Abstract
Bacterial translation is thought to initiate by base pairing of the 16S rRNA and the Shine–Dalgarno sequence in the mRNA’s 5′ untranslated region (UTR). However, transcriptomics has revealed that leaderless mRNAs, which completely lack any 5′ UTR, are broadly distributed across bacteria and can initiate translation in the absence of the Shine–Dalgarno sequence. To investigate the mechanism of leaderless mRNA translation initiation, synthetic in vivo translation reporters were designed that systematically tested the effects of start codon accessibility, leader length, and start codon identity on leaderless mRNA translation initiation. Using these data, a simple computational model was built based on the combinatorial relationship of these mRNA features that can accurately classify leaderless mRNAs and predict the translation initiation efficiency of leaderless mRNAs. Thus, start codon accessibility, leader length, and start codon identity combine to define leaderless mRNA translation initiation in bacteria.
INTRODUCTION
Translation initiation is a critical step for fidelity of gene expression in which the ribosome initiation complex is formed on the start codon of the mRNA. Since the canonical start codon, AUG, complements both initiator and elongator methionyl-tRNAs, the ribosome must distinguish the start AUG codon from elongator AUG codons. Incorrect initiation at an elongator AUG can lead to non-functional products that can be detrimental to cellular fitness (1–3). Canonical start codon selection is thought to occur by the base pairing of the 16S rRNA with a Shine–Dalgarno (SD) sequence in the mRNA located 5 nt upstream of the start codon (4–6). The base pairing between the 16S rRNA and mRNA was shown to be critical for initiation since mutation of the anti-SD (aSD) in the 16S rRNA is lethal (7), and translation of a gene lacking a canonical SD sequence could be restored when the 16S of the rRNA was mutated to a complementary sequence (8). While the SD–aSD pairing clearly impacts translation initiation efficiency (TIE) in Escherichia coli, other studies have found that the SD:aSD interaction is not essential for correct selection of the start codon (9,10). Indeed, ‘orthogonal’ ribosomes with altered 16S rRNA aSD sequences were found to initiate at the normal start codons throughout the transcriptome (11). Interestingly, E. coli lacks SD sites within its genome in ∼30% of its translation initiation regions (TIRs) with other species of bacteria containing SD sites in as few as 8% of their TIRs (12,13). Indeed, RNA-seq-based transcription mapping experiments have found that many bacterial mRNAs are ‘leaderless’ and begin directly at the AUG start codon (14–16), and that these mRNAs are abundant in pathogens such as Mycobacterium tuberculosis and in the mammalian mitochondria (17).
To account for the lack of essentiality of the SD site, a ‘unique accessibility model’ was proposed that posited that start codon selection occurs due to the TIR being accessible to initiating ribosomes, while elongator AUGs are physically inaccessible due to RNA secondary structures (18). This model was based upon a strong negative correlation observed between mRNA secondary structure content in the TIR and TIE (19–21). This model is further supported by genomic analysis of RNA secondary structure prediction of mRNA TIRs in which there is a lower amount of secondary structure in the TIR compared to elongator regions, which is conserved across all domains of life (22). While the unique accessibility model is overly simplistic, more advanced computational approaches have been able to combine TIR accessibility with SD strength, spacing and standby sites to more accurately predict TIE of leadered mRNAs (23). While TIR accessibility has been shown to be critical in many leadered mRNAs, it has not yet been systematically tested for leaderless mRNAs.
Genome-wide RNA-seq transcript mapping experiments have revealed that leaderless mRNAs are widespread across bacteria (14), yet little is known about their mechanism of translation initiation. While very few leaderless mRNAs have been identified in E. coli [0.7% leaderless mRNAs (24)], other bacteria and archaea contain a large majority of their transcripts as leaderless mRNAs [up to 72% leaderless mRNAs (14,25)]. Additionally, sizable proportions of leaderless mRNAs have been identified in bacteria of clinical significance, such as M. tuberculosis, and of industrial significance, such as Corynebacterium glutamicum (15,26). In the model bacterium Caulobacter crescentus, ∼17% of mRNAs are leaderless (27), with the fastest doubling time known of any bacterium with large numbers of leaderless mRNAs. In addition, C. crescentus has good genetic tools, making it an ideal model to study translation initiation of leaderless mRNAs.
Importantly, the role of TIR accessibility has not been systematically tested for leaderless mRNAs; however, some aspects of their initiation have been identified that are distinct from leadered mRNAs. Mitochondrial leaderless mRNAs have been found to lack 5′ secondary structure (28), in support of a TIR accessibility model. Additionally, mutagenesis of the Mycobacterium smegmatis pafA leaderless mRNA to perturb its secondary structure showed that secondary structure content negatively correlated with these translation levels (29). However, the changes in codon usage across the mutants make the relative impact of secondary structure and codon usage unknown for this mRNA. In opposition to the canonical initiation mechanism, leaderless mRNAs can initiate with 70S ribosomes where IF2 is known to stimulate their translation, and IF3 can inhibit leaderless translation (30,31). Additionally, AUG is the most efficient start codon in leaderless mRNAs in E. coli or Haloarchaea (32–36), while AUG and GUG are both efficient leaderless mRNA start codons in M. smegmatis (16). In E. coli, suppressor tRNAs could restore initiation on non-AUG codons for leadered RNAs, but not for leaderless RNAs (32), suggesting that for leaderless mRNAs an AUG start codon has unique initiation properties independent of perfect codon–anticodon base pairing. Indeed, genomic prediction of leaderless mRNAs suggests a very high preference of AUG (79%) at the 5′ end of leaderless mRNAs, with a smaller percentage of GUG (10%), UUG (6%) and others (3%) (13). In addition to the start codon identity, TIE of mRNAs with short leaders (<5 nt) is significantly lower as compared to their fully leaderless counterparts (34,35,37–39). Altogether, this suggests that leaderless mRNAs strongly prefer AUG and are inhibited by having short leaders.
In order to understand the mRNA sequence features needed for leaderless translation initiation, we systematically measured the effect of TIR accessibility, start codon identity and leader length on leaderless mRNA translation initiation in C. crescentus. Using synthetic in vivo translation initiation reporters, we show that TIR accessibility, start codon identity and leader length all dramatically affect leaderless mRNA TIE. The dependences of each mRNA feature on TIE were then built into a simple computational model (TIEleaderless model) that accurately predicts which RNAs in the C. crescentus transcriptome would be initiated as leaderless RNAs with an area under the curve (AUC) of a receiver operator characteristic (ROC) curve of 0.99. The TIEleaderless model also accurately predicts the TIE of in vivo leaderless mRNA reporters (R2 = 0.87). This therefore provides the first systematic analysis of mRNA features required for leaderless initiation and the C. crescentus TIEleaderless model will likely provide a foundation for our understanding of leaderless mRNA translation initiation across bacteria.
MATERIALS AND METHODS
Computational predictions of start codon accessibility
Retrieving transcript sequences
All the RNA sequences were retrieved from transcription start site and translation start site data available from RNA-seq and ribosome profiling, respectively (27,40), using the C. crescentus NA1000 genome sequence (41). For M. smegmatis, RNA-seq and ribosome profiling data were downloaded from the European Nucleotide Archive (https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-2929/), and for M. tuberculosis, RNA-seq data were obtained from Gene Expression Omnibus accession number GSE62152 and analyzed using the CP000480.1 and NC_000962 genome sequences, respectively (16). For Haloferax volcanii, RNA-seq and ribosome profiling data were provided by the DiRuggiero Lab, and analyzed with the H. volcanii NCBI RefSeq genome (taxonomy identification 2246; one chromosome, four plasmids) (42). For Mus musculus mitochondria, RNA-seq and ribosome profiling data were downloaded from (43) and analyzed with the NC_005089 genome sequence. The TIR sequences were then extracted from all open reading frames (ORFs) using 50 nt (25 nt upstream of start codon and 25 nt downstream from start codon). If the 5′ upstream untranslated region (UTR) was <25 nt, then 50 nt from transcription start site was used for all TIR calculations. Classification of mRNA type (leaderless, non-SD or SD) was obtained from (27).
Translation efficiency data
Ribosome profiling and RNA-seq translation efficiency (TE) data for C. crescentus were obtained from (27). Ribosome profiling and RNA-seq TE data for H. volcanii were obtained from the group of Prof. Jocelyn DiRuggiero (44). Ribosome profiling and RNA-seq sequencing data for M. smegmatis were obtained from the European Nucleotide Archive (https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-2929/) (16). For both ribosome profiling and RNA-seq data, the sequencing reads were downloaded as fastq files and the adapter polyA sequences were trimmed using a custom python script. Trimmed reads were then depleted of rRNA and tRNA reads by alignment with bowtie (45), and the remaining non-rRNA/tRNA reads were then aligned to the M. smegmatis MC2 155 genome. RPKM values were then calculated based upon the CP009494.1 annotation. To avoid confounding effects from initiating or terminating ribosomes, the first 15 nt and last 15 nt of ORFs were omitted from the RPKM calculations. ORFs with <50 reads in a given sample were omitted from the RPKM calculation. The TE was then calculated as the ratio of the RPKMribosome profiling/RPKMRNA-seq.
Calculation of ΔGunfold
Start codon accessibility was computed similar to (46) by comparing the native TIR RNA structure (ΔGmRNA) to that of the same TIR bound by an initiating ribosome (ΔGinit). Since ribosome binding requires a single-stranded region of the mRNA, we approximated this by forcing the TIR to be single stranded. The overall calculation was performed in three steps:
Calculation of ΔGmRNA: The minimum free energy (mfe) labeled as ΔGmRNA was calculated using RNAfold web server of the Vienna RNA websuite (47) at the growth temperature of each organism by inputting all the TIR sequences in a text file using command line function ‘RNAfold --temp “temp” <input_sequences.txt >output.txt’. The output file was in the default RNAfold format with each new sequence on one line followed by dot–bracket notation (Vienna format) in the next line. RNAstructure (48) was used to generate ct files for each of the mfe structures predicted in RNAfold that contained all the base-pair indexes for each sequence.
Calculation of ΔGinit: The base pairs in the TIR (from up to 12 nt upstream of the start codon to 13 nt downstream of the start codon) were broken and forced to be single stranded, including any pairs formed from the TIR and outside. If the 5′ UTR length was ≥25 nt, then the RBS was selected from −12 to +13 nt (25 nt). If the 5′ UTR length was <25 nt, then the TIR comprised of the entire 5′ UTR to +13 nt. A new dot–bracket file with these base-pairing constraints was then used in the RNAfold program (47) with the same RNA sequence to calculate the ΔGinit.
- Calculation of ΔGunfold: Lastly, ΔGunfold was calculated by subtracting ΔGmRNA (mfe of mRNA in native state) from ΔGinit (mfe of mRNA after ribosome binding):
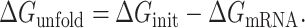
1
Cell growth and media
E. coli culture
For cloning, plasmids with the reporter gene were transformed in E. coli top 10 competent cells using the heat shock method for 50–55 s at 42°C. Luria–Bertani (LB) liquid medium was used for outgrowth and the colonies were plated on LB/kanamycin (50 μg/mL) agar plates. For miniprep, the E. coli cultures were inoculated overnight (O/N) in liquid LB/kanamycin (30 μg/mL).
C. crescentus culture
For cloning, plasmids were transformed in NA1000 C. crescentus cells after sequence verification using electroporation. The C. crescentus NA1000 cells were grown in peptone yeast extract (PYE) liquid medium. After transformation, for the outgrowth liquid PYE medium was used (2 mL) and then plated on PYE/kanamycin (25 μg/mL) agar plates. For imaging, the C. crescentus cultures were grown O/N at different dilutions in liquid PYE/kanamycin (5 μg/mL). Next day, the cultures growing in log phase were diluted and induced in liquid PYE with kanamycin (5 μg/mL) and xylose (final concentration of 0.2%) such that the optical density (OD) was ∼0.05–0.1.
Design and generation of translation reporters
Oligos and plasmid design
For the design and generation of reporter assay, a plasmid with a reporter gene [yellow fluorescent protein (YFP)], under the control of an inducible xylose promoter, was used. The pBYFPC-2 plasmid containing the kanamycin-resistant gene was originally generated from (49). A list of oligos used for generating plasmids with different 5′ UTRs of YFP is provided in Supplementary Table S6.
Inverse PCR mutagenesis and ligation
The 5′ UTR region and start codon of the YFP reporter protein were replaced with other TIR sequences. This was done by inverse PCR, in which the leaderless TIR is attached to the reverse primer as an overhang. Initial denaturation was done at 98°C for 5 min, followed by 30 cycles of denaturation at 98°C for 10 s, annealing at 60°C for 10 s and extension at 72°C for 7 min and 20 s. After 30 cycles, final extension was done at 72°C for 5 min. The polymerase used was Phusion (Thermo Scientific, 2 U/μL). The PCR product was then DPNI treated to cut the template DNA using DPNI enzyme (Thermo Scientific, 10 U/μl). The DPNI-treated sample was then purified using Thermo Fisher GeneJET PCR Purification Kit. The purified sample (50 ng) was then used for blunt-end ligation using T4 DNA Ligase (Thermo Scientific, 1 Weiss U/μL).
Transcription reporter design
For the design of transcription reporter assay, a plasmid with a 28 nt mutant version of 5′ UTR (CCNA_03971) in front of reporter gene (YFP) was used. This reporter gene had the nucleotide A at its +1 position and the reporter gene was under the control of an inducible xylose promoter. The pBYFPC-2 plasmid containing the kanamycin-resistant gene was originally generated from (49). The +1 nucleotide was mutated to all other nucleotides (G, C or T) and these three mutant plasmids were synthesized into DNA oligos and cloned by Genscript.
The insertion sequence from the +1 nt (underlined) to 28 nt including start codon (atg) to the RBS for each construct is shown below:
A. accgattaacgatggtggttgttctggc
C. cccgattaacgatggtggttgttctggc
G. gccgattaacgatggtggttgttctggc
T. tccgattaacgatggtggttgttctggc
Transformation in E. coli cells
Five microliters of the ligation reaction was then added to 50 μL of E. coli top 10 competent cells. Then, the mixture was incubated in ice for 30 min, heat shocked for 50–55 s in the water bath at 42°C and immediately kept in ice for 5 min, after which 750 μL of LB liquid medium was added to the cells for outgrowth and kept for incubation at 37°C for 1 h at 200 rpm. After this, 200–250 μL of the culture was plated on LB/kanamycin (50 μg/mL) agar plates.
Colony screening and sequence verification
The colonies grown on LB/kanamycin plates were screened by colony PCR to first screen for the presence of the new TIR insert. The cloning results in the replacement of the larger 5′ UTR region of YFP with a smaller region containing a leaderless TIR, thus distinguished easily on an analytical gel. The forward and reverse primers used for the screening result in ∼180 bp, whereas the original fragment amplified with the same oligos is 245 bp. The forward oligo used was pxyl-for: cccacatgttagcgctaccaagtgc and reverse oligo was eGYC1: gtttacgtcgccgtccagctcgac. Upon verification, a small aliquot (4 μl) of the colony saved in Taq polymerase buffer was inoculated in 5 mL of liquid LB/kanamycin (30 μg/mL) and incubated O/N at 37°C at 200 rpm. Next day, the culture was miniprepped using Thermo Fisher GeneJET Plasmid Miniprep Kit. The concentration of DNA in the miniprepped samples was measured using Nanodrop 2000C from Thermo Scientific. DNA samples were sent to Genewiz for Sanger sequencing to verify the correct insert DNA sequences using the DNA primer eGYC1: gtttacgtcgccgtccagctcgac (49).
Transformation in C. crescentus NA1000 cells
After the sequences were verified, the plasmids were transformed in C. crescentus NA1000 cells. For transformation, the NA1000 cells were grown O/N at 28°C in PYE liquid medium at 200 rpm. The next day, 5 mL of cells were harvested for each transformation, centrifuged and washed three times with autoclaved Milli-Q water. Then, 1 μL of sequence verified plasmid DNA was mixed with the cells and electroporated using Bio-Rad Micropulser (program Ec1 set at voltage of 1.8 kV). Then, the electroporated cells were immediately inoculated in 2 mL of PYE for 3 h at 28°C at 200 rpm. Then, 10–20 μL of culture was plated on PYE/kanamycin agar plates. Kanamycin-resistant colonies were grown in PYE/kanamycin media O/N and then stored as a freezer stock in the −80°C freezer.
Cellular assay of translation reporters
C. crescentus cells harboring reporter plasmids were serially diluted and grown O/N in liquid PYE/kanamycin medium (5 μg/mL). The next day, cells in the log phase were diluted with fresh liquid PYE/kanamycin (5 μg/ml) to have an OD of 0.05–0.1. The inducer xylose was then added in the medium such that the final concentration of xylose is 0.2%. The cells were grown for 6 h at 28°C at 200 rpm. After this, 2–5 μL of the culture was spotted on M2G 1.5% agarose pads on a glass slide. After the spots soaked into the pad, a coverslip was placed on the pads and the YFP level was measured using fluorescence microscopy using a Nikon eclipse NI-E with CoolSNAP MYO-CCD camera and 100× Oil CFI Plan Fluor (Nikon) objective. Image was captured using Nikon Elements software with a YFP filter cube with exposure times of 30 ms for phase-contrast images and 300 ms for YFP images. The images were then analyzed using a plug-in of software ImageJ (50) called MicrobeJ (51).
Three-component model calculations and leader length/identity analysis
For all RNA transcripts in the C. crescentus genome identified in (27,40), we computed their capacity to initiate as a leaderless mRNA using the following equation:
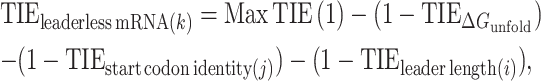 |
(2) |
where k is a given RNA transcript, j is start codon identity and i is leader length (nt). To identify putative leaderless mRNA TIRs, we first asked whether the 5′ end contained an AUG or near-cognate start codon, and if not, we scanned successively from the 5′ end for AUG trinucleotides within the first 8 nt. Near-cognate start codons were omitted from positions containing leader nucleotides since AUG codons yielded higher TIE values even in the presence of a leader. We next asked whether there is an AUG or near-cognate start codon further downstream by scanning 5′ to 3′ through the first 18 nt. If found, we calculated TIEleaderless mRNA with all different possible cognate/near-cognate start codons along the TIR. Then, of all the different possibilities, the one having the highest TIEleaderless score was selected for further analysis (Figure 7A).
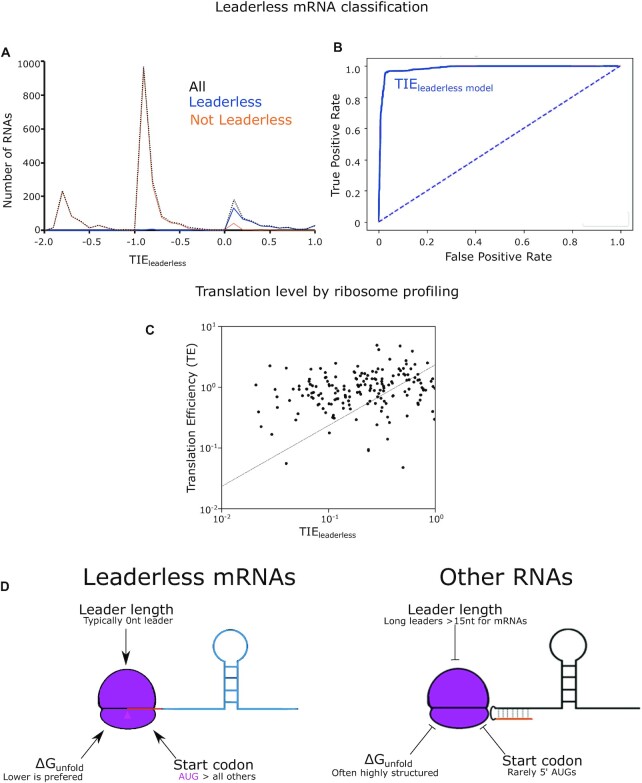
Figure 7.
A combinatorial model accurately predicts translation of leaderless mRNAs. (A) Line graph showing the predicted TIEleaderless scores on the X-axis and the number of RNAs on the Y-axis. The solid blue line represents natural leaderless mRNAs. The orange line represents the RNAs that are not leaderless RNAs. The black dotted line represents all RNAs. RNAs with short leaders are shown in Supplementary Figure S2. (B) ROC curve (shown in solid blue, with ‘random’ shown as a dotted line) with true positive rate on the Y-axis and false positive rate on the X-axis. The AUC was calculated to be 0.99 for classification based on the TIEleaderless score and 0.68 for classification based solely on presence of a 5′ AUG (Supplementary Figure S3). (C) TE of leaderless mRNAs (Y-axis) is plotted compared to TIEleaderless (X-axis). The trendline is the result of a least-squares fit yielding a slope of 0.71 and R2 = 0.06. (D) Model design showing ribosome binding to the AUG trinucleotide (pink triangle) at the 5′ end when it is highly accessible as shown in the left. The ribosome binding is prevented when the region becomes more structured and the accessibility decreases.
To utilize TIEleaderless mRNA for classification, each RNA was then categorized into two different classes based on 5′ end sequencing data and ribosome profiling-based global assays (27,40): true leaderless, RNAs that are known to initiate directly at a 5′ start codon [judged by a complete lack of a 5′ UTR and a ribosome density >1/20 the downstream CDS (27)]; and false leaderless, RNAs that are not initiated at a 5′ start codon. A small subset was classified as ‘unknown’, as they contain very short leaders and lack SD sites, making their mode of translation initiation ambiguous. Using these TIEleaderless mRNA values, an ROC curve was plotted using scikit-learn library in python (52) with the ‘true leaderless’ and ‘false leaderless’ RNAs (TIEleaderless mRNA values for the C. crescentus transcriptome can be found in Supplementary Table S1).
To utilize TIEleaderless mRNA for prediction of translation initiation reporter levels, we first converted all negative TIEleaderless mRNA scores to zero. Next, we compared the TIEleaderless mRNA scores to the YFP levels of the translation initiation and performed a linear regression calculation using the linest function in Microsoft Excel and LibreOffice Calc. For prediction of native leaderless mRNA translation levels, TE measurements from ribosome profiling experiments (27) were compared to the TIEleaderless mRNA scores.
RESULTS
Computational prediction of C. crescentus start codon accessibility
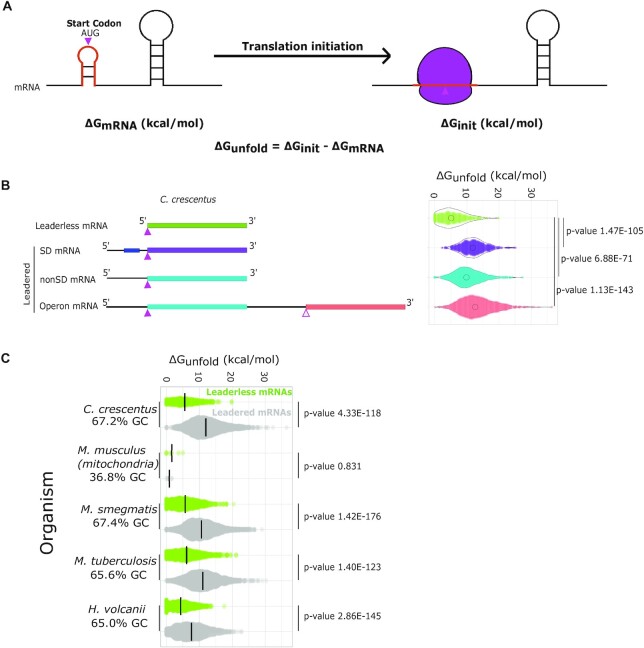
To assess the role of mRNA accessibility across mRNA types, ΔGunfold calculations were performed on all C. crescentus TIRs. ΔGunfold represents the amount of energy required by the ribosome to unfold the mRNA at the TIR and has been identified as a metric that correlates with TE in E. coli (46). ΔGunfold was calculated for all TIRs by first predicting the mfe of the 50 nt region of the mRNA (ΔGmRNA) around the start codon using RNAfold (47). ΔGinit was then calculated in which the TIR (25 nt surround the start codon), roughly equivalent to a ribosome footprint, was constrained to be single stranded to approximate accessibility for the ribosome to initiation. ΔGunfold was then calculated using Equation (1), which represents the energy required to open the TIR to facilitate translation initiation (Figure 1A). ΔGunfold calculations were performed on all the CDSs in the genome (Figure 1B) and classified into mRNA types based on transcriptome and ribosome profiling maps of the C. crescentus genome (27). The transcripts were categorized into two major classes: leaderless (no 5′ UTR) and leadered (those containing a 5′ UTR). Leadered mRNAs were further categorized into subclasses based upon the presence of the SD sequence (27): SD (containing an SD sequence in the 5′ UTR) and non-SD (lacking an SD sequence in the 5′ UTR). Since it is also known that some polycistronic operons reinitiate translation between CDSs without dissociation of the ribosomal subunits, we also examined the ΔGunfold of TIRs occurring downstream of the first CDS in polycistronic mRNAs (operons). The average ΔGunfold value of leaderless mRNAs (5.6 kcal/mol) was significantly lower than those of SD (11.9 kcal/mol, P = 1.5E−105), non-SD (10.3 kcal/mol, P = 6.9E−71) and internal operon TIRs (13.2 kcal/mol, P = 1.1E−143) as calculated by pairwise two-sided t-tests with unequal variance (Figure 1B). The lower ΔGunfold values of non-SD TIRs may be due to the loss of stabilization of TIRs from base pairing between the anti-SD site in the 16S rRNA and the SD site in the mRNA. We also observed that average ΔGunfold of non-SD TIRs was significantly lower than those of SD TIRs (P = 1.8E−14) and operon TIRs (P = 1.4E−44). The difference between the average ΔGunfold of SD and operon genes was also significant (P = 2.1E−09). Because the ribosome is an efficient RNA helicase, it is possible that the increased ΔGunfold of operon TIRs may be tolerated by the ribosome’s ability to unwind such structures when terminating on the previous CDSs. We hypothesized that the low ΔGunfold observed for leaderless mRNAs was due to an intrinsic requirement for their initiation; however, because the size of the leaderless mRNA footprint is significantly smaller than a leadered mRNA footprint, the low ΔGunfold observed for leaderless mRNAs could potentially be explained by the smaller ribosome footprint size. To explore this possibility, we analyzed the ΔGunfold of leadered mRNA TIRs using the same footprint size and region as leaderless mRNAs (13 nt) in the ΔGunfold calculation (Supplementary Figure S1). We observed that the ΔGunfold was still significantly lower for leaderless mRNA TIRs, suggesting that the low ΔGunfold for leaderless mRNA TIRs is not simply an artifact of the smaller mRNA footprint size.
Figure 1.
Leaderless mRNA TIRs are more accessible than leadered mRNAs. (A) Predicted unfolding energy of mRNAs. The predicted mRNA mfe (ΔGmRNA) is represented on the left. The orange TIR indicates a ribosome footprint surrounding the start codon (pink). The image on the right represents the mRNA upon initiation (ΔGinit) where the orange initiation region is unfolded. The ΔGunfold represents the amount of energy required by the ribosome to unfold the TIR of the mRNA. (B) Violin plots of ΔGunfold (right) calculated for all the mRNAs of each class (left) in the C. crescentus genome based on the transcript architecture (27,40). P-values were calculated based on a t-test (two-tailed, unequal variance). (C) Violin plots of ΔGunfold calculated for leaderless (green) and leadered (gray) mRNAs for various organisms with ribosome profiling data. ΔGunfold values were calculated using the optimal growth temperature of each microorganism and 37°C for M. musculus mitochondria. P-values were calculated based on a t-test (two-tailed, unequal variance).
To explore whether low ΔGunfold for leaderless mRNA TIRs is a species-specific property of C. crescentus, or a general property of leaderless mRNA TIRs, we calculated ΔGunfold for TIRs in other organisms identified to contain a significant number of leaderless mRNAs. We identified two additional bacteria (M. smegmatis and M. tuberculosis) (16), one archaeal species (H. volcanii) (25,44) and one mitochondrial genome (M. musculus) (43) that had transcriptome information and ribosome profiling or mass spec data supporting a significant number of leaderless mRNAs. Across bacteria and archaea, the leaderless mRNA ΔGunfold remained quite low as compared to their leadered mRNAs counterparts (leaderless average 5.6–7.6 kcal/mol, leadered average 12.0–12.9 kcal/mol) (Figure 1C). In M. musculus mitochondria, however, leaderless mRNAs and leadered mRNAs were both observed to have low ΔGunfold (Figure 1C), perhaps in part due to the relatively low GC percentage. Since leaderless mRNAs showed a rather low ΔGunfold, and lack complexities associated with leadered mRNAs, such as SD or standby sites that are important for leadered initiation (23), we further explored the functional role of ΔGunfold in C. crescentus leaderless mRNAs.
Systematic analysis of C. crescentus leaderless mRNA TIR determinants using in vivo translation reporters
Leaderless mRNA initiation is known to be strongly influenced by addition of nucleotides prior to the start codon (leader nts) and by start codon identity (32–39); however, the role of TIR accessibility has been poorly described in this class of mRNAs. To understand the role of these three mRNA features, we systematically tested each feature using in vivo leaderless mRNA translation initiation reporters. Translation initiation reporters were designed in which the start codon of plasmid pBXYFPC-2 was replaced with an AUG fused directly to the +1 nt of the xylose promoter (49). The xylose promoter was chosen because it is one of the best characterized promoters in Caulobacter and its TSS was mapped to the same nt by two independent methods (40). An additional 15–24 nt after the 5′ AUG was added to allow complete replacement of the 5′ leader and start codon in pBXYFPC-2 with a leaderless TIR. Since only the first six to nine codons are altered across leaderless mRNA mutants, and the vast majority of the YFP CDS is unaltered, this allows a sensitive system to measure changes in translation initiation. As leaderless TIR mutants may also alter the amino acid sequence, additional care was also taken to ensure that mutations would not alter the N-end rule amino acid preferences of the resulting proteins (53). Using this in vivo translation initiation system, we generated three different sets of leaderless TIR reporters to test the effect of ΔGunfold, start codon identity and additional leader length on C. crescentus translation initiation.
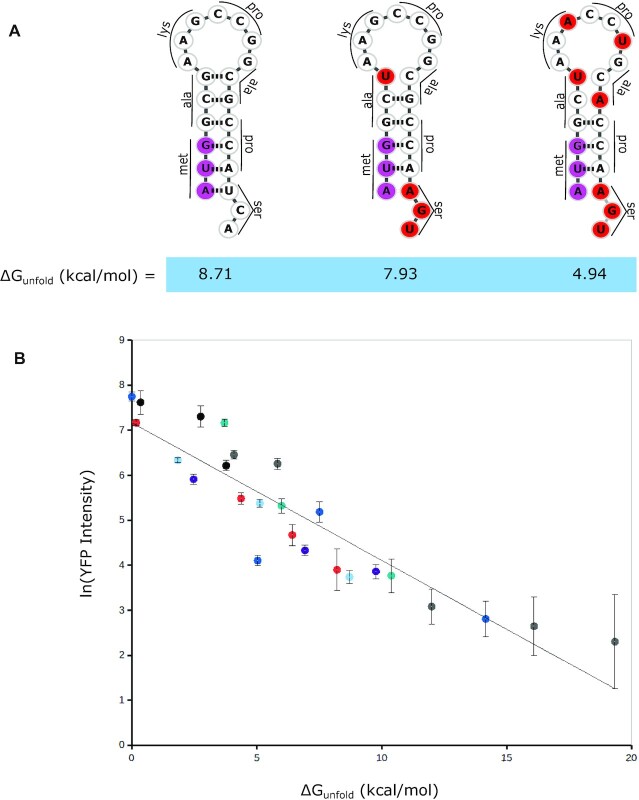
As leaderless mRNAs were predicted to have TIRs with low ΔGunfold values, we engineered several RNA hairpins in the TIR to assess the role of ΔGunfold on translation initiation (Figure 2A). Since very few natural C. crescentus mRNAs contained RNA structure content in their TIRs (Figure 1B), six synthetic hairpins were designed, varying in stem and loop sizes (Supplementary Table S2). Into each construct, we also introduced synonymous codon mutations designed to alter the secondary structure content, yielding a range of ΔGunfold values without altering the amino acid sequence within a given hairpin (Supplementary Table S2). Importantly, the entire range of ΔGunfold values across the synthetic hairpins spans the entire range calculated for natural leaderless mRNAs (Figure 1, Supplementary Table S2). For all hairpins, we observed that lowering ΔGunfold and thereby increasing the accessibility of the start AUG led to an increase in the level of YFP production (Figure 2B). Since six of the seven hairpin mutant sets showed a relationship in which hairpin codon usage frequency positively correlated with ΔGunfold (Supplementary Table S2), it is most likely that the observed reduction in YFP reporter levels is a result of increased structure content and is not likely to be caused by faster elongation of common codons in the TIR. Additionally, across all mutant hairpin sets generated, we observed a strong negative correlation between the YFP reporter level and the ΔGunfold across a vast range of values with a linear correlation R2 value of 0.84 (Figure 2B). These data suggest that accessibility of the start codon is a critical feature for leaderless mRNA translation initiation.
Figure 2.
ΔGunfold strongly influences leaderless mRNA translation. (A) A representative TIR synthetic stem loop synonymous mutation set with varying ΔGunfold values. The bases in the start codon are colored pink, and red bases highlight where mutations were introduced to disrupt base pairing. (B) In vivo translation reporter levels for the various leaderless RNA mutants. Each hairpin and its synonymous codon mutant set are shown with the same color (raw data can be found in Supplementary Table S1). Black points, leaderless set 1; gray points, leaderless set 2; dark blue points, leaderless set 3; purple points, leaderless set 4; light blue points, leaderless set 5; red points, leaderless set 6; and teal points, leaderless set 7. The natural log of the average YFP intensity per cell is shown and error bars represent the standard deviation of three biological replicates. The dotted blue line represents a linear curve fit with an R2 value of 0.84 and a slope of −0.3.
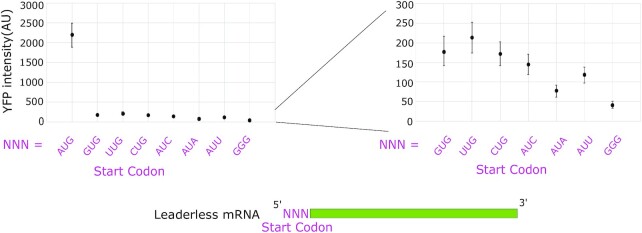
Next, we systematically tested the effect of the start codon identity on the in vivo translation initiation reporters. In C. crescentus, natural leaderless mRNAs initiate with an AUG, GUG or UUG start codon (27,40). Since it is well established that start codon identity can affect leaderless mRNA translation initiation (32–36), we generated variants with different start codon identities. Here, AUG was mutated to other near-cognate start codons GUG, CUG, UUG, AUC, AUU and AUA that are known to be the start codons of other leadered mRNAs in C. crescentus (27). We also included a non-cognate GGG codon as a negative control since no GGG start codons are known to occur in C. crescentus. The results showed that replacing the original AUG codon with any of the other near-cognate codons drastically decreased the translation initiation reporter levels, while the GGG codon yielded the lowest translation initiation reporter levels (Figure 3). To examine whether the mutation in +1 nt resulted from lower transcription or from lower translation, we generated leadered mRNA reporters with all four possible +1 nucleotides and tested their in vivo reporter levels similarly to that in M. smegmatis (16). A +1 G led to a mild reduction in reporter activity compared to a +1 A (Supplementary Figure S5), suggesting that most of the observed changes in the leaderless mRNA reporter levels likely come from translation; however, it remains possible that mutants containing 5′ YTG could have an altered TSS due to the favored +2 pyrimidine. A TSS shift upstream would introduce a leader reducing translation, while a shift downstream would cause a loss of the start codon. These data show that the AUG triplet is by far the preferred start codon for C. crescentus leaderless mRNAs and this may be due to factors affecting both transcription and translation.
Figure 3.
Leaderless mRNAs have a strong preference for AUG start codons. Leaderless mRNA in vivo translation reporters were generated with the start codons listed on the X-axis and their average YFP intensity per cell was measured. On the right is a zoomed-in view of all non-AUG codons tested. Error bars represent the standard deviation from three biological replicates.
Finally, we systematically tested the role of additional leader length in C. crescentus leaderless mRNAs. In E. coli, even a single nucleotide before the AUG is known to inhibit initiation of leaderless mRNAs (37). To test whether C. crescentus leaderless mRNAs were negatively impacted by leader nucleotides, we generated a set of reporters with 0, 1, 2, 3, 5, 10 or 20 5′ adenosines before the AUG start codon (Figure 4). An A-rich sequence was chosen as it lacks any possible SD sites and is unlikely to form secondary structure, and ΔGunfold values were not altered upon addition of these 5′ bases to the leaderless translation initiation reporter (Supplementary Table S1). Across this set of mutants, additional nucleotides showed a strong decrease in translation initiation reporter levels with increasing leader length (Figure 4). The translation initiation reporter levels dropped by ∼2-fold for each additional A that was added to the 5′ end [TIEleader length = 0.45 × i–0.91, R2 = 0.92, i = leader length (nt)]. This confirms that even a short leader can lead to a significant reduction in translation initiation of C. crescentus leaderless mRNAs. Importantly, it is possible that the addition of extra A nucleotides may lead a downstream shift in the TSS position; in such a case, this would imply that additional leader nucleotides may be even more potent inhibitors of leaderless initiation than indicated by the reporter assay.
Figure 4.
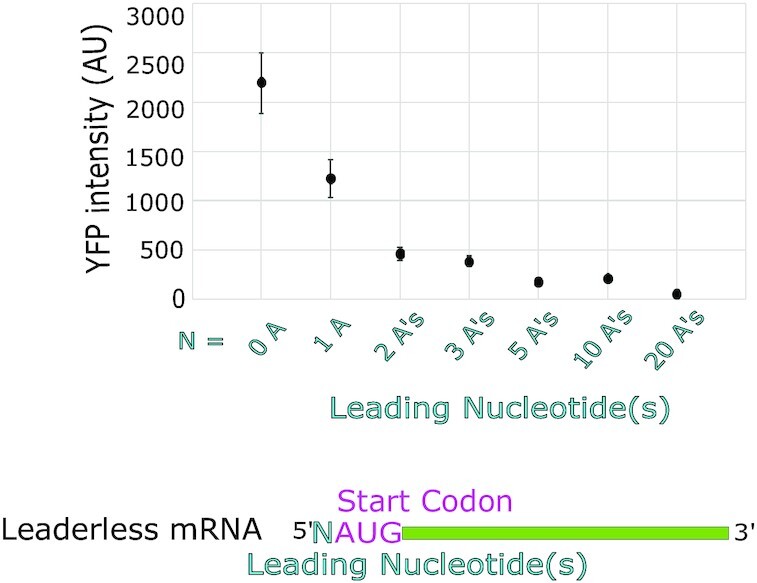
Leaderless mRNAs are inhibited by additional upstream nucleotides. Leaderless mRNA in vivo translation reporters were generated with variable number of leading nucleotides on the X-axis and their average YFP intensity per cell was measured (raw data can be found in Supplementary Table S1). Error bars represent three biological replicates.
Leaderless mRNA TIR determinants affect TE of natural leaderless mRNAs
Because the in vivo translation initiation reporters were all synthetic constructs, we explored the extent to which each mRNA feature (ΔGunfold, start codon identity and leader length) occurs in natural C. crescentus leaderless mRNAs. As noted previously, ΔGunfold is significantly lower for leaderless mRNAs than for other mRNA types (Figure 1B). To analyze the role of start codon selection, we calculated the fraction of AUGs at the 5′ end of all C. crescentus leaderless mRNAs and of the random chance of finding each start codon based on the genomes’ GC percentage. This analysis revealed a strong enrichment of AUGs at the 5′ end of C. crescentus leaderless mRNAs as compared to random, and a slight enrichment of the GUG near-cognate start codons (Figure 5A). While GUG TIR reporters yielded similar TIR reporter levels to UUG and CUG, it is possible that the lack of U and C at the +1 of C. crescentus leaderless mRNAs is due to their low abundance in TSSs (40). Of all the leaderless mRNAs, only 4.4% (17/385) are initiating with non-AUG start codons as compared to the leadered mRNAs of which 27.23% (989/3632) of genes initiate with non-AUG start codons (Supplementary Table S3). Since these near-cognate start codons were translated much more poorly than AUG in our translation initiation reporters, it is possible that for leaderless mRNAs there is a positive selection for the AUG start codon and a negative selection for near-cognate start codons. Additionally, by exploring the length of mRNAs, we noticed that there was a much greater occurrence of leaderless mRNAs than mRNAs with short leaders <10 nt (Figure 5B). Additional leader nucleotides were strongly inhibitory of leaderless translation, and only eight contain SD motifs, suggesting some of these short-leadered mRNAs may be poorly initiated.
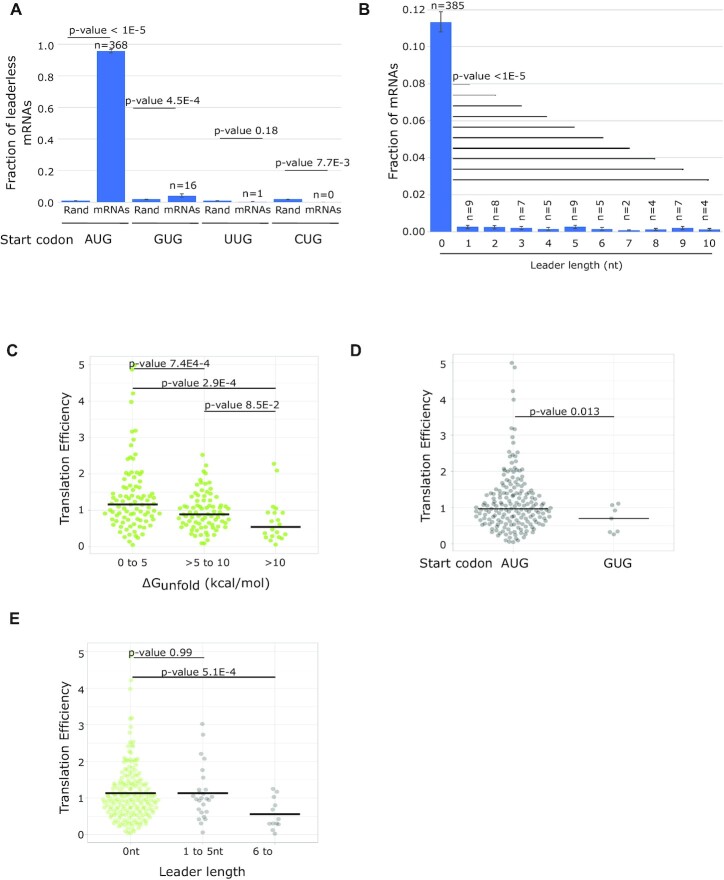
Figure 5.
ΔGunfold, start codon identity and leader length correlate with TE across native leaderless mRNAs. (A) Bar graph showing the fraction of leaderless mRNAs starting with AUG, GUG, UUG and CUG start codons. Also shown are the random chances of trinucleotides being AUG, GUG, UUG and CUG calculated based on GC content (67%) of C. crescentus genome. P-values were calculated based on a two-tailed Z-test. (B) Bar graph showing the fraction of leaderless mRNAs and mRNAs with 5′ UTR of length 1–10 [as determined in (40)]. mRNAs containing SD sites were excluded from this analysis. P-values were calculated based on a two-tailed Z-test of each leader length compared to leader length 0. (C) Violin plot of TE as measured by ribosome profiling (54) of natural leaderless mRNAs binned in three groups depending on ΔGunfold values (0–5, 5–10 and >10 kcal/mol). P-values based on a t-test (two-tailed, unequal variances). (D) Violin plot of TE as measured by ribosome profiling (54) of natural leaderless mRNAs starting with AUG and GUG. P-values were calculated based on a t-test (two-tailed, unequal variance). (E) Violin plot showing the TE as measured by ribosome profiling (54) on the Y-axis of leaderless mRNAs (green) and with leaders of varying length (1–10) shown in gray. P-values were calculated based on a t-test (two-tailed, unequal variance).
To estimate the effects of each mRNA feature (ΔGunfold, start codon identity and leader length) on natural leaderless mRNA translation, we next analyzed ribosome profiling data of the C. crescentus mRNAs (27). Here, we utilized TE measurements that approximate the relative number of ribosome footprints to mRNA fragments from the same cell samples (54). In total, TE data for 191 leaderless mRNAs and 38 short-leadered mRNAs (1–10 leader length) were obtained for cells grown in PYE media (27). We separated leaderless mRNAs into three groups based upon their ΔGunfold values (0–5, 5–10 and >10 kcal/mol) and compared their TE. The median TE was reduced as the ΔGunfold increased (Figure 5C) (median = 1.2 for 0–5 kcal/mol, median = 0.89 for 5–10 kcal/mol, median = 0.54 for >10 kcal/mol), similar to the dependence observed in the synthetic translation reporters (Figure 2B). For start codon identity, we noticed that a majority of leaderless mRNAs with near-cognate start codons had translation efficiencies that were not measurable because their genes contained an additional upstream TSS. However, for the seven GUG mRNAs whose TE was measured, the median (0.70) was lower than that of the AUG initiated leaderless mRNAs (0.97) (Figure 5D), in line with the findings of the synthetic reporters (Figure 3). Finally, we compared the TE of leaderless mRNAs with those with very short leaders (Figure 5E). Since eight of these mRNAs with short leaders contain SD sequences in the leader, we removed these RNAs from the analysis because we expect them to initiate translation by the canonical mechanism. As leader length increases, we generally observed that the TE tends to decrease (Figure 5E), again in line with the synthetic reporters (Figure 4). Overall, these data suggest that the effects of ΔGunfold, start codon identity and leader length observed in the synthetic translation initiation reporters are also observed across natural C. crescentus leaderless mRNAs.
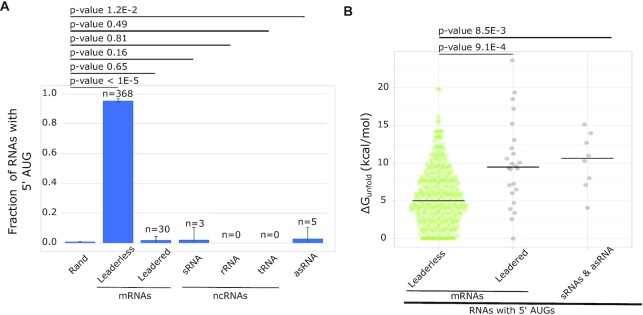
Many RNAs present in the C. crescentus transcriptome are not initiated as leaderless mRNAs, so we explored the relative fraction of 5′ AUG trinucleotides in all classes of RNAs (Figure 6A). As noted previously, leaderless mRNAs are highly enriched in AUG codons (Figure 5A). Surprisingly, leadered mRNAs contain a similar fraction of 5′ AUGs as would be predicted from the genome’s GC percentage, which is also observed in small non-coding RNAs (sRNAs) and antisense RNAs (asRNAs). Conversely, tRNAs and rRNAs contain zero cases with a 5′ AUG. To explore why these RNAs are not initiated as leaderless mRNAs, we calculated the ΔGunfold of each class of 5′ AUG containing RNA (Figure 6B). If these 5′ AUGs found in non-leaderless RNAs were inaccessible to ribosomes, it would be permissible for this sequence to be present at the 5′ end without causing aberrant initiation. Indeed, for the RNAs with 5′ AUGs, we observe that leaderless mRNAs have a low ΔGunfold (median = 5.0), while leadered mRNAs (median = 9.5), sRNAs (median = 14) and asRNAs (median = 9.0) all contain significantly higher ΔGunfold values. This suggests that RNAs with inaccessible 5′ AUGs are blocked from leaderless mRNA initiation.
Figure 6.
Non-coding RNAs with 5′ AUGs are rare and have higher ΔGunfold. (A) Bar graph showing the fraction of natural leaderless mRNAs starting with trinucleotide AUG and other types of RNAs starting with trinucleotide AUG, but not initiated at that AUG (leadered mRNAs, sRNAs, rRNAs, tRNAs and asRNAs). Also shown is the random chance of trinucleotide being AUG out of 10 000 nucleotides, calculated based on GC content of C. crescentus genome. P-values were calculated using a two-tailed Z-test with each RNA class compared to the random probability of 5′ AUG. (B) Violin plot showing ΔGunfold of natural leaderless mRNAs starting with AUG (green) and other types of RNAs starting with AUG, but not initiated at that AUG (leadered mRNAs, RNAs and asRNAs) (shown in gray). P-values were calculated based on a t-test (two-tailed, unequal variance).
Due to the strong involvement of ΔGunfold, start codon identity and leader length in C. crescentus leaderless mRNA TIRs, we also explored these features across organisms. As already shown in Figure 1C, the ΔGunfold for leaderless mRNAs is markedly lower than leadered mRNAs in all species analyzed with the exception of M. musculus mitochondria. The low ΔGunfold in the mitochondria may be due to alterations in the translation initiation mechanism of leadered mRNAs by their highly proteinaceous ribosomes (55). Across these organisms, ribosome profiling and total RNA-seq were performed in M. smegmatis (16) and H. volcanii (44), allowing the comparison of how ΔGunfold tracks with TE. As observed for C. crescentus, as ΔGunfold increases, a drop is observed in the TE in leaderless mRNAs in both M. smegmatis and H. volcanii (Supplementary Figure S4). We hypothesize that the low overall ΔGunfold observed for leaderless mRNAs across all species suggests that ribosome accessibility is a key feature for leaderless mRNAs across organisms. In M. smegmatis and H. volcanii, the observed drop in TE from the 0–5 and 5–10 bins was smaller than that observed for C. crescentus, which may be due to the higher growth temperatures of these organisms. Next, we explored the distributions of leader lengths across species (Supplementary Table S4). As observed in C. crescentus, mRNAs with short leaders have a highly skewed distribution across species, with leaderless mRNAs showing the largest peak, with a small fraction of mRNAs containing a 1 nt leader, and a much smaller population of mRNAs observed with a ≥2 nt leader (Supplementary Table S4). The lower abundance of mRNAs with very short leaders is likely to be due to their poorer translation levels observed across organisms (Supplementary Figure S4) as this has even been observed with E. coli leaderless TIR reporters (37). To explore this possibility, we compared TE across organisms. As observed in C. crescentus, M. smegmatis and H. volcanii, TE is also markedly decreased in mRNAs containing short leaders (Supplementary Figure S4). While C. crescentus leaderless mRNA TE was not significantly lower than the 1–5 nt bin, both M. smegmatis and H. volcanii showed sharper drops in TE. While C. crescentus mRNAs with 6–10 nt leaders showed a significant drop in TE, neither M. smegmatis nor H. volcanii showed a significant decrease. This discrepancy may be explained by a low sample size in M. smegmatis, where only three mRNAs were identified in the 5–10 nt bin, while in H. volcanii the 5–10 nt bin distribution contained a single outliner whose TE was measured to be >30. Finally, we examined start codon identities for leaderless mRNAs across species (Supplementary Table S5). Here, only H. volcanii was similar to C. crescentus with a strong bias in the AUG start codon for leaderless mRNAs (Supplementary Table S5). M. tuberculosis and M. smegmatis both contained GUG start codons in leaderless mRNAs with similar abundance to AUG (Supplementary Table S5). In the M. musculus mitochondria, AUG is the most common start codon across mRNAs; however, AUG is less common in leaderless mRNAs, and the summed use of GUG, AUC, AUU and AUA makes near-cognate start codons more abundant than AUG initiated leaderless mRNAs (Supplementary Table S5). Interestingly, mitochondrial RNAP has been found to initiate transcription efficiently with NAD+ and NADH (56), which has the potential to alter start codon selection by the translation machinery. As observed in C. crescentus, M. smegmatis and H. volcanii both showed a lower TE for leaderless mRNAs starting with GUG as compared to AUG (Supplementary Figure S4). The magnitude of the reduced TE for GUG initiated leaderless mRNAs is significantly smaller in M. smegmatis (1.1 AUG, 0.91 GUG) as compared to H. volcanii (2.5 AUG, 0.82 GUG), which is in line with previous data showing that GUG initiates with similar efficiency to AUG in leaderless mRNAs in mycobacteria (16). Overall, these data suggest that ΔGunfold, start codon identity and leader length have similar effects on leaderless mRNA translation across species. However, minor idiosyncratic differences in the frequency and magnitude of each leaderless mRNA feature on TE were observed across species, likely arising from differences in the translation machinery.
Three-component model describes leaderless mRNA start codon selection
In order to understand how the mRNA determinants combine to dictate leaderless mRNA translation, we built a computational model based upon the three features (ΔGunfold, start codon identity and leader length) and explored its ability to describe leaderless mRNA start codon selection and efficiency of leaderless mRNA translation initiation. From our synthetic in vivo translation initiation reporters, we performed curve fitting to assess the relative effect of each feature on TIE. For each feature (ΔGunfold, start codon identity and leader length), the highest reporter level measured in each mutant set was normalized to 1 before curve fitting. ΔGunfold data were fit to an exponential equation [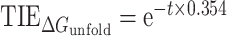 , where t is ΔGunfold (kcal/mol) and R2 = 0.78], leader length data were fit to a power equation (TIEleader length = 0.45 × i–0.92, where i is leader length >0, R2 = 0.92 and TIEleader length = 1 for i = 0), TIEstart codon was based directly on reporter levels for each near-cognate start codon (Figure 3) and all other codons were given a value of 0 (Supplementary Table S1). For each mRNA feature, we therefore generated a function that could calculate the relative TIE of any RNA in C. crescentus based upon the mRNA sequence. We then built a computational model in which the three features were assumed to be independent from each other to calculate a summed TIE. In this model, we set the maximum TIE to 1, and then subtracted the effects of the sequence feature as measured from the in vivo translation reporters in Equation (2). Using Equation (2), we predicted the TIE for each RNA in the C. crescentus transcriptome (Figure 7A). For all RNAs, we successively scanned for the closest AUG or near-cognate start codon to the 5′ end and used this for the TIE calculation. RNAs known to be initiated as leaderless mRNAs (27,40) yielded higher TIE scores (median = 0.15, σ = 0.35), while TIE scores for all other RNAs were typically lower (median = −0.95, σ = 0.45). To estimate the utility of this model at classifying leaderless mRNAs, we used an ROC analysis (Figure 7B). The ROC AUC for the TIEleaderless model was equal to 0.99, which significantly outperforms identifying RNAs with 5′ AUGs (ROC AUC 0.68) suggesting the TIEleaderless model can accurately classify those RNAs that are initiated as leaderless mRNAs with high accuracy and precision. The success of this simple TIEleaderless model to classify leaderless mRNAs based on the combinations of ΔGunfold, start codon identity and leader length suggests that these mRNA features combinatorically control translation initiation on leaderless mRNAs.
, where t is ΔGunfold (kcal/mol) and R2 = 0.78], leader length data were fit to a power equation (TIEleader length = 0.45 × i–0.92, where i is leader length >0, R2 = 0.92 and TIEleader length = 1 for i = 0), TIEstart codon was based directly on reporter levels for each near-cognate start codon (Figure 3) and all other codons were given a value of 0 (Supplementary Table S1). For each mRNA feature, we therefore generated a function that could calculate the relative TIE of any RNA in C. crescentus based upon the mRNA sequence. We then built a computational model in which the three features were assumed to be independent from each other to calculate a summed TIE. In this model, we set the maximum TIE to 1, and then subtracted the effects of the sequence feature as measured from the in vivo translation reporters in Equation (2). Using Equation (2), we predicted the TIE for each RNA in the C. crescentus transcriptome (Figure 7A). For all RNAs, we successively scanned for the closest AUG or near-cognate start codon to the 5′ end and used this for the TIE calculation. RNAs known to be initiated as leaderless mRNAs (27,40) yielded higher TIE scores (median = 0.15, σ = 0.35), while TIE scores for all other RNAs were typically lower (median = −0.95, σ = 0.45). To estimate the utility of this model at classifying leaderless mRNAs, we used an ROC analysis (Figure 7B). The ROC AUC for the TIEleaderless model was equal to 0.99, which significantly outperforms identifying RNAs with 5′ AUGs (ROC AUC 0.68) suggesting the TIEleaderless model can accurately classify those RNAs that are initiated as leaderless mRNAs with high accuracy and precision. The success of this simple TIEleaderless model to classify leaderless mRNAs based on the combinations of ΔGunfold, start codon identity and leader length suggests that these mRNA features combinatorically control translation initiation on leaderless mRNAs.
In addition to the classification of RNAs as leaderless mRNAs, we also explored how well the TIEleaderless model predicted TIE. Here, the translation initiation reporters generated were all scored with the TIEleaderless model and compared to their YFP fluorescence. Since TIEleaderless scores below zero are not physically possible, those with negative TIEleaderless values were set to zero to signify they are not predicted to be translated. As expected, the TIEleaderless score correlates strongly to the YFP reporter levels (R2 = 0.87) with a slope of 2050 A.U. (Supplementary Figure S6). We then compared the TIEleaderless scores to the TE as measured by ribosome profiling of the natural leaderless mRNAs. Since natural leaderless mRNAs encode many genes with diverse codon usages, a poorer correlation was obtained with TE (R2 = 0.06, slope = 0.71 A.U.) than with the TIE reporters (Figure 7C). The correlation of the TIEleaderless model at predicting ribosome profiling TE (R = 0.25) is the same as observed for the RBS calculator model of initiation and E. coli ribosome profiling data (R = 0.25) (57). Since the TIE reporters all code for YFP with near-identical codon usage, and the natural mRNAs have variable codon usage frequencies, it is possible that translation elongation differences between natural ORFs also impact TE. Indeed, translation elongation rates have been estimated to be rate limiting in vivo in other bacteria (58,59). In addition, ribosome occupancy of stalled ribosomes can complicate the analysis of ribosome profiling data, making the interpretation rather difficult. While it is objectively harder to quantitatively predict translation levels, the TIEleaderless model performs rather well.
DISCUSSION
Here, we provide the first systematic analysis of mRNA structure content, start codon identity and leader length on the initiation of leaderless mRNAs (Figure 7D). Importantly, this study was performed using the bacterium C. crescentus that is adapted to efficient leaderless mRNA initiation (27). As has been observed for leadered mRNAs (19,46), mRNA structure content at the leaderless TIR hinders leaderless mRNA translation initiation, suggesting that ribosome accessibility is a key feature for leaderless mRNAs. As previously observed in E. coli, changes in start codon identity from the preferred ‘AUG’ and presence of leader nucleotides lead to a significant reduction of TIE for C. crescentus leaderless mRNAs. Using these quantitative data, we generated a combinatorial TIEleaderless model that predicts the ability of an RNA to initiate as a leaderless mRNA from the individual effects of these features that can be computed for any RNA in the transcriptome. This TIEleaderless model both accurately and sensitively predicts the ability of all RNAs in the C. crescentus transcriptome to initiate as leaderless mRNAs. While a 5′ AUG is highly enriched in leaderless mRNAs and only rarely observed in non-coding RNAs (Figure 6A), non-coding RNAs containing 5′ AUGs utilize a high ΔGunfold to prevent aberrant translation initiation (Figure 6B). Additionally, very short leaders that were found to inhibit leaderless mRNA initiation are selected against in leaderless mRNAs and are common in 5′ regions of non-coding RNAs containing non-initiating AUGs. Finally, leaderless mRNAs are much more selective for AUG start codons than are leadered mRNAs, suggesting that the additional stabilization of the translation initiation complex provided by the SD–aSD base pairing helps facilitate initiation on near-cognate start codons.
Leaderless mRNAs have been found to initiate translation in bacterial, archaeal, and both cytoplasmic and mitochondrial eukaryotic ribosomes (17,28,60) suggesting that leaderless initiation is an ancestral initiation mechanism. It is therefore possible that the TIEleaderless model generated here in C. crescentus may also perform well across organisms. Indeed, even a few nucleotides preceding the AUG inhibit leaderless mRNA translation initiation in C. crescentus, E. coli and mammalian mitochondria (37,39). The strong inhibition of leaderless mRNA translation by TIR secondary structure is likely why leaderless mRNAs in mitochondria have been found to lack 5′ secondary structures (28). C. crescentus shares a similar preference for 5′ AUGs to E. coli for leaderless mRNA initiation (33). Interestingly, in the mycobacteria, GUG start codons are much more abundant in leaderless mRNAs and tend to be initiated more similarly to AUG codons in this organism (16). Mycobacterium GUG initiated leaderless mRNAs tend to code for short regulatory ORFs (16), as opposed to ORFs encoding functional genes in C. crescentus. This suggests that there are likely to be some species-specific differences in leaderless mRNA features arising from the differences in the translation initiation machinery. Indeed, across prokaryotes, 79% of predicted leaderless genes contain AUG as the start codon, whereas GUG, UUG and others are found with an average of 10%, 6% and 3%, respectively (13). Surprisingly, leaderless mRNAs across organisms appear to initiate with assembled 70S/80S ribosomes (31,61–63), further suggesting a conserved mechanism of initiation. Therefore, an important goal moving forward will be to determine how broadly across organisms this TIEleaderless model might apply. Based upon the observations described here, it is likely that these features (ΔGunfold, start codon identity and leader length) will combine similarly across species to define leaderless mRNA TIRs. However, due to differences in the translation initiation machinery across organisms, the specific hierarchy of mRNA features will need to be experimentally determined for a given species in order to generate a TIEleaderless model that accurately classifies leaderless mRNAs.
DATA AVAILABILITY
Transcript architecture for the C. crescentus genome (updated operon map, updated 24 April 2020) was obtained from figshare (https://doi.org/10.6084/m9.figshare.12919208.v3). Data and code for the described analyses are available in GitHub (https://github.com/schraderlab/A-combination-of-mRNA-features_data).
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Schrader lab for critical feedback.
Contributor Information
Mohammed-Husain M Bharmal, Department of Biological Sciences, Wayne State University, Detroit, MI 48202, USA.
Alisa Gega, Department of Biological Sciences, Wayne State University, Detroit, MI 48202, USA.
Jared M Schrader, Department of Biological Sciences, Wayne State University, Detroit, MI 48202, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NARGAB Online.
FUNDING
National Institutes of Health [R35GM124733 to J.M.S.]; Wayne State University [start-up funds to J.M.S.]. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1.Drummond D.A., Wilke C.O.. The evolutionary consequences of erroneous protein synthesis. Nat. Rev. Genet. 2009; 10:715–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kurland C.G., Ehrenberg M.. Growth-optimizing accuracy of gene expression. Annu. Rev. Biophys. Biophys. Chem. 1987; 16:291–317. [DOI] [PubMed] [Google Scholar]
- 3.Rodnina M.V., Wintermeyer W.. Fidelity of aminoacyl-tRNA selection on the ribosome: kinetic and structural mechanisms. Annu. Rev. Biochem. 2001; 70:415–435. [DOI] [PubMed] [Google Scholar]
- 4.Steitz J.A., Jakes K.. How ribosomes select initiator regions in mRNA: base pair formation between 3′ terminus of 16S rRNA and the mRNA during initiation of protein synthesis in Escherichia coli. Proc. Natl Acad. Sci. U.S.A. 1975; 72:4734–4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen H., Bjerknes M., Kumar R., Jay E.. Determination of the optimal aligned spacing between the Shine–Dalgarno sequence and the translation initiation codon of Escherichia coli mRNAs. Nucleic Acids Res. 1994; 22:4953–4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shine J., Dalgarno L.. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc. Natl Acad. Sci. U.S.A. 1974; 71:1342–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacob W.F., Santer M., Dahlberg A.E.. A single base change in the Shine–Dalgarno region of 16S rRNA of Escherichia coli affects translation of many proteins. Proc. Natl Acad. Sci. U.S.A. 1987; 84:4757–4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hui A., de Boer H.A.. Specialized ribosome system: preferential translation of a single mRNA species by a subpopulation of mutated ribosomes in Escherichia coli. Proc. Natl Acad. Sci. U.S.A. 1987; 84:4762–4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calogero R.A., Pon C.L., Canonaco M.A., Gualerzi C.O.. Selection of the mRNA translation initiation region by Escherichia coli ribosomes. Proc. Natl Acad. Sci. U.S.A. 1988; 85:6427–6431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Melancon P., Leclerc D., Destroismaisons N., Brakiergingras L.. The anti-Shine–Dalgarno region in Escherichia coli 16S ribosomal RNA is not essential for the correct selection of translational starts. Biochemistry. 1990; 29:3402–3407. [DOI] [PubMed] [Google Scholar]
- 11.Saito K., Green R., Buskirk A.R.. Translational initiation in E. coli occurs at the correct sites genome-wide in the absence of mRNA–rRNA base-pairing. eLife. 2020; 9:e55002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang B., Halgamuge S., Tang S.L.. Analysis of SD sequences in completed microbial genomes: non-SD-led genes are as common as SD-led genes. Gene. 2006; 373:90–99. [DOI] [PubMed] [Google Scholar]
- 13.Srivastava A., Gogoi P., Deka B., Goswami S., Kanaujia S.P.. In silico analysis of 5′-UTRs highlights the prevalence of Shine–Dalgarno and leaderless-dependent mechanisms of translation initiation in bacteria and archaea, respectively. J. Theor. Biol. 2016; 402:54–61. [DOI] [PubMed] [Google Scholar]
- 14.Beck H.J., Moll I.. Leaderless mRNAs in the spotlight: ancient but not outdated!. Microbiol. Spectr. 2018; 6: 10.1128/microbiolspec.RWR-0016-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cortes T., Schubert O.T., Rose G., Arnvig K.B., Comas I., Aebersold R., Young D.B.. Genome-wide mapping of transcriptional start sites defines an extensive leaderless transcriptome in Mycobacterium tuberculosis. Cell Rep. 2013; 5:1121–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shell S.S., Wang J., Lapierre P., Mir M., Chase M.R., Pyle M.M., Gawande R., Ahmad R., Sarracino D.A., Ioerger T.R.et al.. Leaderless transcripts and small proteins are common features of the mycobacterial translational landscape. PLoS Genet. 2015; 11:e1005641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Montoya J., Ojala D., Attardi G.. Distinctive features of the 5′-terminal sequences of the human mitochondrial mRNAs. Nature. 1981; 290:465–470. [DOI] [PubMed] [Google Scholar]
- 18.Nakamoto T.A unified view of the initiation of protein synthesis. Biochem. Biophys. Res. Commun. 2006; 341:675–678. [DOI] [PubMed] [Google Scholar]
- 19.de Smit M.H., van Duin J.. Secondary structure of the ribosome binding site determines translational efficiency: a quantitative analysis. Proc. Natl Acad. Sci. U.S.A. 1990; 87:7668–7672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Smit M.H., van Duin J.. Control of translation by mRNA secondary structure in Escherichia coli: a quantitative analysis of literature data. J. Mol. Biol. 1994; 244:144–150. [DOI] [PubMed] [Google Scholar]
- 21.Skripkin E.A., Adhin M.R., de Smit M.H., van Duin J.. Secondary structure of the central region of bacteriophage MS2 RNA: conservation and biological significance. J. Mol. Biol. 1990; 211:447–463. [DOI] [PubMed] [Google Scholar]
- 22.Gu W., Zhou T., Wilke C.O.. A universal trend of reduced mRNA stability near the translation-initiation site in prokaryotes and eukaryotes. PLoS Comput. Biol. 2010; 6:e1000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salis H.M.The ribosome binding site calculator. Methods Enzymol. 2011; 498:19–42. [DOI] [PubMed] [Google Scholar]
- 24.Romero D.A., Hasan A.H., Lin Y.F., Kime L., Ruiz-Larrabeiti O., Urem M., Bucca G., Mamanova L., Laing E.E., van Wezel G.P.et al.. A comparison of key aspects of gene regulation in Streptomyces coelicolor and Escherichia coli using nucleotide-resolution transcription maps produced in parallel by global and differential RNA sequencing. Mol. Microbiol. 2014; 94:963–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Babski J., Haas K.A., Nather-Schindler D., Pfeiffer F., Forstner K.U., Hammelmann M., Hilker R., Becker A., Sharma C.M., Marchfelder A.et al.. Genome-wide identification of transcriptional start sites in the haloarchaeon Haloferax volcanii based on differential RNA-Seq (dRNA-Seq). BMC Genomics. 2016; 17:629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pfeifer-Sancar K., Mentz A., Ruckert C., Kalinowski J.. Comprehensive analysis of the Corynebacterium glutamicum transcriptome using an improved RNAseq technique. BMC Genomics. 2013; 14:888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schrader J.M., Zhou B., Li G.W., Lasker K., Childers W.S., Williams B., Long T., Crosson S., McAdams H.H., Weissman J.S.et al.. The coding and noncoding architecture of the Caulobacter crescentus genome. PLoS Genet. 2014; 10:e1004463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones C.N., Wilkinson K.A., Hung K.T., Weeks K.M., Spremulli L.L.. Lack of secondary structure characterizes the 5′ ends of mammalian mitochondrial mRNAs. RNA. 2008; 14:862–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Korman M., Schlussel S., Vishkautzan M., Gur E.. Multiple layers of regulation determine the cellular levels of the pup ligase PafA in Mycobacterium smegmatis. Mol. Microbiol. 2019; 112:620–631. [DOI] [PubMed] [Google Scholar]
- 30.Tedin K., Moll I., Grill S., Resch A., Graschopf A., Gualerzi C.O., Blasi U.. Translation initiation factor 3 antagonizes authentic start codon selection on leaderless mRNAs. Mol. Microbiol. 1999; 31:67–77. [DOI] [PubMed] [Google Scholar]
- 31.O’Donnell S.A., Janssen G.R.. Leaderless mRNAs bind 70S ribosomes more strongly than 30S ribosomal subunits in Escherichia coli. J. Bacteriol. 2002; 184:6730–6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Etten W.J., Janssen G.R.. An AUG initiation codon, not codon-anticodon complementarity, is required for the translation of unleadered mRNA in Escherichia coli. Mol. Microbiol. 1998; 27:987–1001. [DOI] [PubMed] [Google Scholar]
- 33.O’Donnell S.M., Janssen G.R.. The initiation codon affects ribosome binding and translational efficiency in Escherichia coli of cI mRNA with or without the 5′ untranslated leader. J. Bacteriol. 2001; 183:1277–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hering O., Brenneis M., Beer J., Suess B., Soppa J.. A novel mechanism for translation initiation operates in Haloarchaea. Mol. Microbiol. 2009; 71:1451–1463. [DOI] [PubMed] [Google Scholar]
- 35.Chen W.C., Yang G.P., He Y., Zhang S.M., Chen H.Y., Shen P., Chen X.D., Huang Y.P.. Nucleotides flanking the start codon in hsp70 mRNAs with very short 5′-UTRs greatly affect gene expression in Haloarchaea. PLoS One. 2015; 10:e0138473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brock J.E., Pourshahian S., Giliberti J., Limbach P.A., Janssen G.R.. Ribosomes bind leaderless mRNA in Escherichia coli through recognition of their 5′-terminal AUG. RNA. 2008; 14:2159–2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krishnan K.M., Van Etten W.J. 3rd, Janssen G.R. Proximity of the start codon to a leaderless mRNA’s 5′ terminus is a strong positive determinant of ribosome binding and expression in Escherichia coli. J. Bacteriol. 2010; 192:6482–6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones R.L. 3rd, Jaskula J.C., Janssen G.R.. In vivo translational start site selection on leaderless mRNA transcribed from the Streptomyces fradiaeaph gene. J. Bacteriol. 1992; 174:4753–4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Christian B.E., Spremulli L.L.. Preferential selection of the 5′-terminal start codon on leaderless mRNAs by mammalian mitochondrial ribosomes. J. Biol. Chem. 2010; 285:28379–28386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou B., Schrader J.M., Kalogeraki V.S., Abeliuk E., Dinh C.B., Pham J.Q., Cui Z.Z., Dill D.L., McAdams H.H., Shapiro L.. The global regulatory architecture of transcription during the Caulobacter cell cycle. PLoS Genet. 2015; 11:e1004831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marks M.E., Castro-Rojas C.M., Teiling C., Du L., Kapatral V., Walunas T.L., Crosson S.. The genetic basis of laboratory adaptation in Caulobacter crescentus. J. Bacteriol. 2010; 192:3678–3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gelsinger D.R., DiRuggiero J.. Transcriptional landscape and regulatory roles of small noncoding RNAs in the oxidative stress response of the haloarchaeon Haloferax volcanii. J. Bacteriol. 2018; 200:e00779-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rudler D.L., Hughes L.A., Perks K.L., Richman T.R., Kuznetsova I., Ermer J.A., Abudulai L.N., Shearwood A.M.J., Viola H.M., Hool L.C.et al.. Fidelity of translation initiation is required for coordinated respiratory complex assembly. Sci. Adv. 2019; 5:eaay2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gelsinger D.R., Dallon E., Reddy R., Mohammad F., Buskirk A.R., DiRuggiero J.. Ribosome profiling in archaea reveals leaderless translation, novel translational initiation sites, and ribosome pausing at single codon resolution. Nucleic Acids Res. 2020; 48:5201–5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mustoe A.M., Corley M., Laederach A., Weeks K.M.. Messenger RNA structure regulates translation initiation: a mechanism exploited from bacteria to humans. Biochemistry. 2018; 57:3537–3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gruber A.R., Lorenz R., Bernhart S.H., Neubock R., Hofacker I.L.. The Vienna RNA websuite. Nucleic Acids Res. 2008; 36:W70–W74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reuter J.S., Mathews D.H.. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 2010; 11:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thanbichler M., Iniesta A.A., Shapiro L.. A comprehensive set of plasmids for vanillate- and xylose-inducible gene expression in Caulobacter crescentus. Nucleic Acids Res. 2007; 35:e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B.et al.. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012; 9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ducret A., Quardokus E.M., Brun Y.V.. MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat. Microbiol. 2016; 1:16077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V.et al.. Scikit-learn: machine learning in python. J. Mach. Learn. Res. 2011; 12:2825–2830. [Google Scholar]
- 53.Tobias J.W., Shrader T.E., Rocap G., Varshavsky A.. The N-end rule in bacteria. Science. 1991; 254:1374–1377. [DOI] [PubMed] [Google Scholar]
- 54.Ingolia N.T., Ghaemmaghami S., Newman J.R.S., Weissman J.S.. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009; 324:218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Amunts A., Brown A., Toots J., Scheres S.H.W., Ramakrishnan V.. Ribosome: the structure of the human mitochondrial ribosome. Science. 2015; 348:95–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bird J.G., Basu U., Kuster D., Ramachandran A., Grudzien-Nogalska E., Towheed A., Wallace D.C., Kiledjian M., Temiakov D., Patel S.S.et al.. Highly efficient 5′ capping of mitochondrial RNA with NAD+ and NADH by yeast and human mitochondrial RNA polymerase. eLife. 2018; 7:e42179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li G.W., Burkhardt D., Gross C., Weissman J.S.. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell. 2014; 157:624–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Racle J., Picard F., Girbal L., Cocaign-Bousquet M., Hatzimanikatis V.. A genome-scale integration and analysis of Lactococcus lactis translation data. PLoS Comput. Biol. 2013; 9:e1003240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vieira J.P., Racle J., Hatzimanikatis V.. Analysis of translation elongation dynamics in the context of an Escherichia coli cell. Biophys. J. 2016; 110:2120–2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baltz R.H., Hegeman G., Skatrud P.L.. Industrial Microorganisms: Basic and Applied Molecular Genetics. 1993; Washington, DC: American Society for Microbiology. [Google Scholar]
- 61.Moll I., Hirokawa G., Kiel M.C., Kaji A., Blasi U.. Translation initiation with 70S ribosomes: an alternative pathway for leaderless mRNAs. Nucleic Acids Res. 2004; 32:3354–3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Udagawa T., Shimizu Y., Ueda T.. Evidence for the translation initiation of leaderless mRNAs by the intact 70S ribosome without its dissociation into subunits in eubacteria. J. Biol. Chem. 2004; 279:8539–8546. [DOI] [PubMed] [Google Scholar]
- 63.Andreev D.E., Terenin I.M., Dunaevsky Y.E., Dmitriev S.E., Shatsky I.N.. A leaderless mRNA can bind to mammalian 80S ribosomes and direct polypeptide synthesis in the absence of translation initiation factors. Mol. Cell. Biol. 2006; 26:3164–3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Transcript architecture for the C. crescentus genome (updated operon map, updated 24 April 2020) was obtained from figshare (https://doi.org/10.6084/m9.figshare.12919208.v3). Data and code for the described analyses are available in GitHub (https://github.com/schraderlab/A-combination-of-mRNA-features_data).