Abstract
Rationale
Prognostic tools for aiding in the treatment of hospitalized COVID-19 patients could help improve outcome by identifying patients at higher or lower risk of severe disease. The study objective was to develop models to stratify patients by risk of severe outcomes during COVID-19 hospitalization using readily available information at hospital admission.
Methods
Hierarchical ensemble classification models were trained on a set of 229 patients hospitalized with COVID-19 to predict severe outcomes, including ICU admission, development of acute respiratory distress syndrome, or intubation, using easily attainable attributes including basic patient characteristics, vital signs at admission, and basic lab results collected at time of presentation. Each test stratifies patients into groups of increasing risk. An additional cohort of 330 patients was used for blinded, independent validation. Shapley value analysis evaluated which attributes contributed most to the models’ predictions of risk.
Main results
Test performance was assessed using precision (positive predictive value) and recall (sensitivity) of the final risk groups. All test cut-offs were fixed prior to blinded validation. In development and validation, the tests achieved precision in the lowest risk groups near or above 0.9. The proportion of patients with severe outcomes significantly increased across increasing risk groups. While the importance of attributes varied by test and patient, C-reactive protein, lactate dehydrogenase, and D-dimer were often found to be important in the assignment of risk.
Conclusions
Risk of severe outcomes for patients hospitalized with COVID-19 infection can be assessed using machine learning-based models based on attributes routinely collected at hospital admission.
Keywords: Machine learning, COVID-19, Prognostic models, Risk assessment, Clinical decision support systems
1. Introduction
The coronavirus disease (COVID-19) pandemic continues to place world-wide health systems under pressure. The ability to identify patients at greatest risk of developing severe disease would help inform discussions of the risks and benefits of treatments [1], [2], [3], target higher risk populations for clinical trials of therapeutic agents, compare outcomes between patients with similar risk, and prioritize limited resources.
Multiple prognostic factors have been identified, including age, certain comorbidities [4], [5], [6], neutrophil-to-lymphocyte ratio (NLR), D-dimer and C-reactive protein (CRP) [4], [7], [8]. However, the relative influence of these factors on prognosis has been difficult to elucidate.
While traditional statistical modelling has been used to combine multiple attributes to predict outcomes for patients with COVID-19 [9], [10], modern machine learning (ML) has added advantage in its capability to discover more complex interactions between correlated attributes. Such methods have shown promise for predicting acute respiratory distress syndrome (ARDS) in diseases other than COVID-19 [11], [12] and demonstrated potential utility for predicting poor outcomes in COVID-19 [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29].
Many patients hospitalized with COVID-19 provide limited medical history due to not engaging in routine care, not having easily accessible comorbidities data, or being intubated shortly after arrival. Hence, the goal of this study was to create and validate tests to risk stratify COVID-19 patients for progression to severe disease using only readily available clinical, demographic, and laboratory attributes collected at the point of hospital admission. Given the nature of these attributes, the tests stratifying risk of admission to the intensive care unit (ICU), intubation, or progression to ARDS could be validated, blinded to outcome, on an independent patient cohort using data drawn from electronic health records (EHRs).
2. Methods
2.1. Data extraction and patient cohorts
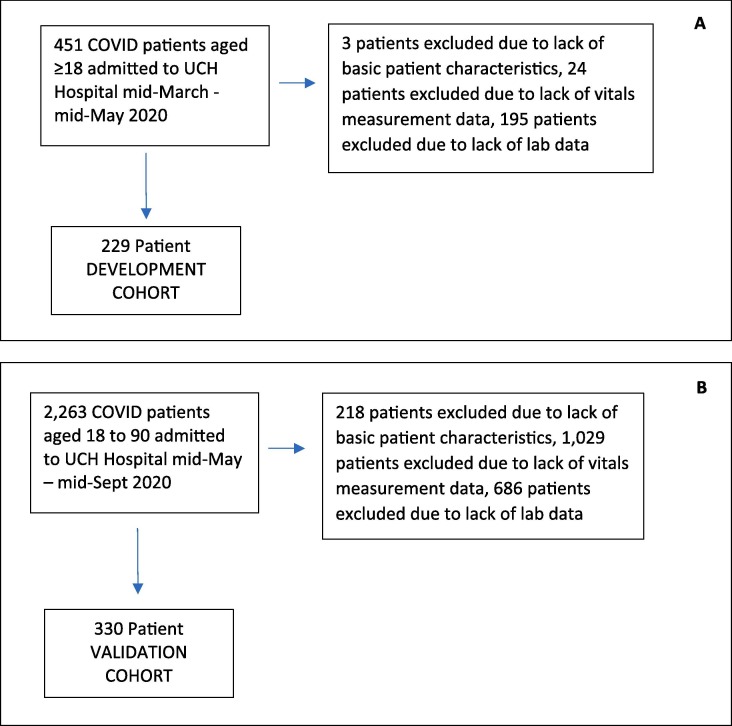
For test development, data was collected for all patients 18 years or older admitted to the University of Colorado Health (UCH) hospital in Aurora, CO with a positive COVID-19 nasopharyngeal polymerase chain reaction test between mid-March and mid-May 2020. Attributes deemed potentially useful at the time were extracted from the EHR by medical students and stored in a REDCap database [30]. Patient data was restricted to first recorded admission and observations for laboratory values and vital measures. ARDS occurrence was defined by any mention of ARDS in the critical care progress notes.
The deidentified dataset was transferred to Biodesix for test development. A set of 26 attributes (indicated with * in Table 1 ) to be used in classifier training was selected, limited to attributes routinely collected at hospital admission and available in the EHR, where inclusion of an attribute did not reduce the size of the cohort with complete data by more than 10%. The development cohort included patients with complete information for the selected attributes.
Table 1.
Patient characteristics for the development and independent validation cohorts.
| Development Cohort (N = 229) | Validation Cohort (N = 330) | ||
|---|---|---|---|
| Categorical Attribute | Class | n (%) | n (%) |
| Race* | White | 41 (17.9) | 113 (34.2) |
| Black/African American | 52 (22.7) | 23 (7.0) | |
| Hispanic/Latino | 94 (41.0) | 164 (49.7) | |
| Other | 31 (13.5) | 25 (7.6) | |
| Unknown | 11 (4.8) | 5 (1.5) | |
| Sex* | Male | 124 (54.1) | 172 (52.1) |
| Female | 105 (45.9) | 158 (47.9) | |
| eGFR* | ≥60 mL/min/1.73 m2 | 180 (78.6) | 244 (73.9) |
| 30 ≥ but < 60 mL/min/1.73 m2 | 34 (14.8) | 61 (18.5) | |
| <30 mL/min/1.73 m2 | 15 (6.6) | 25 (7.6) | |
| Hypertension | Yes | 100 (43.7) | NA |
| No | 126 (56.3) | NA | |
| Diabetes | Yes | 82 (35.8) | NA |
| No | 147 (64.2) | NA | |
| Continuous Attribute | Median (Q1-Q3) | Median (Q1-Q3) | |
| BMI**, kg/m2 | 30 (27–36) | ||
| Age*, years | 57 (43–68) | 57 (44–70) | |
| Temperature*, °C | 37 (37–38) | 37 (37–38) | |
| Heart Rate*, beats/minute | 98 (84–110) | 98 (85–110) | |
| Systolic*, mm Hg | 130 (120–150) | 130 (120–140) | |
| Diastolic BP*, mm Hg | 74 (65–83) | 74 (65–84) | |
| Respiratory Rate*, breaths/minute | 20 (18–24) | 20 (18–24) | |
| Oxygen Saturation*, % | 92 (87–94) | 92 (88–95) | |
| Weight*, kg | 82 (72–100) | 85 (73–100) | |
| QTc*, | 440 (420–460) | 440 (420–470) | |
| Sodium*, mmol/L | 140 (130–140) | 140 (130–140) | |
| Potassium*, mmol/L | 3.8 (3.4–4.0) | 3.9 (3.6–4.2) | |
| Carbon dioxide* mmol/L | 23 (21–25) | 23 (21–25) | |
| BUN* mg/dL | 13 (10–20) | 15 (10–22) | |
| Creatinine* mg/dL | 0.94 (0.69–1.2) | 0.87 (0.72–1.2) | |
| Anion Gap*, mmol/L | 12 (10–13) | 11 (9–13) | |
| WBC Count* ×109 cells/L | 6.8 (5.3–8.9) | 7 (5.5–9.1) | |
| Hemoglobin* g/dL | 15 (13–16) | 14 (13–15) | |
| Hematocrit*, % | 44 (40–47) | 42 (38–46) | |
| Platelet Count* ×109 cells/L | 210 (160–260) | 200 (160–260) | |
| LDH* U/L | 320 (260–420) | 450 (290–750) | |
| D-Dimer* ng/mL | 860 (530–1500) | 740 (410–1500) | |
| CRP*, mg/L | 83 (41–150) | 79 (37–160) | |
| Ferritin*, ng/mL | 360 (170–730) | 310 (130–600) | |
Definition of abbreviations: eGFR = estimated glomerular filtration rate; BP = blood pressure; QTc = corrected QT interval; BUN = blood urea nitrogen; CO2 = carbon dioxide; WBC = white blood cell; LDH = lactate dehydrogenase; CRP = C-reactive protein; BMI = body mass index.
Used for classification.
Only complete for 205 out 229 patients.
An independent validation cohort was derived from data provided by UCH’s EHR data warehouse service for blinded validation of the tests. The validation cohort included all patients aged between 18 and 90 years, hospitalized in any UCH system facility with a positive COVID-19 test or diagnosis, with first admission between mid-May 2020 and mid-September 2020 with complete data on the 26 required attributes (Fig. 1 ). Deidentified data for the validation cohort were transferred to Biodesix for blinded test classification generation. Outcome data for test performance evaluation for the validation cohort was only shared after test classifications were returned to UCH investigators. ARDS occurrence was identified by SNOMED code. More details on data curation and validation are provided in the supplement. The study was reviewed and approved by the Colorado Multiple Institutional Review Board.
Fig. 1.
Consort diagrams of patient selection down to development (A) and validation (B) cohorts.
2.2. Test development
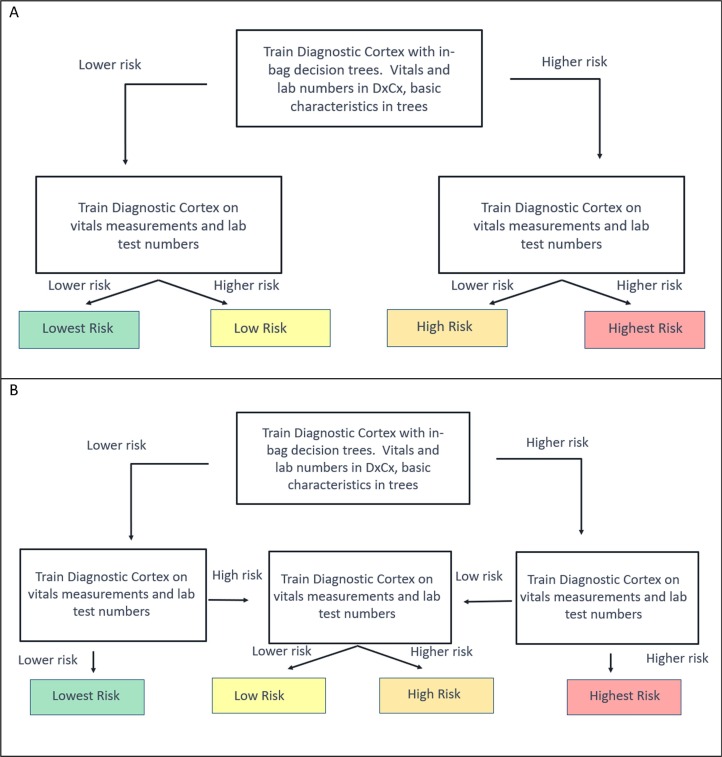
A series of classifiers were trained to classify patients as higher or lower risk for each endpoint (ICU admission, intubation, ARDS) using the same set of 26 attributes that were all used in all final tests to predict risk of outcome. These classifiers were arranged in a hierarchy shown in Fig. 2 to give a final classification of up to four possible risk groups. The fundamental model behind each component classifier was the Diagnostic Cortex® platform, a strongly dropout-regularized, feature abstracted, ensemble logistic regression [31]. The classifier at the top of each test’s hierarchy was augmented with an additional decision tree model. Hyperparameters for the models were picked on heuristic considerations as in [31] and are given in Table E2, and further optimization was not performed to avoid overfitting. Bootstrap validation results in the development cohort were obtained using out-of-bag estimates [32]. ML methods are described fully in the supplement, and source code demonstrating their application here is provided at [33].
Fig. 2.
Hierarchical Configuration of Classifiers used for Risk Assessment for Each Endpoint. A Diagnostic Cortex model with in-bag decision tree model (represented by the top box) was used to stratify the entire development cohort into a higher and lower risk group for each endpoint. Diagnostic Cortex models (middle boxes) without trees were used to split the resulting two groups further according to one of the two schemas. (Schema A was used for the tests predicting risk of any complication and intubation. Schema B was used for the tests predicting risk of ARDS and admission to the ICU.)
Relative attribute importance for individual patient classification was evaluated using Shapley Values [34], [35] for 50 patients in the validation cohort chosen to span different test classification groups and is described in detail in the supplement.
2.3. Statistical methods
The validation cohort was analyzed following a prespecified statistical analysis plan, included in the supplement, using SAS Enterprise Guide 8.2 (SAS 9.4) (SAS Institute, Cary, NC). Performance is presented in terms of positive-predictive-value (PPV), specificity, and F1 scores [36], [37], [38]. Cochran-Armitage test and Fisher’s exact test were used to assess trends and differences in proportions. Confidence intervals (CIs) for proportions were calculated using the Clopper-Pearson or bootstrap methods.
3. Results
Patients included in the development cohort were generally similar to those excluded due to lack of complete data, although rate of intubation and systolic blood pressure were slightly higher and oxygen saturation and D-dimer level lower. Patients included in the validation cohort exhibited higher rates of severe outcomes than patients excluded due to lack of complete data. The included and excluded cohorts are compared in the supplement.
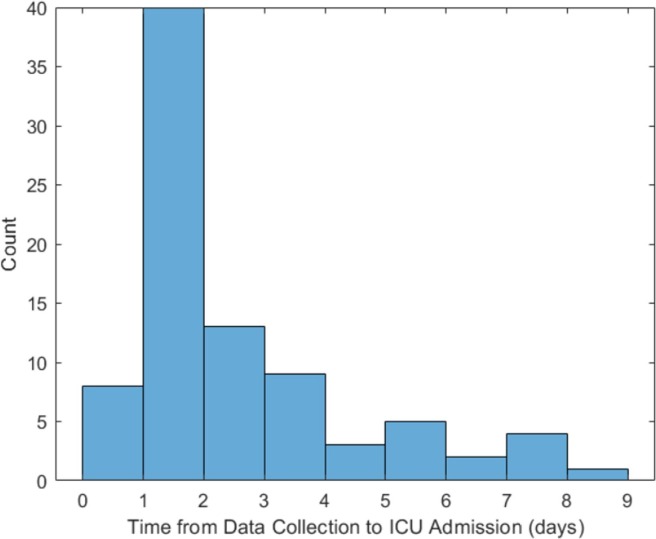
Two-hundred twenty-nine patients were included in the development cohort: 77 (34%) were admitted to the ICU, 53 (23%) were intubated, and 45 (20%) developed ARDS. The proportions of patients experiencing poor outcomes were smaller in the 330 patients in the validation cohort, of whom 85 (26%) were admitted to the ICU, 42 (13%) were intubated, and 35 (11%) developed ARDS. The patient characteristics of the two cohorts are summarized in Table 1. The racial/ethnic composure of both cohorts were skewed compared with overall Colorado demographics [39] with White being significantly underrepresented in both sets, and Black being overrepresented only in the development cohort. All attributes used in classification are contained in Table 1 and denoted with an asterisk. The time from data collection to ICU admission was estimated in the validation cohort and is summarized in Fig. 3 .
Fig. 3.
Time from data collection to admission to the ICU for the 85 patients admitted to the ICU in the validation cohort indicating potential utility for ICU admission risk assessment at hospital admission. A time in the [0, 1] bin indicates the patient was admitted on the same day as the data was collected.
3.1. Predicting ICU admission
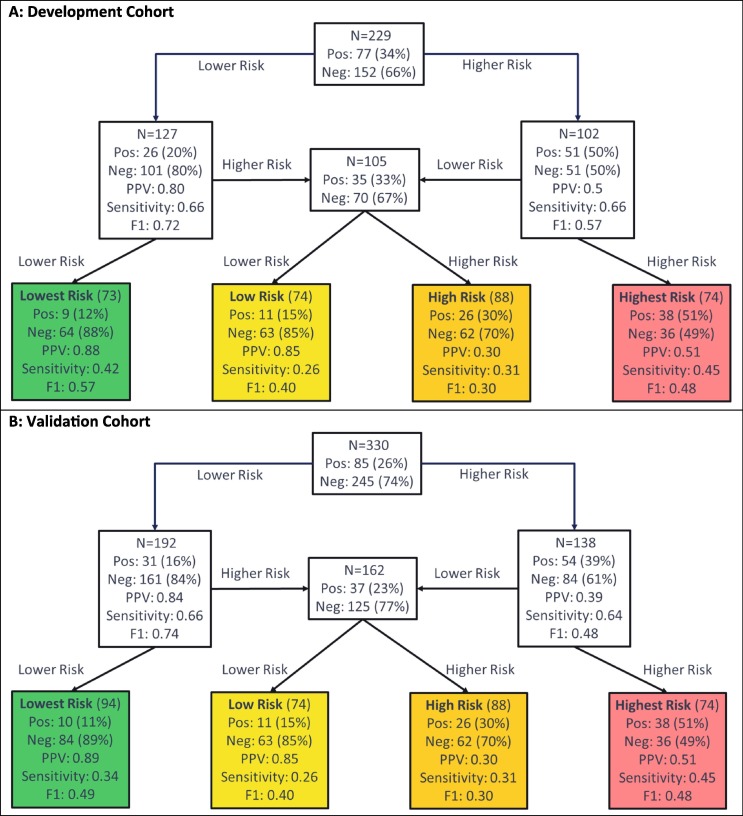
Test development for prediction of ICU admission used the schema in Fig. 2B, with the flow of patients through the hierarchical classification scheme shown in Fig. 3. In Fig. 4 , we see the test classified 73 (32%), 54 (24%), 51 (22%), and 51 (22%) of the development cohort to the lowest, low, high, and highest risk groups, respectively. Only 12% of patients in the lowest risk group were admitted to the ICU, whereas 65% of the highest risk group were admitted. Performance was similar in the validation cohort, of which 28%, 22%, 27%, and 22% were classified to the lowest, low, high, and highest risk groups, respectively. The proportions of patients that were admitted to the ICU were significantly associated with increasing risk subgroup (Cochran-Armitage p < 0.0001). The proportion of patients admitted to the ICU in the lowest risk group was 11% (95% CI: 5–19%), significantly lower than that in the other groups (Fisher’s exact p < 0.0001). The proportion of patients admitted to the ICU in the highest risk group was 51% (95% CI: 39–63%), Fisher’s exact p (highest risk vs other) < 0.0001).
Fig. 4.
Performance Flow Chart for the Risk Assessment Test for ICU Admission for (A) the Development Cohort and (B) the Validation Cohort. Each uncolored box represents a classifier with the contents reflecting the set of patients to be classified by the classifier. The colored boxes represent the final risk groups with the contents reflecting composition of the groups and test performance. Bootstrap 95% confidence intervals for performance metrics are given in the supplement. Pos = Positive (Admitted to ICU); Neg = Negative (Not Admitted to ICU), PPV = Positive Predictive Value.
Test classification was associated with heart rate, respiratory rate, oxygen saturation, sodium, blood urea nitrogen (BUN), anion gap, white blood cell (WBC) count, lactate dehydrogenase (LDH), D-dimer, CRP, and ferritin. Shapley Value analysis showed that the attributes most important for classification generation varied by patient, but that lowest risk classification was most often explained by CRP and D-dimer, with ferritin, LDH, platelet count, heart rate, respiratory rate, oxygen saturation, and WBC also of relatively higher importance. In highest risk patients, classification was related to LDH and D-dimer, although oxygen saturation, respiratory rate, CRP, ferritin, and weight were also important for some patients.
3.2. Predicting ARDS
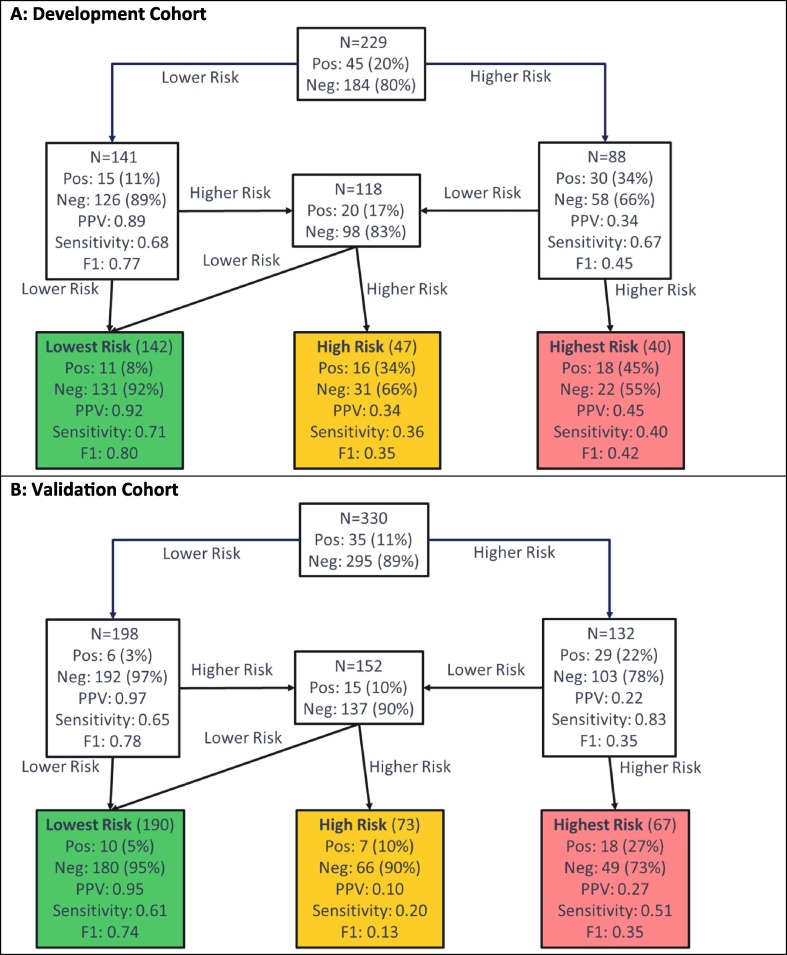
Test development for prediction of ARDS used the schema of Fig. 2B, with the flow of patients through the hierarchical classification scheme shown in Fig. 5 . Note that, for this test, the lowest and lower risk groups were combined into a single lowest risk group, as the proportion of patients with ARDS was found to be similar in both subgroups during test development. In Fig. 4, we see the test predicting risk of developing ARDS classified 142 (62%), 47 (21%), and 40 (17%) patients to the lowest, low, and highest risk groups, respectively, in the development cohort. In the lowest risk group, only 8% of patients developed ARDS, while in the highest risk group 45% of patients developed ARDS. The proportions of patients assigned to the lowest, high, and highest risk groups were similar in the validation cohort: 190 (58%), 73 (22%), and 67 (20%), respectively. The proportion of patients that developed ARDS increased with increasing risk group (Cochran-Armitage p = 0.02). Although the lowest risk group contained more than half of the patients, only 5% (95% CI: 3–9%) of them developed ARDS (Fisher’s exact p (lowest vs other) < 0.0001). In contrast, the percentage of patients developing ARDS in the highest risk group was 27% (95% CI: 17–39%), Fisher’s exact p (highest vs other) = 0.0004.
Fig. 5.
Performance Flow Chart for the Test Predicting Risk of Developing ARDS in (A) the Development Cohort, (B) the Validation Cohort. Each uncolored box represents a classifier with the contents reflecting the set of patients to be classified by the classifier. The colored boxes represent the final risk groups with the contents reflecting composition of the groups and test performance. Bootstrap 95% confidence intervals for performance metrics are given in the supplement. Pos = Positive (Developed ARDS); Neg = Negative (Did not develop ARDS), PPV = Positive Predictive Value.
Test classification was associated with age, blood pressure, oxygen saturation, sodium, BUN, WBC, LDH, D-dimer, CRP, and ferritin. Shapley value analysis again showed that the attributes most important for classification generation varied by patient, but that CRP and D-dimer contributed substantially to a classification of lowest risk, with race and age also contributing for some patients. Highest risk classification was related to LDH, CRP, and BUN in most patients and oxygen saturation in some patients.
3.3. Predicting intubation
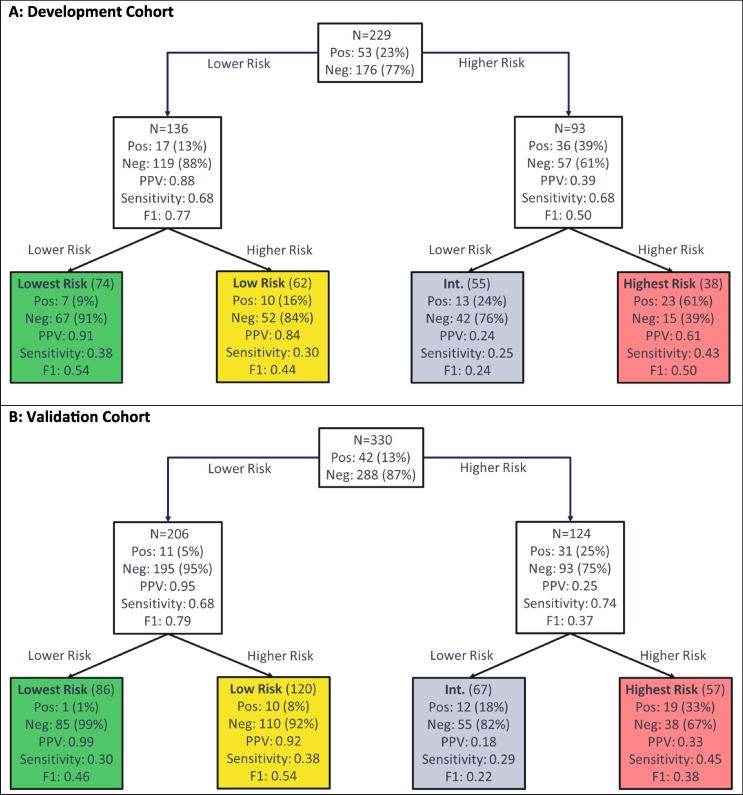
Test development to predict intubation used the schema of Fig. 2A, with the flow of patients through the hierarchical classification scheme shown in Fig. 5. In Fig. 6 , we see the test predicting risk of intubation classified 74 (32%), 62 (27%), 55 (24%), and 38 (17%) patients of the development cohort to the lowest, low, intermediate, and highest risk groups, respectively and 86 (26%), 120 (36%), 67 (20%), and 57 (17%), respectively, in the validation cohort. The patients of the validation cohort were assigned 86 (26%):120 (36%):67 (20%): 57 (17%) to the lowest, low, high and highest risk groups. Only 1% (95% CI: 0–6%) of patients in the lowest risk group were intubated (Fisher’s exact p (vs other <0.0001), while 33% (95 %CI: 21–47%) of patients in the highest risk group were intubated (Fisher’s exact p (vs other) < 0.0001). There was a significant association of increase in intubation with increasing risk group (Cochran-Armitage p < 0.0001).
Fig. 6.
Performance Flow Chart for the Test Assessing Risk of Intubation for (A) the Development Cohort and (B) the Validation Cohort. Each uncolored box represents a classifier with the contents reflecting the set of patients to be classified by the classifier. The colored boxes represent the final risk groups with the contents reflecting composition of the groups and test performance. Bootstrap 95% confidence intervals for performance metrics are given in the supplement. Pos = Positive (Intubated); Neg = Negative (Not intubated), PPV = Positive Predictive Value.
Test classification was associated with respiratory rate, oxygen saturation, BUN, creatinine, anion gap, WBC, LDH, D-dimer CRP, and ferritin. Shapley value analysis revealed that multiple attributes, varying by patient, were of increased relevance for classification. However, CRP was found to be generally most relevant for generation of a lowest risk classification, with D-dimer, LDH, BUN, race, and age also important factors. A highest risk classification was related to LDH, oxygen saturation, CRP, and BUN.
3.4. Comparison with other tests
An attempt was made to compare the performance of the tests to similar published classifiers [8], [10], [15]. The classifier described in Xiao et al. [12] applied on the development cohort yielded very similar areas under the receiver-operating characteristic (ROC) curves to those for the first classifiers in our hierarchies for each endpoint. Using the specified cutoff [10], relatively small, but pure lowest risk groups were observed for all four endpoints. While the purity was similar to that of our tests, the lowest risk group sizes were roughly half the size. The classifier described by Liu et al. [8] used age and NLR to stratify patients into four risk groups. Across all four endpoints, purities in the lowest risk groups were similar to those presented here but were roughly half the size. The highest risk groups were nearly twice the size of those for our tests but had purities a factor of 2–3 smaller. Performance for predicting ICU admission was poor, with the low risk group containing a similar proportion of patients admitted to the ICU as the highest risk group. Many studies have looked at binary classification to similar prognostic endpoints [20], [21], [22], [23], [24], [25], [26], [27]. While our tests stratify patients into more than 2 groups, the top classifier in each hierarchy achieved comparable performance in terms of AUROC.
4. Discussion
Using EHR-derived data recorded at the time of hospital admission, we developed and blindly validated a clinical decision support system composed of three different ML-based risk tests predicting three measures of progression to severe disease in patients hospitalized with COVID-19; namely risk of progression to ICU admission, intubation, and ARDS diagnosis. Given that many patients admitted with COVID-19 may not be in regular care and aware of comorbidities or may be critically ill upon presentation and unable to provide a detailed medical history, we utilized only attributes readily accessible in an emergency care setting.
Multiple previous studies have used ML-based algorithms to predict COVID-19 prognosis by combining basic patient demographics, vital signs, laboratory measurements, and comorbidities [14], [15], [16], sociodemographic information, comorbidities and current medications [19], or chest computed tomography (CT) scan alone [18], [19] or with patient demographics [15]. The performance of our tests compared favorably with that of two existing risk assessment models for which we were able to generate classifications for our development set, with our test achieving similar negative predictive values in the lowest risk groups but superior specificity. However, for other simple nomogram-based models, we were not able to compare performance due to lack of information for required attributes, as these were not obtainable from collected EHR data.
We validated three tests by generating test classifications blinded to all outcome data and analyzing the results according to a prespecified statistical analysis plan. This validation study was possible using data directly extracted from the EHR, without any further curation, demonstrating that the data required for risk assessment was easily and readily accessible. Test performance was similar between the development and validation cohorts, illustrating the ability of the ML platform to generalize to unseen datasets. The successful validation of the performance of our models observed in test development shows that our selection of minimal attributes was rich in predictive information. Fig. 4, Fig. 5, Fig. 6 demonstrate significant enhancement in the outcome of interest from the baseline in each cohort to the final risk groups.
As with all modern ML-based modeling, it was not a priori obvious which attributes would most inform the results for an individual patient. Recent work using Shapley values has directly addressed model explainability at an individual patient level [40]. Our ML architecture made the calculation of exact Shapley values for all classifiers for 50 patients from the validation cohort computationally feasible without the need for commonly made approximations [41], [42], supplement). The Shapley value analysis found that, when used in combination by the ML, simple clinical measures of risk and disease severity (age, weight, oxygen saturation) and general biomarkers of inflammation and acute phase reactants (CRP, D-dimer, LDH) were important for risk assessment. CRP, D-dimer, and LDH have been linked to COVID-19 severity in other studies, likely reflective of a pronounced inflammatory, systemic immune response with heightened thromboembolic risk [43], [44]. Increasing age and body mass index are well-established risk factors for influenza severity [45], and may, in part, exacerbate the inflammatory response [46], [47]. While race/ethnicity was not found to play a key role in explaining risk classification in the Shapley value analysis, a white race was observed to contribute towards a lower risk classification, while all other choices did not contribute substantially to risk category (see Fig. E22 in supplement). These observations were consistent with the associations we observed when we compared patient characteristics between our risk subgroups (see supplement). However, the Shapley values also revealed that our tests combined information across the 26 attributes in non-trivial ways and the attributes that explained patient classification differed from patient to patient. For some patients, all attributes had similar importance, while for others, some attributes pointed towards higher risk, while others pointed towards lower risk.
In contrast to the single center development cohort, the validation cohort included patients from the statewide UC health system. However, the cohort size and restriction to Colorado are limitations of this study. While patients excluded from the development cohort due to missing data were generally similar to those included, in validation, patients with complete data (only 15% of the total available) exhibited higher rates of severe disease and generally worse prognostic factors (laboratory and vital signs) than those without. Further validation of the tests in larger cohorts derived from other health systems and geographic areas is necessary.
The clinical utility of these tests will depend on their application. All tests achieved good performance in identifying patients who did not experience adverse outcome in the lowest risk groups in development and validation. Performance in identifying patients who experienced adverse outcomes in the highest risk groups was promising, but the risks of false positives and negatives would need to be carefully considered in an application where identifying patients likely to experience adverse outcome is desired.
In summary, we have developed and validated a suite of tests able to assess the risk of a poor outcome for patients hospitalized with COVID-19 based on information easily and routinely collected at time of hospital admission. Additional validation, preferably in a prospective setting, is required to further demonstrate the clinical utility of this risk assessment tool beyond clinical assessment alone. However, with readily-derived and quickly-available EHR data, a risk assessment at or near the time of admission can inform prognosis, guide discussions on the risks and benefits of treatments (including intubation), or identify low or high-risk patients for limited resources or enrollment in clinical trials. Furthermore, the methods here may be implemented in the care of future patients with novel viral infections.
5. Summary Table
What is already known?
|
What does this study add?
|
CRediT authorship contribution statement
Thomas W. Campbell: Conceptualization, Formal analysis, Methodology, Software, Writing – original draft, Writing – review and editing. Melissa P. Wilson: Data Curation. Heinrich Roder: Supervision, Methodology, Software, Writing – original draft, Writing – review and editing. Samantha MaWhinney: Supervision, Writing - review & editing. Robert W. Georgantas: Supervision, Writing – review and editing. Laura K. Maguire: Formal analysis, Writing – review and editing. Joanna Roder: Supervision, Methodology, Formal analysis, Writing – original draft, Writing – review and editing. Kristine M. Erlandson: Conceptualization, Resources, Writing - review & editing.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: ‘The authors have no major conflicts of interest to disclose. TC, HR, RG, LM, and JR are named as inventors on a provisional patent assigned to Biodesix relevant to the work and hold stock and/or stock options in Biodesix. KE reports grant funding from the NIH during the conduct of the study and grant funding from Gilead and personal fees from Theratechnologies and ViiV outside of the conduct of the study’.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijmedinf.2021.104594.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Mahase E. Covid-19: FDA authorises neutralising antibody bamlanivimab for non-admitted patients. BMJ. 2020;11(371) doi: 10.1136/bmj.m4362. PMID: 33177042. [DOI] [PubMed] [Google Scholar]
- 2.Elsawah H.K., Elsokary M.A., Abdallah M.S., ElShafie A.H. Efficacy and safety of remdesivir in hospitalized Covid-19 patients: Systematic review and meta-analysis including network meta-analysis. Rev. Med. Virol. 2020 doi: 10.1002/rmv.2187. Epub ahead of print. PMID: 33128490. [DOI] [PubMed] [Google Scholar]
- 3.Tuccori M., Ferraro S., Convertino I., Cappello E., Valdiserra G., Blandizzi C., Maggi F., Focosi D. Anti-SARS-CoV-2 neutralizing monoclonal antibodies: clinical pipeline. MAbs. 2020;12(1):1854149. doi: 10.1080/19420862.2020.1854149. PMID: 33319649; PMCID: PMC7755170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou F., Yu T., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., Guan L., Wei Y., Li H., Wu X., Xu J., Tu S., Zhang Y., Chen H., Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-10 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ciceri F., Castagna A., Rovere-Querini P., De Cobelli F., Ruggeri A., Galli L., Conte C., De Lorenzo R., Poli A., Ambrosio A., Signorelli C., Bossi E., Fazio M., Tresoldi C., Colombo S., Monti G., Fominskiy E., Franchini S., Spessot M., Martinenghi C., Carlucci M., Beretta L., Scandroglio A.M., Clementi M., Locatelli M., Tresoldi M., Scarpellini P., Martino G., Bosi E., Dagna L., Lazzarin A., Landoni G., Zangrillo A. Early predictors of clinical outcomes of COVID-19 outbreak in Milan. Italy. Clin Immunol. 2020;217 doi: 10.1016/j.clim.2020.108509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ioannou G.N., Locke E., Green P., Berry K., O’Hare A.M., Shah J.A., Crothers K. Eastment MC< Dominitz JA, Fan VS. Risk Factors for Hospitalziation, Mechanical Ventilation, or Death Among 10 131 US Veterans with SARS-CoV-2 Infection. JAMA Netw. Open. 2020;3(9):2022310. doi: 10.1001/jamanetworkopen.2020.22310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng B., Hu J., Zuo X., Chen J., Li X. Chen y, Yang G, Shi X, Deng A. Predictors of progression from moderate to severe coronavirus disease 2019: a retrospective cohort. Clin. Microbiol. Infect. 2020;26(10):1400–1405. doi: 10.1016/j.cmi.2020.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu J., Liu Y., Xiang P., Pu L., Xiong H., Li C., Zhang M., Tan J., Xu Y., Song R., Song M., Wang L., Zhang W., Han B., Yang L., Wang X., Zhou G., Zhang T., Li B., Wang Y., Chen Z., Wang X. Neutrophil-to-lymphocyte ratio predicts critical illness patients with 2019 coronavirus disease in the early stage. J. Trans. Med. 2020;18:206. doi: 10.1186/s12967-020-02374-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng Z., Yu Q., Yao S., Luo L., Zhou W., Mao X., Li J., Duan J., Yan Z., Yang M., Tan H., Ma M., Li T., Yi D., Mi Z., Zhao H., Jiang Y., He Z., Li H., Nie W., Liu Y., Zhao J., Luo M., Liu X., Rong P., Wang W. Early prediction of disease progression in COVID-19 pneumonia patients with chest CT and clinical characteristics. Nat. Commun. 2020;11(1):4968. doi: 10.1038/s41467-020-18786-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao L., Zhang W., Gong M., Zhang Ym Chen L., Zhu H., Hu C., Kang P., Liu L., Zhu H. Development and validation of the HNC-LL score for predicting the severity of coronavirus disease 2019. EBioMedicine. 2020;57:102880. doi: 10.1016/j.ebiom.2020.102880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sinha P., Churpek M.M., Calfee C.S. Machine Learning Classifier Models Can Identify Acute Respiratory Distress Syndrome Phenotypes Using Readily Available Clinical Data. Am. J. Respir. Crit. Care Med.. 2020;202(7):996–1004. doi: 10.1164/rccm.202002-0347OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McNicholas B., Madden M.G., Laffey J.G. Machine Learning Classifier Models: The Future for Acute Respiratory Distress Syndrome Phenotyping? Am. J. Respir. Crit. Care Med. 2020;202(7):919–920. doi: 10.1164/rccm.202006-2388ED. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wynants L., Van Calster B., Collins G.S., Riley R.D., Heinze G., Schuit E., Bonten M.M.J., Damen J.A.A., Debray T.P.A., De Vos M., Dhiman P., Haller M.C., Harhay M.O., Henckaerts L., Kreuzberger N., Lohman A., Luijken K., Ma J., Andaur C.L., Reitsma J.B., Sergeant J.C., Shi C., Skoetz N., Smits L.J.M., Snell K.I.E., Sperrin M., Spijker R., Steyerberg E.W., Takada T., van Kuijk S.M.J., van Royen F.S., Wallisch C., Hooft L., Moons K.G.M., van Smeden M. Prediction models for diagnosis and prognosis of covid-19 infection: systematic review and critical appraisal. BMJ. 2020;7(369) doi: 10.1136/bmj.m1328. Erratum. In: BMJ. 2020 Jun 3;369:m2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castro V.M., McCoy T.H., Perlis R.H. Laboratory Findings Associated With Severe Illness and Mortality Among Hospitalized Individuals With Coronavirus Disease 2019 in Eastern Massachusetts. JAMA Netw. Open. 2020;3(10) doi: 10.1001/jamanetworkopen.2020.23934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu G., Yang P., Xie Y., Woodruff H.C., Rao X., Guiot J., Frix A., Loius R., Moutschen M., Li J., Li J., Yan C., Du D., Zhao S., Ding Y., Liu B., Sun W., Albarello F., d’Ambramo A., Schinina V., Nicastri E., Occhipinti M., Barisionse E., Halilaj I., Lovinfosse P., Wang X., Wu J., Lambin P. Development of a clinical decision support system for severity risk prediction and triage of COVID-19 patients at hospital admission: an international multicenter study. Eur. Respir. J. 2020;56:2001104. doi: 10.1183/13993003.01104-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vaid A., Somani S., Russak A.J., De Freitas J.K., Chaudhry F.F., Paranjpe I., Johnson K.W., Lee S.J., Miotto R., Richter F., Zhao S., Beckmann N.D., Naik N., Kia A., Timsina P., Lala A., Paranjpe M., Golden E., Danieletto M., Singh M., Meyer D., O'Reilly P.F., Huckins L., Kovatch P., Finkelstein J., Freeman R.M., Argulian E., Kasarskis A., Percha B., Aberg J.A., Bagiella E., Horowitz C.R., Murphy B., Nestler E.J., Schadt E.E., Cho J.H., Cordon-Cardo C., Fuster V., Charney D.S., Reich D.L., Bottinger E.P., Levin M.A., Narula J., Fayad Z.A., Just A.C., Charney A.W., Nadkarni G.N., Glicksberg B.S. Machine Learning to Predict Mortality and Critical Events in a Cohort of Patients With COVID-19 in New York City: Model Development and Validation. J. Med. Internet Res. 2020;22(11) doi: 10.2196/24018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.An C., Lim H., Kim D.W., Chang J.H., Choi Y.J., Kim S.W. Machine learning prediction for mortality of patients diagnosed with COVID-19: a nationwide Korean cohort study. Sci. Rep. 2020;10(1):18716. doi: 10.1038/s41598-020-75767-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghosh B., Kumar N., Singh N., Sadhu A.K., Ghosh N., Mirta P., Chatterjee I. A Quantitative Lung Computed Tomography Image Feature for Multi-Center Severity Assessment of COVID-19. medRxiv. 2020 doi: 10.1101/2020.07.13.20152231. [DOI] [Google Scholar]
- 19.Wang S., Zha Y., Li W., Wu Q., Li X., Niu M., Wang M., Qiu X., Li H., Yu H., Gong W., Bai Y., Li L., Zhu Y., Wang L., Tian J. A fully automatic deep learning system for COVID_19 diagnostic and prognostic analysis. Eur. Respir. J. 2020;56:2000775. doi: 10.1183/13993003.00775-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marcos M., Belhassen-García M., Sánchez-Puente A., Sampedro-Gomez J., Azibeiro R., Dorado-Díaz P.-I., et al. Development of a severity of disease score and classification model by machine learning for hospitalized COVID-19 patients. PLoS ONE. 2021;16(4) doi: 10.1371/journal.pone.0240200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heldt F.S., Vizcaychipi M.P., Peacock S., et al. Early risk assessment for COVID-19 patients from emergency department data using machine learning. Sci. Rep. 2021;11:4200. doi: 10.1038/s41598-021-83784-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu L., Halalau A., Dalal B., Abbas A.E., Ivascu F., Amin M., et al. Machine learning methods to predict mechanical ventilation and mortality in patients with COVID-19. PLoS ONE. 2021;16(4) doi: 10.1371/journal.pone.0249285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Estiri H., Strasser Z.H., Murphy S.N. Individualized prediction of COVID-19 adverse outcomes with MLHO. Sci. Rep. 2021;11:5322. doi: 10.1038/s41598-021-84781-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodriguez V.A., Bhave S., Chen R., Pang C., Hripcsak G., Sengupta S., Elhadad N., Green R., Adelman J., Metitiri K.S., Elias P., Groves H., Mohan S., Natarajan K., Perotte A. Development and validation of prediction models for mechanical ventilation, renal replacement therapy, and readmission in COVID-19 patients. J. Am. Med. Inform. Assoc. 2021:ocab029. doi: 10.1093/jamia/ocab029. Epub ahead of print. PMID: 33706377; PMCID: PMC7989331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bolourani S., Brenner M., Wang P., McGinn T., Hirsch J., Barnaby D., Zanos T. A Machine Learning Prediction Model of Respiratory Failure Within 48 Hours of Patient Admission for COVID-19: Model Development and Validation. J. Med. Internet Res. 2021;23(2) doi: 10.2196/24246. e24246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aljouie Abdulrhman Fahad, et al. Early Prediction of COVID-19 Ventilation Requirement and Mortality from Routinely Collected Baseline Chest Radiographs, Laboratory, and Clinical Data with Machine Learning. J. Multidisciplinary Healthcare. 2021;14:2017–2033. doi: 10.2147/JMDH.S322431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jakob C.E.M., Mahajan U.M., Oswald M., et al. Prediction of COVID-19 deterioration in high-risk patients at diagnosis: an early warning score for advanced COVID-19 developed by machine learning. Infection. 2021 doi: 10.1007/s15010-021-01656-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Espinosa-Gonzalez A.B., Neves A.L., Fiorentino F., Prociuk D., Husain L., Ramtale S.C., Mi E., Mi E., Macartney J., Anand S.N., Sherlock J., Saravanakumar K., Mayer E., de Lusignan S., Greenhalgh T., Delaney B.C. Predicting Risk of Hospital Admission in Patients With Suspected COVID-19 in a Community Setting: Protocol for Development and Validation of a Multivariate Risk Prediction Tool. JMIR Res. Protocols. 2021:29072. doi: 10.2196/29072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kar S., Chawla R., Haranath S.P., et al. Multivariable mortality risk prediction using machine learning for COVID-19 patients at admission (AICOVID) Sci. Rep. 2021;11:12801. doi: 10.1038/s41598-021-92146-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harris P.A., Taylor R., Thielke R., Payne J., Gonzalez N., Conde J.G. Research electronic data capture (REDCap) - A metadata-driven methodology and workflow process for providing translational research informatics support. J. Biomed. Inform. 2009 Apr;42(2):377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roder J., Oliveira C., Net L., Tsypin M., Linstid B., Roder H. A dropout-regularized classifier development approach optimized for precision medicine test discovery from omics data. BMC Bioinf. 2019;20(1):325. doi: 10.1186/s12859-019-2922-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Breiman L. Department of Statistics, University of California; 1996. Out-of-bag estimation. Technical Report. [Google Scholar]
- 33.https://github.com/Biodesix/dxCortex-forest-COVID19.
- 34.Shapley L.S. A Value for N-Person Games. Contrib. Theory Games. 1953;2:307–317. [Google Scholar]
- 35.Jia R., Dao D., Wang B., Hubis F.A., Hynes N., Gurel N.M., et al. Towards efficient data valuation based on the Shapley value. Proceedings of the 22nd International Conference on Artificial Intelligence and Statistics (AIS-TATS) 2019;vol. 89 [Google Scholar]
- 36.Murphy K.P. Machine Learning, a probabilistic perspective. MIT. 2012:182–186. [Google Scholar]
- 37.Le Nguyen Quoc Khanh, Ho Quang-Thai, Yapp Edward Kien Yee, Yu-Yen Ou, Yeh Hui-Yuan. DeepETC: A deep convolutional neural network architecture for investigating and classifying electron transport chain's complexes. Neurocomputing. 2020;375 [Google Scholar]
- 38.Nguyen Quoc Khanh Le, Truong Nguyen Khanh Hung, Duyen Thi Do, Luu Ho Thanh Lam, Luong Huu Dang, Tuan-Tu Huynh. Radiomics-based machine learning model for efficiently classifying transcriptome subtypes in glioblastoma patients from MRI. Comput. Biol. Med. 132 (2021). [DOI] [PubMed]
- 39.[accessed 2020 Dec 14] https://www.census.gov/quickfacts/CO.
- 40.Lundberg S.M., Lee S. 31st Conference on Neural Information Processing Systems (NIPS) 2017. A unified approach to interpreting model predictions. [Google Scholar]
- 41.E. Kumar, S. Venkatasubramanian, C. Scheidegger, S. Friedler, Problems with Shapley-value-based explanations as feature importance measures. arXiv:2002.11097v1 [cs.AI], 25 Feb 2020.
- 42.K. Aas, M. Jullum, A. Løland, Explaining Individual Predictions When Features are Dependent: More Accurate Approximations to Shapley Values. arXiv:1903.10464v3 [stat.ML], 2020.
- 43.Danwang C., Endomba F.T., Nkeck J.R., Wouna D.L.A., Robert A., Noubiap J.J. A meta-analysis of potential biomarkers associated with severity of coronavirus disease 2019 (COVID-19) Biomark. Res. 2020;31(8):37. doi: 10.1186/s40364-020-00217-0. PMID: 32879731; PMCID: PMC7456766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.García L.F. Immune Response, Inflammation, and the Clinical Spectrum of COVID-19. Front Immunol. 2020;16(11):1441. doi: 10.3389/fimmu.2020.01441. PMID: 32612615; PMCID: PMC7308593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Louie J.K., Acosta M., Samuel M.C., Schechter R., Vugia D.J., Harriman K., Matyas B.T. California Pandemic (H1N1) Working Group. A novel risk factor for a novel virus: obesity and 2009 pandemic influenza A (H1N1) Clin. Infect. Dis. 2011;52(3):301–312. doi: 10.1093/cid/ciq152. Epub 2011 Jan 4. PMID: 21208911. [DOI] [PubMed] [Google Scholar]
- 46.Morais A.H.A., Passos T.S., de Lima Vale S.H., da Silva Maia J.K., Maciel B.L.L. Obesity and the increased risk for COVID-19: mechanisms and nutritional management. Nutr. Res. Rev. 2020:1–13. doi: 10.1017/S095442242000027X. Epub ahead of print. PMID: 33183383; PMCID: PMC7737140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Petrakis D., Margină D., Tsarouhas K., Tekos F., Stan M., Nikitovic D., Kouretas D., Spandidos D.A., Tsatsakis A. Obesity – a risk factor for increased COVID–19 prevalence, severity and lethality (Review) Mol. Med. Rep. 2020;22(1):9–19. doi: 10.3892/mmr.2020.11127. Epub 2020 May 5. PMID: 32377709; PMCID: PMC7248467. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.