Abstract
A key driver of patients’ well-being and clinical trials for Parkinson’s disease (PD) is the course disease takes over time (progression and prognosis). To assess how genetic variation influences the progression of PD over time to dementia (PDD), a major determinant for quality of life, we performed a genome-wide survival study (GWSS) of 11.2 million variants in 3,821 PD patients over 31,053 longitudinal visits. We discover and replicate RIMS2 as a progression locus (P = 2.78 × 10−11; hazard ratio (HR) = 4.77), identify suggestive evidence for TMEM108 (HR = 2.86, P = 2.09 × 10−8) and WWOX (HR = 2.12, P = 2.37 × 10−8), and confirm associations for GBA (HR = 1.93, P = 0.0002) and APOE (HR = 1.48, P = 0.001). Polygenic progression scores exhibit a substantial aggregate association with dementia risk, while polygenic susceptibility scores are not predictive. This study identifies a novel synaptic locus and polygenic score for cognitive disease progression in PD and proposes diverging genetic architectures of progression and susceptibility.
The past decade has seen success in identifying genetic variants linked to susceptibility for common disease from genome-wide association studies (GWAS) through time-static, two-group comparisons of unaffected controls and cases captured at one single snapshot of time1,2. The genetic architecture of progression and prognosis, which are fundamental for patients, has not been established. For Parkinson’s disease (PD), the pace of deterioration varies dramatically between patients3, but most genome-wide analyses do not capture the time dimension. Which genes determine whether a patient will have an aggressive or benign course, and which variants influence who will develop dementia? To shift from the genetics of susceptibility to precision medicine, longitudinal designs4 are needed that examine the critical time dimension and provide information about individual change5.
Patients with PD are projected to double to 14 million worldwide by 20406. The pace of progression varies considerably between patients3,7–9. Parkinson’s disease dementia (PDD) is one of the most debilitating manifestations of disease progression in PD10 with greatest influence on quality of life10, caregivers and health costs11. In clinical trials, the heterogeneity of progression rates obfuscates drug effects. None of the existing PD therapies slow the underlying neuropathology, which relentlessly advances from brainstem to cortex12 and clinically correlates with progression from motor to cognitive symptoms13.
Limited evidence exists on the genetic architecture of cognitive decline in PD beyond the GBA locus established by us7,8,14 and others15. APOE is implicated chiefly based on cross-sectional studies16. Evidence for other candidate genes and GWAS-derived susceptibility variants is controversial (e.g. LRRK217, SNCA18,19, MAPT14,20 and others21,22).
We determined the effects of 11.2 million deeply imputed variants on cognitive decline in 4,872 patients with PD who were prospectively assessed with 36,123 study visits in 15 cohorts14,23–31 from North America and Europe between 1986 and 2017 (Supplementary Fig. 1 and Supplementary Table 1). We evaluated thousands more patients, tens of thousands more follow-up visits, and millions more SNPs than previous longitudinal explorations8,21,22, and confirmed associations in an independent replication population. We performed whole genome genotyping on our cohorts with the new generation, high-density Illumina Infinium Multi-Ethnic Global Array that harnesses content from Phase 3 of the 1000 Genomes Project32 and transethnic tagging strategies to maximize imputation accuracy for low-frequency variants. Imputation33–36 provides power of detection comparable to whole genome sequencing (WGS) for low frequency (minor allele frequency (MAF) ≥ 1% but < 5%)36 and common variants36. We genotyped 1.8 million variants and imputed 11.2 million variants (Methods and Supplementary Fig. 2). Concordance of imputation compared to WGS was 99.4% based on 562 samples probed with both methods (Supplementary Fig. 3).
4,491 samples passed genotyping quality control. Patients were left-censored, and those with missing or non-quality clinical data were excluded (n = 670, Extended Data Fig. 1). To identify genetic variants associated with progression from PD to Parkinson’s disease dementia (PDD; Supplementary Table 2), we performed a longitudinal GWSS (Fig. 1 and Methods) on the remaining 3,821 patients. We assigned 2,650 patients and 11,744 visits to the discovery population. 1,171 patients and 19,309 visits comprised the replication population. We employed GWSS to estimate the influence of common and low-frequency genetic variants on time from the onset of PD to progression to the endpoint of PD dementia (PDD). We performed Cox proportional hazards analyses adjusting for age at onset, gender, years of education at enrollment, ten principal components of genetic population substructure, and a “cohort” term as a random effect (frailty model37). Physicians recruited and longitudinally assessed the participants without knowledge of their genotypes.
Fig. 1 |. Within-cases longitudinal genome-wide survival study identifies three loci associated with progression to Parkinson’s disease dementia (PDD).
a, Manhattan plot of the genome-wide survival analyses. -log10(P value) from the Cox proportional hazards (Cox PH) model with two-sided Wald test for 12-year survival free of dementia are plotted against chromosomal position for the combined population (n = 3,821 cases with PD tracked in 31,053 longitudinal visits for up to 12 years). Each point represents a SNP. The dashed red line corresponds to the genome-wide significance threshold. b, Covariate-adjusted survival curves for PD patients without the RIMS2 rs182987047 variant (cyan line) and for those carrying the variant (magenta dashed line). Cox PH model with two-sided Wald test. c, Adjusted mean MMSE scores across time predicted from the estimated fixed-effect parameters in the LMM analysis are shown for cases carrying the RIMS2 rs182987047 variant (magenta) and cases without the variant (non-carriers; cyan) adjusting for covariates. Shaded ribbons indicate +/− standard error of the mean (s.e.m.) across time. P values from LMM.
An association signal in the RIMS2 locus reached genome-wide significance in the discovery population and was confirmed in the replication population (Fig. 1 and Table 1). The genomic control inflation factor (𝛌GC) was 1.067 in the combined analysis (Supplementary Fig. 4), and the LD Score regression intercept was lower (1.057)38, consistent with a contribution from polygenicity to inflation38,39. The lead variant rs182987047 in the RIMS2 locus (NC_000008.10:g.105249272A>T; Extended Data Fig. 2) was associated with progression to PDD with a hazard ratio (HR) = 4.74 (95% CI 2.87–7.83) and P = 1.16 × 10−9 in the discovery cohort. This was replicated in the replication population with a HR = 6.2 (95% CI 1.78–21.29) with P = 0.004). In the combined analysis, the lead RIMS2 variant showed HR = 4.77 (95% CI 3.01–7.56) with P = 2.78 × 10−11 (Fig. 1a,b). Another linked variant in this locus (rs116918991; NC_000008.10:g.105158401G>A; correlated with r2 = 0.49) also achieved genome-wide significance in the combined analysis with P = 5.21 × 10−9 (Extended Data Fig. 3). We next investigated whether a different measure of longitudinal cognitive function would confirm this association. Generalized linear mixed model meta-analysis (LMM) of serial Mini Mental State Exam (MMSE) scores, a measure of global cognitive function in PD40, in carriers compared to non-carriers confirmed these results. Serial MMSE scores in patients carrying the lead RIMS2 variant declined more rapidly over time compared to patients who were non-carriers with P = 0.0014 (Fig. 1c) in the LMM adjusting for fixed covariates of age, gender, disease duration upon enrollment, years of education, ten principal components, and random effects (Methods). The RIMS2 variant was not associated with motor progression (Supplementary Fig. 5), possibly due to power and design limitations (confounding from PD medications, which treat motor symptoms53, but not dementia).
Table 1 |.
Variants linked to progression from PD to PDD
| Chr. | Position (Mb) | SNP | Risk allele | RAF | HR | 95% CI | P discovery | P replication | P combined | Nearest gene |
|---|---|---|---|---|---|---|---|---|---|---|
| 8 | 105.25 | rs182987047 | T | 0.013 | 4.77 | 3.01–7.56 | 1.16 × 10 −9 | 4.14 × 10 −3 | 2.78 × 10 −11 | RIMS2 |
| 3 | 132.99 | rs138073281 | C | 0.017 | 2.86 | 1.98–4.13 | 3.43 × 10−5 | 4.23 × 10−5 | 2.09 × 10−8 | TMEM108 |
| 16 | 78.28 | rs8050111 | G | 0.066 | 2.12 | 1.63–2.75 | 1.08 × 10−6 | 0.01 | 2.37 × 10−8 | WWOX |
Hazard ratio for developing PDD in PD patients carrying a risk allele. The three variants were imputed; imputation accuracy was confirmed by whole genome sequencing. Chr, chromosome; RAF, risk allele frequency; HR, hazard ratio from the combined analysis; 95% CI, 95% confidence interval of HR; P values from Cox PH models with two-sided Wald test.
RIMS2 (chr. 8) encodes the Regulating Synaptic Membrane Exocytosis 2 protein, a RIM family member, involved in docking and priming of presynaptic vesicles41,42. Mutations in RIMS2 cause cone-rod synaptic disorder syndrome (MIM 618970)43. In mice, knockout of the RIMS2 ortholog leads to critical defects in memory44. The paralog RIMS1 (chr. 6) is a PD susceptibility locus45 that was not associated with progression. Human RIMS2 showed preferential expression in brain compared to 53 tissues (GTEx46 V7; Extended Data Fig. 4) with high expression in dopamine and pyramidal neurons laser-captured from 86 and 13 human brains, respectively (BRAINcode47; Extended Data Fig. 5).
Two suggestive association signals were located in Transmembrane Protein 108 (TMEM108; NC_000003.11:g.132985956A>C) and WW Domain Containing Oxidoreductase (WWOX; NC_000016.9:g.78281160A>G) loci, respectively (Fig. 1a). These loci achieved genome-wide significance (P < 5 × 10−8) in the combined analysis of discovery and replication populations with suggestive P < 5 × 10−5 in discovery and P < 0.05 in the replication cohort (Table 1). These two loci can now be prioritized for further evaluation. Six additional loci reached genome-wide significance in the discovery cohort but were not replicated; Supplementary Table 3 and Supplementary Fig. 4). The rs138073281 variant in the TMEM108 locus, which is implicated in synaptic spine formation48 and cognition48, was associated with progression to PDD with HR = 2.86 (95% CI 1.98–4.13) and P = 2.09 × 10−8 in the combined Cox analysis (Table 1). LMM meta-analysis confirmed that patients carrying the TMEM108 variant had a more rapid decline in serial MMSE scores compared to non-carriers with P = 0.0019 (Extended Data Fig. 3). The WWOX locus had a HR = 2.12 (95% CI 1.63–2.75) and P = 2.37 × 10-8. WWOX is mutated in autosomal recessive ataxia with mental retardation and epilepsy49 and was associated with Alzheimer’s disease50 while this manuscript was in preparation. Patients carrying the WWOX variant had a more rapid longitudinal cognitive decline in MMSE scores compared to non-carriers with P = 0.009 in the LMM analysis (Extended Data Fig. 3). TMEM108 and WWOX are both expressed in human brain (Extended Data Fig. 4) and specifically in dopamine and pyramidal neurons47 (Extended Data Fig. 5).
The RIMS2 locus and the suggestive prognosis-associated loci have not been associated with PD susceptibility in any case-control GWAS, including large meta-analyses, which reported non-significant P values for the three variants45. Thus, if they were to modulate disease susceptibility, their effect sizes would likely be very modest. Because sub-threshold variants may contribute to genetic architecture51, we examined 505 sub-threshold progression variants (P <10−5 and > 5 × 10−8 in the combined analysis) for overlap with susceptibility variants45. None of the sub-threshold progression variants was significantly associated with susceptibility considering multiple testing (e.g. 497 had P values > 0.05; eight had P values between 0.01 and 0.05). Thus, lead variants associated with cognitive progression differed from susceptibility variants.
We next evaluated the effects of two previously nominated candidate prognostic genes, GBA7,8,14,15 and APOE16, on risk of dementia in patients with PD (Supplementary Table 4) in the combined population. Patients carrying a pathogenic mutation for Gaucher’s disease or protein-coding variants associated with PD (as defined previously7) in GBA had a HR of 1.93 (95% CI 1.36–2.73) for dementia with P = 0.0002 (Fig. 2a) in the Cox analysis extending previous results7,8,15. They had a more rapid longitudinal decline in MMSE scores compared to non-carriers in LMM analysis (β = −0.087, P = 0.011, Fig. 2b). Cases carrying the APOE ε4 allele had a HR of 1.48 (95% CI 1.17–1.87) for PDD with P = 0.001 (Fig. 2c) and a more rapid decline in MMSE scores (β = −0.078, P = 0.0003) compared to non-carriers (Fig. 2d). The RIMS2 variant was a 2.5x to 3x-stronger predictor of PD dementia than GBA and than APOE, respectively.
Fig. 2 |. GBA and APOE ε4 accelerate cognitive decline in individuals with Parkinson’s disease.
a, Covariate-adjusted survival curves for PD patients without GBA mutation (cyan line) and those carrying GBA mutation (orange dashed line). b, Adjusted mean MMSE scores across time predicted from the estimated fixed-effect parameters in the LMM for carriers (orange) and non-carriers (cyan) of a GBA variant. c, Covariate adjusted survival curves for patients with PD without an APOE ε4 allele (cyan line), carriers of one APOE ε4 allele (red line) and carriers of two APOE ε4 alleles (purple line). Cox PH model with two-sided Wald test. d, Adjusted mean MMSE scores across time predicted from the estimated fixed-effect for non-carriers (cyan line), APOE ε4 heterozygous (red line) and APOE ε4 homozygous (purple line) carriers. a,c, Cox PH model with two-sided Wald test; b-d, shaded ribbons indicate +/− s.e.m. across time; P values from LMM.
It has been assumed that GWAS-derived susceptibility variants constitute progression drivers with limited evidence (e.g. ref. 21). The aggregate effect of 90 GWAS-derived susceptibility loci45 can be captured in a polygenic risk score (PRS) (Methods) that estimates the cumulatively genetic susceptibility for PD52. We tested this PRS for association with dementia prognosis in our longitudinal PD cohorts. Contrary to expectations, no statistically significant association between PRS and progression to PDD was found in the Cox analysis (HR = 0.95; 95% CI, 0.80–1.13; P = 0.57). The AUC for 10-year prediction of PDD was 0.496 (95% CI 0.444–0.548; Table 2 and Fig. 3a), which was not different from chance. Furthermore, we compared patients in the highest PRS quartile to those in the lowest PRS quartile using survival curves (Fig. 3b, P = 0.91) and LMM (Extended Data Fig. 6) and detected no appreciable differences. Individually, none of the 90 susceptibility variants achieved multiple-testing-corrected significance thresholds for predicting PDD (Supplementary Table 5). They were also not significantly linked to motor progression in PD as measured by transition to HY stage 3 using Cox model analysis and change in MDS-UPDRS III subscale score by LMM model analysis, respectively, adjusting for covariates (Supplementary Data 1). There was no correlation between the statistical power to detect effects at these SNPs and the observed P values (Pearson correlation r2 = 0.016, P = 0.88). This suggests that genetic variants and scores linked to susceptibility are not significantly associated with cognitive progression.
Table 2 |.
Performance of different genetic Cox PH models for predicting PDD
| Genetic factor* | PHS stage# | HR | 95% CI | P | Concordance of model | AUC## | 95% CI |
|---|---|---|---|---|---|---|---|
| PRS: PD GWAS (90 SNPs) | Development | 0.95 | 0.80–1.13 | 0.57 | 0.510 | 0.496 | 0.444–0.548 |
| PHS: Novel 3 variants | Development | 2.54 | 2.10–3.08 | 4.51 × 10−20 | 0.597 | 0.589 | 0.552–0.626 |
| PHS: Novel 3 variants + GBA | Development | 2.49 | 2.08–2.98 | 1.30 × 10−21 | 0.601 | 0.611 | 0.569–0.652 |
| PHS: Novel 3 variants + APOE ε4 | Development | 2.47 | 2.06–2.96 | 5.12 × 10−21 | 0.616 | 0.604 | 0.559–0.649 |
| PHS: Novel 3 variants + GBA + APOE ε4 | Development | 2.42 | 2.04–2.88 | 2.68 × 10−22 | 0.618 | 0.623 | 0.576–0.670 |
| PRS: PD GWAS (90 SNPs) | Validation | 0.60 | 0.25–1.42 | 0.25 | 0.551 | 0.588 | 0.399–0.778 |
| PHS: Novel 3 variants + GBA + APOE ε4 | Validation | 2.05 | 1.16–3.61 | 0.01 | 0.617 | 0.668 | 0.519–0.817 |
Exclusively genetic factors were used in these Cox PH models. HR, hazard ratio and 95% CI, 95% confidence interval.
The PHS Development Stage comprises the discovery and replication cohorts; the PHS Validation Stage comprises three additional, independent cohorts that were not available and not used during the variant discovery, variant replication, and PHS development stages.
AUC, indicates the area under the 10-year cumulative/dynamic receiver operating characteristic curves for PHS development cohorts (e.g. combined discovery and replication cohorts); and the area under the 6-year receiver operating characteristic curves for the independent PHS validation cohorts; 95% confidence interval was estimated using a simulation method. P values from Cox PH models with two-sided Wald test.
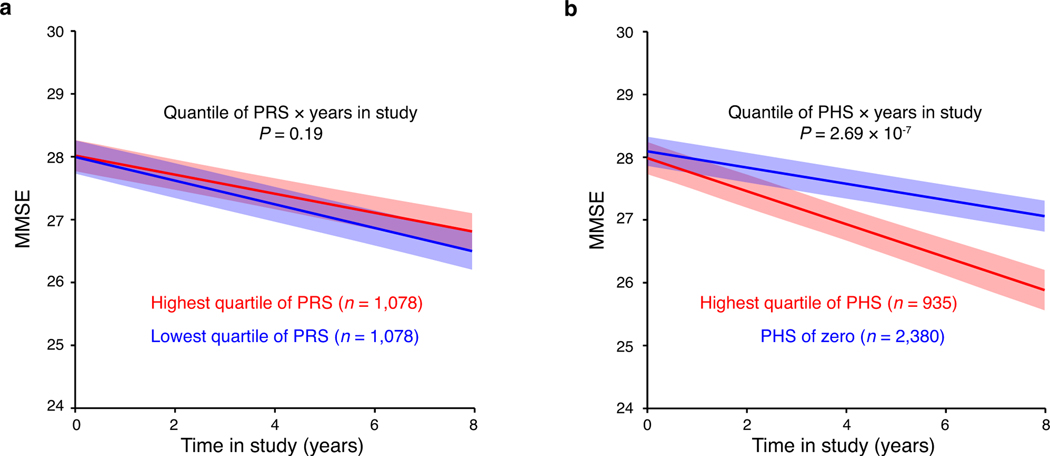
Fig. 3 |. A polygenic hazard score outperforms polygenic risk scores in dementia prediction.
a, Comparison of polygenic Cox PH models for predicting progression to PDD in cases with PD (n = 3,821 with 31,053 longitudinal visits). Data are visualized as the 10-years cumulative AUC (bars) and the 95% CI (error bars), which was estimated as described59 implemented in the timeROC package. P values of the AUC of individual polygenic hazard score models (based on prognosis variants) compared to the AUC of a polygenic risk score (PRS) model (based on 90 susceptibility variants2) are shown; * indicates P < 0.05 (i.e. exact P values of 0.006, 0.002 and 0.003, respectively) and ** indicates P = 0.0009; two-sided z-tests. b, Cox-adjusted survival curves for survival free of PDD for cases scoring in the highest quartile of PRS (orange) compared to cases scoring in the lowest quartile of PRS (cyan) are shown. c, Cox-adjusted survival curves for survival free of PDD for cases scoring in the highest quartile of PHS (magenta) compared to cases scoring zero on the PHS (cyan) are shown for the combined dataset. d, Cox-adjusted survival curves for survival free of PDD for cases scoring in the highest quartile of PHS (magenta) compared to cases scoring zero on the PHS (cyan) are shown for the new PHS validation dataset. b-d, Cox PH model with two-sided Wald test; stratified analyses’ results (HR, 95% CI, P values) are shown; non-stratified analyses’ results are in Table 2. Patients assigned to the highest quartile of PRS or PHS were those with a score greater than the score separating the fourth (highest) and third quartile of values.
We then used the lead variant from each of the three prognosis loci to develop an innovative cognitive polygenic hazard score (PHS) to predict PD dementia (Methods). The HR was 2.54 (95% CI 2.10–3.08) with P = 4.51 × 10−20 for a one unit value increase in PHS. The PHS was associated with prediction of PDD with a 10-year cumulative AUC of 0.589 (95% CI 0.552–0.626; Fig. 3a). Out of 3,821 cases with PD, 688 (18%) carried at least one of the three novel progression alleles (rs182987047, rs138073281, rs8050111), of which 639 cases carried only one progression allele, 47 cases carried two risk alleles, and two cases carried three risk alleles. Cox PH analysis stratified for carriers of 1, 2 (either homozygous or heterozygous for two loci), and 3 risk alleles compared to non-carrier cases indicated an additive effect with HRs of 2.79 (95% CI 2.12–3.67) with P of 2.70 × 10−13, 5.65 (95% CI 3.27–9.74) with P = 4.81 × 10−10, and 30.4 (95% CI 3.77–245.4), respectively.
We evaluated different genetic Cox PH models for prediction of PDD in the combined population (Table 2, Fig. 3a, and Methods). The most robust genetic hazard model included the three new prognosis loci plus GBA and APOE (model concordance = 0.618). This PHS was a significant predictor of PDD with an AUC of 0.623 (95% CI 0.576–0.670). It was significantly more accurate in estimating whether a patient will develop dementia within ten years from disease onset than chance alone (P = 2.68 × 10−22) or compared to the PRS (P = 0.0009). The Cox-adjusted survival curves of patients (Fig. 3c) showed that 89.6% of patients with a low (zero) PHS survived for 10 years since onset of PD without dementia. By contrast, only 73.3% of patients in the highest quartile of the PHS remained free of dementia for 10 years since onset of PD.
To further test the performance of the PHS score in independent patients (that were not previously used to discover and replicate the progression variants, nor to build and optimize the PHS), we analyzed three new independent cohorts (EPIPARK, DeNoPa; HBS2). The association of the PHS score with PDD prediction was replicated with P = 0.01 and HR = 2.05 (1.16–3.61; Table 2) across the three new cohorts. Areas under the curve (AUCs) in the independent development and validation stages were consistent, e.g. 0.623 (0.576–0.670) and 0.668 (0.519–0.817), respectively (Table 2). Similarly, stratified covariate-adjusted survival analysis comparing cases scoring in the highest quartile of PHS vs. cases scoring zero on the PHS were consistent in development and validation stages with HR = 2.82 (2.14–3.72) and HR = 3.20 (1.26–8.11), respectively (Fig. 3c,d; longitudinal follow-up period was considerably shorter in the new cohorts). The PRS score was again not predictive of PDD in the three new cohorts (P = 0.25).
This study uncovered genetic variation linked to cognitive progression in PD with substantial effect sizes. These progression variants were not associated with susceptibility. Susceptibility variants and scores did not appear to predict progression. This is consistent with the hypothesis that disease initiation and progression may, in part, be governed by diverging genetics and mechanisms9,54,55. Cognitive progression in PD strongly correlates with cortical spread of Lewy bodies and neurites56,12. Furthermore, amyloid plaques and tangles present in up to a third of patients56,57. Our study indicates that genetic drivers of PD progression may comprise PD loci (e.g. RIMS2 and potentially TMEM108), loci shared with Dementia with Lewy Bodies (e.g. APOE and GBA58), and, possibly, loci shared with Alzheimer’s disease (e.g. APOE and WWOX50). Analyses of larger longitudinal populations will be required to detect variants with small effect sizes, to increase statistical power for motor phenotypes confounded by PD medications, and to systematically decode the divergent and convergent features of the genetic architecture underlying susceptibility, progression, and dementias.
These results suggest a new paradigm for drug development. Disease-modifying drugs that target the genetic drivers of disease progression could potentially turn fast progressors into slow progressors and substantially improve quality of life. Clinically, this study provides a polygenic score that could be used to enrich trials with patients who have a more aggressive disease course and thus are likely to show the greatest benefits from interventions. This may be useful because ascertaining therapeutic efficacy in patients who naturally progress slowly is exceedingly difficult.
Methods
Study participants.
Supplementary Table 1 describes the cohorts included in this work. Discovery, replication and PHS development stages. We used 15 cohorts14,23–31,60–65 from North America and Europe to discover and replicate progression variants and to build the PHS score. These comprised 4,872 patients with PD (with available genotyping data), which were longitudinally assessed with 36,123 study visits between 1986 and 2017 (Supplementary Fig. 1).
Written informed consent for DNA collection and phenotypic data collection for secondary research use for each cohort was obtained from the participants with approval from the local ethics committees. The Institutional Review Board of Partners HealthCare approved the current genotyping and analyses. For PPMI, approval was obtained to download and analyze the publicly accessible WGS and clinical data. 13 cohorts enrolled patients with a diagnosis of PD established according to modified UK PD Society Brain Bank diagnostic criteria as previously reported1,23,26,29,61,62,64–67. In DATATOP, the eligibility criteria required a clinical diagnosis of early, idiopathic PD (HY stages 1 or 2) with patients not on anti-parkinsonian medications66. Arizona Study of Aging/Brain and Body Donation Program: all subjects have come to autopsy and have had full neuropathological examinations with neuropathological diagnosis63. Diagnostic certainty was increased by confirming the clinical diagnosis of PD during longitudinal follow-up visits68 in all cohorts. Patients whose longitudinal follow-up evaluations were not consistent with a diagnosis of PD were excluded. Cohorts were a priori assigned to discovery or replication cohorts in order to achieve an approximately two-thirds to one-third split among the two stages (while considering fixed cohort sizes) and to achieve an balanced distribution of the distinct types of cohorts (e.g. purpose-designed biomarkers studies, phase 3 clinical trials, population-based cohorts) across the two stages.
Serial Mini Mental State Exam (MMSE) scores69 were longitudinally collected in 10 cohorts. Montreal Cognitive Assessment (MoCA)70 scores were collected in PDBP30, PPMI61 study and converted to MMSE scores according to a published formula71. SCOPA-COG were collected in PROPARK62, PROPARK-C (PROPARK-Cross sectional cohort) and NET-PD Long term Study-1 (LS1)31 cohort and converted to MMSE scores. Cohort-specific definitions of Parkinson’s disease dementia was used (Supplementary Table 2). For seven cohorts, operationalized level 1 diagnostic criteria for Parkinson’s disease dementia (PDD) according to the Movement Disorders Society Task Force40 were available; PreCEPT and DATATOP used distinct definitions. PreCEPT defined PDD as a score of 4 on the UPDRS subscale 1 item 1 defined as “cognitive dysfunction [that] precludes the patient’s ability to carry out normal activities and social interactions”. For DATATOP published criteria for cognitive impairment leading to functional impairment were used72. Depression status were distinct defined according to different cohort. Ethnicity was self-reported. For several cohorts in this analysis, we evaluated previously collected longitudinal phenotypic data; for the active HBS, PDBP and DIGPD cohorts, both retro- and prospectively collected longitudinal data elements were included. HBS, Arizona Study of Aging/Brain and Body Donation Program, NET-PD LS1, CamPaIGN, PICNICS, DIGPD, PDBP, PIB, ParkWest, PROPARK and PROPARK-C cohorts comprised the discovery population and DATATOP, PPMI, PreCEPT and Tartu the replication population.
PHS validation stage.
To avoid overfitting, we tested the performance of the pre-specified polygenic hazard score (PHS) score in 520 patients from three, independent cohorts with detailed longitudinal clinical phenotyping, De Novo Parkinson Cohort (DeNoPa)73, EPIPARK74, and HBS2 (Supplementary Table 1), from Germany and the US. These three longitudinal PD cohorts were not used to discover and replicate progression variants, nor to build the PHS. PD was diagnosed in these cohorts according to modified UK PD Society Brain Bank diagnostic criteria. Cohort-specific definitions of PDD are listed in Supplementary Table 2.
Genotyping and data quality control and processing.
Quality control steps are shown in Extended Data Figure 1. Briefly, the DNA of patients with PD was quality controlled on an Agilent 2100 Bioanalyzer. DNA was quantitated against an 8 point standard curve using Quant-iT Picogreen dsDNA Assay Kit (Life Technologies, P7589) using a SpectraMax Gemini plate reader from Molecular Devices. Sample were genotyped at Translational Genomics Core of Partners HealthCare using the Illumina Multi-Ethnic Genotyping Array (MEGA A1, Illumina)75, which includes 1,779,819 markers (MEG array kit, Illumina, WG-316–1001). DNA was amplified using a whole genome amplification process. After fragmentation of the DNA, the sample was hybridized to 50-mer probes attached to the Beadchips, stopping one base before the interrogated base. Single base extension was then carried out to incorporate a labeled nucleotide. Dual color (Cy3 and Cy5) staining allowed the nucleotide to be detected by the iSCAN reader. Data from the iSCAN were collected in the Illumina LIMS and automated conversion to genotype occured using Autocall v2.0.1. In total, 4,510 PD sample were genotyped with the MEGA array; 512 PD samples from PPMI had whole genome sequences available.
We employed PLINK76 (version 1.90beta) and in-house scripts to conduct genotyping data processing and preform rigorous subject and SNP quality control (Extended Data Figure 1). SNPs with overall missingness > 0.05 were excluded. Samples with mismatched sex were excluded. Samples with a genotype missingness > 0.05 or heterozygosity rate greater than 4 s.d. from the mean were also excluded. To check relatedness among samples, 279,933 LD-independent SNPs were selected and pairwise identity by descent was estimated using PLINK routine “--indep 50 5 2”. For any related sample (pi-hat between 0.1875 to 0.9)77, one case with higher genotyping call rate was selected and kept and the others were excluded. For those sample pairs with pi-hat > 0.9, both cases were excluded. To identify geographical outliers, a pruned data set containing 86,998 LD-independent SNPs were merged with the 1000 Genomes Project data set78. Principal component analysis (smartpca)79 was used to identify and exclude the geographical outliers.
For 4,491 patients with PD, 31,885 (95.4%) of visits occurred within 12 years of longitudinal follow-up from disease onset, with a median follow-up time of 6.7 years (interquartile range, 4.2 years). We therefore focused our survival analyses on the 12-year time frame from disease onset. 3,821 samples passed genotyping quality control (Extended Data Figure 1). Patients were left-censored and those with missing or non-quality clinical data were excluded (n = 670; Extended Data Figure 1). Specifically, 24 were excluded clinical data were not available. Another 646 patients were removed due to missing critical individual data points or left censoring (Extended Data Figure 1) (e.g. 138 participants already had PDD at the baseline visit and were left censored; 39 subjects missed age at onset or age at the baseline visit data; 238 subjects, whose first study visit occurred more than 12 years from disease onset; 231 missed dementia ascertainment data). Thus, a total of 3,821 participants passed rigorous genotyping and clinical data requirements (Extended Data Figure 1). To identify genetic variants associated with progression from PD to Parkinson’s disease dementia (PDD), we performed a longitudinal genome-wide survival study (GWSS) on these 3,821 patients, of whom 2,650 patients (and 11,744 visits) were assigned to the discovery population, and 1,171 patients (and 19,309 visits) were assigned to the replication population.
For 520 independent patients from DeNoPa, EPIPARK and HBS2, the same genotyping quality control was performed, and 425 samples passed quality control, 21 patients were removed due to left censoring. Thus, a total of 404 patients with 1,028 visits were used in the PHS validation stage.
Genotype imputation.
Genotype imputation was performed using Minimac3 (v2.0.1) on the Michigan online imputation server80. The haplotype reference consortium (HRC version r1.1)81 was selected as reference panel, which consists of 64,940 haplotypes of predominantly European ancestry with ~39.2 million SNPs, all with an estimated minor allele count of ≥ 5. Eagle2 (v2.3)82 with 20-Mb chunk size was used to estimate haplotype phasing; pipeline details, including quality check, phasing and imputation, are available at https://imputationserver.sph.umich.edu. Samples from all studies were prephased and imputed in a single batch to avoid batch effects attributable to the imputation process: Multi-Ethnic Genotyping Array (MEGA) data of 4,020 subjects with PD with 1,635,580 SNPs at autosomes were used as input for the online server. To estimate imputation accuracy, imputed genotype calls for 1,052,012 SNPs were compared with directly genotyped data using EmpR to calculate the correlation between the true genotyped values and the imputed values from the output of Minimac3. Mean R2 was 0.996 and EmpR was 0.979 for variants with MAF ≥ 0.1% (Supplementary Fig. 2). Imputed variants with MAF < 0.1% and/or R2 < 0.3 were excluded. In total 11,220,132 imputed SNPs remained for further analysis (Supplementary Fig. 2). In addition, we removed 26,785 variants with discordant minor allele frequency (with Fisher’s exact test FDR < 0.05) observed with MEGA array plus imputation (14 cohorts) compared to whole genome sequencing (PPMI cohort). In total, 7,741,751 variants with MAF ≥ 1% remained for further analysis. Imputation for the PHS validation cohorts was performed separately.
PPMI and HBS whole genome sequencing datasets.
The PPMI and HBS data consists of 512 and 699 individuals with PD, respectively, who passed quality control. Whole-genome sequencing was performed by Macrogen, Inc. under the direction of Andrew Singleton (NIA). Samples were prepared according to the Illumina TruSeq PCR Free DNA sample Preparation Guide. The libraries were sequenced using Illumina HiSeq X Ten Sequencer. Detailed methods are available at https://ida.loni.usc.edu/pages/access/geneticData.jsp.
Evaluating the concordance between imputed genotypes and sequencing.
We employed the SnpSift tool (http://snpeff.sourceforge.net/SnpSift.version_4_0.html) to evaluate the concordance between imputed SNPs (based on the MEGA array) and SNPs directly called from whole genome sequencing in 562 individuals from HBS for which both assays were available (Supplementary Fig. 3). The percentage concordance between 10,421,270 imputed SNPs and whole genome sequencing was 99.4% (standard error 0.0006%). For the three SNPs associated with PD dementia (whose genotypes came from imputation), we observed high concordance rates of 99.5%, 99.6%, and 98.9% for rs182987047, rs138073281, and rs8050111, respectively. Imputation average call rates (AvgCall) and imputation R2 values were for rs182987047, 0.998 and 0.98, respectively; for rs138073281, 0.996 and 0.888; for rs8050111, 0.997 and 0.961.
Candidate loci GBA and APOE.
β-glucocerebrosidase gene (GBA) variants were defined as described7 and included pathogenic mutations associated with Gaucher’s disease as well as the PD-associated, coding risk variants (E326K, T369M and E388K). We previously reported7 GBA genotypes (largely based on targeted or Sanger sequencing of the locus) for 2,625 of the 4,491 patients with PD here included. For the remaining 1,866 PD patients, GBA variants and mutations were identified based on the MEGA array. Participants were classified as carriers (with one or more GBA mutations) or non-carriers (no GBA mutation) as reported7.
Apolipoprotein E (APOE) alleles ε2 ε3 ε4 were identified based on rs7412 and rs429358 from MEGA chip plus imputation data (14 cohorts) or whole genome sequencing (WGS) (PPMI cohort). We compared imputed APOE alleles of 531 HBS PD patients to the results of a TaqMan SNP genotyping assay for the two SNPs. The concordance rate was 98.7%. We classified the 4,491 PD patients into three groups for downstream analysis: 81 homozygous ε4 carriers (ε4/ε4), 1,068 heterozygous ε4 carriers (ε2/ε4, ε3/ε4), and 3,342 non-ε4 carriers (ε2/ε2, ε2/ε3, ε3/ε3).
Statistical analysis.
The Cox proportional hazards statistic was used to estimate the influence of each genotype on time (years from onset of PD) to reaching the endpoint of PDD. Age at onset of PD, gender, years of education, and the top ten principal components of population substructure were included as covariates in the Cox analyses. For the meta-analyses across cohorts, a “cohort” term was included as a random effect (a random effects Cox model is often termed a “frailty” model). Regarding “cohort” as a random term will permit inferences about study level variance among a hypothetical universe of studies in the referent population. 31,885 (95.4%) of visits from 4,467 PD patients occurred within 12 years of longitudinal follow-up from disease onset with a median follow-up time of 6.7 years (inter-quartile range, 4.2 years). We thus focused our survival analyses on the 12-year time frame from disease onset. Cox proportional hazards analyses were performed using the coxph function in the Survival package (Version 2.38–1) in R, and the “Breslow” method was used for handling observations that have tied survival times. P values less than or equal to 5 × 10−8 were considered as indicative of genome-wide significance.
Generalized longitudinal mixed fixed and random effects analysis (LMM)5 of cognitive decline was performed using serial Mini Mental State Exam (MMSE) scores longitudinally assessed (enrollment visit and multiple longitudinal follow-up visits) in the combined data set. The PROPARK-C and Tartu cohorts were excluded from the LMM because no longitudinal MMSE scores were available. The MMSE score was the dependent variable and the primary predictors were group status (e.g. genotype carrier status or alleles), time in the study (years), and their interaction. An intercept term and linear rate of change across time per subject were the random terms (permitted to be correlated). Subject level fixed covariates were age at baseline, gender, years of education, duration of PD illness at baseline, as well as ten principal components. A study term was included as a random effect. The significance, direction and effect size of the group x time terms answers the question of differential progression for the carriers as compared to the non-carrier group. This analysis was performed using the glmmPQL function in the MASS package (version 7.3–37). All analyses were conducted in the R statistical environment version 3.3.1. Nominal P values (not adjusted for multiple testing) were shown except were indicated otherwise. Evidence for genome-wide significance in the discovery population was defined as P ≤ 5 × 10−8; P values ≤ 0.05 were considered evidence of significance in the replication population and in the PHS validation population. Associations for previously established candidate loci were considered significant if they met Bonferroni-adjusted significance thresholds (e.g. 0.05/number of established candidates evaluated).
Polygenic risk score (PRS).
A PRS was calculated as the weighted sum of the number of risk alleles possessed by an individual, in which the weight was taken as the natural log of the odds ratio (OR) associated with each individual SNP. We used 90 lead GWAS variants associated with susceptibility for PD and the ORs from a recent meta-analysis study45 to calculate the PRS (Supplementary Table 5).
Polygenic hazard score (PHS).
For each patient in this study, we calculated an individual polygenic hazard score (PHS) similar to what was described83. We used the hazard ratios of the lead associated SNPs (from the combined data set) in each of the three prognosis loci to calculate the PHS. Briefly, we added the number of risk alleles (0, 1 or 2) for a lead variant multiplied by the effect size (natural log of hazard ratio from combined dataset) for that variant. In other versions of the PHS, we included additionally one or both of the candidate cognitive prognosis genes (GBA mutation status and APOE ε4 allele haplotype). To evaluate the performance of the PHS models, the cumulative/dynamic receiver operating characteristic (ROC) curves, area under curves (AUC), confidence intervals of the AUC (simulation method), and comparisons between two AUCs were calculated using the timeROC package (Version 0.2)59 in R with the inverse probability of censoring weights (IPCW) method used to compute the weights.
Characterization of genomic risk loci.
We used FUMA (http://fuma.ctglab.nl) to characterize the cognitive prognosis loci. Tag SNPs with suggestive P < 1 × 10−5 were inputted; additional SNPs in high linkage disequilibrium (LD) with a tag SNP (with r2 > 0.6 and independent from each other with r2 < 0.6) were identified using the 1000 Genomes Phase 3 reference panel for Europeans. If LD blocks of independent significant SNPs were closely located to each other (< 250 kb based on the most right and left SNPs from each LD block), they were merged into one genomic locus.
Gene expression analysis.
Gene expression profiles of the three significant loci in human tissues was downloaded directly from GTEx Portal V7 (https://gtexportal.org/). Downloaded gene expression profiles were normalized. Detailed processing methods can be found in the GTEx portal V7. Human brain cell type-specifc expression of the three cognitive prognosis loci was evaluated using the BRAINcode dataset47 and portal (http://www.humanbraincode.org).
Data Availability Statement
A Life Sciences Reporting Summary for this paper is available. Human brain cell type-specific expression data from BRAINcode47 RNA-seq data are accessible through a user-friendly webportal at www.humanbraincode.org and individual-level data through dbGAP (acc. Number phs001556.v1.p1). The gene expression profiles of human tissues used in this study can be downloaded from the GTEx Portal V7 (https://gtexportal.org/). The GWSS summary statistics for the combined analysis of discovery and replication populations are publicly accessible through the EGA database https://www.ebi.ac.uk/ega/ (acc. Number EGAS00001005110). Individual-level genetic data for the NIH-funded Illumina Multi-Ethnic Genotyping Array analyses of the HBS2 and EPIPARK cohorts are accessible in dbGAP with accession number phs002328.v1.p1 in accordance with NIH Genomic Data Sharing Policy. The whole genome sequencing and clinical data for PPMI included in this study are publicly available through a PPMI Whole Genome Sequencing Data Agreement at ppmi@loni.usc.edu. Clinical data for PDBP included in this study are publicly available through https://pdbp.ninds.nih.gov. Clinical longitudinal data for the other cohorts included are accessible through appropriate data sharing agreements that protect patient privacy with the institutions which conducted or are conducting study consents and clinical assessments under local IRB approvals.
Code availability
Analysis code is made available at https://github.com/sixguns1984/GWSS.PDD.
Extended Data
Extended Data Fig. 1. Genotyping pipeline for discovery and replication cohorts.
Quality control (QC) steps outlined in blue were performed using PLINK v1.90beta76.
Extended Data Fig. 2. Characteristics of loci associated with cognitive progression in PD.
a, RIMS2 locus. b, TMEM108 locus. c, WWOX locus. Top, chromosomal position; middle, -log10(P values) for individual SNPs at each locus (left y-axis) with the rate of recombination indicated by the red line (right y-axis); bottom, gene positions with the locus. Each point represents a SNP colored according to LD with the lead associated variant. Figure panels were generated with LocusTrack84 and r2 values were calculated based on CEU population in the 1000 Genomes Project data set78.
Extended Data Fig. 3. Associations between a second RIMS2 variant rs116918991, TMEM108 rs138073281, and WWOX rs8050111 with cognitive PD progression.
a,c,e, Covariate-adjusted survival curves for PD patients without the indicated variant (blue line) and for those carrying the indicated variant (heterozygotes and homozygotes; red dashed line) are shown. P values Cox PH models with two-sided Wald test and were not corrected for multiple hypothesis testing. b,d,f, Adjusted mean MMSE scores across time predicted from the estimated fixed-effect parameters of the LMM analysis are shown for cases carrying the variant (heterozygotes and homozygotes; red) and cases without the variant (non-carriers; blue) adjusting for covariates. Shaded ribbons indicate +/− s.e.m. around predicted MMSE scores across time. Note that a second RIMS2 variant rs116918991 (correlated with r2 = 0.49 with the lead variant rs182987047; Fig. 1) is shown in a and b, and that the HR and P values shown here for TMEM108 rs138073281 and WWOX rs8050111 are different from the HR and P values from the main analysis (Table 1), where variant alleles were coded as 0, 1, 2. P values from LMM analysis with two-sided t-test and were not corrected for multiple hypothesis testing
Extended Data Fig. 4. RIMS2, TMEM108, and WWOX are expressed in human brain.
Gene expression profiles were downloaded directly from the GTEx Portal V746. Expression values are shown in Transcript per Million (TPM), calculated from a gene model with isoforms collapsed to a single gene. Box plots visualize first, third quartiles and medians; the ends of the whiskers represent the lowest (or highest) value still within 1.5-times the interquartile range. Outliers are displayed as dots, if they are above or below 1.5-times the interquartile range. n indicates number of individuals for each tissue analyzed in GTEx V7.
Extended Data Fig. 5. Cell-type specific expression of RIMS2, TMEM108, and WWOX in human brain.
Cell type-specific transcriptomes were assayed using laser-capture RNA sequencing (lcRNAseq) as reported47. Gene expression (FPKM) profiles of RIMS2, TMEM108, and WWOX are from BRAINcode consortium (http://www.humanbraincode.org). n indicates the number of individuals assayed for each cell type. SNDA, indicates dopamine neurons laser-captured from human substantial nigra pars compacta; MCPY, pyramidal neurons from human motor cortex; TCPY, pyramidal neurons from human temporal cortex; PBMC, human peripheral blood mononuclear white cells; FB, primary human fibroblasts. Box plots visualize first, third quartiles, and medians; the ends of the whiskers represent the lowest (or highest) value still within 1.5-times the interquartile range. Each dot represents a sample.
Extended Data Fig. 6. The polygenic hazard score (PHS) is associated with decline in serial MMSE scores.
a, PD cases scoring in the highest quartile (red) of a polygenic risk score (PRS based on 90 susceptibility variants45) compared to PD cases scoring in the lowest quartile of the PRS (blue) are shown. b, PD cases scoring in the highest quartile (red) of the PHS (comprising GBA + APOE ε4 + the 3 novel progression variants) compared to PD cases scoring zero on the PHS (blue) are shown. For a and b, adjusted mean MMSE scores across time predicted from the estimated fixed-effect parameters in the LMM analysis for the combined data set comprising discovery and replication populations are shown. The shaded ribbons indicate +/− s.e.m. around predicted MMSE scores across time. The P values from LMM analysis with two-sided t-tests and were not corrected for multiple hypothesis testing.
Supplementary Material
Acknowledgements
We are grateful to Ofer Nemirovsky for his valuable philanthropic support, encouragement, and outstanding insights. We thank all study participants, their families, and friends for their support and participation, and our study coordinators for making this work possible. We thank Alison Brown at Partners HealthCare Personalized Medicine and Yuliya Kuras at Brigham and Women’s Hospital for excellent technical assistance. The study was funded in part by a philanthropic support (to Brigham & Women’s Hospital and C.R.S.) for Illumina MEGA chip genotyping; and in part by NINDS and NIA R01NS115144 (to C.R.S.), which funded genotyping for the PHS validation cohorts EPIPARK and HBS2. C.R.S.’s work was supported by NIH grants NINDS/NIA R01NS115144, U01NS095736, U01NS100603, and the American Parkinson Disease Association Center for Advanced Parkinson Research. T.M.H. is funded by NIH grant K23NS099380. While this manuscript was in revision, G.L. transferred to a new position at Sun Yat-sen University, where he received support from the National Natural Science Foundation of China (Project no. 31900475), the Fundamental Research Funds for the Central Universities (Project no. 19ykpy146), Young Talent Recruitment Project of Guangdong (Project No. 2019QN01Y139) and Shenzhen Basic Research Project (Project No. JCYJ20190807161601692). For each individual cohort, acknowledgements are listed in the Supplementary Note.
International Genetics of Parkinson Disease Progression (IGPP) Consortium
Ganqiang Liu1,2,3, Jiajie Peng1,2,4, Zhixiang Liao1,2, Joseph J. Locascio1,2,5, Jean-Christophe Corvol6, Xianjun Dong1,2, Jodi Maple-Grødem7,8, Meghan C. Campbell9, Alexis Elbaz10, Suzanne Lesage6, Alexis Brice6, Graziella Mangone6, Bernard Ravina12, Ira Shoulson13, Pille Taba14, Sulev Kõks15,16, Thomas G. Beach17, Florence Cormier-Dequaire6, Guido Alves7,8,18, Ole-Bjørn Tysnes19,20, Joel S. Perlmutter9,21,22, Peter Heutink23, Jacobus J. van Hilten25, Meike Kasten26, Brit Mollenhauer27, Claudia Trenkwalder28, Christine Klein29, Roger A. Barker30,31, Caroline H. Williams-Gray30, Johan Marinus25, and Clemens R. Scherzer1,2,5,11*
Footnotes
Competing Interests Statement
Brigham and Women’s Hospital holds a US provisional patent application on the polygenic hazard score for predicting PD progression, on which C.R.S. is named as inventor. Outside this work, C.R.S. has served as consultant, scientific collaborator or on scientific advisory boards for Sanofi, Berg Health, Pfizer, Biogen, and has received grants from NIH, U.S. Department of Defense, American Parkinson Disease Association, and the Michael J Fox Foundation (MJFF).
G.L., J.J.L., J.M., A.E., J.H.G., A.Y.H., S.K., P.T., S.S.A., J.S.P., and M.C.C. report no relevant financial or other conflicts of interest in relation to this study.
M.A.S. has no conflict of interest related to this work. Outside this work, M.A.S. has received grants from NINDS, DoD, MJFF, Farmer Family Foundation, and has served as a consultant to commercial programs: Eli Lilly & Co (data monitoring committee), Prevail Therapeutics (scientific advisory board), Denali Therapeutics (scientific advisory board), nQ Medical (scientific advisory board), Chase Therapeutics (scientific advisory board) and Partner Therapeutics (scientific advisory board).
A.-M.W. has received research funding from the ALS Association, the Parkinson’s Foundation, has participated in clinical trials funded by Acorda, Biogen, Bristol-Myers Squibb, Sanofi/Genzyme, Pfizer, Abbvie, and received consultant payments from Mitsubishi Tanabe and from Accordant.
T.M.H. has no conflict of interest related to this work. Outside this work, he received honoraria for consulting in advisory boards for Boston Scientific and Medtronic.
J.-C.C. has no conflict of interest related to this work. Outside this work, J.C.C. received honoraria for consulting in advisory boards for Abbvie, Actelion, Air Liquide, Biogen, BMS, BrainEver, Clevexel, Denali, Pfizer, Theranexus, and Zambon.
B.R. is an employee of and holds equity in Praxis Precision Medicines and is advisor for Caraway Therapeutics and Brain Neurotherapy Bio.
I.S. is Principal investigator of a MJFF Computational Science Grant (2017–19).
S.K. is supported by Multiple Sclerosis of Western-Australia (MSWA) and the Perron Institute.
P.H. is a Scientific Advisor of Neuron23.
J.J.v.H. has no conflict of interest related to this work. Outside this work, J.J.v.H. has received grants from the Alkemade-Keuls Foundation, Stichting Parkinson Fonds, Parkinson Vereniging, The Netherlands Organisation for Health Research and Development, The Netherlands Organisation for Scientific Research, Hersenstichting, AbbVie, Michael J Fox Foundation, and research support from Hoffmann-La-Roche, Lundbeck and the Centre of Human Drug Research.
B.M. has no conflict related to this work. Outside this work, B.M. has received honoraria for consultancy from Roche, Biogen, AbbVie, Servier and Amprion. B.M. is member of the executive steering committee of the Parkinson Progression Marker Initiative and PI of the Systemic Synuclein Sampling Study of the Michael J. Fox Foundation for Parkinson’s Research and has received research funding from the Deutsche Forschungsgemeinschaft (DFG), EU (Horizon2020), Parkinson Fonds Deutschland, Deutsche Parkinson Vereinigung, Parkinson’s Foundation and MJFF.
R.A.B. has no conflict of interest related to this work. Outside this work, R.A.B. received consultancy monies from LCT; FCDI; Novo Nordisk; Cellino; Sana; UCB; received royalties from Wiley and Springer-Nature; grant funding from CPT; NIHR Cambridge Biomedical Research Centre (146281); MRC; Wellcome Trust (203151/Z/16/Z) and Rosetrees Trust (A1519 M654).
C.H.W.-G. has no conflict of interest related to this work. C.H.W.-G. is supported by a RCUK/UKRI Research Innovation Fellowship awarded by the Medical Research Council (MR/R007446/1) and the NIHR Cambridge Biomedical Research Centre, and receives grant support from MJFF, the Evelyn Trust, the Cure Parkinson’s Trust, Parkinson’s UK, the Rosetrees Trust and the Cambridge Centre for Parkinson=Plus. C.H.W.-G. has received honoraria from Lundbeck and consultancy payments from Modus Outcomes and Evidera.
C.T. is supported by EU Grant Horizon 2020/propag-ageing and the MJFF.
C.K. serves as a medical advisor to Centogene for genetic testing reports in the fields of movement disorders and dementia, excluding Parkinson’s disease.
Reference
- 1.Nalls MA et al. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat. Genet. 46, 989–993 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang D. et al. A meta-analysis of genome-wide association studies identifies 17 new Parkinson’s disease risk loci. Nat. Genet. 49, 1511–1516 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greenland JC, Williams-Gray CH & Barker RA The clinical heterogeneity of Parkinson’s disease and its therapeutic implications. Eur. J. Neurosci. 49, 328–338 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Wijmenga C. & Zhernakova A. The importance of cohort studies in the post-GWAS era. Nat. Genet. 50, 322–328 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Locascio JJ & Atri A. An overview of longitudinal data analysis methods for neurological research. Dement. Geriatr. Cogn. Dis. Extra 1, 330–357 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dorsey ER & Bloem BR The Parkinson pandemic—a call to action. JAMA Neurol. 75, 9–10 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Liu G. et al. Specifically neuropathic Gaucher’s mutations accelerate cognitive decline in Parkinson’s. Ann. Neurol. 80, 674–685 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu G. et al. Prediction of cognition in Parkinson’s disease with a clinical-genetic score: a longitudinal analysis of nine cohorts. Lancet Neurol. 16, 620–629 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aarsland D. et al. Cognitive decline in Parkinson disease. Nat. Rev. Neurol. 13, 217–231 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schrag A, Jahanshahi M. & Quinn N. What contributes to quality of life in patients with Parkinson’s disease? J. Neurol. Neurosurg. Psychiatry 69, 308–312 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Svenningsson P, Westman E, Ballard C. & Aarsland D. Cognitive impairment in patients with Parkinson’s disease: diagnosis, biomarkers, and treatment. Lancet Neurol. 11, 697–707 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Braak H. et al. Staging of brain pathology related to sporadic Parkinson’s disease. Neurobiol. Aging 24, 197–211 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Langston JW The Parkinson’s complex: parkinsonism is just the tip of the iceberg. Ann. Neurol. 59, 591–596 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Williams-Gray CH et al. The CamPaIGN study of Parkinson’s disease: 10-year outlook in an incident population-based cohort. J. Neurol. Neurosurg. Psychiatry 84, 1258–1264 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Cilia R. et al. Survival and dementia in GBA-associated Parkinson’s disease: The mutation matters. Ann. Neurol. 80, 662–673 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Pang S, Li J, Zhang Y. & Chen J. Meta-analysis of the relationship between the APOE gene and the onset of Parkinson’s disease dementia. Parkinsons Dis. 2018, 9497147 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Healy DG et al. Phenotype, genotype, and worldwide genetic penetrance of LRRK2-associated Parkinson’s disease: a case-control study. Lancet Neurol. 7, 583–90 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guella I. et al. alpha-synuclein genetic variability: A biomarker for dementia in Parkinson disease. Ann. Neurol. 79, 991–999 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Markopoulou K. et al. Does alpha-synuclein have a dual and opposing effect in preclinical vs. clinical Parkinson’s disease? Parkinsonism Relat. Disord. 20, 584–589 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mata IF et al. APOE, MAPT, and SNCA genes and cognitive performance in Parkinson disease. JAMA Neurol. 71, 1405–1412 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paul KC, Schulz J, Bronstein JM, Lill CM & Ritz BR Association of polygenic risk score with cognitive decline and motor progression in Parkinson disease. JAMA Neurol. 75, 360–366 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mata IF et al. Large-scale exploratory genetic analysis of cognitive impairment in Parkinson’s disease. Neurobiol. Aging 56, 211 e1–211 e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Locascio JJ et al. Association between alpha-synuclein blood transcripts and early, neuroimaging-supported Parkinson’s disease. Brain 138, 2659–2671 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pankratz N. et al. Meta-analysis of Parkinson’s disease: identification of a novel locus, RIT2. Ann. Neurol. 71, 370–84 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jankovic J. et al. Variable expression of Parkinson’s disease: a base-line analysis of the DATATOP cohort. The Parkinson Study Group. Neurology 40, 1529–1534 (1990). [DOI] [PubMed] [Google Scholar]
- 26.Ravina B. et al. A longitudinal program for biomarker development in Parkinson’s disease: a feasibility study. Mov. Disord. 24, 2081–2090 (2009). [DOI] [PubMed] [Google Scholar]
- 27.Winder-Rhodes SE et al. Glucocerebrosidase mutations influence the natural history of Parkinson’s disease in a community-based incident cohort. Brain 136, 392–399 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Marinus J. et al. A short scale for the assessment of motor impairments and disabilities in Parkinson’s disease: the SPES/SCOPA. J. Neurol. Neurosurg. Psychiatry 75, 388–395 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Breen DP, Evans JR, Farrell K, Brayne C. & Barker RA Determinants of delayed diagnosis in Parkinson’s disease. J. Neurol. 260, 1978–1981 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Rosenthal LS et al. The NINDS Parkinson’s disease biomarkers program. Mov. Disord. 31, 915–923 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Writing Group for the NINDS Exploratory Trials in Parkinson Disease (NET-PD) Investigators et al. Effect of creatine monohydrate on clinical progression in patients with Parkinson disease: a randomized clinical trial. JAMA 313, 584–593 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.1000 Genomes Project Consortium et al. A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Browning SR & Browning BL Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 81, 1084–1097 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y, Willer CJ, Ding J, Scheet P. & Abecasis GR MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol. 34, 816–834 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marchini J, Howie B, Myers S, McVean G. & Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 39, 906–913 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Visscher PM et al. 10 years of GWAS discovery: biology, function, and translation. Am. J. Hum. Genet. 101, 5–22 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ripatti S. & Palmgren J. Estimation of multivariate frailty models using penalized partial likelihood. Biometrics 56, 1016–1022 (2000). [DOI] [PubMed] [Google Scholar]
- 38.Bulik-Sullivan BK et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang J. et al. Genomic inflation factors under polygenic inheritance. Eur. J. Hum. Genet. 19, 807–812 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dubois B. et al. Diagnostic procedures for Parkinson’s disease dementia: recommendations from the movement disorder society task force. Mov. Disord. 22, 2314–2324 (2007). [DOI] [PubMed] [Google Scholar]
- 41.Kaeser PS et al. RIM proteins tether Ca2+ channels to presynaptic active zones via a direct PDZ-domain interaction. Cell 144, 282–295 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu C, Kershberg L, Wang J, Schneeberger S. & Kaeser PS Dopamine secretion is mediated by sparse active zone-like release sites. Cell 172, 706–718 e15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mechaussier S. et al. Loss of function of RIMS2 causes a syndromic congenital cone-rod synaptic disease with neurodevelopmental and pancreatic involvement. Am. J. Hum. Genet. 106, 859–871 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Powell CM et al. The presynaptic active zone protein RIM1alpha is critical for normal learning and memory. Neuron 42, 143–153 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nalls MA et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 18, 1091–1102 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Consortium GTEx et al. Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dong X. et al. Enhancers active in dopamine neurons are a primary link between genetic variation and neuropsychiatric disease. Nat. Neurosci. 21, 1482–1492 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiao HF et al. Transmembrane protein 108 is required for glutamatergic transmission in dentate gyrus. Proc. Natl. Acad. Sci. USA 114, 1177–1182 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mallaret M. et al. The tumour suppressor gene WWOX is mutated in autosomal recessive cerebellar ataxia with epilepsy and mental retardation. Brain 137, 411–419 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kunkle BW et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Abeta, tau, immunity and lipid processing. Nat. Genet. 51, 414–430 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Khera AV et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet. 50, 1219–1224 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nalls MA et al. Diagnosis of Parkinson’s disease on the basis of clinical and genetic classification: a population-based modelling study. Lancet Neurol. 14, 1002–1009 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Armstrong MJ & Okun MS Diagnosis and treatment of Parkinson disease: a review. JAMA 323, 548–560 (2020). [DOI] [PubMed] [Google Scholar]
- 54.Lee JK, Tran T. & Tansey MG Neuroinflammation in Parkinson’s disease. J. Neuroimmune Pharmacol. 4, 419–429 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Johnson ME, Stecher B, Labrie V, Brundin L. & Brundin P. Triggers, Facilitators, and Aggravators: Redefining Parkinson’s Disease Pathogenesis. Trends Neurosci. 42, 4–13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Irwin DJ et al. Neuropathologic substrates of Parkinson disease dementia. Ann. Neurol. 72, 587–598 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Irwin DJ et al. Neuropathological and genetic correlates of survival and dementia onset in synucleinopathies: a retrospective analysis. Lancet Neurol. 16, 55–65 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guerreiro R. et al. Investigating the genetic architecture of dementia with Lewy bodies: a two-stage genome-wide association study. Lancet Neurol. 17, 64–74 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Blanche P, Dartigues JF & Jacqmin-Gadda H. Estimating and comparing time-dependent areas under receiver operating characteristic curves for censored event times with competing risks. Stat. Med. 32, 5381–5397 (2013). [DOI] [PubMed] [Google Scholar]
Methods-only References
- 60.Alves G. et al. Incidence of Parkinson’s disease in Norway: the Norwegian ParkWest study. J. Neurol. Neurosurg. Psychiatry 80, 851–857 (2009). [DOI] [PubMed] [Google Scholar]
- 61.Parkinson Progression Marker I. The Parkinson Progression Marker Initiative (PPMI). Prog. Neurobiol. 95, 629–635 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Verbaan D. et al. Patient-reported autonomic symptoms in Parkinson disease. Neurology 69, 333–341 (2007). [DOI] [PubMed] [Google Scholar]
- 63.Beach TG et al. Arizona Study of Aging and Neurodegenerative Disorders and Brain and Body Donation Program. Neuropathology 35, 354–389 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lucero C. et al. Cognitive reserve and beta-amyloid pathology in Parkinson disease. Parkinsonism Relat. Disord. 21, 899–904 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Corvol JC et al. Longitudinal analysis of impulse control disorders in Parkinson disease. Neurology 91, e189–e201 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.DATATOP: a multicenter controlled clinical trial in early Parkinson’s disease. Parkinson Study Group. Arch. Neurol. 46, 1052–60 (1989). [DOI] [PubMed] [Google Scholar]
- 67.Williams-Gray CH et al. The distinct cognitive syndromes of Parkinson’s disease: 5 year follow-up of the CamPaIGN cohort. Brain 132, 2958–2969 (2009). [DOI] [PubMed] [Google Scholar]
- 68.Hughes AJ, Daniel SE, Ben-Shlomo Y. & Lees AJ The accuracy of diagnosis of parkinsonian syndromes in a specialist movement disorder service. Brain 125, 861–870 (2002). [DOI] [PubMed] [Google Scholar]
- 69.Goetz CG et al. Movement Disorder Society Task Force report on the Hoehn and Yahr staging scale: status and recommendations. Mov. Disord. 19, 1020–1028 (2004). [DOI] [PubMed] [Google Scholar]
- 70.Hoops S. et al. Validity of the MoCA and MMSE in the detection of MCI and dementia in Parkinson disease. Neurology 73, 1738–1745 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.van Steenoven I. et al. Conversion between mini-mental state examination, montreal cognitive assessment, and dementia rating scale-2 scores in Parkinson’s disease. Mov. Disord. 29, 1809–1815 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Uc EY et al. Incidence of and risk factors for cognitive impairment in an early Parkinson disease clinical trial cohort. Neurology 73, 1469–1477 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mollenhauer B. et al. Baseline predictors for progression 4 years after Parkinson’s disease diagnosis in the De Novo Parkinson Cohort (DeNoPa). Mov. Disord. 34, 67–77 (2019). [DOI] [PubMed] [Google Scholar]
- 74.Kasten M. et al. Cohort Profile: a population-based cohort to study non-motor symptoms in parkinsonism (EPIPARK). Int. J. Epidemiol. 42, 128–128k (2013). [DOI] [PubMed] [Google Scholar]
- 75.Bien SA et al. Strategies for enriching variant coverage in candidate disease loci on a multiethnic genotyping array. PLoS One 11, e0167758 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Purcell S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Anderson CA et al. Data quality control in genetic case-control association studies. Nat. Protoc. 5, 1564–1573 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.1000 Genomes Project Consortium et al. A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Price AL et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006). [DOI] [PubMed] [Google Scholar]
- 80.Das S. et al. Next-generation genotype imputation service and methods. Nat. Genet. 48, 1284–1287 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McCarthy S. et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 48, 1279–1283 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Loh PR et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nat. Genet. 48, 1443–1448 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Desikan RS et al. Genetic assessment of age-associated Alzheimer disease risk: Development and validation of a polygenic hazard score. PLoS Med. 14, e1002258 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cuellar-Partida G, Renteria ME & MacGregor S. LocusTrack: Integrated visualization of GWAS results and genomic annotation. Source Code Biol. Med. 10, 1 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
A Life Sciences Reporting Summary for this paper is available. Human brain cell type-specific expression data from BRAINcode47 RNA-seq data are accessible through a user-friendly webportal at www.humanbraincode.org and individual-level data through dbGAP (acc. Number phs001556.v1.p1). The gene expression profiles of human tissues used in this study can be downloaded from the GTEx Portal V7 (https://gtexportal.org/). The GWSS summary statistics for the combined analysis of discovery and replication populations are publicly accessible through the EGA database https://www.ebi.ac.uk/ega/ (acc. Number EGAS00001005110). Individual-level genetic data for the NIH-funded Illumina Multi-Ethnic Genotyping Array analyses of the HBS2 and EPIPARK cohorts are accessible in dbGAP with accession number phs002328.v1.p1 in accordance with NIH Genomic Data Sharing Policy. The whole genome sequencing and clinical data for PPMI included in this study are publicly available through a PPMI Whole Genome Sequencing Data Agreement at ppmi@loni.usc.edu. Clinical data for PDBP included in this study are publicly available through https://pdbp.ninds.nih.gov. Clinical longitudinal data for the other cohorts included are accessible through appropriate data sharing agreements that protect patient privacy with the institutions which conducted or are conducting study consents and clinical assessments under local IRB approvals.