemiological role to carry and circulate resistant bacteria during their migration. Due to the paucity of information on MDR bacteria of migratory bird origin, in this study, we screened fecal materials of migratory birds travelling to Bangladesh to detect MDR Enterococcus spp., Salmonella spp., and Vibrio spp.
In this study among the three bacterial species (Enterococcus spp., Salmonella spp., and Vibrio spp.), Enterococcus spp. were detected as most frequently - in 60.61% (40/66) of the samples. The higher detection of Enterococcus spp. in the fecal samples of migratory birds is not unusual, as they are commensal bacteria in the intestine of many avian species (Silva et al., 2012). Our findings are lined with the detection rate of previous studies conducted in Portugal (63.30%) (Santos et al., 2013), and Northern Tunisia (65.8%) (Klibi et al. 2015). Conversely, a higher prevalence rate was reported from Poland (100%) (Stepien-Pysniak et al., 2017), and comparatively lower was also reported from Poland (51.92%) (Stępień-Pyśniak et al., 2018). In this study, all the Enterococcus isolates were found resistant to ampicillin and frequently resistant to streptomycin, meropenem, erythromycin, and gentamicin (Table 3) which are linked with the previous findings (Santos et al., 2013, Klibi et al., 2015; Stępień-Pyśniak et al., 2018; Yahia et al., 2018). Our findings revealed that vancomycin, ceftriaxone, and chloramphenicol were highly sensitive against all Enterococcus isolates. But surprisingly, a high proportion of meropenem resistant (50%) Enterococcus isolates were also detected- which is alarming for human health. Meropenem is under the carbapenem group of antibiotics which are only used in serious health conditions of humans (Islam et al., 2021). However, before drawing any final conclusion, these resistance need to be further confirmed by MIC and molecular techniques.
Salmonella spp. are ubiquitously distributed in nature, and are part of intestinal microbiota (Tsiodras et al., 2008). Salmonellosis caused by Salmonella spp., is one of the cognizant causes of death in wild migratory birds, however, apparently healthy birds can also carry these types of bacteria (Najdenski et al., 2018). In this study, a total of 21.21% (14/66) fecal samples of migratory birds were found positive for Salmonella spp. which is close to the previous studies conducted in Southern Europe (20.8%) (Antilles et al., 2021) and Czech Republic (24%) (Dolejska et al., 2009), but higher than the findings from Wei et al., 2020, Callaway et al., 2014. Salmonella spp. is an important pathogen that causes serious infections in both humans and animals. The detection of Salmonella spp. from migratory birds in our study, therefore, fortifies the importance of monitoring those birds to forecast the epizootic condition of the Salmonella to one-health components. Although the antibiotic resistance to Salmonella spp. is an ecological event, it mainly develops with the spontaneous competition among pathogens (Matias et al., 2016). In line with previous studies (Matias et al., 2016, Literak et al., 2007), we found Salmonella spp. were frequently resistant against chloramphenicol, ampicillin, tetracycline, streptomycin, vancomycin, and erythromycin; whereas 100% sensitive to ceftriaxone. Alike Enterococcus spp., Salmonella spp. isolates (three) also showed resistance to meropenem. It is noted that 11.54% of Salmonella isolates were resistant against ceftriaxone- a 3rd generation cephalosporin antibiotic that is used for severe Salmonella infections in humans (Lamb et al., 2002).
Vibrio spp. are frequently found in aquatic environments (Fernández-Delgado et al., 2016). Many of the Vibrio spp. such as V. cholera, V. parahaemolyticus are highly pathogenic in nature (Fu et al., 2019, Cardoso et al., 2018). In the present study, Vibrio spp. were detected in 39.40% (26/66) fecal samples of migratory birds which is very much similar to an earlier report in Venezuela (40%) (Fernández-Delgado et al. 2016). The presence of Vibrio spp. in fecal materials of migratory birds may be due to their feeding practices leading to the source of bacteria in their gut (Fernández-Delgado et al., 2016). Migratory birds can easily pick up Vibrio spp. from aquatic contamination during their feeding and moving behavior in the coastal areas. The continuous rising of antibiotic resistance reveals the Vibrio spp. as a notorious pathogen. It is highly alarming that, 34.62% Vibrio spp. isolates were resistant to tetracycline which also impact negatively on healthcare facilities. Tetracycline is an important antibiotic that is commonly and effectively used in cholera treatment (Cardoso et al., 2018). With resistance against these important antibiotics, isolated Vibrio spp. showed frequent resistance to ampicillin, erythromycin, and streptomycin. Previously, Cardoso et al. (2018) detected resistant Vibrio spp. in migratory birds from the north-central coast of the state of Rio de Janeiro, Brazil.
Observed variations in the detection rate of isolated bacteria and their resistance profiles compared to other studies could be due to spatial and temporal variations (geographical and seasonal distribution), types and species of migratory birds, sample size compared to previous studies. In addition, stressful conditions of migratory birds acquired during migration may vary the shedding rate of bacteria (Hubálek, 2004).
By bivariate analysis, strong significant correlations were observed between any of the two antibiotics which were resistant to isolated bacteria i.e. ciprofloxacin-tetracycline and gentamicin-streptomycin in Enterococcus spp. isolates; ciprofloxacin-gentamicin, tetracycline-streptomycin, and erythromycin-streptomycin in Salmonella spp. isolates; and ciprofloxacin-chloramphenicol and gentamicin-ceftriaxone in Vibrio spp. isolates (Fig. 1, Fig. 2, Fig. 3). The observed significant correlations among different antibiotics might be due to the misuse and overuse of antibiotics in humans, animals, and avian species in such areas where migratory birds inhabit (Islam et al., 2021). In addition, cross-contamination facilitates the dissemination of bacteria resistance against multifarious antibiotics in different environmental components.
Multidrug resistant bacteria are major health concern since often they are very difficult to treat. In addition, their treatment is expensive and lengthy. Surprisingly, among the isolates obtained in this study, 85.71% Salmonella spp., 76.92% Vibrio spp., and 60% Enterococcus spp. isolates were MDR in nature (Table 4). The results from our study suggested that migratory birds may convey and transport MDR bacteria from their habitats to far regions where there is no traditionally public awareness to prevent and control infectious diseases (Islam et al., 2021). In addition, migratory birds harboring these with MDR bacteria may carry and circulate them across long distances (Stępień-Pyśniak et al., 2018).
The aquatic environments are deemed as hotspots to circulate MDR bacteria. Human activities can contaminate the natural water bodies by disseminating antibiotic resistant and/or MDR bacteria. For example, the poor drainage system in animal farms, human settlements, healthcare facilities, pharmaceutical companies generates a huge amount of wastewater which can contaminate rivers and other water bodies where migratory birds inhabit. Natural water sources can also be affected by contaminated soil. Migratory birds can easily pick up resistant bacteria from those water bodies, spread them to surrounding aquatic environmental components, and ultimately impact negatively one-health components. In addition, they can also contaminate the environment with MDR bacteria harboring through fecal droppings. There are possibilities for entering these MDR bacteria from contaminated environments into the food chain.
5. Conclusion
AMR is a major global health crisis of the 21st century. This is the first study to detect MDR Enterococcus spp., Salmonella spp., and Vibrio spp. from migratory birds that seasonally travelled to Bangladesh. Our present study suggests that these migratory birds have the potential to carry and circulate MDR bacteria along their flyway. Detection of MDR bacteria in migratory birds is of great public health concern because of their potentiality to contaminate environments and transmit to one-health components. These birds, therefore, need to be kept under active AMR surveillance with strong one-health approach as a crucial step in combating AMR-related hazards.
CRediT authorship contribution statement
Md. Saiful Islam: Conceptualization, Methodology, Software, Formal analysis, Investigation, Data curation, Writing - original draft, Writing - review & editing, Visualization. Anamika Paul: Methodology, Investigation, Writing - original draft. Mithun Talukder: Investigation. Krishna Roy: Investigation. Md. Abdus Sobur: Formal analysis, Writing - review & editing, Visualization. Samina Ievy: Investigation. Md. Mehedi Hasan Nayeem: . Saifur Rahman: Conceptualization. K.H.M. Nazmul Hussain Nazir: Supervision. Muhammad Tofazzal Hossain: Writing - review & editing. Md. Tanvir Rahman: Conceptualization, Methodology, Validation, Writing - original draft, Writing - review & editing, Visualization, Supervision, Project administration.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
We are grateful to the Ministry of Science and Technology, Government of the People’s Republic of Bangladesh, for facilitating the research.
Funding
This research was funded by Bangladesh Agricultural University Research System (BAURES; 2019/8/BAU) and University Grants Commission (2020/28/UGC) of Bangladesh.
Footnotes
Peer review under responsibility of King Saud University.
References
- Akter M., Islam M.S., Islam M.A., Sobur M.A., Jahan M.S., Rahman S., Nazir K.N.H., Rahman M.T. Migratory birds as the potential source for the transmission of Aspergillus and other fungus to Bangladesh. J. Adv. Vet. Anim. Res. 2020;7:338–344. doi: 10.5455/javar.2020.g427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antilles N., García-Bocanegra I., Alba-Casals A., López-Soria S., Pérez-Méndez N., Saco M., González-Solís J., Cerdà-Cuéllar M. Occurrence and antimicrobial resistance of zoonotic enteropathogens in gulls from southern Europe. Sci. Total Environ. 2021;763 doi: 10.1016/j.scitotenv.2020.143018. [DOI] [PubMed] [Google Scholar]
- Atterby C., Ramey A.M., Hall G.G., Järhult J., Börjesson S., Bonnedahl J. Increased prevalence of antibiotic-resistant E. coli in gulls sampled in Southcentral Alaska is associated with urban environments. Infect. Ecol. Epidemiol. 2016;6:32334. doi: 10.3402/iee.v6.32334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer A.T., Kirby W.M.M., Sherris J.C., Turck M. Antibiotic susceptibility testing by a standardized single disc method. Am. J. Clin. Pathol. 1966;45:149–158. [PubMed] [Google Scholar]
- Brown L.D., Cai T.T., DasGupta A. Interval estimation for a binomial proportion. Stat. Sci. 2001;16:101–117. [Google Scholar]
- Callaway T.R., Edrington T.S., Nisbet D.J. Isolation of Escherichia coli O157: H7 and Salmonella from migratory brown-headed cowbirds (Molothrus ater), common grackles (Quiscalus quiscula), and cattle egrets (Bubulcus ibis) Foodborne Pathog. Dis. 2014;11:791–794. doi: 10.1089/fpd.2014.1800. [DOI] [PubMed] [Google Scholar]
- Cardoso M.D., Lemos L.S., Roges E.M., de Moura J.F., Tavares D.C., Matias C.A.R., Rodrigues D.P., Siciliano S. A comprehensive survey of Aeromonas sp. and Vibrio sp. in seabirds from southeastern Brazil: outcomes for public health. J. Appl. Microbiol. 2018;124:1283–1293. doi: 10.1111/jam.13705. [DOI] [PubMed] [Google Scholar]
- Chadfield M.S., Christensen J.P., Christensen H., Bisgaard M. Characterization of streptococci and enterococci associated with septicaemia in broiler parents with a high prevalence of endocarditis. Avian Pathol. 2004;33:610–617. doi: 10.1080/03079450400013089. [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI), 2018. Performance Standards for Antimicrobial Susceptibility Testing, M100-S28. Clinical and Laboratory Standards Institute, Wayne, PA.
- Díaz-Sánchez S., Sánchez S., Ewers C., Höfle U. Occurrence of avian pathogenic Escherichia coli and antimicrobial-resistant E. coli in red-legged partridges (Alectoris rufa): sanitary concerns of farming. Avian Pathol. 2012;41:337–344. doi: 10.1080/03079457.2012.687101. [DOI] [PubMed] [Google Scholar]
- Dolejska M., Bierošová B., Kohoutova L., Literak I., Čížek A. Antibiotic-resistant Salmonella and Escherichia coli isolates with integrons and extended-spectrum beta-lactamases in surface water and sympatric black-headed gulls. J. Appl. Microbiol. 2009;106:1941–1950. doi: 10.1111/j.1365-2672.2009.04155.x. [DOI] [PubMed] [Google Scholar]
- Facklam R.R., Carvalho M.D.G.S., Teixeira L.M. In: The Enterococci: pathogenesis, molecular biology, and antibiotic resistance. Gilmore M.S., Clewell D.B., Courvalin P., Dunny G.M., Murray B.E., Rice L.B., editors. ASM Press; Washington, D.C.: 2002. History, Taxonomy, Biochemical Characteristics, and Antibiotic Susceptibility Testing of Enterococci; pp. 1–54. 10.1128/9781555817923.ch1. [Google Scholar]
- Fernández-Delgado M., Sanz V., Giner S., Suárez P., Contreras M., Michelangeli F. Prevalence and distribution of Vibrio spp. in wild aquatic birds of the Southern Caribbean Sea, Venezuela, 2011–12. J. Wildl. Dis. 2016;52:621–626. doi: 10.7589/2015-06-154. [DOI] [PubMed] [Google Scholar]
- Fisher K., Phillips C. The ecology, epidemiology and virulence of Enterococcus. Microbiology. 2009;155:1749–1757. doi: 10.1099/mic.0.026385-0. [DOI] [PubMed] [Google Scholar]
- Fratamico P.M. Comparison of culture, polymerase chain reaction (PCR), TaqMan Salmonella, and Transia Card Salmonella assays for detection of Salmonella spp. in naturally-contaminated ground chicken, ground turkey, and ground beef. Mol. Cell. Probes. 2003;17:215–221. doi: 10.1016/S0890-8508(03)00056-2. [DOI] [PubMed] [Google Scholar]
- Fu S., Hao J., Yang Q., Lan R., Wang Y., Ye S., Liu Y., Li R. Long-distance transmission of pathogenic Vibrio species by migratory waterbirds: a potential threat to the public health. Sci. Rep. 2019;9:1–11. doi: 10.1038/s41598-019-52791-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther S., Aschenbrenner K., Stamm I., Bethe A., Semmler T., Stubbe A., Stubbe M., Batsajkhan N., Glupczynski Y., Wieler L.H., Ewers C. Comparable high rates of extended-spectrum-beta-lactamase-producing Escherichia coli in birds of prey from Germany and Mongolia. PloS One. 2012;7 doi: 10.1371/journal.pone.0053039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatha A.M., Divya P.S., Saramma A.V., Rahiman M., Krishnan K.P. Migratory bird, Branta leucopis (Barnacle goose), a potential carrier of diverse Escherichia coli serotypes into pristime Arctic environment. Curr. Sci. 2013;104:1078–1080. [Google Scholar]
- Hubálek Z. An annotated checklist of pathogenic microorganisms associated with migratory birds. J. Wildl. Dis. 2004;40:639–659. doi: 10.7589/0090-3558-40.4.639. [DOI] [PubMed] [Google Scholar]
- Ievy S., Islam M., Sobur M., Talukder M., Rahman M., Khan M.F.R., Rahman M. Molecular detection of avian pathogenic Escherichia coli (APEC) for the first time in layer farms in Bangladesh and their antibiotic resistance patterns. Microorganisms. 2020;8:1021. doi: 10.3390/microorganisms8071021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam M., Nayeem M., Hasan M., Sobur M., Ievy S., Rahman S., Kafi M., Ashour H.M., Rahman M. Virulence Determinants and Multidrug Resistance of Escherichia coli Isolated from Migratory Birds. Antibiotics. 2021;10:190. doi: 10.3390/antibiotics10020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.J., Ryu J.O., Lee S.Y., Kim E.S., Kim H.Y. Multiplex PCR for detection of the Vibrio genus and five pathogenic Vibrio species with primer sets designed using comparative genomics. BMC Microbiol. 2015;15:1–11. doi: 10.1186/s12866-015-0577-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klibi N., Amor I.B., Rahmouni M., Dziri R., Douja G., Said L.B., Lozano C., Boudabous A., Slama K.B., Mansouri R., Torres C. Diversity of species and antibiotic resistance among fecal enterococci from wild birds in Tunisia. Detection of vanA-containing Enterococcus faecium isolates. Eur. J. Wildl. Res. 2015;61:319–323. doi: 10.1007/s10344-014-0884-2. [DOI] [Google Scholar]
- Lamb H.M., Ormrod D., Scott L.J., Figgitt D.P. Ceftriaxone. Drugs. 2002;62:1041–1089. doi: 10.2165/00003495-200262070-00005. [DOI] [PubMed] [Google Scholar]
- Lin Y., Dong X., Sun R., Wu J., Tian L., Rao D., Zhang L., Yang K. Migratory birds-one major source of environmental antibiotic resistance around Qinghai Lake. China. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Literak I., Vanko R., Dolejska M., Čížek A., Karpíšková R. Antibiotic resistant Escherichia coli and Salmonella in Russian rooks (Corvus frugilegus) wintering in the Czech Republic. Lett. Appl. Microbiol. 2007;45:616–621. doi: 10.1111/j.1472-765X.2007.02236.x. [DOI] [PubMed] [Google Scholar]
- Matias, C.A.R., Pereira, I.A., de Araújo, M.D.S., Santos, A.F.M., Lopes, R.P., Christakis, S., Rodrigues, D.D.P., Siciliano, S., 2016. Characteristics of Salmonella spp. isolated from wild birds confiscated in illegal trade markets, Rio de Janeiro, Brazil. Biomed Res. Int. 2016, e3416864. DOI: 10.1155/2016/3416864. [DOI] [PMC free article] [PubMed]
- Najdenski H., Dimova T., Zaharieva M.M., Nikolov B., Petrova-Dinkova G., Dalakchieva S., Popov K., Hristova-Nikolova I., Zehtindjiev P., Peev S., Trifonova-Hristova A. Migratory birds along the Mediterranean-Black Sea Flyway as carriers of zoonotic pathogens. Can. J. Microbiol. 2018;64:915–924. doi: 10.1139/cjm-2017-0763. [DOI] [PubMed] [Google Scholar]
- Rahman M., Sobur M., Islam M., Ievy S., Hossain M., El Zowalaty M.E., Rahman A.M.M., Ashour H.M. Zoonotic Diseases: Etiology, Impact, and Control. Microorganisms. 2020;8:1405. doi: 10.3390/microorganisms8091405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramey A.M., Hernandez J., Tyrlöv V., Uher-Koch B.D., Schmutz J.A., Atterby C., Järhult J.D., Bonnedahl J. Antibiotic-resistant Escherichia coli in migratory birds inhabiting remote Alaska. EcoHealth. 2018;15:72–81. doi: 10.1007/s10393-017-1302-5. [DOI] [PubMed] [Google Scholar]
- Rashid M., Rakib M.M., Hasan B. Antimicrobial-resistant and ESBL-producing Escherichia coli in different ecological niches in Bangladesh. Infect. Ecol. Epidemiol. 2015;5:26712. doi: 10.3402/iee.v5.26712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos T., Silva N., Igrejas G., Rodrigues P., Micael J., Rodrigues T., Resendes R., Gonçalves A., Marinho C., Gonçalves D., Cunha R. Dissemination of antibiotic resistant Enterococcus spp. and Escherichia coli from wild birds of Azores Archipelago. Anaerobe. 2013;24:25–31. doi: 10.1016/j.anaerobe.2013.09.004. [DOI] [PubMed] [Google Scholar]
- Silva N., Igrejas G., Gonçalves A., Poeta P. Commensal gut bacteria: distribution of Enterococcus species and prevalence of Escherichia coli phylogenetic groups in animals and humans in Portugal. Ann. Microbiol. 2012;62:449–459. doi: 10.1007/s13213-011-0308-4. [DOI] [Google Scholar]
- Stępień-Pyśniak D., Hauschild T., Nowaczek A., Marek A., Dec M. Wild birds as a potential source of known and novel multilocus sequence types of antibiotic-resistant Enterococcus faecalis. J. Wildl. Dis. 2018;54:219–228. doi: 10.7589/2017-05-118. [DOI] [PubMed] [Google Scholar]
- Stepien-Pysniak D., Hauschild T., Rozanski P., Marek A. MALDI-TOF mass spectrometry as a useful tool for identification of Enterococcus spp. from wild birds and differentiation of closely related species. J. Microbiol. Biotechnol. 2017;27:1128–1137. doi: 10.4014/jmb.1612.12036. [DOI] [PubMed] [Google Scholar]
- Sweeney M.T., Lubbers B.V., Schwarz S., Watts J.L. Applying definitions for multidrug resistance, extensive drug resistance and pandrug resistance to clinically significant livestock and companion animal bacterial pathogens. J. Antimicrob. Chemother. 2018;73:1460–1463. doi: 10.1093/jac/dky043. [DOI] [PubMed] [Google Scholar]
- Tawyabur M., Islam M., Sobur M., Hossain M., Mahmud M., Paul S., Hossain M.T., Ashour H.M., Rahman M. Isolation and Characterization of Multidrug-Resistant Escherichia coli and Salmonella spp. from Healthy and Diseased Turkeys. Antibiotics. 2020;9:770. doi: 10.3390/antibiotics9110770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The European Committee on Antimicrobial Susceptibility Testing, 2019. Break point tables for interpretation of MICs and zone diameters, version 9.0 Available online: http://www.eucast.org (Accessed 31st March, 2021).
- Tsiodras S., Kelesidis T., Kelesidis I., Bauchinger U., Falagas M.E. Human infections associated with wild birds. J. Infect. 2008;56:83–98. doi: 10.1016/j.jinf.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei B., Shang K., Cha S.Y., Zhang J.F., Kang M., Jang H.K. Prevalence and potential risk of Salmonella enterica in migratory birds from South Korea. Vet. Microbiol. 2020;249 doi: 10.1016/j.vetmic.2020.108829. [DOI] [PubMed] [Google Scholar]
- Yahia H.B., Chairat S., Hamdi N., Gharsa H., Sallem R.B., Ceballos S., Torres C., Slama K.B. Antimicrobial resistance and genetic lineages of faecal enterococci of wild birds: Emergence of vanA and vanB2 harbouring Enterococcus faecalis. Int. J. Antimicrob. Agents. 2018;52:936–941. doi: 10.1016/j.ijantimicag.2018.05.005. [DOI] [PubMed] [Google Scholar]
- Zereen F., Akter S., Sobur M.A., Hossain M.T., Rahman M.T. Molecular detection of Vibrio cholerae from human stool collected from SK Hospital, Mymensingh, and their antibiogram. J. Adv. Vet. Anim. Res. 2019;6:451–455. doi: 10.5455/javar.2019.f367. [DOI] [PMC free article] [PubMed] [Google Scholar]