Abstract
The increasing development of antibiotic resistance in bacteria has been a major problem for years, both in human and veterinary medicine. Prophylactic measures, such as the use of vaccines, are of great importance in reducing the use of antibiotics in livestock. These vaccines are mainly produced based on formaldehyde inactivation. However, the latter damages the recognition elements of the bacterial proteins and thus could reduce the immune response in the animal. An alternative inactivation method developed in this work is based on gentle photodynamic inactivation using carbon nanodots (CNDs) at excitation wavelengths λex > 290 nm. The photodynamic inactivation was characterized on the nonvirulent laboratory strain Escherichia coli K12 using synthesized CNDs. For a gentle inactivation, the CNDs must be absorbed into the cytoplasm of the E. coli cell. Thus, the inactivation through photoinduced formation of reactive oxygen species only takes place inside the bacterium, which means that the outer membrane is neither damaged nor altered. The loading of the CNDs into E. coli was examined using fluorescence microscopy. Complete loading of the bacterial cells could be achieved in less than 10 min. These studies revealed a reversible uptake process allowing the recovery and reuse of the CNDs after irradiation and before the administration of the vaccine. The success of photodynamic inactivation was verified by viability assays on agar. In a homemade flow photoreactor, the fastest successful irradiation of the bacteria could be carried out in 34 s. Therefore, the photodynamic inactivation based on CNDs is very effective. The membrane integrity of the bacteria after irradiation was verified by slide agglutination and atomic force microscopy. The method developed for the laboratory strain E. coli K12 could then be successfully applied to the important avian pathogens Bordetella avium and Ornithobacterium rhinotracheale to aid the development of novel vaccines.
1. Introduction
Antibiotics have been used for the treatment of bacterial infectious diseases in human and veterinary medicine for more than 80 years.1 They are mainly applied when the pathogen has not yet been precisely identified, but the infection must be treated quickly. Antibiotics are generally well tolerated by humans and animals, and are thus widely used in medicine. However, antibiotics do not affect certain bacteria. The survival of these antibiotic- or antimicrobial-resistant bacteria has been favored for years by the extensive and global use of antibiotics. In consequence, the World Health Organization (WHO) has issued warnings that the rapid spread of antibiotic resistance is a major public health challenge because it endangers the ability to treat potentially life-threatening infections for years.2 In Europe, more than 33 000 deaths a year have been reported due to infection with multiresistant bacteria, and the numbers are rising.3
In the 2014 WHO global report on surveillance “antimicrobial resistance”, the authors reported the risk of resistances occurring in different bacteria all over the world.4 This was updated in 2020 and supplemented by an Immunization Agenda 2030.5 Among others, the bacteria of the species Escherichia coli already show a resistance to the last generation drugs (cephalosporins and fluoroquinolones) commonly used to treat serious infections of 50% or more in five out of six investigated regions.4 Potential human infections from E. coli can be urinary tract infections, bloodstream infections, and meningitis in neonates. Resistance in E. coli readily develops either through mutations or by acquisition of mobile genetic elements against broad-spectrum penicillins (e.g., ampicillin or amoxicillin). The spread of transmissible antibiotic-resistant bacteria between humans and livestock can take different paths (meat, surface water, wastewater, or manure fertilization used for edible plants).6 Similar resistant plasmids or similar E. coli strains have been found in chicken meat products and human infections.7 Since the beginning of the use of antibiotics in medicine in the 1940s, resistances have steadily grown and current research shows an increase of individual resistance genes by a factor of more than 15.8
While antibiotics or for example bacteriophages are only used after the host (human, animal) has been infected, vaccines can be used to prevent infections, create herd immunity (indirect protection), or even eradicate diseases. These prophylactic measures are of great importance in reducing the use of antibiotics in livestock. Vaccines are mainly produced based on formaldehyde inactivation. However, the latter damages the recognition elements of the bacterial membrane and thus could reduce the immune response in the animal. One alternative to formaldehyde is the photolytic inactivation with ultraviolet (UV) light.9 Due to the absorption of low-UV-wavelength radiation, the bacterial cell walls can be damaged as well. Thus, lower exposure times or intermediary additives (e.g., photosensitizer) were used to increase the integrity of the bacteria to trigger stronger immune responses. One possible gentle inactivation method is the photodynamic inactivation using amorphous carbon nanodots (CNDs)10 at wavelengths λex > 290 nm.11 The inactivation is based on photoinduced formation of reactive oxygen species by CNDs in the bacterium to avoid damage of the outer membrane.11,12
CNDs, due to their promising photophysical and chemical properties (e.g., high water solubility, chemical inertness, resistance to photobleaching, high biocompatibility, and ease of functionalization13) have emerged as very promising nanomaterials for a broad range of applications (e.g., wastewater treatment,14 (photo)catalysis,15 and optoelectronic devices13). In particular, the ability to prepare CNDs from a wide range of accessible organic materials makes them a potential alternative for conventional organic dyes and semiconductor quantum dots (QDs) in various applications. In contrast to QDs, which often consist of toxic elements such as cadmium and other rare earth metals,16,17 the CNDs (cyto-)toxicity is typically lower but still under debate and is strongly influenced by the concentration of the CNDs, their surface, and their application.18−21 Various studies concerning the (cyto-)toxicity of different CNDs types in various model organisms, e.g., cell viability assays,22−24 zebrafish,25 and various rodents,26,27 so far suggest that CNDs of any type appear to have low (cyto-)toxicity in vitro and in vivo, and that they possess excellent biocompatibility.28−31 Thus, CNDs can be efficient photosensitizers for photodynamic inactivation which simultaneously reduce unwanted side effects both in the target bacteria and the vaccine recipient. For the photodynamic inactivation the CNDs must pass through the outer membrane of the bacteria.32,33 The detailed uptake mechanism of the CNDs into bacteria depends among others on the size, shape, degree of dispersion, and surface charge of the CNDs and is part of current studies and reviews.12,34−36
In this work, CNDs synthesized according to previously published protocols37,38 were used to find the smallest irradiation time for the inactivation of the nonvirulent laboratory strain E. coli K12 using UV and visible light radiation. The verification of the integrity of the inactivated E. coli via different methods was the focus. The lowest membrane damage will result in the highest immune response in animals and thus in a very effective vaccine. The method developed for the E. coli was subsequently applied to the important avian pathogens Bordetella avium and Ornithobacterium rhinotracheale (ORT). These pathogens are mainly responsible for respiratory diseases in turkeys.39−42 Infections with these bacterial pathogens lead to high economic losses in the poultry sector and therefore generally require antimicrobial treatment.
2. Results and Discussion
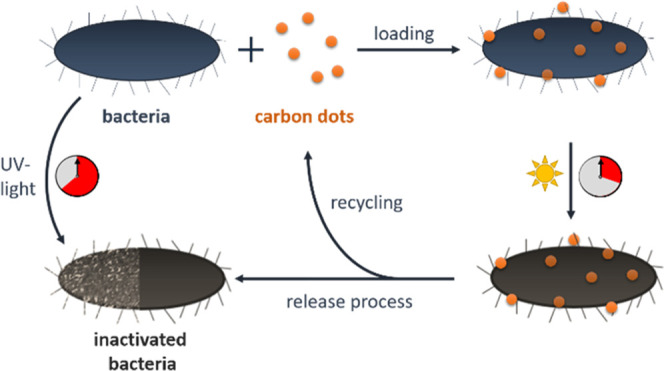
A self-made flow photoreactor was used to show the potential of the gentle photodynamic inactivation of bacteria using CNDs at wavelengths λex > 290 nm. Two batch and one flow inactivation experiments were chosen as reference methods. The batch inactivations were performed in cuvettes with two different light sources (calibration lamp: λex = 230–2250 nm and UV-A diode: λex = 365 ± 40 nm). For the flow inactivation reference, the bacteria without CNDs were irradiated in the photoreactor with UV light (λex ≈ 254 nm).
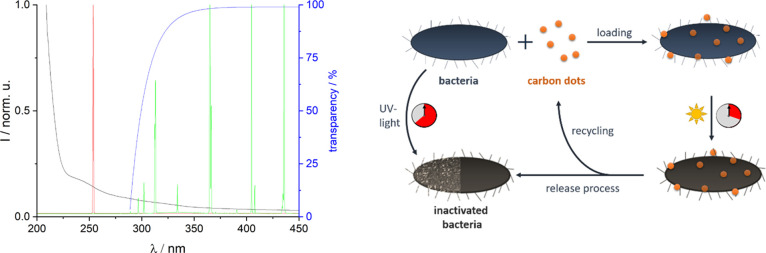
The different radiation wavelengths in the photoreactor were chosen based on the absorption spectra of the E. coli (see Figure 1 left). While 254 nm (low-pressure Hg radiator, red curve) is strongly absorbed directly by the E. coli, the high wavelengths above 290 nm (mid-pressure Hg radiator + borosilicate glass as high-pass filter, green curve) are used to excite the CNDs and thus gently inactivate the E. coli. For photodynamic inactivation, the preceding incorporating process of the CNDs in the bacteria was controlled by fluorescence microscopy (loading process). After irradiation of the CNDs–bacteria mixture in the flow reactor with the mid-pressure radiator, the CNDs were removed from the bacteria via diffusion with a liquid flow of phosphate-buffered saline (PBS) (release process, Figure 1 right).
Figure 1.
Left: normalized extinction spectrum of E. coli (black) and emission spectra of a Hg low-pressure (red) and a Hg mid-pressure radiator (green) in a glass envelope of borosilicate glass with given transparency (blue); right: scheme of the flow inactivation with CNDs and the reference method with UV light.
For monitoring the process of loading and releasing of the CNDs in the bacteria, a fluorescence microscope was used. Changes in the absorption and emission behavior of the CNDs due to their surrounding environment made it possible to monitor their depopulation in the surrounding PBS and their population in the E. coli separately. Both the absorption and emission bands of the CNDs shift to higher wavelengths (around + 100 nm) when CNDs are internalized by the bacteria. This behavior was to be expected, as it has previously been shown that the dipole–dipole interactions between CND surface groups and the surrounding medium can modify the emission properties of CNDs.43 Hence, pH, polarity, and temperature of the surrounding medium can lead to a shift of CNDs absorption- and emission behavior, while their characteristic offset is maintained.44 Two different optical filter cubes, depending on the CND-containing medium, were applied to the microscope to separate the excitation and emission light (see Section 3) of the CNDs in the different surroundings.
2.1. Loading Process
For monitoring the incorporating process of the CNDs into the bacteria, E. coli were fixed with agarose on a coverslip. A bright-field image of the bacteria was recorded via a charge-coupled device (CCD) camera to locate the position of the bacteria in the focus (Figure 2 left, 0 min). Afterward, a defined volume (10 or 100 μL) of a diluted CND stock solution was added and a series of fluorescence images were taken to monitor the uptake process of the CNDs into the E. coli (Figure 2 left, 2 to 9 min). Different bacteria were selected as regions of interest (ROIs). These ROIs were separately integrated to observe the fluorescence intensity over time at 630 nm (CND emission in E. coli—Figure 2 right, Figure 3 left). The emission at this wavelength was mainly induced by the CNDs, the weak autofluorescence (emission) of the bacteria was removed through background subtraction. The process was repeated at different volumes of CND solution (Figure 3 left).
Figure 2.
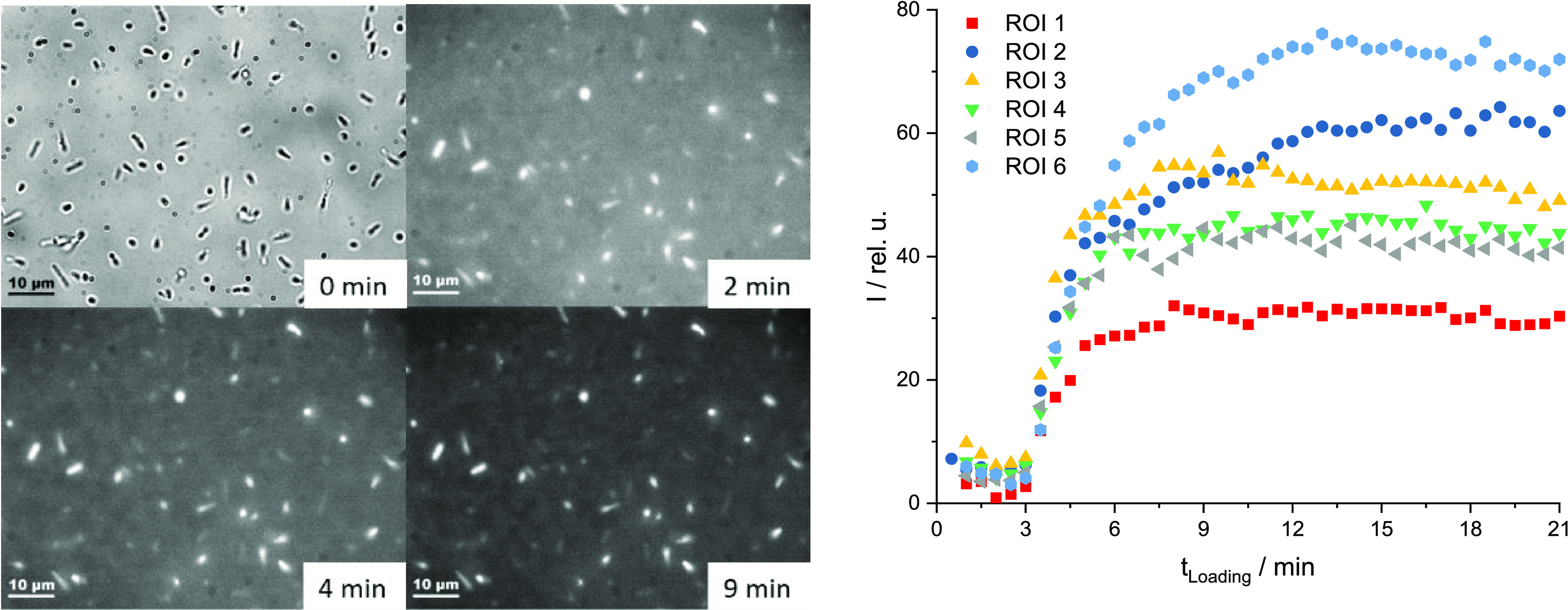
Loading of the E. coli bacteria via adding of 100 μL of diluted CND stock solution to a volume of 1 mL of E. coli in PBS at 0 min—left: microscopic images (0 min = bright-field image, 2/4/9 min = fluorescence images, X-Cite 25%, λex = 565 ± 15 nm, λem = 630 ± 37.5 nm), E. coli (rod) bacteria were fixed with 1 M agarose solution—right: fluorescence intensity over time after loading of different regions of interest (ROIs = clear shaped E. coli bacteria in the focal plane of the microscope), intensities are background-corrected, background = region without loaded bacteria, image acquisition rate was 2 min–1.
Figure 3.
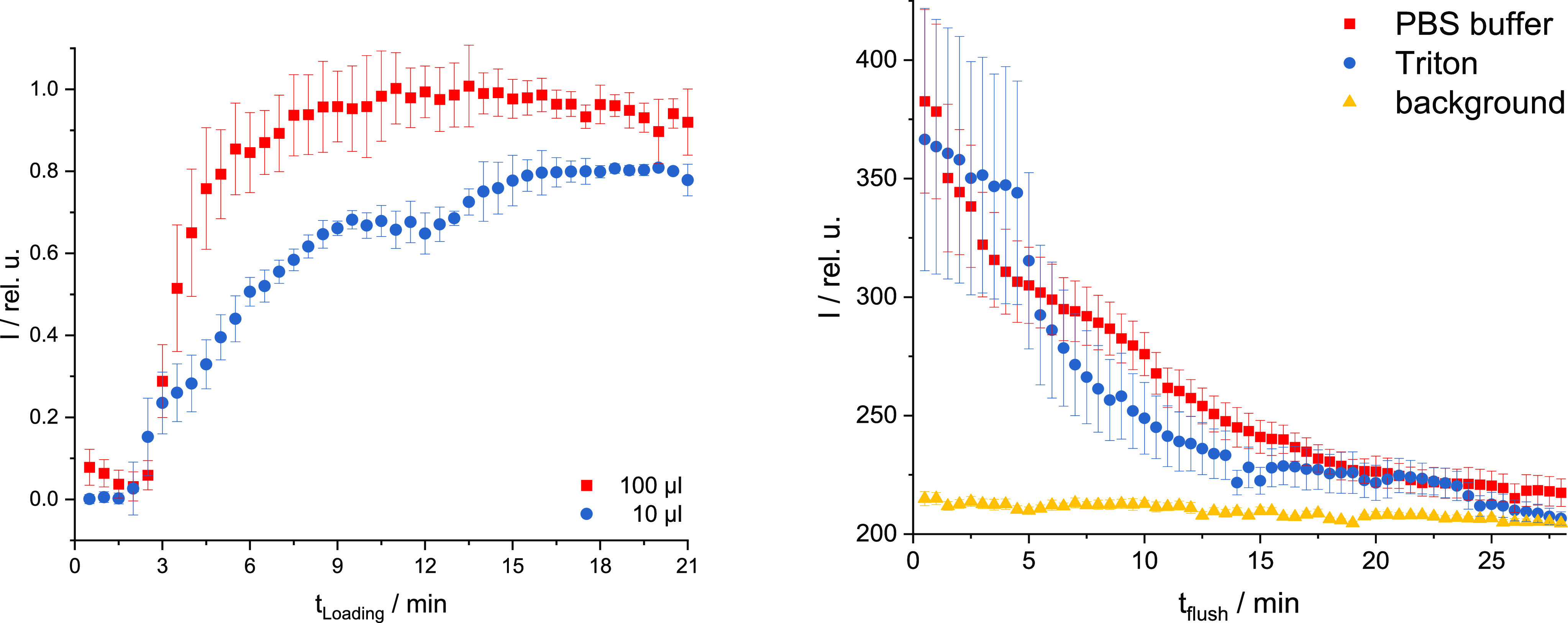
Left: loading of the E. coli bacteria with 10 or 100 μL of CND solution and their normalized averaged ROIs—right: washing out process of CND-loaded E. coli—flow rate 1 mL/min of either 0.5% Triton solution (v/v) or PBS with respect to the background (= region without E. coli).
The uptake of the CNDs into the E. coli begins within several seconds after adding the CND solution to the E. coli regardless of the volume used. This is reflected by the fast increase of the fluorescence intensity within the first minutes (see Figure 2 right and Figure 3 left). By adding 100 μL of the CND solution, the emission intensity reaches a plateau after 7–8 min (Figure 2 right). Smaller CND solution volumes lead to a more undefined result with various changes in the emission, probably due to varying CND concentrations in the bacteria. Larger volumes of added CNDs result in higher average fluorescence emission intensities than smaller volumes (Figure 3 left) due to higher numbers of incorporated CNDs in one bacterium.
2.2. Release Process
To remove the CNDs from the E. coli, the CND-doped E. coli fixed on the coverslip were continuously washed with PBS (1 mL/min). To monitor this process, the fluorescence intensity at λem ≈ 630 nm (CND emission in E. coli) was measured over the washing out time (Figure 3 right). To verify that all CNDs had left the bacteria, the same procedure was done with a 0.5% Triton X-100 solution (v/v). Triton X-100 is a nonionic surfactant that is used in protein purification to detach membrane proteins in their native conformation from membranes. In contrast to the respective background, the emissions of a ROI that was flushed with PBS or Triton decreased significantly over 15 min flushing time. Thereafter, the fluorescence of all three curves behaved similarly with comparable intensities (see Figure 3 right). It can therefore be concluded that the CNDs can be washed out of the bacteria using PBS and thus can be reused for the further photodynamic inactivation process.
2.3. Photodynamic Inactivation
The direct photolytic inactivation with UV light allows a rather targeted damage of biological functions of the cell due to the absorption by the bacteria.45 In contrast, photodynamic inactivation generates reactive oxygen species mainly within the bacteria (e.g., peroxides or singlet oxygen), which attack biologically important molecules in the immediate vicinity of the used photosensitizer. A well-explored photosensitizer which generates reactive oxygen species is methylene blue (MB).46 The photosensitizer is effective against multiresistant bacteria and can be used in high concentration (1% v/v) without human toxicity, but with low efficiency as well as low selectivity to bacteria cell.47 In the present study, CNDs were applied as an alternative photosensitizer due to their special luminescence properties.23
The photodynamic inactivation starts with the doping of the bacteria with a photosensitizer, such as MB or CNDs (Figure 1 right; for details, see Section 3). To find the shortest irradiation time, each bacteria–photosensitizer mixture was irradiated in the flow reactor. To verify the influence of daylight while doping the bacteria with CNDs, the doped E. coli were irradiated additionally with a calibration lamp and a diode in a cuvette (for details, see Section 3). The calibration lamp shows broad emission (λex = 230–2250 nm) nearby the sunlight spectral range, while the diode emits only UV-A wavelengths around λex = 365 nm.
The high emissions at λem > 290 nm of the mid-pressure Hg irradiator leads to a fast inactivation of the doped E. coli independent of the used photosensitizer (CNDs or MB). Thus, our CNDs give comparable results to the well-established photosensitizer MB. Additionally, CNDs have a wide absorption range. Therefore, inactivation due to daylight (calibration lamp and UV-A light) of the CND-doped E. coli was tested. It takes place but needs significantly longer irradiation times (≈30 min, see Table 1) due to the lower energy densities.
Table 1. Indirect Photo-Inactivation of Doped E. coli (Batch Inactivation).
| lamp source | mid-pressure Hg radiatora | calibration lampbT | UV-A lightbT | |
|---|---|---|---|---|
| doping | MB | CNDs | CNDs | CNDs |
| initial sample [108 CFU/mL] | 4 | 4 | 1 | 2 |
| min radiation time | 34 s | 34 s | ≈30 min | ≈30 min |
| sterile test | negative | negative | negative | negative |
This irradiation was performed with the flow reactor (perfluoroalkoxy alkane (PFA) tube with ID = 0.762 mm, water cooled).
hese irradiations were done in cuvettes in front of either the calibration lamp or (365 ± 40) nm diode both without cooling.
The potential of the custom-made flow reactor while using CND-doped bacteria for inactivation was verified with further experiments. First, the number of bacteria strains was increased by Bordetella avium and Ornithobacterium rhinotracheale. Second, the influence of the CNDs on the inactivation process was tested by direct inactivation of the bacteria with UV light. The goal of all experiments was to find the highest possible flow rate or the shortest possible irradiation time for each bacterial strain at which the sterile test is showing a repeatable negative result (inactivated bacteria). To that end, the bacteria solutions in PBS (zero sample) were irradiated by both Hg radiation sources (low and mid-pressures) at different flow rates.
A summary of these results is given in Table 2. The short irradiation time results in low damage to the bacteria, which is reflected in the high recovery rates (all >94%) of the irradiated bacteria compared to untreated bacteria using a matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS). Additionally, the antigen–antibody reaction—clumping of particulate antigens (agglutination)—showed that the recognition elements on the bacteria surface were not damaged significantly during the irradiation. The minimal possible irradiation time varies greatly from strain to strain. While the mid-pressure Hg radiator allows considerably shorter irradiation times for the E. coli, the low-pressure radiator shows a more effective inactivation for the B. avium and ORT. However, all photodynamic inactivations were significantly faster than the reference method without CNDs. The acceleration factor of the inactivation for the mid-pressure Hg irradiator, by adding CNDs, varied depending on the bacteria strains (E. coli = 1.6, B. avium = 2.6, and ORT = 3.7).
Table 2. Results of the Flow Inactivation (with/without CNDs) of the Bacteria E. coli, B. avium, and ORT in the Flow Reactor Using Different Hg Lamps (Upper Part of the Table)a.
| E. coli (K12) | B. avium | ORT | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Hg radiator (pressure) | low | mid | mid + CNDs | low | mid | mid + CNDs | low | mid | mid + CNDs |
| initial sample [108 CFU/mL] | 1 | 4 | 4 | 9 | 9 | 9 | 3 | 3 | 3 |
| min radiation time [s] | 336 | 56 | 34 | 22 | 31 | 12 | 49 | 84 | 23 |
| Verification of integrity | |||||||||
| sterile test | neg. | neg. | neg. | neg. | neg. | neg. | neg. | neg. | neg. |
| agglutination | yes | yes | yes | yes | yes | yes | yes | yes | yes |
| MALDI-TOF recovery rate | >94% | >95% | not tested | >96% | >99% | not tested | >99% | >99% | not tested |
For the possible minimum irradiation time needed for a complete inactivation, the results of used methods to verify the integrity are given (lower part of the table).
2.4. Verification of Integrity
The success of the direct photolytic and the photodynamic inactivation of E. coli was verified by viability assays on agar and sterile tests. If the number of colony-forming units (CFU) on agar was smaller than 10 for the stock solution of the irradiated sample and the sterile tests of the same sample were negative, the sample was defined as inactivated.
Additionally, MALDI mass spectra were recorded and compared to the spectra before irradiation. The high recovery rates of at least 94% (Table 2) give a good indication of the integrity of the bacteria and proteins. Furthermore, the integrity of proteins and lipids was verified by slide agglutination with positive results. For a good immune response, the most important parts of the bacterium are an intact shell and membrane. To verify their integrity, atomic force microscopy (AFM, see Figure 4) and fluorescence lifetime imaging microscopy (FLIM, see Figure 5) were used.
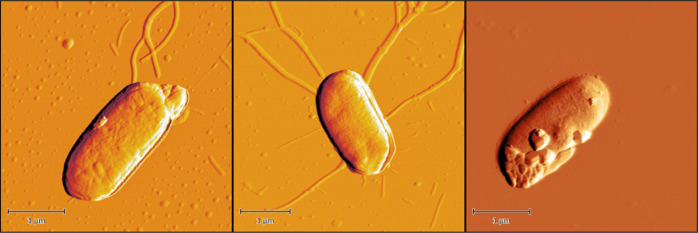
Figure 4.
Examples of AFM images of E. coli bacteria before (left) and after treatment (middle) with radiation or 0.4% formaldehyde (right)—preparation: a solution of E. coli in PBS 107 CFU/mL was added dropwise on a slate plat and dried with air.
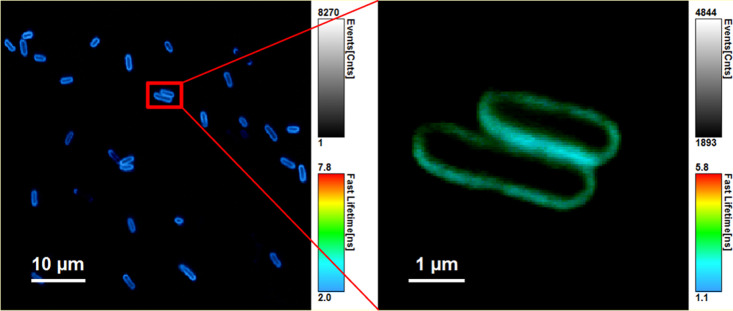
Figure 5.
Representative false color-coded FLIM images of E. coli with the fluorescence marker FM1-43, which is selective for bacteria membrane, λex = 475 nm, λem > 500 nm after irradiation with low-pressure Hg radiator (colder colors indicate lower decay time)—preparation: a solution of E. coli in PBS 107 CFU/mL and FM1-43 (c = 1 μM) was fixed on the coverslip with Vectabond and the staining was stopped after 2 min incubation time with 100 μL of 1% aqueous agarose solution.
In AFM images, the exterior shape of the bacteria can be visualized. The surface of the E. coli bacteria treated with 0.4% formaldehyde showed significant damage. In contrast, the untreated and the irradiated E. coli were undamaged. The FLIM images allow for an additional evaluation of the integrity of the outer E. coli membrane using the membrane selective fluorescence marker FM1-43. The irradiated E. coli showed no leakage of the marker; therefore, an intact membrane can be assumed.
2.5. Application
In summary, the verification of bacterial integrity shows that after irradiation, all tested external parts of the inactivated bacteria show no significant difference to the bacterial integrity before irradiation. The optimization of the photolytic inactivation by the addition of CNDs has not been tested in animals so far. However, since the photodynamically inactivated bacteria can be separated from the CNDs, there is no argument against their future use in animals. Thus, the self-synthesized CNDs could be a good alternative photosensitizer for future work with different bacterial strains.
3. Experimental Section
3.1. Materials
Water was purified in a Milli-Q Advantage A10. Phosphate-buffered saline (PBS) was acquired from Sigma-Aldrich (tablets, pH 7.4, P4417 Sigma). The laboratory strain E. coli (K12), B. avium, and O. rhinotracheale were prepared by RIPAC LABOR GmbH. The CNDs used in this study were custom-made via microwave-assisted hydrothermal treatment at the University of Potsdam (designated as HAc/N-CNDs).37 Our synthesized CNDs have a narrow size distribution of (2.40 ± 0.25) nm, a good solubility in water, and a high hygroscopy.37,38 Accordingly, the determination of the dry weight to specify concentrations was not possible for these CNDs.
3.2. Methods
3.2.1. Sample Preparation for the Inactivation
The photodynamic inactivation starts with the doping of the bacteria with a photosensitizer (methylene blue (MB) or CNDs). For a 1 mL of stock solution with E. coli (≈108 CFU/mL) in agarose (0.6 g agarose in 30 mL PBS), 30 μL of MB (1 mm stock solution in PBS) or 20 μL CND (stock solution), respectively, was added to the E. coli solution and shaken for minimum 10 min to fully load the bacteria with the photosensitizer (verified by fluorescence microscopy, data not shown).
3.2.2. Flow Inactivation
The inactivation of the bacteria was performed with a custom-made flow photoreactor with two different light sources (Hg low- or mid-pressure radiation sources), based on the literature.48 Both are surrounded by an immersion solution and a cooling tube (UV Laboratory Reactor System 2, Peschl Ultraviolet). The mid-pressure Hg radiator (TQ 150, Peschl Ultraviolet) is combined with a cooling tube made of borosilicate glass while the low-pressure Hg radiator (TNN 15/32, ozone-free, Peschl Ultraviolet) quartz glass is used for the cooling tube. The bacteria suspension is pumped with a flow rate between 0.6 and 90 mL/min via a piston pump (REGLO-CPF Digital with RH00 pump head, ISMATEC) through a perfluoroalkoxy alkane (PFA) tube (ID = 0.762 mm, IDEX Health & Science) wrapped around the surface of the cooling tube. The irradiated volume was 5600 μL with 12.5 mm between suspension and radiator surface. All inactivation runs were performed between 25 and 28 °C.
3.2.3. Batch Inactivation
In addition to the flow inactivation in the photoreactor, the CND-doped bacteria were also irradiated in fused silica cuvettes (thickness 5 mm) that were located 2.5 cm in front of a calibration lamp (halogen lamp LSB020 LOT-Quantum Design GmbH, Darmstadt, Germany for sunlight emission) or (365 ± 40) nm diode (OTLH 0480UV, OPTO-Technology, Inc., IL, for UV light emission).
3.2.4. Fluorescence Microscopy
To monitor the process of loading and releasing the CNDs in and from the bacteria, a fluorescence microscope (Zeiss Axio Observer Z1 inverted microscope, Carl Zeiss, Jena, Germany) was equipped with the objective Zeiss Fluar 20×/NA 0.75. To separate the excitation and emission light, two different filter cubes operating at λex = 485 ± 15 nm and λem = 535 ± 25 nm, or at λex = 565 ± 15 nm and λem = 630 ± 37.5 nm were used. The images were taken by a CCD camera (CoolSnap HQ2, Photometrics, Arizona). A short-arc lamp (X-Cite 120, Visitron Systems, Purchheim, Germany) was used for excitation. The data collection was done with the software MetaMorph ver. 7.1 (Molecular Devices, Sunnyvale). Data analysis was performed using Origin 9.1 G (Origin Lab Corp., Northampton).
For monitoring the loading process of the CNDs into the bacteria, 100 μL of E. coli from a stock solution (≈108 colony-forming units per milliliter [CFU/mL]) was fixed with agarose on a coverslip. To minimize the background in the fluorescence microscope images, E. coli bacteria and CNDs out of the focus were removed using a peristaltic pump (1 mL/min PBS flow for 6 min through a slide perfusion chamber). Afterward, a defined volume (10 or 100 μL) of the diluted CND stock solution (10 μL of stock in 5 mL of PBS) was added to the coverslip.
3.2.5. Atomic Force Microscopy (AFM)
The exterior shapes of the E. coli bacteria were analyzed using a Flex-Axiom AFM (Nanosurf GmbH, Langen, Germany). The Flex-Axiom AFM includes CameraView for displaying video output and PicoView for controlling all aspects of alignment, calibration, and imaging.
3.2.6. Fluorescence Lifetime Imaging Microscopy (FLIM)
Measurements of the fluorescence decay times were realized with the MicroTime 200 fluorescence lifetime microscope system (PicoQuant, Berlin, Germany) coupled with the time-correlated single photon counting (TCSPC) module Pico-Harp 300 exhibiting a time resolution of 8 ps. For excitation, a supercontinuum laser source (SC-400-2, Fianium) was used at λex = 475 nm with a repetition rate of 20 MHz. The excitation light was coupled via the dichroic mirror z467/638 rpc (AHF Analysentechnik, Tübingen, Germany) into the inverted microscope (IX71, Olympus, Hamburg, Germany) equipped with a Plan Apo ×100/NA 1.4 oil immersion objective (Olympus). The fluorescence signal passed a 30 μm pinhole and a long-pass filter (LP500, AHF Analysentechnik, Tübingen, Germany) and was detected by a single-photon avalanche diode (SPAD, SPCM-AGR-13, PerkinElmer, Waltham). Time-resolved fluorescence images were acquired by scanning the sample with an xy-piezoelectric scanner. Thus, full frame images of 80 × 80°μm were recorded with a resolution of 150 pixel × 150 pixel and a pixel dwell time of 0.6 ms/pixel. The images shown were recorded via the SymPhoTime64 software version 2.0 (PicoQuant, Berlin, Germany).
3.2.7. Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS)
The MALDI-TOF MS experiments were performed with the standard VITEK-MS RUO System (Biomerieux Deutschland GmbH) with the standard protocol. The cells (before and after photolytic inactivation) were harvested by centrifugation (2 min, 1.500 rpm) from cell suspension. The cell pellet was directly transferred on the MALDI-TOF MS target and mixed with matrix. The MALDI-TOF MS measurement and identification were performed with all manufacturer settings. Identification with 99,9% illustrate the presence of whole intact proteins. After degradation of proteins, like covalent formalin inactivation, the protein peaks are lost the spectra and the identification is negative. The cells after photodynamic inactivation were not tested by MALDI-TOF MS because the interaction of CND with the high voltage system in the instrument (strict warning from manufacturer).
4. Conclusions
The present work shows the potential of a gentle photodynamic inactivation of bacteria using carbon nanodots (CNDs) at wavelengths λex > 290 nm. During the implementation of this alternative inactivation method, the loading and the release process of the CNDs in the representative bacterium E. coli were monitored via fluorescence microscopy. After adding sufficient concentrations of the CND solution to the E. coli, all observed bacteria were fully loaded after 5–10 min. The release process took place in less than 25 min. The photodynamic inactivation with CNDs addition was up to 10 times faster than the reference method without CNDs with a UV lamp. After irradiation, the membrane integrity of the bacteria was successfully verified by agglutination and atomic force microscopy (AFM).
Acknowledgments
The authors gratefully acknowledge the financial support for this research received from the German Federal Ministry for Food and Agriculture (BMEL) (Grant No. 2817600712). We acknowledge the support of the Deutsche Forschungsgemeinschaft and Open Access Publishing Fund of University of Potsdam.
The authors declare no competing financial interest.
References
- D’Costa V. M.; King C. E.; Kalan L.; Morar M.; Sung W. W. L.; Schwarz C.; Froese D.; Zazula G.; Calmels F.; Debruyne R.; et al. Antibiotic resistance is ancient. Nature 2011, 477, 457–461. 10.1038/nature10388. [DOI] [PubMed] [Google Scholar]
- Radke V. Antimicrobial Resistance. J. Environ. Health 2019, 81, 6–7. [Google Scholar]
- Cassini A.; Högberg L. D.; Plachouras D.; Quattrocchi A.; Hoxha A.; Simonsen G. S.; Colomb-Cotinat M.; Kretzschmar M. E.; Devleesschauwer B.; Cecchini M.; et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis. Lancet Infect. Dis. 2019, 19, 56–66. 10.1016/S1473-3099(18)30605-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison L.; Zembower T. R. Antimicrobial Resistance. Gastrointest. Endoscopy Clin. North Am. 2020, 30, 619–635. 10.1016/j.giec.2020.06.004. [DOI] [PubMed] [Google Scholar]
- World Health Organization (WHO). Leveraging Vaccines to Reduce Antibiotic Use and Prevent Antimicrobial Resistance: An Action Framework; CC BY-NC-S, 2020. [DOI] [PMC free article] [PubMed]
- Witte W. Medical Consequences of Antibiotic Use in Agriculture. Science 1998, 279, 996–997. 10.1126/science.279.5353.996. [DOI] [PubMed] [Google Scholar]
- Leverstein-van Hall M. A.; Dierikx C. M.; Cohen Stuart J.; Voets G. M.; van den Munckhof M. P.; van Essen-Zandbergen A.; Platteel T.; Fluit A. C.; van de Sande-Bruinsma N.; Scharinga J.; et al. Dutch patients, retail chicken meat and poultry share the same ESBL genes, plasmids and strains. Clin. Microbiol. Infect. 2011, 17, 873–880. 10.1111/j.1469-0691.2011.03497.x. [DOI] [PubMed] [Google Scholar]
- Bundesministerium für Gesundheit; Bundesministerium für Ernährung und Landwirtschaft; Bundesministerium für Bildung und Forschung. Dart 2020 -Antibiotika-Resistenzen bekämpfen zum Wohl von Mensch und Tier; 2020.
- Wang C.; Lu S.; Zhang Z. Inactivation of airborne bacteria using different UV sources: Performance modeling, energy utilization, and endotoxin degradation. Sci. Total Environ. 2019, 655, 787–795. 10.1016/j.scitotenv.2018.11.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao P.; Han K.; Tang Y.; Wang B.; Lin T.; Cheng W. Recent advances in carbon nanodots: Synthesis, properties and biomedical applications. Nanoscale 2015, 7, 1586–1595. 10.1039/C4NR05712K. [DOI] [PubMed] [Google Scholar]
- Bing W.; Sun H.; Yan Z.; Ren J.; Qu X. Programmed Bacteria Death Induced by Carbon Dots with Different Surface Charge. Small 2016, 12, 4713–4718. 10.1002/smll.201600294. [DOI] [PubMed] [Google Scholar]
- Dong X.; Liang W.; Meziani M. J.; Sun Y. P.; Yang L. Carbon dots as potent antimicrobial agents. Theranostics 2020, 10, 671–686. 10.7150/thno.39863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C. X.; Yu C.; Wang C. F.; Chen S. Facile plasma-induced fabrication of fluorescent carbon dots toward high-performance white LEDs. J. Mater. Sci. 2013, 48, 6307–6311. 10.1007/s10853-013-7430-6. [DOI] [Google Scholar]
- Saud P. S.; Pant B.; Alam A.-M.; Ghouri Z. K.; Park M.; Kim H.-Y. Carbon quantum dots anchored TiO2 nanofibers: Effective photocatalyst for waste water treatment. Ceram. Int. 2015, 41, 11953–11959. 10.1016/j.ceramint.2015.06.007. [DOI] [Google Scholar]
- Li H.; He X.; Kang Z.; Huang H.; Liu Y.; Liu J.; Lian S.; Tsang C. H. A.; Yang X.; Lee S. T. Water-soluble fluorescent carbon quantum dots and photocatalyst design. Angew. Chem., Int. Ed. 2010, 49, 4430–4434. 10.1002/anie.200906154. [DOI] [PubMed] [Google Scholar]
- Derfus A. M.; Chan W. C. W.; Bhatia S. N. Probing the Cytotoxicity of Semiconductor Quantum Dots. Nano Lett. 2004, 4, 11–18. 10.1021/nl0347334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S. Y.; Shen W.; Gao Z. Carbon quantum dots and their applications. Chem. Soc. Rev. 2015, 44, 362–381. 10.1039/C4CS00269E. [DOI] [PubMed] [Google Scholar]
- Tabish T. A.; Zhang S.. Graphene Quantum Dots: Syntheses, Properties, and Biological Applications. In Comprehensive Nanoscience and Nanotechnology; Elsevier, 2016; Vol. 3, pp 171–192. [Google Scholar]
- Wang L.; Zhu S.; Lu T.; Zhang G.; Xu J.; Song Y.; Li Y.; Wang L.; Yang B.; Li F. The effects of a series of carbon dots on fibrillation and cytotoxicity of human islet amyloid polypeptide. J. Mater. Chem. B 2016, 4, 4913–4921. 10.1039/C6TB00921B. [DOI] [PubMed] [Google Scholar]
- Wright P. C.; Qin H.; Choi M. M. F.; Chiu N. H. L.; Jia Z. Carbon nanodots interference with lactate dehydrogenase assay in human monocyte THP-1 cells. Springerplus 2014, 3, 615 10.1186/2193-1801-3-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.; He X.; Li Y.; Zhang Z.; Ma Y.; Li F.; Liu J. A Cytotoxicity Study of Fluorescent Carbon Nanodots Using Human Bronchial Epithelial Cells. J. Nanosci. Nanotechnol. 2013, 13, 5254–5259. 10.1166/jnn.2013.7528. [DOI] [PubMed] [Google Scholar]
- Goh E. J.; Kim K. S.; Kim Y. R.; Jung H. S.; Beack S.; Kong W. H.; Scarcelli G.; Yun S. H.; Hahn S. K. Bioimaging of hyaluronic acid derivatives using nanosized carbon dots. Biomacromolecules 2012, 13, 2554–2561. 10.1021/bm300796q. [DOI] [PubMed] [Google Scholar]
- Wang B.; Wang Y.; Wu H.; Song X.; Guo X.; Zhang D.; Ma X.; Tan M. A mitochondria-targeted fluorescent probe based on TPP-conjugated carbon dots for both one- and two-photon fluorescence cell imaging. RSC Adv. 2014, 4, 49960–49963. 10.1039/C4RA07467J. [DOI] [Google Scholar]
- Wang L.; Yin Y.; Jain A.; Susan Zhou H. Aqueous phase synthesis of highly luminescent, nitrogen-doped carbon dots and their application as bioimaging agents. Langmuir 2014, 30, 14270–14275. 10.1021/la5031813. [DOI] [PubMed] [Google Scholar]
- Ke Y.; Garg B.; Ling Y. C. Waste chicken eggshell as low-cost precursor for efficient synthesis of nitrogen-doped fluorescent carbon nanodots and their multi-functional applications. RSC Adv. 2014, 4, 58329–58336. 10.1039/C4RA10178B. [DOI] [Google Scholar]
- Fowley C.; Nomikou N.; McHale A. P.; McCaughan B.; Callan J. F. Extending the tissue penetration capability of conventional photosensitisers: A carbon quantum dot-protoporphyrin IX conjugate for use in two-photon excited photodynamic therapy. Chem. Commun. 2013, 49, 8934–8936. 10.1039/c3cc45181j. [DOI] [PubMed] [Google Scholar]
- Yang S. T.; Cao L.; Luo P. G.; Lu F.; Wang X.; Wang H.; Meziani M. J.; Liu Y.; Qi G.; Sun Y. P. Carbon dots for optical imaging in vivo. J. Am. Chem. Soc. 2009, 131, 11308–11309. 10.1021/ja904843x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L.; Wang X.; Meziani M. J.; Lu F.; Wang H.; Luo P. G.; Lin Y.; Harruff B. A.; Veca L. M.; Murray D.; et al. Carbon dots for multiphoton bioimaging. J. Am. Chem. Soc. 2007, 129, 11318–11319. 10.1021/ja073527l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y.; Wang R.; Li G.; Chen C.; Chi Y.; Chen G. Polyamine-functionalized carbon quantum dots as fluorescent probes for selective and sensitive detection of copper ions. Anal. Chem. 2012, 84, 6220–6224. 10.1021/ac3012126. [DOI] [PubMed] [Google Scholar]
- Liu C.; Zhang P.; Zhai X.; Tian F.; Li W.; Yang J.; Liu Y.; Wang H.; Wang W.; Liu W. Nano-carrier for gene delivery and bioimaging based on carbon dots with PEI-passivation enhanced fluorescence. Biomaterials 2012, 33, 3604–3613. 10.1016/j.biomaterials.2012.01.052. [DOI] [PubMed] [Google Scholar]
- Zhu S.; Zhang J.; Qiao C.; Tang S.; Li Y.; Yuan W.; Li B.; Tian L.; Liu F.; Hu R.; et al. Strongly green-photoluminescent graphene quantum dots for bioimaging applications. Chem. Commun. 2011, 47, 6858–6860. 10.1039/c1cc11122a. [DOI] [PubMed] [Google Scholar]
- Li H.; Huang J.; Song Y.; Zhang M.; Wang H.; Lu F.; Huang H.; Liu Y.; Dai X.; Gu Z.; et al. Degradable Carbon Dots with Broad-Spectrum Antibacterial Activity. ACS Appl. Mater. Interfaces 2018, 10, 26936–26946. 10.1021/acsami.8b08832. [DOI] [PubMed] [Google Scholar]
- Sharma A.; Das J. Small molecules derived carbon dots: Synthesis and applications in sensing, catalysis, imaging, and biomedicine. J. Nanobiotechnol. 2019, 17, 92 10.1186/s12951-019-0525-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.; Lu F.; Ma C.; Ma Y.; Zhang M.; Wang B.; Zhang Y.; Liu Y.; Huang H.; Kang Z. Carbon dots with positive surface charge from tartaric acid and m-aminophenol for selective killing of Gram-positive bacteria. J. Mater. Chem. B 2021, 9, 125–130. 10.1039/D0TB02332A. [DOI] [PubMed] [Google Scholar]
- Cui F.; Ye Y.; Ping J.; Sun X. Carbon dots: Current advances in pathogenic bacteria monitoring and prospect applications. Biosens. Bioelectron. 2020, 156, 112085 10.1016/j.bios.2020.112085. [DOI] [PubMed] [Google Scholar]
- Liang X.-J.; Chen C.; Zhao Y.; Jia L.; Wang P. Biopharmaceutics and Therapeutic Potential of Engineered Nanomaterials. Curr. Drug Metab. 2008, 9, 697–709. 10.2174/138920008786049230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiling T. T.; Schürmann R.; Vogel S.; Ebel K.; Nicolas C.; Milosavljević A. R.; Bald I. Photophysics and Chemistry of Nitrogen-Doped Carbon Nanodots with High Photoluminescence Quantum Yield. J. Phys. Chem. C 2018, 122, 10217–10230. 10.1021/acs.jpcc.8b00748. [DOI] [Google Scholar]
- Meiling T. T.; Cywiński P. J.; Bald I. White carbon: Fluorescent carbon nanoparticles with tunable quantum yield in a reproducible green synthesis. Sci. Rep. 2016, 6, 28557 10.1038/srep28557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz K.-H.Chapter 15 - Avian bordetellosis (turkey coryza), InPattison M.; McMullin P. F.; Bradbury J. M.; Alexander D. J. B. T., Eds.; W.B. Saunders, Edinburgh, 2008; pp 176–180. [Google Scholar]
- Hinz K. H.; Gluender G. Prevalence of Bordetella avium sp. nov. and Bordetella bronchiseptica in birds. Berl. Muench. Tieraerztl. Wochenschr. 1985, 369–373. [PubMed] [Google Scholar]
- Van Empel P. C. M.; Hafez H. M. Ornithobacterium rhinotracheale: A review. Avian Pathol. 1999, 28, 217–227. 10.1080/03079459994704. [DOI] [PubMed] [Google Scholar]
- Refai M.; El-Gohary A.; Attia S. A.; Khalifa R. A. Diagnosis of Ornithobacterium rhinotracheale infection in chickens by ELISA. Egypt. J. Immunol. 2005, 12, 87–93. [PubMed] [Google Scholar]
- Cushing S. K.; Ding W.; Chen G.; Wang C.; Yang F.; Huang F.; Wu N. Excitation wavelength dependent fluorescence of graphene oxide controlled by strain. Nanoscale 2017, 9, 2240–2245. 10.1039/C6NR08286F. [DOI] [PubMed] [Google Scholar]
- Cushing S. K.; Li M.; Huang F.; Wu N. Origin of strong excitation wavelength dependent fluorescence of graphene oxide. ACS Nano 2014, 8, 1002–1013. 10.1021/nn405843d. [DOI] [PubMed] [Google Scholar]
- Ragàs X.; Dai T.; Tegos G. P.; Agut M.; Nonell S.; Hamblin M. R. Photodynamic inactivation of Acinetobacter baumannii using phenothiazinium dyes: In vitro and in vivo studies. Lasers Surg. Med. 2010, 42, 384–390. 10.1002/lsm.20922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecchio D.; Gupta A.; Huang L.; Landi G.; Avci P.; Rodas A.; Hamblina M. R. Bacterial photodynamic inactivation mediated by methylene blue and red light is enhanced by synergistic effect of potassium iodide. Antimicrob. Agents Chemother. 2015, 59, 5203–5212. 10.1128/AAC.00019-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rienstra-Kiracofe J. C.; Tschumper G. S.; Schaefer H. F.; Nandi S.; Ellison G. B. Atomic and molecular electron affinities: Photoelectron experiments and theoretical computations. Chem. Rev. 2002, 102, 231–282. 10.1021/cr990044u. [DOI] [PubMed] [Google Scholar]
- Shen B.; Bedore M. W.; Sniady A.; Jamison T. F. Continuous flow photocatalysis enhanced using an aluminum mirror: Rapid and selective synthesis of 2′-deoxy and 2′,3′-dideoxynucleosides. Chem. Commun. 2012, 48, 7444–7446. 10.1039/c2cc33356b. [DOI] [PubMed] [Google Scholar]