ABSTRACT
FleQ plays a crucial role in motility and biofilm formation by regulating flagellar and exopolysaccharide biosynthesis in Pseudomonas aeruginosa. It has been reported that the expression of FleQ is transcriptionally downregulated by the virulence factor regulator Vfr. Here, we demonstrated that a LysR-type transcriptional regulator, OsaR, is also capable of binding to the promoter region of fleQ and repressing its transcription. Through gel shift and DNase I footprinting assays, the OsaR binding site was identified and characterized as a dual LysR-type transcriptional regulator box (AT-N11-AT-N7-A-N11-T). Mutation of the A-T palindromic base pairs in the fleQ promoter not only reduced the binding affinity of OsaR in vitro but also derepressed fleQ transcription in vivo. The OsaR binding site was found to cover the Vfr binding site; knockout of osaR or vfr separately exhibited no effect on the transcriptional level of fleQ; however, fleQ expression was repressed by overexpression of osaR or vfr. Furthermore, simultaneously deleting both osaR and vfr resulted in an upregulation of fleQ, but it could be complemented by the expression of either of the two repressors. In summary, our work revealed that OsaR and Vfr function as two transcriptional repressors of fleQ that bind to the same region of fleQ but work separately.
IMPORTANCEPseudomonas aeruginosa is a widespread human pathogen, which accounts for serious infections in the hospital, especially for lung infection in cystic fibrosis and chronic obstructive pulmonary disease patients. P. aeruginosa infection is closely associated with its motility and biofilm formation, which are both under the regulation of the important transcription factor FleQ. However, the upstream regulatory mechanisms of fleQ have not been fully elucidated. Therefore, our research identifying a novel regulator of fleQ as well as new regulatory mechanisms controlling its expression will be significant for better understanding the intricate gene regulatory mechanisms related to P. aeruginosa virulence and infection.
KEYWORDS: Pseudomonas aeruginosa, transcriptional regulation, OsaR, fleQ
INTRODUCTION
Pseudomonas aeruginosa is a ubiquitous Gram-negative pathogenic bacterium that causes acute and chronic infections in immunocompromised individuals, such as burn victims and cystic fibrosis patients (1–3). P. aeruginosa pathogenesis is determined by its production of various virulence factors, for example, flagella, motility, and biofilm formation (4). The expression of these traits is under complex gene regulation, which involves numerous transcriptional regulators, regulatory RNAs, and σ factors (5).
The NtrC family transcription factor FleQ is a major regulator in the process of flagellar biosynthesis (6), as it works together with the alternate sigma factor σ54 (RpoN) to activate the transcription of most flagellar genes (7–9). The fleQ mutant exhibits loss of motility and the ability to synthesize flagellin or assemble a flagellum (7).
FleQ activity is posttranscriptionally downregulated by its antiactivator FleN, which binds to FleQ to inhibit FleQ ATPase activity (10, 11). Sigma factor σ70 is involved in the activation of fleQ transcription (12). AmrZ and Vfr have been reported as two transcriptional repressors of fleQ. The alginate and motility regulator AmrZ functions globally in the regulation of various genes, such as the psl operon, involved in Psl exopolysaccharide synthesis (13), and algD, involved in alginate production (14). fleQ promoter activity was approximately 4-fold higher in an amrZ mutant (15), while transcriptome sequencing (RNA-seq) analysis indicated that fleQ transcription was repressed 8-fold by AmrZ (16). The virulence regulator Vfr is another global transcriptional regulator of a number of genes, such as toxA and recA, involved in exotoxin A production, lasR and rhlR, involved in quorum sensing, and exsA, involved in the type III secretion system (17–19). Overexpression of Vfr downregulates fleQ, but, peculiarly, fleQ promoter activity was not upregulated in the vfr mutant compared with the wild-type (WT) strain (12).
OsaR was previously identified by our group as a transcription factor belonging to the family of LysR-type transcriptional regulators (LTTRs) (20). A number of LTTRs function as global regulators of the genes involved in metabolism, cell division, quorum sensing, virulence, motility, nitrogen fixation, oxidative stress responses, toxin production, attachment, and secretion (21). We performed chromatin immunoprecipitation sequencing (ChIP-seq) to identify genes affected directly by OsaR; the bioinformatic analysis yielded many peaks, and genes were selected by assignment to these peaks. fleQ was one of the selected genes that was putatively regulated by OsaR. In this report, we confirmed the interaction between OsaR and fleQ and ultimately identified a novel regulatory mechanism involving two transcriptional repressors, OsaR and Vfr, that coregulate the expression of fleQ.
RESULTS
OsaR binds to the intergenic region upstream of fleQ in vivo and in vitro.
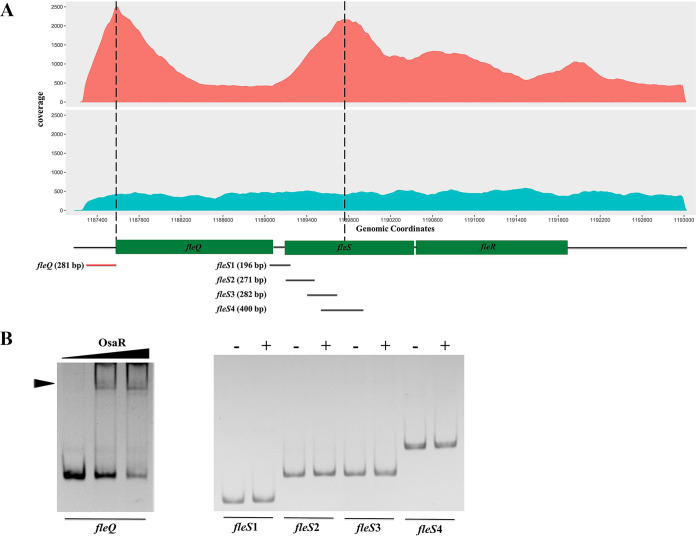
ChIP-seq was conducted to explore the putative interaction of OsaR and its target operons or genes (Seqhealth, China). A set of gene loci was preliminarily identified by peak calling, and those with fold enrichment above 2.0 were selected for further verification (see the supplemental material). Data analyses indicated two peak regions that may contain an OsaR binding site that were relevant to the gene fleQ and the fleSR operon, respectively (Fig. 1A). fleQ and fleSR are both crucial in the biosynthesis of flagella, and their locations in the P. aeruginosa genome are adjacent (6, 7, 22). Therefore, we performed gel shift assays to validate the binding of OsaR to the two regions. DNA fragments representing the whole intergenic region upstream of fleQ were mixed with OsaR protein, followed by polyacrylamide gel electrophoresis. Considering that the center of the fleSR peak region was actually within the coding sequence of the fleS gene, as shown in Fig. 1A, 4 overlapping DNA fragments (fleS1, fleS2, fleS3, and fleS4) covering both the intergenic region and the coding region were assayed. As shown in Fig. 1B, OsaR caused a modest motility shift of the fleQ intergenic fragment, while no shifted band was observed for fleSR, probably due to nonspecific binding that causes false positives in the ChIP-seq. Together, the ChIP-seq data and the gel shift assay results demonstrated that OsaR is capable of binding to the intergenic region upstream of fleQ both in vivo and in vitro.
FIG 1.
OsaR binds to the intergenic region upstream of fleQ in vivo and in vitro. (A) OsaR immunoprecipitation reads (in red) were plotted against the number of reads from the nonimmunoprecipitated DNA (in blue). The ordinate indicates the coverage depth of the reads at a certain genomic location. The region represented by the abscissa corresponds to the genomic location from coordinates 1187000 to 1193000. The locations of the fleQ and fleSR genes are represented in boxes below the abscissa. Peaks (reads, >1,500) represented by DNA accumulations are marked with dashed lines. (B) Gel shift assays of OsaR and the intergenic DNA upstream of fleQ or the DNA fragments covering both the intergenic region and coding region of fleS (fleS1 to fleS4). The protein-DNA complex is indicated by an arrowhead. The length and location of the fragments are shown in panel A; red and black colors indicate shift or no shift, respectively. Purified OsaR was used in a concentration gradient of 0, 20 nM, and 40 nM for fleQ or 0 (−) and 40 nM (+) for fleS1 to fleS4; DNA fragments of 100 ng were used.
osaR overexpression downregulates fleQ and swimming motility.
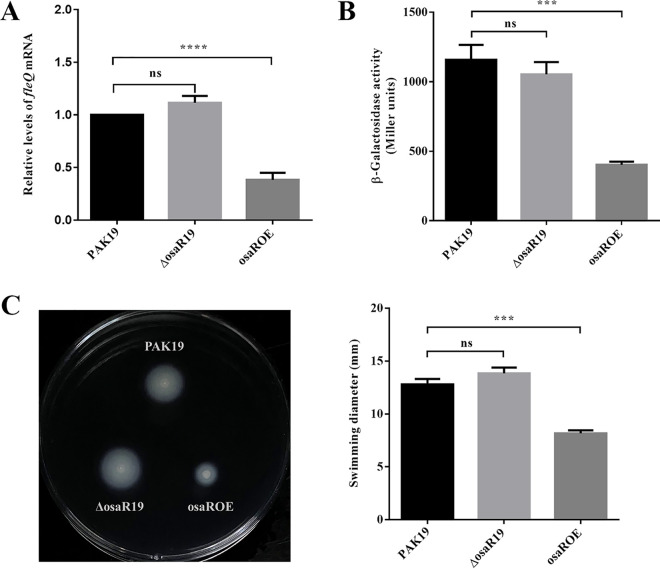
Our previous study found that OsaR, as a typical LTTR, autoregulates itself positively (20). To test whether OsaR regulates fleQ transcription and, if it does, whether the regulation is positive or negative, we detected the mRNA levels and promoter activities of fleQ in a WT strain (PAK19), osaR mutant (ΔosaR19), and osaR overexpressing strain (osaROE). By reverse transcription-quantitative PCR (RT-qPCR), we found that fleQ mRNA level was not affected by osaR knockout but significantly decreased when osaR was overexpressed (Fig. 2A). This result was consistent with that of the reporter assay. We constructed a reporter plasmid, PfleQ-lacZ, by fusing the fleQ promoter region to a promoterless lacZ gene. We then transformed the reporter plasmid into PAK19, ΔosaR19, and osaROE strains. The β-galactosidase assays suggested that fleQ promoter activity was not significantly different in the osaR mutant but obviously reduced in the osaR-overexpressing strain compared with that in the WT strain (Fig. 2B). Since fleQ is essential to flagellar biosynthesis and fleQ expression is closely associated with the flagellum-mediated motility (6, 12), we tested the effect of deleting or overexpressing osaR on swimming motility. As shown in Fig. 2C, deleting osaR did not influence swimming, while overexpressing osaR resulted in a significant loss of swimming ability. These results indicated that OsaR was a repressor of fleQ, but it remained unknown that why the absence of osaR has no effect on fleQ transcription.
FIG 2.
osaR overexpression downregulates fleQ and swimming motility. (A) The effect of OsaR on the transcriptional level of fleQ. The mRNA levels of fleQ were determined by qRT-PCR. (B) The effect of OsaR on the promoter activity of fleQ. WT strain (PAK19), osaR mutant (ΔosaR19), and osaR overexpressing strain (osaROE) were transformed with the reporter plasmid PfleQ-lacZ, followed by β-galactosidase assays when cultured to an OD600 of ∼0.6. (C) The effect of OsaR on swimming motility. Motility was detected on M63 glucose/CAA plates containing carbenicillin (left), and the diameters of the swimming zone were measured from three independent replicates (right). ns, not significant; ***, P < 0.001; ****, P < 0.0001, as determined by Student's t test. Error bars represent standard deviations.
OsaR binding site in fleQ promoter is a dual LTTR box that covers the −10 region and the transcription sites.
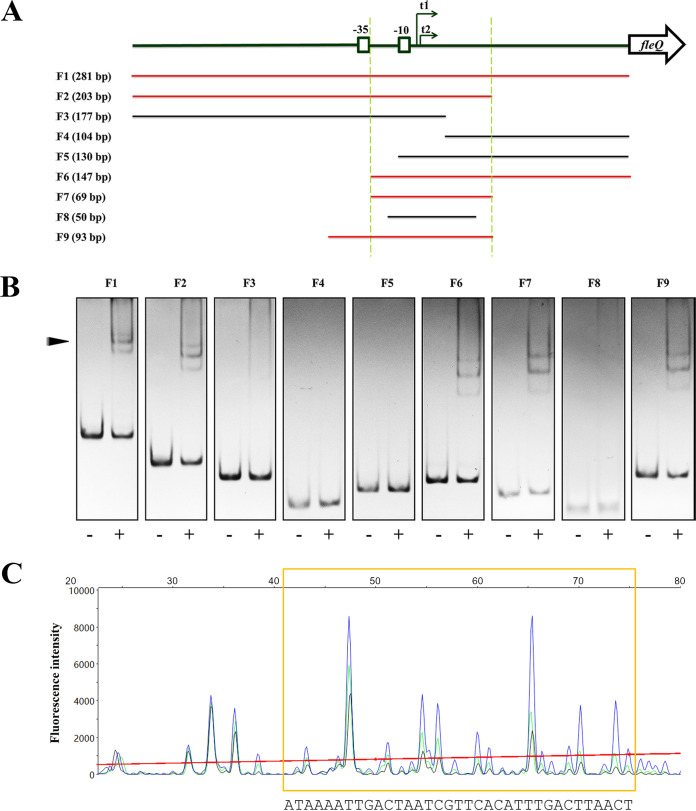
To investigate the detailed mechanism by which OsaR represses fleQ, we further conducted gel shift assays and DNase I footprinting assay to identify the OsaR binding site in the intergenic region upstream of fleQ. DNA fragments that represent different parts of the whole intergenic region were amplified and tested for their affinity to OsaR (Fig. 3A). As shown in Fig. 3A and B, fragments F1, F2, and F6 were able to bind, while F3, F4, and F5 were unable to bind to OsaR, indicating that the overlapping region of F2 and F6, which was named F7, is the shortest fragment containing the OsaR binding site. In fact, a strong and clear motility shift was detected for F7 incubated with OsaR, whereas no shift was observed for a further shortened fragment, F8 (Fig. 3B). To determine the exact binding site, a 93-bp DNA fragment (F9) relative to the first base of the fleQFP-F-FAM primer was used for DNase I footprinting assays. Based on F7, F9 was an extension that covered the −35 box for ensuring that the whole promoter was tested (Fig. 3A). The binding of OsaR to F9 was also verified (Fig. 3B). As shown in Fig. 3C, the fluorescence signal of fragments in lengths of 41 to 75 bp were reduced by addition of OsaR protein. It indicated that a region of approximately 35 bp in length was protected from DNase I digestion by OsaR, and the protection got stronger when the amount of OsaR was increasing, which implied this region was the OsaR binding site in the fleQ promoter. Based on the sequence of F9, the nucleotide sequence of the OsaR binding site was found to be 5′-ATAAAATTGACTAATCGTTCACATTTGACTTAACT-3′.
FIG 3.
Identification of the exact binding sites of OsaR in the fleQ promoter. (A) Schematic diagram of the fragments used for gel shift assays and DNase I footprinting assays. The fleQ gene is represented by an open arrow. The −35 and −10 boxes of the fleQ promoter are boxed. The two transcription start sites, t1 and t2, are indicated by two arrows. The designation and length of each of the fragments, F1 to F9, are shown on the left; red and black colors indicate shift or no shift, respectively. The overlaps of F2 and F6 are defined between the two dashed lines. (B) Gel shift assays of OsaR and DNA fragments F1 to F9. The protein-DNA complex is indicated by an arrowhead. DNA fragments of 100 ng were used and incubated with (+) or without (−) OsaR at a concentration of 40 nM. All fragments were assayed on 5% polyacrylamide gels. F1 to F3, F4 to F6, and F7 to F9 were assayed on independent gels by electrophoresis for 1 h 50 min, 1 h 35 min, and 1 h 20 min, respectively. (C) A DNase I footprinting assay revealed the OsaR binding site. 5′-FAM-labeled DNA fragment F9 (500 ng) was incubated with 80 nM (black), 40 nM (green), or 0 nM OsaR (blue) and then submitted to a 1-min DNase I treatment (0.5 U) and analyzed by capillary electrophoresis. The fluorescence intensity (arbitrary units, ordinate) is plotted against the sequence length (bases, abscissa) of the fragment. Peaks are superimposed, and the differentiated region of the three electropherograms is marked by a yellow box.
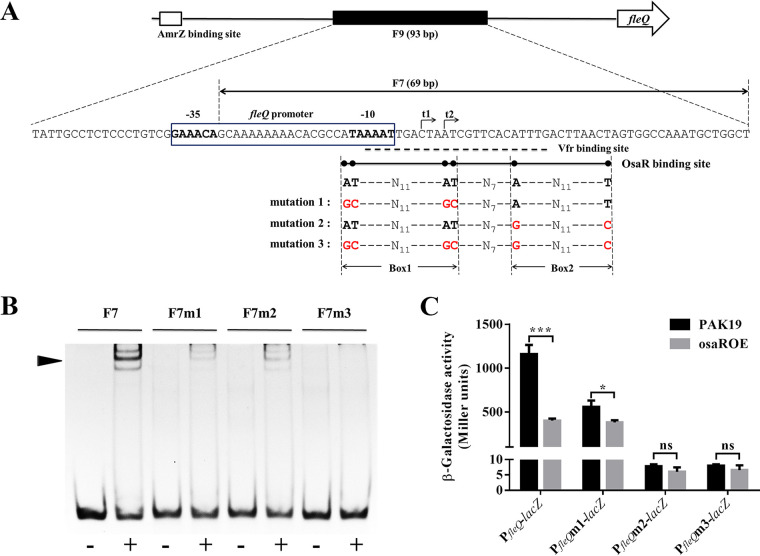
The generally accepted LTTR box consists of the sequence T-N11-A, but this sequence can vary in both base pair composition and length in the reverse complementary region (21, 23). As indicated in Fig. 4A, the OsaR binding site in the fleQ promoter region is located from position −13 to +22, counting from the transcription start site, t2, that was previously reported (12). Examining the base sequence of this region, we found that two typical LTTR boxes, AT-N11-AT (Box1) and A-N11-T (Box2), were presented and distributed on either side of this region (Fig. 4A). We assumed that these two LTTR boxes were the core of the OsaR binding site. Based on the base sequence of fragment F7 (Fig. 3A), complementary oligonucleotides were synthesized (Genewiz) with base mutations corresponding to mutation 1, mutation 2, and mutation 3 in Fig. 4A. Fragments F7m1, F7m2, and F7m3 were then generated by annealing the corresponding complementary oligonucleotides (shown in Table 2), thereby mutating the first, second, and both pairs of A-T palindromic bases of the OsaR binding site in F7, respectively. The binding affinity of OsaR to F7 and its base mutants was tested by gel shift assay, as shown in Fig. 4B. We also quantitated the percentage of each fragment that was shifted by using ImageJ software to measure the gray values. F7m1 and F7m2 both exhibited an obviously attenuated shift compared with the distinct shift of F7, while barely any shift was detected for F7m3 (Fig. 4B; see also Fig. S1 in the supplemental material). This result supports the specificity of the OsaR binding site. More importantly, it demonstrated that both of the A-T palindromic base pairs in Box1 and Box2 are vital for OsaR DNA recognition and affinity, which are weakened or even eliminated when they are altered. To further verify the importance of these two A-T pairs, we performed reporter assays with the three types of mutations indicated in Fig. 4A. As shown in Fig. 4C, mutation 1 lowered the promoter activity for PAK19, because two bases in the −10 box of the promoter were altered, but not for osaROE, due to the derepression caused by affecting the binding of OsaR. Overexpression of osaR decreased the activity by more than half (∼750 Miller units reduced) and only about 1/3 (∼180 Miller units reduced) of WT fleQ promoter and mutated fleQ promoter, respectively. Unexpectedly and intriguingly, mutation 2 and mutation 3 led to extremely low expression of lacZ for both strains. We then examined the transcription of lacZ by RT-qPCR and found that the mRNA of lacZ did not plunge to such a low level (Fig. S2). In addition, as shown in Fig. S2, lacZ transcription was even increased by ∼2-fold when the promoter was mutated in the osaR overexpressing strain (osaROE) but still lower than that in the WT strain (PAK19), which supports the result that mutation 1 detracts but does not abolish OsaR binding. We speculate that the common mutated bases of mutation 2 and mutation 3, that is, the A-T pair of Box2, plays a crucial role in the translation of the gene downstream of the fleQ promoter. Further investigations are ongoing now.
FIG 4.
Characteristics of the OsaR binding site and their implications. (A) Schematic representation of the fleQ promoter region. The fleQ gene is represented by an open arrow. In the upstream intergenic region, the putative AmrZ binding site is represented by an open box, the F9 fragment used in the DNase I footprinting assay is represented by a solid box, and the base sequence matched to it is presented below. The F7 fragment is indicated by a double arrow. The fleQ promoter is enclosed in a box, and the −35 and −10 regions are boldfaced. The transcription start sites, t1 and t2, are indicated by arrows. The Vfr binding site is underlined with a dashed line. The OsaR binding site is underlined with a full line, in which the two pairs of the A-T palindromic bases are indicated by six dots. The sequence features of the OsaR binding site and the three types of base mutations are shown below; mutated bases are marked in red. (B) The binding affinity of OsaR for the F7 DNA fragment and its three different base mutants. F7m1, F7m2, and F7m3 correspond to the three base mutations in panel A. DNA fragments of 100 ng were used and incubated with (+) or without (−) OsaR at a concentration of 40 nM, followed by a 1.5-h electrophoresis on a 8% polyacrylamide gel. (C) The effect of key mutations in OsaR binding site on the promoter activity of fleQ. PfleQm1, PfleQm2, and PfleQm3 correspond to the three base mutations indicated in panel A. β-Galactosidase assays were conducted in WT strain and osaR overexpressing strain. ns, not significant; *, P < 0.05; ***, P < 0.001; determined by Student's t test. Error bars represent standard deviations.
TABLE 2.
Primers used in this study
| Oligonucleotide | Sequence (5′–3′)b | Purpose |
|---|---|---|
| PfleQ-Fa | CCGGAATTCGTTTTTCATGGCTTTGTGCCG | Construction of PfleQ-lacZ |
| PfleQ-R | CGCGGATCCTTTGATCAGCTGCCTTGCATC | Construction of PfleQ-lacZ |
| pKF912-F | CCCAAGCTTGAAGGCTTCGCAGCTCTC | Construction of pKF912 |
| pKF912-R | GGAATTCCTGAAGCGCCTGTTCTTCC | Construction of pKF912 |
| Δvfr-F | CGATGATGTCGACAACATCG | Identification of vfr deletion |
| Δvfr-R | TCAAGAGCCAGACCCTGATG | Identification of vfr deletion |
| fleS1-F | GGCATGAGCCGGCGTGAC | Amplification of fleS1 |
| fleS1-R | CGTATCGGCTGGCTGCTC | Amplification of fleS1 |
| fleS2-F | ATGCAACCAGCCCTCAACGC | Amplification of fleS2 |
| fleS2-R | TGACGATGACGCCGCCGG | Amplification of fleS2 |
| fleS3-F | GCGGAAAAGGAGCGCCTGG | Amplification of fleS3 |
| fleS3-R | GTTGAGCAGGATCAGTTGCC | Amplification of fleS3 |
| fleS4-F | AGCCGTTGGTGGGCATGC | Amplification of fleS4 |
| fleS4-R | CGGGCGAACACCAGCATG | Amplification of fleS4 |
| fleQF123-F | GTTTTTCATGGCTTTGTGCC | Amplification of F1, F2, F3 |
| fleQF1456-R | TTTGATCAGCTGCCTTGCATC | Amplification of F1, F4, F5, F6 |
| fleQF2-R | AGCCAGCATTTGGCCACTA | |
| fleQF3-R | CAAATGTGAACGATTAGTCA | Amplification of F2 |
| fleQF4-F | ACTTAACTAGTGGCCAAATGC | Amplification of F3 |
| fleQF5-F | TAAAATTGACTAATCGTTCAC | Amplification of F4 |
| fleQF6-F | GCAAAAAAAACACGCCATAA | Amplification of F5 |
| fleQF7-F | GCAAAAAAAACACGCCATAAAATTGACTAATCGTTCACATTTGACTTAACTAGTGGCCAAATGCTGGCT | Amplification of F6 |
| fleQF7-R | AGCCAGCATTTGGCCACTAGTTAAGTCAAATGTGAACGATTAGTCAATTTTATGGCGTGTTTTTTTTGC | Annealing of F7 |
| fleQF8-F | ACACGCCATAAAATTGACTAATCGTTCACATTTGACTTAACTAGTGGCCA | Annealing of F7 |
| fleQF8-R | TGGCCACTAGTTAAGTCAAATGTGAACGATTAGTCAATTTTATGGCGTGT | Annealing of F8 |
| fleQF7m1-F | GCAAAAAAAACACGCCGCAAAATTGACTAGCCGTTCACATTTGACTTAACTAGTGGCCAAATGCTGGCT | Annealing of F8 |
| fleQF7m1-R | AGCCAGCATTTGGCCACTAGTTAAGTCAAATGTGAACGGCTAGTCAATTTTGCGGCGTGTTTTTTTTGC | Annealing of F7m1 |
| fleQF7m2-F | GCAAAAAAAACACGCCATAAAATTGACTAATCGTTCACGTTTGACTTAACCAGTGGCCAAATGCTGGCT | Annealing of F7m1 |
| fleQF7m2-R | AGCCAGCATTTGGCCACTGGTTAAGTCAAACGTGAACGATTAGTCAATTTTATGGCGTGTTTTTTTTGC | Annealing of F7m2 |
| fleQF7m3-F | GCAAAAAAAACACGCCGCAAAATTGACTAGCCGTTCACGTTTGACTTAACCAGTGGCCAAATGCTGGCT | Annealing of F7m3 |
| fleQF7m3-R | AGCCAGCATTTGGCCACTGGTTAAGTCAAA | Annealing of F7m3 |
| fleQFP-F-FAM | CGTGAACGGCTAGTCAATTTTGCGGCGTGTTTTTTTTGC | DNase I footprinting assay |
| fleQFP-R | TTTATTGCCTCTCCCTGTCG | DNase I footprinting assay |
| fleQ-qRT-F | AGCCAGCATTTGGCCACTA | qRT-PCR |
| fleQ-qRT-R | ACTACCGCCTCAACGTATTCC | qRT-PCR |
| rpsL-qRT-F | CGCTTCTCATGCTCCATCC | qRT-PCR |
| rpsL-qRT-R | ACGTGCCTGCGCTGCAAAACCCCGAGGTGTCCAGCGAACC | qRT-PCR |
F, forward; R, reverse; FAM, 6-carboxyfluorescein phosphoramidate labeled at the 5′end.
The underlines are the sites of restriction enzymes.
Therefore, we can conclude that the OsaR binding site in the fleQ promoter is a characteristic sequence, AT-N11-AT-N7-A-N11-T. It is highly similar to the consensus binding sequence of another well-studied LTTR, OxyR (24, 25) (Fig. S3A). We have also identified it in the promoter region of other genes that are regulated or putatively regulated by OsaR, such as PA0057 (Fig. S3B) (20).
OsaR and Vfr function as redundant repressors of fleQ.
It was previously reported that the virulence factor regulator Vfr was also able to repress the transcription of fleQ; overexpression of Vfr resulted in a reduction in fleQ promoter activity and motility, whereas no effect was shown with vfr deletion (12). Our study identified that OsaR represses the transcription of fleQ and binds to a promoter region overlapping the Vfr binding site (Fig. 2A and B and 4A). Two transcription factors binding to the same position implies a coregulatory mechanism. We first confirmed the function of Vfr. As shown in Fig. S4, deleting vfr did not affect fleQ mRNA level and promoter activity or bacterial motility; however, they were prominently reduced in the vfr-overexpressing strain.
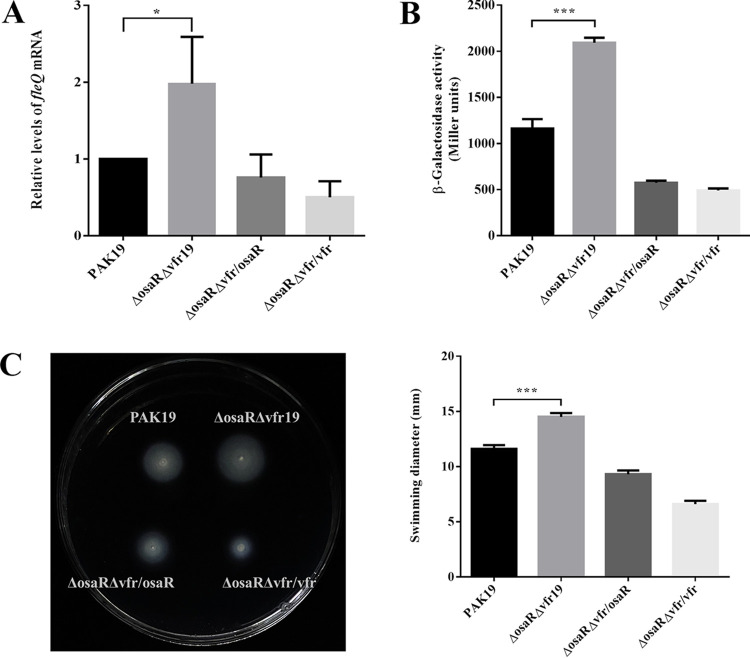
Therefore, OsaR and Vfr display an identical role in regulating fleQ. In particular, either of the two repressors being absent has no significant effect on fleQ transcription, suggesting that their function is redundant. To validate this assumption, we knocked out both osaR and vfr in PAK and then tested the influence on fleQ expression. As shown in Fig. 5, the double-knockout strain exhibited upregulated mRNA levels and promoter activity of fleQ and increased swimming ability. In addition, these phenotypes could be restored with the expression of either osaR or vfr alone (Fig. 5). It is worth noting that the level of fleQ transcription in the ΔosaRΔvfr strain was only approximately 2-fold upregulated compared with the wild-type strain (Fig. 5). AmrZ is likely responsible for this. In the absence of AmrZ, fleQ promoter activity and mRNA levels were upregulated by approximately 4-fold and 8-fold, respectively (15, 16). This suggests that AmrZ is a more effective repressor of fleQ than OsaR and/or Vfr.
FIG 5.
fleQ and swimming motility are upregulated when osaR and vfr are both deleted. (A) The effect on fleQ transcription by deleting osaR and vfr simultaneously. The mRNA levels were determined by RT-qPCR. (B) The effect on the promoter activity of fleQ by deleting both osaR and vfr. Strains were transformed with the reporter plasmid PfleQ-lacZ, followed by β-galactosidase assays when cultured to an OD600 of ∼0.6. (C) The effect on swimming motility by deleting both osaR and vfr. Motility was detected and quantitated by three independent replicates. ns, not significant; *, P < 0.05; ***, P < 0.001; determined by Student's t test. Error bars represent standard deviations.
In conclusion, our results demonstrated that OsaR and Vfr separately bind to the same position covering the entire (OsaR) or partial (Vfr) −10 box and the two transcription start sites in the fleQ promoter. The redundant repression would be attenuated only when the two repressors were simultaneously removed, because either one of them could retain the repression when the other one was absent.
DISCUSSION
The regulator gene fleQ is vital for the expression of almost all known flagellar genes in P. aeruginosa (6–8). Mechanisms that inactivate or downregulate fleQ are sufficient to inhibit flagellum biosynthesis as well as flagellum-mediated motility (7, 12, 26). Overexpression of fleQ in a nonmotile P. aeruginosa strain increased flagellar numbers and restored flagellum-mediated motility (26). It was reported that fleQ was downregulated by overexpression of the virulence factor regulator Vfr (12); however, it was perplexing that knocking out vfr had no effect on fleQ (12). In this study, we unraveled this conundrum and provided updated demonstrations about the upstream regulation of fleQ. We identified a protein-DNA interaction between the LysR-type transcriptional regulator OsaR and the promoter of fleQ. Further experimental results revealed that OsaR can bind to the −10 region and retain the transcriptional repression when Vfr is absent and vice versa.
Many bacterial promoters are controlled by multiple transcription factors with positive or negative interplay, which illustrates the complexity, diversity, and versatility of bacterial gene regulation mechanisms (27–30). The binding sites of competitive repressors partially overlap the control region, and a substantial boost of expression of the controlled gene will only be generated by removing the repressors simultaneously (27, 28, 30). The regulatory pattern of OsaR and Vfr repressing fleQ identified by our study is compatible with the canonical mode of competitive repression, except fleQ only displays modest upregulation in the ΔosaRΔvfr double mutant. We suppose that another repressor, AmrZ, could still repress fleQ to a certain extent when OsaR and Vfr are both absent. Depending on the alternative sigma factor AlgT (31), AmrZ directly interacts with the fleQ promoter and effectively suppresses fleQ transcription (15, 26). Dissimilar to the OsaR/Vfr binding sites, however, the putative AmrZ binding site is located further upstream of the −35 region (Fig. 4A) according to previous ChIP-seq analysis (16). Although further investigation is needed for identifying the specific relationship among the regulation by the three repressors, our discovery indicates the potential of distinct manners in which OsaR/Vfr and AmrZ repress fleQ independently.
Our previous study found that OsaR was involved in bacterial tolerance to antibiotics and oxidative stress (20). OsaR protein undergoes a change in aggregation state between its oxidative state and reductive state and shows different DNA affinity under different redox conditions (20). Therefore, oxidative stresses, including those generated by antibiotics, might be the signaling factor that controls OsaR activity, which is similar to another LTTR, OxyR, that senses reactive oxygen species (ROS) and regulates target genes in a redox-dependent manner (24, 32). Hence, our study might provide critical clues for understanding the motility changes of P. aeruginosa mutants that display altered susceptibility to antibiotics or oxidative stress (33–35).
Members of the LTTR family have a highly conserved amino acid composition and secondary structure in their DNA-binding domain as well as a characteristic binding sequence, called the LTTR box, which consists of the sequence T-N11-A, which can vary in both base pair composition and length (21, 23). The OxyR binding sites in antioxidant genes exhibit typical characteristics of the LTTR box (24, 25), including two LTTR boxes with a spacer composed of 7/8 random bases (see Fig. S3A in the supplemental material). In this report, the OsaR binding site we identified in fleQ was AT-N11-AT-N7-A-N11-T (Fig. 4A), which is highly similar to the OxyR binding sites. In addition, our previous work indicated that an approximately 40-bp section of DNA within the intergenic region of PA0056-PA0057 was protected from DNase I digestion by the OsaR protein (20). The features of a dual LTTR box were found in the base sequence, which were the same as those of the fleQ-bound site (Fig. S3B). The available evidence suggests that AT-N11-AT-N7-A-N11-T, or its variant, is the OsaR consensus sequence, and, based on this, further investigation of the global function of OsaR could be conducted. Moreover, because OsaR and OxyR both transform between oxidation and reduction states (20, 24, 32) and share a binding sequence with high similarity, the potential for cross talk between these two LTTRs appears to have research prospects. Intriguingly, in a study by Panmanee et al. (36), bioinformatics analysis was performed to screen OxyR binding sites in P. aeruginosa based on similarity to the well-characterized Escherichia coli OxyR-regulated promoter sequences (ATAG-N7-CTAT-N7-ATAG-N7-CTAT), and fleQ was identified as one of the OxyR-dependent gene candidates. There was no further evidence supporting the OxyR-fleQ interaction in the aforementioned study; however, our study demonstrated that fleQ is under the regulation of OsaR.
Classical LTTR regulation has been described as transcriptional activation and negative autoregulation, and there are relatively fewer reports of LTTRs acting as transcriptional repressors that positively autoregulate (21). LrhA was first identified in E. coli as a LysR homolog (37), and, with its homologs HexA and PecT in Erwinia spp. (38), subsequent studies revealed that these three LTTRs positively autoregulate themselves and negatively regulate the expression of genes required for flagellation, motility, and chemotaxis (21, 37–39). In E. coli, the flhDC operon encodes the master regulator in the hierarchical regulation of flagellar biogenesis (40); it is activated by the cyclic AMP (cAMP) receptor protein CRP (41) and negatively regulated by LrhA (39). In P. aeruginosa, we previously identified that osaR expression is positively autoregulated by OsaR (20); here, we demonstrated that the top-level regulator in the flagellar biogenesis hierarchy, FleQ, is repressed redundantly by the LTTR OsaR and the cAMP receptor protein Vfr, which is actually a homolog of E. coli CRP (42). We speculate that coregulation mechanisms of the flagellation-related master regulator involving a positively autoregulated LTTR and a cAMP receptor protein is conserved in multiple bacterial species.
In addition to P. aeruginosa, FleQ appears to be at the top level of the hierarchical regulation of flagellar biogenesis in all of the examined pseudomonads, such as P. putida and P. fluorescens (43). FleQ homologs that regulate flagellar biosynthesis are also widely distributed in other bacterial species, including FlrA in Vibrio cholerae (44), FlaK in Vibrio parahaemolyticus (45), and FlgR in Helicobacter pylori (46). Therefore, any further elucidation of the upstream regulatory mechanisms of fleQ will be remarkably significant to the study of all of the related flagellated bacteria, especially because, to the best of our knowledge, OsaR and its homologs in other species have not been reported yet.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
P. aeruginosa and Escherichia coli strains were routinely grown on LB medium at 37°C. Additionally, M63 glucose/CAA medium (M63 minimal medium supplemented with 0.2% glucose, 1 mM MgSO4, and 0.5% Casamino Acids) was used to detect motility (47–49). Antibiotics were added at the following concentrations: 50 μg/ml tetracycline or 150 μg/ml carbenicillin for P. aeruginosa; 10 μg/ml tetracycline, 25 μg/ml kanamycin, or 50 μg/ml carbenicillin for E. coli.
The bacterial strains and plasmids used in this study are listed in Table 1. The primers used in this study are listed in Table 2. The plasmids were constructed based on standard DNA manipulations with the E. coli DH5α strain and confirmed by sequencing (Genewiz), after which they were transformed into P. aeruginosa strains by electroporation (50). The construction of the ΔosaRΔvfr mutant was based on the ΔosaR mutant and the S17/pEX18Tc-Δvfr strain; the conjugation and the selection of double-crossover mutants were conducted according to manipulations previously reported (51).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source |
|---|---|---|
| E. coli | ||
| DH5α | F− ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK− mK+) phoA supE44 λ− thi-1 gyrA96 relA1 | CWBIO |
| BL21 | F− ompT hsdS(rBB− mB−) gal dcm (DE3) | CWBIO |
| S17-1 | RP4-2 Tc::Mu Km::Tn7 Tpr Smr Pro Res− Mod+ | R. Ramphal |
| S17/pEX18Tc-Δvfr | S17-1 harboring the pEX18Tc-Δvfr vector; Tcr | 58 |
| BL21/pETMAL-osaR | BL21 harboring the pETMAL-osaR vector; Kanr | 20 |
| P. aeruginosa | ||
| PAK | Wild type | David Bradley |
| PAK19 | PAK harboring the pUCP19 vector; Cbr | This study |
| ΔosaR | PAK with osaR gene deleted | 20 |
| ΔosaR19 | ΔosaR harboring the pUCP19 vector; Cbr | This study |
| Δvfr | PAK with vfr gene deleted | 58 |
| Δvfr19 | Δvfr harboring the pUCP19 vector; Cbr | This study |
| ΔosaRΔvfr | PAK with both osaR and vfr genes deleted | This study |
| ΔosaRΔvfr19 | ΔosaRΔvfr harboring the pUCP19 vector; Cbr | This study |
| osaROE | PAK harboring the pUCP19-osaR vector for overexpression of osaR; Cbr | 20 |
| vfrOE | PAK harboring the pKF912 vector for overexpression of vfr; Cbr | This study |
| ΔosaRΔvfr/osaR | ΔosaRΔvfr mutant harboring the pUCP19-osaR vector for complement of osaR; Cbr | This study |
| ΔosaRΔvfr/vfr | ΔosaRΔvfr mutant harboring the pKF912 vector for complement of vfr; Cbr | This study |
| Plasmids | ||
| pEX18Tc-Δvfr | vfr gene deletion on pEX18Tc; Tcr | 58 |
| pETMAL-osaR | pETMALc-H vector carrying the malE-OsaR fusion; Kanr | 20 |
| pUCP19 | Multicopy E. coli-P. aeruginosa shuttle vector; Apr/Cbr | 59 |
| pUCP19-osaR | pUCP19 carrying the intact osaR gene; Apr/Cbr | 20 |
| pKF912 | pUCP19 carrying vfr as a 1.2-kb XhoI fragment; Apr/Cbr | 42 |
| pDN19lacΩ | Broad-host-range plasmid containing a promoterless lacZ gene; Tcr | 60 |
| PfleQ-lacZ | pDN19lacΩ carrying the promoter region of fleQ in the EcoRI/BamHI sites; Tcr | This study |
| PfleQm1-lacZ | PfleQ-lacZ with the first A-T pair of the OsaR binding site mutated; Tcr | This study |
| PfleQm2-lacZ | PfleQ-lacZ with the second A-T pair of the OsaR binding site mutated; Tcr | This study |
| PfleQm3-lacZ | PfleQ-lacZ with the two A-T pairs of the OsaR binding site mutated; Tcr | This study |
Protein expression and purification.
Expression and purification of OsaR were conducted as previously described (52), with slight modifications. Briefly, the pETMALc-H vector was used to construct an MBP-OsaR fusion expression plasmid to enhance the solubility of the expressed protein. The BL21/pETMAL-osaR strain was cultured in LB medium supplemented with 0.2% glucose overnight, followed by a 100-fold dilution into fresh medium. The culture was grown at 37°C until the optical density at 600 nm (OD600) reached 0.6, and then 0.3 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was added for induction for another 2 h at 28°C. The cells then were harvested and suspended in PB buffer (20 mM NaH2PO4, 20 mM Na2HPO4, pH 7.4) supplemented with 500 mM NaCl and 1 mM EDTA. The OsaR protein was acquired and purified based on the pMAL protein fusion and purification system manual (NEB E8200), in which amylose resin (NEB) was used for affinity chromatography and thrombin (Macklin) was used for the cleavage of the fusion protein.
Gel shift assay.
To assess the ability of OsaR to bind to the promoter of fleQ in gel shift assays, various desired fragments of the intergenic region upstream of the translation start site were amplified by PCR using different pairs of primers (primers shown in Table 2). Site-directed mutagenesis fragments were obtained by annealing complementary oligonucleotides (base sequences shown in Table 2). All of the DNA fragments used for the gel shift assay were purified with the MiniBEST agarose gel DNA extraction kit (TaKaRa). The binding and shift assays were performed as previously published (53, 54). Purified protein and DNA fragments were mixed in a 25-μl mixture containing the gel-shift binding buffer (20 mM Tris-HCl, 50 mM KCl, 50 mM MgCl2, 10% glycerol, pH 7.5) and subsequently incubated at 37°C for 30 min. The samples were resolved on a 5% or 8% polyacrylamide native gel in Tris-borate-EDTA buffer (0.044 M Tris, 0.044 M boric acid, 0.001 M EDTA, pH 8.0) on ice. Gels were soaked in 0.5 μg/ml ethidium bromide (EB), and then the DNA was visualized under UV light and imaged in a MultiImage light cabinet filter position molecular imager (Alpha Innotech Corporation).
DNase I footprinting assay.
DNase I footprint analysis was performed using a nonradiochemical capillary electrophoresis method as previously described (55, 56). A 93-bp DNA fragment was generated by PCR using a 6-FAM (6-carboxyfluorescein phosphoramidate) primer, fleQFP-F-FAM, labeled at the 5′ end and paired with the primer fleQFP-R (primers are shown in Table 2). The labeled DNA fragment was purified with the MiniBEST agarose gel DNA extraction kit (TaKaRa) and then mixed with purified OsaR protein in 100-μl reaction mixtures containing the same binding buffer as that used in the gel shift assays. After incubation for 30 min at room temperature, 0.5 U of DNase I (Invitrogen) was added for 1 min of further incubation. The samples were then placed at 70°C in a water bath for 10 min to end the reaction, followed by purification utilizing the UNIQ-10 spin column Oligo DNA purification kit (Sangon Biotech). Fragments were analyzed by capillary electrophoresis, and the sizes were determined using ABI Peak Scanner software v2.0.
RNA extraction and RT-qPCR.
Overnight culture of P. aeruginosa strains was diluted 1/100 into fresh LB medium, followed by subculturing until the OD600 reached 1.0. The bacterial cells were subsequently harvested, and total RNA was isolated utilizing the SPARKeasy bacterial RNA kit (SparkJade). cDNA was synthesized with the PrimeScript RT reagent kit (TaKaRa), including a genomic DNA erasing step followed by reverse transcription-PCR with random primers. Real-time PCR was performed using the cDNA as the template, which was mixed with the indicated primers (shown in Table 2) and qPCR SYBR green master mix (Yeasen). The gene rpsL encoding the 30S ribosomal protein was used as internal control.
PfleQ-lacZ reporter assay.
PfleQ, which represents the intergenic region upstream of fleQ or its three variants, PfleQm1, PfleQm2 and PfleQm3, were inserted upstream of the promoter-less lacZ gene in the pDN19lacΩ vector, generating PfleQ-lacZ, PfleQm1-lacZ, PfleQm2-lacZ, and PfleQm3-lacZ, respectively. PfleQ was amplified by PCR; PfleQm1, PfleQm2, and PfleQm3 were synthesized by Genewiz. The transcriptional fusion plasmids were transformed into specific strains for conducting β-galactosidase assays, and the β-galactosidase assay was carried out as previously described (57).
Motility assay.
Motility was assessed as previously described; M63 glucose/CAA medium with 0.3% agar was used for the swimming motility assay (47–49). Fresh bacterial colonies were inoculated onto the surface of the plate with sterile toothpicks, followed by 12 to 16 h of incubation at 30°C.
ACKNOWLEDGMENTS
This work was supported by the Sino-Swiss Scientific and Technological Cooperation Project, supported by the Ministry of Science and Technology of China (no. 2015DFG32140) and the National Natural Science Foundation of China (no. 31770102).
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material is available online only.
Contributor Information
Haijin Xu, Email: xuhaijin@aliyun.com.
Mingqiang Qiao, Email: qiaomq@nankai.edu.cn.
George O'Toole, Geisel School of Medicine at Dartmouth.
REFERENCES
- 1.Church D, Elsayed S, Reid O, Winston B, Lindsay R. 2006. Burn wound infections. Clin Microbiol Rev 19:403–434. 10.1128/CMR.19.2.403-434.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lyczak JB, Cannon CL, Pier GB. 2000. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect 2:1051–1060. 10.1016/s1286-4579(00)01259-4. [DOI] [PubMed] [Google Scholar]
- 3.Sousa AM, Pereira MO. 2014. Pseudomonas aeruginosa diversification during infection development in cystic fibrosis lungs-a review. Pathogens 3:680–703. 10.3390/pathogens3030680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sadikot RT, Blackwell TS, Christman JW, Prince AS. 2005. Pathogen-host interactions in Pseudomonas aeruginosa pneumonia. Am J Respir Crit Care Med 171:1209–1223. 10.1164/rccm.200408-1044SO. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balasubramanian D, Schneper L, Kumari H, Mathee K. 2013. A dynamic and intricate regulatory network determines Pseudomonas aeruginosa virulence. Nucleic Acids Res 41:1–20. 10.1093/nar/gks1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dasgupta N, Wolfgang MC, Goodman AL, Arora SK, Jyot J, Lory S, Ramphal R. 2003. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol Microbiol 50:809–824. 10.1046/j.1365-2958.2003.03740.x. [DOI] [PubMed] [Google Scholar]
- 7.Arora SK, Ritchings BW, Almira EC, Lory S, Ramphal R. 1997. A transcriptional activator, FleQ, regulates mucin adhesion and flagellar gene expression in Pseudomonas aeruginosa in a cascade manner. J Bacteriol 179:5574–5581. 10.1128/jb.179.17.5574-5581.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jyot J, Dasgupta N, Ramphal R. 2002. FleQ, the major flagellar gene regulator in Pseudomonas aeruginosa, binds to enhancer sites located either upstream or atypically downstream of the RpoN binding site. J Bacteriol 184:5251–5260. 10.1128/JB.184.19.5251-5260.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Totten PA, Lara JC, Lory S. 1990. The rpoN gene product of Pseudomonas aeruginosa is required for expression of diverse genes, including the flagellin gene. J Bacteriol 172:389–396. 10.1128/jb.172.1.389-396.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baraquet C, Harwood CS. 2013. Cyclic diguanosine monophosphate represses bacterial flagella synthesis by interacting with the Walker A motif of the enhancer-binding protein FleQ. Proc Natl Acad Sci USA 110:18478–18483. 10.1073/pnas.1318972110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dasgupta N, Ramphal R. 2001. Interaction of the antiactivator FleN with the transcriptional activator FleQ regulates flagellar number in Pseudomonas aeruginosa. J Bacteriol 183:6636–6644. 10.1128/JB.183.22.6636-6644.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dasgupta N, Ferrell EP, Kanack KJ, West SE, Ramphal R. 2002. fleQ, the gene encoding the major flagellar regulator of Pseudomonas aeruginosa, is sigma70 dependent and is downregulated by Vfr, a homolog of Escherichia coli cyclic AMP receptor protein. J Bacteriol 184:5240–5250. 10.1128/JB.184.19.5240-5250.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones CJ, Ryder CR, Mann EE, Wozniak DJ. 2013. AmrZ modulates Pseudomonas aeruginosa biofilm architecture by directly repressing transcription of the psl operon. J Bacteriol 195:1637–1644. 10.1128/JB.02190-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baynham PJ, Brown AL, Hall LL, Wozniak DJ. 1999. Pseudomonas aeruginosa AlgZ, a ribbon-helix-helix DNA-binding protein, is essential for alginate synthesis and algD transcriptional activation. Mol Microbiol 33:1069–1080. 10.1046/j.1365-2958.1999.01550.x. [DOI] [PubMed] [Google Scholar]
- 15.Tart AH, Blanks MJ, Wozniak DJ. 2006. The AlgT-dependent transcriptional regulator AmrZ (AlgZ) inhibits flagellum biosynthesis in mucoid, nonmotile Pseudomonas aeruginosa cystic fibrosis isolates. J Bacteriol 188:6483–6489. 10.1128/JB.00636-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones CJ, Newsom D, Kelly B, Irie Y, Jennings LK, Xu B, Limoli DH, Harrison JJ, Parsek MR, White P, Wozniak DJ. 2014. ChIP-Seq and RNA-Seq reveal an AmrZ-mediated mechanism for cyclic di-GMP synthesis and biofilm development by Pseudomonas aeruginosa. PLoS Pathog 10:e1003984. 10.1371/journal.ppat.1003984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davinic M, Carty NL, Colmer-Hamood JA, San Francisco M, Hamood AN. 2009. Role of Vfr in regulating exotoxin A production by Pseudomonas aeruginosa. Microbiology 155:2265–2273. 10.1099/mic.0.028373-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Croda-Garcia G, Grosso-Becerra V, Gonzalez-Valdez A, Servin-Gonzalez L, Soberon-Chavez G. 2011. Transcriptional regulation of Pseudomonas aeruginosa rhlR: role of the CRP orthologue Vfr (virulence factor regulator) and quorum-sensing regulators LasR and RhlR. Microbiology 157:2545–2555. 10.1099/mic.0.050161-0. [DOI] [PubMed] [Google Scholar]
- 19.Marsden AE, Intile PJ, Schulmeyer KH, Simmons-Patterson ER, Urbanowski ML, Wolfgang MC, Yahr TL. 2016. Vfr directly activates exsA transcription to regulate expression of the Pseudomonas aeruginosa type III secretion system. J Bacteriol 198:1442–1450. 10.1128/JB.00049-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y, Ma Y, Ma Z, Han X, Qi H, Andersen JB, Xu H, Tolker-Nielsen T, Qiao M. 2020. Redox protein OsaR (PA0056) regulates dsbM and the oxidative stress response in Pseudomonas aeruginosa. Antimicrob Agents Chemother 65:e01771-20. 10.1128/AAC.01771-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maddocks SE, Oyston PCF. 2008. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology 154:3609–3623. 10.1099/mic.0.2008/022772-0. [DOI] [PubMed] [Google Scholar]
- 22.Ritchings BW, Almira EC, Lory S, Ramphal R. 1995. Cloning and phenotypic characterization of fleS and fleR, new response regulators of Pseudomonas aeruginosa which regulate motility and adhesion to mucin. Infect Immun 63:4868–4876. 10.1128/iai.63.12.4868-4876.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goethals K, Van Montagu M, Holsters M. 1992. Conserved motifs in a divergent nod box of Azorhizobium caulinodans ORS571 reveal a common structure in promoters regulated by LysR-type proteins. Proc Natl Acad Sci USA 89:1646–1650. 10.1073/pnas.89.5.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei Q, Minh PN, Dotsch A, Hildebrand F, Panmanee W, Elfarash A, Schulz S, Plaisance S, Charlier D, Hassett D, Haussler S, Cornelis P. 2012. Global regulation of gene expression by OxyR in an important human opportunistic pathogen. Nucleic Acids Res 40:4320–4333. 10.1093/nar/gks017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ochsner UA, Vasil ML, Alsabbagh E, Parvatiyar K, Hassett DJ. 2000. Role of the Pseudomonas aeruginosa oxyR-recG operon in oxidative stress defense and DNA repair: oxyR-dependent regulation of katB-ankB, ahpB, and ahpC-ahpF. J Bacteriol 182:4533–4544. 10.1128/JB.182.16.4533-4544.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tart AH, Wolfgang MC, Wozniak DJ. 2005. The alternative sigma factor AlgT represses Pseudomonas aeruginosa flagellum biosynthesis by inhibiting expression of fleQ. J Bacteriol 187:7955–7962. 10.1128/JB.187.23.7955-7962.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Compan I, Touati D. 1993. Interaction of six global transcription regulators in expression of manganese superoxide dismutase in Escherichia coli K-12. J Bacteriol 175:1687–1696. 10.1128/jb.175.6.1687-1696.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Torres Montaguth OE, Bervoets I, Peeters E, Charlier D. 2019. Competitive repression of the artPIQM operon for arginine and ornithine transport by arginine repressor and leucine-responsive regulatory protein in Escherichia coli. Front Microbiol 10:1563. 10.3389/fmicb.2019.01563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bervoets I, Charlier D. 2019. Diversity, versatility and complexity of bacterial gene regulation mechanisms: opportunities and drawbacks for applications in synthetic biology. FEMS Microbiol Rev 43:304–339. 10.1093/femsre/fuz001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tardat B, Touati D. 1993. Iron and oxygen regulation of Escherichia coli MnSOD expression: competition between the global regulators Fur and ArcA for binding to DNA. Mol Microbiol 9:53–63. 10.1111/j.1365-2958.1993.tb01668.x. [DOI] [PubMed] [Google Scholar]
- 31.Garrett ES, Perlegas D, Wozniak DJ. 1999. Negative control of flagellum synthesis in Pseudomonas aeruginosa is modulated by the alternative sigma factor AlgT (AlgU). J Bacteriol 181:7401–7404. 10.1128/JB.181.23.7401-7404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jo I, Chung IY, Bae HW, Kim JS, Song S, Cho YH, Ha NC. 2015. Structural details of the OxyR peroxide-sensing mechanism. Proc Natl Acad Sci USA 112:6443–6448. 10.1073/pnas.1424495112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wongsaroj L, Saninjuk K, Romsang A, Duang-Nkern J, Trinachartvanit W, Vattanaviboon P, Mongkolsuk S. 2018. Pseudomonas aeruginosa glutathione biosynthesis genes play multiple roles in stress protection, bacterial virulence and biofilm formation. PLoS One 13:e0205815. 10.1371/journal.pone.0205815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coleman SR, Smith ML, Spicer V, Lao Y, Mookherjee N, Hancock REW. 2020. Overexpression of the small RNA PA0805.1 in Pseudomonas aeruginosa modulates the expression of a large set of genes and proteins, resulting in altered motility, cytotoxicity, and tobramycin resistance. mSystems 5:e00204-20. [PMC]. 10.1128/mSystems.00204-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi N, Gao Y, Yin D, Song Y, Kang J, Li X, Zhang Z, Feng X, Duan J. 2019. The effect of the sub-minimal inhibitory concentration and the concentrations within resistant mutation window of ciprofloxacin on MIC, swimming motility and biofilm formation of Pseudomonas aeruginosa. Microb Pathog 137:103765. 10.1016/j.micpath.2019.103765. [DOI] [PubMed] [Google Scholar]
- 36.Panmanee W, Charoenlap N, Atichartpongkul S, Mahavihakanont A, Whiteside MD, Winsor G, Brinkman FSL, Mongkolsuk S, Hassett DJ. 2017. The OxyR-regulated phnW gene encoding 2-aminoethylphosphonate:pyruvate aminotransferase helps protect Pseudomonas aeruginosa from tert-butyl hydroperoxide. PLoS One 12:e0189066. 10.1371/journal.pone.0189066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gibson KE, Silhavy TJ. 1999. The LysR homolog LrhA promotes RpoS degradation by modulating activity of the response regulator sprE. J Bacteriol 181:563–571. 10.1128/JB.181.2.563-571.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harris SJ, Shih YL, Bentley SD, Salmond GP. 1998. The hexA gene of Erwinia carotovora encodes a LysR homologue and regulates motility and the expression of multiple virulence determinants. Mol Microbiol 28:705–717. 10.1046/j.1365-2958.1998.00825.x. [DOI] [PubMed] [Google Scholar]
- 39.Lehnen D, Blumer C, Polen T, Wackwitz B, Wendisch VF, Unden G. 2002. LrhA as a new transcriptional key regulator of flagella, motility and chemotaxis genes in Escherichia coli. Mol Microbiol 45:521–532. 10.1046/j.1365-2958.2002.03032.x. [DOI] [PubMed] [Google Scholar]
- 40.Chilcott GS, Hughes KT. 2000. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar typhimurium and Escherichia coli. Microbiol Mol Biol Rev 64:694–708. 10.1128/MMBR.64.4.694-708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Soutourina O, Kolb A, Krin E, Laurent-Winter C, Rimsky S, Danchin A, Bertin P. 1999. Multiple control of flagellum biosynthesis in Escherichia coli: role of H-NS protein and the cyclic AMP-catabolite activator protein complex in transcription of the flhDC master operon. J Bacteriol 181:7500–7508. 10.1128/JB.181.24.7500-7508.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.West SE, Sample AK, Runyen-Janecky LJ. 1994. The vfr gene product, required for Pseudomonas aeruginosa exotoxin A and protease production, belongs to the cyclic AMP receptor protein family. J Bacteriol 176:7532–7542. 10.1128/jb.176.24.7532-7542.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blanco-Romero E, Redondo-Nieto M, Martinez-Granero F, Garrido-Sanz D, Ramos-Gonzalez MI, Martin M, Rivilla R. 2018. Genome-wide analysis of the FleQ direct regulon in Pseudomonas fluorescens F113 and Pseudomonas putida KT2440. Sci Rep 8:13145. 10.1038/s41598-018-31371-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klose KE, Mekalanos JJ. 1998. Distinct roles of an alternative sigma factor during both free-swimming and colonizing phases of the Vibrio cholerae pathogenic cycle. Mol Microbiol 28:501–520. 10.1046/j.1365-2958.1998.00809.x. [DOI] [PubMed] [Google Scholar]
- 45.Kim YK, McCarter LL. 2000. Analysis of the polar flagellar gene system of Vibrio parahaemolyticus. J Bacteriol 182:3693–3704. 10.1128/JB.182.13.3693-3704.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spohn G, Scarlato V. 1999. Motility of Helicobacter pylori is coordinately regulated by the transcriptional activator FlgR, an NtrC homolog. J Bacteriol 181:593–599. 10.1128/JB.181.2.593-599.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vallet I, Olson JW, Lory S, Lazdunski A, Filloux A. 2001. The chaperone/usher pathways of Pseudomonas aeruginosa: identification of fimbrial gene clusters (cup) and their involvement in biofilm formation. Proc Natl Acad Sci USA 98:6911–6916. 10.1073/pnas.111551898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O'Toole GA, Kolter R. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol 30:295–304. 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 49.O'Toole GA, Kolter R. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol 28:449–461. 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 50.Choi KH, Kumar A, Schweizer HP. 2006. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J Microbiol Methods 64:391–397. 10.1016/j.mimet.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 51.Hmelo LR, Borlee BR, Almblad H, Love ME, Randall TE, Tseng BS, Lin C, Irie Y, Storek KM, Yang JJ, Siehnel RJ, Howell PL, Singh PK, Tolker-Nielsen T, Parsek MR, Schweizer HP, Harrison JJ. 2015. Precision-engineering the Pseudomonas aeruginosa genome with two-step allelic exchange. Nat Protoc 10:1820–1841. 10.1038/nprot.2015.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pryor KD, Leiting B. 1997. High-level expression of soluble protein in Escherichia coli using a His6-tag and maltose-binding-protein double-affinity fusion system. Protein Expr Purif 10:309–319. 10.1006/prep.1997.0759. [DOI] [PubMed] [Google Scholar]
- 53.Zheng R, Feng X, Wei X, Pan X, Liu C, Song R, Jin Y, Bai F, Jin S, Wu W, Cheng Z. 2018. PutA is required for virulence and regulated by PruR in Pseudomonas aeruginosa. Front Microbiol 9:548. 10.3389/fmicb.2018.00548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fan Z, Xu C, Pan X, Dong Y, Ren H, Jin Y, Bai F, Cheng Z, Jin S, Wu W. 2019. Mechanisms of RsaL mediated tolerance to ciprofloxacin and carbenicillin in Pseudomonas aeruginosa. Curr Genet 65:213–222. 10.1007/s00294-018-0863-3. [DOI] [PubMed] [Google Scholar]
- 55.Baraquet C, Murakami K, Parsek MR, Harwood CS. 2012. The FleQ protein from Pseudomonas aeruginosa functions as both a repressor and an activator to control gene expression from the pel operon promoter in response to c-di-GMP. Nucleic Acids Res 40:7207–7218. 10.1093/nar/gks384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilson DO, Johnson P, McCord BR. 2001. Nonradiochemical DNase I footprinting by capillary electrophoresis. Electrophoresis 22:1979–1986. . [DOI] [PubMed] [Google Scholar]
- 57.Weng Y, Chen F, Liu Y, Zhao Q, Chen R, Pan X, Liu C, Cheng Z, Jin S, Jin Y, Wu W. 2016. Pseudomonas aeruginosa enolase influences bacterial tolerance to oxidative stresses and virulence. Front Microbiol 7:1999. 10.3389/fmicb.2016.01999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jin Y, Zhang M, Zhu F, Peng Q, Weng Y, Zhao Q, Liu C, Bai F, Cheng Z, Jin S, Wu W. 2019. NrtR regulates the type III secretion system through cAMP/Vfr pathway in Pseudomonas aeruginosa. Front Microbiol 10:85. 10.3389/fmicb.2019.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schweizer HP. 1991. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene 97:109–121. 10.1016/0378-1119(91)90016-5. [DOI] [PubMed] [Google Scholar]
- 60.Totten PA, Lory S. 1990. Characterization of the type a flagellin gene from Pseudomonas aeruginosa PAK. J Bacteriol 172:7188–7199. 10.1128/jb.172.12.7188-7199.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Sets S1 to S4. Download JB.00145-21-s0001.xlsx, XLSX file, 0.1 MB (103.9KB, xlsx)
Fig. S1 to S5. Download JB.00145-21-s0002.pdf, PDF file, 0.4 MB (433.8KB, pdf)