Abstract
The Notch signaling pathway is a fundamental regulator of cell fate determination in homeostasis and regeneration. In this work, we aimed to determine how Notch signaling mediates the interactions between perivascular stem cells and their niches in human dental mesenchymal tissues, both in homeostatic and regenerative conditions. By single cell RNA sequencing analysis, we showed that perivascular cells across the dental pulp and periodontal human tissues all express NOTCH3, and that these cells are important for the response to traumatic injuries in vivo in a transgenic mouse model. We further showed that the behavior of perivascular NOTCH3‐expressing stem cells could be modulated by cellular and molecular cues deriving from their microenvironments. Taken together, the present studies, reinforced by single‐cell analysis, reveal the pivotal importance of Notch signaling in the crosstalk between perivascular stem cells and their niches in tissue homeostasis and regeneration.
Keywords: dental pulp stem cells, carious teeth, mesenchymal stem cells, microenvironment, Notch ligands, Notch signaling, Notch3, pericytes, periodontal stem cells, periodontium, perivascular niche, pulp, single‐cell RNA sequencing, stem cell niches, stem cells, tooth
In the dental pulp and the periodontium, mesenchymal stem cells (MSCs) are highly similar, are mostly located around blood vessels, and express NOTCH3. These NOTCH3 + MSCs are important for the reaction of dental tissues to injury. In both tissues, endothelial cells adjacent to NOTCH3 + MSCs express NOTCH ligands, which could thus mediate the molecular crosstalk between MSCs and their perivascular niche.
Significance statement.
The present results highlight the importance of Notch signaling in the interaction between mesenchymal stem cells (MSCs) and their niches. Endothelial cells in dental tissues express all five NOTCH ligands and constitute conserved perivascular niches, which can regulate the activation of NOTCH3‐expressing perivascular MSCs. This fundamental information establishes a new conceptual framework that could be the basis for future research focusing towards MSCs‐based regenerative approaches.
1. INTRODUCTION
The Notch pathway constitutes a fundamental cell communication mechanism that enables neighboring cells to adopt different fates and regulates stem cell function in many organs and tissues such as intestine, brain, eye, and skin.1, 2, 3 Four Notch receptors (ie, Notch1‐4) that interact with five trans‐membrane‐bound ligands (ie, Jagged1,2; Delta1, Delta‐like3,4) have been identified in vertebrates. Ligand‐receptor interactions trigger the cleavage of the receptor and the subsequent nuclear translocation of its intracellular domain. This leads to the activation of Notch downstream genes, which include Hes1, Hes5, Myc, and CCND1.2, 4, 5 Notch signaling is fundamental in the interactions between stem cells and their surrounding microenvironment, also called the stem cell niche, mediating stem cells responses to injury or stress.6, 7, 8 Our previous studies in injured teeth have shown that the Notch pathway plays a role in regulating the behavior of stem cells in specific niches within the dental pulp tissue.9, 10, 11
The development of the tooth results from sequential and reciprocal interactions between cells of the oral epithelium and the cranial neural crest‐derived mesenchyme.12, 13 Dental epithelial cells give rise initially to ameloblasts, which produce the hardest tissue of the human body that covers the crown of the tooth, the enamel, and then drive the development of the dental roots and ensure the production of the root‐specific hard matrix, the cementum.14, 15, 16 Dental mesenchymal cells form two highly vascularized and innervated tissues, the dental pulp and the periodontium.14, 15 The central portion of the tooth is occupied by the dental pulp, which contains mesenchymal stem cells (MSCs), named dental pulp stem cells (DPSCs).17, 18 DPSCs are mainly located in perivascular areas, where they closely interact with endothelial cells.19, 20, 21 Indeed, our previous results and studies by others showed that perivascular cells express Notch receptors and possess MSCs properties.22, 23, 24 Odontoblasts at the borders of the dental pulp form the dentin, which is another tooth‐specific, mineralized tissue supporting the enamel. The periodontium anchors the tooth to the surrounding alveolar bone and provides tooth stability by continuously remodeling its extracellular matrix, the periodontal ligament.14 The periodontium, upon root completion, contains perivascular MSCs,18 periodontal stem cells (PDSCs), and an additional population of epithelial stem cells formed by the root epithelial remnants.25
All these stem cell populations are multipotent and guarantee homeostasis and regeneration of dental tissues.26 MSCs behavior is subject to their crosstalk with elements of their niches and the influence of molecular cues produced by stromal cells, as well as by physical factors such as stiffness, topography, and shear stress.27 In vivo studies aiming at the regeneration of dental tissues were not fully satisfactory,28, 29, 30 mainly due to the limited information that is currently available concerning the interactions between the various MSC populations and their corresponding niches.
Our previous studies have shown that Notch signaling constitutes a fundamental signaling hub throughout tooth development.31, 32, 33 More recent studies have demonstrated that in adult teeth, Notch signaling is activated in dental tissues, and more specifically in perivascular regions, upon injury,10, 11, 34 where it regulates the fate of MSCs in the dental pulp and the periodontium.33, 35, 36 The actual role of Notch signaling in dental tissues homeostasis and regeneration is however far from being fully understood.
In this work, we investigated the role of Notch signaling in the interactions of the various dental MSC populations with their respective niches based on their composition and gene expression patterns at single‐cell resolution. We observed that dental pulp and periodontal MSCs both express NOTCH3, and that NOTCH ligands expressed by endothelial cells are conserved, potential perivascular niche‐derived modulators of their behavior. We also showed that NOTCH3 expression in cultured dental MSCs significantly decreases upon differentiation. Furthermore, we exploited a transgenic mouse model to characterize the behavior of perivascular Notch3‐expressing MSCs in homeostatic conditions and in response to injuries in vivo. Finally, we demonstrated the distribution of NOTCH3 in the pulp of human carious teeth.
2. MATERIALS AND METHODS
2.1. Data and code availability
Data can be found at GEO; Access number: GSE161267. All code is publicly available at: https://github.com/TheMoorLab/Tooth. Single cell RNA sequencing data are described in detail in Reference 37.
2.2. Human samples
The procedure for the collection of anonymized healthy and carious human teeth at the Center of Dental Medicine (ZZM) of the University of Zurich was approved by the Ethic Commission of the Kanton of Zurich (reference number 2012‐0588) and the patients gave their written informed consent. Samples were obtained in fully anonymized form from patients of 18 to 35 years of age, according to the cantonal regulations. The health status of the tooth was determined by dental thorough examination by specialized dentists. Tissue dissociation and single cell RNA sequencing was performed as described previously.37
2.3. Animals
All experiments were performed according to the guidelines of the Swiss Animal Welfare Law and in compliance with the regulations of the Cantonal Veterinary Office, Zurich (License for animal experimentation ZH018/17). The animal facility provided standardized housing conditions, with a mean room temperature of 21°C ± 1°C, relative humidity of 50% ± 5%, and 15 complete changes of filtered air per hour (HEPA H 14filter); air pressure was controlled at 50 Pa. The light/dark cycle in the animal rooms was set to a 12 hours/12 hours cycle (lights on at 07:00, lights off at 19:00) with artificial light of approximately 40 Lux in the cage. The animals had unrestricted access to sterilized drinking water, and ad libitum access to a pelleted and extruded mouse diet in the food hopper (Kliba No. 3436; Provimi Kliba/Granovit AG, Kaiseraugst, Switzerland). Mice were housed in a barrier‐protected specific pathogen‐free unit and were kept in groups of maximum five adult mice per cage in standard IVC cages (Allentown Mouse 500; 194 mm × 181 mm × 398 mm, floor area 500 cm2; Allentown, New Jersey) with autoclaved dust‐free poplar bedding (JRS GmbH + Co KG, Rosenberg, Germany). A standard cardboard house (Ketchum Manufacturing, Brockville, Canada) served as a shelter, and tissue papers were provided as nesting material. Additionally, crinklets (SAFE crinklets natural, JRS GmbH + Co KG) were provided as enrichment and further nesting material. The specific pathogen‐free status of the animals was monitored frequently and confirmed according to FELASA guidelines by a sentinel program. The mice were free of all viral, bacterial, and parasitic pathogens listed in FELASA recommendations.38 To generate Notch3 CreER R26 mT/m mice, we crossed R26 mT/mG (Gt(ROSA)26Sor tm4(ACTB‐tdTomato,‐EGFP)Luo; Jax: 007576) mice with Notch3 CreER (Notch3 tm1.1[cre/ERT2]Sat; MGI: 5304914) mice. The double‐fluorescent Cre reporter R26 mT/mG mice constitutively express membrane‐targeted tandem dimer Tomato (mT). Upon Cre recombinase activation, mT is excised and membrane‐targeted green fluorescent protein (mGFP) is expressed. In the Notch3 CreER; R26 mT/mG mice, the Cre recombinase activity is triggered upon exposure to 4‐hydroxytamoxifen (4‐OHT) solely in Notch3‐expressing cells, leading to the expression of mGFP in these cells as well as in their progeny. Pain management was ensured via subcutaneous injection of Temgesic (2 mg/kg body weight) 2 hours before surgery, followed by postoperatory injections with 6 hours intervals for a period of 24 hours and completed by supplying Temgesic in drinking water for 3 days postsurgery. We investigated whether Notch3‐expressing cells contribute to pulp homeostasis using the Notch3 CreER; R26 mT/mG transgenic mice. 8‐ to 10‐week‐old mice were injected intraperitoneally with 4‐OHT (H7904, Sigma‐Aldrich, Buchs, Switzerland), dissolved in corn oil (100 mg/kg body weight). One day upon 4‐OHT administration, animals were anesthetized by intraperitoneal injection of a solution consisting of Ketamine (65 mg/kg body weight) and Xylazine (13 mg/kg body weight) dissolved in 0.9% NaCl. Cavity preparations were performed at the occlusal part of the upper first molar teeth, accessing the pulp mesio‐buccal horns, using a slow‐speed dental drill and a round bur size 008 (Brassler, USA) irrigated with a saline solution. Upon operation, direct dental pulp capping was performed with calcium hydroxide (Dycal Cement, LD Caulk Company, Del) followed by further cavity protection filling with AH Plus Root canal sealer (Dentsply, DeTrey, Germany). A total of six treated mice and nine control mice were analyzed.
2.4. Processing of human teeth and mouse tissues for immunofluorescent staining
Human healthy and carious teeth used for histological analysis and immunostaining were immediately fixed by immersion in paraformaldehyde 4% (PFA 4%) for 24 hours, and then decalcified in Morse's solution for 8 weeks, dehydrated, embedded in paraffin, and serially sectioned at 5 μm. From a subset of teeth, the dental pulp was immediately extracted and fixed in PFA 4% for 2 hours. The specimens were then incubated in Sucrose 30%, embedded in Tissue Tek O.C.T.™ (4583, Sakura, Alphen aan den Rijn, the Netherlands), and serially sectioned at 10 μm. For animal experimentation, mice were anaesthetized with Ketamine/Xylazine, and sacrificed at 3, 10, and 28 days postinjection, by intracardiac perfusion with Phosphate Buffer Saline (PBS) followed by PFA 4%. Thereafter, the heads of the animals were postfixed by overnight incubation in PFA 4% at 4°C. Subsequently, specimens were decalcified in 10% Ethylenediamine Tetraacetic Acid (Titriplex, 1084211000, Merck, Zug, Switzerland) for up to 8 to 10 weeks, dehydrated, embedded in paraffin, and serially sectioned at 5 μm. Sections were then used for further immunofluorescent staining.
2.5. Immunostaining
Paraffin sections were rehydrated by incubation in Xylol followed by a series of Ethanol solutions (100% ‐ 30%) and distilled H2O. Cryosections were let dry at room temperature for 1 hour and then washed with PBS before immunostaining. Cells used for immunofluorescent staining were first fixed in PFA 4% for 15 minutes at 4°C. Thereafter, specimens were blocked with PBS supplemented with 2% fetal bovine serum (FBS) and incubated with primary antibodies for 1 hour at room temperature. The following primary antibodies were used: Rabbit anti‐Thy1/CD90 (1:100; 555595, BD, Eysin, Switzerland), biotinylated Rat anti‐CD31 (1:200; 553371; BD Biosciences, San Jose, California), Mouse anti‐Vimentin (1:100; M0725, Dako, Baar, Switzerland), Rabbit, anti‐Smooth Muscle Actin (1:100; MS‐113‐P0, Thermo Fisher Scientific, Reinach, Switzerland), anti‐Notch3,22 Rabbit anti‐GFP (1:100, A‐11122, Thermo Fisher Scientific), Goat anti‐GFP (1:100; ab6673, Abcam, Cambridge, UK), Goat anti‐Jagged1 (1:100, AF1277, R&D Systems, Minneapolis, Minnesota), Rabbit anti‐Laminin (1:20; ab11575, Abcam, Cambridge, United Kingdom), Rat anti‐Endomucin (1:20, ab106100, Abcam), anti‐CD45 (1:50, 103 104, BioLegend, San Diego, California), anti‐Fibronectin (1:100, ab2413, Abcam), anti‐MBP (1:200, MAB386, Millipore). The sections were then incubated with Fluorochrome‐conjugated secondary antibodies for 1 hour at room temperature at dark. The following secondary antibodies were used: Alexa‐568 Donkey anti‐Rabbit (1:500; A10042, Thermo Fisher Scientific), Alexa‐488 Chicken anti‐Goat (1:500; A‐21467, Thermo Fisher Scientific), Alexa‐488 Goat anti‐Rabbit (1:500; A32731, Thermo Fisher Scientific), Alexa‐568 Goat anti‐Rat (1:500; A‐11077, Thermo Fisher Scientific). DAPI (4′,6‐diamidino‐2‐Phenylindole; D1306, Thermo Fisher Scientific) was then used for nuclear staining. After immunofluorescent staining, samples were mounted in ProLong Diamond Antifade Mountant (P36965, Thermo Fisher Scientific), and imaged with a Leica SP8 Inverted Confocal Laser Scanning Microscope (Leica Microsystems‐ Schweiz AG, Heerbrugg, Switzerland).
2.6. Isolation, culture, and differentiation of human dental pulp MSCs
The dental pulps of healthy teeth were enzymatically digested for 1 hour at 37°C in a solution of collagenase (3 mg/mL; Life Technologies Europe BV, Zug ZG, Switzerland) and Dispase (4 mg/mL; Sigma‐Aldrich Chemie GmbH, Buchs SG, Switzerland). A filtered single‐cell suspension was plated in a 40 mm Petri dish with growth medium containing DMEM/F12 (Sigma‐Aldrich Chemie GmbH) with 10% FBS (PAN Biotech GmbH, Aidenbach, Germany), 1% penicillin/streptomycin (P/S) (Sigma‐Aldrich Chemie GmbH), 1% L‐glutamine (Sigma‐Aldrich Chemie GmbH), and 0.5 μg/mL fungizone (Life Technologies Europe BV) after washing away the enzyme solution. Cells were passaged at 80% to 90% confluence and expanded in the same growth medium. The osteogenic differentiation medium consisted of DMEM supplemented with Ascorbic Acid (200 μM), β‐Glycerolphosphate (10 mM), Dexamethasone (10 nM) (Sigma‐Aldrich/Merck, Darmstadt, Germany), and Amphotericin B 0.25 μg/μL (Thermo Fisher Scientific, Switzerland). The adipogenic differentiation medium consisted of DMEM (1 mL) supplemented with Dexamethasone (1 μM), IBMX (0.5 mM), Indomethacin (200 μM), Insulin (10 μM) (Sigma‐Aldrich/Merck) and Amphotericin B 0.25 μg/μL. For chondrogenic differentiation, 5 × 105 cells were pelleted and resuspended in 0.5 mL of StemPro© chondrogenic differentiation medium (A10071‐01, Thermo Fisher Scientific). Cells were cultured for 21 days in osteogenic, adipogenic, and chondrogenic medium. Cells were collected on day 0 (plating day), 7, 14, and 21 and used for RNA extraction. An additional set of cells was cultured for 21 days, stained (see following paragraph) and examined under a bright‐field microscope. Alizarin Red S staining was performed to identify extracellular calcium deposits of cells differentiated into osteoblasts. Alizarin Red S powder was dissolved in distilled water, pH 4.2. Cells were washed with PBS, fixed with 4% PFA for 30 minutes, washed with distilled water and finally Alizarin Red S staining solution was added to each well for 45 minutes at room temperature in the dark. Thereafter, wells were washed with deionized water and then PBS was added. The cells were viewed under a bright‐field microscope, where calcium deposits exhibited a bright orange‐red color. Oil Red O staining was performed to identify lipids in cells differentiated into adipocytes. A 300 mg of Oil Red O powder was added to 100 mL of 99% isopropanol and then mixed with deionized water and filtered through a funnel. Cells were washed with PBS, fixed with 4% PFA for 30 minutes, washed again with deionized water, 60% isopropanol was added for 2 to 5 minutes and after isopropanol aspiration Oil Red O was added for 5 minutes. Thereafter, the wells were rinsed with tap water, hematoxylin counterstain was performed for 1 minute and cells were rinsed with warm tap water. The cells were viewed under a bright‐field microscope, where lipids exhibited a red color while the nuclei of cells were blue. Alcian blue staining was performed on cells pellets subjected to chondrogenic differentiation. Pellets were washed in PBS, fixed in 4% PFA for 30 minutes at room temperature, washed in PBS, and embedded in Cryotek. Ten micrometer sections were cut, stained with 1% Alcian Blue, and counterstained with 0.1% Fast Red. Cells for gene expression analysis were collected by trypsinization at day 0, 7, 14, and 21, snap‐frozen in liquid nitrogen and stored at −80°C. RNA was isolated with the RNeasy Plus Universal Mini Kit (Qiagen AG, Hombrechtikon ZH, Switzerland). Reverse transcription of the isolated RNA was performed using the iScript cDNA synthesis Kit (Bio‐Rad Laboratories AG, Cressier FR, Switzerland). The three‐step quantitative real‐time PCRs were performed using an Eco Real‐Time PCR System (Illumina Inc, San Diego, California). Expression level analysis of S18 (housekeeping gene), ALP, OSX/SP7, DSPP, and NOTCH3 were carried out using the SYBR Green PCR Master Mix (Applied Biosystems, Carlsbad, California) in combination with oligonucleotide primers. Gene expression analysis was performed on six independent samples per condition. Samples were always compared one‐vs‐one using the Mann‐Whitney U/Wilcoxon rank‐sum test (Graph Pad Prism 8.0).
3. RESULTS
3.1. Human dental MSCs express NOTCH3
Recently, we performed a thorough single cell RNA sequencing analysis of the human dental pulp and periodontium.37 We here interrogated this dataset to determine the expression of NOTCH receptors and ligands in dental pulp and periodontal MSCs (Figures 1 and 2). In the dental pulp, MSCs were characterized by the higher expression of NOTCH3 (log‐fold change of 1.24 and adjusted P value <.001, compared to other cell types in the pulp) (Figure 1A). NOTCH3 + MSCs also expressed the dental MSCs markers THY1, MYH11, and ACTA2 (Figure 1A).20, 37, 39 NOTCH3‐expressing MSCs were mainly localized around the vessels, where the perivascular niches are formed21, 22 (Figure 1C‐G). Double staining against Notch3 and the endothelial cell marker CD31, followed by cell surface analysis, showed that the Notch3‐positive MSCs (pericytes) are in direct contact with the CD31‐positive endothelial cells (Figure 1H,I). Similar to what observed in the dental pulp, MSCs in the periodontium expressed high levels of NOTCH3 (Figure 2A). NOTCH3‐expressing MSCs were particularly enriched in proximity to the tooth roots (Figure 2B‐D) and often in direct contact with CD31‐positive endothelial cells (Figure 2E).
FIGURE 1.
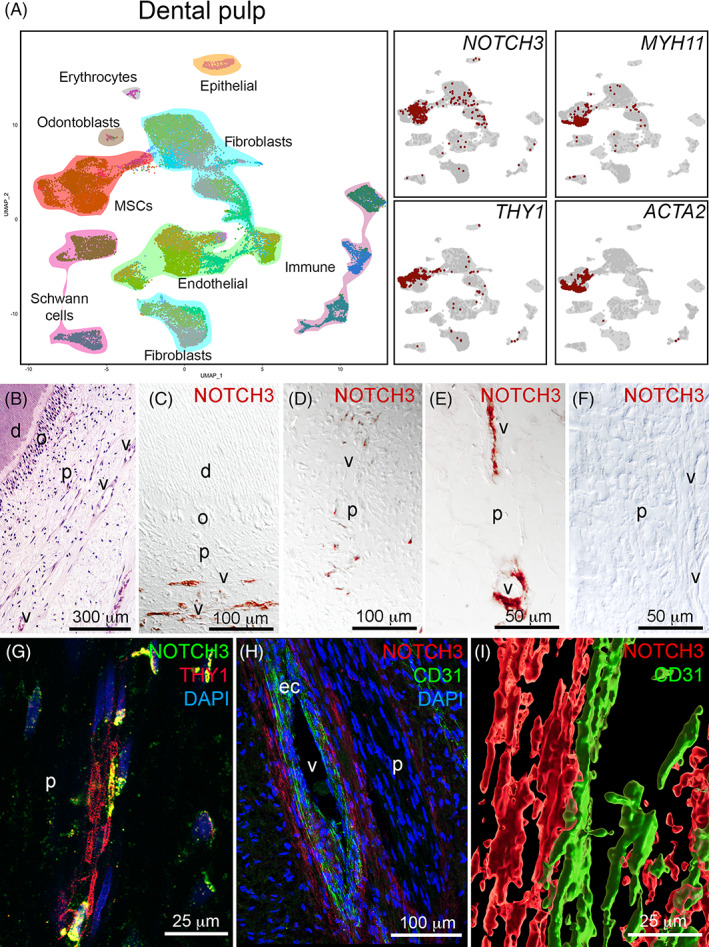
Expression of NOTCH3 in the human dental pulp. A, UMAP visualization of color‐coded clustering of the dental pulp (n > 32 000 cells).37 B, Hematoxylin‐eosin staining showing the structure of the dental pulp. C‐E, Immunohistochemistry against NOTCH3 (red color) in the human dental pulp showing the localization of NOTCH3 in pericytes. F, Negative control for the NOTCH3 staining. G, Immunofluorescent staining showing localization of NOTCH3+ (green color) and THY1+ (red color) mesenchymal stem cells (MSCs). Blue color: DAPI. H, Immunofluorescent staining in the human dental pulp showing NOTCH3 staining in the pericytes (red color) and CD31 staining in endothelial cells (green color). Blue color: DAPI. I, Surface rendering of the relative localization of NOTCH3+ MSCs (red color) and CD31+ endothelial cells (green color). d, dentin; ec, endothelial cells; o, odontoblasts; p, pulp; v, vessels. Scale bars = B = 300 μm; C and D = 100 μm; E and F = 50 μm; G = 25 μm; H = 100 μm; I = 25 μm
FIGURE 2.
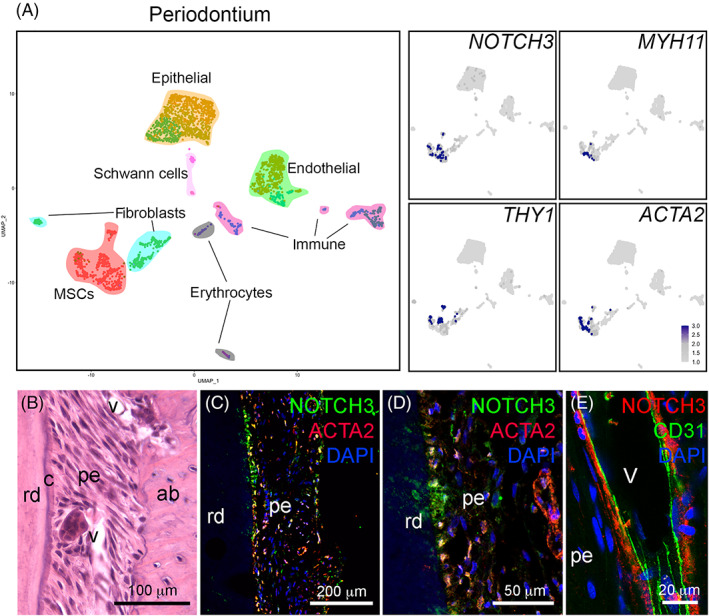
Expression of NOTCH3 in the human periodontium. A, UMAP visualization of color‐coded clustering of the periodontium (n > 2800 cells).37 B, Hematoxylin‐eosin staining showing the structure of the periodontium. C,D, Immunofluorescent staining showing localization of NOTCH3+ (green color) and ACTA2+ (red color) mesenchymal stem cells (MSCs). Blue color: DAPI. E, Immunofluorescent staining showing localization of NOTCH3+ MSCs (red color) and CD31+ endothelial cells (green color). Blue color: DAPI. ab, alveolar bone; c, cementum; pe, periodontium; rd, root dentin; v, vessels. Scale bars = B = 100 μm; C = 200 μm; D = 50 μm; E = 10 μm
3.2. Identification of potential NOTCH signaling activators in MSCs niches
We analyzed the potential interactions between NOTCH ligands and receptors in cells from different clusters. Activation of the NOTCH signaling in human MSCs was indicated by the expression of the NOTCH downstream gene HES1. Expression of HES1 in MSCs was higher than in non‐endothelial cell types in the periodontium and the dental pulp (logFC = 0.77 and P value <.001). Additionally, periodontal MSCs showed a higher expression of MYC and CCND1 (logFC = 0.64 and 0.27, respectively, and P values <.001; Figure 3A), which have been shown to be direct target genes of Notch signaling in specific cell contexts and inducers of cell proliferation.4, 5 Endothelial cells in both the human dental pulp and periodontal tissues expressed all five NOTCH ligands JAG1, JAG2, DLL1, DLL3, DLL4, as well as the NOTCH receptors NOTCH1 and NOTCH4 (Figure 3B,C; dataset). In addition, we observed expression of JAG2, DLL1, and DLL4 in dental pulp fibroblasts, and expression of JAG1 and JAG2 in periodontal fibroblasts and epithelial cells (Figure 3B,C).
FIGURE 3.
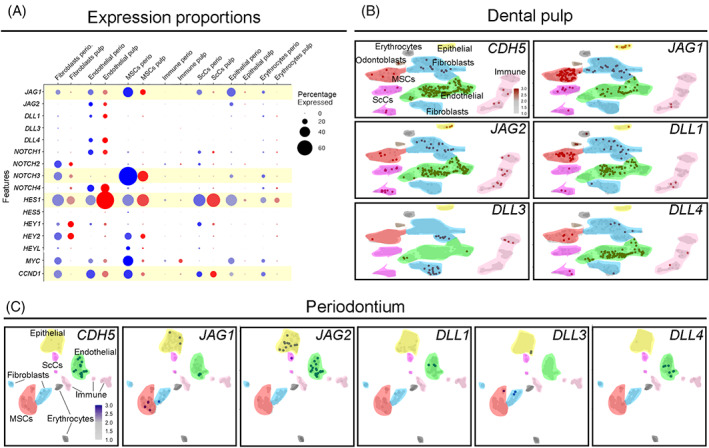
Analysis of the expression of NOTCH receptors and ligands in mesenchymal stem cells (MSCs) and endothelial cells of human teeth. A, Dot‐plot showing the proportion of cells expressing NOTCH ligands, receptors, and target genes. Light yellow highlights genes of interest discussed in detail in the text. The size of the circles is proportional to the percentage of cells expressing the indicated genes. Blue circles represent periodontal cells; red circles represent dental pulp cells. B,C, Feature plots showing expression of Notch ligands in endothelial cells (green color) in the dental pulp (B) and in the periodontium (C) of human teeth
3.3. NOTCH3 expression is associated with MSCs stemness in vitro
We further observed that human dental pulp MSCs maintain NOTCH3 expression when cultured in vitro (Figure 4A). Isolated dental pulp MSCs were multipotent, as they could undergo osteogenic (showed by Alizarin Red staining; Figure 4B), adipogenic (showed by Oil Red O staining; Figure 4C), and chondrogenic differentiation (showed by Alcian Blue staining; Figure 4D). Differentiation was accompanied by upregulation of differentiation markers, and by the significant decrease of NOTCH3 expression (Figure 4E, Fc = −0.86; P‐value <.01). The expression of NOTCH3 significantly decreased already 7 days after the onset of differentiation (Figure 4E). These results indicate that NOTCH3 expression is associated with stemness of human dental MSCs, and that its expression is downregulated early upon MSCs differentiation.
FIGURE 4.
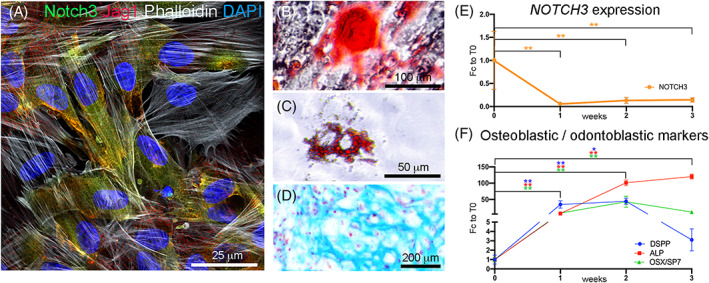
NOTCH3 expression in human dental pulp mesenchymal stem cells (MSCs) in vitro. A, Immunofluorescent staining showing distribution of NOTCH3 (green color), JAG1 (red color), Phalloidin (white color) and DAPI (blue color) in cultured dental pulp MSCs. B‐D, Staining showing multilineage differentiation potential of human dental pulp MSCs. Differentiation was obtained by culturing human dental pulp cells in osteogenic, adipogenic, and chondrogenic media for 3 weeks. B, Alizarin Red staining performed on dental pulp MSCs cultured in osteogenic conditions; C, Oil Red O staining performed on dental pulp MSCs cultured in adipogenic conditions; D, Alcian Blue staining performed on dental pulp MSCs cultured in chondrogenic conditions. E, Real‐time PCR analysis of the expression of NOTCH3 in dental pulp MSCs undergoing osteogenic/odontoblastic differentiation. F, Real‐time PCR analysis of the expression of the osteogenic and odontoblastic differentiation markers DSPP (Dentin Sialophosphoprotein), ALP (Alkaline Phosphatase), and OSX/SP7 (Osterix) in dental pulp MSCs undergoing osteogenic/odontoblastic differentiation. **P‐value <.01; *P‐value <.05. N = 4 for each time point and condition. Error bars indicate standard deviation. Scale bars = A = 25 μm; B = 100 μm; C = 50 μm; D = 200 μm
3.4. Notch3+MSCs are involved in tissue homeostasis in vivo
Based on the identification of NOTCH3 as a general marker for MSCs in the human dental tissues, we investigated the behavior and fate of Notch3‐expressing populations in vivo in a transgenic mouse model. We analyzed Notch3‐expressing cells, as well as their progeny, in homeostatic conditions in intact molars of Notch3‐Cre ER ; R26 mT/mG mice (Figure 5). Notch3‐expressing cells were detected by immunofluorescent staining against GFP. Four days after 4‐OHT induction, Notch3‐expressing cells in the dental pulp were localized around blood vessels and in the subodontoblastic layer (Figure 5C,G). In the periodontium, Notch3‐GFP staining was localized in perivascular cells and in cells close to the dental root (Figure 5C,K), a pattern similar to that already observed in human tissues (Figure 2C,D). A higher number of Notch3‐GFP cells were found 10 days after 4‐OHT induction in both dental pulp and periodontal tissues, indicating the involvement of the Notch3‐GFP cells in tooth homeostasis (Figure 5D,H,L). Notch3‐GFP cells were still detected in the perivascular areas in both dental pulp and periodontal tissues 28 days 4‐OHT postinduction (Figure 5E,I,M).
FIGURE 5.
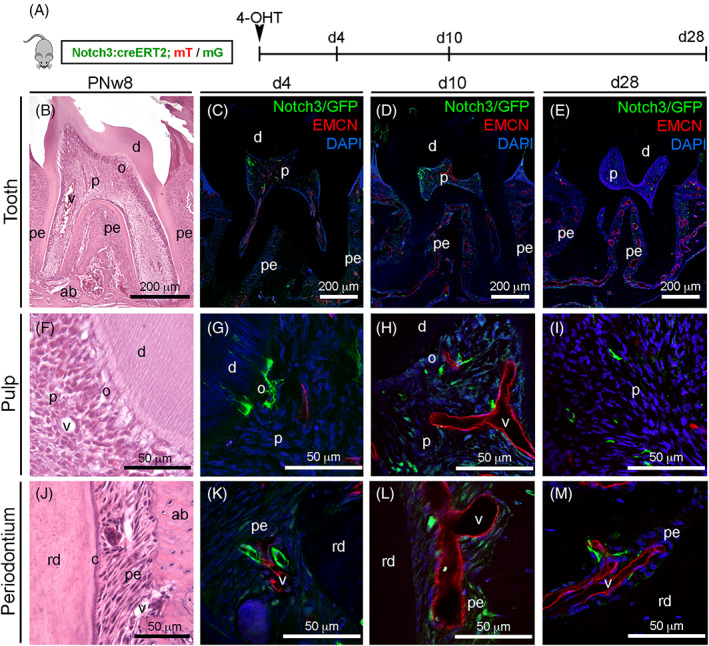
Lineage tracing of Notch3‐expressing cells and their progeny in intact mouse molars. A, Experimental design for lineage tracing in homeostatic conditions. B, Hematoxylin‐Eosin staining showing the first upper molar tooth. C‐E, Immunofluorescent staining showing Notch3‐GFP cells (green color) and endomucin‐positive blood vessels (red color) 4 days (C), 10 days (D), and 28 days (E) after 4‐OHT induction. F, Hematoxylin‐eosin staining showing the dentin‐pulp complex. G‐I, Immunofluorescent staining showing Notch3‐GFP cells (green color) and endomucin‐positive blood vessels (red color) in the dental pulp 4 days (G), 10 days (H) and 28 days (I) after 4‐OHT induction. J, Hematoxylin‐Eosin staining showing the structure of the periodontium. K‐M, Immunofluorescent staining showing Notch3‐GFP cells (green color) and endomucin‐positive blood vessels (red color) in the periodontium 4 days (K), 10 days (L), and 28 days (M) after 4‐OHT induction. ab, alveolar bone; c, cementum; d, dentin; o, odontoblasts; p, pulp; pe, periodontium; rd, root dentin; v, vessels. Scale bars = B‐E = 200 μm; F‐M = 50 μm
3.5. Notch3 + MSCs contribute to tissue regeneration upon injury in vivo
To determine whether Notch3‐expressing MSCs could participate in dental pulp remodeling upon injury, we first injected 4‐OHT to Notch3‐Cre ER ; R26 mT/mG mice, and the following day we performed cavity preparations in the first molars (Figure 6A‐C), and finally we analyzed the distribution of Notch3‐GFP cells in the dental pulp, 4, 10, and 28 days postsurgery. Injuries linked to tooth cavity preparations are associated with activation of the various MSCs populations.10, 40 Significant damage of the dental pulp tissue located under the cavity was visible 4 days after the injury (Figure 6B). Notch3‐GFP+ cells could be already found in pulp regions underlying the injury (Figure 6D). Ten days post‐injury, the number of Notch3‐GFP+ cells increased, with 50% more GFP+ cells compared to the 4 days postinjured dental pulp and 400% more GFP+ cells (P < .05) compared to the dental pulp of intact teeth (Figure 6D,K). At that time, Notch3‐GFP+ cells were not found only in perivascular regions but also in other pulp areas, which spanned from the cavity site to regions of the dental pulp more distant from the injury (Figure 6E,G‐J). Twenty‐eight days postinjury, the total number of Notch3‐GFP+ cells slightly decreased (Figure 6K), with GFP signal still detected in proximity to the injured dental pulp region as well as in perivascular sites (Figure 6C,F). To assess the in vivo differentiation potential of Notch3‐expressing MSCs upon tooth injury, we performed double immunofluorescent staining against molecules specific for the different dental pulp cell types, such as DSPP (a marker of dentinogenic and osteogenic cells), Fibronectin (a marker of fibroblasts), CD45 (a general marker for immune cells), and Myelin Basic Protein (MBP, a marker for Schwann cells). Ten days postinjury, we observed a gradient of Notch3‐GFP distribution within the pulp where the highest signal was detected nearby the injury site and the lowest signal far away in the intact pulp region (Figure 6G, white arrows and arrowheads). We detected several cells that were costained with Notch3‐GFP and DSPP adjacent to the lesion area (Figure 6G, white arrows). In contrast, Notch3‐GFP+ cells did not express markers for fibroblasts (Figure 6H), immune cells (Figure 6I), and glial cells (Figure 6J).
FIGURE 6.
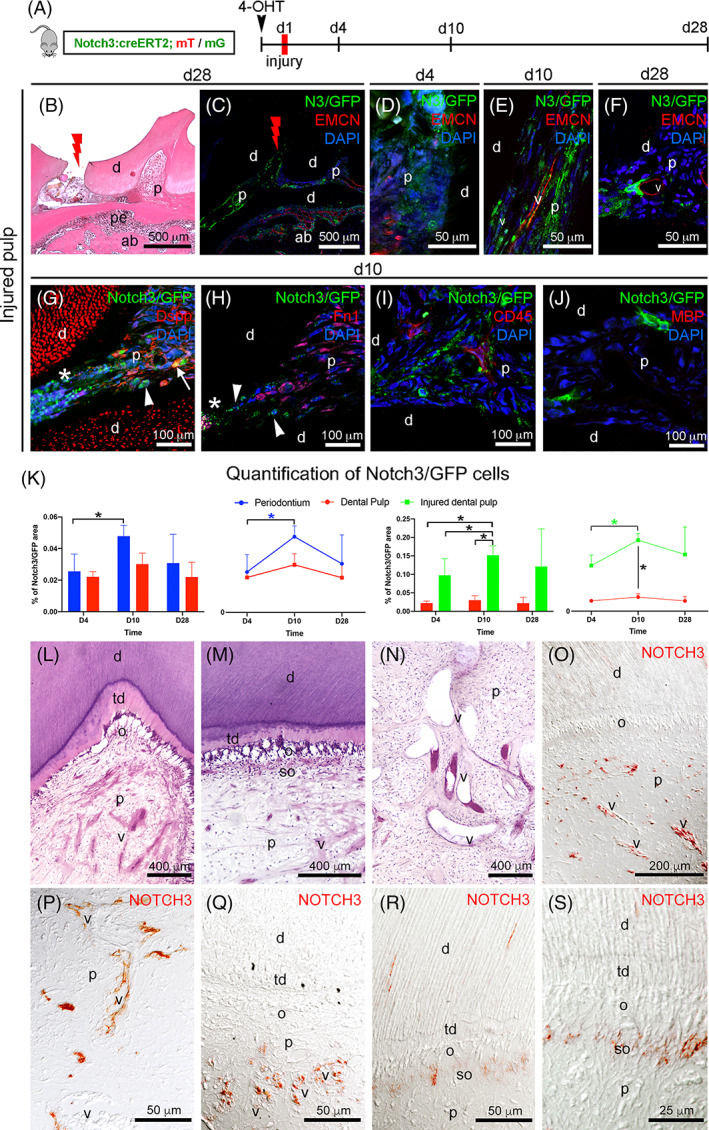
Lineage tracing of Notch3‐expressing cells and their progeny in injured mouse molars, and NOTCH3 distribution in the dental pulp of carious human teeth. A, Experimental design for lineage tracing upon cavity preparation in the first upper molar of transgenic mice. B, Hematoxylin‐eosin staining showing the cavity preparation in the molar. Red lightning marks the area of the cavity preparation. C, Immunofluorescent staining showing an overview of the distribution of Notch3‐GFP (green color) signal in the dental pulp of injured teeth 28 days after cavity preparation (indicated by the red lightning symbol). Red color: endomucin; blue color: DAPI. D, Immunofluorescent staining showing Notch3‐GFP (green color) signal in the region immediately below the cavity, 4 days after injury. Red color: endomucin; blue color: DAPI. E, Immunofluorescent staining showing Notch3‐GFP cells (green color) 10 days after injury. Red color: endomucin; Blue color: DAPI. F, Immunofluorescent staining showing Notch3‐GFP cells (green color) 28 days after injury. Red color: endomucin; Blue color: DAPI. G, Immunofluorescent staining showing Notch3‐GFP cells (green color) and DSPP+ cells (red color) in the injured dental pulp. Arrowheads indicate Notch3‐GFP+ cells; arrows indicate double‐positive Notch3‐GFP+/DSPP+ cells. White asterisk marks the damaged dental pulp. H, Immunofluorescent staining showing Notch3‐GFP cells (green color) and Fn1+ fibroblasts (red color) in the injured dental pulp. Arrowheads indicate Notch3‐GFP+ cells. White asterisk marks the damaged dental pulp. Blue color: DAPI. I, Immunofluorescent staining showing Notch3‐GFP cells (green color) and CD45‐expressing immune cells (red color) in the injured dental pulp. Blue color: DAPI. J, Immunofluorescent staining showing Notch3‐GFP+ cells (green color) and MBP+ Schwann cells (red color) in the injured dental pulp. No MBP+ cells were observed in the injured pulp. Blue color: DAPI. K, Quantification of Notch3‐GFP+ cells in the periodontium, in the healthy dental pulp and in the injured dental pulp 4 days, 10 days and 28 days 4‐OHT postinjection (N = 3 for each time point and condition). L‐N, Hematoxylin‐eosin staining showing different areas of the dentin‐pulp complex in carious human teeth. L, Overview of the dentin‐pulp complex underneath the carious lesion. Notice the deposition of tertiary dentin, the disorganization of the odontoblasts, and the presence of a rich vascular network. M, Picture showing the deposition of tertiary dentin beneath the carious lesion site, the disorganized odontoblasts, and a rich layer of subodontoblastic cells. N, Blood vessels are strongly dilatated in the dental pulp of carious teeth. O‐S, Immunohistochemistry against NOTCH3 (red color) in the carious human dental pulp. O, Overview showing NOTCH3 expression in the dental pulp underneath the carious lesion. P,Q, NOTCH3 is highly expressed by cells located in proximity of dilatated blood vessels. R,S, NOTCH3 is strongly expressed in the subodontoblastic layer underneath the carious front. ab, alveolar bone; d, dentin; o, odontoblasts; p, dental pulp; pe, periodontium; so, subodontoblasts; td, tertiary dentin; v, vessels. Scale bars = B and C = 500 μm; D‐F, P‐R = 50 μm; G‐J = 100 μm; L‐N = 400 μm; O = 200 μm; S = 25 μm. *P‐value <.05. Error bars in J indicate standard deviation
3.6. NOTCH3 distribution in the pulp of carious human teeth
We further analyzed the expression of NOTCH3 in carious human teeth (Figure 6L‐S). In the dentin‐pulp complex area underlying the carious lesion, tertiary dentin was deposited, odontoblasts were strongly disorganized and blood vessels were significantly dilatated (Figure 6L‐N). We observed that NOTCH3 was more widely distributed in the pulps of carious teeth (Figure 6O‐S) when compared to the healthy dental pulps (Figure 1C‐E). NOTCH3 protein was not exclusively detected around blood vessels (Figure 6P,Q), but also in the subodontoblastic cell layer of cells underlying the lesion site (Figure 6R,S) and sometimes in the odontoblastic processes within dentinal tubules (Figure 6R).
4. DISCUSSION
Understanding the mechanisms that mediate the interaction between stem cells and their niches is of paramount importance for the development of stem cell‐based regenerative therapies. We showed that MSCs in human dental tissues are characterized by the expression of NOTCH3, which makes these cells responsive to niche cells expressing NOTCH ligands. Previous studies have shown that the expression of NOTCH3 in perivascular MSCs is not limited to teeth,22, 23, 41 since its expression has been reported in many other tissues such as the brain,42 the bone marrow,43 and joints.44 Lineage tracing experiments in transgenic mice showed that Notch3‐expressing perivascular MSCs cells are important in both dental tissue homeostasis and regeneration. From our data, it appears that Notch3‐GFP cells are relatively quiescent in homeostatic conditions, at least on a time span of 28 days, while they are moderately active in the periodontium, a tissue subject to continuous remodeling in response to various stresses associated with mastication.18, 45, 46, 47, 48 However, in the injured teeth, Notch3‐expressing MSCs are strongly activated in the dental pulp. Dental injuries can lead to the odontoblastic death, thus inducing a regenerative pulp reaction involving the generation of new odontoblasts (also called odontoblast‐like cells), which are derived from MSCs, are responsible for the formation of the tertiary dentin, and express dentin sialophosphoprotein (DSPP), a key component of the dentin matrix.49, 50 Indeed, we observed that, upon tooth injury in mice, Notch3‐expressing cells accumulate close to the injured site and give rise to DSPP‐producing odontoblast‐like cells. Conversely, Notch3‐expressing MSCs did not appear to give rise to other dental pulp cell types, such as activated fibroblasts (marked by the expression of Fn1), immune cells (CD45+), or Schwann cells (MBP+). Similar results obtained in carious human teeth, where distribution of the NOTCH3 protein was correlated with vascular structures and subodontoblastic cells, which substitute the damaged odontoblasts and differentiate into odontoblast‐like cells. Therefore, Notch3‐expressing MSCs are essentially predisposed to differentiate into hard tissue forming cells, contributing to the remodeling and healing of the injured pulp‐dentin complex. In our recent work, we showed that the similarity of MSCs in the dental pulp and the periodontium is counteracted by a great divergence in the composition of their niches.37 However, previous studies from other laboratories had identified dental pulp and periodontal MSCs as divergent cell populations with distinctive properties.51, 52, 53 Therefore, we investigated how tissue‐specific niches could modulate the behavior and fates of similar dental MSCs. Our analysis showed that endothelial cells from the dental pulp and periodontal tissues expressed the NOTCH1 receptor and all the NOTCH ligands (JAG1, JAG2, DLL1, DLL3, DLL4; Figure 7). The expression of the NOTCH3 receptor by perivascular cells, combined with the high expression of NOTCH ligands by endothelial cells, indicate the existence of a Notch‐dependent perivascular MSC niche in dental tissues, as already suggested in previous studies in injured rodent teeth.10, 22 The crosstalk between endothelial and perivascular cells is a conserved feature within different organs and tissues. Signals originating from endothelial cells have been shown to direct the fate of perivascular stem cell populations in diverse tissues such as the brain, bone, bone marrow, and liver.54 Among these signals, Notch ligands expressed by endothelial cells play a primary role in modulating the activity of MSCs localized within perivascular niches.6, 55, 56 NOTCH3 in particular is a primary receptor involved in the crosstalk between endothelial cells and MSCs. Signals derived from endothelial cells maintain the quiescence of perivascular neural stem cells in the brain via Notch3 activation.57 Similarly, endothelial cells‐derived JAG1 is a key modulator of NOTCH3‐expressing MSCs differentiation in the bone marrow.43 Conversely, NOTCH3 activity in perivascular cells is necessary for the proper maturation of arteries,58 and mutations affecting NOTCH3 function are associated with CADASIL, a genetic diseases characterized by degeneration of small brain capillaries and multiple small infarcts.59 It is therefore clear that the Notch‐mediated crosstalk between endothelial and perivascular cells possesses bidirectional functions important for proper vessels formation and the activation of perivascular MSCs. Relevant differences exist however within the human dental pulp and periodontal tissues in their cellular and molecular composition,37 which could represent important cues guiding MSCs activation in various physiological or pathological stimuli (Figure 7).
FIGURE 7.
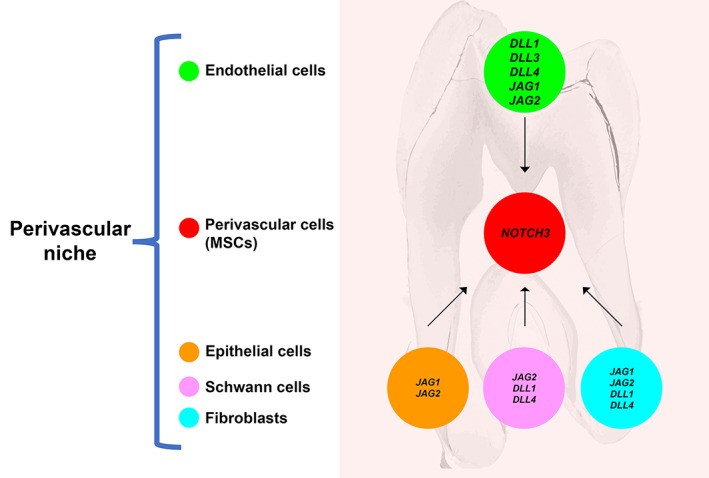
Model showing how Notch signaling is involved in the interactions between mesenchymal stem cells (MSCs) and their niches in dental tissues. MSCs in the human dental pulp and the periodontium express NOTCH3, are highly similar, and are composed by identical subpopulations. In both tissues, NOTCH ligands expressed by endothelial cells establish a communication mechanism for NOTCH3‐expressing pericytes. Among the few genes differentially expressed between dental pulp and periodontal MSCs, HES1 and MYC indicate different activation of Notch signaling and cell cycle. This can be due to the differences in the cellular compositions of the dental pulp and periodontal microenvironment
Previous in vitro studies support the functional importance of Notch signaling in human dental MSCs behavior.36, 60, 61, 62 Indeed, inhibition of Notch signaling leads to a loss of their stemness,61 while JAG1‐dependent activation of the Notch signaling maintains their undifferentiated status.36 On the contrary, DLL1‐dependent Notch signaling activation induces MSCs differentiation into odontoblasts.60 The diverse effects exerted by the different Notch ligands highlight the great importance of this pathway in the interplay between MSCs and their niches. Divergences in the expression of NOTCH ligands in the various niches may privilege the differentiation of human MSCs toward an osteogenic fate in the dental pulp, and a fibroblastic fate in the periodontium. Previous studies have shown that pharmacological blockage of Notch signaling in MSCs induces loss of their stemness, and that Notch signaling activation via Jag1 blocks their differentiation.36 These data point toward the conclusion that inhibition of Notch signaling facilitates MSCs differentiation. Nevertheless, in vivo experiments demonstrating the effects of Notch signaling, upon either its stimulation or inhibition, on dental MSCs behavior are still missing, and therefore no evidence exists to definitely prove that Notch downregulation is required to lead to MSCs differentiation.
In conclusion, the present results highlight the importance of Notch signaling in the interaction between MSCs and their niches. Endothelial cells in human dental tissues express all five NOTCH ligands and constitute conserved perivascular niches, which can regulate the activation of NOTCH3‐expressing perivascular MSCs. This fundamental information establishes a new conceptual framework that could be the basis for future research focusing toward MSCs‐based regenerative approaches.
AUTHOR CONTRIBUTIONS
P.P.: conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript; L.d.V.R., A.E.M.: collection and/or assembly of data, data analysis and interpretation, final approval of manuscript; B.S.: provision of study material from patients, data analysis and interpretation, final approval of manuscript; T.A.M.: conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing, financial support, administrative support, provision of study material or patients, final approval of manuscript.
CONFLICT OF INTERESTS
The authors declared no potential conflicts of interest.
ACKNOWLEDGMENTS
We thank Prof. Spyros Artavanis‐Tsakonas (Harvard Medical School, Harvard University, Boston, Massachusetts) and Dr. Silvia Fre (Institut Curie, CNRS, Paris, France) for providing the Notch3 creER mice. We thank Prof. Giovanna Orsini (Polytechnic University of Marche, Italy) for the technical assistance in tooth cavity preparations and Dr. Cristina Porcheri (Institute for Oral Biology, University of Zurich) for assistance in imaging. Imaging was performed with equipment maintained by the Center for Microscopy and Image Analysis, University of Zurich. This work was supported by institutional funds from the University of Zurich and by the Swiss National Science Foundation (310030_197782).
Pagella P, de Vargas Roditi L, Stadlinger B, Moor AE, Mitsiadis TA. Notch signaling in the dynamics of perivascular stem cells and their niches. STEM CELLS Transl Med. 2021;10(10):1433–1445. 10.1002/sctm.21-0086
Funding information Swiss National Science Foundation, Grant/Award Number: 310030_197782; University of Zurich
DATA AVAILABILITY STATEMENT
Data can be found at GEO: GSE161267. All code is publicly available at: https://github.com/TheMoorLab/Tooth. Single cell RNA sequencing data are described previously.37
REFERENCES
- 1.Ho DM, Artavanis‐Tsakonas S. The notch‐mediated proliferation circuitry. Curr Top Dev Biol. 2016;116:17‐33. [DOI] [PubMed] [Google Scholar]
- 2.Guruharsha KG, Kankel MW, Artavanis‐Tsakonas S. The notch signalling system: recent insights into the complexity of a conserved pathway. Nat Rev Genet. 2012;13:654‐666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siebel C, Lendahl U. Notch signaling in development, tissue homeostasis, and disease. Physiol Rev. 2017;97:1235‐1294. [DOI] [PubMed] [Google Scholar]
- 4.Cohen B, Shimizu M, Izrailit J, et al. Cyclin D1 is a direct target of JAG1‐mediated notch signaling in breast cancer. Breast Cancer Res Treat. 2010;123:113‐124. [DOI] [PubMed] [Google Scholar]
- 5.Palomero T, Lim WK, Odom DT, et al. NOTCH1 directly regulates c‐MYC and activates a feed‐forward‐loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci USA. 2006;103:18261‐18266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tikhonova AN, Dolgalev I, Hu H, et al. The bone marrow microenvironment at single‐cell resolution. Nature. 2019;569:222‐228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koch U, Lehal R, Radtke F. Stem cells living with a notch. Development. 2013;140:689‐704. [DOI] [PubMed] [Google Scholar]
- 8.Andersson ER, Sandberg R, Lendahl U. Notch signaling: simplicity in design, versatility in function. Development. 2011;138:3593‐3612. [DOI] [PubMed] [Google Scholar]
- 9.Mitsiadis TA, Feki A, Papaccio G, et al. Dental pulp stem cells, niches, and notch signaling in tooth injury. Adv Dent Res. 2011;23:275‐279. [DOI] [PubMed] [Google Scholar]
- 10.Mitsiadis TA, Caton J, Pagella P, et al. Monitoring notch signaling‐associated activation of stem cell niches within injured dental pulp. Front Physiol. 2017;8:372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitsiadis TA, Roméas A, Lendahl U, Sharpe PT, Farges JC. Notch2 protein distribution in human teeth under normal and pathological conditions. Exp Cell Res. 2003;282(2):101‐109. 10.1016/s0014-4827(02)00012-5. [DOI] [PubMed] [Google Scholar]
- 12.Kollar EJ. Tissue interactions in development of teeth and related ectodermal derivatives. Dev Biol (N Y 1985). 1986;4:297‐313. [DOI] [PubMed] [Google Scholar]
- 13.Pagella P, Porcheri C, Mitsiadis TA. Exploiting teeth as a model to study basic features of signaling pathways. Biochem Soc Trans. 2020;48:2729‐2742. [DOI] [PubMed] [Google Scholar]
- 14.Nanci A. Ten Cate's Oral Histology. St. Louis, Missouri: Elsevier; 2013:379. [Google Scholar]
- 15.Mitsiadis TA, Graf D. Cell fate determination during tooth development and regeneration. Birth Defects Res C Embryo Today. 2009;87:199‐211. [DOI] [PubMed] [Google Scholar]
- 16.Diekwisch TG. The developmental biology of cementum. Int J Dev Biol. 2001;45:695‐706. [PubMed] [Google Scholar]
- 17.Gronthos S, Mankani M, Brahim J, et al. Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proc Natl Acad Sci USA. 2000;97:13625‐13630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roguljic H, Matthews BG, Yang W, et al. In vivo identification of periodontal progenitor cells. J Dent Res. 2013;92:709‐715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oh M, Zhang Z, Mantesso A, et al. Endothelial‐initiated crosstalk regulates dental pulp stem cell self‐renewal. J Dent Res. 2020;99:1102‐1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vidovic I, Banerjee A, Fatahi R, et al. alphaSMA‐expressing perivascular cells represent dental pulp progenitors in vivo. J Dent Res. 2017;96:323‐330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi S, Gronthos S. Perivascular niche of postnatal mesenchymal stem cells in human bone marrow and dental pulp. J Bone Miner Res. 2003;18:696‐704. [DOI] [PubMed] [Google Scholar]
- 22.Lovschall H, Mitsiadis TA, Poulsen K, et al. Coexpression of Notch3 and Rgs5 in the pericyte‐vascular smooth muscle cell axis in response to pulp injury. Int J Dev Biol. 2007;51:715‐721. [DOI] [PubMed] [Google Scholar]
- 23.Jamal M, Chogle SM, Karam SM, et al. NOTCH3 is expressed in human apical papilla and in subpopulations of stem cells isolated from the tissue. Genes Dis. 2015;2:261‐267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaltz N, Ringe J, Holzwarth C, et al. Novel markers of mesenchymal stem cells defined by genome‐wide gene expression analysis of stromal cells from different sources. Exp Cell Res. 2010;316:2609‐2617. [DOI] [PubMed] [Google Scholar]
- 25.Athanassiou‐Papaefthymiou M, Papagerakis P, Papagerakis S. Isolation and characterization of human adult epithelial stem cells from the periodontal ligament. J Dent Res. 2015;94:1591‐1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Orsini G, Pagella P, Mitsiadis TA. Modern trends in dental medicine: an update for internists. Am J Med. 2018;131:1425‐1430. [DOI] [PubMed] [Google Scholar]
- 27.Lane SW, Williams DA, Watt FM. Modulating the stem cell niche for tissue regeneration. Nat Biotechnol. 2014;32:795‐803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu XY, Li X, Wang J, et al. Concise review: periodontal tissue regeneration using stem cells: strategies and translational considerations. Stem Cells Translational Medicine. 2019;8:392‐403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen H, Fu H, Wu X, et al. Regeneration of pulpo‐dentinal‐like complex by a group of unique multipotent CD24a(+) stem cells. Sci Adv. 2020;6:eaay1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xuan K, Li B, Guo H, et al. Deciduous autologous tooth stem cells regenerate dental pulp after implantation into injured teeth. Sci Transl Med. 2018;10(455):eaaf3227. 10.1126/scitranslmed.aaf3227. [DOI] [PubMed] [Google Scholar]
- 31.Mitsiadis TA, Lardelli M, Lendahl U, et al. Expression of notch 1, 2 and 3 is regulated by epithelial‐mesenchymal interactions and retinoic acid in the developing mouse tooth and associated with determination of ameloblast cell fate. J Cell Biol. 1995;130:407‐418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitsiadis TA, Henrique D, Thesleff I, et al. Mouse Serrate‐1 (Jagged‐1): expression in the developing tooth is regulated by epithelial‐mesenchymal interactions and fibroblast growth factor‐4. Development. 1997;124:1473‐1483. [DOI] [PubMed] [Google Scholar]
- 33.Mitsiadis TA, Hirsinger E, Lendahl U, et al. Delta‐notch signaling in odontogenesis: correlation with cytodifferentiation and evidence for feedback regulation. Dev Biol. 1998;204:420‐431. [DOI] [PubMed] [Google Scholar]
- 34.Mitsiadis TA, Fried K, Goridis C. Reactivation of delta‐notch signaling after injury: complementary expression patterns of ligand and receptor in dental pulp. Exp Cell Res. 1999;246:312‐318. [DOI] [PubMed] [Google Scholar]
- 35.Denes BJ, Bolton C, Illsley CS, et al. Notch coordinates periodontal ligament maturation through regulating Lamin A. J Dent Res. 2019;98:1357‐1366. [DOI] [PubMed] [Google Scholar]
- 36.Zhang C, Chang J, Sonoyama W, et al. Inhibition of human dental pulp stem cell differentiation by notch signaling. J Dent Res. 2008;87:250‐255. [DOI] [PubMed] [Google Scholar]
- 37.Pagella P, de Vargas Roditi L, Stadlinger B, et al. A single cell atlas of human teeth. iScience. 2021;24:102405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mahler Convenor M, Berard M, Feinstein R, et al. FELASA recommendations for the health monitoring of mouse, rat, hamster, guinea pig and rabbit colonies in breeding and experimental units. Lab Anim. 2014;48:178‐192. [DOI] [PubMed] [Google Scholar]
- 39.Dominici M, Le Blanc K, Mueller I, et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315‐317. [DOI] [PubMed] [Google Scholar]
- 40.Mitsiadis TA, De Bari C, About I. Apoptosis in developmental and repair‐related human tooth remodeling: a view from the inside. Exp Cell Res. 2008;314:869‐877. [DOI] [PubMed] [Google Scholar]
- 41.Wang Y, Pan L, Moens CB, et al. Notch3 establishes brain vascular integrity by regulating pericyte number. Development. 2014;141:307‐317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vanlandewijck M, He L, Mäe MA, et al. A molecular atlas of cell types and zonation in the brain vasculature. Nature. 2018;554:475‐480. [DOI] [PubMed] [Google Scholar]
- 43.Blache U, Vallmajo‐Martin Q, Horton ER, et al. Notch‐inducing hydrogels reveal a perivascular switch of mesenchymal stem cell fate. EMBO reports. 2018;19(8):e45964. 10.15252/embr.201845964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei K, Korsunsky I, Marshall JL, et al. Notch signalling drives synovial fibroblast identity and arthritis pathology. Nature. 2020;582:259‐264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Howard PS, Kucich U, Taliwal R, et al. Mechanical forces alter extracellular matrix synthesis by human periodontal ligament fibroblasts. J Periodontal Res. 1998;33:500‐508. [DOI] [PubMed] [Google Scholar]
- 46.Smith PC, Martinez C, Martinez J, et al. Role of fibroblast populations in periodontal wound healing and tissue remodeling. Front Physiol. 2019;10:270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sodek J, Overall CM. Matrix metalloproteinases in periodontal tissue remodelling. Matrix Suppl. 1992;1:352‐362. [PubMed] [Google Scholar]
- 48.Xu Q, Yuan X, Zhang X, et al. Mechanoadaptive responses in the periodontium are coordinated by Wnt. J Dent Res. 2019;98:689‐697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.D'Souza RN, Cavender A, Sunavala G, et al. Gene expression patterns of murine dentin matrix protein 1 (Dmp1) and dentin sialophosphoprotein (DSPP) suggest distinct developmental functions in vivo. J Bone Miner Res. 1997;12:2040‐2049. [DOI] [PubMed] [Google Scholar]
- 50.Liang T, Zhang H, Xu Q, et al. Mutant dentin sialophosphoprotein causes dentinogenesis imperfecta. J Dent Res. 2019;98:912‐919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Otabe K, Muneta T, Kawashima N, et al. Comparison of gingiva, dental pulp, and periodontal ligament cells from the standpoint of mesenchymal stem cell properties. Cell Med. 2012;4:13‐21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gopinathan G, Foyle D, Luan X, et al. The Wnt antagonist SFRP1: a key regulator of periodontal mineral homeostasis. Stem Cells Dev. 2019;28:1004‐1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamada S, Tomoeda M, Ozawa Y, et al. PLAP‐1/asporin, a novel negative regulator of periodontal ligament mineralization. J Biol Chem. 2007;282:23070‐23080. [DOI] [PubMed] [Google Scholar]
- 54.Rafii S, Butler JM, Ding BS. Angiocrine functions of organ‐specific endothelial cells. Nature. 2016;529:316‐325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Poulos MG, Guo P, Kofler NM, et al. Endothelial Jagged‐1 is necessary for homeostatic and regenerative hematopoiesis. Cell Rep. 2013;4:1022‐1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ottone C, Krusche B, Whitby A, et al. Direct cell‐cell contact with the vascular niche maintains quiescent neural stem cells. Nat Cell Biol. 2014;16:1045‐1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kawai H, Kawaguchi D, Kuebrich BD, et al. Area‐specific regulation of quiescent neural stem cells by Notch3 in the adult mouse subependymal zone. J Neurosci. 2017;37:11867‐11880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Domenga V, Fardoux P, Lacombe P, et al. Notch3 is required for arterial identity and maturation of vascular smooth muscle cells. Genes Dev. 2004;18:2730‐2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Prakash N, Hansson E, Betsholtz C, et al. Mouse notch 3 expression in the pre‐ and postnatal brain: relationship to the stroke and dementia syndrome CADASIL. Exp Cell Res. 2002;278:31‐44. [DOI] [PubMed] [Google Scholar]
- 60.He F, Yang Z, Tan Y, et al. Effects of notch ligand Delta1 on the proliferation and differentiation of human dental pulp stem cells in vitro. Arch Oral Biol. 2009;54:216‐222. [DOI] [PubMed] [Google Scholar]
- 61.Uribe‐Etxebarria V, Luzuriaga J, Garcia‐Gallastegui P, et al. Notch/Wnt cross‐signalling regulates stemness of dental pulp stem cells through expression of neural crest and core pluripotency factors. Eur Cell Mater. 2017;34:249‐270. [DOI] [PubMed] [Google Scholar]
- 62.Osathanon T, Ritprajak P, Nowwarote N, et al. Surface‐bound orientated Jagged‐1 enhances osteogenic differentiation of human periodontal ligament‐derived mesenchymal stem cells. J Biomed Mater Res A. 2013;101:358‐367. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data can be found at GEO; Access number: GSE161267. All code is publicly available at: https://github.com/TheMoorLab/Tooth. Single cell RNA sequencing data are described in detail in Reference 37.
Data can be found at GEO: GSE161267. All code is publicly available at: https://github.com/TheMoorLab/Tooth. Single cell RNA sequencing data are described previously.37


