ABSTRACT
Swimming motility is a critical virulence factor in pathogenesis for numerous Vibrio species. Vibrio campbellii DS40M4 is a wild-type isolate that has been recently established as a highly tractable model strain for bacterial genetics studies. We sought to exploit the tractability and relevance of this strain for characterization of flagellar gene regulation in V. campbellii. Using comparative genomics, we identified homologs of V. campbellii flagellar and chemotaxis genes conserved in other members of the Vibrionaceae and determined the transcriptional profile of these loci using differential RNA-seq. We systematically deleted all 63 predicted flagellar and chemotaxis genes in V. campbellii and examined their effects on motility and flagellum production. We specifically focused on the core regulators of the flagellar hierarchy established in other vibrios: RpoN (σ54), FlrA, FlrC, and FliA. Our results show that V. campbellii transcription of flagellar and chemotaxis genes is governed by a multitiered regulatory hierarchy similar to other motile Vibrio species. However, there are several critical differences in V. campbellii: (i) the σ54-dependent regulator FlrA is dispensable for motility; (ii) the flgA, fliEFGHIJ, flrA, and flrBC operons do not require σ54 for expression; and (iii) FlrA and FlrC coregulate class II genes. Our model proposes that the V. campbellii flagellar transcriptional hierarchy has three classes of genes, in contrast to the four-class hierarchy in Vibrio cholerae. Our genetic and phenotypic dissection of the V. campbellii flagellar regulatory network highlights the differences that have evolved in flagellar regulation across the Vibrionaceae.
IMPORTANCEVibrio campbellii is a Gram-negative bacterium that is free-living and ubiquitous in marine environments and is an important global pathogen of fish and shellfish. Disruption of the flagellar motor significantly decreases host mortality of V. campbellii, suggesting that motility is a key factor in pathogenesis. Using this model organism, we identified >60 genes that encode proteins with predicted structural, mechanical, or regulatory roles in function of the single polar flagellum in V. campbellii. We systematically tested strains containing single deletions of each gene to determine the impact on motility and flagellum production. Our studies have uncovered differences in the regulatory network and function of several genes in V. campbellii compared to established systems in Vibrio cholerae and Vibrio parahaemolyticus.
KEYWORDS: motility, flagellar gene regulation, Vibrio, Vibrio campbellii, flagellar motility
INTRODUCTION
Swimming motility is an important behavior that is common among aquatic bacteria. By using a rotating molecular nanomachine called a flagellum, bacteria can propel themselves through fluid environments as a resource-scavenging strategy (1, 2). For many bacteria swimming motility is also critical for host interactions. Vibrio species are a highly motile group of marine bacteria that closely associate with a diverse range of marine hosts (3–6). Some vibrios form symbiotic pairings, such the mutualism between Vibrio fischeri and the Hawaiian bobtail squid, Euprymna scolopes (3, 7). Other vibrios are pathogenic and can infect humans (e.g., Vibrio cholerae and Vibrio parahaemolyticus) and/or are pathogens of fish and shellfish (e.g., Vibrio harveyi and Vibrio campbellii) (8–11). Flagellum-mediated motility plays a critical role for each of these interactions between Vibrio species and their hosts; both colonization and lethality of these strains in their respective hosts decreases when motility is inhibited (4, 12, 13). For this reason, vibrios have served as key models for study of flagellar regulation and its role in colonization.
Of the various Vibrio species, V. cholerae has been the most studied Vibrio species regarding flagellar regulation and has thus served as the predominant model for polar flagellar regulation in other vibrios (14–17). In V. cholerae, flagellar regulation is organized stepwise into a four-tiered transcriptional hierarchy, whereby expression of downstream flagellar components is activated after expression of upstream components (16, 17). Under this model, the sole class I gene encodes the σ54-dependent transcriptional activator FlrA. Once FlrA is expressed, class II gene expression is activated, which includes several structural components of the flagellum (flagellar export machinery, early basal body genes, and ATPase), some chemotaxis genes, and the flrBC and fliA regulatory genes whose products regulate expression of classes III and IV, respectively (16, 18, 19). Class III is activated by FlrC, another σ54-dependent transcriptional regulator that belongs to a two-component system, along with its cognate histidine kinase FlrB (19, 20). Class III genes include those for intermediate structural components, such as the distal and proximal rod, hook proteins, and the essential flagellin FlaA (16). Once the flagellum has reached a certain length after class III expression, the anti-sigma factor FlgM is exported from the cell, leading to activation of FliA (the alternative sigma factor σ28) that activates class IV genes (21). These include those for late structural components such as the flagellar stator, four alternative flagellins, and the T ring, as well as other chemotaxis proteins, including the methyl-accepting chemoreceptors. The synthesis of a functional flagellum in V. cholerae depends on this stepwise regulatory hierarchy, and deletion of any of the four regulators disrupts the subsequent steps and results in complete loss of motility (19).
The regulatory network driving flagellar motility has also been studied in other Vibrio species, including V. fischeri, V. parahaemolyticus, and Vibrio vulnificus (22). The polar flagellar genes are highly conserved between Vibrio species and are organized into similar genetic clusters in each genome (16, 22, 23). Despite this, flagellar phenotypes differ among each species. While most Vibrios produce a sodium-driven monotrichous polar sheathed flagellum, some species, such as V. fischeri, exhibit peritrichous polar sheathed flagella (24, 25). V. parahaemolyticus and Vibrio alginolyticus encode a secondary set of flagellar genes that allow them to produce proton-driven peritrichous lateral unsheathed flagella in addition to their polar flagellum, enabling them to undergo swarming motility over surfaces (26, 27). These differences in motile phenotypes suggest that there is much diversity in the number, type, and regulation of flagella among Vibrio species.
V. campbellii is a marine bacterium belonging to the Harveyi clade that shares similar free-living and pathogenic life cycles with other members of the Vibrionaceae (28–30). V. campbellii has served as an important organism for the study of pathogenesis because it infects many different organisms critical to aquaculture, including shrimp, oysters, and fish (8–10). V. campbellii uses a monotrichous sheathed polar flagellum to propel itself. As with other vibrios, swimming motility plays an important role in pathogenesis for V. campbellii, and inhibition of motility significantly decreases host mortality during infection (13). However, while V. campbellii encodes flagellar and chemotaxis genes conserved by other Vibrio species, the expression and function of these genes have not been characterized. In addition, the V. campbellii genome encodes genes with homology to V. parahaemolyticus lateral flagellar genes for swarming motility. However, only a few strains appear to express these genes with little phenotypic or genetic characterization (31, 32). Thus, there are several gaps in our understanding of V. campbellii flagellum-based motility.
Recently, we established the wild isolate V. campbellii DS40M4 as a new model strain for V. campbellii research because it is capable of natural transformation, thus facilitating powerful genetic manipulations such as MuGENT (33–35). The genetic tractability of DS40M4 enabled us to use it as a model system to study flagellar motility in V. campbellii. We systematically deleted all 63 predicted polar flagellar and chemotaxis genes in V. campbellii and characterized the swimming phenotypes of each mutant. We used RNA-seq to identify the genes controlled by canonical flagellar regulators in V. campbellii. With these data, we propose a model for the transcriptional regulation program that governs expression of polar flagellar genes in V. campbellii.
RESULTS
Identification of the flagellar gene operons in V. campbellii.
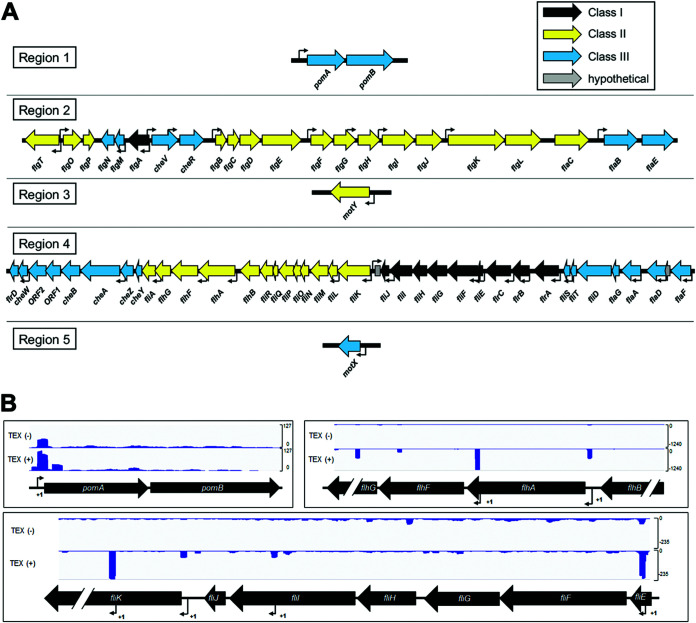
Using reciprocal BLASTp analyses based on known flagellar homologs characterized in other vibrios, we identified 63 distinct polar flagellar genes organized across five regions within chromosome I of V. campbellii DS40M4 (Fig. 1A), as well as 41 distinct lateral flagellar genes organized across two regions within chromosome II (see Fig. S1 in the supplemental material) (22, 26). In our previous work, we conducted differential RNA-seq (dRNA-seq) to identify transcription start sites across the entire DS40M4 genome (34). We used the dRNA-seq data to identify transcriptional start sites in these five loci, allowing us to predict corresponding operons for each flagellar gene (Fig. 1A). Examples of the dRNA-seq transcript data are shown in Fig. 1B for the fliEFGHIJ, pomAB, and flhAFG genes (Fig. 1B). Thus, Fig. 1 presents a diagram showing the predicted organization of the core polar flagellar and chemotaxis genes in V. campbellii and respective transcriptional start sites for each.
FIG 1.
Organization of the polar flagellar gene system in V. campbellii DS40M4. (A) Diagram showing the five regions of polar flagellar genes in chromosome I (GenBank accession number CP030788.1) in V. campbellii. Transcriptional start sites (TSS) are mapped to each region (bent arrows) based on the dRNA-seq results. Flagellar genes are color coded based on their classification in the transcriptional hierarchy via our RNA-seq data. (B) Data shown are cDNA reads from dRNA-seq data mapped to three genetic loci in V. campbellii DS40M4 for samples untreated [TEX(–)] or treated [TEX(+)] with terminator 5′-phosphate-dependent endonuclease. Bent arrows labeled “+1” indicate predicted TSS for flagellar genes.
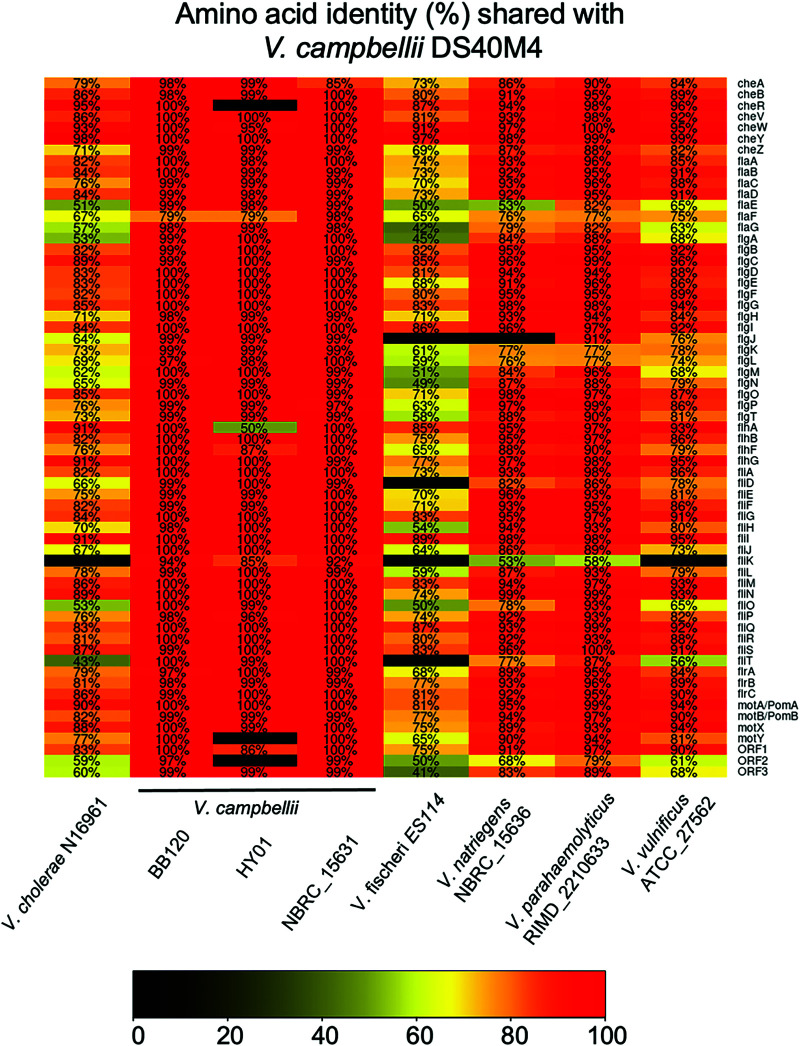
We next used comparative genomics to investigate the conservation of each of the known polar flagellar genes between V. campbellii and other known polar flagellated Vibrio species (Fig. 2). We used a minimum amino acid percent identity requirement of 40% (the lowest possible value for clustering allowed by cd-hit) in this analysis to increase the number of functional homologs identified in our results. For the flagellins that are highly similar to each other, we also independently examined their similarity to flagellins in other Vibrio species using BLASTp. Our genomic data suggest that the genome of V. campbellii encodes homologs that match the complete set of known polar flagellar genes established in Vibrio species thus far (Fig. 2). These genes are highly conserved in the V. campbellii clade. The flagellar gene system in V. campbellii shares amino acid identity most closely with V. parahaemolyticus, whereas most polar flagellar genes in V. campbellii are more divergent compared to other members of the Vibrionaceae, such as V. cholerae or V. fischeri.
FIG 2.
Polar flagellar gene homology across Vibrionaceae. Comparative genomics show the amino acid identity of known V. cholerae polar flagellar genes. The chart indicates the amino acid identity shared with V. campbellii DS40M4 proteins, as indicated by the color scale.
Deletion and characterization of all known flagellar/chemotaxis genes in V. campbellii.
Few studies have characterized the complete set of flagellar genes in a Vibrio species, likely owing to the time and labor associated with genetic mutation of dozens of genes (22, 23). Recently, we showed that genetic manipulation of V. campbellii DS40M4 is highly efficient due to inducible natural transformation via overexpression of the master competence regulator Tfox (33, 34). To automate primer design for constructing deletion mutants in V. campbellii, we programmed an algorithm to generate a list of primers for deletion of every annotated open reading frame (ORF) in the DS40M4 genome. Candidate primers were designed so that overlapping genes were not disrupted and followed a specific set of criteria for validity, including preference for a shorter length (25 to 30 nucleotides), no homopolymers of five or more bases, a melting temperature of 58 to 63°C, and the 3′ end of each primer ends in a cytosine or guanine. We used this algorithm to design primers to construct unmarked deletions of each predicted flagellar or chemotaxis gene in V. campbellii.
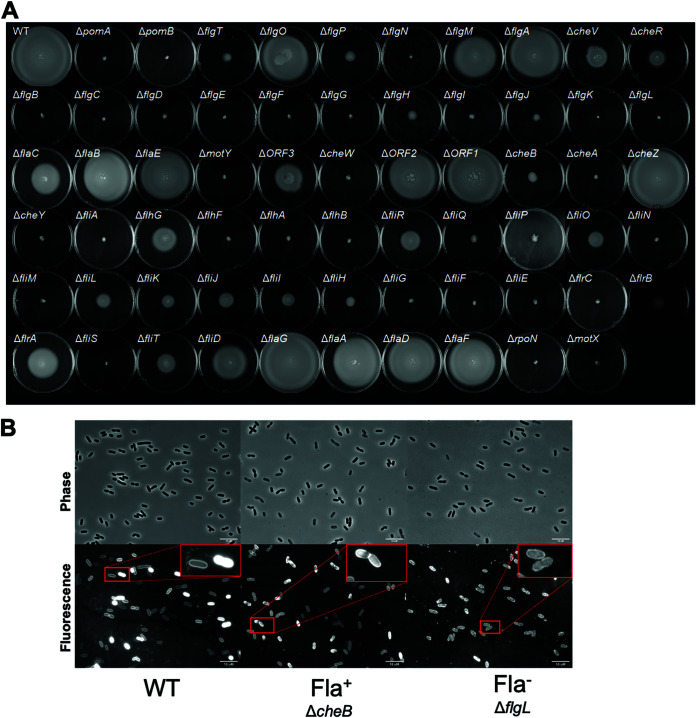
Altogether, we constructed 64 mutants, including each of the 63 flagellar and chemotaxis genes spread across the five flagellar regions in chromosome I of DS40M4, as well as rpoN, which encodes σ54. We classified mutants into groups based on their capacity to swim via soft agar swim plates (Fig. 3A) and the presence or absence of flagella observed via microscopy using the fluorescence stain NanoOrange, a reagent which is virtually nonfluorescent in aqueous solution that fluoresces brightly when it binds hydrophobic regions of proteins and lipids, allowing visualization of cell bodies and flagella (Fig. 3B) (36, 37). Mutants that deviated from wild-type were classified into three phenotypic classes: (i) nonmotile (Mot–) mutants are inhibited for motility on swim plates but produce a detectable flagellum; (ii) aflagellate (Fla–) mutants are inhibited for motility on swim plates and produce no detectable flagellum; and (iii) semimotile (Mot+/–) mutants demonstrate reduced motility (smaller swim halos) on swim plates (Table 1). Of these 64 mutants, 34 are still capable of producing a swim halo (ranging from significantly reduced to wild-type levels), whereas 30 are completely inhibited for swimming motility on swim plates (Fig. 3A). The reduced motility phenotypes in some of these mutants may be explained by a chemotaxis defect such as in che genes (i.e., cheABRWVYZ), which were not studied further within the scope of this work. Overall, we identified a total of 7 Mot– mutants, 25 Fla– mutants, and 20 Mot+/– mutants, with 12 mutants that have phenotypes indistinguishable from the wild type.
FIG 3.
Swim phenotypes of DS40M4 polar flagellar mutants. (A) Soft agar (0.3%) swim plates showing swim halo phenotypes for wild-type and mutant strains of DS40M4. (B) Fluorescence microscopy images of wild-type, ΔcheB (Fla+), and ΔflgL (Fla–) strains using the fluorescent protein stain NanoOrange to observe the presence of flagella.
TABLE 1.
Phenotypic classification of V. campbellii DS40M4 strains containing deletions of predicted flagellar or chemotaxis genes
| Gene | Predicted function | Locus tag | Swim plates | Flagella | Mutant class |
|---|---|---|---|---|---|
| pomA | Flagellar motor protein PomA | DSB67_03225 | – | + | Mot– |
| pomB | Flagellar motor protein PomB | DSB67_03230 | – | + | Mot– |
| flgT | Hypothetical protein (flagellum basal-body protein) | DSB67_03720 | + | + | Swm– |
| flgO | Membrane protein | DSB67_03725 | +++ | + | WT |
| flgP | Flagellar assembly lipoprotein FlgP | DSB67_03730 | + | + | Mot+/– |
| flgN | Molecular chaperone (for FlgK and FlgL) | DSB67_03735 | – | – | Fla– |
| flgM | Flagellar biosynthesis anti-sigma factor FlgM | DSB67_03740 | +++ | + | WT |
| flgA | Flagella basal body P-ring formation protein FlgA | DSB67_03745 | +++ | + | WT |
| cheV | Chemotaxis protein CheV | DSB67_03750 | + | + | Mot+/– |
| cheR | Protein-glutamate O-methyltransferase | DSB67_03755 | + | + | Mot+/– |
| flgB | Flagellar basal body rod protein FlgB | DSB67_03760 | – | – | Fla– |
| flgC | Flagellar basal body rod protein FlgC | DSB67_03765 | – | – | Fla– |
| flgD | Flagellar hook assembly protein FlgD | DSB67_03770 | – | – | Fla– |
| flgE | Flagellar hook protein FlgE | DSB67_03775 | – | – | Fla– |
| flgF | Flagellar basal body rod protein FlgF | DSB67_03780 | – | – | Fla– |
| flgG | Flagellar basal-body rod protein FlgG | DSB67_03785 | – | – | Fla– |
| flgH | Flagellar basal body L-ring protein FlgH | DSB67_03790 | + (fuzzy) | + | Mot+/– |
| flgI | Flagellar basal body P-ring protein FlgI | DSB67_03795 | + (fuzzy) | + | Mot+/– |
| flgJ | Flagellar assembly peptidoglycan hydrolase FlgJ | DSB67_03800 | + (fuzzy) | + | Mot+/– |
| flgK | Flagellar hook-associated protein FlgK | DSB67_03805 | – | – | Fla– |
| flgL | Flagellar hook-associated protein FlgL | DSB67_03810 | – | – | Fla– |
| flaC | Flagellin | DSB67_03815 | + | + | Mot+/– |
| flaB | Flagellin | DSB67_03820 | +++ | + | WT |
| flaE | Flagellin | DSB67_03825 | +++ | + | WT |
| motY | Motor protein (OmpA family protein) | DSB67_10725 | – | + | Mot- |
| flrD | DUF2802 domain-containing protein | DSB67_11255 | + | + | Swm– |
| cheW | Chemotaxis protein CheW | DSB67_11260 | – | + | Mot– |
| ORF2 | Chemotaxis protein CheW | DSB67_11265 | +++ | + | WT |
| ORF1 | ParA family protein | DSB67_11270 | +++ | + | WT |
| cheB | Chemotaxis response regulator protein-glutamate methylesterase | DSB67_11275 | + | + | Mot+/– |
| cheA | Chemotaxis protein CheA | DSB67_11280 | + (fuzzy) | + | Mot+/– |
| cheZ | Protein phosphatase | DSB67_11285 | +++ | + | WT |
| cheY | Chemotaxis protein CheY | DSB67_11290 | – | + | Mot- |
| fliA | RNA polymerase sigma factor FliA | DSB67_11295 | – | – | Mot- |
| flhG | MinD/ParA family protein | DSB67_11300 | + | + | Mot+/– |
| flhF | Flagellar biosynthesis regulator FlhF | DSB67_11305 | + (fuzzy) | + | Swm– |
| flhA | Flagellar biosynthesis protein FlhA | DSB67_11310 | – | – | Fla– |
| flhB | Flagellar biosynthesis protein FlhB | DSB67_11315 | – | – | Fla– |
| fliR | Flagellar type III secretion system protein FliR | DSB67_11320 | + | + | Mot+/– |
| fliQ | Flagellar biosynthetic protein FliQ | DSB67_11325 | + (fuzzy) | + | Mot+/– |
| fliP | Flagellar biosynthetic protein FliP | DSB67_11330 | + (fuzzy) | + | Mot+/– |
| fliO | Flagellar biosynthetic protein FliO | DSB67_11335 | + | + | Swm– |
| fliN | Flagellar motor switch protein FliN | DSB67_11340 | – | – | Fla– |
| fliM | Flagellar motor switch protein FliM | DSB67_11345 | – | – | Fla– |
| fliL | Flagellar basal body-associated protein FliL | DSB67_11350 | + | + | Mot+/– |
| fliK | Flagellar hook-length control protein FliK | DSB67_11355 | + | + | Mot+/– |
| fliJ | Flagellar biosynthesis chaperone FliJ | DSB67_11365 | + | + | Swm– |
| fliI | Flagellum-specific ATP synthase FliI | DSB67_11370 | + | + | Swm– |
| fliH | Flagellar assembly protein FliH | DSB67_11375 | + | + | Swm– |
| fliG | Flagellar motor switch protein FliG | DSB67_11380 | – | – | Fla– |
| fliF | Flagellar basal body M-ring protein FliF | DSB67_11385 | – | – | Fla– |
| fliE | Flagellar hook-basal body complex protein FliE | DSB67_11390 | – | – | Fla– |
| flrC | σ54-dependent Fis family transcriptional regulator | DSB67_11395 | – | – | Fla– |
| flrB/flaL | PAS domain-containing protein | DSB67_11400 | – | – | Fla– |
| flrA | σ54-dependent Fis family transcriptional regulator | DSB67_11405 | + | + | Mot+/– |
| fliS | Flagellar export chaperone FliS | DSB67_11410 | – | – | Fla– |
| fliT | Flagellar protein FliT | DSB67_11415 | + | + | Mot+/– |
| fliD | Flagellar cap protein FliD | DSB67_11420 | + | + | Mot+/– |
| flaG | Flagellar protein flag | DSB67_11425 | +++ | + | WT |
| flaA | Flagellin | DSB67_11430 | +++ | + | WT |
| flaD | Flagellin | DSB67_11435 | +++ | + | WT |
| flaF | Flagellin | DSB67_11445 | +++ | + | WT |
| rpoN | RNA polymerase σ54 factor | DSB67_13575 | – | – | Fla– |
| motX | Motor protein (Sel1 repeat family protein) | DSB67_14290 | – | + | Mot– |
Among the 7 Mot– mutants from our pool are the four motor/stator genes pomAB, motX, and motY, which are crucial for flagellar rotation but not assembly, as well as fliA (σ28; class IV regulator) and cheYW (chemotaxis) (38, 39). The 25 Fla– mutants included flgN (hook chaperone), flgBCF (proximal rod), flgDE (hook), flgG (distal rod), flgH (L ring), flgI (P ring), flgJ (rod cap), flgKL (hook), flhBA (exporter apparatus), fliEF (MS ring), fliGMN (C ring), fliPQ (exporter apparatus), fliS (flagellin chaperone), flrBC (class III regulators), and σ54. Among the mutants in our pool of 20 Mot+/– mutants, we identified flgT and flgP (H ring), cheVR (chemotaxis), flaC (flagellin), flrD (unknown), cheAB (chemotaxis), flhFG (flagellar number/localization), fliHIJ (ATPase), fliK (hook length), fliL (torque generator), fliOR (export apparatus), flrA (class II regulator), fliT (chaperone), and fliD (filament cap). Finally, we found little to no change in motility in flgM (anti-σ28), flgO (H ring), flgA, flaABDEF (flagellins), cheZ (chemotaxis), and ORF1/ORF2 (unknown/chemotaxis) mutants compared to the wild-type strain.
Most mutants within our pool of 64 were sorted into phenotypic classes that agree with previous observations in other vibrios or flagellate bacterial species. However, some discrepancies were found compared to these findings. Notable examples include mutants in the flagellar hook length control gene (fliK), as well as the flagellar export apparatus (fliOR) and the filament cap (fliD and fliT), both of which are required for motility in other bacteria such as Salmonella enterica but resulted in Mot+/– phenotypes for our mutants (40–45). Although soft agar plates are often used to assess swimming motility, the interpretation of these can be misleading because the observed phenotypes are the result of a combination of factors, including but not limited to flagellar synthesis, chemotactic sensing, growth rates, biofilm formation, and sensing of metabolites. Thus, in addition to using swim plates we used phase-contrast microscopy to directly observe swimming in all semimotile mutants. We distinguished semimotile mutants by creating a fourth phenotypic class that we designated nonswimmers (Swm–), which produce swim halos on soft agar plates but show no detectable swimming cells when observed by live cell microscopy. While we observed swimming in many of the Mot+/– mutants, we were surprised to identify several mutants that were semimotile on soft agar plates appear nonmotile by microscopy, including flgT (H ring), flrD (regulatory protein), flhF (flagellar placement), fliO (export apparatus), and fliHIJ (ATPase) mutants. These results corroborate previous observations of flagellar mutants in other bacteria, in which deletion of these genes results in nonmotile and/or aflagellate cells (40, 46–48). This suggests an important difference between the behaviors observed on swim plates versus microscope that may be explained by defects in flagellar assembly, structure, or function.
Effects of core flagellar transcriptional regulators on swimming motility and flagellum synthesis.
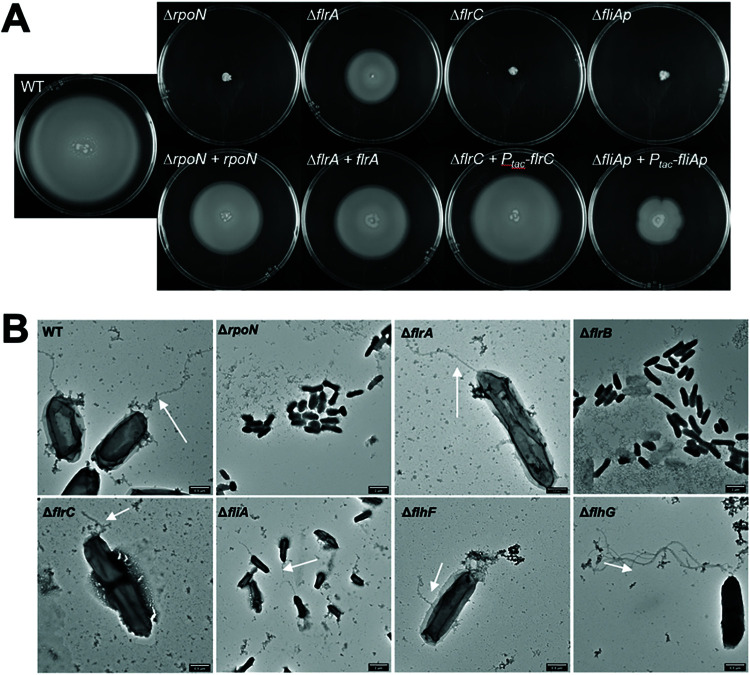
In vibrios with a defined polar flagellar gene system, including V. cholerae, V. fischeri, V. vulnificus, V. alginolyticus, and V. parahaemolyticus, previous studies have shown that the flagellum is regulated by a four-tiered transcriptional hierarchy that includes σ54, FlrA (or the homolog FlaK), FlrC (or the homolog FlaM), and FliA (σ28). Deletion of any one of these transcriptional regulators results in complete loss of motility in most vibrios, with the exception of V. parahaemolyticus that also produces a compensating regulator of lateral flagella, LafK (18, 19, 49–52). To determine whether the V. campbellii flagellar genes are regulated in a similar transcriptional hierarchy, we examined the four predicted regulators: σ54, FlrA (annotated as FlaK), FlrC (annotated as FlaM), and FliA. While the annotation of V. campbellii DS40M4 shares the same naming scheme for FlaKLM with V. parahaemolyticus, we refer to them as FlrABC, respectively, henceforth for consistency with the naming in other vibrios and due to the meaning behind the prefix Flr (flagellar regulatory protein) (49). We constructed isogenic strains with a single deletion of each of the flagellar transcriptional regulator genes in DS40M4 and tested for the presence or absence of motility in each mutant on soft agar plates (Fig. 3A). Similar to other vibrios, deletion of rpoN (that encodes σ54), flrC, and fliA results in the complete abrogation of motility on swim plates (Fig. 4A). However, the ΔflrA mutant demonstrates swimming motility, albeit partially reduced compared to the wild-type strain. We complemented each mutant at an ectopic locus in the genome. For rpoN and flrA, our transcriptional start site (TSS) analyses indicated that these genes are likely the first gene in the operon; thus, we complemented these using their cognate promoter. However, for flrC and fliA, it was unclear from our dRNA-seq data which promoter(s) controls expression of these genes, so we complemented flrC and fliA using an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible promoter. In each case, complementation results in increased levels of motility compared to the deletion strain (Fig. 4A).
FIG 4.
Swimming and flagellar phenotypes of V. campbellii flagellar regulator mutants. (A) Soft agar (0.3%) swim plates showing swim halo phenotypes for wild-type and mutant DS40M4 strains. The top panel shows deletion mutants. The bottom panel shows deletion mutants that were complemented at an ectopic locus (luxB). (B) TEM of wild-type and mutant DS40M4 strains grown in LM liquid culture. Arrows point to flagellar protrusions in strains.
Because our results showed that DS40M4 FlrA is dispensable for motility, we sought to test the role of FlrA in swimming motility in another Vibrio campbellii strain: BB120 (also known as ATCC BAA-1116, previously classified as Vibrio harveyi) (28). We constructed deletions of flrA, flrC, and fliA in BB120, and we observed the same results as in DS40M4. The ΔflrC and ΔfliA strains are nonmotile, whereas the ΔflrA strain retains motility that is slightly diminished compared to wild-type BB120 (see Fig. S2). From these data, we conclude that FlrA is dispensable for swimming motility in V. campbellii DS40M4 and BB120.
We also directly observed the presence or absence of flagella via transmission electron microscopy (TEM) to compare the structural effects of deletion of flagellar regulators (Fig. 4B). We chose to assess deletion strains for each of the four flagellar transcriptional regulators (ΔrpoN, ΔflrA, ΔflrC, and ΔfliA), as well as ΔflrB, ΔflhF, and ΔflhG strains, which act within the four-tiered transcriptional hierarchy and have observable flagellar patterns in other vibrios (46, 53, 54). When rpoN and flrB are deleted, no flagella are detectable via TEM. The ΔflrC cells are also largely aflagellate; however, some observable short cable structures did protrude from some of the cells (Fig. 4B). When flhF is deleted, the cells were largely aflagellate except for a few that produce short and/or nonpolar cable-like structures that appear to be nonlocalized flagella. Conversely, ΔflhG mutants demonstrate hyperflagellation. These findings correlate with prior studies in Vibrio species indicating FlhF’s role in flagellar localization and FlhG’s role in regulating flagellar number (46, 53). Deletion of either flrA or fliA had no observable impact on detectable flagella via TEM (Fig. 4B). In the ΔfliA mutant this may be explained by the role of FliA in primarily regulating chemotaxis and motor genes rather than structural genes, though FliA is typically required for expression of alternate flagellins in vibrios (19). However, it is notable that the ΔflrA mutant produces intact flagella, conflicting with the models in other Vibrio species in which it is one of the earliest required transcriptional regulators for expression of flagellar genes (18, 19). Altogether, these results largely support previous findings in other vibrios with one major exception: we conclude that FlrA is not required for flagellar production or motility in V. campbellii.
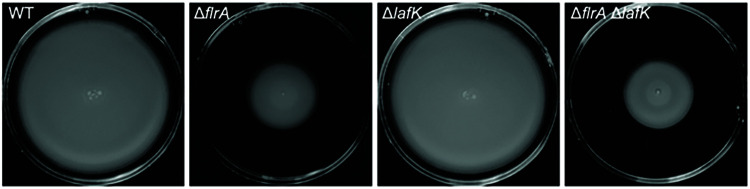
The lateral flagellar gene lafK does not compensate for the absence of flrA.
In addition to encoding the polar flagellar gene system, V. campbellii also encodes a set of genes homologous to known V. parahaemolyticus lateral flagellar genes (26). The lateral flagellar gene system is a separate set of similar but distinct genes which produce numerous, nonsheathed lateral flagella and which are encoded in chromosome II rather than I. Historically, lateral flagella have been characterized in V. parahaemolyticus and V. alginolyticus, but other vibrios, such as V. campbellii, have been found to either produce or encode these genes as well (26, 31, 32, 55–57). Importantly, the lateral flagellar σ54-dependent transcriptional regulator LafK cross-regulates class II polar flagellar genes in the absence of the FlrA homolog (FlaK) in V. parahaemolyticus (51). V. campbellii encodes a lafK homolog in its genome. We hypothesized that V. campbellii LafK regulation facilitates the partially motile phenotype in the ΔflrA mutant strain. The V. campbellii ΔlafK mutant demonstrates no change in swimming motility compared to wild-type, mimicking the results in V. parahaemolyticus (Fig. 5). However, unlike V. parahaemolyticus, the ΔlafK ΔflrA mutant shows no further decrease in motility compared to the ΔflrA mutant (Fig. 5). We also tested the role of LafK in V. campbellii BB120 and likewise saw that the ΔlafK ΔflrA mutant does not show a decrease in motility compared to the ΔflrA strain (see Fig. S2). Finally, the dRNA-seq analysis shows that the level of transcription of the lafK locus is very low, much like most of the lateral flagellar genes (34). Thus, we conclude that V. campbellii LafK does not compensate for FlrA.
FIG 5.
LafK does not regulate swimming motility in V. campbellii. Soft agar (0.3%) swim plates show swim halo phenotypes for wild-type and mutant strains of DS40M4.
Due to the lack of observed LafK-mediated polar flagellar regulation, we hypothesized that V. campbellii may not have an active lateral flagellar system for swarming motility. Although V. campbellii encodes lateral flagella, few studies have characterized these flagella or the capacity for swarming motility in V. campbellii (31, 32). We tested to see whether V. campbellii was capable of swarming motility by performing swarm plate assays with various agar concentrations. V. parahaemolyticus is capable of swarming at agar concentrations of 1.5%, while other swarming bacteria such as B. subtilis swarm at agar concentrations between 0.3 and 1.0% (58). Under each condition tested, we were unable to observe swarming in V. campbellii DS40M4 (see Fig. S3). We also assayed for swarming motility in three additional V. campbellii strains but no swarming was observed, whereas we were able to visualize swarming in V. parahaemolyticus as a positive control (see Fig. S3). Finally, all known lateral flagellar are either extremely lowly expressed or are not expressed at all when V. campbellii is grown in liquid media according to both dRNA-seq and RNA-seq data sets (see Data Set S1) (34). Together, our data suggest that despite carrying lateral flagellar genes, V. campbellii DS40M4 has an inactive lateral flagellar system under the conditions we tested. In addition, we conclude that the lateral flagellar regulator LafK does not compensate for FlrA activity in a ΔflrA mutant.
Transcriptome profiling of flagellar regulators.
To identify the V. campbellii genes that are controlled by the transcriptional regulators σ54, FlrA, FlrC, and FliA, we compared the transcription profiles between wild-type and isogenic mutant strains lacking one of these four genes (rpoN, flrA, flrC, and fliA, respectively) using RNA-seq (see Data Set S1). Our goal was to sort genes into different classes in the regulatory hierarchy based on gene expression relative to wild-type similar to previous studies (16). We predicted that genes expressed earlier in the transcriptional hierarchy are regulated by only the earliest flagellar regulator(s) and not any downstream regulators, whereas genes expressed later in the hierarchy will be affected by all upstream regulators.
Figure 6 shows the expression profiles of flagellar genes for ΔrpoN, ΔflrA, ΔflrC, and ΔfliA strains compared to the wild type, as determined by RNA-seq (Fig. 6; differential gene expression data, statistical analyses, and fold changes in transcript levels for each mutant compared to the wild type are presented in Data Set S1). We compared gene expression by examining genes that were differentially regulated 1.5-fold or more (P < 0.05) in the mutant compared to the wild type. These data revealed that much of the downstream hierarchy is organized similarly to the transcriptional hierarchy established in other vibrios (16, 27). Specifically, FliA-dependent expression patterns are very similar to class IV flagellar genes in other vibrios, where we observed that transcription decreases in the absence of FliA and/or upstream regulators for pomAB, flgMN, cheVR, flaBE, flrD, cheW, ORF1, ORF2, cheBAZY, fliS, flaI, fliD, flaG, flaA, flaDF, and motX. Similarly, FlrC-dependent expression patterns are comparable to class III in other vibrios as well, since we identified several genes clusters that were decreased in expression in the absence of FlrC: flgT, flgOP, flgBCDE, flgFGHIJ, flgKL, flaC, fliKLMNOPQR, flhB, and motY.
FIG 6.
Expression patterns of class I to III polar flagellar genes. A heat map shows the expression levels of polar flagellar genes (the color key indicates the log2 fold change) determined by RNA-seq analysis in the wild type (DS40M4) compared to ΔrpoN (BDP029), ΔflrA (BDP030), ΔflrC (BDP031), and ΔfliAp (BDP046) strains. Flagellar gene names and locus tags are indicated.
The expression patterns for FlrA showed the largest deviation from studies of other vibrios. In V. cholerae, FlrA is required for regulation of several flagellar operons, including fliEFGHIJ, flhAFG, flrBC, and fliA (16, 19). Deletion of flrA also results in decreased expression of downstream genes controlled by either FlrC or FliA, such as hook, rod, chemotaxis, and flagellin genes. Conversely, our data show that FlrA is not required for expression of fliEFGHIJ or flrBC, which are expressed at levels similar to wild type in the absence of either FlrA or σ54 (Fig. 6; see also Data Set S1). The only unique set of genes solely affected by the ΔflrA mutation are flhAFG. This result correlates with the swim plate results in which flrA is not required for swimming motility, but deletion of flrA reduces swim halo size. Notably, however, fliK, fliLMNOPQR, and flhB expression patterns are decreased in both a ΔflrA mutant and a ΔflrC mutant, which suggests that both FlrA and FlrC regulate these genes. From these data, we conclude that FlrA-dependent regulatory patterns in V. campbellii DS40M4 is distinct from that of FlrA in V. cholerae. In addition, transcription of genes—fliEFGHIJ, flrA, and flrBC—classified as class II in V. cholerae is independent of the four regulators studied here in V. campbellii.
FlrA and FlrC coregulate class II genes.
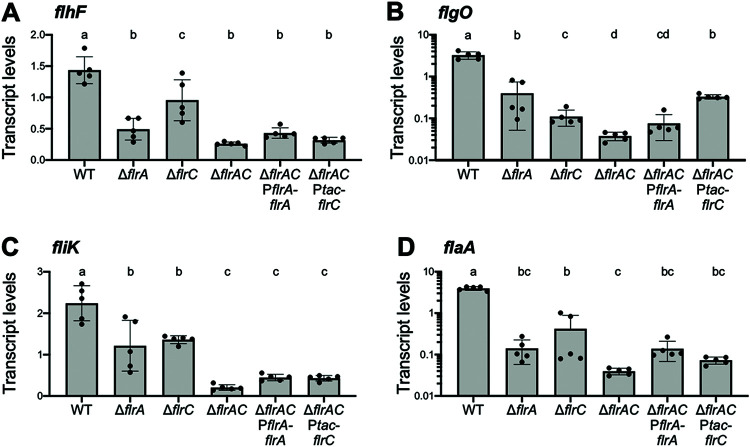
The results of our RNA-seq analysis indicate that some flagellar genes are decreased in expression in both the ΔflrA and ΔflrC backgrounds compared to wild type. FlrA and FlrC are both bacterial enhancer binding proteins (bEBPs) that share 39.79% identity and serve as σ54-dependent transcriptional regulators (49, 59, 60). Thus, we hypothesized that FlrA shares a coregulon with FlrC. To test this, we performed qPCR on wild-type, ΔflrA, ΔflrC, and ΔflrA ΔflrC strains. In addition, we complemented flrA and flrC in the double ΔflrA ΔflrC mutant background. We chose to examine genes from each class based on our RNA-seq data: flhF appears to be regulated by FlrA, flgO appears to be regulated by FlrC, and fliK appears to be coregulated by FlrA and FlrC. As a control, we assessed gene expression of flaA, a gene controlled by FliA late in the hierarchy, which is thus affected by all upstream regulators. Similar to our RNA-seq results, we found that flhF is more strongly decreased in expression in the ΔflrA mutant, whereas flgO is more strongly decreased in the ΔflrC mutant compared to the wild-type strain (Fig. 7A and B). However, deletion of either flrA or flrC has a significant effect on both genes compared to wild type. In addition, deletion of either flrA or flrC partially decreases the expression of fliK compared to the wild type, whereas the deletion of both flrA and flrC together further decreases fliK expression compared to the single mutants (Fig. 7C). Complementation of flrA or flrC in the ΔflrA ΔflrC background restores flgO levels to the single-mutant level (Fig. 7B), whereas complementation did not affect flhF or fliK levels (Fig. 7A and B). As predicted, we also find that deletion of either flrA or flrC significantly impairs flaA expression (Fig. 7D). We observe additive effects on regulation of fliK and flgO, suggesting that both FlrA and FlrC control expression of the fliK and flgO operons. Although deletion of flrC does significantly decrease flhF expression as measured by qPCR, the deletion of flrA and flrC is not additive, suggesting that FlrA is the dominant regulator (Fig. 7A). From these data, we conclude that FlrA and FlrC have compensatory roles and coregulate the class II genes in the flagellar hierarchy.
FIG 7.
FlrA and FlrC coregulate class II genes. Data shown are absolute transcripts quantified by qPCR of genes from representative flhF (a), flgO (b), fliK (c), and flaA (d) operons regulated by FlrA and FlrC in the wild type and in ΔflrA, ΔflrC, ΔflrA ΔflrC, ΔflrA ΔflrC PflrA-flrA, and ΔflrA ΔflrC Ptac-flrC strains. Different lowercase letters indicate significant differences in ANOVA (on log-transformed data for flgO and flaA), followed by Tukey’s multiple-comparison test (n = 5, P < 0.05). Error bars represent the means and standard deviations from five biological replicates.
DISCUSSION
Swimming motility is employed by many different Vibrio species for taxis toward or away from stimuli. The genetic and regulatory networks underlying flagellar swimming motility have primarily been studied in V. cholerae, V. parahaemolyticus, and V. fischeri by testing individual flagellar mutants or by impairing specific flagellar gene intervals (16, 22, 23, 50). By exploiting the power of natural transformation, we were able to efficiently delete every known gene in the five flagellar and chemotaxis genetic loci in V. campbellii. Our work here is both the first broad examination of the polar flagellar genetic network in V. campbellii and one of the first studies to characterize the complete set of known genes in the flagellar and chemotaxis genomic regions in a Vibrio species using reverse genetics.
Of the 64 genes we examined in this study, the phenotypes of most mutants are consistent with predicted outcomes based on prior studies in other bacterial species. The majority of mutants we characterized are semimotile, as has been observed in previous flagellar systems as well, suggesting that these strains are still capable of synthesizing a partially functional flagellum (22). These strains include those impaired in components of the basal body or the outer flagellar rings (L ring, P ring, and H ring), though notably hook and rod mutants were not observed in this mutant class. We found that flagellar hook and rod mutants were nonmotile, likely because these structures are critical for downstream flagellar assembly steps or flagellar joint functionality, which aligns with previous findings in other bacteria (22, 61). We were surprised to find that flagellar filament cap mutants lacking either fliT or fliD demonstrated detectable motility, though this result does support previous findings in V. parahaemolyticus in which absence of fliD does not completely inhibit motility unlike in S. enterica (62, 63). In addition, mutants lacking the internal ruler for flagellar hook length control, FliK, were semimotile on soft agar plates, which is notably different compared to findings in other bacteria (44, 45, 64, 65). However, previous research has shown that ΔfliK mutants can quickly produce suppressor mutants that restore partial motility compared to wild-type cells, most notably in flhB, a component of the flagellar type III secretion system (66, 67). Because motility provides a strong selective pressure for gain-of-function suppressor mutants, we cannot exclude the possibility that some of the phenotypes that we observed in our flagellar mutants arose from suppressor mutations (68). Most of the nonmotile mutants we identified were also aflagellate, except for the motor mutants, due to the stepwise nature of assembly of the flagellum in bacteria.
Notably, prior studies that characterized presence or absence of a flagellum did so using Western immunoblots to measure the presence of polar flagellin production (22). Rather, in this study we directly observed the presence or absence of a flagellum using fluorescence staining and microscopy. However, this approach excludes cells that continue to produce and secrete downstream flagellar components (such as flagellins), regardless of whether these secreted products correlate with an intact flagellum. Both approaches have their advantages, and this distinction could account for differences we observed between flagellate and aflagellate mutants in this study versus previous work. We also noted that several mutants (ΔfliO, ΔflgT, ΔfliHIJ, ΔflhF, and ΔflrD) produce swim flares at the edges of the inoculum center. These are likely suppressor mutants derived from the strong selection for motility in this assay, and future examination of these mutants may enable a broader understanding of the flagellar system in V. campbellii.
Importantly we found that none of the flagellin genes (flaABCDEF) are absolutely required for motility in V. campbellii. Only deletion of flaC impairs motility compared to the other mutants, which all resemble the wild type. FlaC is most closely related to FlaA in V. cholerae, which is the only filament protein required and sufficient for motility (69). The reduced motility in the V. campbellii ΔflaC mutant correlates with most other vibrios that also encode a predominant flagellin subunit that impairs (but does not eliminate) motility when deleted (62, 70–72).
A few Vibrio species such as V. parahaemolyticus, V. alginolyticus, and V. campbellii also encode lateral flagellar genes in addition to their polar flagellar system, which provide them the capacity for swarming motility over surfaces (26, 31, 32). While characterizing the polar flagellar system in V. campbellii in this study, we also studied the lateral flagella and swarming motility in an attempt to describe this behavior as well. When comparing several different strains of V. campbellii using different hard agar concentrations, we were unable to observe swarming motility in any of these strains. We also did not observe additional flagella beyond the presence of the polar flagellum using TEM of cells prepared from hard agar or soft agar plates in the BB120 strain (Fig. 4B; data not shown). Notably, the first study to identify lateral flagella in V. campbellii (then called Beneckea campbellii) observed that only some strains produced both polar and lateral flagella, while many of the strains only produced a single polar flagellum (32). Taking our observations here and the results found in previous studies of V. campbellii, we believe it is likely that the V. campbellii strains we examined have an inactive lateral flagellar system under our laboratory conditions.
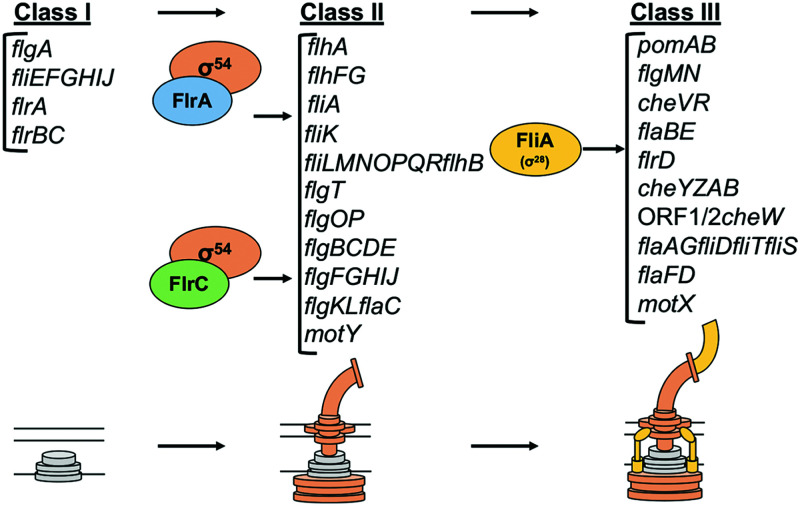
Our transcriptome analysis of the flagellar regulatory mutants in V. campbellii highlighted some unique features of the flagellar regulatory hierarchy, which allowed us to construct a model for the flagellar regulatory hierarchy in V. campbellii (Fig. 8). First, we note that while FlrA plays an observable role in the flagellar regulatory hierarchy, it is also not necessary for V. campbellii cells to be motile on swim plates, which we tested in two strains (DS40M4 and BB120). Genes that were classified as class II in V. cholerae are mostly expressed independently of both σ54 and FlrA in V. campbellii, except for flhAFG and fliA (16). This presents two possibilities for the flagellar regulatory hierarchy in V. campbellii. One possibility is that the four-tiered flagellar hierarchy is actually a three-tiered hierarchy in V. campbellii, where some previously identified class II genes such as fliEFGHIJ and the downstream flagellar regulators like flrB and flrC are expressed constitutively and independently of other regulators like σ54. Taking the data we have presented here into account, without bias from previously defined hierarchies from other Vibrio flagellar species, this is our proposed model for flagellar transcriptional regulation in V. campbellii (Fig. 8). However, we recognize that without a stepwise checkpoint system in place, if multiple class I operons were expressed constitutively in V. campbellii, this could cause a competitive disadvantage because cells would needlessly waste resources in expressing flagellar gene products when swimming motility is not required. Previous reports show that flagellar synthesis is a costly behavior in bacteria, and expression of flagellar genes can impair growth (73–75). Thus, an alternative hypothesis to explain our data are that an unidentified regulator controls the expression of these independently expressed class I genes in V. campbellii. While this model would preserve the four-tiered stepwise mechanism of flagellar regulation conserved by other vibrios, our current data suggest the class I genes are independent of σ54 regulation, and we have not found any evidence of a novel regulator for these genes in V. campbellii. Thus, the first hypothesis is our proposed model that we present in Fig. 8. We anticipate that future work will uncover the presence or absence of a potential novel class I flagellar regulator.
FIG 8.
Transcriptional hierarchy of polar flagellar genes in V. campbellii. A model of the transcriptional hierarchy for polar flagellar genes in V. campbellii DS40M4 is shown. Class I encodes early structural components of the flagella, as well as the genes encoding class II regulators FlrA and FlrC. Class II genes depend on regulation by σ54 and one or both σ54-dependent transcriptional activators, FlrA and FlrC. These genes include most structural components of the flagellum, including the export apparatus, hook, rod, ATPase, flagellin, flagellar rings, and more. Finally, FliA (σ28) activates class III genes, which encode additional flagellins, chemotaxis proteins, and the motor/stator.
Another critical difference in our proposed hierarchy model is how class II genes are regulated in V. campbellii. In our RNA-seq analysis, FlrC in V. campbellii showed very similar expression profiles to those observed in V. cholerae; thus, our model groups class IIV.camp and class IIIV.chol genes very similarly between both species (16). However, as we noted earlier, FlrA still plays an observable role in regulation of downstream flagellar genes such as class II and class III, and we found that fliKLMNOPQR and flhB showed stronger decreases in expression in a ΔflrA mutant compared to a ΔflrC mutant. When testing these further with qPCR in both single mutants and a ΔflrA ΔflrC double mutant, we found that deletion of either FlrA or FlrC significantly impaired expression of genes from the opposite regulon. In addition, a subset of genes classified as FlrC-dependent in both our V. campbellii RNA-seq data and previous V. cholerae models (17) showed additive downregulation when both σ54-dependent flagellar regulators were deleted. Based on these findings, we propose that FlrA and FlrC are both partially interchangeable σ54-dependent transcriptional regulators and that class II flagellar genes are coregulated by both FlrA and FlrC. Future work will more carefully tease apart the mechanisms behind regulation by both bEBPs. Finally, we note that fliS, flaI, fliD, flaG, flaA, ORF1, ORF2, and flrD display FliA-dependent regulation, as opposed to the FlrC-dependent regulation that is observed in V. cholerae (16). Thus, we group these genes with other FliA-regulated genes (class III) in the V. campbellii regulatory hierarchy.
Although the flagellar genes in V. campbellii are conserved and share similar chromosomal organization with other Vibrio species, we have shown that the regulation of the polar flagellum in V. campbellii is distinct in several ways. Furthering our understanding of the regulation of motility in different Vibrio species is important because motility and taxis in response to external signals are key behaviors of bacteria in various niches. In brief, our work here has established tools and broadened our understanding of the flagellar gene network in V. campbellii that will be valuable in future work both in studying swimming motility in V. campbellii and in comparing flagellar regulation across the Vibrionaceae.
MATERIALS AND METHODS
Strains and growth conditions.
All bacterial strains and plasmids are listed in Tables S1 and S2 in the supplemental material. V. campbellii strains were grown at 30°C on Luria marine (LM) medium (lysogeny broth supplemented with an additional 10 g NaCl per liter). Instant ocean water (IOW) medium was used in chitin-independent transformations; it consists of Instant Ocean sea salts (Aquarium Systems, Inc.) diluted in sterile water (2× = 28 g/liter). Transformations were outgrown in LBv2 (lysogeny broth medium supplemented with additional 200 mM NaCl, 23.14 mM MgCl2, and 4.2 mM KCl). When necessary, strains were supplemented with kanamycin (100 μg/ml), spectinomycin (200 μg/ml), or trimethoprim (10 μg/ml).
Analysis of Vibrio flagellar genes.
Identification of Vibrio flagellar and chemotaxis genes was performed as described previously (33). A data set comprising all of the protein sequences from various Vibrio species (including DS40M4) was clustered into groups based on sequence similarity using cd-hit version v4.8.1-2019-0228 (76) (parameters: -M 0 -g 1 -s 0.8 -c 0.4 -n 2 -d 500). The protein sequences from the polar flagellar gene clusters were compared against the corresponding DS40M4 flagellar gene using BLASTp and the percent identity of the best scoring homolog from each Vibrio species was recorded and plotted in the heat map. The six flagellins overlapped in a single cluster due to high sequence identity and were resolved manually by comparing synteny of these genes with other Vibrio flagellin genes.
Primer design and strain construction.
For constructing deletion strains in V. campbellii DS40M4, natural transformation or MuGENT was used as previously described (33–35). A candidate primer set was generated for deletion of each gene consisting of two primer pairs chosen to maximize the portion of the gene removed such that (i) any overlapping genes were left complete and (ii) all four primers met our criteria for validity. A valid primer is between 25 and 30 bp with a preference for shorter primers, contains no homopolymers of 5 or more bases, ends in cytosine or guanine, and has a Tm between 58 and 63°C as determined by the BioPerl implementation of Allawi and SantaLucia (77).
All primers are listed in Table S3 in the supplemental material. These primers were used to generate linear transforming DNAs (tDNA) by splicing-by-overlap extension (SOE) PCR as previously described (35). Briefly, an UP arm containing ∼3-kb homology of the area upstream of the gene to be deleted was PCR amplified via a F1 and R1 primer set. Similarly, a DOWN arm containing ∼3-kb homology of the area downstream of the gene to be deleted was PCR amplified via a F2 and R2 primer set. For unselected tDNA products, the UP and DOWN arms were used as the template and amplified via the F1 and R2 primers to make a SOE product that lacked an antibiotic marker. Unselected tDNA products were cotransformed with a selected tDNA product, which targets the nonfunctional luxB site in DS40M4 and inserts an antibiotic resistance cassette (for either spectinomycin or trimethoprim). These tDNAs were used to construct V. campbellii DS40M4 strains using previously published methods (78). Briefly, DS40M4 were grown overnight in LBv2 at 30°C, with shaking and with antibiotic and 100 μM IPTG. The next day, the culture was back-diluted into IOW containing 100 μM IPTG and tDNA. The tDNA marked with an antibiotic resistance cassette was added, and cells were incubated statically at 30°C for 4 to 6 h. LBv2 was then added, and the cells were incubated shaking at 30°C for an additional 1 to 2 h before streaking cultures onto LM plates with respective selective antibiotics. After natural transformation, strains containing the correct target mutation were identified via colony PCR with a forward and reverse detection primer and further confirmed by sending the products to Eurofins Scientific for sequencing.
V. campbellii BB120 gene deletions were made as previously described (79). Briefly, plasmids to make unmarked gene deletions were constructed using the suicide vector pRE112. Genetic regions flanking genes of interest (1,000 bp) were PCR amplified and inserted into pRE112, and the plasmid was then introduced to BB120 by conjugation and selection on chloramphenicol. Integration of the plasmid was verified by PCR, and each strain was plated on 20% sucrose for counterselection. Colonies that were sucrose resistant were screened for chloramphenicol sensitivity, indicating a crossover event had undergone to delete the gene of interest. Chloramphenicol-sensitive mutants were sequenced to confirm gene deletions.
Motility assays.
Swimming motility was measured by performing swimming assays on soft agar (medium containing 0.3% agar). V. campbellii strains were grown overnight in LM medium and then diluted to an optical density at 600 nm (OD600) of 0.5 the following day. Then, 5-μl portions of the diluted cultures were stabbed and pipetted into the centers of soft agar LM plates and grown in a humid container at 30°C for 24 h. After growth, plates were imaged via AlphaImager HP imaging system (ProteinSimple) and compared for swim halo size.
Swarm assays were performed using LM hard agar plates (0.7, 1.0, and 1.5% agar). V. campbellii strains were grown overnight in LM medium and concentrated to an OD600 of 10 the following day. Next, 5-μl portions of the cultures were spotted onto the surfaces of LM swarm plates and grown in a humid container at 30°C for 72 h. After growth, the plates were measured for swarming radius.
RNA extraction, quantitative reverse transcription-PCR (qRT-PCR), and RNA-seq.
Strains were inoculated in 5 ml of LM medium and grown overnight shaking at 30°C at 275 rpm. Each strain was back diluted to 1:1,000 in 5 ml of LM medium and grown with shaking at 30°C at 275 rpm until they reached an OD600 of ∼0.5. Cells were collected by centrifugation at 3,700 rpm at 4°C for 10 min, the supernatant was removed, and the cell pellets were flash frozen in liquid N2 and stored at −80°C. Triplicate biological samples were collected for RNA-seq and dRNA-seq. RNA was isolated from pellets using a TRIzol-chloroform extraction protocol as described previously (80) and treated with DNase by using a DNA-free DNA removal kit (Invitrogen). RNA-seq was performed as described previously (79).
Real-time qRT-PCR was used to quantify transcript levels from samples collected as described above using a SensiFast SYBR Hi-ROX One-Step kit (Bioline) according to the manufacturer’s guidelines. First, 1 μg of each RNA template was converted to cDNA using SuperScript IV reverse transcriptase (Invitrogen). All reactions were then performed using a LightCycler 4800 II (Roche) with a 0.4 μM concentration of each primer and 5 ng of template RNA (10 μl total volume). Primers were designed to have the following parameters: amplicon size of 100 bp, a primer size of 20 to 28 nucleotides, and a melting temperature of 55 to 60°C. qRT-PCR was performed with five biological replicates and two technical replicates. Data were analyzed using the standard curve method, which was assessed by qPCR on a quantified cDNA dilution series (see Data Set S3). After normalization to the standard curve for each gene, all crossing point values for target genes were normalized to hfq Cp values. Statistical analyses were performed using GraphPad Prism software to conduct one-way analysis of variance (ANOVA), followed by Tukey’s multiple-comparison test. The qRT-PCR data passed normal distribution tests (Shapiro-Wilk and Kolmogorov-Smirnov). We include the raw data for qRT-PCR in Data Set S3.
dRNA-seq analysis.
cDNA libraries from RNA (isolated as described above) were constructed as described previously (81) by Vertis Biotechnology AG (Freising, Germany) and sequenced using an Illumina NextSeq 500 machine in single-read mode (75-bp read length).
Transcriptional start site analysis.
TSS prediction was performed using the program TSSpredator on normalized coverage data (see Data Set S2). TSS classified as primary or secondary are located upstream of a gene, not further apart than the chosen untranslated region (UTR) length of 300 bp. Of these, the primary TSS is the one with the strongest expression. The ones within the annotated gene located on the sense strand are classified as internal, while the ones which are on the antisense strand not farther than the chosen UTR length are classified as antisense. Finally, those which are not within the vicinity of any of the annotated genes are classified as orphans. The TSS data are presented in Data Set S2.
Flagellar labeling and microscopy.
Fluorescence microscopy was performed with a Nikon Eclipse 80i microscope equipped with a Plan Apo 100× phase-contrast objective. Cells were grown in LM medium to an OD600 of ∼0.5, near mid-log phase, and 1 ml of culture volume was concentrated by centrifugation at 4,000 × g for 1 min at room temperature and resuspended in 50 μl of phosphate buffer saline (PBS). Then, 1 μl of NanoOrange dye (ThermoFischer Scientific) was mixed with the cells, followed by 15 min for staining to occur. NanoOrange-stained cells and flagella were visualized with a FITC HYQ filter cube (excitation filter, 460 to 500 nm; barrier filter, >590 nm). Images were captured with a Photometrics Coolsnap HQ2 camera and processed using MetaMorph 7.7.9.0 image software (Universal Imaging Corp.).
For TEM, strains were inoculated in 5 ml of LM medium and grown overnight shaking at 30°C at 275 rpm. Each strain was back diluted at 1:1,000 in 5 ml of LM medium and grown with shaking at 30°C at 275 rpm until it reached an OD600 of ∼0.5, near mid-log phase. Cells were collected by centrifugation at 4,000 × g for 1 min at room temperature and then washed with PBS. Cells were collected on grids and stained in 2% uranyl acetate. The grids were commercially prepared continuous carbon films on 400 mesh copper grids obtained from Ted Pella, Inc. Electron microscopy was performed at nominal magnifications ranging from 2,500× to 50,000× using a JEOL JEM 14000plus TEM operating at 120 kV. Images were recorded with a Gatan OneView camera using 0.5 s exposures and Gatan’s built-in motion correction.
Data availability.
The raw, demultiplexed reads and coverage files from RNA-seq and dRNA-seq were deposited in the National Center for Biotechnology Information Gene Expression Omnibus (NCBI GEO) under accession numbers GSE167483 and GSE147616. Custom scripts are available on GitHub (https://github.com/Juliacvk/vibrio_primer_design).
ACKNOWLEDGMENTS
We thank Victoria Lydick for providing excellent technical support in the lab. We also thank David Gene Morgan and the IU Electron Microscopy Center for their help with imaging vibrio flagella by TEM. Finally, we thank Daniel Kearns for project guidance and comments on the manuscript.
This study was supported by the National Institutes of Health grant R35GM124698 to J.C.V.K.
Footnotes
Supplemental material is available online only.
Contributor Information
Julia C. van Kessel, Email: jcvk@indiana.edu.
Michael Y. Galperin, NCBI, NLM, National Institutes of Health
REFERENCES
- 1.Boin MA, Austin MJ, Häse CC. 2004. Chemotaxis in Vibrio cholerae. FEMS Microbiol Lett 239:1–8. 10.1016/j.femsle.2004.08.039. [DOI] [PubMed] [Google Scholar]
- 2.Ottemann KM, Miller JF. 1997. Roles for motility in bacterial-host interactions. Mol Microbiol 24:1109–1117. 10.1046/j.1365-2958.1997.4281787.x. [DOI] [PubMed] [Google Scholar]
- 3.Norsworthy AN, Visick KL. 2013. Gimme shelter: how Vibrio fischeri successfully navigates an animal’s multiple environments. Front Microbiol 4:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reidl J, Klose KE. 2002. Vibrio cholerae and cholera: out of the water and into the host. FEMS Microbiol Rev 26:125–139. 10.1111/j.1574-6976.2002.tb00605.x. [DOI] [PubMed] [Google Scholar]
- 5.Letchumanan V, Chan KG, Lee LH. 2014. Vibrio parahaemolyticus: a review on the pathogenesis, prevalence, and advance molecular identification techniques. Front Microbiol 5:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pujalte MJ, Sitjà-Bobadilla A, Macián MC, Belloch C, Álvarez-Pellitero P, Pérez-Sánchez J, Uruburu F, Garay E. 2003. Virulence and molecular typing of Vibrio harveyi strains isolated from cultured dentex, gilthead sea bream, and European sea bass. Syst Appl Microbiol 26:284–292. 10.1078/072320203322346146. [DOI] [PubMed] [Google Scholar]
- 7.Ruby EG. 1996. Lessons from a cooperative, bacterial-animal association: the Vibrio fischeri-Euprymna scolopes light organ symbiosis. Annu Rev Microbiol 50:591–624. 10.1146/annurev.micro.50.1.591. [DOI] [PubMed] [Google Scholar]
- 8.Austin B, Zhang XH. 2006. Vibrio harveyi: a significant pathogen of marine vertebrates and invertebrates. Lett Appl Microbiol 43:119–124. 10.1111/j.1472-765X.2006.01989.x. [DOI] [PubMed] [Google Scholar]
- 9.Zhang X-H, He X, Austin B. 2020. Vibrio harveyi: a serious pathogen of fish and invertebrates in mariculture. Mar Life Sci Technol 2:1–245. 10.1007/s42995-020-00037-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haldar S, Chatterjee S, Sugimoto N, Das S, Chowdhury N, Hinenoya A, Asakura M, Yamasaki S. 2011. Identification of Vibrio campbellii isolated from diseased farm-shrimps from south India and establishment of its pathogenic potential in an Artemia model. Microbiology (Reading) 157:179–188. 10.1099/mic.0.041475-0. [DOI] [PubMed] [Google Scholar]
- 11.Daniels NA, Mackinnon L, Bishop R, Altekruse S, Ray B, Hammond RM, Thompson S, Wilson S, Bean NH, Griffin PM, Slutsker L. 2000. Vibrio parahaemolyticus infections in the United States, 1973–1998. J Infect Dis 181:1661–1666. 10.1086/315459. [DOI] [PubMed] [Google Scholar]
- 12.Lee JH, Rho JB, Park KJ, Kim CB, Han YS, Choi SH, Lee KH, Park SJ. 2004. Role of flagellum and motility in pathogenesis of Vibrio vulnificus. Infect Immun 72:4905–4910. 10.1128/IAI.72.8.4905-4910.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Q, Defoirdt T. 2015. Quorum sensing positively regulates flagellar motility in pathogenic Vibrio harveyi. Environ Microbiol 17:960–968. 10.1111/1462-2920.12420. [DOI] [PubMed] [Google Scholar]
- 14.Rugel AR, Klose KE. 2011. Vibrio cholerae flagellar synthesis and virulence. Epidemiol Mol Asp Cholera :203–212. [Google Scholar]
- 15.Zhu S, Kojima S, Homma M. 2013. Structure, gene regulation, and environmental response of flagella in Vibrio. Front Microbiol 4:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Syed KA, Beyhan S, Correa N, Queen J, Liu J, Peng F, Satchell KJF, Yildiz F, Klose KE. 2009. The Vibrio cholerae flagellar regulatory hierarchy controls expression of virulence factors. J Bacteriol 191:6555–6570. 10.1128/JB.00949-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Echazarreta MA, Klose KE. 2019. Vibrio flagellar synthesis. Front Cell Infect Microbiol 9:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Millikan DS, Ruby EG. 2003. FlrA, a σ54-dependent transcriptional activator in Vibrio fischeri, is required for motility and symbiotic light-organ colonization. J Bacteriol 185:3547–3557. 10.1128/JB.185.12.3547-3557.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prouty MG, Correa NE, Klose KE. 2001. The novel σ54- and σ28-dependent flagellar gene transcription hierarchy of Vibrio cholerae. Mol Microbiol 39:1595–1609. 10.1046/j.1365-2958.2001.02348.x. [DOI] [PubMed] [Google Scholar]
- 20.Burnham PM, Kolar WP, Hendrixson DR. 2020. A polar flagellar transcriptional program mediated by diverse two-component signal transduction systems and basal flagellar proteins is broadly conserved in polar flagellates. mBio 11:e03107-19. 10.1128/mBio.03107-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Correa NE, Barker JR, Klose KE. 2004. The Vibrio cholerae FlgM homologue is an anti-σ28 factor that is secreted through the sheathed polar flagellum. J Bacteriol 186:4613–4619. 10.1128/JB.186.14.4613-4619.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim Y, McCarter LL. 2000. Analysis of the polar flagellar gene system of Vibrio parahaemolyticus. J Bacteriol 182:3693–3704. 10.1128/JB.182.13.3693-3704.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brennan CA, Mandel MJ, Gyllborg MC, Thomasgard KA, Ruby EG. 2013. Genetic determinants of swimming motility in the squid light-organ symbiont Vibrio fischeri. Microbiologyopen 2:576–594. 10.1002/mbo3.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Millikan DS, Ruby EG. 2002. Alterations in Vibrio fischeri motility correlate with a delay in symbiosis initiation and are associated with additional symbiotic colonization defects. Appl Environ Microbiol 68:2519–2528. 10.1128/AEM.68.5.2519-2528.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCarter LL. 2006. Motility and chemotaxis, p 113–132. In Thompson FL, Austin B, Swings J (ed), The biology of vibrios. ASM Press, Washington, DC. [Google Scholar]
- 26.McCarter LL. 2004. Dual flagellar systems enable motility under different circumstances. J Mol Microbiol Biotechnol 7:18–29. 10.1159/000077866. [DOI] [PubMed] [Google Scholar]
- 27.Gode-Potratz CJ, Kustusch RJ, Breheny PJ, Weiss DS, McCarter LL. 2011. Surface sensing in Vibrio parahaemolyticus triggers a programme of gene expression that promotes colonization and virulence. Mol Microbiol 79:240–263. 10.1111/j.1365-2958.2010.07445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin B, Wang Z, Malanoski AP, O’Grady EA, Wimpee CF, Vuddhakul V, Alves N, Thompson FL, Gomez-Gil B, Vora GJ. 2010. Comparative genomic analyses identify the Vibrio harveyi genome sequenced strains BAA-1116 and HY01 as Vibrio campbellii. Environ Microbiol Rep 2:81–89. 10.1111/j.1758-2229.2009.00100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Urbanczyk H, Ogura Y, Hayashi T. 2013. Taxonomic revision of Harveyi clade bacteria (family Vibrionaceae) based on analysis of whole-genome sequences. Int J Syst Evol Microbiol 63:2742–2751. 10.1099/ijs.0.051110-0. [DOI] [PubMed] [Google Scholar]
- 30.Sawabe T, Kita-Tsukamoto K, Thompson FL. 2007. Inferring the evolutionary history of vibrios by means of multilocus sequence analysis. J Bacteriol 189:7932–7936. 10.1128/JB.00693-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shinoda S, Nakahara N, Uchida E, Hiraga M. 1985. Lateral flagellar antigen of Vibrio alginolyticus and Vibrio harveyi: existence of serovars common to the two species. Microbiol Immunol 29:173–182. 10.1111/j.1348-0421.1985.tb00817.x. [DOI] [PubMed] [Google Scholar]
- 32.Allen RD, Baumann P. 1971. Structure and arrangement of flagella in species of the genus Beneckea and Photobacterium fischeri. J Bacteriol 107:295–302. 10.1128/jb.107.1.295-302.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simpson CA, Podicheti R, Rusch DB, Dalia AB, van Kessel JC. 2019. Diversity in natural transformation frequencies and regulation across Vibrio species. mBio 10:e02788-19. 10.1128/mBio.02788-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simpson CA, Petersen BD, Haas NW, Geyman LJ, Lee AH, Podicheti R, Pepin R, Brown LC, Rusch DB, Manzella MP, Papenfort K, van Kessel JC. The quorum-sensing systems of Vibrio campbellii DS40M4 and BB120 are genetically and functionally distinct. Environ Microbiol, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dalia AB, McDonough E, Camilli A. 2014. Multiplex genome editing by natural transformation. Proc Natl Acad Sci USA 111:8937–8942. 10.1073/pnas.1406478111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen M, Zhao Z, Yang J, Peng K, Baker MAB, Bai F, Lo CJ. 2017. Length-dependent flagellar growth of Vibrio alginolyticus revealed by real time fluorescent imaging. Elife 6:1–16. 10.7554/eLife.22140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grossart H-P, Steward GF, Martinez J, Azam F. 2000. A simple, rapid method for demonstrating bacterial flagella. Appl Environ Microbiol 66:3632–3636. 10.1128/AEM.66.8.3632-3636.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kojima S, Blair D-I. 2004. The bacterial flagellar motor: structure and function of a complex molecular machine, p 93–134. Academic Press, New York, NY. [DOI] [PubMed] [Google Scholar]
- 39.Parkinson JS. 1978. Complementation analysis and deletion mapping of Escherichia coli mutants defective in chemotaxis. J Bacteriol 135:45–53. 10.1128/jb.135.1.45-53.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barker CS, Meshcheryakova IV, Kostyukova AS, Samatey FA. 2010. FliO regulation of FliP in the formation of the Salmonella enterica flagellum. PLoS Genet 6:e1001143. 10.1371/journal.pgen.1001143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Arnam JS, McMurry JL, Kihara M, Macnab RM. 2004. Analysis of an engineered salmonella flagellar fusion protein, FliR-FlhB. J Bacteriol 186:2495–2498. 10.1128/JB.186.8.2495-2498.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fraser GM, Bennett JCQ, Hughes C. 1999. Substrate-specific binding of hook-associated proteins by FlgN and FliT, putative chaperones for flagellum assembly. Mol Microbiol 32:569–580. 10.1046/j.1365-2958.1999.01372.x. [DOI] [PubMed] [Google Scholar]
- 43.Chilcott GS, Hughes KT. 2000. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar Typhimurium and Escherichia coli. Microbiol Mol Biol Rev 64:694–708. 10.1128/MMBR.64.4.694-708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waters RC, O’Toole PW, Ryan KA. 2007. The FliK protein and flagellar hook-length control. Protein Sci 16:769–780. 10.1110/ps.072785407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shibata S, Takahashi N, Chevance FFV, Karlinsey JE, Hughes KT, Aizawa SI. 2007. FliK regulates flagellar hook length as an internal ruler. Mol Microbiol 64:1404–1415. 10.1111/j.1365-2958.2007.05750.x. [DOI] [PubMed] [Google Scholar]
- 46.Correa NE, Peng F, Klose KE. 2005. Roles of the regulatory proteins FlhF and FlhG in the Vibrio cholerae flagellar transcription hierarchy. J Bacteriol 187:6324–6332. 10.1128/JB.187.18.6324-6332.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Erhardt M, Mertens ME, Fabiani FD, Hughes KT. 2014. ATPase-independent type-III protein secretion in Salmonella enterica. PLoS Genet 10:e1004800. 10.1371/journal.pgen.1004800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martinez RM, Jude BA, Kirn TJ, Skorupski K, Taylor RK. 2010. Role of FlgT in anchoring the flagellum of Vibrio cholerae. J Bacteriol 192:2085–2092. 10.1128/JB.01562-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klose KE, Mekalanos JJ. 1998. Distinct roles of an alternative sigma factor during both free-swimming and colonizing phases of the Vibrio cholerae pathogenic cycle. Mol Microbiol 28:501–520. 10.1046/j.1365-2958.1998.00809.x. [DOI] [PubMed] [Google Scholar]
- 50.Klose KE, Novik V, Mekalanos JJ. 1998. Identification of multiple σ54-dependent transcriptional activators in Vibrio cholerae. J Bacteriol 180:5256–5259. 10.1128/JB.180.19.5256-5259.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim YK, McCarter LL. 2004. Cross-regulation in Vibrio parahaemolyticus: compensatory activation of polar flagellar genes by the lateral flagellar regulator LafK. J Bacteriol 186:4014–4018. 10.1128/JB.186.12.4014-4018.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kawagishi I, Nakada M, Nishioka N, Homma M. 1997. Cloning of a Vibrio alginolyticus rpoN gene that is required for polar flagellar formation. J Bacteriol 179:6851–6854. 10.1128/jb.179.21.6851-6854.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kusumoto A, Shinohara A, Terashima H, Kojima S, Yakushi T, Homma M. 2008. Collaboration of FlhF and FlhG to regulate polar-flagella number and localization in Vibrio alginolyticus. Microbiology (Reading) 154:1390–1399. 10.1099/mic.0.2007/012641-0. [DOI] [PubMed] [Google Scholar]
- 54.Correa NE, Lauriano CM, McGee R, Klose KE. 2000. Phosphorylation of the flagellar regulatory protein FlrC is necessary for Vibrio cholerae motility and enhanced colonization. Mol Microbiol 35:743–755. 10.1046/j.1365-2958.2000.01745.x. [DOI] [PubMed] [Google Scholar]
- 55.Merino S, Shaw JG, Tomás JM. 2006. Bacterial lateral flagella: an inducible flagella system. FEMS Microbiol Lett 263:127–135. 10.1111/j.1574-6968.2006.00403.x. [DOI] [PubMed] [Google Scholar]
- 56.Atsumi T, Maekawa Y, Yamada T, Kawagishi I, Imae Y, Homma M. 1996. Effect of viscosity on swimming by the lateral and polar flagella of Vibrio alginolyticus. J Bacteriol 178:5024–5026. 10.1128/jb.178.16.5024-5026.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kawagishi I, Maekawa Y, Atsumi T, Homma M, Imae Y. 1995. Isolation of the polar and lateral flagellum-defective mutants in Vibrio alginolyticus and identification of their flagellar driving energy sources. J Bacteriol 177:5158–5160. 10.1128/jb.177.17.5158-5160.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kearns DB. 2010. A field guide to bacterial swarming motility. Nat Rev Microbiol 8:634–644. 10.1038/nrmicro2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dey S, Biswas M, Sen U, Dasgupta J. 2015. Unique ATPase site architecture triggers cis-mediated synchronized ATP binding in heptameric AAA+-ATPase domain of flagellar regulatory protein FlrC. J Biol Chem 290:8734–8747. 10.1074/jbc.M114.611434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chakraborty S, Biswas M, Dey S, Agarwal S, Chakrabortty T, Ghosh B, Dasgupta J. 2020. The heptameric structure of the flagellar regulatory protein FlrC is indispensable for ATPase activity and disassembled by cyclic-di-GMP. J Biol Chem 295:16960–16974. 10.1074/jbc.RA120.014083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cohen EJ, Hughes KT. 2014. Rod-to-hook transition for extracellular flagellum assembly is catalyzed by the L-ring-dependent rod scaffold removal. J Bacteriol 196:2387–2395. 10.1128/JB.01580-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McCarter LL. 1995. Genetic and molecular characterization of the polar flagellum of Vibrio parahaemolyticus. J Bacteriol 177:1595–1609. 10.1128/jb.177.6.1595-1609.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Homma M, Fujita H, Yamaguchi S, Iino T. 1984. Excretion of unassembled flagellin by Salmonella Typhimurium mutants deficient in hook-associated proteins. J Bacteriol 159:1056–1059. 10.1128/jb.159.3.1056-1059.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Patterson-Delafield J, Martinez RJ, Stocker BAD, Yamaguchi S. 1973. A new fla gene in Salmonella Typhimurium—flaR—and its mutant phenotype-superhooks. Arch Mikrobiol 90:107–120. 10.1007/BF00414513. [DOI] [PubMed] [Google Scholar]
- 65.Silverman MR, Simon MI. 1972. Flagellar assembly mutants in Escherichia coli. J Bacteriol 112:986–993. 10.1128/jb.112.2.986-993.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hirano T, Yamaguchi S, Oosawa K, Aizawa SI. 1994. Roles of FliK and FlhB in determination of flagellar hook length in Salmonella Typhimurium. J Bacteriol 176:5439–5449. 10.1128/jb.176.17.5439-5449.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kutsukake K, Minamino T, Yokoseki T. 1994. Isolation and characterization of FliK-independent flagellation mutants from Salmonella Typhimurium. J Bacteriol 176:7625–7629. 10.1128/jb.176.24.7625-7629.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Partridge JD, Harshey RM. 2020. Investigating flagella-driven motility in Escherichia coli by applying three established techniques in a series. J Vis Exp 2020:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Echazarreta MA, Kepple JL, Yen LH, Chen Y, Klose KE. 2018. A critical region in the FlaA flagellin facilitates filament formation of the Vibrio cholerae flagellum. J Bacteriol 200:1–11. 10.1128/JB.00029-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim SY, Thanh XTT, Jeong K, Kim S, Pan SO, Jung CH, Hong SH, Lee SE, Rhee JH. 2014. Contribution of six flagellin genes to the flagellum biogenesis of Vibrio vulnificus and in vivo invasion. Infect Immun 82:29–42. 10.1128/IAI.00654-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Millikan DS, Ruby EG. 2004. Vibrio fischeri flagellin A is essential for normal motility and for symbiotic competence during initial squid light organ colonization. J Bacteriol 186:4315–4325. 10.1128/JB.186.13.4315-4325.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Milton DL, O’Toole R, Horstedt P, Wolf-Watz H. 1996. Flagellin A is essential for the virulence of Vibrio anguillarum. J Bacteriol 178:1310–1319. 10.1128/jb.178.5.1310-1319.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Martínez-García E, Nikel PI, Chavarría M, de Lorenzo V. 2014. The metabolic cost of flagellar motion in Pseudomonas putida KT2440. Environ Microbiol 16:291–303. 10.1111/1462-2920.12309. [DOI] [PubMed] [Google Scholar]
- 74.Ni B, Colin R, Link H, Endres RG, Sourjik V. 2020. Growth-rate dependent resource investment in bacterial motile behavior quantitatively follows potential benefit of chemotaxis. Proc Natl Acad Sci USA 117:595–601. 10.1073/pnas.1910849117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ni B, Ghosh B, Paldy FS, Colin R, Heimerl T, Sourjik V. 2017. Evolutionary remodeling of bacterial motility checkpoint control. Cell Rep 18:866–877. 10.1016/j.celrep.2016.12.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li W, Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659. 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 77.Allawi HT, SantaLucia J. 1997. Thermodynamics and NMR of internal G·T mismatches in DNA. Biochemistry 36:10581–10594. 10.1021/bi962590c. [DOI] [PubMed] [Google Scholar]
- 78.Dalia TN, Hayes CA, Stolyar S, Marx CJ, McKinlay JB, Dalia AB. 2017. Multiplex genome editing by natural transformation (MuGENT) for synthetic biology in Vibrio natriegens. ACS Synth Biol 6:1650–1655. 10.1021/acssynbio.7b00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chaparian RR, Olney SG, Hustmyer CM, Rowe-Magnus DA, van Kessel JC. 2016. Integration host factor and LuxR synergistically bind DNA to coactivate quorum-sensing genes in Vibrio harveyi. Mol Microbiol 101:823–840. 10.1111/mmi.13425. [DOI] [PubMed] [Google Scholar]
- 80.Rutherford ST, van Kessel JC, Shao Y, Bassler BL. 2011. AphA and LuxR/HapR reciprocally control quorum sensing in vibrios. Genes Dev 25:397–408. 10.1101/gad.2015011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Papenfort K, Förstner KU, Cong JP, Sharma CM, Bassler BL. 2015. Differential RNA-seq of Vibrio cholerae identifies the VqmR small RNA as a regulator of biofilm formation. Proc Natl Acad Sci USA 112:E766–E775. 10.1073/pnas.1500203112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Set S2. Download JB.00276-21-s0001.xlsx, XLSX file, 11.4 MB (11.4MB, xlsx)
Data Set S3. Download JB.00276-21-s0002.xlsx, XLSX file, 0.06 MB (59.7KB, xlsx)
Legends to Data Sets S1 to S3, Fig. S1 to S3, and Tables S1 to S3. Download JB.00276-21-s0003.pdf, PDF file, 3.9 MB (3.9MB, pdf)
Data Set S1A. Download JB.00276-21-s0004.xlsx, XLSX file, 6.6 MB (6.6MB, xlsx)
Data Set S1B. Download JB.00276-21-s0005.xlsx, XLSX file, 6.6 MB (6.6MB, xlsx)
Data Set S1C. Download JB.00276-21-s0006.xlsx, XLSX file, 6.6 MB (6.6MB, xlsx)
Data Set S1D. Download JB.00276-21-s0007.xlsx, XLSX file, 6.6 MB (6.6MB, xlsx)
Data Availability Statement
The raw, demultiplexed reads and coverage files from RNA-seq and dRNA-seq were deposited in the National Center for Biotechnology Information Gene Expression Omnibus (NCBI GEO) under accession numbers GSE167483 and GSE147616. Custom scripts are available on GitHub (https://github.com/Juliacvk/vibrio_primer_design).