ABSTRACT
Enterococcus faecalis, a multiple antibiotic-resistant Gram-positive bacterium, has emerged as a serious nosocomial pathogen. Here, we used a genetic approach to characterize the strategies used by E. faecalis to fulfill its requirements for endogenous fatty acid (FA) synthesis in vitro and in vivo. The type II fatty acid synthesis (FASII) pathway is encoded by two operons and two monocistronic genes. Expression of all of these genes is repressed by exogenous FAs, which are incorporated into the E. faecalis membrane and modify its composition. Deletion of nine genes of the 12-gene operon abolished growth in an FA-free medium. Addition of serum, which is lipid rich, restored growth. Interestingly, the E. faecalis membrane contains cyclic fatty acids that modify membrane properties but that are unavailable in host serum. The cfa gene that encodes the cyclopropanation process is located in a locus independent of the FASII genes. Its deletion did not alter growth under the conditions tested, but yielded bacteria devoid of cyclic FAs. No differences were observed between mice infected with wild-type (WT) or with FASII or cyclopropanation mutant strains, in terms of bacterial loads in blood, liver, spleen, or kidneys. We conclude that in E. faecalis, neither FASII nor cyclopropanation enzymes are suitable antibiotic targets.
IMPORTANCE Membrane lipid homeostasis is crucial for bacterial physiology, adaptation, and virulence. Fatty acids are constituents of the phospholipids that are essential membrane components. Most bacteria incorporate exogenous fatty acids into their membranes. Enterococcus faecalis has emerged as a serious nosocomial pathogen that is responsible for urinary tract infections, bacteremia, and endocarditis and is intrinsically resistant to numerous antibiotics. E. faecalis synthesizes saturated and unsaturated fatty acids, as well as cyclic fatty acids that are not found in the human host. Here, we characterized mutant strains deficient in fatty acid synthesis and modification using genetic, biochemical, and in vivo approaches. We conclude that neither the fatty acid synthesis pathway nor the cyclopropanation enzyme are suitable targets for E. faecalis antibiotic development.
KEYWORDS: Enterococcus faecalis, FASII pathway, antibiotic target, cyclopropane ring formation, fatty acids, septicemic infection
INTRODUCTION
Bacterial fatty acids (FAs) are usually synthesized by the type II fatty acid synthesis (FASII) pathway. FAs constitute two hydrophobic tails, which, together with a hydrophilic head, comprise phospholipids (PL) that are essential components of cell membranes. Enterococcus faecalis synthesizes both saturated and unsaturated fatty acids (SFA and UFA, respectively), as do numerous streptococci (1–3). Additionally, E. faecalis and numerous other bacteria encode a cyclopropane fatty acid synthase (Cfa), which catalyzes the transfer of a methyl group from S-adenosyl-l-methionine to the double bond of a lipid chain, thereby forming a cyclopropane ring (4). Recent studies uncovered that E. faecalis also synthesizes trans-UFA, further expanding the variety of fatty acids produced by this bacterium (5). Both cyclopropane and trans fatty acids are more rigid than their nonmodified substrates, and they contribute to E. faecalis membrane homeostasis under starvation and stress conditions (6–8).
The FASII pathway defects prevent bacterial growth in vitro in medium not supplemented with FA; this suggests that FASII enzymes may be essential for bacterial viability and are therefore feasible targets for antibiotic design (9–15). However, most bacteria incorporate exogenous FA into their membranes. FA abundance in host compartments and in serum led us to hypothesize and then prove that FA incorporation can bypass FASII-targeted antibiotics in the neonatal pathogen Streptococcus agalactiae, as well as in other Streptococcaceae taxa (1). Nevertheless, antibiotic bypass may differ according to whether bacteria can survive on exclusively exogenous FAs; for example, Mycobacterium tuberculosis remains sensitive to the anti-FASII isoniazid because of its complex lipid requirement (16). In contrast, Staphylococcus aureus, which synthesizes FAs not produced by humans, can nevertheless bypass FASII inhibitors with host-derived fatty acids (17–19).
Enterococcus faecalis is a Gram-positive bacterium that inhabits the gastrointestinal tract of humans and animals (20) and has long been associated with food fermentations. E. faecalis has emerged as a serious nosocomial pathogen and is the second most frequently isolated Gram-positive pathogen isolated in hospitals after S. aureus (21, 22). E. faecalis is responsible for urinary tract infections, bacteremia, and endocarditis (23). It is intrinsically resistant to many antibiotics, and its acquisition of antibiotic resistance genes has made it a public health concern worldwide (23). This situation prompts the search for new antibiotic targets, notably within the FASII pathway (24, 25).
Whether targeting of the FASII pathway or cyclopropanation enzymes could lead to treatment of E. faecalis systemic infection is not resolved. E. faecalis was previously shown to use environmental fatty acids (26). In the presence of 15% human serum, the E. faecalis membrane comprises about 10% vaccenic acid (VA; C18:1Δ11), an FA not present in serum (26). Thus, the E. faecalis FASII pathway is likely active even in the presence of exogenous FAs. The E. faecalis FASII pathway is encoded by the 12-gene cluster fabT to accA, whose genes are present in numerous Gram-positive species, and by a set of three genes, fabO-fabN and the divergent fabI gene, which encode enzymes involved in UFA and SFA synthesis, respectively (14, 27). Finally, cfa encodes the cyclopropanation reaction. In one study, deletion of fabN resulted in E. faecalis UFA auxotrophy (28). In E. faecalis, expression of FASII genes is controlled by FabT, the transcriptional repressor. An auxiliary acyl carrier protein (ACP) carrying long-chain FAs binds to FabT and facilitates repression of a consensus sequence that regulates FASII gene expression (14, 29, 30).
Although E. faecalis incorporates FA from its environment, the FA synthesis requirement in an FA-rich environment cannot be deduced (31, 32). In this study, we characterized the repressive effect of serum on expression of FASII and FA-related genes. We also tested whether FASII pathway and/or cyclopropanation functions are possible antibiotic targets in a murine model of E. faecalis systemic infection.
RESULTS AND DISCUSSION
FASII pathway genetic organization and regulation of expression.
In E. faecalis, a total of 15 genes belonging to the FASII pathway have been previously identified (14, 27, 33). However, a 16th gene, ef1773 (NCBI GenPept accession number WP_010706667), named fabG, is present in the annotated V583 genome. This gene is conserved among E. faecalis isolates. As two genes were assigned the same annotation, we suggest renaming ef2881, which belongs to the 12-gene cluster, to fabGa, and ef1773 to fabGb. The FabGa and FabGb proteins share 32% identity and 65% similarity. The physiological and enzymatic properties of the two FabG homologues remain unknown, and we hypothesized that they may catalyze the same 3-oxoacyl-ACP reduction step but with different substrate preferences or conditions of expression. In silico analysis using the E. faecalis FabT consensus sequence indicated the presence of a putative FabT binding site, AATTTGAAACAAAACG (palindromic nucleotides shown in bold) 10 bp downstream of the fabGb translation start codon (Fig. 1A) (33). FabT belongs to the family of MarR regulators (34). An HpaR binding site, HpaR also belonging to the MarR family, is centered 47 bp downstream of the hpaG to hpaI operon start site, and it was suggested that HpaR interferes with transcription elongation (35). Based on this precedent, we suggest that fabGb is controlled by FabT and, furthermore, that FabGb is involved in FA synthesis.
FIG 1.
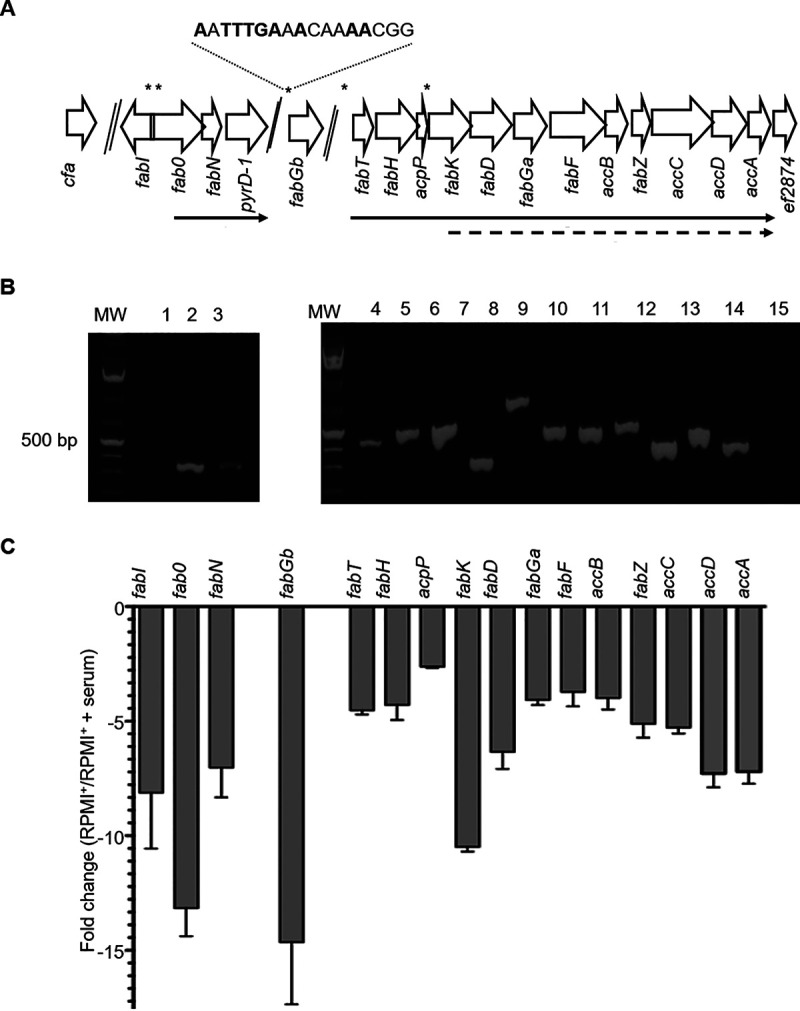
Genetic organization of the Enterococcus faecalis type II fatty acid synthesis (FASII) pathway and impact of exogenous fatty acids (FAs) on E. faecalis FASII gene expression. (A) Schematic representation of E. faecalis VE14089 FASII pathway genes. Gene positions with names below are represented with open arrows. Two genes in the locus, pyrD-1 (ef0285), and ef2874, are unrelated to FASII. Above, asterisks indicate putative FabT binding sites; below, solid arrows indicate transcripts predicted by PCRs, and dotted arrows indicate transcripts identified by predicted FabT binding sequence and expression levels. (B) Agarose gel of the real-time PCR (RT-PCR) amplification products on cDNA using primer pairs from neighboring genes. The order is the same as that in panel A. Lane 1, fabI-fabO; 2, fabO-fabN; 3, fabN-ef0285; 4, fabT-fabH; 5, fabH-acpP; 6, acpP-fabK; 7, fabK-fabD; 8, fabD-fabGa; 9, fabGa-fabF; 10, fabF-accB; 11, accB-fabZ; 12, fabZ-accC; 13, accC-accD; 14, accD-accA; and 15, accA-ef2874. MW, molecular weight standard. (C) FASII gene expression fold change in E. faecalis grown in RPMI+ medium in the absence or the presence of 40% human serum. The means ± standard deviation (SD) from three independent experiments are represented.
Genetic organization of the 12-gene cluster was initially defined in silico on the basis of an upstream FabT consensus binding sequence (27). Depending on the species, an organization in two or three operons was suggested (14, 27, 33). In E. faecalis, FabT consensus binding sequences are present upstream of fabT and fabK (Fig. 1A), suggesting a two-operon organization. Also, the UFA synthesis genes fabO and fabN may constitute another operon (14, 27, 33). This organization was tested by overlapping real-time PCR (RT-PCR) mapping (Fig. 1A and B). Results show that the 12-gene cluster is organized as a single operon that ends before ef2874. A transcription reinitiation site may be linked to the FabT consensus binding sequence upstream of fabK. Furthermore, intense and faint bands, respectively, were obtained for fabO-fabN and fabN-pyrD-1 (= ef0285), suggesting that fabO and fabN constitute an operon and that there may be some read-through downstream of fabN. Together, our data show that E. faecalis FASII genes are organized in two operons (fabT to accA and fabO-fabN) and two monocistronic transcriptional units (fabI and fabGb) (Fig. 1A and B).
The FASII pathway can be strongly repressed by exogenous fatty acids, such that phospholipid synthesis relies on exogenous FA substrates (1, 36). Human serum is rich in lipids comprising long-acyl-chain fatty acids such as 18:1 (15%) and 16:0 (25%), which can act as FabT corepressors (29). To our knowledge, the regulation of E. faecalis FASII gene expression elicited by the addition of FAs has not been studied. To address this question, and after checking that there was no growth difference in RPMI+ medium in the presence or absence of added human serum (data not shown), we used real-time quantitative PCR (qRT-PCR) to analyze the effect of 40% human serum on E. faecalis FASII gene expression (Fig. 1A and C). Serum addition resulted in decreased expression of all FASII genes, ranging from 2.5- to 14-fold depending on the gene. Interestingly, a putative FabT binding site was found close to the most strongly repressed genes, fabGb, fabO, and fabK. This suggests that a FabT-controlled transcription initiation site or a transcription repression site exists upstream of these genes. Together, our data show that the 12-gene operon (from fabT to accA), as defined above, is strongly repressed in the presence of a physiological concentration of serum.
The E. faecalis FASII pathway and cyclopropanation are dispensable for growth in the presence of serum.
Supplementation with serum or unsaturated FA overcomes growth inhibition mediated by cerulenin, a FASII inhibitor (1, 17, 37). We confirmed these results with the wild-type (WT) strain VE14089 (data not shown). The consequences of a nonfunctional FASII pathway were then examined by constructing the EfΔFASII mutant, in which 9 genes of the 12-gene operon, from fabK to accA (ef2883 to ef2875) were deleted. The genes that were removed encode products implicated in the initiation and elongation modules (Fig. 1A). Growth of the WT, the EfΔFASII mutant, and back-to-the-wild-type (BWT-FASII) (see Materials and Methods) strains was first compared in RPMI+ (Fig. 2A). The WT and the BWT-FASII strains behaved similarly. In contrast, growth of the EfΔFASII strain was essentially arrested in RPMI+. To test whether the slight growth observed with the EfΔFASII strain was due to fatty acids carried over from the preculture, we rediluted the cultures to an absorbance at 600 nm (OD600) of 0.01 in RPMI+ when the EfΔFASII culture reached a plateau (Fig. 2B). Although the WT and BWT strains resumed growth, the EfΔFASII strain barely did (OD600 change from 0.01 to 0.015). These results confirm that an active FASII pathway is required for growth in FA-free medium. Addition of 40% human serum to growth medium led to equivalent growth of all strains (Fig. 2A), indicating that serum is sufficient to fully restore growth of the EfΔFASII mutant strain. Together, these results indicate that the exogenous FAs present in serum are necessary and sufficient to restore growth when E. faecalis FASII pathway is abolished, either via the addition of an FASII inhibitor (1) or by deletion of FASII genes.
FIG 2.
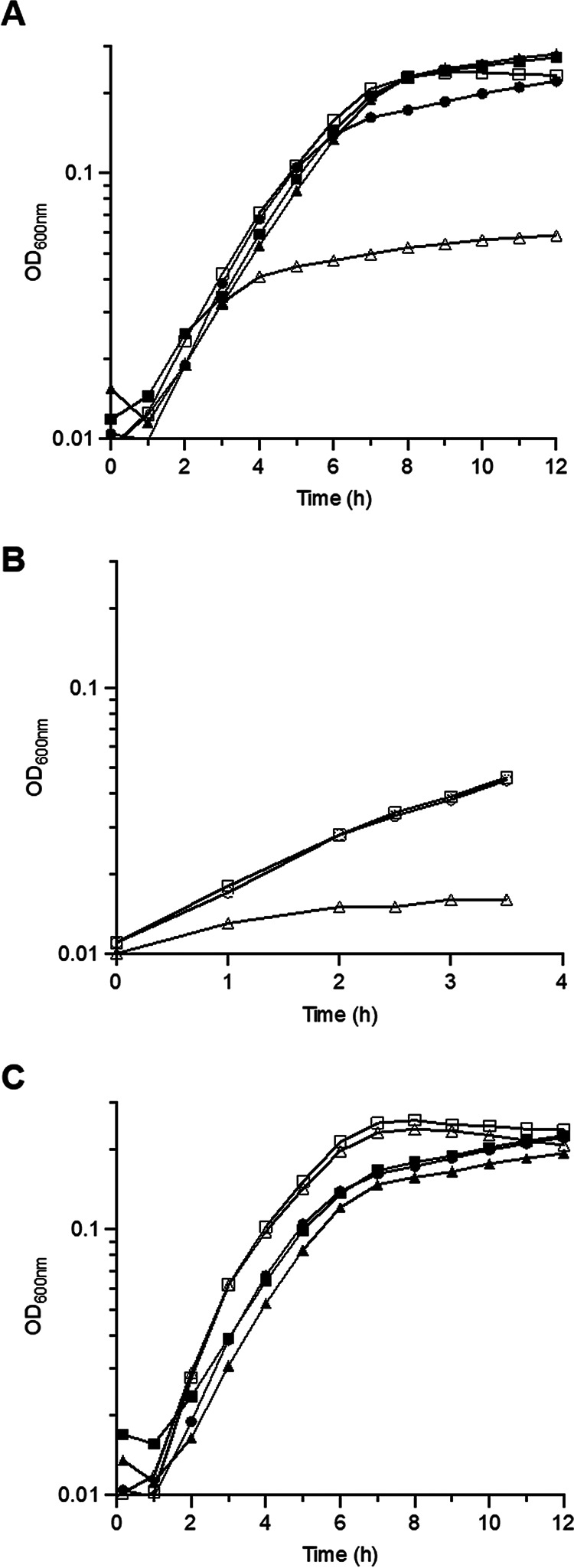
Impact of FASII pathway mutations on E. faecalis growth. (A) Growth curves of VE14089 (WT) (●), EfΔFASII (▲), and BWT-FASII (■) strains in RPMI+ in the absence (empty symbols) or presence (closed symbols) of 40% human serum. (B) Growth curves of VE14089 (WT) (○), EfΔFASII (Δ) and BWT-FASII (□) and diluted in RPMI+ to an absorbance at 600 nm (OD600) of 0.01 when EfΔFASII strain growth reached a plateau. (C) Growth curves of VE14089 (●), EfΔcfa (■), and BWT-cfa (▲) strains in RPMI+ in the absence (empty symbols) or presence (closed symbols) of 40% human serum. Growth curves shown are representative of at least 3 independent experiments. Note change of scales for panel B.
The role of cyclopropanation in E. faecalis growth in the presence of exogenous FAs remained an open question. E. faecalis produces cyclopropane FAs, which are produced from both endogenous and exogenous substrates, presumably to mediate membrane fluidity adjustment by modifying UFA (8). A cfa in-frame deletion mutant was constructed to assess the role of cyclopropane FA synthesis in E. faecalis growth in medium without or with host serum. The mutant strain grew like the WT and BWT-cfa strains in both media, indicating that the presence or absence of cyclic FA does not impact E. faecalis growth in vitro (Fig. 2C).
E. faecalis WT and EfΔFASII strains show identical FA composition upon growth in serum.
Strong repression of E. faecalis FASII genes in the presence of human serum correlates with incorporation of FAs, as previously observed under the same culture conditions (26). We determined the FA compositions of WT VE14089 and BWT-derivative (BWT-FASII) strains grown in the absence or presence of 40% serum (Fig. 3A; see also Tables S2 and S3 in the supplemental material). BWT-FASII profiles were similar to those of the WT; the slight differences probably arose from growth variations that could alter the efficiency of conversion between the UFA and cyclo forms. As previously described, FA of cells grown in FA-free medium (RPMI+) comprised mainly palmitic acid (PA; 16:0) (40%), vaccenic acid (VA; 18:1Δ11) (20%), and cyclopropanated VA form (19Δ11) (20%) (Fig. 3A). In contrast, membrane FA composition of bacteria grown in serum resembled the profiles of serum alone (Table S3 and Fig. 3A) (14, 26, 31). Thus, the proportion of endogenously produced 18:1Δ11 dropped to 1.7%, and that of serum-provided oleic acid (OA; 18:1Δ9) rose to 13.5%. These latter percentages are different from previously reported ones (26, 31); this could be due to the use here of 40% rather than 15% serum. Thus, VE14089, like other E. faecalis isolates, incorporates serum-supplied exogeneous FA (6). Another notable change in the presence of serum was a decrease in the amount of C19Δ11, as reported previously (26), while cyclopropanated OA (C19Δ9), which is not found in serum, rose to 15%, indicating that E. faecalis CFA synthase cyclizes both 18:1Δ9 and 18:1Δ11. This indicates that E. faecalis CFA synthase is active on different substrates, which is in agreement with the fact that bacterial CFAs display lax requirements for positioning of the cis double bond, except for cis double-bond positions at FA extremities, or trans double bonds, which are generally not converted (4).
FIG 3.
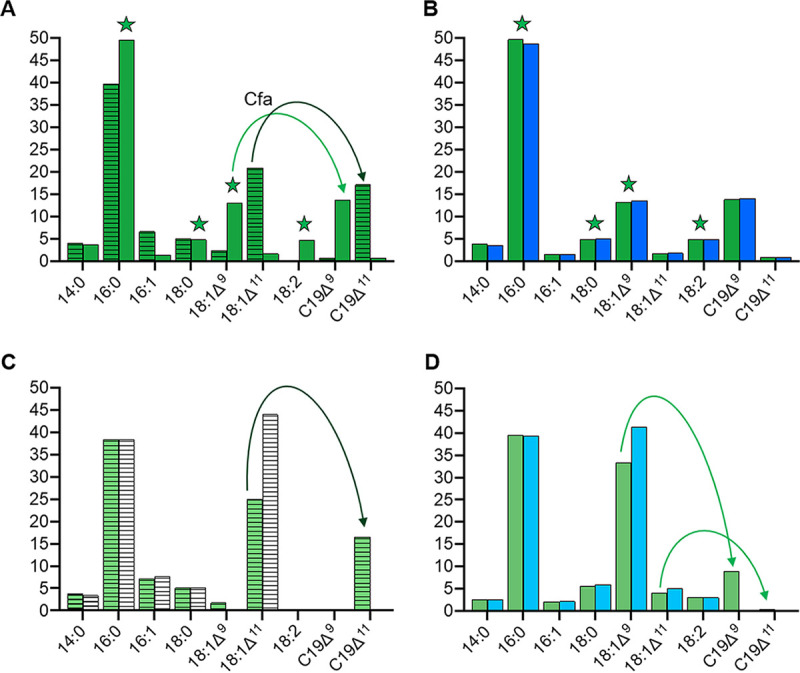
Fatty acid profiles of E. faecalis WT, ΔFASII, and Δcfa strains grown without and with serum. E. faecalis was grown in RPMI+ or in RPMI+ containing 40% human serum to the stationary phase. FA composition was compared to assess the roles of medium FA content and genetic background. The means of the percentages of each FA found in E. faecalis BWT or mutant strains grown to the stationary phase in the presence or absence of 40% serum of three independent experiments are presented (see also Tables S2 to S4 in the supplemental material). (A) BWT-FASII grown in the absence or presence of serum. (B). BWT-FASII and EfΔFASII strains grown in 40% serum. (C) BWT-cfa and EfΔcfa strains grown in RPMI+. (D) BWT-cfa and EfΔcfa strains grown in 40% human serum. Only major fatty acids (>2% of total in at least one profile) are shown. Stars, fatty acids derived from serum. Green arrows, cyclopropanation in the BWT strains of unsaturated fatty acids; dark green, produced by E. faecalis (mainly 18:1Δ11); light green, incorporated from serum (mainly 18:1Δ9). Columns are colored as follows: dark green, BWT-FASII; dark blue, EfΔFASII; light green, BWT-cfa; light blue, EfΔcfa; hatched, strains grown in RPMI+; empty, strains in RPMI+ supplemented with 40% human serum.
We also established the FA content of the EfΔFASII mutant strain grown in 40% human serum (Fig. 3B). The FA profiles of the EfΔFASII and BWT-FASII strains were similar (Fig. 3B). Both strains similarly incorporated serum FAs and generated C19Δ9. These data demonstrate that FASII activity has no impact on membrane composition in the presence of human serum.
E. faecalis cyclopropane fatty acid synthase impacts cellular FA composition.
To characterize the impact of the cfa deletion on E. faecalis membrane FA composition, we compared the FA profiles of EfΔcfa, BWT-cfa, and WT strains grown in FA-free medium and in the presence of human serum (Fig. 3C and D; see also Tables S2 and S4 in the supplemental material). The BWT-cfa profiles were nearly identical to those of the WT (Tables S2 and S4). Compared to the BWT-cfa control strain in RPMI+, the EfΔcfa mutant strain was devoid of C19Δ11 and was enriched in 18:1Δ11 (Fig. 3C). In serum-supplemented medium, the EfΔcfa strain was devoid of C19Δ9 and enriched in 18:1Δ9 (Fig. 3D). These data confirm that the EfΔcfa strain is defective for cyclopropanation of unsaturated fatty acids, mainly 18:1Δ11 in RPMI+ and 18:1Δ9 in the presence of serum. This implies that, in vivo, the fatty acid composition of cfa-defective strains may be different from those of wild-type strains, and this could have physiological consequences.
E. faecalis septicemic infection is independent of both the FASII pathway and fatty acid cyclopropanation.
The WT, EfΔFASII, and BWT-FASII strains grew similarly and displayed the same FA composition when grown in the presence of human serum. During infection, however, E. faecalis infects organs whose FA contents are variable (38, 39). We therefore asked whether exogenous FAs in the host may compensate the loss of the active FASII pathway in vivo.
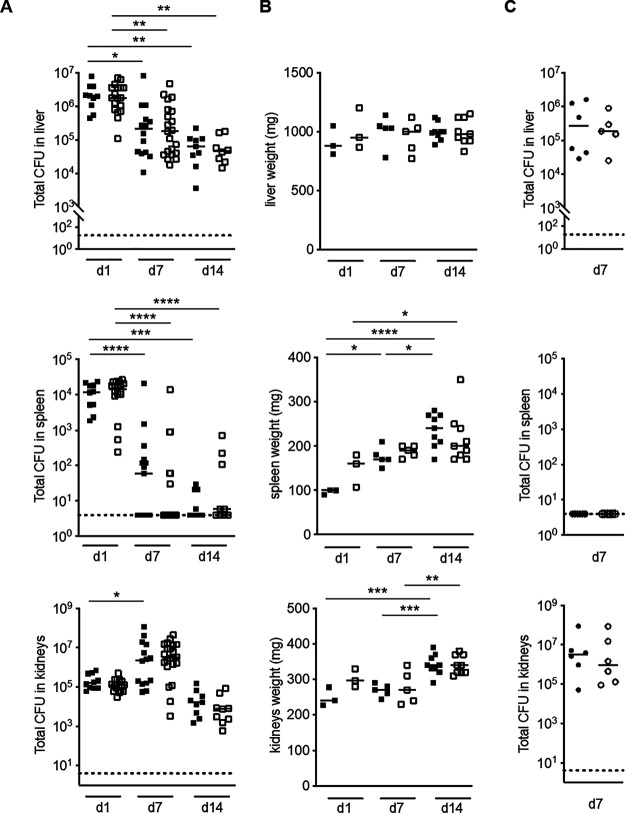
We first followed the infection process, looking at the physiological consequences of intravenous infection with the BWT-FASII strain. CFU counts and organ weights were examined at 1, 7, and 14 days postinfection (dpi) (Fig. 4A and B). Throughout the course of the experiment, the CFU counts in blood were below the detection threshold (data not shown). The CFU counts in the liver diminished 31-fold, at 14 dpi, with no change in the weight of livers throughout the experiment. In the spleen, the bacterial levels strongly decreased over a 2-week period, from approximately 104 CFU 24 h postinfection (hpi) to 66% of spleens being sterile at the end of the experiment. However, day 14 spleens weighed twice as much as day 1 spleens (Fig. 4B). Finally, kidney weights increased between day 7 to day 14 (Fig. 4B). This correlated, albeit with a time lag, to the tendency in the CFU counts to increase at day 7 in BWT-FASII-infected mice (15-fold) and is in agreement with E. faecalis tropism for the kidneys (31, 32).
FIG 4.
EfΔFASII and BWT-FASII infection in a mouse septicemic model. (A) CFU shown by scatter graph plots at 1, 7, and 14 days postinfection (dpi) of bacteria recovered from BWT-FASII- and EfΔFASII-infected mice. Nine to 20 mice were intravenously infected with 6 × 108 to 8 × 108 CFU E. faecalis BWT-FASII (closed squares) or EfΔFASII (open squares) strains. (B) Organ weights shown by scatter graph plots at 1, 7, and 14 dpi of 3 to 9 mice infected by the EfΔFASII or BWT-FASII strains. Symbols are as in panel A. (C) CFU shown by scatter graph plots at 7 dpi of viable bacteria recovered from EfΔcfa- and BWT-cfa-infected mice. Six mice were intravenously infected with 6 × 108 to 8 × 108 CFU E. faecalis BWT-cfa (closed circles) or EfΔcfa (open circles) strains. Upper panels, liver; middle panels, spleen; lower panels, kidneys. Each symbol represents the number of CFU per organ of one infected mouse or the weight of one organ. Median ranges are represented. Statistical analysis was as follows: 2-way analysis of variance (ANOVA) and a Bonferroni posttest (A and B) Mann-Whitney test (C). *, P < 0.05; **, P < 0.01; ***, P < 0.005; ****, P < 0.0001.
We then assessed the consequences of the FASII gene deletion by comparing the BWT-FASII- and EfΔFASII-infected mice. Bacterial counts in liver, spleen, and kidneys, as well as organ weights, were similar for BWT-FASII- and EfΔFASII-infected mice at all time points (Fig. 4A and B). This demonstrates that, as in other species, targeting the FASII pathway would be ineffective for the treatment of E. faecalis septicemic infection (1).
The consequences of a cfa deletion were also examined in the mouse septicemia infection model. cfa is reportedly more expressed in the stationary phase (40). We reasoned that differences, if any, between EfΔcfa and BWT-cfa strains would be more pronounced after CFU counts reached their highest level in the wild-type strain (here BWT)-infected mice, i.e., at day 7. No differences were observed in terms of bacterial loads in the different organs between EfΔcfa- and BWT-cfa-infected mice (Fig. 4C). This indicates that cyclopropanation activity does not confer an advantage to E. faecalis during systemic infection in this model.
In conclusion, our data demonstrate that targeting the FASII pathway or the cyclopropanation process would be ineffective for the treatment of E. faecalis septicemic infection.
MATERIALS AND METHODS
Bacterial strains, media, and culture conditions.
The vancomycin-resistant E. faecalis V583 strain was isolated from a bloodstream infection and sequenced. It was used for the in silico analysis. VE14089, a plasmid-cured derivative of the V583 strain, was the wild type (WT) reference strain used for the construction of mutant and back-to-the-wild-type (BWT [see “Strain construction,” below]) strains (41–43). Two mutants were constructed, EfΔFASII and EfΔcfa, corresponding to the in-frame deletion of nine genes from fabK to accA (7,863 bp) and of cfa (1,176 bp), respectively. The respective isogenic BWT strains BWT-FASII and BWT-cfa were obtained. Escherichia coli TG1 (Invitrogen) was used for cloning experiments. E. faecalis was cultured in Todd-Hewitt (TH) broth or agar (Difco Laboratories, Detroit, MI), supplemented with 0.1% Tween 80 (Sigma) when appropriate, or in RPMI+ medium (RPMI 1640 supplemented with 1× amino acids; Gibco, Life Technologies), 1× vitamins (Sigma-Aldrich), 1% glucose, and 0.1 mM HEPES (Gibco, Life Technologies). E. coli was cultured in trypticase soy (TS) medium (Difco Laboratories,). When needed, antibiotics were used at the following concentrations: for E. coli, ticarcillin, 100 μg/ml, and erythromycin, 160 μg/ml; and for E. faecalis, erythromycin, 50 μg/ml. Cerulenin (Sigma) was added at 8 μg/ml final concentration.
In silico analysis.
The AGTTTGATAATCAAATT sequence was used to seek potential FabT binding sites in the regulatory regions or within the FASII genes (27).
General DNA techniques.
E. coli plasmid DNA was prepared by rapid alkaline lysis using the QIAprep spin miniprep kit (Qiagen). Genomic DNA from E. faecalis was prepared using the DNeasy blood and tissue kit (Qiagen) with pretreatment for Gram-positive bacteria, as recommended by the manufacturer. PCR was carried out with AmpliTaq Gold polymerase as described by the manufacturer (Applied Biosystems). The molecular weight standard used in agarose gels is the 1 kb Plus DNA ladder (Life Technologies). Amplification products were purified using a QIAquick PCR purification kit (Qiagen). PCR products and plasmids used were sequenced with an ABI 310 automated DNA sequencer, using the Prism dye terminator cycle sequencing kit (Applied Biosystems). To determine the genetic and transcriptional organization of VE14089 FASII genes, primer pairs were designed to flank neighboring FASII genes. Primers are listed in Table S1 in the supplemental material.
Strain construction.
The primers used for the construction of the in-frame EfΔFASII and EfΔcfa mutant strains are listed in Table S1. Primers used are based on the published genome sequence of E. faecalis V583 (GenBank accession number NC_004668.1) (41). The ΔFASII deletion was constructed by using splicing by overlap extension PCR as described previously (44). The corresponding PCR fragment was cloned into the thermosensitive shuttle plasmid pG1. The Δcfa (ef0203; NCBI reference sequence NC_004668.1) deletion was constructed using the In-Fusion PCR cloning kit (Clontech), cloning the two PCR-amplified fragments into pG1, previously linearized in a one-step reaction, following the manufacturer’s recommendations. The resulting intermediate plasmids, pG1ΔFASII and pG1Δcfa, were checked by sequencing the inserts. Electroporation of VE14089 and allelic exchange were performed as described previously with minor modifications (44, 45). For each construction, starting from a single clone in which the intermediate plasmid was integrated by homologous recombination, we selected clones in which the second homologous recombination event had taken place. One class of clones was deleted for the considered gene, and the other reverted to the wild-type genotype. An isolated reverted clone was termed back-to-the-wild type (BWT). The BWT strains are isogenic to their deleted counterparts, i.e., they should possess the same secondary mutations if any came up during the construction steps (43). That they displayed the WT phenotype was checked for all in vitro phenotypes. For the EfΔFASII strain construction, 0.1% Tween 80 (Sigma), a rich source of FA containing 70% oleic acid (OA; 18:1Δ9), was added during all steps of the experiment after the electroporation. The in-frame deletion mutants and BWT strains were confirmed by PCR and sequence analysis.
Growth curves.
E. faecalis strains were inoculated into RPMI+ supplemented with Tween 80 (0.1%) and cultured overnight at 37°C. Overnight cultures were washed twice in phosphate-buffered saline (PBS) and subcultured (1:100) in RPMI+ and in RPMI+ supplemented with 40% human serum. In these experiments, the bacterial cultures carried out in RPMI+ were further diluted in RPMI+ before assessing growth. Growth was determined by measuring absorbance at 600 nm (OD600). Experiments were repeated three times.
RNA extraction and RT-PCR.
RNA was extracted from bacteria by the phenol-chloroform technique as described previously (46). RNA (2 to 5 μg) was treated with DNase I (Promega) according to the manufacturer’s recommendations. RNA was quantified by absorbance at 260 and 280 nm. RNA purity and integrity were controlled, and RNA was stored at −80°C until use.
Reverse transcription was carried out using the SuperScript II kit (Invitrogen) and random hexamer primers (Fermentas) according to the manufacturers’ instructions. PCRs were carried out on cDNA, as well as on genomic DNA (gDNA) and RNA as positive and negative controls, using primer pairs flanking neighboring FASII genes (Table S1). PCR amplicons were examined on a 1% agarose gel.
Real-time quantitative PCRs.
Overnight VE14089 RPMI+ cultures were then subcultured (1:100) to the mid-log phase (OD600 = 0.3) in RPMI+ with or without the addition of 40% human serum. RNAs were extracted, and cDNA samples were diluted to 50 ng/μl. qPCR was performed using the LightCycler (Roche) and the SYBR green PCR kits (Applied Biosystems) using primers listed in Table S1. Each assay was performed in duplicate, with two independently prepared total RNA samples. The relative quantification in gene expression was determined by the comparative threshold cycle (2−ΔΔCT) method, using rpoB as the reference gene (47).
Fatty acid analysis.
Strains were inoculated into RPMI+ supplemented with Tween 80 (0.1%) and grown overnight at 37°C. Cells were washed three times in RPMI+ and diluted 100-fold in fresh RPMI+ cultures with or without 40% human serum. Cells were harvested after 14 h of growth (stationary phase) under static conditions at 37°C and washed once in NaCl 0.9% containing 0.01% Triton X-100 and twice in NaCl 0.9%. Cell pellets were stored at −20°C until analysis. The FA composition of serum used for experiments was also determined. Whole-cell esterified fatty acid determinations were done on an AutoSystem XL gas chromatograph (Perkin-Elmer) equipped with a DB-Wax column (30 m × 30.25 mm × 30.25 mm; Agilent, France), for all analyses except those shown in Fig. 3D, or on a ZB-Wax capillary column (30 m × 0.25 mm × 0.25 mm; Phenomenex, France), for that shown in Fig. 3D, as described previously (1, 17). Bacterial fatty acid composition of each sample was analyzed in three independent experiments.
Ethics statement.
All animal experiments described in this study were conducted in accordance with the guidelines of Paris Descartes University, in compliance with the European animal welfare regulation (http://ec.europa.eu/environment/chemicals/lab_animals/home_en.html), and were approved by the Paris Descartes University animal care and use committee and by the Ministère de l’Education Nationale, l’Enseignement Supérieur et la Recherche (2015032714098562_v1, APAFIS no. 390).
Intravenous infection of mice.
All in vivo infections in this study were performed with 6-week-old female BALB/c mice (Charles River, L’Arbresle, France). Bacterial suspensions used for intravenous (i.v.) infection were prepared as follows. Overnight broth cultures of mutant and isogenic BWT strains, grown in TH medium supplemented with 0.1% Tween 80, were diluted 1:100 into 50 ml of TH-Tween 80 medium and grown until an OD600 of 0.6. The bacterial suspensions were washed twice in 0.9% NaCl and resuspended in 0.9% NaCl to obtain a final concentration of 6 × 108 to 8 × 108 CFU/ml in 500 μl for intravenous injections. Mice were injected in the tail vein using a 28-gauge 0.5-in. needle.
Dissemination assay and organ analysis.
Randomized groups of 9 to 21 mice or of 6 mice were infected with the BWT-FASII and EfΔFASII or BWT-cfa and EfΔcfa strains, respectively, and mice were sacrificed by cervical dislocation at days 1, 7, and 14 postinfection or at day 7, postinfection; blood, liver, spleen, and kidneys were harvested. All organs were weighed prior to further experimentation. Liver, spleen, and kidneys were homogenized in 0.9% NaCl. Serial dilutions were immediately plated on TH agar plates containing 0.1% Tween 80 (TH-Tween agar). CFU were enumerated after 24 h of incubation at 37°C.
Statistical analysis.
Statistical analyses were performed using a 2-way analysis of variance (ANOVA) and a Bonferroni posttest or a Mann-Whitney test (Prism 8). A P value of ≤0.05 was considered statistically significant.
ACKNOWLEDGMENTS
We fondly acknowledge our colleague G. Lamberet, who passed away 23 Dec 2019. We thank A. Tazi for helpful discussions.
This work was supported by funding from the Agence Nationale de la Recherche (ANR-13001038), the Fondation pour la recherche Médicale (DBF20161136769), and from INSERM, CNRS, and Université de Paris.
Footnotes
Supplemental material is available online only.
Contributor Information
Claire Poyart, Email: claire.poyart@inserm.fr.
Agnes Fouet, Email: agnes.fouet@inserm.fr.
William W. Metcalf, University of Illinois at Urbana Champaign
REFERENCES
- 1.Brinster S, Lamberet G, Staels B, Trieu-Cuot P, Gruss A, Poyart C. 2009. Type II fatty acid synthesis is not a suitable antibiotic target for Gram-positive pathogens. Nature 458:83–86. 10.1038/nature07772. [DOI] [PubMed] [Google Scholar]
- 2.Altabe S, Lopez P, de Mendoza D. 2007. Isolation and characterization of unsaturated fatty acid auxotrophs of Streptococcus pneumoniae and Streptococcus mutans. J Bacteriol 189:8139–8144. 10.1128/JB.01275-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Montgomerie JZ, Kalmanson GM, Guze LB. 1973. Fatty acid composition of L-forms of Streptococcus faecalis cultured at different osmolalities. J Bacteriol 115:73–75. 10.1128/jb.115.1.73-75.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grogan DW, Cronan JE. 1997. Cyclopropane ring formation in membrane lipids of bacteria. Microbiol Mol Biol Rev 61:429–441. 10.1128/mmbr.61.4.429-441.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kondakova T, Kumar S, Cronan JE. 2019. A novel synthesis of trans-unsaturated fatty acids by the Gram-positive commensal bacterium Enterococcus faecalis FA2–2. Chem Phys Lipids 222:23–35. 10.1016/j.chemphyslip.2019.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y-M, Rock CO. 2008. Membrane lipid homeostasis in bacteria. Nat Rev Microbiol 6:222–233. 10.1038/nrmicro1839. [DOI] [PubMed] [Google Scholar]
- 7.Parsons JB, Rock CO. 2013. Bacterial lipids: metabolism and membrane homeostasis. Prog Lipid Res 52:249–276. 10.1016/j.plipres.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poger D, Mark AE. 2015. A ring to rule them all: the effect of cyclopropane fatty acids on the fluidity of lipid bilayers. J Phys Chem B 119:5487–5495. 10.1021/acs.jpcb.5b00958. [DOI] [PubMed] [Google Scholar]
- 9.Heath RJ, White SW, Rock CO. 2001. Lipid biosynthesis as a target for antibacterial agents. Prog Lipid Res 40:467–497. 10.1016/s0163-7827(01)00012-1. [DOI] [PubMed] [Google Scholar]
- 10.Parsons JB, Rock CO. 2011. Is bacterial fatty acid synthesis a valid target for antibacterial drug discovery? Curr Opin Microbiol 14:544–549. 10.1016/j.mib.2011.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y-M, White SW, Rock CO. 2006. Inhibiting bacterial fatty acid synthesis. J Biol Chem 281:17541–17544. 10.1074/jbc.R600004200. [DOI] [PubMed] [Google Scholar]
- 12.Heath RJ, White SW, Rock CO. 2002. Inhibitors of fatty acid synthesis as antimicrobial chemotherapeutics. Appl Microbiol Biotechnol 58:695–703. 10.1007/s00253-001-0918-z. [DOI] [PubMed] [Google Scholar]
- 13.Lu H, Tonge PJ. 2008. Inhibitors of FabI, an enzyme drug target in the bacterial fatty acid biosynthesis pathway. Acc Chem Res 41:11–20. 10.1021/ar700156e. [DOI] [PubMed] [Google Scholar]
- 14.Zhu L, Bi H, Ma J, Hu Z, Zhang W, Cronan JE, Wang H. 2013. The two functional enoyl-acyl carrier protein reductases of Enterococcus faecalis do not mediate triclosan resistance. mBio 4:e00613-13–e00613. 10.1128/mBio.00613-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parsons JB, Frank MW, Rosch JW, Rock CO. 2013. Staphylococcus aureus fatty acid auxotrophs do not proliferate in mice. Antimicrob Agents Chemother 57:5729–5732. 10.1128/AAC.01038-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhatt A, Molle V, Besra GS, Jacobs WR, Kremer L. 2007. The Mycobacterium tuberculosis FAS-II condensing enzymes: their role in mycolic acid biosynthesis, acid-fastness, pathogenesis and in future drug development: the mycobacterial FAS-II condensing enzymes. Mol Microbiol 64:1442–1454. 10.1111/j.1365-2958.2007.05761.x. [DOI] [PubMed] [Google Scholar]
- 17.Kénanian G, Morvan C, Weckel A, Pathania A, Anba-Mondoloni J, Halpern D, Gaillard M, Solgadi A, Dupont L, Henry C, Poyart C, Fouet A, Lamberet G, Gloux K, Gruss A. 2019. Permissive fatty acid incorporation promotes staphylococcal adaptation to FASII antibiotics in host environments. Cell Rep 29:3974–3982.e4. 10.1016/j.celrep.2019.11.071. [DOI] [PubMed] [Google Scholar]
- 18.Gloux K, Guillemet M, Soler C, Morvan C, Halpern D, Pourcel C, Vu Thien H, Lamberet G, Gruss A. 2017. Clinical relevance of type II fatty acid synthesis bypass in Staphylococcus aureus. Antimicrob Agents Chemother 61:e02515-16. 10.1128/AAC.02515-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morvan C, Halpern D, Kénanian G, Hays C, Anba-Mondoloni J, Brinster S, Kennedy S, Trieu-Cuot P, Poyart C, Lamberet G, Gloux K, Gruss A. 2016. Environmental fatty acids enable emergence of infectious Staphylococcus aureus resistant to FASII-targeted antimicrobials. Nat Commun 7:12944. 10.1038/ncomms12944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fisher K, Phillips C. 2009. The ecology, epidemiology and virulence of Enterococcus. Microbiology (Reading) 155:1749–1757. 10.1099/mic.0.026385-0. [DOI] [PubMed] [Google Scholar]
- 21.Karchmer AW. 2000. Nosocomial bloodstream infections: organisms, risk factors, and implications. Clin Infect Dis 31(Suppl 4):S139–S143. 10.1086/314078. [DOI] [PubMed] [Google Scholar]
- 22.Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis 39:309–317. 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- 23.Arias CA, Murray BE. 2012. The rise of the Enterococcus: beyond vancomycin resistance. Nat Rev Microbiol 10:266–278. 10.1038/nrmicro2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng Z, Chakraborty D, Dewell SB, Reddy BVB, Brady SF. 2012. Environmental DNA-encoded antibiotics fasamycins A and B inhibit FabF in type II fatty acid biosynthesis. J Am Chem Soc 134:2981–2987. 10.1021/ja207662w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gerusz V. 2010. Recent advances in the inhibition of bacterial fatty acid biosynthesis. Annu Rep Med Chem 45:295–311. [Google Scholar]
- 26.Saito HE, Harp JR, Fozo EM. 2014. Incorporation of exogenous fatty acids protects Enterococcus faecalis from membrane-damaging agents. Appl Environ Microbiol 80:6527–6538. 10.1128/AEM.02044-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eckhardt TH, Skotnicka D, Kok J, Kuipers OP. 2013. Transcriptional regulation of fatty acid biosynthesis in Lactococcus lactis. J Bacteriol 195:1081–1089. 10.1128/JB.02043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diederich A-K, Duda KA, Romero-Saavedra F, Engel R, Holst O, Huebner J. 2016. Deletion of fabN in Enterococcus faecalis results in unsaturated fatty acid auxotrophy and decreased release of inflammatory cytokines. Innate Immun 22:284–293. 10.1177/1753425916639669. [DOI] [PubMed] [Google Scholar]
- 29.Jerga A, Rock CO. 2009. Acyl-Acyl carrier protein regulates transcription of fatty acid biosynthetic genes via the FabT repressor in Streptococcus pneumoniae. J Biol Chem 284:15364–15368. 10.1074/jbc.C109.002410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu L, Zou Q, Cao X, Cronan JE. 2019. Enterococcus faecalis encodes an atypical auxiliary acyl carrier protein required for efficient regulation of fatty acid synthesis by exogenous fatty acids. mBio 10:e00577-19. 10.1128/mBio.00577-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harp JR, Saito HE, Bourdon AK, Reyes J, Arias CA, Campagna SR, Fozo EM. 2016. Exogenous fatty acids protect Enterococcus faecalis from daptomycin-induced membrane stress independently of the response regulator LiaR. Appl Environ Microbiol 82:4410–4420. 10.1128/AEM.00933-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kau AL, Martin SM, Lyon W, Hayes E, Caparon MG, Hultgren SJ. 2005. Enterococcus faecalis tropism for the kidneys in the urinary tract of C57BL/6J mice. Infect Immun 73:2461–2468. 10.1128/IAI.73.4.2461-2468.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu Y-J, White SW, Rock CO. 2005. Domain swapping between Enterococcus faecalis FabN and FabZ proteins localizes the structural determinants for isomerase activity. J Biol Chem 280:30342–30348. 10.1074/jbc.M504637200. [DOI] [PubMed] [Google Scholar]
- 34.Osterman A, Overbeek R. 2003. Missing genes in metabolic pathways: a comparative genomics approach. Curr Opin Chem Biol 7:238–251. 10.1016/s1367-5931(03)00027-9. [DOI] [PubMed] [Google Scholar]
- 35.Galán B, Kolb A, Sanz JM, García JL, Prieto MA. 2003. Molecular determinants of the hpa regulatory system of Escherichia coli: the HpaR repressor. Nucleic Acids Res 31:6598–6609. 10.1093/nar/gkg851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yao J, Rock CO. 2017. Exogenous fatty acid metabolism in bacteria. Biochimie 141:30–39. 10.1016/j.biochi.2017.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saito HE, Harp JR, Fozo EM. 2017. Enterococcus faecalis responds to individual exogenous fatty acids independently of their degree of saturation or chain length. Appl Environ Microbiol 84:e01633-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cunnane SC, McAdoo KR, Prohaska JR. 1986. Lipid and fatty acid composition of organs from copper-deficient mice. J Nutr 116:1248–1256. 10.1093/jn/116.7.1248. [DOI] [PubMed] [Google Scholar]
- 39.Gentry-Weeks C, Estay M, Loui C, Baker D. 2003. Intravenous mouse infection model for studying the pathology of Enterococcus faecalis infections. Infect Immun 71:1434–1441. 10.1128/IAI.71.3.1434-1441.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.To TMH, Grandvalet C, Tourdot-Marechal R. 2011. Cyclopropanation of membrane unsaturated fatty acids is not essential to the acid stress response of Lactococcus lactis subsp. cremoris. Appl Environ Microbiol 77:3327–3334. 10.1128/AEM.02518-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paulsen IT, Banerjei L, Myers GSA, Nelson KE, Seshadri R, Read TD, Fouts DE, Eisen JA, Gill SR, Heidelberg JF, Tettelin H, Dodson RJ, Umayam L, Brinkac L, Beanan M, Daugherty S, DeBoy RT, Durkin S, Kolonay J, Madupu R, Nelson W, Vamathevan J, Tran B, Upton J, Hansen T, Shetty J, Khouri H, Utterback T, Radune D, Ketchum KA, Dougherty BA, Fraser CM. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071–2074. 10.1126/science.1080613. [DOI] [PubMed] [Google Scholar]
- 42.Rigottier-Gois L, Alberti A, Houel A, Taly J-F, Palcy P, Manson J, Pinto D, Matos RC, Carrilero L, Montero N, Tariq M, Karsens H, Repp C, Kropec A, Budin-Verneuil A, Benachour A, Sauvageot N, Bizzini A, Gilmore MS, Bessières P, Kok J, Huebner J, Lopes F, Gonzalez-Zorn B, Hartke A, Serror P. 2011. Large-scale screening of a targeted Enterococcus faecalis mutant library identifies envelope fitness factors. PLoS One 6:e29023. 10.1371/journal.pone.0029023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Danne C, Dubrac S, Trieu-Cuot P, Dramsi S. 2014. Single cell stochastic regulation of pilus phase variation by an attenuation-like mechanism. PLoS Pathog 10:e1003860. 10.1371/journal.ppat.1003860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Biswas I, Gruss A, Ehrlich SD, Maguin E. 1993. High-efficiency gene inactivation and replacement system for Gram-positive bacteria. J Bacteriol 175:3628–3635. 10.1128/jb.175.11.3628-3635.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cruz-Rodz AL, Gilmore MS. 1990. High efficiency introduction of plasmid DNA into glycine treated Enterococcus faecalis by electroporation. Mol Gen Genet 224:152–154. 10.1007/BF00259462. [DOI] [PubMed] [Google Scholar]
- 46.Lamy M-C, Zouine M, Fert J, Vergassola M, Couve E, Pellegrini E, Glaser P, Kunst F, Msadek T, Trieu-Cuot P, Poyart C. 2004. CovS/CovR of group B Streptococcus: a two-component global regulatory system involved in virulence: the CovS/CovR regulatory system of Streptococcus agalactiae. Mol Microbiol 54:1250–1268. 10.1111/j.1365-2958.2004.04365.x. [DOI] [PubMed] [Google Scholar]
- 47.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 to S4. Download JB.00221-21-s0001.xlsx, XLSX file, 0.02 MB (16.5KB, xlsx)