Abstract
The coronavirus disease 2019 (COVID-19) pandemic caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has claimed millions of lives to date. Antigenic drift has resulted in viral variants with putatively greater transmissibility, virulence, or both. Early and near real-time detection of these variants of concern (VOC) and the ability to accurately follow their incidence and prevalence in communities is wanting. Wastewater-based epidemiology (WBE), which uses nucleic acid amplification tests to detect viral fragments, is a reliable proxy of COVID-19 incidence and prevalence, and thus offers the potential to monitor VOC viral load in a given population. Here, we describe and validate a primer extension PCR strategy targeting a signature mutation in the N gene of SARS-CoV-2. This allows quantification of B.1.1.7 versus non-B.1.1.7 allele frequency in wastewater without the need to employ quantitative RT-PCR standard curves. We show that the wastewater B.1.1.7 profile correlates with its clinical counterpart and benefits from a near real-time and facile data collection and reporting pipeline. This assay can be quickly implemented within a current SARS-CoV-2 WBE framework with minimal cost; allowing early and contemporaneous estimates of B.1.1.7 community transmission prior to, or in lieu of, clinical screening and identification. Our study demonstrates that this strategy can provide public health units with an additional and much needed tool to rapidly triangulate VOC incidence/prevalence with high sensitivity and lineage specificity.
Keywords: COVID-19, Alpha variant, Variant of concern, WBE, Public health
Graphical abstract
1. Introduction
Since the start of the COVID-19 pandemic, the possibility of SARS-CoV-2 antigenic drift, whereby genetic mutation gives rise to more transmissible and/or virulent viruses was identified as a potential risk to public health (Pachetti et al., 2020). SARS-CoV-2, being a single-stranded RNA (ssRNA) virus, is susceptible to frequent mutation (Duffy, 2018; Holmes and Rambaut, 2004; Kustin and Stern, 2021; Sanjuán et al., 2010). Generally, most viral mutations have a low impact on disease in populations (Grubaugh et al., 2020), but in late 2020, several concurrent mutations emerged in circulating SARS-CoV-2 genomes which increase infectivity (Chen et al., 2020; Korber et al., 2020) and potentially decrease sensitivity to neutralizing antibodies (Gaebler et al., 2021; Wang et al., 2021). As of May 2021, these variants of concern (VOC) as declared by the WHO include B.1.1.7 (α), B.1.351 (β), P.1 (γ), and B.1.617.2 (δ) (Konings et al., 2021). With communities around the world already reeling from the effects of the initial pandemic waves, the putative increase in morbidity and mortality associated with VOC infection relative to that of earlier variants, is affecting the resilience of health care systems and communities.
SARS-CoV-2 wastewater-based epidemiology (WBE) is being adopted as a means of surveillance in many jurisdictions around the world (reviewed in Hamouda et al. 2021). The common method currently used to detect fragments of SARS-CoV-2 viral RNA in wastewater is quantitative RT-PCR (qRT-PCR) utilizing hydrolysis probe and primer sets that were validated for clinical testing (Vogels et al., 2020). The high specificity and sensitivity of the CDC “2019-nCoV N1” (N1) and ”2019-nCoV N2” (N2) probe/primer sets seen in clinical samples (Vogels et al., 2020), appear to be mirrored in wastewater matrices (Hamouda et al., 2021) although relative sensitivities between the sample types haven't been formally investigated. N1 and N2 assays are thus used by many WBE research groups including those participating in the provincial wastewater surveillance initiative in Ontario, Canada. Critically, these qRT-PCR assays currently do not distinguish VOCs. Here, we describe and validate a qRT-PCR assay that targets a B.1.1.7-specific allele that lies adjacent to the N1 region and which employs new forward primers along with the existing N1 probe and reverse primer. This assay uses an allele-specific primer extension strategy capable of discriminating single nucleotide variants in a pooled sample such as wastewater. As such, this strategy can be employed to detect other VOCs as they emerge.
2. Materials and methods
2.1. Water resource recovery facility description and sample collection
24 h composite samples of primary clarified sludge (PCS) were collected from the Ottawa, Ontario (pop. ∼1M) water resource recovery facility (WRRF) between January 16 and April 28, 2021 (n=33). At the initiation of sampling there was no known community transmission of B.1.1.7 in this region. 24 h composite post-grit influent and grab samples of PCS were collected from the Barrie, Ontario (pop. ∼150K) WRRF on January 26, 2021 (n=1 for each sample type) during an institutional B.1.1.7 outbreak. Both sampled WRRFs are designed and operated as conventional treatment trains with influent entering screens and flowing to grit chambers followed by primary clarifiers and then activated sludge units. WRRF characteristics are outlined in Table 1 .
Table 1.
Characteristics of the studied water resource recovery facilities (WRRF) in Ottawa and Barrie. The population serviced and average daily and peak design flow rates in million litres per day (MLD) are indicated together with annual average influent concentrations of total suspended solids (TSS), carbonaceous biological oxygen demand (cBOD5), ammonia-nitrogen (NH3-N) and total phosphorus (TP).
| WRRF Characteristic | Ottawa | Barrie |
|---|---|---|
| Population serviced (approx. number of inhabitants) | 1,000,000 | 135,000 |
| Average (peak) rated design flow (MLD) | 545 (1363) | 76 (156) |
| Annual average influent [TSS] (mg/L) | 323 | 294 |
| Annual average influent [cBOD5] (mg/L) | 171 | 132 |
| Annual average influent [NH3-N] (mg/L) | 30.6 | 24.7 |
| Annual average influent [TP] (mg/L) | 5.8 | 5.3 |
2.2. Isolation of wastewater solids and RNA extraction
The solids in influent collected at the Barrie, Ontario WRRF were first allowed to settle at 4°C for one hour prior to decanting. Settled influent solids or PCS samples collected at Barrie or Ottawa WRRFs were well-mixed and 40 ml transferred to a 50 mL round-bottom centrifuge tube. Samples were centrifuged at 10,000 x g in a fixed angle rotor for 45 min at 4°C. The supernatant was discarded, taking care not to disturb the pellet. Samples were centrifuged a second time at 10,000 x g for 5 min and the remaining supernatant decanted. The resulting pellets were transferred to a RNase-free microfuge tube, the tared weight was recorded, and the RNA was immediately extracted. Total RNA was extracted using the RNeasy PowerMicrobiome Kit (Qiagen, Germantown, MD) on a QIAcube Connect automated extraction platform, with a modified protocol. The following changes to the manufacturer's protocol were performed: (1) 250 ± 15 mg of sample pellet was added to the initial extraction step in place of 200 µL of liquid sample, and (2) the optional phenol-chloroform step was instead performed with Trizol LS reagent (ThermoFisher, Ottawa, Canada) prior to vortexing and centrifugation. The resulting aqueous phase was retained and processed as per the original manufacturer's protocol and included the DNase treatment step. RNA was eluted in 100 µl of RNase-free water.
2.3. Human feces collection and RNA extraction
Feces were collected from confirmed COVID-19 positive in-patients at the Ottawa Hospital. The patient specimens used in this study were anonymous donations, collected with consent. Their collection and use were approved by the Ottawa Health Science Network Research Ethics Board (OHSN-REB) under REB 20200235-01H. For this study, feces from 2 confirmed COVID-19 patients being treated in Intensive Care was pooled and approximately 200 μl of this fecal sample (n=1) was extracted using the same method as for the wastewater solids detailed above.
2.4. qRT-PCR
One-step qRT-PCR was performed with TaqMan® Fast Virus 1-Step Master Mix (ThermoFisher, Ottawa, Canada). The N1 assay was performed using the premixed 2019-nCoV N1 Assay-RUO probe/primers set (500/125 nM, respectively) (IDT, Kanata, Canada). Allele-specific qRT-PCR was performed in parallel to detect non-B.1.1.7 alleles (D3) or only the B.1.1.7 allele (D3L) using newly designed forward primers combined with the N1 probe and reverse primers at 500, 125 and 500 nM, respectively. Sequences of the probes/primers are as follows: 5’-GACCCCAAAATCAGCGAAAT-3’ (US-CDC_2019-nCoV_N1_for); 5’-6-FAM-ACCCCGCAT/ZEN/TACGTTTGGTGGACC-IOWA BLACK FQ-3’ (US-CDC_2019-nCoV_N1_probe); 5’-TCTGGTTACTGCCAGTTGAATCTG-3’ (US-CDC_2019-nCoV_N1_rev); 5’-CATCTAAACGAACAAACTAAAATGTCTGAT-3’ (D3_for); 5’-CATCTAAACGAACAAACTAAATGTCTCTA-3’ (D3L_for). Critically, the D3L_for primer used in this study also incorporates a deletion at nt 28271 (underlined nucleotide in the D3_for primer above) that emerged in GISAID-deposited B.1.1.7 sequences from the UK in January 2021 when we were designing this assay. qRT-PCR reactions were run in triplicate using 1.5 µl of RNA input in a final reaction volume of 10 μl. Cycling was performed on a CFX Connect qPCR thermocycler (Bio-Rad, Hercules, CA) as follows: RT at 50°C, 5 min, followed by RT inactivation, polymerase activation and template denaturation at 95°C for 20 s, and 45 cycles of denaturation (95°C/3s), then annealing/extension (55°C/45s). No-template controls (NTC) showed either no amplification after 45 cycles or, rarely, poor amplification above 40 cycles. Reactions were considered positive when Ct<40. Wastewater samples from the treatment plants studied were routinely tested in our laboratory for RT-PCR inhibition and negative extraction controls as well recovery controls were performed as part of the Ontario Wastewater Surveillance Initiative QA programme (D'Aoust et al., 2021a). RT-PCR inhibition was assessed by diluting samples by a factor of 4 and 10 and comparing the proportionality of resultant N1 assay Cts. No contamination or inhibition was observed during the study period at either location (D'Aoust et al., 2021a).
2.5. RT-ddPCR
Singleplex one-step RT-ddPCR was performed to accurately quantify commercial standards used in generating qRT-PCR standard curves. 1-Step RT-ddPCR Advanced Kit for Probes supermix (Bio-Rad, Hercules, CA) together with CDC_N1 probe and primers (see above) were used to determine absolute copy number (cp) of N RNA in the following SARS-CoV-2 genomic RNA standards: Twist Bioscience Control 2, based on NCBI Genbank accession MN908947.3 (also known as Wuhan-Hu-1 and assigned to Pango lineage B) and Twist Bioscience Control 14, based on GISAID accession EPI_ISL_710528 and assigned to the B.1.1.7 (α) Pango lineage. Reactions consisted of 5 µL of the serial-diluted RNA template, forward and reverse primers (working concentration of 900 nM each), probe (working concentration of 250 nM), 5 µL of supermix, 2 µL of reverse transcriptase, 1 µL of DTT, and PCR-grade water to make up the balance of the 20 µL reaction. The droplet generation, PCR reaction, quantification and data analysis was carried out as described in an earlier study (D'Aoust et al., 2021a).
2.6. Clinical sequencing and case data
To identify a signature mutation that is a good candidate for a single locus, B.1.1.7-specific qRT-PCR assay, we used the global GISAID clinical SARS-CoV-2 genomic database (Elbe and Buckland-Merrett, 2017) to probe for amino acid mutations that were present in >95% of B.1.1.7 sequences and with <0.1% prevalence in other lineages. This was accomplished by manually surveying nucleotide mutation tables across GISAID sequences categorized by Pango lineage using covidcg.org (Chen et al., 2021). We then confirmed and visualized these findings by generating radial plots from subsampled GISAID sequences using nextstrain.org (Hadfield et al., 2018). Specifically, radial plots of amino acid- and nucleotide-level genomic loci were generated using the global database which subsampled 3799 GISAID SARS-CoV-2 sequences on February 12, 2021.
To compare our wastewater findings to the local clinical picture, we obtained case data from Ottawa Public Health and the Simcoe Muskoka District Health Unit. These Ontario Public Health Units (PHUs) track public health in the cities of Ottawa and Barrie, respectively . “Cases” were confirmed cases of COVID-19 attributed to the PHU as determined by the Ministry of Health of Ontario's surveillance case definition (PHO Case definition, 2021). Reported cases were constrained to those residing in the PHU providing data at the time of illness. Confirmed case data was extracted from Ontario's Public Health Case and Contact Management system (CCM), a central data depository for COVID-19 case and contact management and reporting in Ontario, managed by the Ontario Ministry of Health and accessed by PHUs. Data on VOCs are entered into this system from a data stream from the Ontario Lab Integration System (OLIS) or by manual entry by PHU staff. Designation of a confirmed case as having a variant or mutation of interest was done manually for the period of this study. Confirmed COVID-19 cases and confirmed B.1.1.7 lineage infection among people residing in the City of Barrie at the time of illness with accurate episode dates (earliest of onset of symptoms, testing date or date case reported) from January 7, 2021 to March 23, 2021 was extracted from CCM on March 25, 2021. Case data among people residing in Ottawa and testing positive between January 1, 2021 and April 28, 2021 was extracted on April 28, 2021. It is important to note that practices related to routine screening or whole genome sequencing for VOC including B.1.1.7 vary over time in Ontario with testing practices scaling up and down in response to changing public health priorities and capacity.
2.7. Statistical tests and plots
Spearman's rank correlation was determined between D3L wastewater allele frequency and clinical case proportion over time. A two-tailed Student's t-test was used to determine the significance of the correlation relative to the null hypothesis of zero correlation. Both were calculated in the “Analysis:Correlation” dialogue within GraphPad Prism v. 9.1.2 for Mac OS (Motulsky). Radial plots were generated on nextstrain.org (Hadfield et al., 2018). Other plots were generated with GraphPad Prism v. 9.1.2 for Mac OS.
2.8. VOC allele frequency
The proportion of SARS-CoV-2 RNA signal in wastewater that can be attributed to D3L alleles – the D3L allele frequency – was determined based on equivalent primer efficiencies and y-intercepts (i.e., assay sensitivities) observed for both D3 and D3L standard curves. These equivalent assay properties allow direct comparison of Ct values that fall along the qRT-PCR standard curves derived from RNA templates. Thus, a perceived advantage of this method is that it doesn't require absolute quantification via standard curves to determine the B.1.1.7-specific (D3L) signal as a proportion of the total SARS-CoV-2 (D3L+D3) signal. D3 and D3L Ct values were transformed into linear space and allele frequency was calculated as the ratio of D3L and the sum of D3 and D3L allele amounts. These operations are defined in the formula: 2−Ct[D3L] / (2−Ct[D3L] + 2−Ct[D3]).
3. Results and discussion
3.1. Assay design strategy
Clinical diagnosis of a B.1.1.7 case, with its multiple mutations, requires a highly specific assay with a very high positive predictive value. This is difficult to achieve in a singleplex qRT-PCR assay, as targeting multiple, co-occurring mutations at different loci may be needed to unambiguously define a VOC. Multiplexed assays or secondary screens incorporating Sanger sequencing and/or whole-genome sequencing are therefore required to confirm the genetic lineage of a virus isolated from clinical specimens (PHO VOC testing descriptions, 2021). Unlike testing of individuals, wastewater samples include genomes (and fragments thereof) sourced from multiple infections and can be collected at high frequency (e.g., daily) over continuous periods. The additional longitudinal granularity provided by wastewater sampling allows passive, population-level probing of COVID-19 infection courses. This property provides substantial power to probe and follow the dynamics of allele frequencies in a population as infections wax and wane.
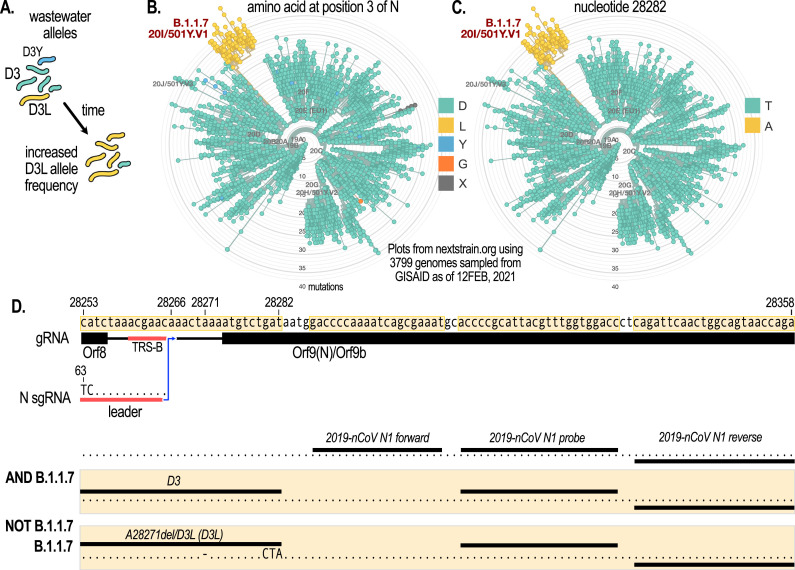
We hypothesized that following a single, signature mutation in this context would yield sufficient power to distinguish VOC lineages. This is because other variants carrying a mutation at the same locus are likely to be rare and below detection limits as they are believed to be less transmissible and thus have a very low prevalence which does not change over time (Fig. 1 A). We found that one such signature mutation on the N gene at the amino acid level (D3L) is present in the B.1.1.7 lineage (Fig. 1B). Moreover, a single nucleotide variant (SNV) in the wobble position of N:D3, T28282A appears to be unique to the B.1.1.7 lineage (Fig. 1C). Serendipitously, D3L lies only 5 bp upstream from the start of the CDC 2019-nCoV N1 amplicon region, which we currently target to detect SARS-CoV-2 in wastewater (D'Aoust et al., 2021a, 2021b, 2021c). Taking advantage of the high sensitivity of the N1 target, we designed forward primers with mutations at the 3’ end complementary to the B.1.1.7 signature allele at this locus (Fig. 1D). This allele-specific primer extension strategy works by virtue of the inability of DNA polymerase to extend mismatched nucleotides at the 3’ end of the primer (Lefever et al., 2013). This method affords high allele specificity, while obviating the complexities and high cost associated with the development of competition probe assays. We then combined this forward primer with the existing N1 probe and reverse primer (Fig. 1D, and Materials and Methods). We call these two new assays “D3” and “D3L”, distinguishing non-B.1.1.7 from B.1.1.7 viral lineages, respectively, when applied in the wastewater context. Coincident with the development of the assay described in this work, Lee et al also demonstrated the feasibility of the allele-specific primer extension strategy to monitor VOC mutations in wastewater with assays designed to detect 3 different mutations on the SARS-CoV-2 S protein (Lee et al., 2021). Additional VOC-specific qRT-PCR assays, such as those that are based on allele-specific competition probes have also been described and validated in wastewater samples (Peterson et al., 2021).
Fig. 1.
The N gene D3L mutation serves as a signature target to detect VOC of the B.1.1.7 lineage. (A) D3L allele frequency is hypothesized to rapidly increase in wastewater in communities where B.1.1.7 has been introduced. Other mutations at the same genomic locus which are at low prevalence would not be detected. (B) A radial plot of mutations amongst globally sampled GISAID SARS-CoV-2 sequences at the N:D3 locus. The D3L mutation is a signature of the B.1.1.7 lineage. (C) A radial plot as in B but at the nucleotide level showing that the T28282A SNV is unique to B.1.1.7. (D) Genomic neighborhood and primer/probe locations for the 3 different assays discussed in the text: N1 which amplifies 99+% of viral variants (AND B.1.1.7), D3 which amplifies alleles other than the B.1.1.7-specific allele (NOT B.1.1.7), and D3L which target B.1.1.7-specific alleles. The D3L primer also incorporates a deletion at A28271 which is present in the vast majority of B.1.1.7 genomes. TRS-B=Transcriptional regulatory site-body. Radial plots were generated on nextstrain.org (Hadfield et al., 2018) with sequences subsampled global sequencing data subsampled from GISAID (Elbe and Buckland-Merrett, 2017).
While the strategy of using a single genomic locus signature might give sufficient resolution in the wastewater context, it is important to note that this assay may not have a high specificity in clinical samples for viruses of the B.1.1.7 lineage as a rare mutation may occur at this location in individual cases. Another caveat of the proposed assay is that emerging viral variants might accumulate additional mutations in the probe and primer region of the assay that would affect sensitivity. Indeed, partway into designing and testing this new assay at the end of December 2020 when B.1.1.7 was becoming prevalent in the United Kingdom, we noted the emergence of a SNV in B.1.1.7 sequences deposited at GISAID that co-occurred with the D3L mutation and is located in the middle of the D3L forward primer. This deletion mutant (A28271del) quickly became the dominant SNV in the B.1.1.7 lineage, co-occurring with D3L at similar frequencies in the United Kingdom (Fig. 1D). The D3L forward primer used in this study therefore incorporates this SNV. Together, this design strategy (i.e., targeting a signature mutation that lies close to existing sensitive qPCR probe/primer sets) has the potential to create specific and sensitive qRT-PCR assays for determining the proportion of VOC-specific RNA signal in wastewater samples.
3.2. Sensitivity and allele-specificity on synthetic and patient feces-derived RNA templates
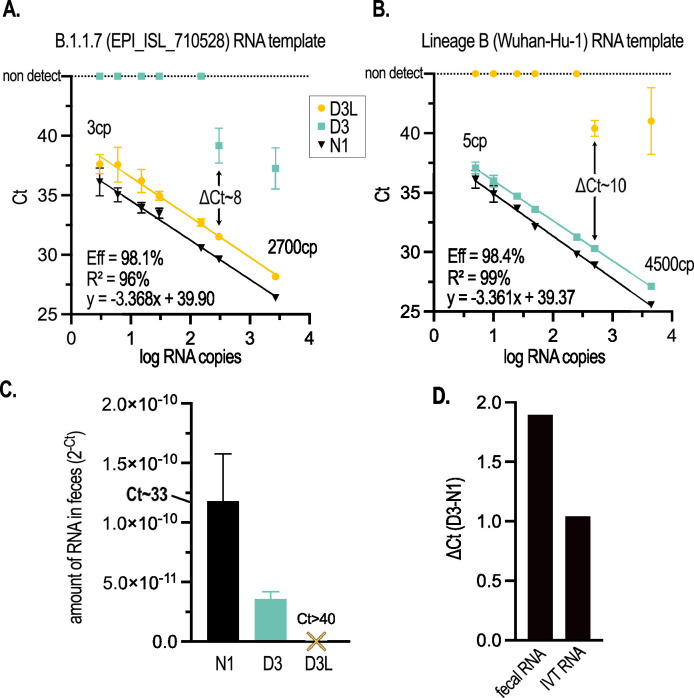
We performed D3L, D3 and N1 qRT-PCR assays on serial dilutions of B.1.1.7 or Lineage B (Wuhan-Hu-1) in vitro transcribed RNA templates obtained from a commercial supplier (Twist Bioscience, San Francisco, CA) to generate standard curves (Fig. 2 A, B). To compare similar template inputs, we determined the copy number (cp) of the RNA templates based on N1 RT-ddPCR (indicated in Figs. 2A, B and S1). qRT-PCR for N1 exhibited excellent efficiency (>98%) and analytical sensitivity (4/4 wells detected) down to ∼5 cp with either the B.1.1.7 or Lineage B templates (Fig. 2A, B), consistent with our previous data (D'Aoust et al., 2021b) and that of others (Ahmed et al., 2021). Importantly, both D3L and D3 assays on their respective complementary RNA templates demonstrated excellent efficiency (98.1 and 98.4%), linearity (96 and 99%), and analytical sensitivity (4/4 wells detected) comparable to N1, although y-intercepts tended to be approximately 2 cycles higher, indicating that the limit of detection for D3 and D3L is higher than N1 under these conditions.
Fig. 2.
The D3L and D3 qRT-PCR assays are highly sensitive and sufficiently discriminate targeted alleles on IVT RNA templates and in COVID-19 patient fecal samples. (A) Standard curve of N1, D3L, and D3 assays performed with serial dilutions of a synthetic B.1.1.7 genomic RNA template (GISAID accession ID EPI_ISL_710528, Twist Biosciences). (B) Standard curve of N1, D3L, and D3 assays performed with serial dilutions of a synthetic non-B1.1.7 genomic RNA template (Wuhan-Hu-1 isolate). For both standard curves, N1 RNA copy number (cp) determined by RT-ddPCR is indicated. In panels (A) and (B), error bars represent the standard deviation of the mean of 4 qRT-PCR replicates. Calculated primer efficiencies ([10-1/m -1]*100), linear fit, and equation of the line of regression are indicated for D3L (A) and D3 (B). (C) The B.1.1.7 allele (D3L) is not detectable in RNA extracted from the pooled feces of 2 human COVID-19 patients admitted to an Ottawa hospital prior to the emergence of B.1.1.7 in Canada; while both N1 and D3 (non-B.1.1.7) alleles are detected. Error bars represent the standard deviation of the mean of 2-3 qRT-PCR replicates. (D) D3 alleles amplify approx. 1 Ct later than the N1 allele on IVT RNA, with data derived from (B); and approx. 2 Cts later in patient feces, with data derived from (C).
An allele-specific assay must be able to discriminate the targeted alleles without cross-talk (i.e., it must not amplify the non-targeted allele). To this end, we assessed the D3L and D3 assays on serial dilutions of their orthogonal templates (Lineage B and B.1.1.7, respectively, Fig. 2A, B). We observed excellent allele specificity, with no detectable cross-talk (no amplification at 45 cycles) until template copy number approached 300 (Ct∼31) at which point weak detections were observed (differences of 8-10 cycles between complementary and orthogonal templates; compare ΔCt in Fig. 2A, B). Importantly, this cycle difference was maintained when template copies were an order of magnitude higher. To independently confirm our observations, the laboratory of Dr. Mark Servos, a participant in the Ontario Wastewater Surveillance Initiative (Manuel et al., 2021) assessed the D3 and D3L primer efficiencies and analytical sensitivity on the same RNA templates but using a different one-step qRT-PCR supermix and found similar performance (Fig. S1).
We next assessed the sensitivity and specificity of the D3L assay by probing fecal RNA extracted from COVID-19 hospital patients that had been admitted in Fall 2020 to the Ottawa Hospital, prior to the introduction of B.1.1.7 into the community. SARS-CoV-2 RNA isolated from human feces could be partially degraded/fragmented and contains both sub-genomic (sg) and genomic (g) RNAs. The D3L and D3 forward primers span an intergenic region of the genome and the two 5’ terminal nucleotides of the forward primers are a mismatch for the leader sequence that is present on intact N sgRNA transcripts (Fig. 1D). Thus, D3L and D3 qRT-PCR sensitivity may be lower in viral RNA extracted from COVID-19 patient feces compared to N1. Although we readily detected N1 amplicons (Ct∼33) in COVID-19 patient feces (pooled from 2 patients), we observed that D3 signal in the same sample was ∼30% of N1 (Fig. 2C). Critically, we did not observe any amplification of the D3L allele in patient feces after 45 cycles, consistent with the allele-specificity observed with the synthetic RNA template and the putative B.1.1.7-negative status of the patients (Fig. 2C). Using N1 to normalize any RT and/or PCR inhibition that might be present in RNA extracted from feces, we compared D3 sensitivity in patient fecal RNA vs. that obtained from IVT RNA. Comparing the difference in Cts (ΔCt) between D3 and N1 signals (Fig. 2D), we found that the D3 assay was approximately 50% less sensitive (amplified ∼1 cycle later) with a fecally-extracted vs. IVT viral RNA template. Although this data was derived from a single pooled fecal sample from two patients, it nonetheless suggests some form of RT and/or PCR inhibition that is specific to the D3 assay (as we are controlling for general RT and/or PCR inhibition by normalizing to N1 which shares a common probe and reverse primer with the D3 assay). This could be due to a number of properties that are inherent to the fecal matrix including: (1) increased competition from other RNAs present, (2) increased D3 RNA fragmentation, (3) sgRNA-derived cDNAs that are amplified less efficiently due to mismatches at the 5’ end of the forward primer, (4) Impaired reverse transcription of the D3 region, or 5) PCR inhibitors that differentially affect D3 cDNA amplification. Further investigation is needed to understand this phenomenon. Together, these data demonstrate the sensitivity and allelic specificity of D3L and D3 assays in COVID-19 patient fecal samples and support the applicability of this assay to the wastewater context.
3.3. Application of assay to influent and primary sludge wastewaters
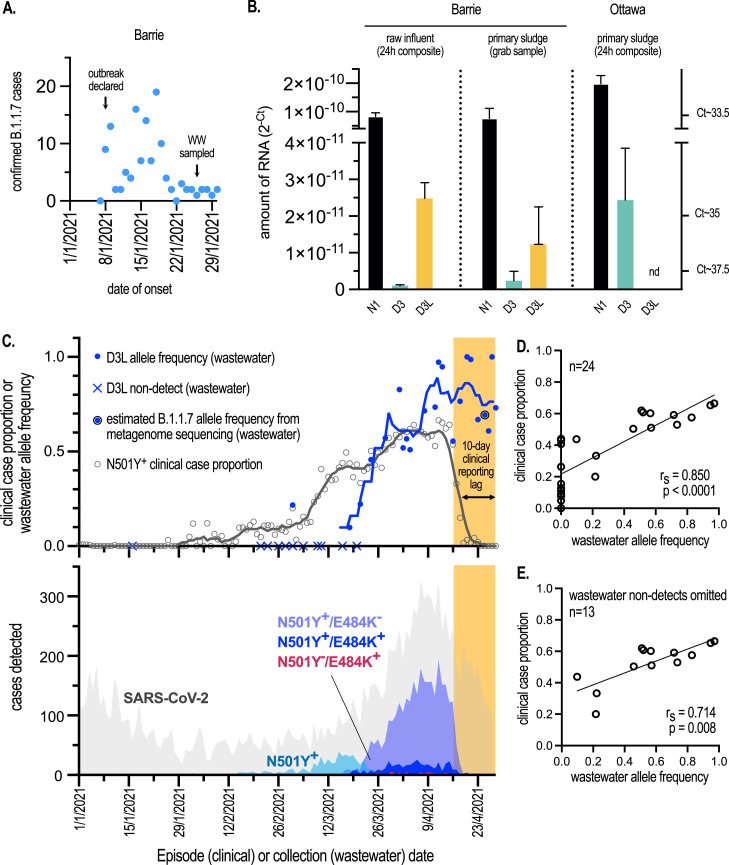
We obtained 24h composite influent and grab primary sludge samples (n=1 for each sample type) collected on January 26, 2021 from a WRRF serving the mid-size (pop. ∼135K) community of Barrie, Ontario. More than two weeks prior to sampling, on January 8, a COVID-19 outbreak was declared at a long-term care facility (LTCF) within the WRRF catchment. More than 100 B.1.1.7 cases associated with this outbreak were later confirmed by whole genome sequencing of clinical samples at the provincial laboratories. Most cases were likely active at the time of sampling (Fig. 3 A). We readily detected N1 amplicons in both samples, indicating that SARS-CoV-2 was prevalent in the community at the time of sampling and served as a positive control (Fig. 3B). Critically both samples from Barrie yielded detectable signal with the D3L assay (Fig. 3B). D3, targeting non-B.1.1.7 was at very low levels compared to D3L. Given that primer efficiencies of D3 and D3L are virtually identical (Fig. 2A, B), this allowed us to estimate their relative abundance and suggested that B.1.1.7 was the dominant viral lineage in the community at the time of sampling. To assess the sensitivity and specificity of the D3L assay in a background of non-B.1.1.7 SARS-CoV-2 in a wastewater context, we sampled a 24h composite of primary sludge obtained on January 16, 2021, from a wastewater resource recovery facility (WRRF) serving a large Canadian urban center (Ottawa, Ontario; pop. ∼1M). Although we could detect both N1 and D3 alleles, the D3L assay failed to detect B.1.1.7-specific RNA fragments (Fig. 3B). According to Ottawa Public Health, fewer than 5 clinically verified and travel-related B.1.1.7 cases were active in this community at the time of the wastewater sampling.
Fig. 3.
Rapid detection and quantification of B.1.1.7 allele frequency in wastewater samples following community outbreaks. (A) The number of B.1.1.7 cases by date of symptom onset that were associated with an isolated outbreak at a congregate living setting in Barrie, Ontario in January 2021. Cases were retrospectively assigned to the B.1.1.7 lineage by sequencing performed at Public Health Laboratories in Ontario. Wastewater (WW) was sampled from the community treatment plant serving approx. 150K inhabitants on January 26, 2021. (B) Amount of alleles in wastewater determined by qRT-PCR and expressed as linearly transformed Ct values (2-Ct). Raw Ct values are indicated on the right axis. N1, D3, D3L assays were performed on raw influent solids or primary sludge samples collected from WRRFs in (i) Barrie, Ontario on January 26, 2021 and experiencing an outbreak of B.1.1.7 COVID-19 (approx. 100-200 cases), and (ii) Ottawa on January 16, 2021 and with <5 known and active B.1.1.7 cases. Mean of qRT-PCR replicates (n=3) from each of the 3 indicated samples is presented with error bars representing s.d. (C, top) The proportion of N501Y+ clinical cases from all tested SARS-CoV-2 cases by episode date compared to D3L allele frequency in wastewater. Trend lines represent 7-day midpoint rolling averages. “N501Y+” includes positive cases detected with the singleplex N501Y assay, as well as N501Y+/E484+ and N501Y+/E484K- genotypes assigned with the multiplex assay. Metagenomic sequencing on April 25 confirmed the major proportion of wastewater SARS-CoV-2 RNA was of B.1.1.7 origin. See text for details. (C, bottom) SARS-CoV-2 case distributions in Ottawa, Ontario by episode date and assigned genotypes. Cases initially screened positive by qRT-PCR (SARS-CoV-2) were subsequently screened using qRT-PCR assays targeting either N501Y beginning in February, or N501Y/E484K beginning in March. The estimated reporting lag period between the time a case is detected and the reported results of the N501Y/E484K assay is indicated by the yellow shaded region. Clinical data were provided by Ottawa Public Health and Simcoe Muskoka District Health Unit. (D) Correlation of D3L allele frequency in wastewater vs. clinical N501Y+ case proportion in Ottawa. 24 paired clinical and wastewater observations falling in the time period between January 1 and April 15, 2021, prior to the clinical reporting lag, were included from the data presented in (C). A line of regression was computed, and Spearman's rank correlation coefficient (rs) is indicated. A two-tailed t-test indicated this correlation deviated significantly from zero. (E) Analysis as in (D), but with wastewater qRT-PCR non-detects omitted (n=13). A significant correlation was still observed (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
To validate the D3L qRT-PCR assay, we sent the same Barrie raw influent sample to be sequenced at the National Microbiology Laboratory (NML) in Winnipeg, Manitoba. In a companion report, by Landgraff et al, a consensus metagenome was sequenced from this sample that was consistent with viruses of the B.1.1.7 lineage, thus confirming both our qRT-PCR data and the contemporaneous clinical landscape in the community (Landgraff et al., 2021).
3.4. Measurement of community incidence
Passive and near-real-time monitoring tools to assess disease incidence and prevalence within communities and across larger political jurisdictions have transformative potential for public health. In the context of the current pandemic where an already highly transmissible respiratory virus has undergone antigenic drift that has led to higher attack rates, rapid intelligence and action is required to effectively curtail new outbreaks. Where the immunized predominate, breakthrough events resulting from infections with emerging viral variants must be rapidly identified. Clinical COVID-19 surveillance of populations continues to be hampered in many parts of the world due to low resources and inadequate supply chain networks (Vandenberg et al., 2021). Even in jurisdictions with excellent clinical surveillance systems, socioeconomic barriers and hesitancy to testing make it difficult for epidemiologists and governments to maintain a clear picture of COVID-19 in their communities (Rubin, 2020). Complementary approaches are therefore highly desirable.
We aimed to assess the feasibility of quantifying the proportion of SARS-CoV-2 RNA signal in wastewater that can be attributed to a VOC in a community using the D3L/D3 assays. Twice weekly measurements for N1, D3L and D3 in Ottawa primary sludge were undertaken beginning February 21, 2021. N1 served as a VOC-agnostic positive control. We did not reliably detect (>1 of 3 replicates under 40 Cts) D3L alleles until March 2, 2021 at which point we computed the D3L allele frequency making up the total SARS-CoV-2 allelic signature in wastewater; a ratio of D3L (B.1.1.7) signal to total SARS-CoV-2 signal (D3+D3L). See Materials and Methods for more detail. Additional non-detects were observed until the middle of March at which point D3L allele concentrations consistently increased and with which we were able to calculate allele frequencies (Fig. 3C, top). To quantify the functional sensitivity of the D3/D3L vs. N1 assays in situ (i.e., the relative ability to detect fecally shed viral RNA fragments present in the primary sludge) vs. analytical sensitivity derived in vitro (i.e., on synthetic RNA template) we measured the distribution of N1 as well as the sum of D3 and D3L signals, the latter of which is an estimate of the total signal from this genomic locus in wastewater. We observed a nearly 1 log difference in the mean of the two distributions (Fig. S2A), indicating that amplification from the D3 and D3L loci is significantly impaired in the wastewater context compared to that on synthetic RNA or template extracted from patient feces. Possible explanations include differential inhibition of RT and/or PCR, increased fragmentation in the locus targeted by the forward primers, increased competition of forward primers for other nucleic acid templates, or the fact that they overlap an intergenic region that might differentially detect genomic vs. sub-genomic RNA concentrations.
We next determined if the D3L allele frequency observed in wastewater correlated with VOC-specific clinical surveillance efforts. SARS-CoV-2 genomic surveillance in Ontario could not process the volume of cases during the Winter and Spring COVID-19 waves in Ottawa which precluded unbiased sampling and reporting. The SARS-CoV-2 case distribution across this period (January-April, 2021) in Ottawa is shown in grey in Fig. 3C, bottom. Coincident with our assay development in January/February 2021, and in an effort to minimize sampling bias and reporting lags, the PHO laboratories developed, validated and deployed (February/March, 2021) a clinical qRT-PCR test targeting S:N501Y; this mutation was at first deemed diagnostic for the B.1.1.7 lineage (PHO VOC testing descriptions, 2021). However, the emergence of additional variants with N501Y in Ontario in February/March 2021, namely B.1.351 and P.1, necessitated a new multiplexed PCR test targeting both N501Y and E484K. This replaced the singleplex N501Y test in late March and N501Y+/E484K− genotypes were considered probable B.1.1.7 (PHO VOC testing descriptions, 2021). Only a very small number of P.1 and B.1.351 cases were reported in Ottawa during this study period and contributed to N501Y+ and N501Y+/E484+ but not N501Y+/E484− or N501Y−/E484+ counts (Fig. 3C, bottom, and personal communication from Ottawa Public Health). Therefore, due to the low prevalence of P.1 and B.1.351 in Ottawa before April 2021, we assigned N501Y+ cases as probable B.1.1.7 in this study. Retrospective analysis of the number of clinical cases plotted as a function of presumed date of onset of symptoms or test (episode date) between January 1 and April 28, 2021 showed a rapid and sustained increase in the number of N501Y+ or N501Y+/E484K− (probable B.1.1.7) cases beginning in the middle of March and peaking by the beginning of April, consistent with the general increase in SARS-CoV-2 cases in Ottawa observed in this period (Fig. 3C, bottom). Note the significant time-lag in clinical VOC reporting (approx.10 days; denoted by the orange shaded regions in Fig. 3C) due to bottlenecks in the primary and secondary screening of nasopharyngeal swabs, due to both lab and data entry delays during a period of high disease incidence in the community (personal communication, Ottawa Public Health).
We calculated the proportion of N501Y+ (N501Y+ + N501Y+/E484− + N501Y+/E484K+) cases relative to all cases and plotted this based on episode date (Fig. 3C, top). For simplicity, we included N501Y+/E484K+ cases (i.e. not B.1.1.7) as we could not distinguish B.1.1.7, B.1.351, P.1 cases from the earlier singleplex-derived N501Y+ counts. Moreover, N501Y+/E484K+ cases did not materially contribute to case proportion in the study window (Fig. S2B). Thus, considering all N501Y+ cases derived from both singleplex and duoplex testing simplified our analysis and interpretation. A cursory examination shows that the distribution of clinical N501Y+ case proportion generally followed that of the wastewater D3L allele frequency over time (Fig. 3C, top). Exceptions include in the first half of March when a bump in clinical N501Y+ cases was observed that wasn't matched in the wastewater data. We speculate that this might have been caused by sampling bias (i.e., overrepresentation of travelers or outbreaks) and non-B.1.1.7 VOCs that may have been captured by the N501Y singleplex assay. Additionally, we observed a suppression of wastewater N1 signal due to snowmelt in early March that may have resulted in false negatives for D3L. We also observed a loss of correlation due to clinical reporting time-lags towards the end of the study period in late April.
Notwithstanding these biases associated with both the clinical and wastewater data, when we considered 21 paired clinical-wastewater observations in the period of January 1-April 15, 2021 (omitting the clinical reporting lag period of April 16-28, 2021) we found a high correlation that deviated significantly from zero (rs=0.861, p<0.0001) between B.1.1.7-specific clinical case proportions and wastewater allele frequencies (Fig. 3D). During the initial phase of the B.1.1.7 epidemic in Ottawa, when clinical cases were low, the wastewater assay had insufficient sensitivity leading to 11 non-detect observations between January 16 and March 20. These zero values skew the distribution, and we therefore repeated our correlation analysis, omitting the non-detected observation pairs (Fig. 3E). This 2nd analysis also showed a strong correlation that deviates significantly from zero (rs=0.714, p=0.008).
Finally, to again validate the reliability and specificity of the D3L qRT-PCR assay in reporting B.1.1.7 viral RNA in wastewater, an influent sampled collected at Ottawa WRRF on April 25, 2021 was sent for metagenomic sequencing to NML (timepoint indicated in Fig. 3C) (Landgraff et al., 2021). As was seen with the Barrie sample, the Ottawa wastewater sample yielded a consensus B.1.1.7 metagenome.
4. Conclusion
These data support the use of wastewater-based allele-specific qRT-PCR strategies to follow the emergence and proportion of VOC-specific RNA signal that makes up total wastewater SARS-CoV-2 RNA signal in populations served by WRRFs. We demonstrate that an allele-specific qRT-PCR assay targeting a single locus on the SARS-CoV-2 genome can provide sufficiently high lineage specificity and detection sensitivity in the wastewater context. We also demonstrate good correlation between two independent measures of VOC (B.1.1.7) infection in a population, namely clinical case proportion and wastewater allele frequency, which together can be used to triangulate incidence and prevalence of VOC-specific infections. The latter method to monitor VOC appears less susceptible to data inaccuracies and bottlenecks associated with scaling that occurs during times of elevated disease incidence in the community. Moreover, the flexibility of this proxy measure of COVID-19 incidence allows rapid and parallel implementation of multiple assays targeting other VOC-specific alleles within existing WBE surveillance architectures. In conclusion, our study demonstrates that targeting signature mutations in the N gene with allele-specific primer extension qRT-PCR represents a viable strategy to quantitatively monitor emerging VOC in wastewater over time with high sensitivity, specificity, and speed.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors wish to acknowledge the leadership of Dr. Alex MacKenzie (CHEORI) in this continuing endeavour. We are grateful to Dr. Chrystal Landgraff and her team at National Microbiology Laboratory for performing the metagenomic sequencing and to Dr. Kyrylo Bessonov and Mr. Cameron McDermaid for their helpful comments in reviewing the manuscript. The authors acknowledge the crucial participation and fruitful collaborations with Water Services at the Cities of Ottawa and Barrie; public health agencies (Ottawa Public Health, Simcoe Muskoka District Health Unit, Public Health Ontario), along with funding provided through the Ontario Wastewater Surveillance Initiative (Manuel et al., 2021). Their respective employees’ time, facilities, resources, and assistance provided throughout the study greatly contributed to this work. We gratefully acknowledge the originating and submitting laboratories responsible for sharing genetic sequence data via the GISAID Initiative, which was used in this project.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.watres.2021.117681.
Appendix. Supplementary materials
References
- Ahmed W., Bertsch P.M., Angel N., Bibby K., Bivins A., Dierens L., Edson J., Ehret J., Gyawali P., Hamilton K.A., Hosegood I., Hugenholtz P., Jiang G., Kitajima M., Sichani H.T., Shi J., Shimko K.M., Simpson S.L., Smith W.J.M., Symonds E.M., Thomas K.v., Verhagen R., Zaugg J., Mueller J.F. Detection of SARS-CoV-2 RNA in commercial passenger aircraft and cruise ship wastewater: A surveillance tool for assessing the presence of COVID-19 infected travellers. J. Travel Med. 2021;27 doi: 10.1093/JTM/TAAA116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A.T., Altschuler K., Zhan S.H., Chan Y.A., Deverman B.E. Covid-19 CG enables sars-cov-2 mutation and lineage tracking by locations and dates of interest. eLife. 2021;10:1–15. doi: 10.7554/eLife.63409. e63409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Wang R., Wang M., Wei G.W. Mutations strengthened SARS-CoV-2 infectivity. J. Mol. Biol. 2020;432:5212–5226. doi: 10.1016/j.jmb.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Aoust P.M., Towhid S.T., Mercier É., Hegazy N., Tian X., Bhatnagar K., Zhang Z., Naughton C.C., MacKenzie A.E., Graber T.E., Delatolla R. COVID-19 wastewater surveillance in rural communities: comparison of lagoon and pumping station samples. Sci. Total Environ. 2021;801 doi: 10.1016/j.scitotenv.2021.149618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Aoust Patrick M., Mercier E., Montpetit D., Jia J.J., Alexandrov I., Neault N., Baig A.T., Mayne J., Zhang X., Alain T., Langlois M.A., Servos M.R., MacKenzie M., Figeys D., MacKenzie A.E., Graber T.E., Delatolla R. Quantitative analysis of SARS-CoV-2 RNA from wastewater solids in communities with low COVID-19 incidence and prevalence. Water Res. 2021;188 doi: 10.1016/j.watres.2020.116560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Aoust P.M., Graber T.E., Mercier E., Montpetit D., Alexandrov I., Neault N., Baig A.T., Mayne J., Zhang X., Alain T., Servos M.R., Srikanthan N., MacKenzie M., Figeys D., Manuel D., Jüni P., MacKenzie A.E., Delatolla R. Catching a resurgence: Increase in SARS-CoV-2 viral RNA identified in wastewater 48 h before COVID-19 clinical tests and 96 h before hospitalizations. Sci. Total Environ. 2021;770 doi: 10.1016/j.scitotenv.2021.145319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy S. Why are RNA virus mutation rates so damn high? PLoS Biol. 2018;16 doi: 10.1371/journal.pbio.3000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbe S., Buckland-Merrett G. Data, disease and diplomacy: GISAID's innovative contribution to global health. Glob. Chall. 2017;1:33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaebler C., Wang Z., Lorenzi J.C.C., Muecksch F., Finkin S., Tokuyama M., Cho A., Jankovic M., Schaefer-Babajew D., Oliveira T.Y., Cipolla M., Viant C., Barnes C.O., Bram Y., Breton G., Hägglöf T., Mendoza P., Hurley A., Turroja M., Gordon K., Millard K.G., Ramos V., Schmidt F., Weisblum Y., Jha D., Tankelevich M., Martinez-Delgado G., Yee J., Patel R., Dizon J., Unson-O’Brien C., Shimeliovich I., Robbiani D.F., Zhao Z., Gazumyan A., Schwartz R.E., Hatziioannou T., Bjorkman P.J., Mehandru S., Bieniasz P.D., Caskey M., Nussenzweig M.C. Evolution of antibody immunity to SARS-CoV-2. Nature. 2021;591:639–644. doi: 10.1038/s41586-021-03207-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubaugh N.D., Petrone M.E., Holmes E.C. We shouldn’t worry when a virus mutates during disease outbreaks. Nat. Microbiol. 2020;5:529–530. doi: 10.1038/s41564-020-0690-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield J., Megill C., Bell S.M., Huddleston J., Potter B., Callender C., Sagulenko P., Bedford T., Neher R.A. NextStrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamouda M., Mustafa F., Maraqa M., Rizvi T., Aly Hassan A. Wastewater surveillance for SARS-CoV-2: Lessons learnt from recent studies to define future applications. Sci. Total Environ. 2021;759:143493. doi: 10.1016/j.scitotenv.2020.143493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes E.G., Rambaut A. Viral evolution and the emergence of SARS coronavirus. Philos. Trans. R. Soc. B. 2004;359:1059–1065. doi: 10.1098/rstb.2004.1478. Biological Sciences. Royal Society. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konings F., Perkins M.D., Kuhn J.H., Pallen M.J., Alm E.J., Archer B.N., Barakat A., Bedford T., Bhiman J.N., Caly L., Carter L.L., Cullinane A., de Oliveira T., Druce J., el Masry I., Evans R., Gao G.F., Gorbalenya A.E., Hamblion E., Herring B.L., Hodcroft E., Holmes E.C., Kakkar M., Khare S., Koopmans M.P.G., Korber B., Leite J., MacCannell D., Marklewitz M., Maurer-Stroh S., Rico J.A.M., Munster V.J., Neher R., Munnink B.O., Pavlin B.I., Peiris M., Poon L., Pybus O., Rambaut A., Resende P., Subissi L., Thiel V., Tong S., van der Werf S., von Gottberg A., Ziebuhr J., van Kerkhove M.D. SARS-CoV-2 Variants of Interest and Concern naming scheme conducive for global discourse. Nat. Microbiol. 2021;6:821–823. doi: 10.1038/s41564-021-00932-w. [DOI] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., Hastie K.M., Parker M.D., Partridge D.G., Evans C.M., Freeman T.M., de Silva T.I., Angyal A., Brown R.L., Carrilero L., Green L.R., Groves D.C., Johnson K.J., Keeley A.J., Lindsey B.B., Parsons P.J., Raza M., Rowland-Jones S., Smith N., Tucker R.M., Wang D., Wyles M.D., McDanal C., Perez L.G., Tang H., Moon-Walker A., Whelan S.P., LaBranche C.C., Saphire E.O., Montefiori D.C. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kustin T., Stern A. Biased Mutation and Selection in RNA Viruses. Mol. Biol. Evol. 2021;38:575–588. doi: 10.1093/molbev/msaa247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraff, C., Ya, L., Wang, R., Buchanan, C., Wells, M., Schonfeld, J., Bessonov, K., Ali, J., Robert, E., Nadon, C., 2021. Metagenomic sequencing of municipal wastewater provides a near-complete SARS-CoV-2 genome sequence identified as the B.1.1.7 variant of concern from a Canadian municipality concurrent with an outbreak. medRxiv. doi: 10.1101/2021.03.11.21253409. [DOI]
- Lee W.L., Imakaev M., Armas F., McElroy K.A., Gu X., Duvallet C., Chandra F., Chen H., Leifels M., Mendola S., Floyd-O'sullivan R., Powell M.M., Wilson S.T., Berge K.L.J., Lim C.Y.J., Wu F., Xiao A., Moniz K., Ghaeli N., Matus M., Thompson J., Alm E.J. Quantitative SARS-CoV-2 alpha variant B.1.1.7 tracking in wastewater by allele-specific RT-qPCR. Environ. Sci. Technol. Lett. 2021;8:675–682. doi: 10.1021/acs.estlett.1c00375. [DOI] [Google Scholar]
- Lefever S., Pattyn F., Hellemans J., Vandesompele J. Single-nucleotide polymorphisms and other mismatches reduce performance of quantitative PCR assays. Clin. Chem. 2013;59:1470–1480. doi: 10.1373/clinchem.2013.203653. [DOI] [PubMed] [Google Scholar]
- Motulsky, H., accessed Sept 11, 2021 Graphpad statistics guide: spearman rank correlation and t-test. https://www.graphpad.com/guides/prism/latest/statistics/stat_interpreting_results_correlati.htm.
- Manuel DG, Delatolla R, Fisman DN, Fuzzen M, Graber T, Katz GM, Kim J, Landgraff C, MacKenzie A, Maltsev A, Majury A, McKay RM, Minnery J, Servos M, Weese JS, McGeer A, Born KB, Barrett K, Schwarz B, Jüni P. The role of wastewater testing for SARS-CoV-2 surveillance. Science Briefs of Ontario COVID-19 Science Advisory Table. 2021;2(40) doi: 10.47326/ocsat.2021.02.40.1.0. [DOI] [Google Scholar]
- Pachetti M., Marini B., Benedetti F., Giudici F., Mauro E., Storici P., Masciovecchio C., Angeletti S., Ciccozzi M., Gallo R.C., Zella D., Ippodrino R. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J. Transl. Med. 2020;18 doi: 10.1186/s12967-020-02344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson S.W., Lidder R., Daigle J., Wonitowy Q., Mulvey M.R., Mangat C.S. RT-qPCR detection of SARS-CoV-2 mutations S 69-70 del, S N501Y and N D3L associated 1 with variants of concern in Canadian wastewater samples. medRxiv preprint server. 2021 doi: 10.1101/2021.05.20.21257536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PHO Case definition 2021 [WWW Document], n.d. URL https://www.publichealthontario.ca/-/media/documents/i/2020/iphis-bulletin-client-responsibility.pdf (accessed 6.16.21).
- PHO VOC testing descriptions 2021 [WWW Document], n.d. URL https://www.publichealthontario.ca/en/about/blog/2021/lab-voc-testing-updates (accessed 6.16.21).
- Rubin R. First it was masks; Now some refuse testing for SARS-CoV-2. JAMA. 2020;324:2015–2016. doi: 10.1001/jama.2020.22003. [DOI] [PubMed] [Google Scholar]
- Sanjuán R., Nebot M.R., Chirico N., Mansky L.M., Belshaw R. Viral mutation rates. J. Virol. 2010;84:9733–9748. doi: 10.1128/jvi.00694-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenberg O., Martiny D., Rochas O., van Belkum A., Kozlakidis Z. Considerations for diagnostic COVID-19 tests. Nat. Rev. Microbiol. 2021;19:171–183. doi: 10.1038/s41579-020-00461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogels C.B.F., Brito A.F., Wyllie A.L., Fauver J.R., Ott I.M., Kalinich C.C., Petrone M.E., Casanovas-Massana A., Catherine Muenker M., Moore A.J., Klein J., Lu P., Lu-Culligan A., Jiang X., Kim D.J., Kudo E., Mao T., Moriyama M., Oh J.E., Park A., Silva J., Song E., Takahashi T., Taura M., Tokuyama M., Venkataraman A., Weizman O., el Wong P., Yang Y., Cheemarla N.R., White E.B., Lapidus S., Earnest R., Geng B., Vijayakumar P., Odio C., Fournier J., Bermejo S., Farhadian S., dela Cruz C.S., Iwasaki A., Ko A.I., Landry M.L., Foxman E.F., Grubaugh N.D. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT–qPCR primer–probe sets. Nat. Microbiol. 2020;5:1299–1305. doi: 10.1038/s41564-020-0761-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C.O., Finkin S., Schaefer-Babajew D., Cipolla M., Gaebler C., Lieberman J.A., Oliveira T.Y., Yang Z., Abernathy M.E., Huey-Tubman K.E., Hurley A., Turroja M., West K.A., Gordon K., Millard K.G., Ramos V., Silva J.da, Xu J., Colbert R.A., Patel R., Dizon J., Unson-O’Brien C., Shimeliovich I., Gazumyan A., Caskey M., Bjorkman P.J., Casellas R., Hatziioannou T., Bieniasz P.D., Nussenzweig M.C. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592:616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.