Abstract

(E)-9-Oxooctadec-10-en-12-ynoic acid is found to mediate its antidiabetic activity by increasing insulin-stimulated glucose uptake in L6 myotubes by activating the phosphoinositide 3-kinase (PI3K) pathway. A simultaneous study of site-specific modification followed by structure–activity relationship provides a tremendous scope for exploiting the bioactivity of the parent molecule. Therefore, in the present study, we focused on site-specific modification of (E)-9-oxooctadec-10-en-12-ynoic acid (1) to generate multiple derivatives and extensive structure–activity relationship (SAR) studies. We have done structural base design and synthesized a series of amides from acid compound 1. Compound 1 consists of an acid functionality, which is known for its metabolism-related liabilities. The SAR has been generated using scaffolds of different antidiabetic drugs such as biguanides, sulfonylureas, thiazolidinediones/glitazones, peroxisome proliferator-activated receptors, K + ATP, α-glucosidase inhibitors, and others. Furthermore, the study demonstrates and explains the promising derivatives and importance of SAR of the compound (E)-9-oxooctadec-10-en-12-ynoic acid. In order to gain mechanistic insights, a molecular docking study was performed against PI3K, which could identify the binding modes and thermodynamic interactions governing the binding affinity. According to our research, compounds 5, 6, 27, 28, 31, 32, and 33 are the best compounds from the series having EC50 values of 15.47, 8.89, 7.00, 13.99, 8.70, 12.27, and 16.14 μM, respectively.
Introduction
Diabetes mellitus (DM) is a chronic metabolic disorder occurring in humans that is caused by the inability to produce enough insulin by the islet cells in the pancreas (insulin insufficiency) or when the cells in the body cannot effectively utilize the insulin it produces (insulin resistance). It is a metabolic disorder where disrupted glucose homeostasis results in high circulating levels of glucose in the blood.1 Insulin plays an important role in maintaining glucose homeostasis by facilitating glucose uptake, largely in adipocytes and skeletal muscles of healthy individuals.2 Diabetic patients are not able to metabolize glucose in their bodies and are categorized mainly as having type I (T1DM) or type II (T2DM). T2DM is the major type of diabetes and accounts for around 90% of all diabetes cases.
According to the World Health Organization (WHO), 422 million people worldwide are suffering from diabetes, which is the leading cause of death from the years 2000 to 2019. The disease has severe metabolic and multi-organ complications such as kidney failure, heart disease, vascular disease, and retinal disease due to sustained high blood glucose levels for a long period of time.3 The available treatment options have limited efficacy, significant mechanism-based side effects, and high cost.4 Hence, it is an urgent and unmet need to explore new drugs and treatment modalities with better safety and efficacy profiles.
Historically, plants have been a source of developing low-cost drugs that have little potential side effects in many countries and cultures. These medicinal plants have been known and reported for their antidiabetic activity. Natural products from plants have an excellent history in ancient medicine and a vast potential for the discovery of active pharmacophores.5,6
The aqueous extract of the leaves of Ixora coccinea Linn showed a significant reduction in blood glucose levels and serum lipid profile levels in alloxan-induced diabetic rats.7 There have been literature reports that the compound (E)-9-oxooctadec-10-en-12-ynoic acid (1) mediates its effects on glucose uptake activity. It was demonstrated that this compound increases insulin-stimulated glucose uptake in L6 myotubes by activating the phosphoinositide 3-kinase (PI3K) pathway.8 Based on this reported finding, we have designed a series of amides of (E)-9-oxooctadec-10-en-12-ynoic acid (1).
Compound 1 consists of an acid functionality, which is associated with metabolite liabilities. An acid moiety can also be responsible for metabolic instability and toxicity.9 To counter such problems in metabolism, biotransformation studies are becoming significant in guiding the modification of a lead series during drug discovery and in characterizing lead candidates.10 One of the commonly used strategies in medicinal chemistry is the use of bioisosteres to improve the drug-like properties and resolve such issuses.11 Bioisosterism is used in the modification of lead compounds that eliminate toxicity and enhance the potency and pharmacokinetic properties of the molecule.
Therefore, we planned to design and synthesize various amides as bioisosteres of acid compound 1. Furthermore, the amide group has a significant role in forming hydrogen bond interactions, which may act as a hydrogen bond acceptor or hydrogen bond donor. The amide functional group enhances the conformational rigidity.12 The amide functional group plays an important role in the design of drug molecules; hence, it appears in many clinically approved drugs.
Due to the widespread occurrence of amides in natural products, pharmaceuticals, and biologically active compounds, various synthetic methods are available for the synthesis of amides. The common method for the synthesis of amides involves the reaction of activated carboxylic acid derivatives, such as chlorides, anhydrides, or esters, with amines, or, alternatively, the direct union of the carboxylic acids with amines assisted by stoichiometric amounts of coupling reagents.13
Experimental Section
General Information
1H, 13C, DEPT, and 2D NMR spectra were recorded on Bruker-Avance 300 MHz and 500 MHz instruments. The NMR experiments were carried out in the indicated deuterated solvent. Chemical shift data for protons are reported in parts per million (ppm) downfield from TMS and are referenced to the residual proton in the NMR solvent [(CD3)2SO, 2.50; CDCl3, 7.28 ppm]. 13C NMR spectra were recorded at 125 and 75 MHz; chemical shift data for carbons are reported in parts per million (ppm, δ scale) downfield from TMS and are referenced to the carbon resonance of the solvent [(CD3)2SO, 39.52; CDCl3, 77.16 ppm]. ESI-MS and HRMS spectra were recorded on a Bruker Daltonics MicroTOFq instrument. All the solvents used were of analytical grade and purchased from Merck.
Plant Materials
The aerial parts of I. brachiata were collected from the southern parts of the Maharashtra state in India in the months of March 2008 and taxonomically classified. The voucher specimens (collection no. 01650), for future reference, are deposited to the herbarium of the Department of Natural Products Botany, Piramal Enterprises Limited, Mumbai.
Isolation of (E)-9-Oxooctadec-10-en-12-ynoic Acid (1)
The chopped pieces of the collected plant were dried in the drying room with the help of a dehumidifier. The completely dried materials were then coarsely ground with the help of a “Bulani pulverizer” to a mesh size of 8 and taken for extraction. The pulverized plant material (500 g) was taken in a 5 L flat-bottom flask for extraction and soaked in 3.5 L of petroleum ether and stirred for 3 h in a water bath at 40 ± 5 °C. After 3 h, the soaked material was filtered using a Whatman filter paper. The same process was repeated twice, and the collected filtrates were concentrated with a rotary evaporator at 45 °C under line vacuum until it is completely dried. The concentrated material was in the form of thick dark green paste. The yield of the extract was 25 g (5.0%). The cold preserved petroleum ether extract was subjected to fractionation and purification using normal-phase flash chromatography. The fractions were eluted with a petroleum ether and ethyl acetate gradient. Further, the pure compound was isolated by reverse-phase semipreparative HPLC using a C18 (250 mm × 21.5 mm, 10 μm) HPLC column; the mobile phase was a water and acetonitrile premix in a ratio of 30:70 and detection of (λmax) = 280 nm.
General Methods for Preparation of Compounds 4–43
To a stirred solution of (E)-9-oxooctadec-10-en-12-ynoic acid (25 mg) in dichloromethane (2 mL), DIPEA (1.5 equiv), HATU (1.5 equiv) and 1 equiv of amine were added. The reaction was then stirred for 12 h at room temperature. After completion of the reaction, the solvent was removed under a high vacuum and again dissolved in 50 mL of ethyl acetate washed with water and brine. The solvent was removed and purified by a semi-preparative column to get the purified final compounds (yield: 60–95%). The data sets supporting the results of this article are available in the Supporting Information.
(E)-9-Oxooctadec-10-en-12-ynoic Acid (1)
White solid; HPLC purity: 99.72% (tR = 13.08 min); 1H NMR (500 MHz, CDCl3, ppm): δ 6.67 (d, 1H, J = 16.0 Hz), 6.46 (d, 1H J = 16.0 Hz), 2.53 (t, 2H, J = 7.5 Hz), 2.40 (t, 2H, J = 7.0 Hz), 2.36 (t,2H, J = 8.0 Hz), 1.63 (m, 4H), 1.56 (m, 2H), 1.63–1.34 (m, 10H), 0.92 (t, 3H, J = 7.0 Hz); 13C NMR (125 MHz, CDCl3, ppm): δ 199.63 (C=O), 178.24 (Cx1), 136.51 (1xCH), 124.07 (1xCH), 101.88 (Cx1), 78.48 (1xC), 29.01 (2xCH2), 40.81, 34.01, 31.04, 28.88, 28.02, 24.70, 23.99, 22.15, 19.83 (9xCH2), 13.94 (1xCH3); HRMS–ESI m/z: calcd for [C18H27O3]− [M – H]−: 291.1996; found: 291.1955.
(E)-9-Oxo-N-phenyloctadec-10-en-12-ynamide (4)
Yield: 79%; brown solid; HPLC purity: 98.63% (tR = 14.10 min); 1H NMR (500 MHz, CDCl3, ppm): δ 7.53 (d, 2H, J = 8.0 Hz), 7.43 (bs, −NH), 7.32 (t, 2H, J = 7.5 Hz), 7.12 (t, 2H, J = 7.0 Hz), 6.66 (d, 1H, J = 16.0 Hz), 6.46 (d, 1H, J = 16.0 Hz), 2.54 (t, 2H, J = 7.5 Hz), 2.40 (t, 2H, J = 7.5 Hz), 2.35 (t, 2H, J = 8.0 Hz), 1.74 (t, 2H, J = 7.0 Hz), 1.60 (m, 2H), 1.41–1.27 (m, 10H), 0.92 (t, 3H, J = 7.0 Hz); 13C NMR (125 MHz, CDCl3, ppm): δ 199.72 (C=O), 171.45 (1xC), 138.05 (1xC), 136.52 (1xCH), 128.96 (3xCH), 124.14 (1xCH), 119.76 (2xCH), 101.98 (1xC), 78.48 (1xC), 40.79, 38.63, 37.70, 31.05, 29.02, 28.95, 28.03, 25.47, 23.96, 22.17, 19.85 (11xCH2), 13.96 (1xCH3); HRMS-ESI m/z: calcd for [C24H32NO2]−[M – H]−: 366.2439; found: 366.2461.
(E)-N-(Cyclohexylmethyl)-9-oxooctadec-10-en-12-ynamide (5)
Yield: 89%; white solid; HPLC purity: 98.09% (tR = 14.88 min); 1H NMR (500 MHz, CDCl3, ppm): δ 6.67 (d, 1H, J = 16.0 Hz), 6.46 (d, 1H, J = 16.0 Hz), 5.50 (bs, −NH), 3.10 (t, 2H, J = 6.0 Hz), 2.52 (t, 2H, J = 7.0 Hz), 2.39 (t, 2H, J = 7.5 Hz), 2.16 (t, 2H, J = 7.5 Hz), 1.70–1.17 (m, 27H), 0.92 (t, 3H, J = 7.0 Hz); 13C NMR (100 MHz, CDCl3, ppm): δ 199.61 (C=O), 173.04 (1xC), 136.53 (1xC), 124.06 (1xC), 101.88 (1xC), 78.48 (1xC), 30.84 (2xCH2), 29.05 (2xCH2), 25.81 (2xCH2), 37.95 (1xCH), 45.67, 40.82, 36.89, 31.05, 29.01, 28.03, 26.40, 25.74, 23.98, 22.16, 19.84 (11xCH2), 13.94 (1xCH3); HRMS-ESI m/z: calcd for [C25H40NO2]− [M – H]−: 386.3065; found: 386.3101.
(R,E)-N-(1-Cyclohexylethyl)-9-oxooctadec-10-en-12-ynamide (6)
Yield: 90%; off-white solid; yield, 90%; HPLC purity: 99.15% (tR = 15.24 min); 1H NMR (500 MHz, CDCl3, ppm): δ 6.66 (d, 1H, J = 16.0 Hz), 6.45 (d, 1H, J = 16.0 Hz), 5.32 (d, −NH, J = 8.0 Hz), 3.87 (q, 1H, J = 7.0 Hz), 2.51 (t, 2H, J = 7.3 Hz), 2.38 (t, 2H, J = 6.5 Hz), 2.14 (t, 2H, J = 7.5 Hz), 1.17–1.14 (m-27H), 1.08 (d, 3H, J = 7.0 Hz), 0.91 (t, 3H, J = 7.0 Hz); 13C NMR (125 MHz, CDCl3, ppm): δ 199.61 (C=O), 172.29 (1xC), 136.52 (1xCH), 124.05 (1xCH), 101.88 (1xC), 78.48 (1xC), 49.18 (1xCH), 43.08 (1xCH), 29.03 (5xCH2), 26.17 (2xCH2), 40.81, 37.06, 31.05, 28.02, 26.41, 25.78, 23.99, 22.15, 19.83 (9xCH2), 18.02 (1xCH3), 13.93 (1xCH3); HRMS-ESI m/z: calcd for [C26H42NO2]− [M – H]−: 400.3221; found: 400.3304.
(S,E)-N-(1-Cyclohexylethyl)-9-oxooctadec-10-en-12-ynamide (7)
Yield: 65.3%; off-white solid; yield: 65%; HPLC purity: 98.85% (tR = 15.25 min); 1H NMR (500 MHz, CDCl3, ppm): δ 6.66 (d, 1H, J = 16.0 Hz), 6.45 (d, 1H, J = 16.0 Hz), 5.28 (d, −NH, J = 8.0 Hz), 3.87 (q, 1H, J = 7.5 Hz), 2.52 (t, 2H, J = 7.5 Hz), 2.39 (t, 2H, J = 6.5 Hz), 2.15 (t, 2H, J = 7.5 Hz), 1.74–1.11 (m-27H), 1.08 (d, 3H, J = 6.8 Hz), 0.92 (t, 3H, J = 7.0 Hz); 13C NMR (125 MHz, CDCl3, ppm): δ 199.59 (C=O), 172.26 (1xC), 136.53 (1xCH), 124.04 (1xCH), 101.86 (1xC), 78.49 (1xC), 49.17 (1xCH), 43.09 (1xCH), 29.04 (5xCH2), 26.18 (2xCH2), 40.81, 37.08, 31.05, 28.03, 26.42, 25.78, 23.99, 22.16, 19.84 (9xCH2), 18.07 (1xCH3), 13.94 (1xCH3); HRMS-ESI m/z: calcd for [C26H42NO2]− [M – H]−: 400.3221; found: 400.3297.
(E)-N-Cyclohexyl-N-isopropyl-9-oxooctadec-10-en-12-ynamide (8)
Yield: 74.2%; brown solid; HPLC purity: 98.67% (tR = 16.86 min); 1H NMR (500 MHz, CDCl3, ppm): δ 6.66 (d, 1H, J = 16.0 Hz), 6.45 (d, 1H, J = 16.0 Hz), 3.96 (t, 1H, J = 6.0 Hz), 3.44 (bm, 1H), 2.58 (t, 2H, J = 7.0 Hz), 2.38 (t, 2H, J = 6.0 Hz), 2.27 (q, 2H, J = 7.0 Hz), 1.86–1.32 (m-32H), 1.20 (d, 6H, J = 8.0 Hz), 0.91 (t, 3H, J = 7.0 Hz); 13C NMR (125 MHz, CDCl3, ppm): δ 199.71 (C=O), 172.21 (1xC), 136.54 (1xCH), 124.04 (1xCH), 101.84 (1xC), 78.49 (1xC), 54.98 (1xCH), 54.70 (1xCH), 29.24 (2xCH2), 40.90, 35.39, 31.04, 30.18, 29.07, 28.02, 26.63, 26.07, 25.44, 25.33, 24.57, 24.04, 22.15, 19.83 (14xCH2), 20.74 (2xCH3), 13.93 (1xCH3); HRMS-ESI m/z: calcd for [C27H46NO2]+ [M + H]+: 416.3523; found: 416.3359.
(E)-N-(Naphthalen-1-ylmethyl)-9-oxooctadec-10-en-12-ynamide (9)
Yield: 92%; brown solid; HPLC purity: 99.09% (tR = 14.46 min); 1H NMR (500 MHz, CDCl3, ppm): δ 8.01 (d, 1H, J = 8.0 Hz), 7.89 (d, 1H, J = 8.0 Hz), 7.83 (t, 1H, J = 5.0 Hz), 7.53 (t, 2H, J = 8.0 Hz), 7.43 (d, 2H, J = 5.0 Hz), 6.66 (d, 1H, J = 16.0 Hz), 6.43 (d, 1H, J = 16.0 Hz), 5.86 (bs, 1H, −NH), 4.88 (d, 2H, J = 5.0 Hz), 2.57 (t, 2H, J = 7.0 Hz), 2.39 (t, 2H, J = 7.0 Hz), 2.17 (t, 2H, J = 7.0 Hz), 1.62–1.17 (m-16H), 0.92 (t, 3H, J = 7.0 Hz); 13C NMR (125 MHz, CDCl3, ppm): δ 199.66 (C=O), 172.83 (1xC), 136.54 (1xC),133.88 (1xC), 133.65 (1xC), 131.39 (1xCH), 128.76 (1xCH), 128.61 (1xCH), 126.75 (1xCH), 126.61 (1xCH), 126.02 (1xCH), 125.40 (1xCH), 124.09 (1xCH), 123.57 (1xCH), 101.94 (1xC), 78.49 (1xC), 28.97 (2xCH2), 41.75, 40.76, 36.64, 31.05, 28.95, 28.03, 25.61, 23.92, 22.16, 18.94 (10xCH2), 13.95 (1xCH3); HRMS-ESI m/z: calcd for [C29H36NO2]− [M – H]−: 430.2752; found: 430.2719.
(E)-N-(3,5-Difluorobenzyl)-9-oxooctadec-10-en-12-ynamide (10)
Yield: 77%; brown solid; HPLC purity: 98.45% (tR = 13.95 min); 1H NMR (500 MHz, CDCl3, ppm): δ 6.80 (d, 1H, J = 6.0 Hz), 6.74–6.69 (m, 1H), 6.67 (t, 1H, J = 2.0 Hz), 6.61 (d, 1H, J = 2.0 Hz), 6.43 (d, 1H, J = 16.0 Hz), 6.14 (bs, 1H, −NH), 4.43 (d, 2H, J = 6.0 Hz), 2.51 (t, 2H, J = 7.0 Hz), 2.39 (t, 2H, J = 7.0 Hz), 2.24 (t, 2H, J = 7.0 Hz), 1.62–1.32 (m-16H), 0.92 (t, 3H, J = 7.0 Hz); 13C NMR (125 MHz, CDCl3, ppm): δ 199.69 (C=O), 173.26 (1xC), 164.87 (1xC), 161.19 (1xC), 142.73 (1xC), 136.49 (1xCH), 124.12 (1xCH), 110.40 (1xCH), 110.07 (1xCH), 102.73 (1xCH), 101.99 (1xC), 78.46 (1xC), 28.96 (2xCH2), 28.93 (2xCH2), 40.76, 42.65, 38.8, 36.51, 31.04, 28.02, 25.52, 23.91, 22.15, 19.83, (10xCH2), 13.94 (1xCH3); HRMS-ESI m/z: calcd for [C25H32F2NO2]− [M–H]−: 416.2407; found: 416.2379.
(E)-N-(3,5-Difluorobenzyl)-9-oxooctadec-10-en-12-ynamide (11)
Yield: 93%; brown semi-solid; HPLC purity: 98.11% (tR = 14.55 min); 1H NMR (300 MHz, CDCl3, ppm): δ 6.67 (d, 1H, J = 16.0 Hz), 6.45 (d, 1H, J = 16.0 Hz), 5.54 (bs, 1H, −NH), 3.31–3.21 (m, 2H), 2.51 (t, 2H, J = 7.0 Hz), 2.38 (t, 2H, J = 6.0 Hz), 2.13 (t, 2H, J = 8.0 Hz), 1.59–1.30 (m-18H), 0.92 (s, 12H); 13C NMR (75 MHz, CDCl3, ppm): δ 199.70 (C=O), 173.11 (1xC), 136.50 (1xCH), 124.10 (1xCH), 101.95 (1xCH), 78.46 (1xC), 29.38 (3xCH3), 43.34, 40.79, 36.78, 38.80, 36.17, 31.03, 29.90, 29.21, 29.01, 28.01, 25.65, 23.97, 22.14, 18.82 (13xCH2), 13.93 (1xCH3); HRMS-ESI m/z: calcd for [C24H41NO2Na]+ [M + H + Na]+: 398.3030; found: 398.3035.
(E)-N-Cyclopentyl-9-oxooctadec-10-en-12-ynamide (12)
Yield: 79%; brown solid; HPLC purity: 98.95% (tR = 14.03 min);1H NMR (500 MHz, CDCl3, ppm): δ 6.67 (d, 1H, J = 16.0 Hz), 6.45 (d, 1H, J = 16.0 Hz), 5.48 (bs, 1H, −NH), 4.22–4.17 (m, 1H), 2.52 (t, 2H, J = 7.0 Hz), 2.39 (t, 2H, J = 6.0 Hz), 2.13 (t, 2H, J = 7.0 Hz), 2.0–1.3 (m-24H), 0.92 (t, 3H, J = 7.0 Hz); 13C NMR (75 MHz, CDCl3, ppm): δ 199.98 (C=O), 172.03 (1xC), 136.45 (1xCH), 124.37 (1xCH), 99.61 (1xCH), 78.45 (1xC), 55.31 (1xCH), 40.72, 36.33, 35.09, 33.96, 31.04, 29.69, 29.00, 28.83, 28.01, 25.28, 24.74, 24.01, 23.89, 22.15, 19.84 (15xCH2), 13.93 (1xCH3); HRMS-ESI m/z: calcd for [C23H36NO2]− [M–H]−: 358.2752; found: 358.2709.
(E)-9-Oxo-N-(thiazol-2-yl)octadec-10-en-12-ynamide (13)
Yield: 69.1%; off-white solid; HPLC purity: 99.03% (tR = 12.87 min); 1H NMR (300 MHz, CDCl3, ppm): δ 12.08 (bs, −NH), 7.44 (d, 1H, J = 4.0 Hz), 7.02 (d, 1H, J = 4.0 Hz), 6.68 (d, 1H, J = 16 Hz), 6.46 (d, 1H, J = 16.0 Hz), 2.59–2.49 (m, 4H), 2.39 (t, 2H, J = 7.0 Hz), 1.78 (t, 2H, J = 7.0 Hz) 1.63–1.36 (m-18H), 0.92 (t, 3H, J = 7.0 Hz); 13C NMR (75 MHz, CDCl3, ppm): δ 199.58 (C=O), 171.23 (1xC), 159.95 (1xC), 136.51 (1xCH), 136.09 (1xCH), 124.10 (1xCH), 113.51 (1xCH), 101.94 (1xCH), 78.48 (1xC), 40.78, 36.16, 31.04, 29.09, 29.02, 28.96, 28.02, 24.99, 23.95, 22.16, 19.84 (11xCH2), 13.94 (1xCH3); HRMS-ESI m/z: calcd for [C21H29N2O2S]− [M–H]−: 373.1955; found: 373.1961.
(E)-N-(2-Methoxyphenyl)-9-oxooctadec-10-en-12-ynamide (14)
Yield: 88%; brown solid; HPLC purity: 98.61% (tR = 14.47 min); 1H NMR (300 MHz, CDCl3, ppm): δ 8.38 (d, 1H, J = 8.0 Hz), 7.78 (bs, −NH), 7.03–6.92 (m, 2H), 6.88 (d, 1H, J = 8.0 Hz), 6.67 (δ, 1H, J = 16.0 Hz), 6.45 (d, 1H, J = 16.0 Hz), 3.88 (s, 3H), 2.51 (t, 2H, J = 7.0 Hz), 2.38 (t, 4H, J = 7.0 Hz), 1.70 (t, 2H, J = 8.0 Hz), 1.60–1.35 (m-14H), 0.91 (t, 3H, J = 7.0 Hz); 13C NMR (75 MHz, CDCl3, ppm): δ 199.63 (C=O), 171.25 (1xC), 147.68 (1xC), 136.51 (1xCH), 127.70 (1xC), 124.06 (1xCH), 123.48 (1xCH), 121.06 (1xCH), 119.74 (1xCH), 109.83 (1xCH), 101.88 (1xCH), 78.48 (1xC), 55.65 (1xCH3), 29.01 (2xCH2), 40.80, 37.99, 31.03, 29.11, 28.01, 25.53, 24.00, 22.15, 19.82 (9xCH2), 13.93 (CH3); HRMS-ESI m/z: calcd for [C25H34NO3]− [M – H]−: 396.2544; found: 396.2546.
(E)-N-(1,3-Dimethyl-1H-pyrazol-5-yl)-9-oxooctadec-10-en-12-ynamide (15)
Yield: 76%; off-white solid; HPLC purity: 99.66% (tR = 12.59 min); 1H NMR (300 MHz, CDCl3, ppm): δ 7.69 (s, −NH), 6.67 (d, 1H, J = 16.0 Hz), 6.45 (d, 1H, J = 16.0 Hz), 6.02 (s, 1H), 3.65 (s, 3H), 2.53 (t, 2H, J = 7.0 Hz), 2.37 (t, 4H, J = 7.0 Hz), 2.20 (s, 3H), 1.73–1.34 (m-16H), 0.91 (t, 3H, J = 7.0 Hz); 13C NMR (75 MHz, CDCl3, ppm): δ 199.95 (C=O), 171.14 (C), 147.18 (C), 136.47 (CH), 135.87 (C), 124.31 (CH), 102.22 (CH), 99.65 (CH), 78.44 (C), 36.29 (CH3), 28.85 (2xCH2), 40.72, 34.14, 31.04, 28.00, 25.31, 24.77, 23.90, 22.15, 19.83 (9 × CH2), 13.93 (CH3), 12.58 (CH3); HRMS-ESI m/z: calcd for [C23H34N3O2]− [M – H]−: 384.2657; found: 384.2656.
(E)-N-(2-Cyanophenyl)-9-oxooctadec-10-en-12-ynamide (16)
Yield: 80%; brown solid; HPLC purity: 99.01% (tR = 12.86 min); 1H NMR (300 MHz, CDCl3, ppm): δ 7.40 (d, 1H, J = 8.0 Hz), 7.30 (t, 1H, J = 7.0 Hz), 6.76 (d, 1H, J = 8.0 Hz), 6.68 (m, 1H), 6.62 (d, 1H, J = 16.0 Hz), 6.46 (d, 1H, J = 16.0 Hz), 2.51 (t, 2H, J = 7.5 Hz), 2.36 (m, 4H), 1.63–1.32 (m-16H), 0.91 (t, 3H, J = 7.0 Hz); 13C NMR (75 MHz, CDCl3, ppm): δ 199.68 (C=O), 178.29 (1 × C), 149.67 (1 × C), 136.51 (1 × CH), 134.00 (1 × CH), 132.34 (1 × CH), 124.10 (1 × CH), 117.93 (1 × CH), 117.63 (1 × C), 115.15 (1 × CH), 101.91 (1 × C), 95.96 (1 × C), 78.49 (1 × C), 29.01 (2 × CH2), 40.81, 33.97, 31.04, 28.87, 28.01, 24.68, 24.00, 22.15, 19.83, (9 × CH2), 13.94 (1 × CH3); HRMS-ESI m/z: calcd for [C25H33N2O2]+ [M + H]+: 393.2537; found: 393.2416.
(E)-9-Oxo-N-(1,3,4-thiadiazol-2-yl)octadec-10-en-12-ynamide (17)
Yield: 50.8%; off-white solid; HPLC purity: 99.19% (tR = 13.09 min); 1H NMR (300 MHz, CDCl3, ppm): δ 13.29 (bs, −NH), 8.82 (s, 1H), 6.67 (d,1H, J = 16.0 Hz), 6.46 (d, 1H, J = 16.0 Hz), 2.79 (t, 2H, J = 7.5 Hz), 2.51 (t, 2H, J = 7.5 Hz), 2.39 (t, 2H, J = 6.5 Hz), 1.81 (t, 2H, J = 7.0), 1.63–1.36 (m-14H), 0.92 (t, 3H, J = 6.5 Hz); 13C NMR (75 MHz, CDCl3, ppm): δ 199.65 (C=O), 172.20 (1 × C), 160.52 (1 × C), 147.34 (1 × CH), 136.52 (1 × CH), 124.08 (1 × CH), 101.93 (1 × C), 78.48 (1 × C), 40.81, 36.13, 31.05, 29.01, 28.96, 28.87, 28.02, 25.13, 23.99, 22.16, 19.84, (11 × CH2), 13.95 (1 × CH3); HRMS-ESI m/z: calcd for [C20H28N3O2S]− [M – H]−: 374.1897; found: 374.1918.
(E)-N-(5-Bromopyrimidin-2-yl)-9-oxooctadec-10-en-12-ynamide (18)
Yield: 63.4%; brown solid; HPLC purity: 98.41% (tR = 13.70 min); 1H NMR (300 MHz, CDCl3, ppm): δ 8.28 (s, 2H), 6.67 (d, 1H, J = 16.0 Hz), 6.46 (d, 1H, J = 16.0 Hz), 5.96 (bs, −NH), 2.51 (t, 2H, J = 7.5 Hz), 2.39 (m, 4H), 1.63–1.26 (m-16H), 0.92 (t, 3H, J = 6.5 Hz); 13C NMR (75 MHz, CDCl3, ppm): δ 199.74 (C=O), 172.16 (C), 158.38 (2 × CH), 149.72 (1 × C), 136.49 (1 × CH), 124.14 (1 × CH), 120.44 (1 × C), 101.97 (1 × C), 78.47 (1 × C), 29.00 (2 × CH2), 40.79, 34.01, 31.03, 28.86, 28.01, 24.67, 23.99, 22.15, 19.82 (9xCH2), 13.93 (1 × CH3); HRMS-ESI m/z: calcd for [C22H30BrN3O2Na]+ [M + H + Na]+: 470.1414; found: 470.1450.
(E)-N-(5-Bromopyrimidin-2-yl)-9-oxooctadec-10-en-12-ynamide (19)
Yield: 91.67%; brown solid; HPLC purity: 99.27% (tR = 13.70 min); 1H NMR (500 MHz, CDCl3, ppm): δ 6.67 (d, 1H, J = 16.0 Hz), 6.46 (d, 1H, J = 16.0 Hz), 5.60 (bs, −NH), 2.51 (t, 2H, J = 7.0 Hz), 2.39 (t, 2H, J = 7.0 Hz), 2.36 (m, 2H), 1.71–1.31 (m,14H), 0.92 (t, 3H, J = 7.0 Hz); 13C NMR (125 MHz, CDCl3, ppm): δ 199.69 (C=O), 172.17 (1xC), 136.53 (1xCH), 124.10 (1xCH), 101.92 (1xC), 78.49 (1xC), 48.05 (1xC), 31.35 (2xCH2), 44.61, 40.80, 36.70, 31.05, 28.99, 28.96, 28.03, 25.57, 23.97, 22.16, 19.84, 15.04 (12xCH2), 13.94 (1xCH3); HRMS-ESI m/z: calcd for [C22H35NO2Na]+: 368.3560; found [M + H + Na]+: 368.2577.
(E)-N,N-Diisobutyl-9-oxooctadec-10-en-12-ynamide (20)
Yield: 85%; brown oil; HPLC purity: 97.82% (tR = 14.72 min); 1H NMR (300 MHz, CDCl3, ppm): δ 6.67 (d, 1H, J = 16.0 Hz), 6.44 (d, 1H, J = 16.0 Hz), 3.18 (d, 2H, J = 8.0 Hz), 3.09 (d, 2H, J = 8.0 Hz), 2.51 (t, 2H, J = 7.5 Hz), 2.37–2.29 (m, 4H), 2.0–1.87 (m, 2H), 1.58–1.30 (m-16H), 0.91–0.83 (m, 15H); 13C NMR (75 MHz, CDCl3, ppm): δ 199.74 (C=O), 175.05 (C), 136.50 (1xCH), 124.14 (1xCH), 101.93 (1xC), 78.47 (1xC), 55.93, 53.55, 40.78, 33.15, 31.04, 29.20, 29.07, 28.99, 28.01, 26.76, 25.83, 23.97, 19.83 (13xCH2), 27.90 (2xCH), 20.10 (2xCH3), 20.04 (2xCH3), 13.93 (1xCH3); HRMS-ESI m/z: calcd for [C26H46NO2]+ [M + H]+: 404.3523; found: 404.3561.
(E)-N-(1-(4-Bromophenyl)ethyl)-9-oxooctadec-10-en-12-ynamide (21)
Yield: 90%; off-white solid; HPLC purity: 98.09% (tR = 13.41 min); 1H NMR (300 MHz, CDCl3, ppm): δ 7.48 (d, 2H, J = 8.0 Hz), 7.21 (d, 1H, J = 8.0 Hz), 6.68 (d, 1H, J = 16.0 Hz), 6.47 (d, 1H, J = 16.0 Hz), 5.72 (d, −NH, J = 7.0 Hz), 5.10 (m,1H), 2.52 (t, 2H, J = 7.5 Hz), 2.40 (m, 4H), 2.17 (d, 2H, J = 7.5 Hz), 1.67–1.60 (m, 6H), 1.48 (d, 3H, J = 7.0 Hz) 1.39–1.31 (m, 10H), 0.93 (m, 3H, J = 7.0 Hz); 13C NMR (75 MHz, CDCl3, ppm): δ 199.61 (C=O), 172.15 (1xC), 142.48 (1xC), 136.52 (1xCH), 131.70 (2xCH), 127.93 (2xCH), 124.08 (1xCH), 121.08 (1xC), 101.94 (1xC), 78.48 (1xC), 28.94 (2xCH2), 48.08 (1xCH), 40.78, 36.71, 31.06, 28.98, 28.03, 25.52, 23.92, 22.17, 19.85 (9xCH2), 21.68 (1xCH3), 13.95 (1xCH3); HRMS-ESI m/z: calcd for [C26H35BrNO2]− [M–H]−: 472.1857; found: 472.1893.
(E)-N-(2-Hydroxyethyl)-9-oxooctadec-10-en-12-ynamide (22)
Yield: 91%; brown solid; HPLC purity: 98.53% (tR = 15.28 min); 1H NMR (300 MHz, CDCl3, ppm): δ 6.69 (d,1H, J = 16.0 Hz), 6.42 (d,1H, J = 16.0 Hz), 6.29 (bs, −NH), 3.78–3.70 (m, 2H), 3.40 (bs, −OH), 3.21–3.14 (m,2H), 2.53 (t, 2H, J = 7.5 Hz), 2.40 (t, 2H, J = 6.5 Hz), 2.18 (t, 2H, J = 7.0 Hz), 1.66–127 (m, 16H), 0.92 (t, 3H, J = 6.5 Hz); 13C NMR (75 MHz, CDCl3, ppm): δ 199.51 (C=O), 174.50 (1xC), 136.74 (1xCH), 123.97 (1xCH), 101.84 (1xC), 78.49 (1xC), 28.97 (2xCH2), 59.47, 42.99, 40.25, 36.66, 31.18, 29.71, 28.03, 25.50, 23.91, 22.17, 19.84 (9xCH2), 13.94 (1xCH3); HRMS-ESI m/z: calcd for [C20H34NO3]+[M + H]+: 336.2533; found: 336.2517.
(E)-N-(3,4-Dihydroxyphenethyl)-9-oxooctadec-10-en-12-ynamide (23)
Yield: 89%; brown solid; HPLC purity: 99.46% (tR = 11.77 min); 1H NMR (300 MHz, CDCl3, ppm): δ 7.52 (bs, −OH), 6.85 (d, 1H, J = 8.0 Hz), 6.75 (bs, −OH), 6.73 (d, 1H, J = 16.0 Hz), 6.60 (d, 1H, J = 8.0 Hz), 6.49 (d, J = 16.0 Hz), 5.6 (bs, −NH), 3.54–3.49 (m, 2H), 2.70 (t, 3H, J = 6.0 Hz), 2.55 (t, 2H, J = 7.0 Hz), 2.41 (t, 2H, J = 6.5 Hz), 2.13 (t, 2H, J = 7.0 Hz), 1.70–1.25 (m, 16H), 0.93 (t, 3H, J = 7.0 Hz); 13C NMR (75 MHz, CDCl3, ppm): δ 200.93 (C=O), 173.58 (1xC), 144.24 (1xC), 142.82 (1xC), 136.33 (1xCH), 130.85 (1xC), 125.09 (1xCH), 120.52 (1xCH), 115.51 (1xCH), 115.30 (1xCH), 102.91 (1xC), 78.46 (1xC), 40.74, 40.39, 36.77, 34.75, 31.06, 28.98, 28.84, 28.63, 28.00, 25.58, 23.95, 22.17, 19.89 (13xCH2), 13.95 (1xCH3); HRMS-ESI m/z: calcd for [C26H37NO4Na]+ [M + H + Na]+: 450.2615; found: 450.2570.
(E)-N-(3-Chloro-4-fluorophenyl)-9-oxooctadec-10-en-12-ynamide (24)
Yield: 78%; pale yellow solid; HPLC purity: 98.70% (tR = 13.59 min); 1H NMR (300 MHz, CDCl3, ppm): δ 7.75 (d, 1H, J = 8.0 Hz), 7.65 (bs, −NH), 7.37 (m, 1H), 7.08 (t, 1H, J = 8.0 Hz), 6.69 (d, 1H, J = 8.0 Hz), 6.47 (d, 1H, J = 16.0 Hz), 2.54 (t, 2H, J = 7.0 Hz), 2.40–2.32 (m, 4H), 1.87–1.27 (m, 18H), 0.92 (t, 3H, J = 6.5 Hz); 13C NMR (75 MHz, CDCl3, ppm): δ 199.95 (C=O), 171.64 (1xC), 152.97 (1xC), 136.45 (1xCH), 134.66 (1xC), 124.35 (1xCH), 122.05 (1xCH), 119.49 (1xCH), 116.66 (1xCH), 116.37 (1xCH), 102.25 (1xC), 78.44 (1xC), 40.73, 37.44, 31.05, 29.69, 28.87, 28.80, 28.01, 25.31, 23.88, 22.16, 19.85 (11xCH2), 13.9 (1xCH3); HRMS-ESI m/z: calcd for [C24H30ClFNO2]− [M – H]−: 418.1955; found: 418.1961.
(E)-3-(9-Oxooctadec-10-en-12-ynoyl)thiazolidine-2,4-dione (25)
Yield: 90%; brown solid; HPLC purity: 99.47% (tR = 11.87 min); 1H NMR (300 MHz, CDCl3, ppm): δ 6.68 (d, 1H, J = 8.0 Hz), 6.44 (d, 1H, J = 16.0 Hz), 3.67 (s, 2H), 2.52 (t, 2H, J = 7.0 Hz), 2.36–2.32 (m, 4H), 1.61–1.33 (m, 16H), 0.92 (m, 3H, J = 7.0 Hz); 13C NMR (75 MHz, CDCl3, ppm): δ 199.85 (C=O), 174.57 (1xC), 169.96 (1xC), 165.76 (1xCH), 136.48 (1xCH), 124.35 (1xCH), 102.06 (1xC), 78.48 (1xC), 28.98 (2xCH2), 40.79, 35.82, 33.73, 31.05, 28.82, 28.02, 24.58, 24.00, 22.15, 19.85 (10xCH2), 13.92 (1xCH3); MS m/z: calcd for [C21H28NO4S]− [M – H]−: 390.18; found 390.29.
(E)-N,N-Dimethyl-9-oxooctadec-10-en-12-ynamide (26)
Yield: 86%; pale yellow semi-solid; HPLC purity: 99.04% (tR = 13.55 min); 1H NMR (500 MHz, CDCl3, ppm): δ 6.67 (d, 1H, J = 16.0 Hz), 6.46 (d, 1H, J = 16.0 Hz), 3.02 (s, 3H), 2.95 (s, 3H), 2.52 (t, 2H, J = 7.0 Hz), 2.39 (t, 2H, J = 7.0 Hz), 1.65 (m, 6H), 1.56 (m, 2H), 1.41–1.27 (m, 10H), 0.92 (t, 3H, J = 7.0 Hz); 13C NMR (125 MHz, CDCl3, ppm): δ 199.64 (C=O), 173.20 (1xC), 136.59 (1xCH), 124.02 (1xCH), 101.84 (1xC), 78.50 (1xC), 40.86 (2xCH2), 37.31 (2xCH3), 35.37, 33.35, 31.06, 29.28, 29.21, 29.08, 28.04, 25.09, 24.04, 22.17, 19.84 (11xCH2), 13.96 (1xCH3); HRMS-ESI: m/z calcd for [C20H34NO2]+ [M + H]+: 320.25895; found 320.25827 .
(E)-9-Oxo-N-(phenylsulfonyl)octadec-10-en-12-ynamide (27)
Yield: 81%; brown solid; HPLC purity: 97.50% (tR = 13.35 min); 1H NMR (300 MHz, CDCl3, ppm): δ 8.72 (bs, −NH), 8.09 (d, 2H, J = 8.0 Hz), 7.67 (t,1H, J = 7.0 Hz), 7.75 (t, 2H, J = 7.0 Hz), 6.69 (d, 1H, J = 8.0 Hz), 6.47 (d, 1H, J = 16.0 Hz), 2.52 (t, 2H, J = 7.0 Hz), 2.42 (t, 2H, J = 7.0 Hz), 2.26 (t, 2H, J = 7.0 Hz), 1.58–1.25 (m, 16H), 0.92 (m, 3H, J = 6.5 Hz); 13C NMR (75 MHz, CDCl3, ppm): δ 199.85 (C=O), 170.79 (1xC), 138.66 (1xC), 136.45 (1xCH), 133.95 (1xC), 128.98 (2xCH), 128.32 (2xCH), 124.34 (1xCH), 102.19 (CH), 78.47 (1xC), 28.72 (2xCH2), 40.70, 36.22, 31.05, 28.43, 28.02, 24.13, 23.82, 22.16, 19.85 (9xCH2), 13.95 (1xCH3); HRMS-ESI m/z: calcd for [C24H32NO4S]− [M–H]−: 430.2058; found: 430.2081.
(E)-1-(2-Mercapto-1H-benzo[d]imidazol-1-yl)octadec-10-en-12-yne-1,9-dione (28)
Yield: 85%; brown solid; HPLC purity: 97.63% (tR = 14.07 min); 1H NMR (300 MHz, CDCl3, ppm): δ 10.47 (bs, −SH), 8.20 (d, 1H, J = 8.0 Hz), 7.18 (d, 3H, J = 8.0 Hz), 6.70 (d, 1H, J = 8.0 Hz), 6.49 (d, 1H, J = 8.0 Hz), 3.63 (t, 2H, J = 7.0 Hz), 2.54 (t, 2H, J = 7.0 Hz), 2.40 (t, 2H, J = 6.0 Hz), 1.84–1.27 (m, 16H), 0.93 (m, 3H, J = 6.5 Hz); 13C NMR (75 MHz, CDCl3, ppm): δ 199.82 (C=O), 175.33 (1xC), 170.18 (1xC), 136.53 (1xC), 131.59 (1xCH), 130.31 (1xC), 125.48 (2xCH), 124.17 (2xCH), 116.26 (1xCH), 109.022 (1xCH), 101.95 (CH), 78.51 (1xC), 40.86, 39.69, 31.05, 29.17, 29.06, 28.81, 28.03, 24.50, 24.05, 22.16, 19.85 (11xCH2), 13.95 (1xCH3); HRMS-ESI m/z: calcd for [C25H33N2O2S]+ [M + H]+: 425.2257; found: 425.2154.
(E)-9-Oxo-N-(2-sulfamoylphenyl)octadec-10-en-12-ynamide (29)
Yield: 92%; brown solid; HPLC purity: 99.33% (tR = 14.30 min); HRMS-ESI m/z: calcd for [C24H33N2O4S]− [M – H]−: 445.2292; found: 445.2339.
(E)-9-Oxo-N-(4-sulfamoylphenethyl)octadec-10-en-12-ynamide (30)
Yield: 94%; pale yellow solid; HPLC purity: 99.41% (tR = 14.29 min); 1H NMR (300 MHz, CDCl3, ppm): δ 7.85 (d, 2H, J = 8.0 Hz), 7.30 (d, 2H, J = 8.0 Hz), 6.68 (dt, 1H, J = 16.0 Hz), 6.46 (d, 1H, J = 16.0 Hz), 5.42 (bs, −NH), 3.52 (m, 2H), 2.96 (s, −NH2), 2.89 (t, 2H, J = 7.0 Hz), 2.53 (t, 2H, J = 7.0 Hz), 2.39 (t, 2H, J = 6.0 Hz), 2.14 (t, 2H, J = 7.0 Hz), 1.57–1.28 (m, 16H), 0.92 (t, 3H, J = 6.5 Hz); 13C NMR (75 MHz, CDCl3, ppm): δ 200.09 (C=O), 173.87 (1xC), 144.29 (1xC), 140.58 (1xC), 136.47 (1xCH), 129.44 (2xCH), 126.63 (2xCH), 124.40 (1xCH), 102.28 (1xC), 78.46 (C), 40,72, 40.62, 40.31 36.51, 35.49, 31.05, 28.97, 28.85, 28.01, 25.57, 23.91, 22.16, 19.85 (13xCH2), 13.95 (1xCH3); HRMS-ESI m/z: calcd for [C26H37N2O4S]− [M – H]−: 473.2469; found: 473.2439.
(E)-N-(4-Chlorophenyl)sulfonyl-9-oxooctadec-10-en-12-ynamide (31)
Yield: 85%; off-white solid; HPLC purity: 99.33% (tR = 13.69 min); 1H NMR (300 MHz, CDCl3, ppm): δ 8.56 (bs, −ΝΗ), 8.04 (d, 2H, J = 8.0 Hz), 7.55 (d, 2H, J = 8.0 Hz), 6.71 (d, 1H, J = 16.0 Hz), 6.48 (d, 1H, J = 16.0 Hz), 2.53 (t, 2H, J = 7.0 Hz), 2.38 (t, 2H, J = 7.0 Hz), 2.28 (t, 2H, J = 7.0 Hz), 1.72–1.27 (m, 16H) 0.93 (t, 3H, J = 6.5 Hz); 13C NMR (75 MHz, CDCl3, ppm): δ 199.88 (C=O), 170.63 (1xC), 140.70 (1xC), 137.01 (1xCH),136.43 (1xCH), 129.94 (2xCH), 129.30 (2xCH), 124.41 (1xCH), 102.24 (1xC), 78.47 (1xC), 28.65 (2xCH2), 40.68, 36.28, 31.06, 28.39, 28.02, 24.09, 23.77, 22.17, 19.86 (9xCH2), 13.95 (1xCH3); HRMS-ESI m/z: calcd for [C24H31ClNO4S]− [M-H]−: 464.1668; found: 464.1695.
(E)-N-Cyclohexyl-N-(2-hydroxyethyl)-9-oxooctadec-10-en-12-ynamide (32)
Yield: 92%; brown oil; HPLC purity: 99.31% (tR = 13.29 min); 1H NMR (300 MHz, CDCl3, ppm): δ 6.69 (d, 1H, J = 16.0 Hz), 6.48 (d, 1H, J = 16.0 Hz), 3.75 (bs, 2H), 3.59 (t, −OH), 3.51 (t, 2H, J = 7.5 Hz), 2.53 (t, 2H, J = 7.5 Hz), 2.40 (t, 4H, J = 6.5 Hz), 1.91–1.27 (m, 26H), 0.92 (m, 3H, J = 7.0 Hz); 13C NMR (75 MHz, CDCl3, ppm): δ 199.68 (C=O), 176.12 (1xC), 136.52 (1xCH), 124.13 (1xCH), 101.94 (1xCH), 78.48 (1xC), 64.42 (1xCH), 29.15 (2xCH2), 25.79 (2xCH2), 57.74, 44.92, 40.80, 33.54, 31.58, 31.05, 29.02, 28.92, 28.02, 25.38, 25.15, 23.97, 22.16, 19.84 (14xCH2), 13.94 (1xCH3); HRMS-ESI m/z: calcd for [C26H44NO3]+ [M + H]+: 418.3316; found: 418.3348.
(E)-N-(1-(3-Ethoxy-4-methoxyphenyl)-2-(methylsulfonyl)ethyl)-9-oxooctadec-10-en-12-ynamide (33)
Yield: 90%; off-white solid; HPLC purity: 99.15% (tR = 12.34 min); 1H NMR (300 MHz, CDCl3, ppm): δ 6.88 (bs, 3H), 6.73 (d, 1H, J = 8.0 Hz), 6.67 (d, 1H, J = 16.0 Hz), 6.46 (d, 1H, J = 16.0 Hz), 5.54–5.48 (m, 1H), 4.13–4.06 (m, 2H), 3.02 (s, 3H), 3.69 (dd, 1H, J = 6.0 Hz, J = 15.0 Hz), 3.48 (dd, 1H, J = 5.0 Hz, J = 15.0 Hz), 2.66 (s, 3H), 2.51 (t, 2H, J = 7.0 Hz), 2.39 (t, 2H, J = 7.0 Hz), 2.39 (t, 2H, J = 7.5 Hz), 1.62–1.31 (m, 19H), 0.92 (m, 3H, J = 6.5 Hz); 13C NMR (75 MHz, CDCl3, ppm): δ 199.65 (C=O), 172.81 (1xC), 149.33 (C), 148.77 (1xC), 136.51 (1xCH), 130.96 (1xC), 124.12 (1xCH), 118.54 (1xCH), 111.72 (1xCH), 111.35 (1xCH), 101.98 (1xCH), 78.47 (1xC), 36.57 (1xCH), 56.00 (1xCH3), 48.76 (1xCH3), 28.93 (3xCH2), 64.57, 59.50, 41.90, 40.76, 31.05, 28.02, 25.36, 23.91, 22.16, 19.84 (10xCH2), 14.76 (1xCH3), 13.95 (1xCH3); HRMS-ESI m/z: calcd for [C30H45NO6SNa]+ [M + H + Na]+: 570.2860; found: 570.2848.
(E)-N-Cyclohexyl-9-oxooctadec-10-en-12-ynamide (34)
Yield: 88%; brown solid; HPLC purity: 99.16% (tR = 13.68 min); 1H NMR (300 MHz, CDCl3, ppm): δ 6.68 (d, 1H, J = 16.0 Hz), 6.47 (d, 1H, J = 16.0 Hz), 5.42 (d, −NH, J = 6.0 Hz), 3.79 (m, 1H), 2.51 (t, 2H, J = 7.0 Hz), 2.39 (t, 2H, J = 7.0 Hz), 2.14 (t, 2H, J = 7.5 Hz), 1.93 (d, 2H, J = 12.0 Hz), 1.62–131 (m, 24H), 0.92 (t, 3H, J = 7.0 Hz); 13C NMR (75 MHz, CDCl3, ppm): δ 199.64 (C=O), 172.30 (1xC), 136.52 (1xCH), 124.07 (1xCH), 101.90 (1xCH), 78.48 (1xC), 48.10 (1xCH), 33.23 (2xCH2), 24.87 (2xCH2), 28.98, (2xCH2), 40.80, 36.97, 31.05, 29.03, 28.02, 25.75, 25.53, 23.97, 22.16, 19.84 (10xCH2), 13.94 (1xCH3); HRMS-ESI m/z calcd for [C24H39NO2Na]+: [M + H + Na]+: 396.2873; found: 396.2868.
(E)-9-Oxo-N-tosyloctadec-10-en-12-ynamide (35)
Yield: 92%; off-white solid; HPLC purity: 99.05% (tR = 11.22 min); 1H NMR (300 MHz, CDCl3, ppm): δ 9.71 (s, −NH), 7.60 (bs, 2H), 7.47 (bs, 2H), 6.68 (d, 1H, J = 16.0 Hz), 6.45 (d, 1H, J = 16.0 Hz), 2.53 (t, 2H, J = 7.0 Hz), 2.40–2.31 (m, 5H), 1.59–1.18 (m, 18H), 0.91 (t, 3H, J = 7.0 Hz); 13C NMR (75 MHz, CDCl3, ppm): δ 200.46 (C=O), 174.86 (1xC), 139.82 (1xC), 136.49 (2xCH), 126.73 (2xCH), 124.07 (1xCH), 120.71 (2xCH), 102.33 (1xCH), 78.53 (1xC), 40.80 (2xCH2), 31.08 (2xCH2), 29.07 (2xCH2), 36.60, 25.73, 28.05, 23.98, 19.87 (5xCH2), 22.17 (1xCH3), 13.94 (1xCH3); HRMS-ESI m/z: calcd for [C25H34NO4S]− [M – H]−: 444.2203; found: 444.2219.
(E)-4-(N-Acetyl-9-oxooctadec-10-en-12-ynamido)benzene-1-sulfonyl Chloride (36)
Yield: 87%; off-white solid; HPLC purity: 99.13% (tR = 12.98 min); 1H NMR (300 MHz, CDCl3, ppm): δ 8.45 (s, −NH), 7.97 (d, 2H, J = 8.0 Hz), 7.37 (d, 2H, J = 8.0 Hz), 6.69 (d, 1H, J = 16.0 Hz), 6.47 (d, 1H, J = 16.0 Hz), 2.53 (t, 2H, J = 7.0 Hz), 2.46 (s, 3H, J = 7.0 Hz), 2.25 (t, 2H, J = 7.0 Hz), 1.61–1.25 (m, 16H), 0.91 (t, 3H, J = 6.5 Hz); 13C NMR (75 MHz, CDCl3, ppm): δ 199.76 (C=O), 170.64 (2xC), 145.08 (1xC), 136.45 (1xCH), 135.65 (1xC), 129.61 (2xCH), 128.39 (2xCH), 124.29 (1xCH), 102.12 (1xCH), 78.48 (1xC), 28.73 (2xCH2), 40.71, 36.22, 31.05, 28.46, 28.02, 24.13, 23.82, 22.17, 19.85 (9xCH2), 21.69 (1xCH3), 13.95 (1xCH3); HRMS-ESI m/z: calcd for [C26H33ClNO5S]− [M-H]−: 506.1773; found: 506.1687.
(E)-N-(2-Morpholinoethyl)-9-oxooctadec-10-en-12-ynamide (37)
Yield: 56.1%; pale yellow solid; HPLC purity: 98.55% (tR = 9.74 min); 1H NMR (500 MHz, CDCl3, ppm): δ 6.66 (d, 1Η, J = 16.0 Hz), 6.46 (d, 1H, J = 16.0 Hz), 6.07 (s, −NH), 3.74 (s, 2H), 3.39 (dd, 2H, J = 6.0 Hz, J = 12.0 Hz), 2.52 (m, 6H), 2.39 (t, 2H, J = 7.0 Hz), 2.19 (t, 2H J = 7.0 Hz), 1.61–1.27 (m, 18H), 0.92 (t, 3H J = 7.0 Hz); 13C NMR (125 MHz, CDCl3, ppm): δ 199.72 (C=O), 173.39 (1xC), 136.52 (1xCH), 124.07 (1xCH), 101.92 (1xC), 78.48 (1xC), 66.80 (2xCH2), 57.22 (1xCH2), 53.32 (2xCH2), 40.80 (1xCH2), 36.63, 35.52, 31.05, 29.08, 29.02, 29.01, 28.029, 25.62, 23.98, 22.16, 19.84 (11xCH2), 13.94 (1xCH3); HRMS-ESI m/z: calcd for [C24H39N2O3]− [M-H]−: 403.2966; found: 403.2941.
(E)-N-(1,3-Dihydroxypropan-2-yl)-9-oxooctadec-10-en-12-ynamide (38)
Yield: 80%; yellow solid; HPLC purity: 99.61% (tR = 13.99 min); 1H NMR (300 MHz, CDCl3, ppm): δ 6.69 (d, 1H, J = 16 Hz), 6.47 (d, 1H, J = 16 Hz), 5.42 (bs, −NH), 4.26–4.19 (m, 1H), 2.52 (t, 2H, J = 7.5 Hz), 2.40 (t, 2H, J = 7.0 Hz), 2.14 (t, 2H, J = 7.0 Hz), 2.06–1.99 (m, 2-OH), 1.65–1.27 (m, 20H), 0.93 (t, 3H, J = 6.5 Hz); 13C NMR (75 MHz, CDCl3, ppm): δ 200.09 (C=O), 174.53 (1xC), 136.46 (1xCH), 124.39 (1xCH), 102.23 (1xCH), 78.46 (1xC), 63.07 (2xH2), 52.38 (1xCH), 28.87 (2xCH2), 40.74, 36.59, 31.05, 28.75, 28.01, 25.53, 23.83, 22.16, 19.85 (9xCH2), 13.94 (1xCH3); HRMS-ESI m/z: calcd for [C21H36NO4]+ [M + H]+: 366.26443; found: 366.26309.
(E)-N-(Methylsulfonyl)-9-oxooctadec-10-en-12-ynamide (39)
Yield: 90%; brown solid; HPLC purity: 99.60% (tR = 14.03 min); 1H NMR (300 MHz, CDCl3, ppm): δ 8.64 (bs, 1H), 6.71 (d, 1H, J = 16.0 Hz), 6.48 (d, 1H, J = 16.0 Hz), 3.32 (s, 3H), 2.52 (t, 2H, J = 7.5 Hz), 2.40–2.32 (m,4H), 2.14 (t, 2H, J = 7.0 Hz), 1.65–1.34 (m, 16H), 0.92 (t, 3H, J = 6.5 Hz); 13C NMR (75 MHz, CDCl3, ppm): δ 200.21 (C=O), 172.02 (1xC), 136.38 (1xCH), 124.63 (1xCH), 102.43 (1xCH), 78.46 (1xC), 28.66 (2xCH2), 41.58 (1xCH3), 40.65, 36.45, 31.05, 28.47, 28.00, 24.23, 23.81, 22.16, 19.86 (9xCH2), 13.94 (1xCH3); HRMS-ESI m/z: calcd for [C19H30NO4S]− [M – H]−: 368.1901; found: 368.1925.
(E)-9-Oxo-N-(pyrazin-2-yl)octadec-10-en-12-ynamide (40)
Yield: 82%; brown solid; HPLC purity: 97.66% (tR = 12.47 min); 1H NMR (300 MHz, CDCl3, ppm): δ 8.79 (d, 1H, J = 4.0 Hz), 8.52 (d, 1H, J = 8.0 Hz), 7.53–4.49 (dd, 1H, J = 4.0 Hz, 8.0 Hz), 6.71 (dd, 1H, J = 2.0 Hz, J = 16.0 Hz), 6.48 (dd, 1H, J = 2.0 Hz, J = 16.0 Hz), 2.52 (t, 2H, J = 7.5 Hz), 2.40 (t, 2H, J = 6.5 Hz), 1.63–1.28 (m, 16H), 0.92 (t, 3H, J = 7.0 Hz); 13C NMR (75 MHz, CDCl3, ppm): δ 200.02 (C=O), 169.28 (1xC), 151.44 (1xC), 140.17 (1xCH), 136.43 (1xCH), 135.11 (1xCH), 130.06 (1xCH), 124.49 (1xCH), 102.34 (1xC), 78.47 (1xC), 28.99 (2xCH2), 40.72, 34.07, 31.05, 28.91, 28.01, 24.33, 23.96, 22.16, 19.85 (9xCH2), 13.94 (1xCH3); HRMS-ESI m/z: calcd for [C22H30N3O2]− [M-H]−: 368.2344; found: 368.2100.
(E)-N-(1-Methyl-1H-benzo[d]imidazol-2-yl)-9-oxooctadec-10-en-12-ynamide (41)
Yield: 92%; brown solid; HPLC purity: 98.90% (tR = 14.00 min); 1H NMR (300 MHz, CDCl3, ppm): δ 8.06 (d, 2H, J = 8.0 Hz), 7.30 (d, 1H, J = 8.0 Hz), 7.03 (d, 2H, J = 8.0 Hz), 6.69 (d, 1H, J = 16.0 Hz), 6.48 (d, 1H, J = 16.0 Hz), 5.41 (s, −NH), 3.90 (s, 3H), 2.53 (t, 2H, J = 7.5 Hz), 2.38 (m, 4H), 1.67–1.27 (m, 25H,) 0.93 (t, 3H, J = 7.0 Hz); 13C NMR (75 MHz, CDCl3, ppm): δ 199.61 (C=O), 173.62 (1xC), 167.48 (1xC), 163.28 (1xC), 162.53 (1xC), 136.53 (1xC), 128.46 (2xCH), 128.40 (1xC), 124.06 (1xCH), 114.31 (2xCH), 102.31 (1xC), 101.87 (1xC), 78.49 (1xC), 55.66 (1xCH), 55.47 (1xCH3), 43.64 (1xCH), 29.00 (3xCH3), 28.85 (2xCH2), 40.82, 33.50, 31.05, 30.16, 28.03, 24.64, 23.98, 22.16, 19.84 (9xCH2), 13.95 (1xCH3); HRMS-ESI m/z: calcd for [C33H46N3O3]+ [M + H]+: 532.3534; found: 532.3519.
(E)-N-(1-Methyl-1H-benzo[d]imidazol-2-yl)-9-oxooctadec-10-en-12-ynamide (42)
Yield: 89%; off-white solid; HPLC purity: 99.63% (tR = 14.30 min); 1H NMR (300 MHz, CDCl3, ppm): δ 7.38 (m, 1H),7.29–7.25 (m, 3H), 6.69 (d, 1H, J = 16.0 Hz), 6.48 (d, 1H, J = 16.0 Hz), 3.69 (s, 3H), 5.41 (s, −NH), 2.54 (t, 3H, J = 7.5 Hz), 2.39 (m, 2H), 1.76–1.26 (m, 18H), 0.92 (t, 3H, J = 7.0 Hz); 13C NMR (75 MHz, CDCl3, ppm): δ 199.73 (C=O), 172.89 (1xC), 151.87 (1xC), 136.56 (1xCH), 130.67 (1xC), 129.98 (1xC), 124.01 (1xCH), 123.18 (2xCH), 112.13 (1xCH), 109.22 (1xCH), 101.81 (1xC), 78.50 (1xC), 29.22 (1xCH3), 29.11 (2xCH2), 40.88, 39.70, 31.04, 28.77, 28.03, 25.91, 24.09, 22.16, 19.83 (9xCH2), 13.95 (1xCH3); HRMS-ESI m/z: calcd for [C26H36N3O2]+ [M + H]+: 422.2802; found: 422.2792.
(E)-9-Oxo-N-(4-phenylthiazol-2-yl)octadec-10-en-12-ynamide (43)
Yield: 85%; brown solid; HPLC purity: 99.10% (tR = 14.29 min); 1H NMR (300 MHz, CDCl3, ppm): δ13.07 (bs, −NH), 7.80 (d, 2H, J = 8.0 Hz), 7.48–7.42 (m, 3H), 7.12 (s, 1H), 6.69 (d, 1H, J = 16.0 Hz), 6.48 (d, 1H, J = 16.0 Hz), 2.52 (t, 2H, J = 7.5 Hz), 2.47–2.37 (m, 4H), 1.70–1.30 (m, 18H), 0.92 (t, 3H, J = 7.0 Hz); 13C NMR (75 MHz, CDCl3, ppm): δ 199.58 (C=O), 172.25 (1xC), 161.01 (1xC), 145.79 (1xC), 136.52 (1xCH), 131.10 (1xCH), 129.29 (1xCH), 129.19 (2xCH), 126.28 (2xCH), 124.08 (1xCH), 107.32 (1xC), 101.88 (1xC), 78.50 (1xC), 40.80, 35.94, 31.05, 28.96, 28.93, 28.74, 28.03, 24.60, 23.96, 22.16, 19.84 (11xCH2), 13.94 (1xCH3); HRMS-ESI m/z: calcd for [C27H33N2O2S]− [M-H]−: 449.2268; found: 449.2268.
HPLC Analysis of Compounds 1 and 4–43
All samples were (0.5 mg/mL) prepared in methanol and filtered through a 0.45 μ membrane filter injected (20 μL) in the HPLC system (Waters) using a Unisphere column (150 × 4.6 mm, 3 μm). Mobile phase A (0.1% trifluoric acid) and mobile phase B (acetonitrile) were used. A gradient program was used as follows: time/% A: 0/98, 12/2, 15/2, 16/98, 20/98. The flow rate was 1.0 mL/min. The results were analyzed using Empower software 2.0.
HTS: Glucose Uptake Assay: Glucose Uptake Assay in L6 Myotubes
L6 rat skeletal muscle cells (ATCC, USA) were cultured in 96-well plates (Nunc) in an MEM alpha medium (AMIMED) containing 10% serum and 1% penicillin–streptomycin solution. Differentiation was induced in confluent myoblast cultures by culturing the cells in a medium containing 2% serum. The differentiated fused myotubes were further starved in serum-free media, treated with the samples, and incubated overnight. After 24 h, the cells were incubated with insulin for 20 min and pulsed with C14-tagged deoxyglucose (GE Healthcare, UK) for 10 min in the presence of insulin (Sigma, St. Louis, MO, USA). Glucose uptake was measured by lysing the cells with Microscint PS (Perkin Elmer, USA). The plates were read using a Top Count Reader (Perkin Elmer, USA).
Molecular Docking
A molecular docking study was performed against PI3K using the standard protocol implemented in the GLIDE (grid-based ligand docking with energetics) module of the Schrödinger Molecular modeling suite (Schrödinger, LLC, New York, NY, 2018).15−17 With this objective, the crystal structure of PI3K was retrieved from the Protein Data Bank (PDB ID: 1E7U)18,19 and optimized using the protein preparation wizard. The 3D structures of (E)-9-oxooctadec-10-en-12-ynoic acid analogues were sketched in the builder panel of the Maestro and were subjected to geometry optimization through the ligand preparation tool. With this setup, the molecular docking study was performed to identify the binding modes of (E)-9-oxooctadec-10-en-12-ynoic acid analogues at the active site of PI3K and the various thermodynamic interactions that govern the binding of these molecules.
Results and Discussion
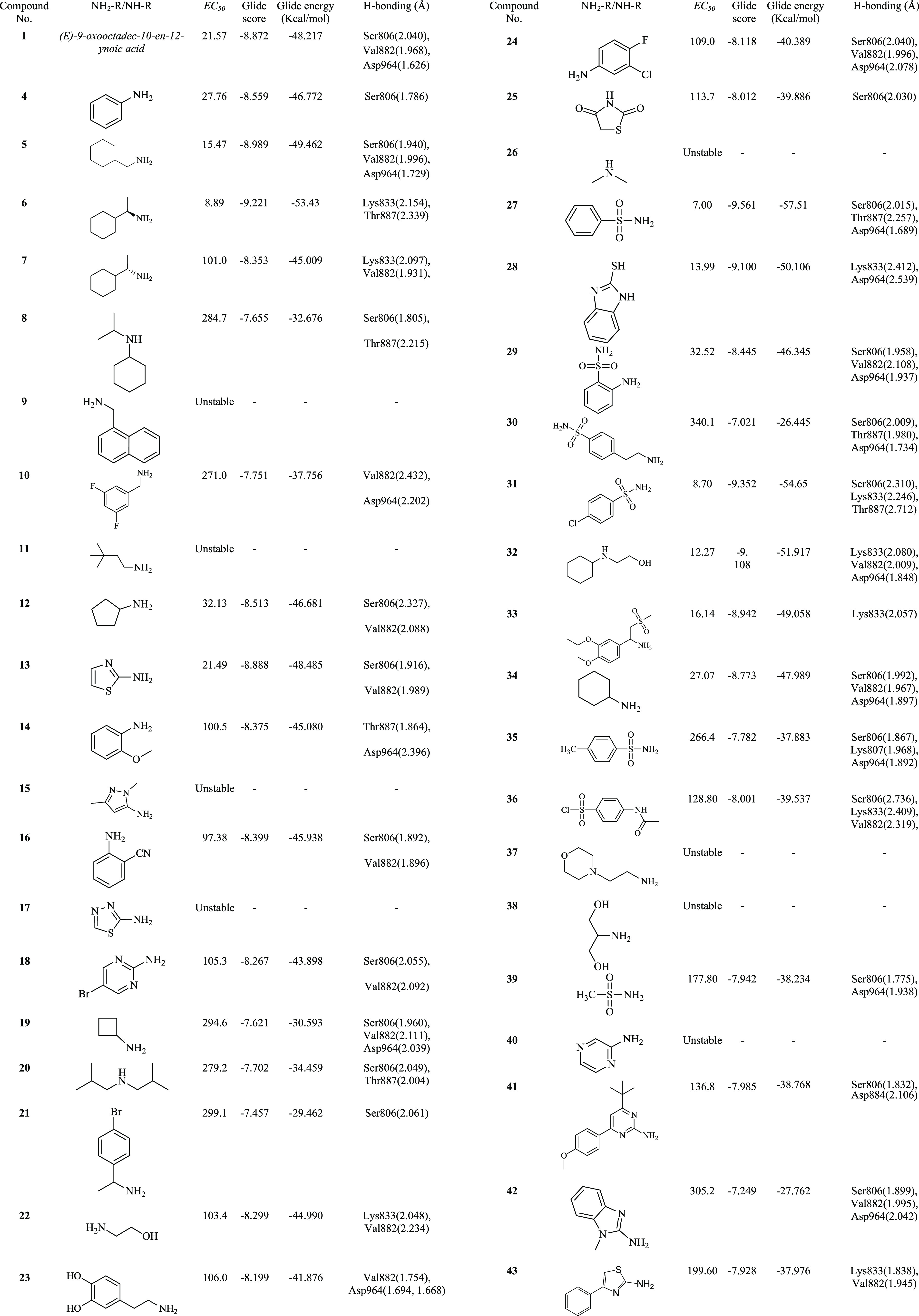
To explore the SAR of compound 1, herein we describe the design and synthesis of various amides derived from compound 1 using HATU as a reagent (Scheme 1). HATU was first reported by Louis A. Carpino in 1993 and is commonly encountered in alcohol and amine acylation reactions.13,14 Various amines were coupled with compound 1 in the presence of HATU and triethylamine in dichloromethane. After completion of the reaction, products were purified with flash column chromatography. The selected amine pharmacophore was from known antidiabetic drugs available as generics sulfonylureas (glimepiride, glipizide, and glyburide), biguanides (metformin), thiazolidinediones (pioglitazone and Actos generic), α-glucosidase inhibitors (acarbose), and meglitinides (nateglinide). All the newly synthesized pure amides were evaluated for their effect on glucose uptake in L6 myotubes. The EC50 values for the effect on glucose uptake in L6 myotubes are presented in Table 1.
Scheme 1. Reaction Scheme for Synthesis of Amides.

Table 1. Effects of Novel Amides to Promote Glucose Uptake with EC50 Values.
Biological Activity
Compounds 1 and 4–43 were evaluated for their effect on glucose uptake in L6 myotubes, and it was demonstrated that some of the compounds derived from (E)-9-oxooctadec-10-en-12-ynoic acid were found to increase insulin-stimulated glucose uptake in L6 myotubes compared with the parent compound by activating the PI3K pathway.8
Acid compound 1 exhibited an EC50 of 21.57 μM in the glucose uptake assay.8 Compound 4, an aniline derivative of compound 1, exhibited poor activity (EC50 of 27.76 μM). Compound 5 is an amide analogue of N-methylamine with acid compound 1, which showed a good activity (EC50 of 15.47 μM). For further improvement in the activity, the following compounds were synthesized and evaluated for glucose uptake. Compound 6 showed an increase in the activity trend, and compound 7 was inactive with an EC50 of 101.0 μM. These compounds were synthesized from cyclohexylmethanamine, (R)-1-cyclohexylethan-1-amine, and (S)-1-cyclohexylethan-1-amine, respectively. Compound 6 showed very good activity with an EC50 of 8.89 μM. This indicates that the stereochemistry of (S)-1-cyclohexylethan-1-amine plays an important role in the activity. Compound 8 was inactive and was synthesized from N-isopropylcyclohexanamine. The bulky substituent in compound 9 amide, i.e., naphthalen-1-ylmethanamine, was not tolerated as such and was an inactive compound.
Compounds 10 and 11 were inactive. Compound 12 with cyclopentanamine shows a poor activity (EC50 of 32.13). Compound 13, with thiazol-2-amine, a five-membered ring, was not tolerated and showed a similar EC50 of 21.49 μM. Compounds 14–25 were unstable and did not show any activity. Compounds 30 and 37 were prepared using 4-(2-aminoethyl)benzenesulfonamide and 2-morpholinoethanamine linkers, respectively, and showed no activity. Similarly, compound 39 was also an inactive compound prepared with methane-sulfonamide substitution.
Compound 26, a dimethylamine analogue, showed decent activity. Compounds 27and 28, amino-benzenesulfonamide and benzo[d]imidazole-2-thiol analogues, showed the best activity of this series (EC50 of 7.00 and 13.99 μM, respectively). However, compound 29 had an EC50 of 32.52 μM. Compounds 31–33 were 4-chloro-N-methylbenzenesulfonamide, 2-(cyclohexyl(methyl)amino)ethanol, and 1-(3-ethoxy-4-methoxyphenyl)-N-methyl-2-(methylsulfonyl)ethanamine analogues and exhibited moderate activity with EC50 values of 8.70, 12.27, and 16.14 μM, respectively. The other compounds of this series from 34 to 43 were inactive compounds.
Molecular Docking
In order to gain an insight into the mechanism of action and to rationalize the promising levels of antidiabetic activity potency demonstrated by the (E)-9-oxooctadec-10-en-12-ynoic acid analogues, a molecular docking study was performed against PI3K using the standard protocol implemented in the GLIDE module of the Schrödinger Molecular modeling suite to identify the binding modes of (E)-9-oxooctadec-10-en-12-ynoic acid analogues at the active site of PI3K and the various thermodynamic interactions that govern the binding of these molecules. The results show that all the docked (E)-9-oxooctadec-10-en-12-ynoic acid analogues could bind to the active site of PI3K with significant binding affinity and could engage in a series of bonded and nonbonded interactions (Table 1) with amino acid residues lining the active site. Also, their docking scores (Glide scores) were found to be in harmony with the observed antidiabetic activity with active compounds showing higher binding affinity than compounds with lower potency. Furthermore, to identify and understand the most significantly interacting residues and the various thermodynamic interactions governing the affinity to PI3K, a detailed analysis of per-residue interactions is elaborated for one of the most active analogue 27 in the next section.
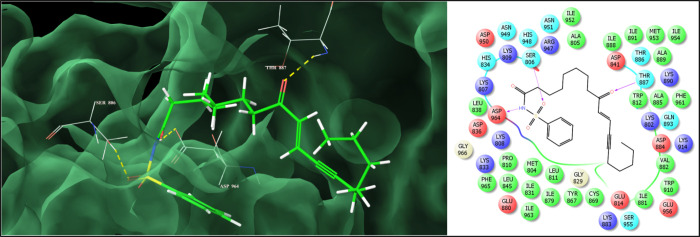
The lowest energy docked conformation of 27 showed that it possesses higher binding affinity (Glide dock score: −9.561, Glide binding energy: −57.51 kcal/mol) to PI3K and could snugly fit into the active site engaging in a network of significant bonded and nonbonded interactions (Figure 1). The major driving factor for the mechanical interlocking of 27 was observed to be a series of significant van der Waals interactions observed with the backbone 9-oxo-octadec-10-ene portion through Met953 (−2.105 kcal/mol), Asn951 (−1.104 kcal/mol), Asp950 (−2.154 kcal/mol), Lys890 (−1.694 kcal/mol), Thr887 (−2.404 kcal/mol), Thr886 (−2.404 kcal/mol), Ala885 (−2.76 kcal/mol), Asp884 (−1.426 kcal/mol), Lys883 (−1.041 kcal/mol), Val882 (−1.711 kcal/mol), Ile881 (−1.036 kcal/mol), Trp812 (−2.064 kcal/mol), and Ala805 (−1.499 kcal/mol) residues, while the N-phenylsulfonamide section was engaged in similar interactions with Asp964 (−2.461 kcal/mol), Ile963 (−1.728 kcal/mol), Phe961 (−1.145 kcal/mol), Glu880 (−1.031 kcal/mol), Ile879 (−1.626 kcal/mol), Tyr867 (−2.098 kcal/mol), Lys833 (−2.045 kcal/mol), Ile831 (−1.365 kcal/mol), Pro810 (−2.026 kcal/mol), Lys808 (−2.238 kcal/mol), Lys807 (−1.64 kcal/mol), Ser806 (−1.738 kcal/mol), and Met804 (−2.226 kcal/mol) residues. Equally significant electrostatic interactions observed with Asp964 (−2.172 kcal/mol), Asp950 (−1.927 kcal/mol), Lys890 (−2.166 kcal/mol), Thr886 (−1.513 kcal/mol), Val882 (−1.888 kcal/mol), Glu880 (−1.083 kcal/mol), Asp841 (−2.133 kcal/mol), Lys833 (−2.591 kcal/mol), Lys809 (−1.361 kcal/mol), Lys808 (−4.444 kcal/mol), Lys807 (−1.853 kcal/mol), and Ser806 (−3.143 kcal/mol) residues also contributed to the enhanced binding affinity of the compound. Furthermore, it also displayed a very close hydrogen bonding interaction with Ser806 (2.015 Å) through the sulfonyl (O=S=O) group, with Asp964 (1.689 Å) through the amide (−NH−) group, and with the Thr887 (2.257 Å) residue through a ketonic (C=O) function, which serves as an “anchor” to guide the 3D orientation of the compound into the active site and further facilitate the steric and electrostatic interactions. Other analogues in the series also exhibited such bonded and nonbonded interactions, which contributed to their antidiabetic potential.
Figure 1.
Binding mode and key interactions of compound 27 into the active site of PI3K (on the right side: the pink lines represent the hydrogen bonding interactions).
Conclusions
In conclusion, the compounds derived from (E)-9-oxooctadec-10-en-12-ynoic acid showed promising results by way of increasing glucose uptake in L6 myotubes, especially compounds 5, 6, 27, 28, and 31–33 with EC50 values of 15.47, 8.89, 7.00, 13.99, 8.70, 12.27, and 16.14 μM, respectively. Molecular docking analysis against PI3K revealed very clear preference for these compounds wherein all of them could snugly fit into the active site of the enzyme through a series of bonded and nonbonded interactions. We have successfully generated the SAR of novel amide compounds. This study opens a new scope for these types of derivatives for antidiabetic medications.
Acknowledgments
The authors wish to thank the management of Piramal Enterprises Limited, India, for supporting this study and providing the infrastructure needed for carrying out the research work reported in this paper. Authors are thankful to Schrödinger Inc. for GLIDE software to perform the molecular docking studies. The authors also thank Dr. Koteppa Pari for his scientific and valuable insights and Chandani Thakkar for preparation of the manuscript.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.1c03600.
Purity by 1H and 13C NMR, HRMS, and HPLC data of compounds 1 and 4–43 (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Reddy B. N.; Ruddarraju R. R.; Kiran G.; Pathak M.; Rama A. R. N. Novel Pyrazolo[3,4-d]pyrimidine-Containing Amide Derivatives: Synthesis, Molecular Docking, In Vitro and In Vivo Antidiabetic Activity. ChemistrySelect 2019, 4, 10072–10078. 10.1002/slct.201900208. [DOI] [Google Scholar]
- Wang Z.; Thurmond D. C. Mechanisms of biphasic insulin-granule exocytosis – roles of the cytoskeleton, small GTPases and SNARE proteins. J. Cell Sci. 2009, 122, 893–903. 10.1242/jcs.034355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.; Jang H. J. Anti-diabetic effects of natural products an overview of therapeutic strategies. Mol. Cell. Toxicol. 2017, 13, 1. 10.1007/s13273-017-0001-1. [DOI] [Google Scholar]
- Moller D. E. New drug targets for type 2 diabetes and the metabolic syndrome. Nature 2001, 414, 821–827. 10.1038/414821a. [DOI] [PubMed] [Google Scholar]
- Chan C.-H.; Ngoh G.-C.; Yusoff R. A Brief Review on Anti Diabetic Plants: Global Distribution, Active ingredients, Extraction Techniques and Acting Mechanisms. Pharmacogn Rev. 2012, 6, 22–28. 10.4103/0973-7847.95854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonio C. M.; Tânia S. F. Isolated Compounds from Natural Products with Potential Antidiabetic Activity - A Systematic Review. Curr. Diabetes Rev. 2017, 14, 106. 10.2174/1573399813666170505120621. [DOI] [PubMed] [Google Scholar]
- Maniyar Y.; Bhixavatimath P. Evaluation of the Hypoglycaemic and Hypolipidaemic Activities of the Aqueous Extract of the leaves of Ixora Coccinea Linn in Diabetic Rats. J. Clin. Diagn. Res. 2011, 5, 1381–1384. [Google Scholar]
- Thakkar C. S.; Kshirsagar R. R.; Naik V. R.; Ghosh A. R.; Vijayakumar V.; Saklani A.; Kulkarni-Almeida A. A. (E)-9-oxooctadec-10-en-12-ynoic acid from Ixora brachiataRoxb. increases glucose uptake in L6 myotubes by activating the PI3K pathway. Fitoterapia 2016, 114, 26–33. 10.1016/j.fitote.2016.08.004. [DOI] [PubMed] [Google Scholar]
- Ballatore C.; Huryn D. M.; Smith A. B. III Carboxylic acid (bio)isosteres in drug design. ChemMedChem 2013, 8, 385–395. 10.1002/cmdc.201200585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y. Z.; Johnson B. M.; Yang T. J. Role of biotransformation studies in minimizing metabolism-related liabilities in drug discovery. AAPS J. 2008, 10, 178–192. 10.1208/s12248-008-9016-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadekar P. K.; Roychowdhury A.; Kharkar P. S.; Khedkar V. M.; Arkile M.; Manek H.; Sarkar D.; Sharma R.; Vijayakumar V.; Sarveswari S. Design, synthesis and biological evaluation of novel azaspiro analogs of linezolid as antibacterial and antitubercular agents. Eur. J. Med. Chem. 2016, 122, 475–487. 10.1016/j.ejmech.2016.07.001. [DOI] [PubMed] [Google Scholar]; Gadekar P. K.; Urunkar G.; Roychowdhury A.; Sharma R.; Bose J.; Khanna S.; Damre A.; Sarveswari S. Design, synthesis and biological evaluation of 2,3-dihydroimidazo[2,1-b]thiazoles as dual EGFR and IGF1R inhibitors. Bioorg. Chem. 2021, 115, 105151. 10.1016/j.bioorg.2021.105151. [DOI] [PubMed] [Google Scholar]
- Kumari S.; Carmona A. V.; Tiwari A. K.; Trippier P. C. Amide Bond Bioisosteres: Strategies, Synthesis, and Successes. J. Med. Chem. 2020, 63, 12290–12358. 10.1021/acs.jmedchem.0c00530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter W. S.; Wilden J. D. The Preparation and Applications of Amides Using Electrosynthesis. Green Chem. 2020, 22, 7737–7759. 10.1039/D0GC02976A. [DOI] [Google Scholar]
- Upare A. A.; Gadekar P. K.; Sivaramakrishnan H.; Naik N.; Khedkar V. M.; Sarkar D.; Choudhari A.; Roopan S. M. Design, synthesis and biological evaluation of (E)-5-styryl-1,2,4-oxadiazoles as anti-tubercular agents. Bioorg. Chem. 2019, 86, 507–512. 10.1016/j.bioorg.2019.01.054. [DOI] [PubMed] [Google Scholar]
- Friesner R. A.; Murphy R. B.; Repasky M. P.; Frye L. L.; Greenwood J. R.; Halgren T. A.; Sanschagrin P. C.; Mainz D. T. Extra Precision Glide: Docking and Scoring Incorporating a Model of Hydrophobic Enclosure for Protein–Ligand Complexes. J. Med. Chem. 2006, 49, 6177–6196. 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- Halgren T. A.; Murphy R. B.; Friesner R. A.; Beard H. S.; Frye L. L.; Pollard W. T.; Banks J. L. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 2. Enrichment Factors in Database Screening. J. Med. Chem. 2004, 47, 1750–1759. 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- Friesner R. A.; Banks J. L.; Murphy R. B.; Halgren T. A.; Klicic J. J.; Mainz D. T.; Repasky M. P.; Knoll E. H.; Shelley M.; Perry J. K.; Shaw D. E. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 1. Method and Assessment of Docking Accuracy. J. Med. Chem. 2004, 47, 1739–1749. [DOI] [PubMed] [Google Scholar]
- Walker E. H.; Pacold M. E.; Perisic O.; Stephens L.; Hawkins P. T.; Wymann M. P.; Williams R. L. Structural Determinants of Phosphoinositide 3-Kinase Inhibition by Wortmannin, LY294002, Quercetin, Myricetin, and Staurosporine. Mol. Cell 2000, 6, 909–919. 10.1016/S1097-2765(05)00089-4. [DOI] [PubMed] [Google Scholar]
- Stalin A.; Irudayaraj S. S.; Gandhi G. R.; Balakrishna K.; Ignacimuthu S.; Al-Dhabi N. A. Hypoglycemic activity of 6-bromoembelin and vilangin in high-fat diet fed-streptozotocin-induced type 2 diabetic rats and molecular docking studies. Life Sci. 2016, 153, 100–117. ISSN 0024-3205 10.1016/j.lfs.2016.04.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.