Abstract
Microbial interactions are increasingly recognized as an integral part of microbial physiology. Cell-cell communication mediated by quorum sensing and metabolite exchange is a formative element of microbial interactions. However, loss-of-function mutations in quorum-sensing components are common across diverse species. Furthermore, quorum sensing is modulated by small molecules and environmental conditions that may be altered in the presence of other microbial species. Recent evidence highlights how strain heterogeneity impacts microbial interactions. There is great potential for microbial interactions to act as selective pressures that influence the emergence of common mutations in quorum-sensing genes across the bacterial and fungal domains.
Keywords: quorum sensing, intraspecies, interspecies, microbial interactions, heterogeneity, mutations, evolution, signaling, cross-feeding
Introduction.
Microbial cells are hardly ever alone. Because microbes almost always live adjacent to kin or different microbial species, microbial interactions are intimately linked with microbial viability, activity and community structure. In fact, it has been proposed that obligate mutualism between microbes is a reason why many remain unculturable [1]. The continuous presence of neighbors necessitates control over inter- and intra-species interactions. Quorum sensing is one important mechanism that governs both interaction types. Although the regulatory components vary in composition and specific gene targets, commonalities exist across species (Fig. 1A). For example, quorum sensing frequently controls the production of catabolic enzymes, small inhibitory molecules, and biofilm related factors via signal-responsive transcription factors. Alongside its use in intraspecies signaling, quorum-sensing systems respond to the signals produced by other species in the local vicinity. Despite the importance of quorum-sensing pathways for intra- and inter-species relationships, loss-of-function mutations in quorum-sensing genes are frequently observed in bacteria and fungi (Fig 1B). At first this appears to present a paradox: since microbial neighbors are present for most microbial life, why might loss-of-function mutations in key mediators of microbial interactions be beneficial? Recent studies provide novel insight into this phenomenon. In this review, we will summarize frequent loss-of-function mutations in quorum sensing-related genes and discuss recent factors that influence the fitness of quorum-sensing mutants. We will review how heterogeneity in quorum sensing affects microbe-microbe interactions and discuss how microbial interactions may influence the rates at which these mutations arise.
Figure 1. Quorum sensing (QS) mediates microbial interactions between gram positives (G+), gram negatives (G−), and fungi.
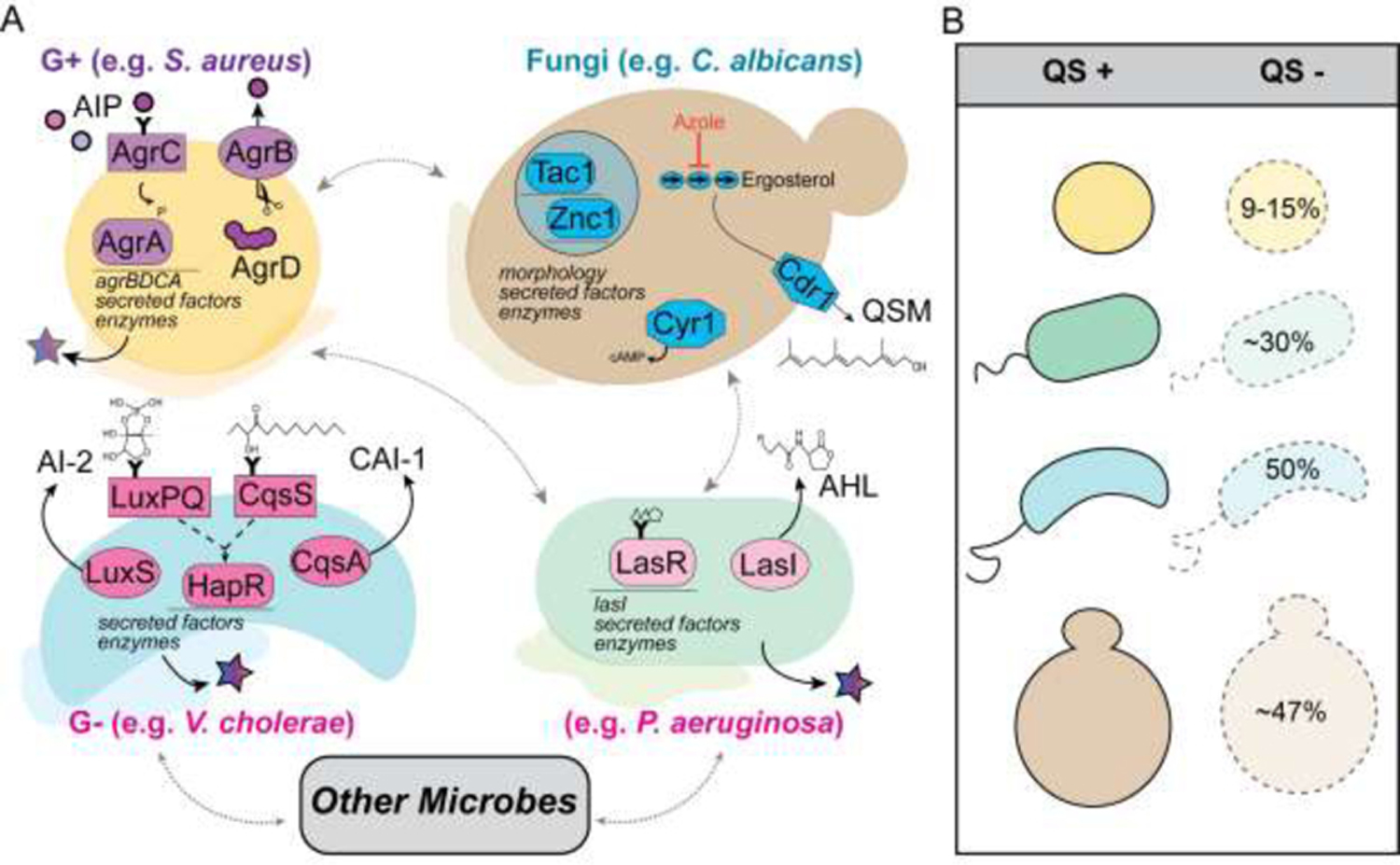
A. Diffusible autoinducers signal within and between species to regulate biofilm formation (colored base) and the production of secreted factors and enzymes (stars) which can modulate the extracellular milieu. Key elements of QS systems in Staphylococcus aureus, Candida albicans, Vibrio cholerae and Pseudomonas aeruginosa whose function is implicated in microbial interactions are shown. Shapes indicate protein activity: histidine kinases, rectangles; transcription factors, rounded rectangles bound to thin line of DNA; and enzymes involved in the synthesis or modification of the autoinducer signal, ovals. A small “Y” amended to a protein structure indicates that it binds the autoinducer. Structures for the autoinducers AI-2, CAI-1, acyl homoserine lactones such as 3OC12HSL, and the QS molecule (QSM) farnesol are shown. Autoinducing peptides (AIPs) from S. aureus and other species are shown as circles with color indicating species specificity. Dashed black lines indicate indirect activity, and dotted grey lines depict microbes with characterized interactions. B. In the species shown in A, strains with non-functional components of the QS system are frequently identified. In S. aureus (yellow) 9–15% of strains have dysfunctional Agr [63,64], in P. aeruginosa (green) roughly 30% have lasR loss-of-function alleles [65], in V. cholerae (blue) as high as 50% of isolates show defective quorum sensing [8], and although less clear in C. albicans (tan), roughly 47% of isolates show reduced farnesol production [66].
Loss-of-function mutations in cell-cell communication genes are common across diverse species.
Numerous studies suggest that the quorum-sensing systems in both gram-positive and gram-negative bacteria are frequently under selection. The agr quorum-sensing locus of Staphylococcus aureus (agrBDCA) exists in several types based on specific single nucleotide polymorphisms in agrC and agrD encoding a histidine sensor kinase and the precursor peptide of its cognate signal respectively, and loss-of-function mutations have been identified in isolates from persistent and chronic infections [2,3]. LasR, a master regulator of Pseudomonas aeruginosa quorum sensing, is selected against in vivo [4–6] and a non-functional lasR allele is present in one outgroup of the P. aeruginosa phylogeny [7]. In a survey of Vibrio cholerae clinical and natural isolates, loss-of-function mutations were prevalent in genes that encode the HapR transcriptional regulator or other quorum-sensing components [8]. Furthermore, experimental evolution selects for quorum sensing-defective mutants in S. aureus [9], P. aeruginosa [10], and the plant pathogen Burkholderia glumae [11]. Although frequent, these mutations do not necessarily abolish expression of the genes controlled by quorum sensing as S. aureus lacking Agr activity and P. aeruginosa lacking LasR function often sustain secondary mutations in other genes that restore downstream activity [3,10,12]. This restorative phenomenon highlights the conditional importance of quorum-sensing regulation.
Compared to bacteria, less is known about fungal quorum sensing though phenotypic evidence suggests that it is also variable. The levels of farnesol, a quorum sensing molecule detected in Candida albicans supernatants, varies between strains and across eight Candida species with at least one strain producing undetectable amounts and others producing ~ 60 µM, a concentration well-above that needed to induce a response in individual cells [13–15]. Sensitivity (or responsiveness) to farnesol (as measured by reduced growth, metabolic inhibition, reduced biofilm formation, or morphology changes) is also variable within and across Candida species including the emerging multi-drug resistant fungal pathogen C. auris [16,17]. Farnesol modulates the Cyr1 adenylate cyclase signaling pathway which influences the activity of the transcription factor Efg1 essential for the control of morphogenesis, and strains frequently lose Efg1 function in vivo [18] to suggest interactions may also be altered. Recently, farnesol was shown to induce the activity of two nuclear transcription factors, Tac1 and Znc1, which upregulate the Cdr1 multidrug efflux pump that may mediate farnesol transport [19]. The Cdr1 transporter sequence or Tac1 transcription factor copy number are often altered in drug resistant isolates suggesting that evolution of drug resistance may also modulate quorum-sensing regulation and vice versa. Repeated loss of quorum-sensing function across species has long been a subject of interest [20], and new findings may provide additional insight into the multitude of ways it both positively and negatively regulates fitness in different contexts.
Small molecules modulate quorum sensing during microbial interactions.
Signal and receptor promiscuity allow for “eavesdropping,” or communication between species and across domains of life. This may provide useful information about the local environment, but the ability of other species to modulate quorum sensing regulation may also make it less useful or even a liability. An investigation into the extent of signal promiscuity in the well-studied quorum sensing receptors of P. aeruginosa (LasR, RhlR, and QscR), Vibrio fischeri (LuxR), Chromobacterim violaceum (CviR), and Burkholderia thailandensis (BtaR1 and BtaR2) revealed most of these signal-activated transcription factors were strongly responsive to at least one non-cognate signal with structurally similar signals exhibiting more activity [21]. Examples of quorum sensing modulation by other species include autoinducing peptides (AIPs) from coagulase-negative staphylococci that compete for binding with S. aureus AgrC signal receptor and inhibit the expression of Agr-regulated targets [22]. Some studies suggest this may also be true across domains of life as C. albicans-produced farnesol has chemical characteristics in common with the LasR-specific acyl-homoserine lactone signal 3-oxo-C12-homoserine lactone (3OC12HSL) produced by P. aeruginosa. 3OC12HSL was able to modulate two farnesol-responsive phenotypes in C. albicans: fluconazole resistance and hyphal formation [23]. Likewise, farnesol has been shown to inhibit the non-acyl homoserine lactone-responsive pathway of the P. aeruginosa quorum-sensing system regulated by the transcription factor PqsR (MvfR) [24]. Some quorum-sensing systems detect both broad-spectrum and species-specific (i.e. cognate) quorum-sensing signals. For example, the species-specific cholera autoinducer-1 signal (CAI-1, (S)-3-hydroxytridecan-4-one) initiates quorum-sensing activation in wild-type V. cholerae but the timing of induction ultimately depends on the production of autoinducer-2, a furanosyl borate diester or tetrahydroxy furan, by neighboring microbes [25]. This coincidence mechanism is possible because of the higher threshold for AI-2-mediated activation compared to the species-specific autoregulatory signal [25]. In another study, exogenous and C. albicans-produced farnesol inhibited S. aureus production of the golden pigment staphyloxanthin, an important S. aureus virulence factor, and this was proposed to be due to competitive inhibition with the pigment precursor farnesyl-diphosphate for an upstream synthase in staphyloxanthin biosynthesis [26]. In turn, farnesol induced a transcriptional response that rendered S. aureus more tolerant to H2O2 stress and resistant to phagocytic killing [26]. These examples illustrate situations where quorum-sensing mutations may escape control from or avoid the effects on other species.
Environmental changes can trigger or repress quorum-sensing activity. C. albicans stimulates S. aureus Agr-dependent alpha hemolysis as a result of amino acid-dependent alkalinization of the medium [27,28]. For C. albicans, acidic conditions lead to transient un-masking of immunogenic beta-glucan in the cell wall, which can be re-masked through a quorum-sensing dependent process [29]. The effects of pH on quorum sensing highlight how physical environments can influence quorum sensing-mediated interactions between microbes. Quorum-sensing mutations may have added benefit if quorum-sensing regulated genes can be spuriously induced by environmental conditions. Activation of alternate regulatory pathways may enable expression of the quorum-sensing regulon in otherwise non-permissive conditions even in mutants lacking the canonical quorum-sensing systems.
What are the consequences of quorum sensing heterogeneity on microbial interactions?
Strains with deficiencies in quorum sensing can have unexpected intra- and inter-species interactions compared to their wild-type counterparts. Because the hapR mutant is often used to study V. cholerae biofilm formation due to its strong preference for biofilm over dispersal, the biofilm behaviors of hapR loss-of-function mutants are perhaps better understood than their HapR+ counterparts. By investigating the wild-type V. cholerae biofilm development over time, it is apparent that hapR loss-of-function mutants display some unique biofilm characteristics, including significantly more biomass [25]. The phenotypic differences observed between wild type and quorum-sensing loss-of-function mutants in V. cholerae speak to the importance of strain diversity. Intra-strain heterogeneity is particularly relevant in chronic conditions where in vivo evolution occurs and closely related isolates can coexist. For example, in mixed colony biofilms with wild-type P. aeruginosa, strains lacking the quorum-sensing regulator LasR hyper-produced the LasR-regulated antimicrobial phenazine pyocyanin [30]. This surprising result was mediated by lasR mutant siderophore production which was shown to promote citrate release particularly by the LasR+ wild type strain. Ultimately, citrate stimulated quorum-sensing activity in the lasR mutant, but not the wild type (Fig. 2). Likewise, C. albicans-produced farnesol has been shown to inhibit P. aeruginosa quorum sensing through Pseudomonas Quinolone Signal (PQS) in wild type whereas it activates PQS signaling in LasR loss-of-function mutants [24]. The loss of LasR function in P. aeruginosa can also lead to reduced rates of oxygen consumption [31], increased expression of microoxic fitness determinants [32], and enhanced catabolism of ethanol [33], which may improve its ability to grow in the presence of oxygen-consuming or ethanol-producing cells. Clinical antibiotic exposure selects for mutations at different rates based on genotype, and this may also reflect differences in susceptibility to antimicrobial compounds produced by other species. As an example, P. aeruginosa lasR mutants did not evolve antibiotic resistance as readily as the wild type [34]. Given the frequency with which quorum-sensing mutants are isolated, an understanding of how these types respond to wild-type strains, to other microbes and to the environment is warranted.
Figure 2. Model of intraspecies interactions between P. aeruginosa wild type (WT) and frequently observed lasR loss-of-function mutants (LasR−).
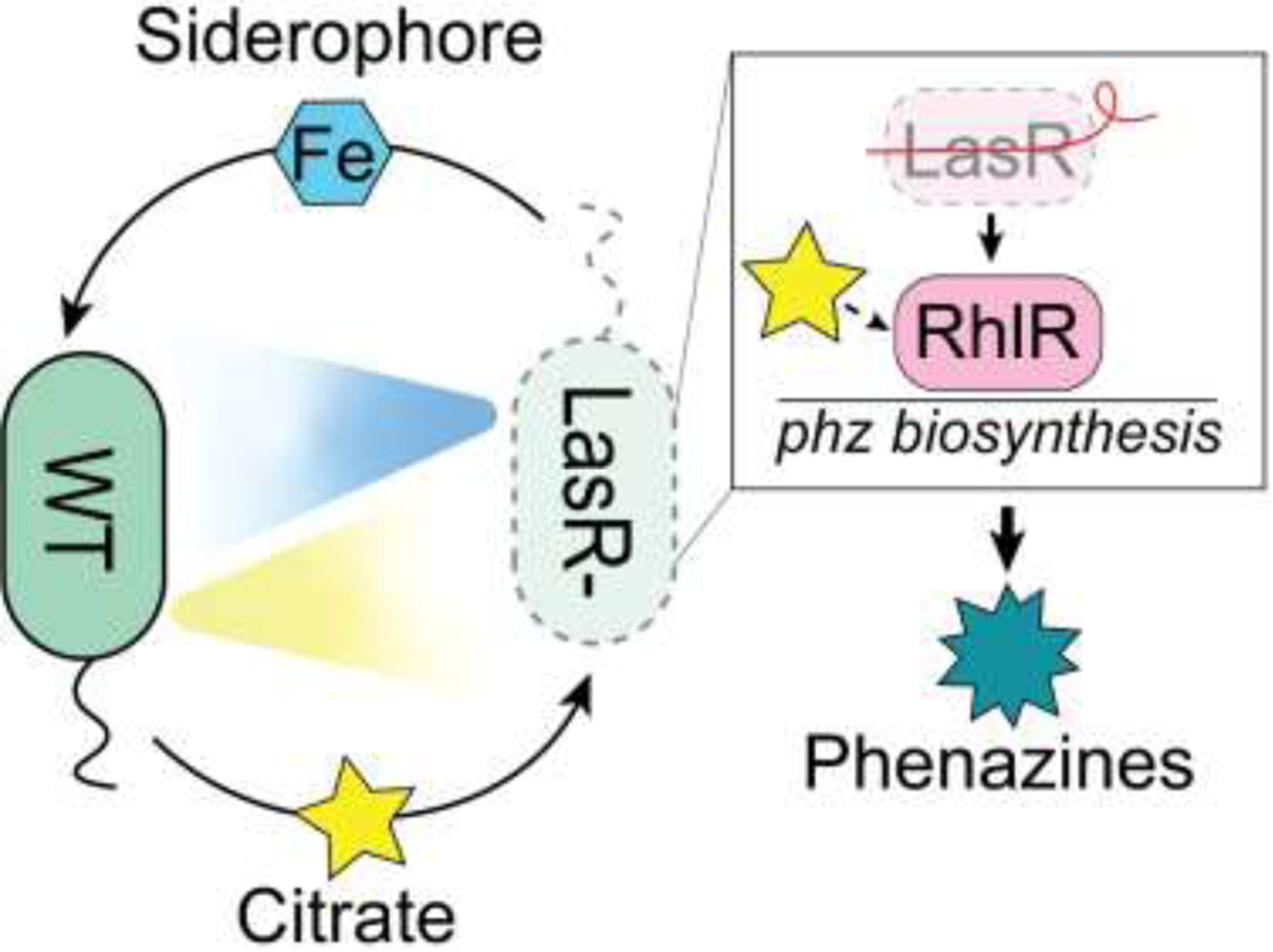
In response to the siderophore pyochelin (blue hexagon) produced by LasR− strains, WT (LasR+) P. aeruginosa releases citrate (star). Citrate preferentially stimulates quorum sensing in LasR− strains (and not WT) by activating the downstream quorum-sensing regulator RhlR resulting in the increased production of phenazines (decagram) and other secreted factors [30].
Do microbial interactions affect the selection for quorum sensing mutations?
In addition to altering composition and community function through mutualism, direct combat, resource competition, and cross-signaling, interactions between organisms can alter evolutionary trajectories [35]. Nearby microbial life can alter many abiotic factors like pH, redox, carbon/nitrogen availability, and metal content—all of which are potential selective pressures. In a cellular automation model investigating a mutualistic interspecies interaction, a mutation with a mildly self-detrimental effect but increasingly cooperative interspecies interaction (i.e. net indirect fitness benefit) rose above kin in four out of ten simulations, but only did so in structured, as opposed to well-mixed, environments [35]. Investigation of soil yeast communities revealed strain fitness was largely determined by the other organisms present [36]. Recent evidence suggests that different quorum-sensing signal variants in the Bacillus cereus group (known as ‘pherotypes’) may reflect environment-specific benefits [37]. Loss-of-function mutants in quorum sensing may provide indirect fitness advantages as a result of social cheating or metabolic differences.
There are a number of recent examples which illustrate how interspecies interactions may place selective pressure on quorum-sensing systems. A study of 23 clinical S. aureus isolates from polymicrobial diabetic ulcers identified one example wherein co-culture with P. aeruginosa significantly altered S. aureus agr copy number thus illustrating P. aeruginosa has the potential to select for differences in S. aureus agr quorum sensing components [38]. Experimental co-evolution of P. aeruginosa in the presence or absence of dense populations of S. aureus revealed differences in the selected outcomes. Specifically, mutations associated with the major outer membrane component, lipopolysaccharide, were only observed when S. aureus was present whereas mutations in the Wsp chemosensory system which controls biofilm formation were observed in both lineages to suggest an adaptation to environmental features rather than S. aureus itself [39]. Interestingly, lasR mutants were only observed in monoculture conditions to suggest they may have been selected against in the presence of S. aureus [39]. For V. cholerae, quorum sensing directly impacts the acquisition of new genetic material by controlling natural competence [40]. When density is low, the transcription factor AphA, well characterized for its induction of V. cholerae virulence and biofilm, inhibits DNA uptake through interactions with the promoter of the gene encoding the master regulator of competence, tfoX. When density is high and dispersal is initiated, the quorum-sensing regulator HapR activates the transcriptional cascade to allow competence [40]. In these cases, intra- and inter-species signaling indirectly and directly impact evolutionary trajectories.
Other common genetic changes may influence and be influenced by microbial interactions.
Like quorum-sensing molecules, metabolite exchange mediates microbial interactions. Often the metabolite pool is a direct result of microbial metabolism mediated by bi-directional or uni-directional exchange. Such exchanges can lead to the emergence of metabolic feedback loops that can drive antagonism or mutual growth. While other interactions increase respiration [41,42], P. aeruginosa antagonizes C. albicans via an antifungal phenazine [43] that inhibits respiration to promote C. albicans fermentation. The fermentation product ethanol stimulates phenazine production particularly when phosphate is limiting [44]. On the other hand, the bi-directional exchange of organic acids observed across three bacterial-fungal pairs of Betaproteobacteria and Mucoromycotina stimulated growth [45]. The recently described mutualistic interaction between the fungus Aspergillus nidulans and the bacterium Bacillus subtilis exemplify the dynamics of access inherent in microbial exchange [46]. B. subtilis supplied nutrition in the form of thiamine to promote A. nidulans hyphal growth, and in return B. subtilis benefited from the access to space on the growing hyphae [46]. Such metabolic differences are a determining factor in interaction outcomes. For example, in a three-species microcosm, P. aeruginosa dominance over S. aureus and Burkholderia cepacia depended on its ability to metabolize secondary metabolites through uni-directional cross-feeding [47]. Siderophores, a common secondary metabolite, are also mediators of bacterial-bacterial [30,48] and bacterial-fungal interactions [49,50] due to their roles in competition for iron, their contribution to ROS generation, and direct and indirect effects on metabolism. Inter-strain variability greatly influences the nature of microbial interactions, and this is most often the product of altered metabolism, including the secretion and uptake of secondary metabolites.
The impact of metabolism on microbial interactions highlights the importance of recognizing the variability of respiration and general metabolism phenotypes among strains within a species. Strain-dependent microbial interactions are reported between S. aureus and P. aeruginosa in which some pairings are capable of antagonism and others are not depending on their secretion profiles [51]. In fungi, mitochondria composition and function are diverse across fungal clades [52,53] and mitochondrial DNA has been proposed to display more heterogeneity than nuclear DNA [54]. Iron availability is a major driver of metabolism in many species, thus it is also important to recognize the variability in iron-acquisition strategies among strains. Iron-scavenging siderophores are the result of diverse biosynthetic gene clusters. The specific gene content and regulation can vary across isolates [55]. Pseudomonads make diverse siderophores, enabling the practice of siderophore typing among Pseudomonas isolates [56]. In a study looking at over 50 clinical isolates of P. aeruginosa, pyoverdine production was found to be very heterogenous [57] and isolates are often observed to lose pyoverdine production altogether [57,58]. The variation observed in siderophore type may be a result of the concurrent gain and loss of functionally redundant biosynthetic clusters through horizontal gene transfer [59] given presence on mobile elements [60]. The nutrient and metabolite spatiotemporal gradients [61] created during microbial interactions increase the opportunity for selection by generating more niches. However, one of the factors that may restrict the rise of metabolic and regulatory mutants from dominance is the dynamic environment itself.
Conclusions.
The diversity in functional genotype, especially with respect to quorum sensing, makes it important to consider the specific physiology of the interacting strains and genotypes when studying an interaction. Given the probability that a microbe is in the presence of others (either non-identical kin or distinct species), the impact of microbial interactions on evolution may be extensive. We posit that microbe-microbe interactions have the potential to drive and prevent selection of many of the mutations that commonly arise. The metabolic capacity of a community can depend on the relative initial ratios of each organism [62] and this likely extends to genotype heterogeneity in cases where the interacting variants have different metabolic properties. Continued investigation of commonly mutated genes or pathways and their roles in microbial interactions may reveal interesting commonalities. This will merit additional studies to understand further the consequences of these mutations on microbial interactions and to assess the contribution of microbial interactions for their selection.
Acknowledgements
Research reported in this publication was supported by grants from the National Institutes of Health to D.A.H. from the Cystic Fibrosis Foundation (HOGAN19G0), STANTO19R0 from the Cystic Fibrosis Foundation, and NIDDK P30-DK117469 (Dartmouth Cystic Fibrosis Research Center). Support for D.L.M came in part from NIH/NIAID T32AI007519 (D.L.M).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- [1].Pande S, Kost C: Bacterial unculturability and the formation of intercellular metabolic networks. Trends in Microbiology 2017, 25:349–361. [DOI] [PubMed] [Google Scholar]
- [2].Suligoy CM, Lattar SM, Noto Llana M, González CD, Alvarez LP, Robinson DA, Gómez MI, Buzzola FR, Sordelli DO: Mutation of agr is associated with the adaptation of Staphylococcus aureus to the host during chronic osteomyelitis. Frontiers in cellular and infection microbiology 2018, 8:18–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Altman DR, Sullivan MJ, Chacko KI, Balasubramanian D, Pak TR, Sause WE, Kumar K, Sebra R, Deikus G, Attie O, et al. : Genome plasticity of agr-defective Staphylococcus aureus during clinical infection. Infect Immun 2018, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Gabrielaite M, Johansen HK, Molin S, Nielsen FC, Marvig RL: Gene loss and acquisition in lineages of Pseudomonas aeruginosa evolving in cystic fibrosis patient airways. mBio 2020, 11:e02359–02320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Persyn E, Sassi M, Aubry M, Broly M, Delanou S, Asehnoune K, Caroff N, Crémet L: Rapid genetic and phenotypic changes in Pseudomonas aeruginosa clinical strains during ventilator-associated pneumonia. Scientific Reports 2019, 9:4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Subedi D, Vijay AK, Kohli GS, Rice SA, Willcox M: Comparative genomics of clinical strains of Pseudomonas aeruginosa strains isolated from different geographic sites. Sci Rep 2018, 8:15668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].García-Reyes S, Soto-Aceves MP, Cocotl-Yañez M, González-Valdez A, Servín-González L, Chávez GS: The outlier Pseudomonas aeruginosa strain ATCC 9027 harbors a defective LasR quorum-sensing transcriptional regulator. FEMS Microbiology Letters 2020, 367. [DOI] [PubMed] [Google Scholar]
- [8].Joelsson A, Liu Z, Zhu J: Genetic and phenotypic diversity of quorum-sensing systems in clinical and environmental isolates of Vibrio cholerae. Infection and immunity 2006, 74:1141–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Gor V, Takemura AJ, Nishitani M, Higashide M, Medrano Romero V, Ohniwa RL, Morikawa K: Finding of Agr phase variants in Staphylococcus aureus. mBio 2019, 10:e00796–00719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kostylev M, Kim DY, Smalley NE, Salukhe I, Greenberg EP, Dandekar AA: Evolution of the Pseudomonas aeruginosa quorum-sensing hierarchy. Proceedings of the National Academy of Sciences 2019, 116:7027–7032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Gnanasekaran G, Lim JY, Hwang I: Disappearance of quorum sensing in Burkholderia glumae during experimental evolution. Microb Ecol 2020, 79:947–959. [DOI] [PubMed] [Google Scholar]
- [12].Cruz RL, Asfahl KL, Van den Bossche S, Coenye T, Crabbé A, Dandekar AA: RhlR-regulated acyl-homoserine lactone quorum sensing in a cystic fibrosis isolate of Pseudomonas aeruginosa. mBio 2020, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hornby JM, Nickerson KW: Enhanced production of farnesol by Candida albicans treated with four azoles. Antimicrobial agents and chemotherapy 2004, 48:2305–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hornby JM, Jensen EC, Lisec AD, Tasto JJ, Jahnke B, Shoemaker R, Dussault P, Nickerson KW: Quorum sensing in the dimorphic fungus Candida albicans is mediated by farnesol. Applied and environmental microbiology 2001, 67:2982–2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Weber K, Sohr R, Schulz B, Fleischhacker M, Ruhnke M: Secretion of E,E-farnesol and biofilm formation in eight different Candida species. Antimicrobial agents and chemotherapy 2008, 52:1859–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Nagy F, Vitális E, Jakab Á, Borman AM, Forgács L, Tóth Z, Majoros L, Kovács R: In vitro and in vivo effect of exogenous farnesol exposure against Candida auris. Front Microbiol 2020, 11:957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Srivastava V, Ahmad A: Abrogation of pathogenic attributes in drug resistant Candida auris strains by farnesol. PLoS One 2020, 15:e0233102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Liang SH, Anderson MZ, Hirakawa MP, Wang JM, Frazer C, Alaalm LM, Thomson GJ, Ene IV, Bennett RJ: Hemizygosity enables a mutational transition governing fungal virulence and commensalism. Cell Host Microbe 2019, 25:418–431 e416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Liu Z, Rossi JM, Myers LC: Candida albicans Zn cluster transcription factors Tac1 and Znc1 are activated by farnesol to upregulate a transcriptional program including the multidrug efflux pump CDR1. Antimicrobial Agents and Chemotherapy 2018, 62:e00968–00918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Whiteley M, Diggle SP, Greenberg EP: Progress in and promise of bacterial quorum sensing research. Nature 2017, 551:313–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wellington S, Greenberg EP: Quorum sensing signal selectivity and the potential for interspecies cross talk. mBio 2019, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Peng P, Baldry M, Gless BH, Bojer MS, Espinosa-Gongora C, Baig SJ, Andersen PS, Olsen CA, Ingmer H: Effect of co-inhabiting coagulase negative Staphylococci on S. aureus agr quorum sensing, host factor binding, and biofilm formation. Front Microbiol 2019, 10:2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Bandara HMHN, Wood DLA, Vanwonterghem I, Hugenholtz P, Cheung BPK, Samaranayake LP: Fluconazole resistance in Candida albicans is induced by Pseudomonas aeruginosa quorum sensing. Scientific Reports 2020, 10:7769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Li W-R, Zeng T-H, Xie X-B, Shi Q-S, Li C-L: Inhibition of the pqsABCDE and pqsH in the pqs quorum sensing system and related virulence factors of the Pseudomonas aeruginosa PAO1 strain by farnesol. International Biodeterioration & Biodegradation 2020, 151:104956. [Google Scholar]
- [25].Bridges AA, Bassler BL: The intragenus and interspecies quorum-sensing autoinducers exert distinct control over Vibrio cholerae biofilm formation and dispersal. PLoS Biol 2019, 17:e3000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Vila T, Kong EF, Ibrahim A, Piepenbrink K, Shetty AC, McCracken C, Bruno V, Jabra-Rizk MA: Candida albicans quorum-sensing molecule farnesol modulates staphyloxanthin production and activates the thiol-based oxidative-stress response in Staphylococcus aureus. Virulence 2019, 10:625–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Todd OA, Noverr MC, Peters BM: Candida albicans impacts Staphylococcus aureus alpha-toxin production via extracellular alkalinization. mSphere 2019, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Todd OA, Fidel PL Jr., Harro JM, Hilliard JJ, Tkaczyk C, Sellman BR, Noverr MC, Peters BM: Candida albicans augments Staphylococcus aureus virulence by engaging the Staphylococcal agr quorum sensing system. mBio 2019, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Cottier F, Sherrington S, Cockerill S, Del Olmo Toledo V, Kissane S, Tournu H, Orsini L, Palmer GE, Pérez JC, Hall RA: Remasking of Candida albicans β-Glucan in response to environmental pH Is regulated by quorum sensing. mBio 2019, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Mould DL, Botelho NJ, Hogan DA: Intraspecies signaling between common variants of Pseudomonas aeruginosa increases production of quorum-sensing-controlled virulence factors. mBio 2020, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hoffman LR, Richardson AR, Houston LS, Kulasekara HD, Martens-Habbena W, Klausen M, Burns JL, Stahl DA, Hassett DJ, Fang FC, et al. : Nutrient availability as a mechanism for selection of antibiotic tolerant Pseudomonas aeruginosa within the CF airway. PLoS Pathog 2010, 6:e1000712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Clay ME, Hammond JH, Zhong F, Chen X, Kowalski CH, Lee AJ, Porter MS, Hampton TH, Greene CS, Pletneva EV, et al. : Pseudomonas aeruginosa lasR mutant fitness in microoxia is supported by an Anr-regulated oxygen-binding hemerythrin. Proceedings of the National Academy of Sciences 2020, 117:3167–3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Crocker AW, Harty CE, Hammond JH, Willger SD, Salazar P, Botelho NJ, Jacobs NJ, Hogan DA: Pseudomonas aeruginosa ethanol oxidation by AdhA in low-oxygen environments. J Bacteriol 2019, 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Hernando-Amado S, Sanz-Garcia F, Martinez JL: Antibiotic resistance evolution is contingent on the quorum-sensing response in Pseudomonas aeruginosa. Mol Biol Evol 2019, 36:2238–2251. [DOI] [PubMed] [Google Scholar]
- [35].Gorter FA, Manhart M, Ackermann M: Understanding the evolution of interspecies interactions in microbial communities. Philosophical Transactions of the Royal Society B: Biological Sciences 2020, 375:20190256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Bleuven C, Nguyen GQ, Després PC, Filteau M, Landry CR: Competition experiments in a soil microcosm reveal the impact of genetic and biotic factors on natural yeast populations. The ISME Journal 2020, 14:1410–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Zhou L, Slamti L, Lereclus D, Raymond B: Optimal response to quorum-sensing signals varies in different host environments with different pathogen group size. mBio 2020, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Matias C, Serrano I, Van-Harten S, Mottola C, Mendes JJ, Tavares L, Oliveira M: Polymicrobial interactions influence the agr copy number in Staphylococcus aureus isolates from diabetic foot ulcers. Antonie Van Leeuwenhoek 2018, 111:2225–2232. [DOI] [PubMed] [Google Scholar]
- [39].Tognon M, Kohler T, Gdaniec BG, Hao Y, Lam JS, Beaume M, Luscher A, Buckling A, van Delden C: Co-evolution with Staphylococcus aureus leads to lipopolysaccharide alterations in Pseudomonas aeruginosa. ISME J 2017, 11:2233–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Haycocks JRJ, Warren GZL, Walker LM, Chlebek JL, Dalia TN, Dalia AB, Grainger DC: The quorum sensing transcription factor AphA directly regulates natural competence in Vibrio cholerae. PLoS Genet 2019, 15:e1008362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Rivett DW, Lilley AK, Connett GJ, Carroll MP, Legg JP, Bruce KD: Contributions of composition and interactions to bacterial respiration are reliant on the phylogenetic similarity of the measured community. Microbial Ecology 2017, 74:757–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Tognon M, Kohler T, Luscher A, van Delden C: Transcriptional profiling of Pseudomonas aeruginosa and Staphylococcus aureus during in vitro co-culture. BMC Genomics 2019, 20:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Morales DK, Grahl N, Okegbe C, Dietrich LEP, Jacobs NJ, Hogan DA: Control of Candida albicans metabolism and biofilm formation by Pseudomonas aeruginosa phenazines. mBio 2013, 4:e00526–00512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Doing G, Koeppen K, Occipinti P, Harty CE, Hogan DA: Conditional antagonism in co-cultures of Pseudomonas aeruginosa and Candida albicans: An intersection of ethanol and phosphate signaling distilled from dual-seq transcriptomics. PLoS Genet 2020, 16:e1008783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Uehling JK, Entler MR, Meredith HR, Millet LJ, Timm CM, Aufrecht JA, Bonito GM, Engle NL, Labbe JL, Doktycz MJ, et al. : Microfluidics and metabolomics reveal symbiotic bacterial-fungal interactions between Mortierella elongata and Burkholderia include metabolite exchange. Front Microbiol 2019, 10:2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Abeysinghe G, Kuchira M, Kudo G, Masuo S, Ninomiya A, Takahashi K, Utada AS, Hagiwara D, Nomura N, Takaya N, et al. : Fungal mycelia and bacterial thiamine establish a mutualistic growth mechanism. Life Sci Alliance 2020, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Morgan BG, Warren P, Mewis RE, Rivett DW: Bacterial dominance is due to effective utilisation of secondary metabolites produced by competitors. Scientific Reports 2020, 10:2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Li K, Gifford AH, Hampton TH, O’Toole GA: Availability of zinc impacts interactions between Streptococcus sanguinis and Pseudomonas aeruginosa in coculture. J Bacteriol 2020, 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Pierce EC, Morin M, Little JC, Liu RB, Tannous J, Keller NP, Pogliano K, Wolfe BE, Sanchez LM, Dutton RJ: Bacterial-fungal interactions revealed by genome-wide analysis of bacterial mutant fitness. Nat Microbiol 2021, 6:87–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Sass G, Nazik H, Penner J, Shah H, Ansari SR, Clemons KV, Groleau MC, Dietl AM, Visca P, Haas H, et al. : Studies of Pseudomonas aeruginosa mutants indicate pyoverdine as the central factor in inhibition of Aspergillus fumigatus biofilm. J Bacteriol 2018, 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Pallett R, Leslie LJ, Lambert PA, Milic I, Devitt A, Marshall LJ: Anaerobiosis influences virulence properties of Pseudomonas aeruginosa cystic fibrosis isolates and the interaction with Staphylococcus aureus. Scientific Reports 2019, 9:6748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].De Chiara M, Friedrich A, Barré B, Breitenbach M, Schacherer J, Liti G: Discordant evolution of mitochondrial and nuclear yeast genomes at population level. BMC biology 2020, 18:49–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Sun N, Parrish RS, Calderone RA, Fonzi WA: Unique, diverged, and conserved mitochondrial functions influencing Candida albicans respiration. mBio 2019, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].De Chiara M, Friedrich A, Barré B, Breitenbach M, Schacherer J, Liti G: Discordant evolution of mitochondrial and nuclear yeast genomes at population level. BMC Biology 2020, 18:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Searle LJ, Méric G, Porcelli I, Sheppard SK, Lucchini S: Variation in siderophore biosynthetic gene distribution and production across environmental and faecal populations of Escherichia coli. PloS one 2015, 10:e0117906–e0117906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Meyer J-M, Geoffroy VA, Baida N, Gardan L, Izard D, Lemanceau P, Achouak W, Palleroni NJ: Siderophore typing, a powerful tool for the identification of fluorescent and nonfluorescent Pseudomonads. Applied and Environmental Microbiology 2002, 68:2745–2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Kang D, Revtovich AV, Chen Q, Shah KN, Cannon CL, Kirienko NV: Pyoverdine-dependent virulence of Pseudomonas aeruginosa isolates from cystic fibrosis patients. Frontiers in Microbiology 2019, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Schick A, Kassen R: Rapid diversification of Pseudomonas aeruginosa in cystic fibrosis lung-like conditions. Proceedings of the National Academy of Sciences 2018, 115:10714–10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Bruns H, Crüsemann M, Letzel A-C, Alanjary M, McInerney JO, Jensen PR, Schulz S, Moore BS, Ziemert N: Function-related replacement of bacterial siderophore pathways. The ISME Journal 2018, 12:320–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Lam MMC, Wick RR, Wyres KL, Gorrie CL, Judd LM, Jenney AWJ, Brisse S, Holt KE: Genetic diversity, mobilisation and spread of the yersiniabactin-encoding mobile element ICEKp in Klebsiella pneumoniae populations. Microbial genomics 2018, 4:e000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Gupta S, Ross TD, Gomez MM, Grant JL, Romero PA, Venturelli OS: Investigating the dynamics of microbial consortia in spatially structured environments. Nat Commun 2020, 11:2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Gao C-H, Cao H, Cai P, Sørensen SJ: The initial inoculation ratio regulates bacterial coculture interactions and metabolic capacity. The ISME Journal 2021, 15:29–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Traber KE, Lee E, Benson S, Corrigan R, Cantera M, Shopsin B, Novick RP: agr function in clinical Staphylococcus aureus isolates. Microbiology (Reading, England) 2008, 154:2265–2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Shopsin B, Drlica-Wagner A, Mathema B, Adhikari RP, Kreiswirth BN, Novick RP: Prevalence of agr dysfunction among colonizing Staphylococcus aureus strains. J Infect Dis 2008, 198:1171–1174. [DOI] [PubMed] [Google Scholar]
- [65].Hoffman LR, Kulasekara HD, Emerson J, Houston LS, Burns JL, Ramsey BW, Miller SI: Pseudomonas aeruginosa lasR mutants are associated with cystic fibrosis lung disease progression. Journal of Cystic Fibrosis 2009, 8:66–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Jung S-I, Shin JH, Kim SH, Kim J, Kim JH, Choi MJ, Chung E-K, Lee K, Koo SH, Chang HH, et al. : Comparison of E,E-farnesol secretion and the clinical characteristics of Candida albicans bloodstream isolates from different multilocus sequence typing clades. PLOS ONE 2016, 11:e0148400. [DOI] [PMC free article] [PubMed] [Google Scholar]


