Abstract
Pandemic dynamics and health care responses are markedly different during the COVID-19 pandemic than in earlier outbreaks. Compared with established infectious disease such as influenza, we currently know relatively little about the origin, reservoir, cross-species transmission and evolution of SARS-CoV-2. Health care services, drug availability, laboratory testing, research capacity and global governance are more advanced than during 20th century pandemics, although COVID-19 has highlighted significant gaps. The risk of zoonotic transmission and an associated new pandemic is rising substantially. COVID-19 vaccine development has been done at unprecedented speed, with the usual sequential steps done in parallel. The pandemic has illustrated the feasibility of this approach and the benefits of a globally coordinated response and infrastructure. Some of the COVID-19 vaccines recently developed or currently in development might offer flexibility or sufficiently broad protection to swiftly respond to antigenic drift or emergence of new coronaviruses. Yet many challenges remain, including the large-scale production of sufficient quantity of vaccines, delivery of vaccines to all countries and ensuring vaccination of relevant age groups. This wide vaccine technology approach will be best employed in tandem with active surveillance for emerging variants or new pathogens using antigen mapping, metagenomics and next generation sequencing.
Keywords: SARS-CoV-2, pandemic, vaccine, COVID-19
Introduction
Since the 1980s, at least 30 new infectious disease threats have emerged (Mukherjee, 2017). Of emerging infectious diseases (EIDs) identified since 1940, 60% were zoonotic in nature, of which 70% originated in wildlife (Jones et al., 2008). This trend is expected to rise because of increased human–animal contact, climate change, land use changes, global population growth, and increased global interconnectedness (Jones et al., 2008, Mukherjee, 2017, Petersen et al., 2018).
Pandemics and epidemics have increased in frequency since the Middle Ages: bubonic plague (started 1347), smallpox (early 1500s), influenza 'Great Pandemic' (1833), cholera (1881), Spanish influenza (1918), Asian influenza (1957), hepatitis C (1960s), Hong Kong influenza (1968), Russian influenza (1977), human immunodeficiency virus (HIV, 1981), severe acute respiratory syndrome coronavirus (SARS-CoV-1, 2003), swine influenza (2009), Middle East respiratory syndrome coronavirus (MERS-CoV, 2012), West Africa Ebola virus (2013), chikungunya virus (2013), Zika virus (2015) and coronavirus disease 2019 (COVID-19) (SARS-CoV-2, 2019). Although the World Health Organization (WHO) includes coronavirus as a priority pathogen, until the COVID-19 pandemic, influenza was considered to be the most likely causal pathogen of the next significant pandemic, and modelling suggested an annual 1% chance of an influenza pandemic that would result in 6 million global deaths (Madhav et al., 2018).
The first difficulty in drawing lessons from previous influenza A pandemics is that the COVID-19 outbreak is the first and only pandemic caused by a coronavirus during the scientific era (CDCP, 2018a, Honigsbaum, 2020, WHO, 2020a). The biological differences between influenza A viruses and coronaviruses are substantial (Abdelrahman et al., 2020). The ultimate goal in declaring a pandemic or a Public Health Emergency of International Concern (PHEIC) is to create a global alert (Durrheim et al., 2020). The term pandemic has been used for approximately 40 years and was not widely adopted during the pandemic events of 1957 and 1968. It is more than 50 years since the occurrence of a pandemic with similar severity and extent to the current COVID-19 pandemic. Mortality in the 1957 Asian and 1968 Hong Kong influenza pandemics was comparable to that of COVID-19 so far (Honigsbaum, 2020). However, severity and mortality were substantially lower in the 2009 influenza A pandemic than in the current COVID-19 pandemic (Faust and Del Rio, 2020, Saunders-Hastings and Krewski, 2016). Human contact has been far greater with the H1N1 influenza A virus subtype than with other influenza viruses (Palese and Wang, 2011). One reason for the mildness of the 2009 pandemic could be the intrinsic pathogenicity and virulence of the H1 subtype (Belser et al., 2010) and the presence of partial immunity against the A/California/04/2009 (H1N1) virus among older adults (Hancock et al., 2009).
While there is a surveillance network made up of 144 national influenza centres all over the world (WHO, 2020b), there is nothing similar for the surveillance of other diseases. In the current COVID-19 pandemic, surveillance has been implemented with the same tools used for influenza surveillance (WHO, 2021a).
Key features of previous influenza pandemics
Features of previous pandemics are summarised in Table 1 .
Table 1.
Features of 20th and 21st century influenza pandemics
| Pandemic | Dates | Influenza virus | No. waves | No. deaths | Age groups with highest mortality |
|---|---|---|---|---|---|
| Spanish influenza | 1918–1920 | A/H1N1 | 3(Barry, 2005) | 20–100 million (Barry, 2005, Johnson and Mueller, 2002, Jordan, 1927, Nicholson et al., 2019) | Infants, young adults (20–40 years), elderly (Taubenberger and Morens, 2006) |
| Asian influenza | 1957–1958 | A/H2N2 | 2(Rogers 2020) | 1–2 million (Saunders-Hastings and Krewski, 2016) | Infants, children (5–14 years), young adults (15–24 years), elderly (Viboud et al., 2016) |
| Hong Kong influenza | 1968–1969 | A/H3N2 | 2(Cockburn et al., 1969, Saunders-Hastings and Krewski, 2016) | 0.5–2 million (Saunders-Hastings and Krewski, 2016) | >65 years (Centers for Disease Control and Prevention 2019b) |
| Russian influenza | 1977–1979 | A/H1N1 | 1(Gregg et al., 1978) | 700,000 (Gregg et al., 1978) | Infants, young adults (<25 years) (Gregg et al., 1978) |
| Swine influenza | 2009–2010 | A/H1N1pdm09 | 2 or 3 depending on location(Jhung et al., 2011, Saunders-Hastings and Krewski, 2016) | 123,000–203,000 (Simonsen et al., 2013) | 5–59 years (Charu et al., 2011) |
Pandemic dynamics and cultural contexts
The spread of the presumed influenza 'Great Pandemic' in 1833 occurred via transatlantic shipping between the Americas and Africa (Patterson, 1986). The first outbreak of the 1918 pandemic was identified in military camps across USA towards the end of World War 1 (Barry, 2005). The pandemic spread globally over nine months (Barry, 2005, Byerly, 2010). War-related factors might have promoted high transmission levels and hampered efforts to contain the pandemic (National Academies of Sciences et al., 2019). In the US, it was illegal to be critical of the government during the war (Nicholson et al., 2019), whilst the UK mounted one of the least effective responses to the pandemic largely because health care professionals underplayed its importance (Tomkins, 1992). The causal agent is now known to be influenza A/H1N1. Influenza A virus was first isolated only in 1933 by Smith, Andrewes and Laidlaw, followed by influenza B virus by Francis in 1936 (Smith et al., 1933, CDCP, 2019a).
In February 1957, a new reassorting A/H2N2 influenza strain (Asian influenza pandemic) emerged in the Yunnan Province of China, and rapidly spread globally (Saunders-Hastings and Krewski, 2016). A second wave affected the Northern hemisphere in November 1957 which was more severe than the first (Rogers, 2020). Most global transmission occurred via land and sea routes (Saunders-Hastings and Krewski, 2016). In 1957, the existence of the avian reservoir and the phenomena of antigenic drift and genetic reassortment were not recognised (Kilbourne, 1980, Kilbourne, 2006).
First reported in July 1968, a new A/H3N2 pandemic strain (a reassortment of the Asian influenza strain with an avian influenza H3 subtype) resulted in the Hong Kong influenza pandemic (Cockburn et al., 1969, Saunders-Hastings and Krewski, 2016). By autumn, the virus had been detected in several Asian countries and the US, before spreading more widely (Cockburn et al., 1969, Saunders-Hastings and Krewski, 2016). The 1968 pandemic was the first in which air travel played a significant role in transmission (Saunders-Hastings and Krewski, 2016). Russian influenza emerged in the former Soviet Union during November 1977 and had spread elsewhere by early 1978 (Gregg et al., 1978). It was caused by an influenza A/H1N1 virus that had circulated worldwide during the 1950s, and mainly affected individuals younger than 25 years of age (Gregg et al., 1978).
The 2009 influenza pandemic was caused by the influenza A/H1N1pdm09 virus (Saunders-Hastings and Krewski, 2016), a triple reassortment virus containing genes from classic swine influenza A viruses, North American lineage avian influenza viruses and human influenza A viruses (Dawood et al., 2009). The first case was recorded in April 2009 in Mexico and the US. It was declared a pandemic by the WHO in June 2009 (Chan, 2009). The pattern of the pandemic varied between countries (Jhung et al., 2011, Saunders-Hastings and Krewski, 2016). School cycles were shown to be important in transmission dynamics in Mexico and Hong Kong (Chowell et al., 2011, Wu et al., 2010).
Morbidity and mortality
Mortality during the ‘Great Pandemic’ in 1833 was up to 80%, with a high death toll for people between 21 and 40 years of age (Patterson, 1986). The Spanish influenza pandemic infected approximately 500 million people, roughly one-third of the world's population at the time, with mortality estimates of 20–100 million deaths worldwide (Barry, 2005, Johnson and Mueller, 2002, Jordan, 1927, Nicholson et al., 2019). The overall mortality was >2.5% (Taubenberger and Morens, 2006), but with wide differences; for example, mortality was high where populations were naive to influenza such as islands in Oceania (e.g. 22% in Western Samoa)(Shanks et al., 2018) or Alaska and Eskimo populations (e.g. up to 38% in Alaska and up to 75% in Labrador)(Mamelund et al., 2013). Age-specific death rates followed a W-shaped curve, with high death rates in the very young and the elderly (as observed with seasonal influenza), but with the addition of a third peak of deaths in adults 20–40 years of age (Taubenberger and Morens, 2006). Nearly half of the influenza deaths during the 1918 pandemic occurred in persons 20–40 years of age (Taubenberger and Morens, 2006). Total mortality in Europe has been estimated at 2.64 million i.e. 1.1% of the total population (Ansart et al., 2009). Mortality varied more than 30-fold across countries, with poorer countries disproportionately affected (Murray et al., 2006). Extrapolating mortality rates to the 2004 population estimated that a similar pandemic would result in 62 million deaths (Murray et al., 2006).
The 1957 Asian influenza was relatively mild and resulted in approximately 1–2 million deaths worldwide (Saunders-Hastings and Krewski, 2016). A study using historical mortality data estimated an average pandemic-associated excess respiratory mortality rate of 1.9 per 10,000 persons (Viboud et al., 2016). The Hong Kong influenza (1968) pandemic is estimated to have caused half a million to 2 million deaths worldwide (Saunders-Hastings and Krewski, 2016), with most deaths occurring in people >65 years of age (CDC, 2019b). All-cause mortality rose by between 3.6% in Canada to 13.0% in England and Wales (Viboud et al., 2005). Mortality in the 1977 Russian influenza pandemic was estimated at 700,000 (Gregg et al., 1978).
Global respiratory deaths from the 2009 A/H1N1 pandemic have been estimated at 123,000–203,000, approximately 10 times as high as the WHO's laboratory-confirmed count (Simonsen et al., 2013). Another modelling study estimated 105,700–395,600 respiratory deaths globally with a further 46,000–179,900 cardiovascular deaths (Dawood et al., 2012); up to 85% of deaths occurred in people under 65 years of age (Dawood et al., 2012, Simonsen et al., 2013). Another study estimated that 71% of deaths in Mexico occurred in people under 60 years of age (Charu et al., 2011). Pregnant women were also identified to be at particular risk (Louie et al., 2010).
Pandemic responses: non-pharmaceutical interventions, treatments and vaccination
Mitigation efforts in the 1918 pandemic relied upon non-pharmaceutical interventions (NPI) such as wearing face masks, quarantines, restriction of activities and gatherings, closures of schools and churches (Nicholson et al., 2019). Concurrent school closures and public gathering bans were the most commonly implemented interventions and resulted in a statistically significant reduction in excess pneumonia and influenza deaths (Markel et al., 2007). Although no antibiotics, antivirals or modern vaccines were available, passive immunisation with immunoglobulins was implemented (Luke et al., 2006). Mortality later in the pandemic is believed to be the result of secondary bacterial infections (Brundage, 2006, Morens et al., 2010).
The 1957 influenza pandemic was the first in which a previous vaccine was available; however, it took 6 months before it could be rolled out because the developers needed to adjust its formulation with the new pandemic strain. Only 30 million vaccine doses were distributed globally and inadequate coverage meant that vaccination had little impact on the pandemic (Henderson et al., 2009, Jensen et al., 1958, Saunders-Hastings and Krewski, 2016). The generally mild nature of the 1968 pandemic again meant that NPIs were not considered necessary (Saunders-Hastings and Krewski, 2016). However, vaccines became available only after the pandemic had peaked (Rogers, 2020).
School closures and other NPIs were implemented swiftly in Mexico after the outbreak of the 2009 A/H1N1 pandemic, followed by similar measures in many other countries (Chowell et al., 2011, Saunders-Hastings and Krewski, 2016). This was the first pandemic to employ both vaccination and use of antivirals (Saunders-Hastings and Krewski, 2016). Data from the first vaccine clinical trial became available in August 2009, 4 months after the outbreak was first identified (WHO, 2012). A month later, the WHO identified that the amount of vaccine that could be produced within a year would be substantially less than the 4.9 billion doses previously estimated (WHO, 2012). However, the vaccine could be administered as a 1-dose instead of 2-dose series (WHO, 2012). Vaccination rates in Europe varied from 0.6% in the Czech Republic to 59% in Sweden (Samanlioglu and Bilge, 2016), while the rate in Canada was estimated at 41% (Gilmour and Hofmann, 2010), in the US at between 20% and 37% depending on age group (CDCP, 2010) and in Brazil at 86% (Domingues and de Oliveira, 2012). The WHO Pandemic Influenza A(H1N1) Vaccine Deployment Initiative facilitated access to the pandemic vaccine in developing countries. However, demand from countries varied considerably and sufficient vaccine was delivered to provide coverage ranging from 0.4% to >100% of the population, with a median coverage of 9.9% (WHO, 2012).
Economic impact
It has been estimated that the Spanish influenza pandemic reduced per capita Gross Domestic Product (GDP) by 6% compared with an 8% reduction resulting from World War 1 (Barro et al., 2020). However, economic recovery was swift, with a 25.5% rebound in industrial production in the US between March 1919 and January 1920 (Anderson, 1979, Grant, 2014, Vernon, 1991). An analysis of the post-1940 US population showed that unborn children at the time of the pandemic had lower educational attainment, income and socioeconomic status compared with other birth cohorts (Almond, 2006). In contrast, the economic impact of the Asian, Hong Kong and 2009 A/H1N1 influenza pandemics was small (Henderson et al., 2009, James and Sargent, 2006, Saunders-Hastings and Krewski, 2016). A systematic review of the cost effectiveness of pandemic interventions concluded that, at a threshold of $45,000 per disability-adjusted life-year (DALY), hospital quarantine and vaccination were cost-effective options, but school closures and social distancing were not (Pasquini-Descomps et al., 2017). Cost-effectiveness ratios for antivirals ranged from $350 to $3500 per DALY or quality-adjusted life-year (QALY) in lethal epidemics but were as high as $40,000 to $250,000 for less severe infections (Pasquini-Descomps et al., 2017).
Coronavirus PHEIC threats (SARS 2003 and MERS 2012) and COVID-19 pandemic
Coronaviruses are positive, single-stranded RNA viruses belonging to the family Coronaviridae (Li, 2016, Su et al., 2016). They are classified into four genera: Alphacoronavirus, Betacoronavirus, Gammacoronavirus and Deltacoronavirus (Li, 2016, Su et al., 2016). There are four endemic coronaviruses that circulate widely in humans. These cause mainly upper and mild respiratory infections such as the common cold, but also less frequently result in more severe respiratory syndromes including pneumonia in vulnerable and immunocompromised hosts (Su et al., 2016). Two of these four coronaviruses are Betacoronavirus (HCoV-OC43 and HCoV-HKU1) and two are Alphacoronavirus (HCoV-229E and HCoV-NL63) (Forni et al., 2017, Greenberg, 2016, Su et al., 2016). There are a further two epidemic coronaviruses: MERS-CoV that produces epidemic outbreaks with short chains of transmission and without clear adaptation to spread widely in humans, and SARS-CoV-1 which has been eradicated by control measures. Both are Betacoronavirus, with MERS-CoV belonging to lineage C (subgenus merbecovirus from etymology “MER” “BEta” “COronavirus”) and SARS-CoV-1 belonging to lineage B (subgenus sarbecovirus “SARs” “BEta” “COronavirus”)(Greenberg, 2016, Tang et al., 2020). Pandemic SARS-CoV-2 also belongs to Betacoronavirus and to the sarbecovirus subgenus (Zhu et al., 2020). Bats or rodents are believed to be the original, natural hosts of all seven human coronaviruses, with cattle, camels, civets and mink implicated as probable intermediate hosts (Forni et al., 2017, Hul et al., 2021, Tang et al., 2020, Tu et al., 2004).
SARS-CoV-1 first emerged in China in November 2002 and spread to 26 countries, although most cases were concentrated in China, Taiwan, Hong Kong, Singapore and Toronto (Canada) (Wilder-Smith et al., 2020). The outbreak was short-lived and was brought under control by July 2003, by which time it had resulted in 8098 cases and 774 deaths (case fatality rate 9.5%) (Wilder-Smith et al., 2020). MERS-CoV emerged in 2012 in Saudi Arabia, but was never defined as having pandemic potential. So far, it has resulted in 2562 laboratory-confirmed cases and 881 deaths (case fatality rate 34.4%) (WHO, 2019). SARS-CoV-2 was first identified in Wuhan, China in December 2019 and has rapidly spread throughout the world. As of 21 March 2021, more than 123 million cases and 2.7 million deaths had been reported globally (Johns Hopkins University, 2020). The death rate per 100,000 population reported varies enormously over time and in different countries, probably because of different testing and reporting policies (Johns Hopkins University, 2020). A systematic review including data up to June 2020 has estimated the infection fatality rate of SARS-CoV-2 at 0.68% (range 0.09–1.60) (Meyerowitz-Katz and Merone, 2020).
Individual and community-based NPIs such as isolation, physical distancing, mask use, and closure of public places were widely implemented during the SARS 2003 outbreak, as were hospital isolation facilities and widespread use of personal protective equipment (Wilder-Smith et al., 2020). There was a strong political will in affected countries to implement effective public health measures (Wilder-Smith et al., 2020). SARS-CoV-1 viral loads in the lungs peaked at 6–11 days after the onset of illness and the number of secondary cases was substantially reduced if an infected individual could be isolated within four days of symptom appearance (Cheng et al., 2004, Li et al., 2004). Fortunately, person-to-person transmission of MERS-CoV was substantially lower than SARS-CoV-1 (Peeri et al., 2020), although hospital transmission was significant (Al-Tawfiq and Auwaerter, 2019).
Some countries, for example Taiwan, Vietnam, South Korea and New Zealand, were successful in controlling COVID-19 (Bacay Watson, 2020). Vietnam closed its border with China and implemented testing, contact tracing and social distancing (Bacay Watson, 2020). New Zealand banned entry from China and mandated a strict quarantine for visitors very early, followed by a stringent national lockdown and extensive testing (Bacay Watson, 2020). Many other countries have relied upon physical distancing, handwashing, mask wearing in public spaces, national lockdowns and contact tracing (Petersen et al., 2020). A study concluded that the most effective measures were closure or restriction of places where people gather, including businesses, educational institutions, bars, restaurants, etc (Haug et al., 2020). However, less intrusive measures such as border restrictions and risk communication strategies were also effective (Haug et al., 2020).
SARS-CoV-2 has spread at a massively higher rate than SARS-CoV-1. Several factors might account for this. Firstly, there was pre-epidemic circulation in the months before the initial outbreak, and the first major outbreak occurred at the time of a Chinese holiday in a city of over 11 million people that is also a large transport hub, facilitating early spread (Wilder-Smith et al., 2020). Secondly, there is a marked difference in virus shedding between SARS-CoV-1 and MERS-CoV versus SARS-CoV-2. In the former two, the virus primarily replicates in the lower airways, but more in the upper airways in the latter (Cheng et al., 2004, Petersen et al., 2020, Wölfel et al., 2020). Peak viral loads in nasopharyngeal aspirates of SARS-CoV-1 occur 6–11 days after symptom onset; in contrast, viral loads of SARS-CoV-2 peak during the first few days of infection, including before the patient becomes symptomatic.(Cheng et al., 2004, To et al., 2020, Wölfel et al., 2020). In addition, many SARS-CoV-2 infections remain asymptomatic (Day, 2020, Li et al., 2020, Petersen et al., 2020). The “serial interval” provides an estimate of the time from symptom onset in a primary case to symptom onset in a secondary case. This has been estimated at 8.4 days for SARS-CoV-1, but only 4.0 days for SARS-CoV-2 (Lipsitch et al., 2003, Nishiura et al., 2020). A third factor is the very high transmissibility of SARS-CoV-2. The basic reproduction number, R0, of SARS-CoV-2 was estimated in the European Union at 0.91–6.33 at the beginning of the outbreak, with a mean of 4.22 (Linka et al., 2020). In contrast, the R0 values for MERS-CoV and SARS-CoV-1 were estimated at 0.6–0.7 and 2–4, respectively, before mitigation measures were put in place (Breban et al., 2013, WHO, 2003).
Pandemics now: where are we in 2021?
The present situation differs significantly from that of 1918. In 2021, we have substantially improved health care and no intense combat or world war. We also have in place the WHO, other international governance mechanisms and the Global Health Security Agenda (GHSA) (Nicholson et al., 2019). In this section, we review the current situation with regard to likely pandemic dynamics and health care responses. We discuss the role of new vaccine technology later in the paper.
Risk of zoonotic transmission
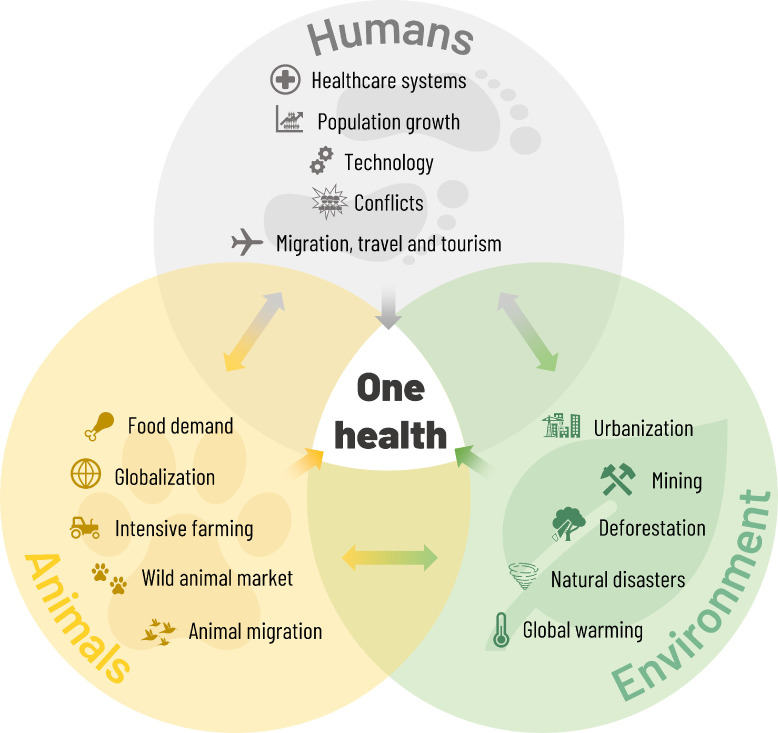
The COVID-19 pandemic has led to increasingly urgent calls to change how humans impact on the environment (Settele et al.). Land use change, climate change, mining, urbanisation, population growth, wild animal markets and modern human-animal interactions, travel and globalisation all act to bring humans into closer contact both with each other and with animals that host novel pathogens (Bedford et al., 2019, Settele et al.). A study of EIDs between 1940 and 2004 showed that human population density was an independent predictor of zoonotic EIDs and wildlife host species richness was an additional predictor of zoonotic EIDs of wildlife origin (Jones et al., 2008). Another study showed that in areas with substantial human activity, wildlife that hosts pathogens shared with humans comprised a higher proportion of local species compared with nearby undisturbed areas (Gibb et al., 2020). Both analyses concluded that conserving wildlife diversity by reducing human activity is likely to contribute to reducing zoonotic disease (Gibb et al., 2020, Jones et al., 2008).
The One Health initiative is a collaborative, transdisciplinary approach with focus on the interconnection between people, animals, plants and their shared environment (Figure 1 ) (CDCP, 2018b). A number of tools and programmes using the One Health approach have been developed (CDCP, 2018b, Rist et al., 2014, Salyer et al., 2017). PREDICT, part of USAID's Emerging Pandemic Threats programme, was established in 2009 to conduct and build capacity for surveillance for zoonotic pathogens with pandemic potential (USAID PREDICT, 2020). However, funding for PREDICT ended in early 2020 (Carlson, 2020). The related Global Virome Project has the ambitious goal of detecting the majority of zoonotic viral threats to human health and food security within 10 years (Global Virome Project, 2020). The project has estimated that there are between 631,000 and 827,000 unknown viruses with zoonotic potential in mammal and bird hosts (Carroll et al., 2018).
Figure 1.
The One Health concept.
Virus variability and the human immune response
Bats, and particularly those of the genus Rhinolophus, are believed to be the main natural reservoir of SARS- and MERS-related coronaviruses (Hu et al., 2015, Li et al., 2005). However, the origin and reservoir of SARS-CoV-2 still remain unclear (Mallapaty, 2020). In contrast, the origin, reservoir, cross-species transmission, genetic and antigenic evolution of influenza viruses are much better understood (Wille and Holmes, 2020). The antigenic drift in influenza virus means that the formulation of seasonal influenza vaccines must be updated regularly (Petrova and Russell, 2018). The SARS-CoV-2 genome experiences lower mutation rates (Alouane et al., 2020) but it is too early to predict whether regular reformulation of COVID vaccines will be necessary. The recent emergence and rapid spread in different parts of the world of SARS-CoV-2 virus variants that harbour point mutations in the spike protein is a matter of concern (Kupferschmidt, 2021, Lauring and Hodcroft, 2021, Tegally et al., 2020), and it has been shown that such mutations are associated with reduced neutralising activity of convalescent serum antibodies (Greaney et al., 2021). Vaccination will progressively increase the population's immunity against the virus but might also contribute to the selection of some specific escape mutants (Kupferschmidt, 2021).
Drugs for pandemic use
Bacterial co-infection, secondary infection and superinfection are common during virus pandemics (Brundage, 2006, Chertow and Memoli, 2013). Studies of autopsy data found bacterial infection in nearly all deaths in the 1918 influenza pandemic (Morens et al., 2008) and up to 55% of deaths in the 2009 A/H1N1 pandemic (CDC, 2009, Gill et al., 2010). Antibiotics are often overused or misused in both humans and animals, leading to development of antimicrobial resistance (Buchy et al., 2020). A recent systematic review of patients with confirmed COVID-19 found that bacterial co-infection was identified in 3.5% of patients and secondary bacterial infection in 14.3%; however, 71.9% received treatment with antibiotics (Langford et al., 2020). The WHO considers antimicrobial resistance as one of the top 10 threats to global health (World Health Organization).
Antivirals are likely to be of significant benefit in reducing morbidity and mortality and are the only specific intervention available before a vaccine is developed, however supplies are limited (WHO, 2018). Traditional antivirals used in influenza infection are the adamantane derivatives, amantadine and rimantadine, and the neuraminidase inhibitors, oseltamivir, zanamivir, peramivir and laninamivir (Principi et al., 2019). However, resistance to amantadine and rimantadine is widespread (Centers for Disease Control and Prevention 2020). A Cochrane review and meta-analysis found that oseltamivir reduced the time to symptom alleviation, although it was not possible to determine whether serious influenza complications such as pneumonia were reduced (Jefferson et al., 2014).
Baloxavir marboxil is a new drug against influenza virus targeting the endonuclease function of the viral PA polymerase subunit. It has been licensed in Japan and the US since 2018 and acts by inhibiting viral replication without cytotoxicity (Principi et al., 2019). It has been shown to be as effective as oseltamivir at alleviating symptoms of uncomplicated influenza and is also effective at preventing influenza infection among household contacts (Hayden et al., 2018, Ikematsu et al., 2020, Ison et al., 2020). The clinical development of pimodivir for the treatment of influenza A infection has recently been halted (Anon, 2020).
Several monoclonal antibodies against the conserved stalk region of the influenza A haemagglutinin (HA) protein are in development and have demonstrated antiviral activity in phase 2 trials (Ali et al., 2018, Hershberger et al., 2019, McBride et al., 2017). A systematic review and meta-analysis concluded that convalescent plasma might reduce mortality in severe influenza and SARS-CoV-1 infection (Mair-Jenkins et al., 2015); however, a meta-analysis of randomised, controlled trials in severe influenza found no benefit (Xu et al., 2020). A Cochrane review of convalescent plasma used in patients with COVID-19 concluded that its benefit is as yet uncertain (Piechotta et al., 2020).
Numerous drugs are being evaluated for the treatment of COVID-19. As of March 2021, the Milken Institute listed over 300 new or repurposed drugs being investigated (Milken Institute, 2021). The WHO SOLIDARITY trial evaluated four repurposed drugs: remdesivir, hydroxychloroquine, lopinavir-ritonavir fixed dose combination and interferon-β1a in patients hospitalised for COVID-19 (Pan et al., 2021). None of the study drugs reduced mortality, initiation of ventilation or duration of hospital stay (Pan et al., 2021). Two previously published randomised trials had shown non-significant improvements with remdesivir versus placebo (Beigel et al., 2020, Wang et al., 2020). The CoDEX randomised trial showed a significant increase in the number of ventilator-free days with dexamethasone compared with standard care, although there was no significant difference in all-cause mortality (Tomazini et al., 2020). The RECOVERY trial showed a significant mortality benefit with dexamethasone versus standard care in patients receiving ventilator support or oxygen, but not in patients receiving no respiratory support (Horby et al., 2020). A number of neutralising monoclonal antibodies are in development for the treatment of patients with mild to moderate disease who are at high risk of progression to severe disease, with promising results (Taylor et al., 2021).
Several guidelines have been published to guide clinical use of treatments for COVID-19 (NIH, 2021, Chalmers et al., 2021, WHO, 2021b, Bhimraj et al., 2021). Neutralising antibodies are recommended for patients with mild to moderate illness who are high risk of progression (NIH, 2021, Bhimraj et al., 2021). Dexamethasone or other systemic corticosteroids are widely recommended in hospitalised patients receiving supplemental oxygen or mechanical ventilation, with the possible additional of remdesivir, baricitinib or tocilizumab (NIH, 2021, Chalmers et al., 2021, WHO, 2021b, Bhimraj et al., 2021). Guidelines do not recommend drugs such as hydroxychloroquine, azithromycin, ivermectin and protease inhibitors such as lopinavir or ritonavir (NIH, 2021, Chalmers et al., 2021, WHO, 2021b, Bhimraj et al., 2021).
Vaccine acceptance and hesitancy
Even though effective vaccines against COVID-19 are now available, it is still unclear how widely accepted they will be by the public. Vaccine hesitancy has become a significant barrier to uptake (Salmon et al., 2015, Shetty, 2010), often fuelled by social media. The Mott Poll Report in the US reported that only 68% of parents intended to have their children vaccinated against influenza in the 2020–21 season (CS Mott Children's Hospital, 2020). Vaccine hesitancy might become a barrier to uptake of COVID-19 vaccines (French et al., 2020, McAteer et al., 2020). A survey in France found that 26% of respondents would be unwilling to receive a SARS-CoV-2 vaccine, rising to 37% among low-income people (COCONEL Group, 2020). Similarly, a survey in the US reported that 32% of adults were unsure whether they would take a vaccine and 11% stated that they would not (Fisher et al., 2020). In contrast, over 90% of adults in China were willing to be vaccinated (Wang et al., 2020).
Multi-country studies of likely acceptance of COVID-19 vaccines have shown variation between countries. In a survey of 12 countries, likely vaccine acceptance ranged from 86% in China to 63% in the US and Sweden (Kerr et al., 2021). A systematic review of survey studies from 33 countries found likely acceptance rates of over 90% in Ecuador, Malaysia, Indonesia and China, while the lowest acceptance rates were found in France (59%), US (57%), Poland (56%), Russia (55%), Italy (54%), Jordan (28%) and Kuwait (24%) (Sallam 2021). Another study found higher COVID-19 vaccine acceptance in 10 low- and middle-income countries in Asia, Africa and South America (80.3%) compared with Russia (30.4%) and the US (64.6%) (Arce et al., 2021). Among the low- and middle-income countries, vaccine acceptance ranged from 66.5% in Pakistan and Burkina Faso to 96.6% in Nepal.
Tracking pandemic death rates
Although there are multiple sources of data on the number of deaths caused by COVID-19, the true number is unknown. The statistics available from different countries can be substantially influenced by different definitions used; for example, some countries include only laboratory-confirmed COVID-19-related deaths, whilst others also include suspected deaths. Limited testing capacity in some countries, particularly in the developing world, can lead to considerable underestimation of the death rate. These issues can be avoided, at least to a degree, by calculation of the number of excess deaths, defined as the increase in all-cause mortality over the expected mortality based on historical data. This method has been used to estimate mortality in previous pandemics and seasonal influenza epidemics. Although it does not provide the exact mortality associated with the pandemic or epidemic, it is considered to provide an objective indicator (Beaney et al., 2020).
The excess mortality resulting from the COVID-19 pandemic is being tracked by several groups. Data from the World Mortality Dataset, which tracks mortality in 103 countries, found the highest excess mortality per 100,000 population in Latin American and Eastern European countries: Peru (590), Bulgaria (460), North Macedonia (420), Serbia (400), Mexico (360), Ecuador (350), Lithuania (350) and Russia (340) (Karlinsky & Kobak 2021). Some countries, including Uruguay, Australia and New Zealand, had negative excess mortality i.e. fewer deaths than expected. Similar results were obtained from other groups (The Economist, 2021, Sanmarchi et al, 2021). The excess mortality data indicated that the official number of COVID-19-related deaths reported by many countries are underestimates (Karlinsky & Kobak, 2021, The Economist, 2021, Sanmarchi et al, 2021).
Health care capacity
The COVID-19 pandemic has highlighted significant gaps in health services. In the US, hospitals were largely operating at full capacity pre-pandemic (Ajao et al., 2015). An adequately staffed health workforce cannot be depended on in a pandemic because of high absentee rates, driven by illness or fear (Madhav et al., 2017 Nov 27). In addition, influenza vaccination coverage among health care professionals continues to be variable and often suboptimal in many countries (CDC, 2021, Haviari et al., 2015). There is a broad recognition among stakeholders that health systems need to change post-COVID-19 to provide a better basis to manage future pandemics. Active debate is taking place regarding initiatives such as improving prevention and early care; strengthening health, public health and social services; integrating elements of global health security with universal health coverage; developing resilience in health care systems; applying equity as a basis for all health care systems; moving from reactive, short-term approaches to planning for longer-term outcomes; maximizing digital health care and information systems; rebuilding trust in governments and healthcare systems; enhancing high-value and eliminating low-value services; (Lal et al., 2021, EFPIA, 2021, Sorensen et al., 2020, Alami et al., 2021).
Risk factors for infectious diseases
In many countries, the population is getting older, and many age-related diseases such as diabetes and renal disease predispose people to infections (Muller et al., 2005, Wang et al., 2011). Treatments for cancer lead to immunosuppression and increased susceptibility to infection, as do immunosenescence and frailty in elderly people (American Cancer Society, 2020, Sadighi Akha, 2018). The COVID-19 pandemic has illustrated the high risk of viral transmission in elderly care facilities (Barnett et al., 2020, Graham et al., 2020). Moreover, immunosenescence leads to a poorer immune response to vaccination, meaning that vaccines are often less effective in the elderly (Crooke et al., 2019); at present, it is unknown whether this will impact upon effectiveness of COVID-19 vaccines. Obesity has become a serious health care concern, even in low- and middle-income countries, and is associated with increased risk for diseases such as cancer, diabetes and cardiovascular disease, as well as with a weaker immune system. Numerous studies have shown that obese patients with COVID-19 have a higher risk of severe disease outcomes and death, and the highest death rates have been seen in countries with the highest proportion of overweight individuals (Public Health England, 2020, World Obesity Federation, 2020, Cai et al., 2021, Gao et al., 2021, Wise, 2021).
Laboratory and research capacity
The genome of the SARS-CoV-2 virus was fully sequenced within a few weeks of identification of the first patients with unidentified pneumonia in China (Zhu et al., 2020). The Global Initiative on Sharing All Influenza Data (GISAID) was launched in 2008 with the aim of sharing influenza genomic data, and the initiative played an important role in the response to the 2009 influenza A/H1N1 pandemic and now to the COVID-19 pandemic (GISAID, 2021, Shu and McCauley, 2017). As of March 2021, nearly one million SARS-CoV-2 genomic sequences have been made available via the GISAID platform. Genomic epidemiology combines genomics data with epidemiological investigations and is able to answer key questions about virus outbreaks more quickly than traditional epidemiological case tracking (Grubaugh et al., 2019, Rockett et al., 2020).
Global research networks can be used to assess pandemic characteristics (Simonsen et al., 2018). For example, data from two cohort studies of influenza conducted by the International Network for Strategic Initiatives in Global HIV Trials (INSIGHT) have been used to estimate the case fatality rate during the 2009 A/H1N1 influenza pandemic (Simonsen et al., 2018). Although this was a retrospective analysis, the same methodology could be used to assess an emerging pandemic (Simonsen et al., 2018). The COVID-19 pandemic has illustrated how rapidly research can be shared, with extensive use of online journal publications and pre-print uploads.
The Coalition for Epidemic Preparedness Innovations (CEPI) was founded in 2017 as a public–private partnership to develop vaccines for future epidemics and enable equitable access to vaccines during outbreaks (CEPI, 2020a). During the COVID-19 pandemic, CEPI has formed the COVAX collaboration with the WHO and Gavi, the Vaccine Alliance; it aims to produce 2 billion SARS-CoV-2 vaccine doses for distribution in 2021 and is providing funding for the development of 11 vaccine candidates (CEPI, 2020b).
Global governance
The International Health Regulations (IHR) 2005 are an international legal agreement between 196 countries, including all WHO Member States, whose implementation is coordinated by the WHO (WHO, 2016). The WHO also coordinates the Global Influenza Programme (GIP) with the aim of providing strategic guidance, technical support and coordination of activities to prepare health systems for seasonal, zoonotic and pandemic influenza threats (WHO, 2020c). Activities include surveillance and monitoring via the Global Influenza Surveillance and Response System (GISRS) and platforms such as FluNet and FluID125. It also includes the Pandemic Influenza Preparedness Framework which came into effect in 2011 and whose goals are to improve sharing of influenza viruses with human pandemic potential and to increase access of developing countries to vaccines and other pandemic-related supplies (WHO, 2020d). Under this framework, an advance supply contract with manufacturers, research institutes and other bodies ensures that the WHO receives supplies of vaccines and other products needed to respond to a pandemic.
A 2011 review of the IHR and the response to the 2009 A/H1N1 pandemic made three summary conclusions: (1) The IHR improved preparedness for public health emergencies, but capacities called for in the IHR were not fully operational nor on a path towards timely implementation; (2) The WHO performed well in many ways, confronted systemic difficulties and demonstrated some shortcomings; (3) The world is ill-prepared for a severe influenza pandemic or similar event (WHO, 2011). The review made a total of 15 recommendations, including to accelerate implementation of core capacities, reinforce evidence-based decisions on international travel and trade, develop and apply measures to assess severity, create a more extensive public health reserve workforce, reach agreement on sharing of viruses and access to vaccines and pursue a comprehensive influenza research programme (WHO, 2011).
Infodemics and fake news
Misinformation about COVID-19 has spread widely, particularly on, but not limited to, social media. Common misinformation has included erroneous health advice, such as self-injection with bleach and use of hydroxychloroquine, as well as conspiracy theories such as bioengineering of the SARS-CoV-2 virus and the role of 5G networks in spreading the virus (Callisher et al., 2020, World Health Organization 2021c). Polls in the UK and US have indicated that almost half of the population have been exposed to misinformation about COVID-19 (Ofcom, 2020, Mitchell & Oliphant, 2020) and several studies have identified a high proportion of false or inaccurate information related to COVID-19 in social media content (Islam et al., 2020, Islam et al., 2021, Marwah et al., 2021) Furthermore, exposure to misinformation appears to be associated with the likelihood of rejecting public health guidelines and vaccine hesitancy (Uscinski et al., 2020, Freeman et al., 2020, Romer and Hall Jamieson, 2021, Romer and Hall Jamieson, 2020). At the same time, the number of scientific papers has proliferated. Unfortunately, the quality of some of the papers is uncertain and the rate of retractions is higher than other related research topics (Tentolouris et al., 2021, Yeo-Teh & Tang, 2021).
Development of pandemic vaccines
The 2009 A/H1N1 pandemic, in which a vaccine did not become available until after the peak of the pandemic had passed, illustrates the problems associated with traditionally delivered influenza vaccines in the pandemic setting (WHO, 2012). A major problem is the difficulty in predicting which virus will cause the next pandemic and the use of vaccines based on influenza virus cultures in eggs. This method of manufacture cannot be upscaled quickly, in contrast to the adenovirus and RNA-based vaccines that have been developed against SARS-CoV-2. Because it was widely believed that the next pandemic would be caused by an influenza virus, significant effort has been invested in development of 'ready to go' pre-pandemic influenza vaccines (Nicholson et al., 2019). As of October 2020, the WHO had identified 41 vaccine candidates against influenza A/H5 viruses, 22 candidates against influenza A/H7 viruses and eight candidates against influenza A/H9N2 virus (WHO, 2020e).
However, it is not sustainable to continuously update multiple candidate vaccines (Marston et al., 2017, Nicholson et al., 2019). Therefore, the focus in more recent years has changed towards development of vaccine platforms; the theory behind this approach is that any platform can be used to present any immunogen from any pathogen (Marston et al., 2017). In addition, for influenza vaccines, substantial work has been done towards development of a universal vaccine capable of eliciting a broad immune response against antigenically diverse influenza viruses (Ostrowsky et al., 2020). It is hoped that a similar approach can be adopted towards vaccines against non-influenza pathogens (Cassone and Rappuoli, 2010).
Vaccine platforms
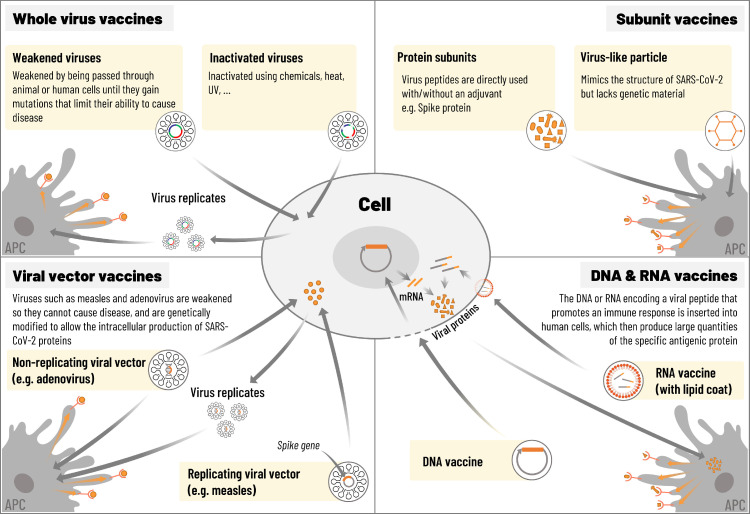
Several vaccine platforms are available or under development (Figure 2 ). Whole virus vaccines include inactivated virus vaccines and live attenuated virus vaccines. Inactivated vaccines are widely used in licensed vaccines such as influenza, polio and hepatitis A (Jeyanathan et al., 2020). Production infrastructure for inactivated vaccines is well established and there are few safety concerns associated with this type of vaccine (Jeyanathan et al., 2020). However, more than one dose is often needed to elicit a robust immune response or an adjuvant might be required (Jeyanathan et al., 2020). An adjuvant is a vaccine component that modifies or enhances the antigen-specific immune response (Di Pasquale et al., 2015). Using an adjuvant allows the dose of antigen to be decreased (antigen-sparing) or the number of doses to be reduced (Cohet et al., 2019, Di Pasquale et al., 2015).
Figure 2.
Platforms for COVID-19 vaccines. Adapted from Callaway, 2020 and Jeyanathan et al., 2020.
Several live attenuated vaccines are also available, including oral polio vaccine, influenza vaccine, measles, mumps and rubella vaccine (MMR), and varicella (chickenpox) vaccine (Jeyanathan et al., 2020, Krammer, 2020, Strugnell et al., 2011). Before use of a live attenuated vaccine, it must be clearly shown that the virus cannot revert genetically to become pathogenic (Jeyanathan et al., 2020).
Viral vector platforms include, among others, adenoviruses, alphaviruses, vesicular stomatitis virus (VSV) and Modified Vaccinia Virus Ankara (MVA, poxvirus) (Rajão and Pérez, 2018, Sebastian and Lambe, 2018). Replicating or non-replicating viral vector vaccines use a viral backbone that is genetically engineered to express antigens from the target virus (Callaway, 2020, Jeyanathan et al., 2020). Replicating viral vector vaccines use a weakened viral backbone such as measles; although they can still replicate within cells, they are unable to cause disease (Callaway, 2020, Jeyanathan et al., 2020).
In non-replicating viral vector vaccines, key replication genes in the backbone are disabled (Callaway, 2020, Jeyanathan et al., 2020). The first adenovirus vaccine vectors were based on human adenoviruses such as HAdV-5 (Colloca et al., 2012). However, a large proportion of adults have pre-existing neutralising antibodies to human adenoviruses (Barouch et al., 2011). High seroprevalence of neutralising antibodies against the vector was largely blamed for the failure of a human adenovirus-based HIV vaccine and the finding that study participants with high titres of anti-adenovirus antibodies were more susceptible to HIV infection than those without such antibodies (Buchbinder et al., 2008, McElrath et al., 2008, Sekaly, 2008). This problem has since been addressed by development of vectors based on viruses to which humans are naive, including chimpanzee adenoviruses (Farina et al., 2001). Licensed viral vector vaccines are available against Ebola, dengue fever and Japanese encephalitis, and the technology for large-scale production already exists (Jeyanathan et al., 2020, Sebastian and Lambe, 2018).
Protein-based subunit vaccines use an isolated protein or protein fragment from the pathogen (Vartak and Sucheck, 2016). They show a strong immunogenicity when administered with an adjuvant, as well as safety in immunocompromised individuals (Vartak and Sucheck, 2016). Virus-like particles (VLPs) comprise empty virus shells that contain no genetic material. Both subunit and VLP technologies are well established, but the vaccines can be poorly immunogenic and might require repeated administration or use of an adjuvant (Callaway, 2020, Jeyanathan et al., 2020).
Nucleic acid vaccines (mRNA or DNA) use only genetic material from the target pathogen, inserted into human cells which then produce copies of the virus protein encoded by the genetic material, leading to acquired immunity (Callaway, 2020, Jeyanathan et al., 2020). RNA vaccines use an RNA-containing vector, such as lipid nanoparticles, while DNA vaccines use a genetically engineered plasmid to deliver the genetic sequence to the cells. The vaccines are non-infectious, with advantages over other platforms in terms of safety, and are also easy to produce and scale up (Callaway, 2020, Jeyanathan et al., 2020).
Development of vaccines against human coronaviruses
Immunological considerations
Studies have shown that SARS-CoV-1, SARS-CoV-2 and MERS-CoV efficiently suppress activation of the innate immune system, which might explain the long pre-symptomatic period observed with SARS-CoV-2 infection (Zhou et al., 2020). Suppression of the innate immune system is likely associated with the dysregulated inflammatory responses observed in severe cases of COVID-19.
The spike protein is the main target for neutralising antibodies, which makes it an essential antigen for vaccine development (Buchholz et al., 2004, Du et al., 2013). Antibodies that bind to the S1 receptor binding domain of SARS-CoV-1 block its interaction with ACE2, while antibodies binding with other regions of S1 inhibit conformational changes of the S protein (Coughlin et al., 2007). Antibodies against the spike protein of SARS-CoV-1 have been shown to be cross-neutralising and to inhibit entry of SARS-CoV-2 into host cells (Walls et al., 2020). Neutralising antibodies to SARS-CoV-1 have remained detectable in patients recovering from natural infection for up to 17 years (Anderson et al., 2020). High antibody titres against the nucleocapsid protein have also been demonstrated following natural infection with SARS-CoV-2 (To et al., 2020), suggesting that this protein could also be a useful target for vaccine development. Currently, no immune correlate of protection has been identified for any of the coronaviruses.
T-cell mediated immunity is also an important element in COVID-19 vaccine design. A study of patients recovering from COVID-19 showed that SARS-CoV-2-specific CD4+ and CD8+ T-cells were detected in 100% and 70% of patients, respectively (Grifoni et al., 2020). Most of the CD4+ T-cell response was targeted against the spike protein (27%), membrane protein (21%) and nucleocapsid protein (11%) (Grifoni et al., 2020). Specific CD4+ T-cell responses against the spike protein correlated highly with anti-spike antibody titres (Grifoni et al., 2020). Other studies have shown that T-cell activation is delayed in SARS-CoV-2 infection (as well as SARS-CoV-1 and MERS-CoV infection), particularly for CD8+ T-cells (Remy et al., 2020, Zhou et al., 2020). Further evidence suggests that patients with less severe COVID-19 illness have higher levels of CD8+ memory T-cells compared with more severe cases (Liao et al., 2020, Peng et al., 2020).
Vaccine-enhanced disease
Two types of vaccine-enhanced disease are currently recognised: antibody-dependent enhancement (ADE) and vaccine-associated enhanced respiratory disease (VAERD) (Graham, 2020). ADE is mediated by the Fc antibody portion, whereby a virus-antibody complex binds more efficiently to cells bearing an Fc receptor, facilitating virus entry to the cell (Graham, 2020). This primarily occurs when vaccination induces neutralising antibodies that are unable to effectively neutralise the virus (Graham, 2020). ADE was reported during preclinical evaluation of a SARS-CoV-1 MVA-based vaccine (Weingartl et al., 2004). VAERD was observed in young children in the 1960s during evaluation of measles and respiratory syncytial virus (RSV) whole inactivated vaccines (Graham, 2020). In the RSV trial, it was observed that a high ratio of binding antibody to neutralising antibody could result in immune complex deposition and complement activation (Graham, 2020).
In the context of COVID-19 vaccine development, the risk of vaccine-enhanced disease can be reduced by demonstrating induction of neutralising antibodies in early clinical studies, as well as protection against virus replication and disease in animal models (Graham, 2020). It is also important to use conformationally correct antigens to induce high quality antibodies and to avoid induction of TH2-biased immune responses (Graham, 2020). Finally, study of lung pathology in animal challenge studies might help to predict abnormal pathology in vaccine recipients (Graham, 2020).
Vaccine candidates against SARS-CoV-1 and MERS-CoV
Several candidate vaccines for SARS-CoV-1 and MERS-CoV were developed, but none reached commercial development, likely because there was little incentive to continue vaccine development for an outbreak that resolved within months (SARS-CoV-1) or resulted in relatively few cases (MERS-CoV) (Padron-Regalado, 2020). For both viruses, vaccines based on inactivated virus, live-attenuated virus, viral vectors, protein-based subunits, VLPs and DNA were evaluated (Padron-Regalado, 2020). The spike protein and nucleocapsid protein were the primary targets for non-whole virus vaccines (Padron-Regalado, 2020).
Vaccine candidates against SARS-CoV-2
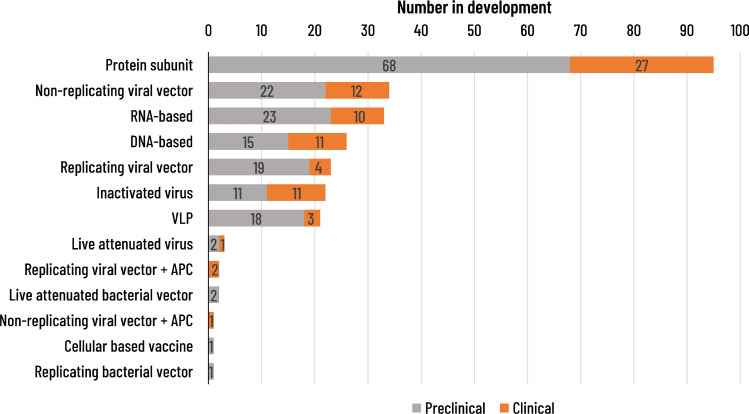
As of 22 March 2021, the WHO listed 82 COVID-19 candidate vaccines in clinical development (Table 2 ) and 182 in preclinical development (WHO, 2021d). The candidate vaccines use a wide range of vaccine platforms (Figure 3 ). Vaccines based on mRNA, adenovirus vector and inactivated virus technologies have advanced most rapidly. The Pfizer/BioNTech vaccine (BNT162b2) is a lipid nanoparticle-formulated, nucleoside-modified RNA vaccine encoding the full-length spike protein (Polack et al., 2020). A phase 2/3 study demonstrated 95.0% (95% CI: 90.0–97.9) vaccine efficacy in preventing COVID-19 disease after two vaccine doses; notably, the cumulative incidence of cases in the vaccine and placebo groups began to diverge by 12 days after the first vaccine dose (Polack et al., 2020). The vaccine was well tolerated (Polack et al., 2020). Another lipid nanoparticle-formulated mRNA vaccine encoding the spike protein (mRNA-1273; Moderna) has reported 94.5% (95% CI: 86.5–97.8) vaccine efficacy against COVID-19 disease after two doses and raised no safety concerns during a phase 3 clinical trial (Baden et al., 2021). A phase 1/2 study of the NVX-CoV2373 vaccine, a nanoparticle vaccine adjuvanted with Matrix-M1 developed by Novavax, has demonstrated a high immune response and good safety profile (Keech et al., 2020). Phase 3 data have shown 89.7% vaccine efficacy against symptomatic disease (Heath et al., 2021).
Table 2.
COVID-19 vaccine candidates in clinical development (WHO draft landscape on 22 March 2021) (World Health Organization, 2020b)
| Type of candidate vaccine | Developer | Dose and route of administration | Phase 1 | Phase 1/2 | Phase 2 | Phase 2/3 | Phase 3 |
|---|---|---|---|---|---|---|---|
| Whole virus vaccines | |||||||
| Inactivated | Sinovac | D0-14, IM | |||||
| Inactivated | Wuhan Institute of Biological Products; Sinopharm; China National Biotech Group | D0-21, IM | |||||
| Inactivated (BBIBP-CorV) | Beijing Institute of Biological Products; Sinopharm; China National Biotech Group | D0-21, IM | |||||
| Inactivated (Vero cells) | Institute of Medical Biology, Chinese Academy of Medical Sciences | D0-28, IM | |||||
| Inactivated QazCovid-in | Research Institute for Biological Safety Problems, Republic of Kazakhstan | D0-21, IM | |||||
| Whole-virion inactivated | Bharat Biotech | D0-14, IM | |||||
| Inactivated SARS-CoV-2 vaccine (Vero cell) | Beijing Minhai Biotechnology Co. | Not disclosed, IM | |||||
| COVI-VAC, live attenuated virus | Codagenix/Serum Institute of India | D0 or D0-28, IN | |||||
| VLA2001 | Valneva, National Institute for Health Research, UK | D0-21, IM | |||||
| ERUCOV-VAC, inactivated virus | Erciyes University | D0-21, IM | |||||
| Inactivated | Shifa Pharmed Industrial Co | D0-14, IM | |||||
| FAKHRAVAC (MIVAC) | Organization of Defensive Innovation and Research | D0-14 or D0-14-21, IM | |||||
| Viral vector vaccines | |||||||
| ChAdOx1-S | University of Oxford; AstraZeneca | D0-28, IM | |||||
| Adenovirus Type 5 Vector | CanSino Biological Inc; Beijing Institute of Biotechnology | IM | |||||
| Adeno-based (rAd26-S+rAd5-S) | Gamaleya Research Institute; Heath Ministry Russian Federation | D0-21, IM | |||||
| Ad26COVS1 | Janssen Pharmaceutical Companies | D0 or D0-56, IM | |||||
| DelNS1-2019-nCoV-RBD-OPT1 (intranasal influenza-based RBD) | University of Hong Kong; Beijing Wantai Biological Pharmacy; Xiamen University | D0, IN | |||||
| MVA-SARS-2-S | Ludwig-Maximilians - University of Munich | D0-28, IM | |||||
| Replication defective Simian adenovirus (GRAd) encoding S | ReiThera; LEUKOCARE; Univercells | D0, IM | |||||
| Ad5 adjuvanted oral vaccine platform | Vaxart | D0-28, oral | |||||
| Measles-vector based | Merck & Co; Institute Pasteur; Themis; University of Pittsburgh | D0-28, IM | |||||
| Covid-19/aAPC vaccine | Shenzhen Geno-Immune Medical Institute | D0-14-28, SC | |||||
| LV-SMENP-DC vaccine | Shenzhen Geno-Immune Medical Institute | D0, SC & IV | |||||
| hAd5-S-Fusion+N-ETSD vaccine | ImmunityBio, Inc. & NantKwest, Inc. | D0, SC or Oral | |||||
| COH04S1 (MVA-SARS-2-S) – Modified vaccinia ankara (sMVA) platform + synthetic SARS-CoV-2 | City of Hope Medical Center + National Cancer Institute | D0-28, IM | |||||
| rVSV-SARS-CoV-2-S Vaccine | Israel Institute for Biological Research | D0, IM | |||||
| Dendritic cell vaccine AV-COVID-19 | Aivita Biomedical, Inc.National Institute of Health Research and Development, Ministry of Health, Republic of Indonesia | D0, IM | |||||
| AdCLD-CoV19 (adenovirus vector) | Cellid Co., Ltd. | D0, IM | |||||
| AdCOVID (adenovirus vector) | Altimmune, Inc. | D0, IN | |||||
| NDV-HXP-S (Newcastle disease virus vector) | Mahidol University; Government Pharmaceutical Organization; Icahn School of Medicine at Mount Sinai | D0-28, IM | |||||
| BBV153 (adenovirus vector) | Bharat Biotech International Ltd | D0, IN | |||||
| Spike or spike plus T-cell epitopes (chimpanzee adenovirus) | Gritstone Oncology | D0-14-28 or D0-28-56 or D0-112, IM | |||||
| mRNA vaccines | |||||||
| mRNA-1273 | Moderna; NIAID | D0-28, IM | |||||
| BNT162 (3 LNP-mRNAs) | Pfizer; BioNTech; Fosun Pharma | D0-28, IM | |||||
| CVnCoV vaccine | Curevac AG | D0-28, IM | |||||
| ARCT-021 | Arcturus Therapeutics | Not disclosed | |||||
| ARCoV | Academy of Military Sciences; Walvax Biotechnology; Suzhou Abogen Biosciences | D0-14 or D0-28, IM | |||||
| LNP-nCoVsaRNA | Imperial College London | D0, IM | |||||
| ChulaCov19 mRNA vaccine | Chulalongkorn University | D0-21, IM | |||||
| m-RNA-1273.351 (B.1.351 variant) | Moderna; NIAID | D0 or D0-28 or D56 | |||||
| PTX-COVID19-B | Providence Therapeutics | D0-28, IM | |||||
| CoV2 self-amplifying RNA LNP | GlaxoSmithKline | D0-28, IM | |||||
| DNA vaccines | |||||||
| DNA plasmid vaccine with electroporation | Inovio Pharmaceuticals; International Vaccine Institute; Advaccine (Suzhou) Biopharmaceutical Co Ltd | D0-28, ID | |||||
| DNA plasmid vaccine | Zydud Cadila | D0-28-56, ID | |||||
| DNA vaccine (GX-19) | Genexine Consortium | D0-28, IM | |||||
| AG0301-COVID19 | AnGes; Takara Bio; Osaka University | D0-14, IM | |||||
| Covigenix VAX-001 + PLV | Entos Pharmaceuticals Inc. | D0-14, IM | |||||
| CORVax – spike protein plasmid DNA vaccine | Providence Health & Services | D0-14, ID | |||||
| bacTRL-Spike oral DNA vaccine | Symvivo Corporation | D0, Oral | |||||
| GLS-5310 | GeneOne Life Science, Inc. | D0-56 or D0-84, ID | |||||
| COVID-eVax | Takis; Rottapharm Biotech | IM or IM + electroporation | |||||
| COVIGEN | University of Sydney; Bionet Co Ltd; Technovalia | D0-28, IM or ID | |||||
| Protein-based vaccines | |||||||
| Full length recombinant SARS CoV-2 glycoprotein nanoparticle vaccine adjuvanted with Matrix M | Novavax | D0-21, IM | |||||
| Recombinant vaccine (CHO cell) | Anhui Zhifei Longcom Biopharmaceutical; Institute of Microbiology, Chinese Academy of Sciences | D0-28 or D0-28-56, IM | |||||
| KBP-COVID-19 (RBD-based protein subunit) | Kentucky Bioprocessing, Inc | D0-21, IM | |||||
| Protein subunit RBD (baculovirus production expressed in Sf9 cells) | West China Hospital, Sichuan University | D0-28, IM | |||||
| S protein subunit (baculovirus production) + AS03 | Sanofi Pasteur; GSK | D0-21, IM | |||||
| SCB-2019 + CpG1018 adjuvant plus alum (native-like trimeric subunit spike protein vaccine) | Clover Biopharmaceuticals Inc; Dynavax | D0-21, IM | |||||
| Recombinant spike protein plus adjuvant | Vaxine Pty Ltd; Medytox | D0, IM | |||||
| MF59 adjuvanted SARS-CoV-2 Sclamp vaccine | University of Queensland; CSL; Seqirus | D0-28, IM | |||||
| MCV-COV1901 (S-2P protein + CpG 1018) | Medigen Vaccine Biologics Corporation; NIAID; Dynavax | D0-28, IM | |||||
| FINLEY-FR1 (RBD + adjuvant) | Instituto Finlay de Vacunas, Cuba | D0-28, IM | |||||
| FINLEY-FR2 (RBD chemically conjugated to tetanus toxoid + adjuvant) | Instituto Finlay de Vacunas, Cuba | D0-28, IM | |||||
| Peptide | Federal Budgetary Research Institution State Research Center of Virology and Biotechnology 'Vector' | D0-21, IM | |||||
| IMP CoVac1 (SARS-CoV-2 HLA-DR peptides) | University Hospital Tuebingen | D0, SC | |||||
| UB612 (multitope peptide-based S1-RBD-protein) | COVAXX; United Biomedical Inc Asia | D0-28, IM | |||||
| AdimrSC-2f (recombinant RBD +/- aluminium) | Adimmune Corporation | Not disclosed | |||||
| CIGB-669 (RBD+AgnHB) | Center for Genetic Engineering and Biotechnology (CIGB) | D0-14-28 or D0-28-56, IN | |||||
| CIGB-66 (RBD+aluminium hydroxide) | Center for Genetic Engineering and Biotechnology (CIGB) | D0-14-28 or D0-28-56, IM | |||||
| BECOV2 | Biological ELimited | D0-28, IM | |||||
| Recombinant SARS-CoV-2 Spike protein, Aluminum adjuvanted | Nanogen Pharmaceutical Biotechnology | D0-21, IM | |||||
| Recombinant protein vaccine S-268019 (baculovirus expression vector system) | Shionogi | D0-21, IM | |||||
| SARS-CoV-2-RBD-Fc fusion protein | University Medical Center Groningen + Akston Biosciences Inc. | SC or IM | |||||
| COVAC-1 and COVAC-2 sub-unit vaccine (spike protein) + SWE adjuvant | University of Saskatchewan | D0-28, IM | |||||
| EuCorVac-19 | POP Biotechnologies; EuBiologics Co Ltd | D0-21, IM | |||||
| SK SARS-CoV-2 recombinant surface antigen (NBP2001) + alum adjuvant | SK Bioscience Co Ltd | D0-28, IM | |||||
| SpFN (spike ferritin nanoparticle) + QS21 adjuvant | Walter Reed Army Institute of Research | D0-28-180, IM | |||||
| Razi Cov Pars | Razi Vaccine and Serum Research Institute | D0-21-51, IM and IN | |||||
| GBP510 recombinant surface protein vaccine + AS03 | SK Bioscience Co Ltd; CEPI; GSK | D0-28, IM | |||||
| VLP-based vaccines | |||||||
| RBD-HBsAg VLPs | SpyBiotech; Serum Institute of India; Accelagen Pty | D0-28, IM | |||||
| Plant-derived VLP adjuvanted with GSK or Dynavax adjuvants | Medicago Inc.; Dynavax; GSK | D0-21, IM | |||||
| VBI-2902a | VBI Vaccines Inc | D0-28, IM | |||||
aAPC: artificial antigen presenting cells; CHO: Chinese hamster ovary; D: Day of vaccination; DC: dendritic cell; HLA: human leukocyte antigen; ID: intradermal; IM: intramuscular; IN: intranasal; LNP: lipid nanoparticle; LV: lentivirus; MVA: Modified Vaccinia Virus Ankara; PLV: proteo lipid vehicle; RBD: receptor binding domain; sa: self-activating; SC: subcutaneous; VLP: virus-like particle; VSV: vesicular stomatitis virus; WHO: World Health Organization
The landscape of candidate vaccines is changing constantly. For the latest WHO updates, refer to (https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines).
Figure 3.
COVID-19 vaccine candidates in clinical development according to vaccine platform (WHO draft landscape on 22 March 2021, 2021b). VLP: virus-like particle; WHO: World Health Organization.
The University of Oxford/AstraZeneca vaccine (ChAdOx1 nCoV-19; AZD-1222) is based on a chimpanzee adenovirus vector that was developed some years ago (Dicks et al., 2012) and contains a codon-optimised full-length spike protein (S1 and S2 subunits) of SARS-CoV-2 (van Doremalen et al., 2020). A pooled analysis of safety from four phase 1/2/3 studies showed that the vaccine is well tolerated (Voysey et al., 2021). Recently, however, concerns over rare reports of coagulation disorders with unexpected deaths have been expressed by some European countries. Efficacy against COVID-19 disease was evaluated in two phase 2/3 studies (Voysey et al., 2021). In one study, the first vaccination was given at a lower dose than planned to some participants and efficacy data have therefore been presented for two study groups: low dose at first vaccination followed by standard dose at the second vaccination (LD/SD) and both vaccinations given at the standard dose (SD/SD). Vaccine efficacy was 90.0% (95% CI: 67.4–97.0) in the LD/SD group and 62.1% (95% CI: 41.0–75.7) in the SD/SD group (Voysey et al., 2021). The mechanism for the higher efficacy in the group who received a lower priming dose is as yet unknown.
The Gam-COVID-Vac combined vector vaccine, Sputnik V, developed by the Russian Gamaleya Center, uses two adenovirus vectors (Ad5 and Ad26) containing the spike protein (Logunov et al., 2021). A phase 3 study reported vaccine efficacy of 91.6% after two vaccine doses, with no moderate or severe cases of COVID-19 reported in the vaccine group (Logunov et al., 2021). A phase 2 study of another adenovirus vector vaccine, Ad26.COV2.S developed by Janssen, has reported a good safety profile and high immune response (Sadoff et al., 2021a). A phase 3 trial demonstrated that a single dose offered 66% protection against moderate to severe disease and 85% against severe disease (Sadoff et al., 2021b).
Inactivated vaccines created from Vero cells inoculated with SARS-CoV-2 are being developed by the Chinese companies, Sinovac and Sinopharm (Xia et al., 2021, Zhang et al., 2021). A phase 2 trial demonstrated that the Sinovac vaccine (CoronaVac) at a dose of 3 µg was well tolerated and resulted in up to 97% seroconversion for neutralising antibodies (Zhang et al., 2021). Interim data from a phase 3 trial in Turkey demonstrated vaccine efficacy of 83.5% against symptomatic COVID-19 (Tanriover et al., 2021). In a phase 2 trial, the Sinopharm vaccine (BBIBP-CorV) was well tolerated and a 4 µg dose administered in a 2-dose schedule (days 0 and 21 or days 0 and 28) resulted in higher neutralising antibody titres than a 4 µg 2-dose schedule at days 0 and 14 or a single administration of a 8 µg dose (Xia et al., 2021). Phase 3 trials of both vaccines are ongoing.
According to Our World in Data, as of 6 September 2021, 40.6% of the world's population and 1.9% of people in low-income countries had received at least one dose of a COVID-19 vaccine (Mathieu et al., 2021). The proportion of the population who had been fully vaccinated was 28% globally, 49% in Europe (59% in the European Union), 43% in North America, 32% in South America, 29% in Asia and 3% in Africa (Mathieu et al., 2021). These values illustrate sharply the global inequity in access to COVID-19 vaccines, which is allowing spread of the virus to continue in unvaccinated populations and increasing the risk of emergence of new variants (Padma, 2021).
Universal influenza vaccines
The HA and neuraminidase (NA) glycoproteins are the major surface proteins of the influenza virus. Current influenza vaccines are designed to induce neutralising antibodies against HA, which consists of a membrane-distal globular head domain and a membrane-proximal stalk domain (Kaminski and Lee, 2011). The head domain is immunodominant, meaning that the immune response is mainly directed towards this part of the glycoprotein (Angeletti et al., 2017). The head domain has high plasticity and undergoes constant antigenic drift (Heaton et al., 2013, Kirkpatrick et al., 2018). This means that antibodies against the head domain are highly strain-specific and become ineffective when the virus mutates. The aim for a universal influenza virus vaccine is to elicit a robust and durable immune response to all influenza virus strains and subtypes.
The Universal Influenza Vaccine Technology Landscape is a publicly available database of universal influenza vaccine technologies that have reached clinical or late pre-clinical development (CIDRAP, 2020, Ostrowsky et al., 2020). It includes 22 vaccine candidates in clinical development and 74 in late pre-clinical development, utilising a variety of vaccine platforms (CIDRAP, 2020, Ostrowsky et al., 2020). Target antigens are aimed at inducing neutralising antibody responses, cross-reactive T-cell responses or a combination of both. Targets for neutralising antibodies include the stalk domain of the HA glycoprotein, the NA glycoprotein and the matrix 2 membrane protein (M2). T-cell targets are mainly focused on internal proteins such as nucleoprotein, matrix protein (M1) and nonstructural proteins (CIDRAP, 2020, Ostrowsky et al., 2020).
Conclusions
For pandemic use, an ideal platform would allow progress within a few weeks or months from viral sequencing to clinical trials, demonstrate consistent immune responses with different pathogens and be suitable for large-scale manufacture regardless of the pathogen included in the vaccine (Lurie et al., 2020). It has been suggested that pandemic vaccines can be developed using a new pandemic paradigm with a rapid start and the usual sequential steps done in parallel (Lurie et al., 2020). The COVID-19 pandemic has illustrated the feasibility of this approach and the benefits of a globally coordinated response and infrastructure (Ball, 2021). Adequate financial support and a system for fair allocation of vaccine is essential for this approach to be successful (Ball, 2021, Lurie et al., 2020).
The current pandemic has also demonstrated these new vaccines can be developed at high speed without compromising safety. Yet, the development of mRNA and viral vector COVID-19 vaccines benefited from years of research on these technologies (e.g., Ebola vaccine) and on other coronaviruses (SARS-CoV-1 and MERS-CoV)(Ball, 2021). Some of the COVID-19 vaccines recently developed or currently in development might offer some flexibility or sufficient broad protection to swiftly respond to antigenic drift or emergence of new coronaviruses. Conventional vaccines have several advantages such as long experience of safety, cost and breadth of immune response with adjuvants. However, vaccines based on cultured virus face problems with rapid upscaling of production; future pandemic vaccines will most probably be fully synthetic, as we are witnessing with the new SARS-CoV-2 vaccines.
This wide vaccine technology approach will be best employed in tandem with active surveillance for emerging variants or new pathogens using antigen mapping, metagenomics and next generation sequencing. New generations of vaccines will certainly be developed (Lavine et al., 2021). In addition, pandemic preparedness needs to take account of challenges such as the large-scale production of sufficient quantity of vaccines, delivery of vaccines to all countries and ensuring vaccination of relevant age groups. Large harmonised networks of vaccination registries, together with robust infrastructure, are needed to ensure that people of all ages can be vaccinated during inter-pandemic periods. A plain language summary of our observations is presented in Supplementary Figure 1.
Conflict of Interest
Philippe Buchy, Otavio Cintra, Michael Nissen are employed by the GSK Group of Companies also have shares in the GSK Group of Companies. They declare financial and non-financial conflict of interest. Dominic E Dwyer, Michael Nissen, Raul Ortiz de Lejarazu, Yves Buisson and Eskild Petersen have nothing to disclose.
Funding
This paper was funded by GlaxoSmithKline Biologicals SA. GlaxoSmithKline Biologicals SA was involved in all stages of the conduct and analysis of the study and covered the costs associated with the development and the publishing of the present manuscript.
Ethical approval
NA.
Contributorship
All authors contributed to study conception, interpretation of the data, and critical review of the paper for important intellectual content. All authors are in agreement with the content of the final article and have approved it for submission and publication.
Acknowledgements
Authors thank Business and Decisions Life Science for editorial assistance and manuscript coordination, on behalf of GSK. Mary Greenacre provided medical writing support, Pierre-Paul Prévot coordinated the manuscript development and provided editorial support.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ijid.2021.09.045.
Appendix. Supplementary materials
References
- Abdelrahman Z, Li M, Wang X. Comparative review of SARS-CoV-2, SARS-CoV, MERS-CoV, and influenza A respiratory viruses. Frontiers in Immunology. 2020;11 doi: 10.3389/fimmu.2020.552909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajao A, Nystrom SV, Koonin LM, et al. Assessing the capacity of the US health care system to use additional mechanical ventilators during a large-scale public health emergency. Disaster Med Public Health Prep. 2015;9:634–641. doi: 10.1017/dmp.2015.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alami H, Lehoux P, Fleet R, et al. How can health systems better prepare for the next pandemic? lessons learned from the management of COVID-19 in Quebec (Canada) Front Public Health. 2021 doi: 10.3389/fpubh.2021.671833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali SO, Takas T, Nyborg A, et al. Evaluation of MEDI8852, an anti-influenza A monoclonal antibody, in treating acute uncomplicated influenza. Antimicrob Agents Chemother. 2018;62:e00694–e00718. doi: 10.1128/aac.00694-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almond D. Is the 1918 influenza pandemic over? Long-term effects of in utero influenza exposure in the post-1940 US population. J Polit Econ. 2006;114:672–712. [Google Scholar]
- Alouane T, Laamarti M, Essabbar A, et al. Genomic diversity and hotspot mutations in 30,983 SARS-CoV-2 genomes: moving toward a universal vaccine for the “confined virus”? Pathogens. 2020;9:829. doi: 10.3390/pathogens9100829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Tawfiq JA, Auwaerter PG. Healthcare-associated infections: the hallmark of Middle East respiratory syndrome coronavirus with review of the literature. J Hosp Infect. 2019;101:20–29. doi: 10.1016/j.jhin.2018.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Cancer Society. Why people with cancer are more likely to get infections. 2020. Available at: https://www.cancer.org/treatment/treatments-and-side-effects/physical-side-effects/low-blood-counts/infections/why-people-with-cancer-are-at-risk.html (accessed December 2020).
- Anderson BW. Liberty Press; 1979. Economics and the public welfare: a financial and economic history of the United States, 1914-1946; pp. 61–69. [Google Scholar]
- Anderson DE, Tan CW, Chia WN, et al. Lack of cross-neutralization by SARS patient sera towards SARS-CoV-2. Emerg Microbes Infect. 2020;9:900–902. doi: 10.1080/22221751.2020.1761267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angeletti D, Gibbs JS, Angel M, et al. Defining B cell immunodominance to viruses. Nat Immunol. 2017;18:456–463. doi: 10.1038/ni.3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anon. Janssen drops pimodivir development program in flu. 2020. Available at: https://www.thepharmaletter.com/article/janssen-drops-pimodivir-development-program-in-flu (accessed December 2020).
- Ansart S, Pelat C, Boelle PY, et al. Mortality burden of the 1918–1919 influenza pandemic in Europe. Influenza Other Respir Viruses. 2009;3:99–106. doi: 10.1111/j.1750-2659.2009.00080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arce JS, Warren SS, Meriggi NF, et al. COVID-19 vaccine acceptance and hesitancy in low- and middle-income countries. Nat Med. 2021;27:1385–1394. doi: 10.1038/s41591-021-01454-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacay Watson V. Five coronavirus success stories: different, but the same. Security Nexus, Daniel K Inouye Asia-Pacific Center for Security Studies. 2020 https://www.jstor.org/stable/pdf/resrep24880.pdf Available at. (accessed October 2020) [Google Scholar]
- Baden LR, El Sahly HM, Essink B, et al. Efficacy ans safety of the mRNA SARS-CoV2 vaccine. N Engl J Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball P. The lightning-fast quest for COVID vaccines – and what it means for other diseases. Nature. 2021;589:16–18. doi: 10.1038/d41586-020-03626-1. [DOI] [PubMed] [Google Scholar]
- Barnett ML, Hu L, Martin T, et al. Mortality, admissions, and patient census at SNFs in 3 US cities during the COVID-19 pandemic. JAMA. 2020;324:507–509. doi: 10.1001/jama.2020.11642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barouch DH, Kik SV, Weverling GJ, et al. International seroepidemiology of adenovirus serotypes 5, 26, 35, and 48 in pediatric and adult populations. Vaccine. 2011;29:5203–5209. doi: 10.1016/j.vaccine.2011.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barro R, Ursua J, Weng J. The coronavirus and the great influenza epidemic: lessons from the “Spanish Flu” for the coronavirus’ potential effects on mortality and economic activity. American Enterprise Institute. 2020 [Google Scholar]
- Barry JM. Penguin Group; New York: 2005. The great influenza: the epic story of the deadliest plague in history. [Google Scholar]
- Beaney T, Clarke JM, Jain V, et al. Excess mortality: the gold standard in measuring the impact of COVID-19 worldwide? J Royal Soc Med. 2020;113:329–334. doi: 10.1177/0141076820956802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford J, Farrar J, Ihekweazu C, et al. A new twenty-first century science for effective epidemic response. Nature. 2019;575:130–136. doi: 10.1038/s41586-019-1717-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beigel JH, Tomashek KM, Dodd LE, et al. Remdesivir for the treatment of Covid-19 – final report. N Engl J Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belser JA, Wadford DA, Pappas C, et al. Pathogenesis of pandemic influenza A (H1N1) and triple-reassortant swine influenza A (H1) viruses in mice. J Virol. 2010;84:4194–4203. doi: 10.1128/jvi.02742-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhimraj A, Morgan R, Hirsch Shumaker A, et al. Infectious Diseases Society of America guidelines on the treatment and management of patients with COVID-19. Infectious Diseases Society of America. 2021 doi: 10.1093/cid/ciaa478. https://www.idsociety.org/practice-guideline/covid-19-guideline-treatment-and-management/ version 4.4.1. 2021. Available from: (accessed September 2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breban R, Riou J, Fontanet A. Interhuman transmissibility of Middle East respiratory syndrome coronavirus: estimation of pandemic risk. Lancet. 2013;382:694–699. doi: 10.1016/s0140-6736(13)61492-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brundage JF. Interactions between influenza and bacterial respiratory pathogens: implications for pandemic preparedness. Lancet Infect Dis. 2006;6:303–312. doi: 10.1016/s1473-3099(06)70466-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchbinder SP, Mehrotra DV, Duerr A, et al. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet. 2008;372:1881–1893. doi: 10.1016/s0140-6736(08)61591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz UJ, Bukreyev A, Yang L, et al. Contributions of the structural proteins of severe acute respiratory syndrome coronavirus to protective immunity. Proc Natl Acad Sci USA. 2004;101:9804–9809. doi: 10.1073/pnas.0403492101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchy P, Ascioglu S, Buisson Y, et al. Impact of vaccines on antimicrobial resistance. Int J Infect Dis. 2020;90:188–196. doi: 10.1016/j.ijid.2019.10.005. [DOI] [PubMed] [Google Scholar]
- Byerly CR. The U.S. military and the influenza pandemic of 1918–1919. Public Health Rep. 2010;125(Suppl 3):82–91. [PMC free article] [PubMed] [Google Scholar]
- Cai Z, Yang Y, Zhang J. Obesity is associated with severe disease and mortality in patients with coronavirus disease 2019 (COVID-19): a meta-analysis. BMC Public Health. 2021;21:1505. doi: 10.1186/s12889-021-11546-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callisher C, Carroll D, Codwell R, et al. Statement in support of the scientists, public health professionals, and medical professionals of China in combatting COVID-19. Lancet. 2020;395:e42–e43. doi: 10.1016/S0140-6736(20)30418-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway E. The race for coronavirus vaccines: a graphical guide. Nature. 2020;580:576–577. doi: 10.1038/d41586-020-01221-y. [DOI] [PubMed] [Google Scholar]
- Carlson CJ. From PREDICT to prevention, one pandemic later. Lancet Microbe. 2020;1:e6–e7. doi: 10.1016/s2666-5247(20)30002-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll D, Daszak P, Wolfe ND, et al. The Global Virome Project. Science. 2018;359:872–874. doi: 10.1126/science.aap7463. [DOI] [PubMed] [Google Scholar]
- Cassone A, Rappuoli R. Universal vaccines: shifting to one for many. mBio. 2010;1:e00042–e000110. doi: 10.1128/mBio.00042-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Center for Infectious Disease Research and Policy (CIDRAP). Universal influenza vaccine technology landscape. 2020. Available at: https://www.cidrap.umn.edu/universal-influenza-vaccine-technology-landscape (acessed October 2020).
- Centers for Disease Control and Prevention Bacterial coinfections in lung tissue specimens from fatal cases of 2009 pandemic influenza A (H1N1) – United States, May–August 2009. MMWR Morb Mortal Wkly Rep. 2009;58:1071–1074. [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention Interim results: state-specific influenza A (H1N1) 2009 monovalent vaccination coverage – United States, October 2009–January 2010. MMWR Morb Mortal Wkly Rep. 2010;59:363–368. [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. Past pandemics. 2018a. Available at: https://www.cdc.gov/flu/pandemic-resources/basics/past-pandemics.html (accessed December 2020).
- Centers for Disease Control and Prevention. One Health Basics. 2018b. Available at: https://www.cdc.gov/onehealth/basics/index.html (accessed October 2020).
- Centers for Disease Control and Prevention. Influenza historic timeline. 2019a. Available at: https://www.cdc.gov/flu/pandemic-resources/pandemic-timeline-1930-and-beyond.htm (accessed December 2020).
- Centers for Disease Control and Prevention. 1968 pandemic (H2N2 virus). 2019b. Available at: https://www.cdc.gov/flu/pandemic-resources/1968-pandemic.html (accessed December 2020).
- Centers for Disease Control and Prevention. Influenza antiviral drug resistance. 2020. Available at: https://www.cdc.gov/flu/treatment/antiviralresistance.htm (accessed October 2020).
- Centers for Disease Control and Prevention. Influenza vaccination information for health care workers. 2021. Available at: https://www.cdc.gov/flu/professionals/healthcareworkers.htm?CDC_AA_refVal=https%3A%2F%2Fwwwcdcgov%2Fflu%2Fhealthcareworkers.htm (accessed March 2021).
- Chalmers JD, Crichton ML, Goeminne PC, et al. Management of hospitalised adults with coronavirus disease 2019 (COVID-19): a European Respiratory Society living guideline. Eur Respir J. 2021;57(4) doi: 10.1183/13993003.00048-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan M. World now at the start of 2009 influenza pandemic. 2009. Available at: https://www.who.int/mediacentre/news/statements/2009/h1n1_pandemic_phase6_20090611/en/ (accessed September 2020).
- Charu V, Chowell G, Palacio Mejia LS, et al. Mortality burden of the A/H1N1 pandemic in Mexico: a comparison of deaths and years of life lost to seasonal influenza. Clin Infect Dis. 2011;53:985–993. doi: 10.1093/cid/cir644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng PK, Wong DA, Tong LK, et al. Viral shedding patterns of coronavirus in patients with probable severe acute respiratory syndrome. Lancet. 2004;363:1699–1700. doi: 10.1016/s0140-6736(04)16255-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chertow DS, Memoli MJ. Bacterial coinfection in influenza: a grand rounds review. JAMA. 2013;309:275–282. doi: 10.1001/jama.2012.194139. [DOI] [PubMed] [Google Scholar]
- Chowell G, Echevarría-Zuno S, Viboud C, et al. Characterizing the epidemiology of the 2009 influenza A/H1N1 pandemic in Mexico. PLoS Med. 2011;8 doi: 10.1371/journal.pmed.1000436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CEPI (Coalition for Epidemic Preparedness Innovations). New vaccines for a safer world. 2020a. Available at: https://cepi.net (accessed October 2020).
- CEPI (Coalition for Epidemic Preparedness Innovations). How COVAX will work. 2020b. Available at: https://cepi.net/COVAX/ (accessed October 2020).
- Cockburn WC, Delon PJ, Ferreira W. Origin and progress of the 1968–69 Hong Kong influenza epidemic. Bull World Health Organ. 1969;41:345–348. [PMC free article] [PubMed] [Google Scholar]
- COCONEL Group A future vaccination campaign against COVID-19 at risk of vaccine hesitancy and politicisation. Lancet Infect Dis. 2020;20:769–770. doi: 10.1016/s1473-3099(20)30426-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohet C, van der Most R, Bauchau V, et al. Safety of AS03-adjuvanted influenza vaccines: a review of the evidence. Vaccine. 2019;37:3006–3021. doi: 10.1016/j.vaccine.2019.04.048. [DOI] [PubMed] [Google Scholar]
- Colloca S, Barnes E, Folgori A, et al. Vaccine vectors derived from a large collection of simian adenoviruses induce potent cellular immunity across multiple species. Sci Transl Med. 2012;4::115ra112. doi: 10.1126/scitranslmed.3002925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coughlin M, Lou G, Martinez O, et al. Generation and characterization of human monoclonal neutralizing antibodies with distinct binding and sequence features against SARS coronavirus using XenoMouse. Virology. 2007;361:93–102. doi: 10.1016/j.virol.2006.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooke SN, Ovsyannikova IG, Poland GA, et al. Immunosenescence and human vaccine immune responses. Immun Ageing. 2019;16:25. doi: 10.1186/s12979-019-0164-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CS Mott Children's Hospital. Mott Poll Report. Flu vaccine for children in the time of COVID. 2020. Available at: https://mottpoll.org/reports/flu-vaccine-children-time-covid (accessed October 2020).
- Dawood FS, Jain S, Finelli L, et al. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med. 2009;360:2605–2615. doi: 10.1056/NEJMoa0903810. [DOI] [PubMed] [Google Scholar]
- Dawood FS, Iuliano AD, Reed C, et al. Estimated global mortality associated with the first 12 months of 2009 pandemic influenza A H1N1 virus circulation: a modelling study. Lancet Infect Dis. 2012;12:687–695. doi: 10.1016/s1473-3099(12)70121-4. [DOI] [PubMed] [Google Scholar]
- Day M. Covid-19: four fifths of cases are asymptomatic, China figures indicate. BMJ. 2020;369:m1375. doi: 10.1136/bmj.m1375. [DOI] [PubMed] [Google Scholar]
- Di Pasquale A, Preiss S, Tavares Da Silva F, et al. Vaccine adjuvants: from 1920 to 2015 and beyond. Vaccines. 2015;3:320–343. doi: 10.3390/vaccines3020320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dicks MD, Spencer AJ, Edwards NJ, et al. A novel chimpanzee adenovirus vector with low human seroprevalence: improved systems for vector derivation and comparative immunogenicity. PloS One. 2012;7:e40385. doi: 10.1371/journal.pone.0040385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingues CM, de Oliveira WK. Uptake of pandemic influenza (H1N1)-2009 vaccines in Brazil, 2010. Vaccine. 2012;30:4744–4751. doi: 10.1016/j.vaccine.2012.05.007. [DOI] [PubMed] [Google Scholar]
- Du L, Zhao G, Kou Z, et al. Identification of a receptor-binding domain in the S protein of the novel human coronavirus Middle East respiratory syndrome coronavirus as an essential target for vaccine development. J Virol. 2013;87:9939–9942. doi: 10.1128/jvi.01048-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrheim DN, Gostin LO, Moodley K. When does a major outbreak become a public health emergency of international concern? Lancet Infect Dis. 2020;20:887–889. doi: 10.1016/s1473-3099(20)30401-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Federation of Pharmaceutical Industries and Associations (EFPIA). Health systems after COVID-19. A perspective on the future of European health systems. 2021. Available at: https://www.google.com/url?sa=t&rct=j&q=&esrc=s&source=web&cd=&cad=rja&uact=8&ved=2ahUKEwjQ-tPdj-3yAhUNHcAKHY0BCgQQFnoECCAQAQ&url=https%3A%2F%2Fwww.efpia.eu%2Fmedia%2F602847%2Fhealth-systems-after-covid-19_en.pdf&usg=AOvVaw374s2OdtBAx9zStoyh-ZSL (accessed September 2021).
- Farina SF, Gao GP, Xiang ZQ, et al. Replication-defective vector based on a chimpanzee adenovirus. J Virol. 2001;75:11603–11613. doi: 10.1128/jvi.75.23.11603-11613.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust JS, Del Rio C. Assessment of deaths from COVID-19 and from seasonal influenza. JAMA Intern Med. 2020;180:1045–1046. doi: 10.1001/jamainternmed.2020.2306. [DOI] [PubMed] [Google Scholar]
- Fisher KA, Bloomstone SJ, Walder J, et al. Attitudes toward a potential SARS-CoV-2 vaccine: a survey of U.S. adults. Ann Intern Med. 2020;173:964–973. doi: 10.7326/m20-3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forni D, Cagliani R, Clerici M, et al. Molecular evolution of human coronavirus genomes. Trends Microbiol. 2017;25:35–48. doi: 10.1016/j.tim.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman D, Waite F, Rosebrock L, et al. Coronavirus conspiracy beliefs, mistrust, and compliance with government guidelines in England. Psychol Med. 2020:1–13. doi: 10.1017/S0033291720001890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French J, Deshpande S, Evans W, et al. Key guidelines in developing a pre-emptive COVID-19 vaccination uptake promotion strategy. Int J Environ Res Public Health. 2020;17:5893. doi: 10.3390/ijerph17165893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Piernas C, Astbury NM, et al. Associations between body-mass index and COVID-19 severity in 6.9 million people in England: a prospective, community-based, cohort study. Lancet Diabetes Endocrinol. 2021;9:350–359. doi: 10.1016/S2213-8587(21)00089-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibb R, Redding DW, Chin KQ, et al. Zoonotic host diversity increases in human-dominated ecosystems. Nature. 2020;584:398–402. doi: 10.1038/s41586-020-2562-8. [DOI] [PubMed] [Google Scholar]
- Gill JR, Sheng ZM, Ely SF, et al. Pulmonary pathologic findings of fatal 2009 pandemic influenza A/H1N1 viral infections. Arch Pathol Lab Med. 2010;134:235–243. doi: 10.1043/1543-2165-134.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour H, Hofmann N. H1N1 vaccination. Health Rep. 2010;21:63–69. [PubMed] [Google Scholar]
- GISAID. 2021. Available at: https://www.gisaid.org (accessed March 2021).
- Global Virome Project. Preparing for the next pandemic. 2020. Available at: www.globalviromeproject.org (accessed October 2020).
- Graham BS. Rapid COVID-19 vaccine development. Science. 2020;368:945–946. doi: 10.1126/science.abb8923. [DOI] [PubMed] [Google Scholar]
- Graham NSN, Junghans C, Downes R, et al. SARS-CoV-2 infection, clinical features and outcome of COVID-19 in United Kingdom nursing homes. J Infect. 2020;81:411–419. doi: 10.1016/j.jinf.2020.05.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant J. Simon and Schuster; 2014. The forgotten depression: 1921, the crash that cured itself. [Google Scholar]
- Greaney A, Loes A, Crawford K, et al. Comprehensive mapping of mutations to the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human serum antibodies. bioRxiv: the preprint server for biology 2021. doi: 10.1101/2020.12.31.425021. [DOI] [PMC free article] [PubMed]
- Greenberg SB. Update on human rhinovirus and coronavirus infections. Semin Respir Crit Care Med. 2016;37:555–571. doi: 10.1055/s-0036-1584797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregg MB, Hinman AR, Craven RB. The Russian flu. Its history and implications for this year's influenza season. JAMA. 1978;240:2260–2263. doi: 10.1001/jama.240.21.2260. [DOI] [PubMed] [Google Scholar]
- Grifoni A, Weiskopf D, Ramirez SI, et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubaugh ND, Ladner JT, Lemey P, et al. Tracking virus outbreaks in the twenty-first century. Nat Microbiol. 2019;4:10–19. doi: 10.1038/s41564-018-0296-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock K, Veguilla V, Lu X, et al. Cross-reactive antibody responses to the 2009 pandemic H1N1 influenza virus. N Engl J Med. 2009;361:1945–1952. doi: 10.1056/NEJMoa0906453. [DOI] [PubMed] [Google Scholar]
- Haug N, Geyrhofer L, Londei A, et al. Ranking the effectiveness of worldwide COVID-19 government interventions. Nat Hum Behav. 2020;4:1303–1312. doi: 10.1038/s41562-020-01009-0. [DOI] [PubMed] [Google Scholar]
- Haviari S, Bénet T, Saadatian-Elahi M, et al. Vaccination of healthcare workers: a review. Hum Vaccin Immunother. 2015;11:2522–2537. doi: 10.1080/21645515.2015.1082014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden FG, Sugaya N, Hirotsu N, et al. Baloxavir marboxil for uncomplicated influenza in adults and adolescents. N Engl J Med. 2018;379:913–923. doi: 10.1056/NEJMoa1716197. [DOI] [PubMed] [Google Scholar]
- Heath PT, Galiza EP, Baxter DN, et al. Safety and efficacy of NVX-CoV2373 Covid-19 vaccine. N Engl J Med. 2021 doi: 10.1056/NEJMoa2107659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton NS, Sachs D, Chen CJ, et al. Genome-wide mutagenesis of influenza virus reveals unique plasticity of the hemagglutinin and NS1 proteins. Proc Natl Acad Sci USA. 2013;110:20248–20253. doi: 10.1073/pnas.1320524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson DA, Courtney B, Inglesby TV, et al. Public health and medical responses to the 1957–58 influenza pandemic. Biosecur Bioterror. 2009;7:265–273. doi: 10.1089/bsp.2009.0729. [DOI] [PubMed] [Google Scholar]
- Hershberger E, Sloan S, Narayan K, et al. Safety and efficacy of monoclonal antibody VIS410 in adults with uncomplicated influenza A infection: results from a randomized, double-blind, phase-2, placebo-controlled study. EBioMedicine. 2019;40:574–582. doi: 10.1016/j.ebiom.2018.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honigsbaum M. Revisiting the 1957 and 1968 influenza pandemics. Lancet. 2020;395:1824–1826. doi: 10.1016/s0140-6736(20)31201-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horby P, Lim WS, Emberson JR, et al. Dexamethasone in hospitalized patients with Covid-19 – preliminary report. N Engl J Med. 2020 doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Ge X, Wang LF, et al. Bat origin of human coronaviruses. Virol J. 2015;12:221. doi: 10.1186/s12985-015-0422-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikematsu H, Hayden FG, Kawaguchi K, et al. Baloxavir marboxil for prophylaxis against influenza in household contacts. N Engl J Med. 2020;383:309–320. doi: 10.1056/NEJMoa1915341. [DOI] [PubMed] [Google Scholar]
- Islam MS, Sarkar T, Khan SH, et al. COVID-19-related infodemic and its impact on public health: a global social media analysis. Am J Trop Med Hyg. 2020;103:1621–1629. doi: 10.4269/ajtmh.20-0812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam MS, Mostofa Kamal A-H, Kabir A, et al. COVID-19 vaccine rumors and conspiracy theories: The need for cognitive inoculation against misinformation to improve vaccine adherence. PLoS One. 2021;16 doi: 10.1371/journal.pone.0251605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ison MG, Portsmouth S, Yoshida Y, et al. Early treatment with baloxavir marboxil in high-risk adolescent and adult outpatients with uncomplicated influenza (CAPSTONE-2): a randomised, placebo-controlled, phase 3 trial. Lancet Infect Dis. 2020;20:1204–1214. doi: 10.1016/s1473-3099(20)30004-9. [DOI] [PubMed] [Google Scholar]
- James S, Sargent T. Working paper 2004–2007. 2006. The economic impact of an influenza pandemic.http://dsp-psd.pwgsc.gc.ca/collection_2008/fin/F21-8-2007-4E.pdf Available at: (accessed September 2020) [Google Scholar]
- Jefferson T, Jones MA, Doshi P, et al. Neuraminidase inhibitors for preventing and treating influenza in healthy adults and children. Cochrane Database Syst Rev. 2014;2014 doi: 10.1002/14651858.CD008965.pub4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen KE, Dunn FL, Robinson RQ. Influenza, 1957: a variant and the pandemic. Prog Med Virol. 1958;1:165–209. [PubMed] [Google Scholar]
- Jeyanathan M, Afkhami S, Smaill F, et al. Immunological considerations for COVID-19 vaccine strategies. Nat Rev Immunol. 2020;20:615–632. doi: 10.1038/s41577-020-00434-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhung MA, Swerdlow D, Olsen SJ, et al. Epidemiology of 2009 pandemic influenza A (H1N1) in the United States. Clin Infect Dis. 2011;52(Suppl 1):S13–S26. doi: 10.1093/cid/ciq008. [DOI] [PubMed] [Google Scholar]
- Johnson NP, Mueller J. Updating the accounts: global mortality of the 1918–1920 "Spanish" influenza pandemic. Bull Hist Med. 2002;76:105–115. doi: 10.1353/bhm.2002.0022. [DOI] [PubMed] [Google Scholar]
- Jones KE, Patel NG, Levy MA, et al. Global trends in emerging infectious diseases. Nature. 2008;451:990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan EO. American Medical Association; Chicago: 1927. Epidemic influenza: a survey. [Google Scholar]
- Kaminski DA, Lee FE. Antibodies against conserved antigens provide opportunities for reform in influenza vaccine design. Front Immunol. 2011;2:76. doi: 10.3389/fimmu.2011.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlinsky A, Kobak D. Tracking excess mortality across countries during the COVID-119 pandemic with the World Mortality Dataset. eLife. 2021;10:e69336. doi: 10.7554/eLife.69336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keech C, Albert G, Cho I, et al. Phase 1-2 trial of a SARS-CoV-2 recombinant spike protein nanoparticle vaccine. N Engl J Med. 2020;383:2320–2332. doi: 10.1056/NEJMoa2026920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr JR, Schneider CR, Recchia G, et al. Correlates of intended COVID-19 vaccine acceptance across time and countries: results from a series of cross-sectional surveys. BMJ Open. 2021;11 doi: 10.1136/bmjopen-2020-048025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilbourne ED. Influenza: viral determinants of the pathogenicity and epidemicity of an invariant disease of variable occurrence. Philos Trans R Soc Lond B Biol Sci. 1980;288:291–297. doi: 10.1098/rstb.1980.0004. [DOI] [PubMed] [Google Scholar]
- Kilbourne ED. Influenza pandemics of the 20th century. Emerg Infect Dis. 2006;12:9–14. doi: 10.3201/eid1201.051254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick E, Qiu X, Wilson PC, et al. The influenza virus hemagglutinin head evolves faster than the stalk domain. Sci Rep. 2018;8:10432. doi: 10.1038/s41598-018-28706-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586:516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- Kupferschmidt K. New coronavirus variants could cause more reinfections, require updated vaccines. Science. 2021 doi: 10.1126/science.abg6028. [DOI] [Google Scholar]
- Lal A, Erondu NE, Heymann DL, Gitahi G, Yates R. Fragmented health systems in COVID-19: rectifying the misalignment between global health security and universal health coverage. Lancet. 2021;397:61–67. doi: 10.1016/S0140-6736(20)32228-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langford BJ, So M, Raybardhan S, et al. Bacterial co-infection and secondary infection in patients with COVID-19: a living rapid review and meta-analysis. Clin Microbiol Infect. 2020;26:1622–1629. doi: 10.1016/j.cmi.2020.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauring AS, Hodcroft EB. Genetic variants of SARS-CoV-2 – what do they mean? JAMA. 2021 doi: 10.1001/jama.2020.27124. [DOI] [PubMed] [Google Scholar]
- Lavine JS, Bjornstad ON, Antia R. Immunological characteristics govern the transition of COVID-19 to endemicity. Science. 2021:eabe6522. doi: 10.1126/science.abe6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Li W, He X, et al. Asymptomatic and presymptomatic infectors: hidden sources of COVID-19 disease. Clin Infect Dis. 2020;71:2018. doi: 10.1093/cid/ciaa418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Shi Z, Yu M, et al. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Li Y, Yu IT, Xu P, et al. Predicting super spreading events during the 2003 severe acute respiratory syndrome epidemics in Hong Kong and Singapore. Am J Epidemiol. 2004;160:719–728. doi: 10.1093/aje/kwh273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao M, Liu Y, Yuan J, et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- Linka K, Peirlinck M, Kuhl E. The reproduction number of COVID-19 and its correlation with public health interventions. medRxiv: the preprint server for health sciences 2020. doi: 10.1101/2020.05.01.20088047. [DOI] [PMC free article] [PubMed]
- Lipsitch M, Cohen T, Cooper B, et al. Transmission dynamics and control of severe acute respiratory syndrome. Science. 2003;300:1966–1970. doi: 10.1126/science.1086616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logunov DY, Dolzhikova IV, Shcheblyakov DV, et al. Safety and efficacy of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine: an interim analysis of a randomised controlled phase 3 trial in Russia. Lancet. 2021 doi: 10.1016/s0140-6736(21)00234-8. S0140–6736(21)00234-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louie JK, Acosta M, Jamieson DJ, et al. Severe 2009 H1N1 influenza in pregnant and postpartum women in California. N Engl J Med. 2010;362:27–35. doi: 10.1056/NEJMoa0910444. [DOI] [PubMed] [Google Scholar]
- Luke TC, Kilbane EM, Jackson JL, et al. Meta-analysis: convalescent blood products for Spanish influenza pneumonia: a future H5N1 treatment? Ann Intern Med. 2006;145:599–609. doi: 10.7326/0003-4819-145-8-200610170-00139. [DOI] [PubMed] [Google Scholar]
- Lurie N, Saville M, Hatchett R, et al. Developing Covid-19 vaccines at pandemic speed. N Engl J Med. 2020;382:1969–1973. doi: 10.1056/NEJMp2005630. [DOI] [PubMed] [Google Scholar]
- Madhav N, Oppenheim B, Gallivan M, et al. In: Disease control priorities: improving health and reducing poverty. Jamison DT, Gelband H, Horton S, Jha P, Laxminarayan R, Mock CN, Nugent R, editors. The International Bank for Reconstruction and Development /The World Bank; Washington (DC): 2017 Nov 27. Pandemics: risks, impacts, and mitigation. Chapter 17. [PubMed] [Google Scholar]
- Mair-Jenkins J, Saavedra-Campos M, Baillie JK, et al. The effectiveness of convalescent plasma and hyperimmune immunoglobulin for the treatment of severe acute respiratory infections of viral etiology: a systematic review and exploratory meta-analysis. J Infect Dis. 2015;211:80–90. doi: 10.1093/infdis/jiu396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallapaty S. Coronaviruses closely related to the pandemic virus discovered in Japan and Cambodia. Nature. 2020;588:15–16. doi: 10.1038/d41586-020-03217-0. [DOI] [PubMed] [Google Scholar]
- Mamelund S, Sattenspiel L, Dimka J. Influenza-associated mortality during the 1918–1919 influenza pandemic in Alaska and Labrador. Soc Sci Hist. 2013;37:177–229. doi: 10.1215/01455532-2074420. [DOI] [Google Scholar]
- Markel H, Lipman HB, Navarro JA, et al. Nonpharmaceutical interventions implemented by US cities during the 1918–1919 influenza pandemic. JAMA. 2007;298:644–654. doi: 10.1001/jama.298.6.644. [DOI] [PubMed] [Google Scholar]
- Marston HD, Paules CI, Fauci AS. The critical role of biomedical research in pandemic preparedness. JAMA. 2017;318:1757–1758. doi: 10.1001/jama.2017.15033. [DOI] [PubMed] [Google Scholar]
- Marwah HK, Carlson K, Rosseau NA, et al. Videos, views, and vaccines: evaluating the quality of COVID-19 communications on YouTube. Disaster Med Public Health Prep. 2021:1–24. doi: 10.1017/dmp.2021.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAteer J, Yildirim I, Chahroudi A. The VACCINES Act: deciphering vaccine hesitancy in the time of COVID-19. Clin Infect Dis. 2020;71:703–705. doi: 10.1093/cid/ciaa433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride JM, Lim JJ, Burgess T, et al. Phase 2 randomized trial of the safety and efficacy of MHAA4549A, a broadly neutralizing monoclonal antibody, in a human influenza A virus challenge model. Antimicrob Agents Chemother. 2017;61:e01154–e01217. doi: 10.1128/aac.01154-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElrath MJ, De Rosa SC, Moodie Z, et al. HIV-1 vaccine-induced immunity in the test-of-concept step study: a case-cohort analysis. Lancet. 2008;372:1894–1905. doi: 10.1016/s0140-6736(08)61592-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerowitz-Katz G, Merone L. A systematic review and meta-analysis of published research data on COVID-19 infection-fatality rates. Int J Infect Dis. 2020;101:138–148. doi: 10.1016/j.ijid.2020.09.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milken Institute. COVID-19 treatment and vaccine tracker. 2021. Available at: https://covid-19tracker.milkeninstitute.org/ (accessed March 2021).
- Mitchell A, Oliphant JB. Americans immersed in COVID-19 news; most think media are doing fairly well covering it. Pew Research Center. 2020 https://www.journalism.org/2020/03/18/americans-immersed-in-covid-19-news-most-think-media-are-doing-fairly-well-covering-it/ Available at: (accessed September 2021) [Google Scholar]
- Morens DM, Taubenberger JK, Fauci AS. Predominant role of bacterial pneumonia as a cause of death in pandemic influenza: implications for pandemic influenza preparedness. J Infect Dis. 2008;198:962–970. doi: 10.1086/591708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morens DM, Taubenberger JK, Harvey HA, et al. The 1918 influenza pandemic: lessons for 2009 and the future. Crit Care Med. 2010;38:e10–e20. doi: 10.1097/CCM.0b013e3181ceb25b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S. Emerging infectious diseases: epidemiological perspective. Indian J Dermatol. 2017;62(5):459–467. doi: 10.4103/ijd.IJD_379_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller LM, Gorter KJ, Hak E, et al. Increased risk of common infections in patients with type 1 and type 2 diabetes mellitus. Clin Infect Dis. 2005;41:281–288. doi: 10.1086/431587. [DOI] [PubMed] [Google Scholar]
- Murray CJ, Lopez AD, Chin B, et al. Estimation of potential global pandemic influenza mortality on the basis of vital registry data from the 1918–20 pandemic: a quantitative analysis. Lancet. 2006;368:2211–2218. doi: 10.1016/s0140-6736(06)69895-4. [DOI] [PubMed] [Google Scholar]
- National Institutes of Health. Coronavirus disease 2019 (COVID-19) treatment guidelines. 2021. Available from: https://www.covid19treatmentguidelines.nih.gov/ (accessed 31 August). Accessed September 2021. [PubMed]
- Nicholson A, Shah CM, Ogawa VA., National Academies of Sciences, Engineering, Medicine . National Academies Press (US); Washington (DC): 2019. Exploring lessons learned from a century of outbreaks: readiness for 2030: proceedings of a workshop. [PubMed] [Google Scholar]
- Nishiura H, Linton NM, Akhmetzhanov AR. Serial interval of novel coronavirus (COVID-19) infections. Int J Infect Dis. 2020;93:284–286. doi: 10.1016/j.ijid.2020.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofcom. Half of UK adults exposed to false claims about coronavirus. 2020. Available at: https://www.ofcom.org.uk/about-ofcom/latest/features-and-news/half-of-uk-adults-exposed-to-false-claims-about-coronavirus (accessed September 2021).
- Ostrowsky J, Arpey M, Moore K, et al. Tracking progress in universal influenza vaccine development. Curr Opin Virol. 2020;40:28–36. doi: 10.1016/j.coviro.2020.02.003. [DOI] [PubMed] [Google Scholar]
- Padma TV. COVID vaccines to reach poorest countries in 2023 - despite recent pledges. Nature. 2021;595:342–343. doi: 10.1038/d41586-021-01762-w. [DOI] [PubMed] [Google Scholar]
- Padron-Regalado E. Vaccines for SARS-CoV-2: lessons from other coronavirus strains. Infect Dis Ther. 2020;9:1–20. doi: 10.1007/s40121-020-00300-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palese P, Wang TT. Why do influenza virus subtypes die out? A hypothesis. mBio. 2011;2:e00150–e00211. doi: 10.1128/mBio.00150-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H, Peto R, Henao-Restrep AM, et al. Repurposed antiviral drugs for COVID-19 - interim WHO SOLIDARITY trial results. N Engl J Med. 2021;384:497–511. doi: 10.1056/NEJMoa2023184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquini-Descomps H, Brender N, Maradan D. Value for money in H1N1 influenza: a systematic review of the cost-effectiveness of pandemic interventions. Value Health. 2017;20:819–827. doi: 10.1016/j.jval.2016.05.005. [DOI] [PubMed] [Google Scholar]
- Patterson KD. Rowman & Littlefield; Lanham, MD: 1986. Pandemic influenza, 1700–1900: a study in historical epidemiology. [Google Scholar]
- Peeri NC, Shrestha N, Rahman MS, et al. The SARS, MERS and novel coronavirus (COVID-19) epidemics, the newest and biggest global health threats: what lessons have we learned? Int J Epidemiol. 2020;49:717–726. doi: 10.1093/ije/dyaa033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Mentzer AJ, Liu G, et al. Broad and strong memory CD4 (+) and CD8 (+) T cells induced by SARS-CoV-2 in UK convalescent COVID-19 patients. 2020. bioRxiv 2020 8;2020.06.05.134551. doi: 10.1101/2020.06.05.134551. [DOI] [PMC free article] [PubMed]
- Petersen E, Petrosillo N, Koopmans M. Emerging infections - an increasingly important topic: review by the Emerging Infections Task Force. Clin Microbiol Infect. 2018;24:369–375. doi: 10.1016/j.cmi.2017.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen E, Koopmans M, Go U, et al. Comparing SARS-CoV-2 with SARS-CoV and influenza pandemics. Lancet Infect Dis. 2020;20:e238–e244. doi: 10.1016/s1473-3099(20)30484-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova VN, Russell CA. The evolution of seasonal influenza viruses. Nat Rev Microbiol. 2018;16:47–60. doi: 10.1038/nrmicro.2017.118. [DOI] [PubMed] [Google Scholar]
- Piechotta V, Chai KL, Valk SJ, et al. Convalescent plasma or hyperimmune immunoglobulin for people with COVID-19: a living systematic review. Cochrane Database Syst Rev. 2020;7 doi: 10.1002/14651858.CD013600.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polack FP, Thomas SJ, Kitchin N, et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N Engl J Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Principi N, Camilloni B, Alunno A, et al. Drugs for influenza treatment: is there significant news? Front Med. 2019;6:109. doi: 10.3389/fmed.2019.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Public Health England. Excess weight and COVID-19: insights from new evidence. 2020. Available at: https://www.gov.uk/government/publications/excess-weight-and-covid-19-insights-from-new-evidence (accessed September 2021).
- Rajão DS, Pérez DR. Universal vaccines and vaccine platforms to protect against influenza viruses in humans and agriculture. Front Microbiol. 2018;9:123. doi: 10.3389/fmicb.2018.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remy KE, Mazer M, Striker DA, et al. Severe immunosuppression and not a cytokine storm characterizes COVID-19 infections. JCI Insight. 2020;5 doi: 10.1172/jci.insight.140329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rist CL, Arriola CS, Rubin C. Prioritizing zoonoses: a proposed one health tool for collaborative decision-making. PloS One. 2014;9 doi: 10.1371/journal.pone.0109986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockett RJ, Arnott A, Lam C, et al. Revealing COVID-19 transmission in Australia by SARS-CoV-2 genome sequencing and agent-based modeling. Nat Med. 2020;26:1398–1404. doi: 10.1038/s41591-020-1000-7. [DOI] [PubMed] [Google Scholar]
- Rogers K. Hong Kong flu of 1968. 2020. Available at: http://www.britannica.com/event/Hong-Kong-flu-of-1968 (accessed 25 September 2020).
- Romer D, Hall Jamieson K. Conspiracy theories as barriers to controlling the spread of COVID-19 in the US. Soc Sci med. 2020;263 doi: 10.1016/j.socscimed.2020.113356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romer D, Hall Jamieson K. Patterns of media use, strength of belief in COVID-19 conspiracy theories, and the prevention of COVID-19 From March to July 2020 in the United States: survey study. J Med Internet Res. 2021;23:e25215. doi: 10.2196/25215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadighi Akha AA. Aging and the immune system: an overview. J Immunol Methods. 2018;463:21–26. doi: 10.1016/j.jim.2018.08.005. [DOI] [PubMed] [Google Scholar]
- Sadoff J, Le Gars M, Shukarev G, et al. Interim results of a phase 1-2a trial of Ad26.COV2.S Covid-19 vaccine. N Engl J Med. 2021;384:1824–1835. doi: 10.1056/NEJMoa2034201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadoff J, Gray G, Vandebosch A, et al. Safety and efficacy of single-dose Ad26CoV2.S vaccine against Covid-19. N Engl J Med. 2021;384:2187–2201. doi: 10.1056/NEJMoa2101544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallam M. COVID-19 vaccine hesitancy worldwide: a concise systematic review of vaccine acceptance rates. Vaccines (Basel) 2021;9:160. doi: 10.3390/vaccines9020160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon DA, Dudley MZ, Glanz JM, et al. Vaccine hesitancy: causes, consequences, and a call to action. Vaccine. 2015;33(Suppl 4):D66–D71. doi: 10.1016/j.vaccine.2015.09.035. [DOI] [PubMed] [Google Scholar]
- Salyer SJ, Silver R, Simone K, et al. Prioritizing zoonoses for global health capacity building – themes from one health zoonotic disease workshops in 7 countries, 2014–2016. Emerg Infect Dis. 2017;23:S55–S64. doi: 10.3201/eid2313.170418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanlioglu F, Bilge AH. An Overview of the 2009 A(H1N1) pandemic in Europe: efficiency of the vaccination and healthcare strategies. J Healthc Eng. 2016;2016 doi: 10.1155/2016/5965836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanmarchi F, Golinelli D, Lenzi J, et al. Exploring the gap between excess mortality and COVID-19 deaths in 67 countries. JAMA Network Open. 2021;4 doi: 10.1001/jamanetworkopen.2021.17359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders-Hastings PR, Krewski D. Reviewing the history of pandemic influenza: understanding patterns of emergence and transmission. Pathogens. 2016;5:66. doi: 10.3390/pathogens5040066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian S, Lambe T. Clinical advances in viral-vectored influenza vaccines. Vaccines. 2018;6:29. doi: 10.3390/vaccines6020029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekaly RP. The failed HIV Merck vaccine study: a step back or a launching point for future vaccine development? J Exp Med. 2008;205:7–12. doi: 10.1084/jem.20072681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settele J, Diaz S, Brondizio E, et al. COVID-19 stimulus measures must save lives, protect livelihoods, and safeguard nature to reduce the risk of future pandemics. IBPES Expert Guest Article, 2020. Available at: https://ipbes.net/covid19stimulus (accessed October 2020).
- Shanks GD, Wilson N, Kippen R, et al. The unusually diverse mortality patterns in the Pacific region during the 1918–21 influenza pandemic: reflections at the pandemic's centenary. Lancet Infect Dis. 2018;18:e323–e332. doi: 10.1016/s1473-3099(18)30178-6. [DOI] [PubMed] [Google Scholar]
- Shetty P. Experts concerned about vaccination backlash. Lancet. 2010;375:970–971. doi: 10.1016/s0140-6736(10)60421-7. [DOI] [PubMed] [Google Scholar]
- Shu Y, McCauley J. GISAID: Global initiative on sharing all influenza data – from vision to reality. Euro Surveill. 2017;22:30494. doi: 10.2807/1560-7917.es.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen L, Spreeuwenberg P, Lustig R, et al. Global mortality estimates for the 2009 influenza pandemic from the GLaMOR project: a modeling study. PLoS Med. 2013;10 doi: 10.1371/journal.pmed.1001558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen L, Higgs E, Taylor RJ, et al. Using clinical research networks to assess severity of an emerging influenza pandemic. Clin Infect Dis. 2018;67:341–349. doi: 10.1093/cid/ciy088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith W, Andrewes C, Laidlaw P. A virus obtained from influenza patients. Lancet. 1933;222:66–68. [Google Scholar]
- Sorensen C, Japinga M, Crook H, McClellan M. Building a better health care system post-COVID-19: steps for reducing low-value and wasteful care. NEJM Catalyst. 2020 doi: 10.1056/CAT.20.0368. [DOI] [Google Scholar]
- Strugnell R, Zepp F, Cunningham A, et al. Vaccine antigens. Perspectives in vaccinology. 2011;1:61–68. doi: 10.1016/j.pervac.2011.05.003. [DOI] [Google Scholar]
- Su S, Wong G, Shi W, et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Comish P, Kang R. The hallmarks of COVID-19 disease. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanriover MD, Doğanay HL, Akova M, et al. Efficacy and safety of an inactivated whole-virion SARS-CoV-2 vaccine (CoronaVac): interim results of a double-blind, randomised, placebo-controlled, phase 3 trial in Turkey. Lancet. 2021;398:213–222. doi: 10.1016/S0140-6736(21)01429-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubenberger JK, Morens DM. 1918 Influenza: the mother of all pandemics. Emerg Infect Dis. 2006;12:15–22. doi: 10.3201/eid1201.050979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor PC, Adams AC, Hufford MM, de la Torre I, Winthrop K, Gottlieb RL. Neutralizing monoclonal antibodies for treatment of COVID-19. Nat Rev Immunol. 2021;21:382–393. doi: 10.1038/s41577-021-00542-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegally H, Wilkinson E, Giovanetti M, et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. 2021. Available at: https://www.medrxiv.org/content/10.1101/2020.12.21.20248640v1 (accessed February 2021).
- Tentolouris A, Ntanasis-Stathopoulos I, Vlachakis PK, Tsilimigras DI, Gavriatopoulou M, Dimopoulos MA. COVID-19: time to flatten the infodemic curve. Clin Exp Med. 2021;1–5 doi: 10.1007/s10238-020-00680-x. (epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Economist. Tracking covid-19 excess deaths across countries. 2021. Available at: https://www.economist.com/graphic-detail/coronavirus-excess-deaths-tracker (accessed September 2021).
- To KK, Tsang OT, Leung WS, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20:565–574. doi: 10.1016/s1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomazini BM, Maia IS, Cavalcanti AB, et al. Effect of dexamethasone on days alive and ventilator-free in patients with moderate or severe acute respiratory distress syndrome and COVID-19: the CoDEX randomized clinical trial. JAMA. 2020;324:1–11. doi: 10.1001/jama.2020.17021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomkins SM. The failure of expertise: public health policy in Britain during the 1918–19 influenza epidemic. Soc Hist Med. 1992;5:435–454. doi: 10.1093/shm/5.3.435. [DOI] [PubMed] [Google Scholar]
- Tu C, Crameri G, Kong X, et al. Antibodies to SARS coronavirus in civets. Emerg Infect Dis. 2004;10:2244–2248. doi: 10.3201/eid1012.040520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- USAID PREDICT. Reducing pandemic risk, promoting global health. 2020. Available at: https://www.usaid.gov/sites/default/files/documents/1864/predict-global-flyer-508.pdf (accessed October 2020).
- Uscinski JE, Enders AM, Klofstad C, et al. Why do people believe COVID-19 conspiracy theories? Harvard Kennedy School (HKS) Misinform Rev. 2020;1:1–12. doi: 10.37016/mr-2020-015. [DOI] [Google Scholar]
- van Doremalen N, Lambe T, Spencer A, et al. ChAdOx1 nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature. 2020;586:578–582. doi: 10.1038/s41586-020-2608-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartak A, Sucheck SJ. Recent advances in subunit vaccine carriers. Vaccines. 2016;4:12. doi: 10.3390/vaccines4020012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernon JR. The 1920-21 Deflation: the role of aggregate supply. Economic Inquiry. 1991;29:572–580. [Google Scholar]
- Viboud C, Grais RF, Lafont BA, et al. Multinational impact of the 1968 Hong Kong influenza pandemic: evidence for a smoldering pandemic. J Infect Dis. 2005;192:233–248. doi: 10.1086/431150. [DOI] [PubMed] [Google Scholar]
- Viboud C, Simonsen L, Fuentes R, et al. Global mortality impact of the 1957–1959 influenza pandemic. J Infect Dis. 2016;213:738–745. doi: 10.1093/infdis/jiv534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voysey M, Clemens SAC, Madhi SA, et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2021;397:99–111. doi: 10.1016/s0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls AC, Park YJ, Tortorici MA, et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HE, Gamboa C, Warnock DG, et al. Chronic kidney disease and risk of death from infection. Am J Nephrol. 2011;34:330–336. doi: 10.1159/000330673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Jing R, Lai X, et al. Acceptance of COVID-19 vaccination during the COVID-19 pandemic in China. Vaccines. 2020;8:482. doi: 10.3390/vaccines8030482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang D, Du G, et al. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet. 2020;395:1569–1578. doi: 10.1016/s0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingartl H, Czub M, Czub S, et al. Immunization with modified vaccinia virus Ankara-based recombinant vaccine against severe acute respiratory syndrome is associated with enhanced hepatitis in ferrets. J Virol. 2004;78:12672–12676. doi: 10.1128/jvi.78.22.12672-12676.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilder-Smith A, Chiew CJ, Lee VJ. Can we contain the COVID-19 outbreak with the same measures as for SARS? Lancet Infect Dis. 2020;20:e102–e107. doi: 10.1016/s1473-3099(20)30129-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wille M, Holmes EC. The ecology and evolution of influenza viruses. Cold Spring Harb Perspect Med. 2020;10 doi: 10.1101/cshperspect.a038489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise J. Covid-19: highest death rate seen in countries with most overweight populations. BMJ. 2021;372:n623. doi: 10.1136/bmj.n623. [DOI] [PubMed] [Google Scholar]
- Wölfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Hul V, Karlsson EA, Hassanin A, et al. A novel SARS-CoV-2 related coronavirus in bats from Cambodia. 2021. bioRxiv preprint doi: 10.1101/2021.01.26.428212 [DOI] [PMC free article] [PubMed]
- World Health Organization. Consensus document on the epidemiology of severe acute respiratory syndrome (SARS). 2003. Available at: https://www.who.int/csr/sars/WHOconsensus.pdf?ua=1 (accessed October 2020).
- World Health Organization. Strengthening response to pandemics and other public-health emergencies: report of the review committee on the functioning of the international health regulations (2005) and on Pandemic Influenza (H1N1) 2009. 2011. https://www.who.int/ihr/publications/RC_report/en/ (accessed October 2020).
- World Health Organization. Report of the WHO pandemic influenza A(H1N1) vaccine deployment initiative. 2012. Available at: https://apps.who.int/iris/handle/10665/44795 (accessed March 2021).
- World Health Organization. About IHR. 2016. Available at: https://www.who.int/ihr/about/en/ (accessed October 2020).
- World Health Organization. Draft thirteenth general programme of work, 2019–2023. 2018. Available at: http://apps.who.int/gb/ebwha/pdf_files/WHA71/A71_4-en.pdf?ua=1 (accessed October 2020).
- World Health Organization. Middle East respiratory syndrome coronavirus (MERS-CoV). MERS monthly summary, November 2019. Available at: https://www.who.int/emergencies/mers-cov/en/ (accessed October 2020).
- World Health Organization. Pandemic influenza. 2020a. Available at: https://www.euro.who.int/en/health-topics/communicable-diseases/influenza/pandemic-influenza (accessed December 2020).
- World Health Organization. Influenza. Global Influenza Surveillance and Response System (GISRS). 2020b. Available at: https://www.who.int/influenza/gisrs_laboratory/en/ (accessed January 2021).
- World Health Organization. Influenza. Surveillance and monitoring. 2020c. Available at: https://www.who.int/influenza/surveillance_monitoring/en/ (accessed October 2020).
- World Health Organization. Pandemic influenza preparedness (PIP) framework. 2020d. Available at: https://www.who.int/influenza/pip/en/ (accessed October 2020).
- World Health Organization. Antigenic and genetic characteristics of zoonotic influenza A viruses and development of candidate vaccine viruses for pandemic preparedness. 2020e. Available at: https://www.who.int/influenza/vaccines/virus/202009_zoonotic_vaccinevirusupdate.pdf?ua=1 (accessed October 2020).
- World Health Organization. Influenza. COVID-19 sentinel surveillance by GISRS. 2021a. Available at: https://www.who.int/influenza/gisrs_laboratory/covid19/en/ (accessed January 2021).
- World Health Organization. Therapeutics and COVID-19 Living Guideline (6 July 2021). 2021b. Available at: https://www.who.int/publications/i/item/WHO-2019-nCoV-therapeutics-2021.2 (accessed September 2021).
- World Health Organization. Coronavirus disease (COVID-19) advice for the public: mythbusters. 2021c. Available at: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/advice-for-public/myth-busters#virus (accessed September 2021).
- World Health Organization. Draft landscape of COVID-19 candidate vaccines. 2021d. Available at: https://www.who.int/who-documents-detail/draft-landscape-of-covid-19-candidate-vaccines2020 (accessed February 2021).
- World Obesity Federation. Obesity and COVID-19: policy statement. 2012. Available at: https://www.worldobesity.org/news/obesity-and-covid-19-policy-statement (accessed September 2021).
- Wu JT, Cowling BJ, Lau EH, et al. School closure and mitigation of pandemic (H1N1) 2009, Hong Kong. Emerg Infect Dis. 2010;16:538–541. doi: 10.3201/eid1603.091216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S, Zhang Y, Wang Y, et al. Safety and immunogenicity of an inactivated SARS-CoV-2 vaccine, BBIBP-CorV: a randomised, double-blind, placebo-controlled, phase 1/2 trial. Lancet Infect Dis. 2021;21:39–51. doi: 10.1016/s1473-3099(20)30831-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Zhou J, Huang Y, et al. Efficacy of convalescent plasma for the treatment of severe influenza. Crit Care. 2020;24:469. doi: 10.1186/s13054-020-03189-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo-Teh NSL, Tang BL. An alrming retraction rate for scientific publications on coronavirus disease 2019 (COVID-19) Account Res. 2021;28:47–53. doi: 10.1080/08989621.2020.1782203. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zeng G, Pan H, et al. Safety, tolerability, and immunogenicity of an inactivated SARS-CoV-2 vaccine in healthy adults aged 18–59 years: a randomised, double-blind, placebo-controlled, phase 1/2 clinical trial. Lancet Infect Dis. 2021;21:181–192. doi: 10.1016/s1473-3099(20)30843-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, To KK, Wong YC, et al. Acute SARS-CoV-2 infection impairs dendritic cell and T cell responses. Immunity. 2020;53:864–877. doi: 10.1016/j.immuni.2020.07.026. e865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.