ABSTRACT
Macromolecular cell-envelope-spanning structures such as the bacterial flagellum must traverse the cell wall. Lytic transglycosylase enzymes are capable of enlarging gaps in the peptidoglycan meshwork to allow the efficient assembly of supramolecular complexes. In the periplasmic space, the assembly of the flagellar rod requires the scaffold protein FlgJ, which includes a muramidase domain in the canonical models Salmonella enterica and Escherichia coli. In contrast, in Rhodobacter sphaeroides, FlgJ and the dedicated flagellar lytic transglycosylase SltF are separate entities that interact in the periplasm. In this study, we show that sltF is expressed, along with the genes encoding the early components of the flagellar hierarchy that include the hook-basal body proteins, making SltF available during the rod assembly. Protein-protein interaction experiments demonstrated that SltF interacts with the rod proteins FliE, FlgB, FlgC, FlgF, and FlgG through its C-terminal region. A deletion analysis that divides the C terminus in two halves revealed that the interacting regions for most of the rod proteins are not redundant. Our results also show that the presence of the rod proteins FliE, FlgB, FlgC, and FlgF displace the previously reported SltF-FlgJ interaction. In addition, we observed modulation of the transglycosylase activity of SltF mediated by FlgB and FlgJ that could be relevant to coordinate rod assembly with cell wall remodeling. In summary, different mechanisms regulate the flagellar lytic transglycosylase, SltF, ensuring a timely transcription, a proper localization and a controlled enzymatic activity.
IMPORTANCE Several mechanisms participate in the assembly of cell-envelope-spanning macromolecular structures. The sequential expression of substrates to be exported, selective export, and a specific order of incorporation are some of the mechanisms that stand out to drive an efficient assembly process. Here, we analyze how the structural rod proteins, the scaffold protein FlgJ and the flagellar lytic enzyme SltF, interact in an orderly fashion to assemble the flagellar rod into the periplasmic space. A complex arrangement of transient interactions directs a dedicated flagellar muramidase toward the flagellar rod. All of these interactions bring this protein to the proximity of the peptidoglycan wall while also modulating its enzymatic activity. This study suggests how a dynamic network of interactions participates in controlling SltF, a prominent component for flagellar formation.
KEYWORDS: Rhodobacter sphaeroides, lytic transglycosylase, flagellar biogenesis, flagellar rod, SltF
INTRODUCTION
The bacterial flagellum is a complex molecular motor embedded in the cell envelope; rotation of this structure and its control by the chemotactic system allows swimming and surveillance of the surroundings, which ultimately directs bacterial cells toward optimal conditions for survival (1). Although the paradigmatic flagellar systems have been those of Salmonella enterica and Escherichia coli, the recent study of several flagellar systems in different proteobacteria has revealed a conserved core structure and numerous accessory components (2).
This organelle can be divided into three major components, namely, the basal body, the hook, and the filament (3–6). The basal body contains the rotor, the export apparatus, several ring structures, and a central rod. The extracellular structures are the hook, formed by approximately 121 to 130 subunits of the protein FlgE (4, 5), and the long helical filament, with more than 20,000 copies of FliC (7, 8). The filament is a long hollow tube that adopts a helical shape and works as a propeller through the rotational motion driven by the flagellar motor (8). These two structures are joined together by hook-associated proteins (HAP1 and -3), and at the tip of the filament the capping protein, HAP2, enables the polymerization of flagellin subunits (9, 10).
The cytoplasmic ring (C-ring) is composed of three proteins: FliG, FliM, and FliN. This structure has been implicated in torque generation and morphogenesis (11–13). FliG interacts with the membrane/supramembrane ring (MS ring) that is formed by several subunits of the protein FliF (14–17). The periplasmic side of the MS ring is connected to the flagellar hook through the rod that is also a helical component of the bacterial flagellum. In contrast to the filament and the hook, which are composed of a single protein, the rod is composed of five different proteins—FliE, FlgB, FlgC, FlgF, and FlgG (18–21).
The structure can be divided into a proximal and a distal rod. The proximal rod is estimated to contain 6 subunits of each FlgB, FlgC, and FlgF, and the distal rod is composed by 26 subunits of FlgG (4, 22, 23). During the assembly of the rod, the peptidoglycan layer (PG) must be penetrated by the growing structure; it has been reported that in betaproteobacteria and most gammaproteobacteria, FlgJ is a bidomain protein composed of an N-terminal scaffolding domain required for the polymerization of the rod and a C-terminal domain with an acetylglucosaminidase activity required to penetrate the peptidoglycan layer (24–26).
The rod spans the cell wall and the outer membrane through the P and L rings that act as bushings during the rotation of this structure (4, 18, 19, 27). The proteins that form the P and L rings are FlgI and FlgH, respectively (4). The L-ring besides acting as a bushing, has been implicated in dislodging the scaffolding protein FlgJ from the tip of the rod and allowing the transition for the polymerization of the hook (28).
The export apparatus is composed by 9 proteins, catalyzes the transport of most of the flagellar axial proteins, and is formed by membrane and cytoplasmic components. The membrane components form an export gate that is housed in the central portion of the MS ring the cytoplasmic portion is projected from the export gate into a large cavity in the central portion of the C ring (29, 30). The axial proteins that conform the rod, the hook, and the scaffolding protein FlgJ have an N-terminal export signal that is recognized by the export apparatus, while FlgI and FlgH are exported trough the general secretion (Sec) system.
Flagellar biogenesis is a highly regulated process that proceeds outwardly in an orchestrated fashion (31, 32); at a certain point during the assembly process, the rod must penetrate the PG layer since the diameter of the growing structure (11 nm) is larger than the mesh pores of the PG layer (4 to 8 nm) (33). The PG mesh is constantly remodeled and reinforced in order to allow cell growth; however, it represents a physical barrier for the assembly of structures that have a diameter greater than its pores (34, 35). Nevertheless peptidoglycan-degrading enzymes are not an absolute requirement for flagellar assembly given that the growth of the axial structures can proceed if growth coincides with the gaps that appear in the structure during growth and/or remodeling of the cell wall (36). The activity of the muramidase enzymes that participate in the assembly of the flagellum is under spatial and temporal control. Most of these enzymes are lytic transglycosylases and are a class of autolysins that rearrange the PG cell wall (34, 37, 38).
In betaproteobacteria and gammaproteobacteria such as Bordetella parapertussis and S. enterica, the bidomain FlgJ protein facilitates the penetration of the PG layer by the axial structures (25, 26, 39). However, in alphaproteobacteria like Caulobacter crescentus and Rhodobacter sphaeroides instead of finding this bidomain protein a short version of FlgJ has been identified. This short FlgJ is characterized by having only the scaffolding domain, and the cell wall hydrolase is a separate polypeptide (40); in these bacteria, the cell wall hydrolase proteins are encoded by genes located in a flagellar context (39, 41).
R. sphaeroides is an alphaproteobacterium that under the standard growth conditions used in the laboratory assembles a single subpolar flagellum. In this microorganism, many of the genes encoding for the components of the flagellum have been identified, and it has been demonstrated that the expression of these genes follows a hierarchical pattern in which the early flagellar components are transcribed by the RNA polymerase associated with a sigma 54 factor (Eσ54) and the late components are dependent on Eσ28 (42).
σ54, also known as RpoN, recognizes promoters with the conserved positions GG and GC that are located –24 and –12 nucleotides upstream the transcription initiation site (43), and open complex formation requires the presence of an activator protein that binds approximately 150 bp upstream of the promoter sequence; this type of protein is known as an enhancer-binding protein (EBP) (44). The R. sphaeroides genome encodes four rpoN genes (rpoN1 to rpoN4) (45). Previous work has demonstrated that RpoN2 is Fla1 specific (45, 46), and one of its EBPs is the protein FleQ (42). Eσ54 and FleQ activate transcription of the genes encoding the MS ring (fliF), and some export apparatus components (fliHIJ), the first rod protein (fliE), and a second EBP (fleT). FleQ, together with FleT and RpoN2, activates the expression of the genes that encode the rest of the export apparatus components, the basal body, the hook, motAB, fliA, and flgM. Finally, once the HBB-hook complex is formed, fliC transcription dependent on Eσ28 proceeds (42, 47).
In this bacterium, the flagellum-specific soluble lytic transglycosylase (SltF) is encoded within the flgG operon, and the protein is exported to the periplasm via the general secretion (Sec) pathway, where it interacts with FlgJ and is directed to open a gap in the PG layer (40, 48, 49). SltF has a conserved enzymatic domain and a C-terminal region that is relevant for flagellar formation but not for activity (41, 49). Thus far, it has been determined that the interaction between SltF and FlgJ depends on the integrity of the C-terminal domain of SltF (41, 49). In addition, it has been shown that SltF can interact with the rod proteins FlgB and FlgF, affecting its hydrolytic activity in opposite ways, i.e., FlgB activates, whereas FlgF inhibits, enzymatic activity (50).
To further understand the role of these proteins during rod biogenesis, we performed an in vitro analysis of the interactions of wild type and three different C-terminal mutant versions of SltF with the rod proteins and examined whether these interactions were modified by the inclusion of FlgJ. A model is proposed in which SltF is preferentially recruited to the forming rod by FliE, FlgB, FlgC, and FlgF over FlgJ, and we postulate how the observed interactions modulate SltF activity to facilitate rod assembly.
RESULTS
Expression of sltF occurs in early stages of flagellar formation.
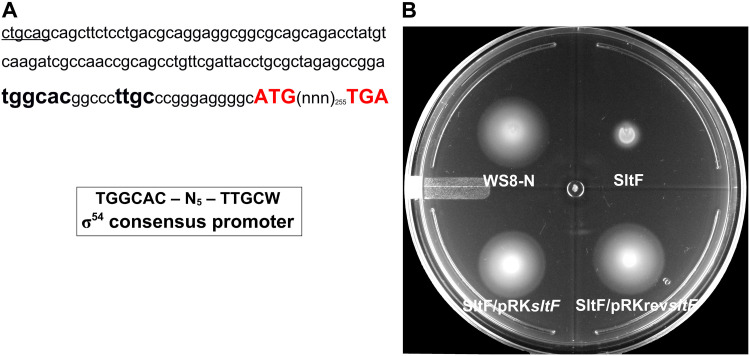
The gene that codes for SltF is positioned in chromosome 1 of the genome of R. sphaeroides in a flagellar genetic context between flgL and fleQ (48). sltF seems to be the last gene of the flgGHIJKLsltF operon (40). Genetic evidence suggests that a σ54-dependent promoter located upstream flgG (pflgG) is responsible for the expression of these genes. Nonetheless, a σ28-dependent promoter sequence was identified upstream of flgK that could express the last three genes of the operon (40). However, SltF activity is required early during flagellar assembly, so it must be transcribed from a σ54-dependent promoter such as pflgG. A close inspection of the 33 bp-intercistronic region between the stop codon of flgL and the start codon of sltF allowed us to identify a sequence similar to a σ54 consensus promoter (Fig. 1A). Therefore, in order to ascertain whether this putative promoter sequence could transcribe sltF, we cloned sltF with a 120-bp sequence upstream of sltF in plasmid pRK415. An sltF mutant was successfully complemented regardless of the orientation of sltF from the lac promoter (plac) present in the plasmid vector (Fig. 1B), suggesting that sltF has its own promoter.
FIG 1.
A functional promoter dependent on a σ54 factor is located upstream of sltF. (A) The 120-nucleotide sequence upstream of sltF is shown. The nucleotides shown in red correspond to the start and stop codons of sltF. The sequence similar to the σ54 binding site is shown in boldface type. (B) Swimming plate for motility assays. The plate shows the swimming phenotype of wild-type WS8N, a sltF mutant strain (SltF), and the mutant strain complemented with two sltF constructs: SltF/pRKsltF, a construct with the gene coding for sltF and 120 bp upstream, and SltF/pRKrevsltF, a construct with the gene coding for sltF and 120 bp upstream cloned in the opposite sense of the vector promoter.
In order to confirm the presence of a promoter in this region, we constructed a transcriptional fusion of the upstream region of sltF with the reporter gene uidA, which encodes the β-glucuronidase enzyme. The plasmid carrying this fusion (up120sltFpfw plasmid) was introduced in the wild-type strain WS8N of R. sphaeroides, and the amount of β-glucuronidase was determined. The results show that this region promotes the expression of the reporter gene, but when this region was cloned in the opposite sense of the reporter gene, the β-glucuronidase activity is lost (Table 1). In addition, as expected for a gene that is expressed within the flagellar gene expression hierarchy, the β-glucuronidase activity was completely dependent on the master regulator FleQ. Overall, these results suggest that sltF is expressed form its own promoter during the early stages of flagellar biogenesis.
TABLE 1.
β-Glucuronidase activity of reporter gene uidA under the control of the upstream region of the sltF gene
| Vector | Mean activity (nmol/min/mg of protein) ± SD |
|
|---|---|---|
| WS8N | SP13 (ΔfleQ) | |
| Empty vector | 0.09395 ± 0.0233 | 0.132 ± 0.0229 |
| Up120sltFprev | 2.6865 ± 0.7248 | NDa |
| Up120sltFpfw | 123.755 ± 25.5741 | 0.11905 ± 0.0204 |
ND, not determined.
Single interactions of SltF with rod proteins and scaffolding protein FlgJ.
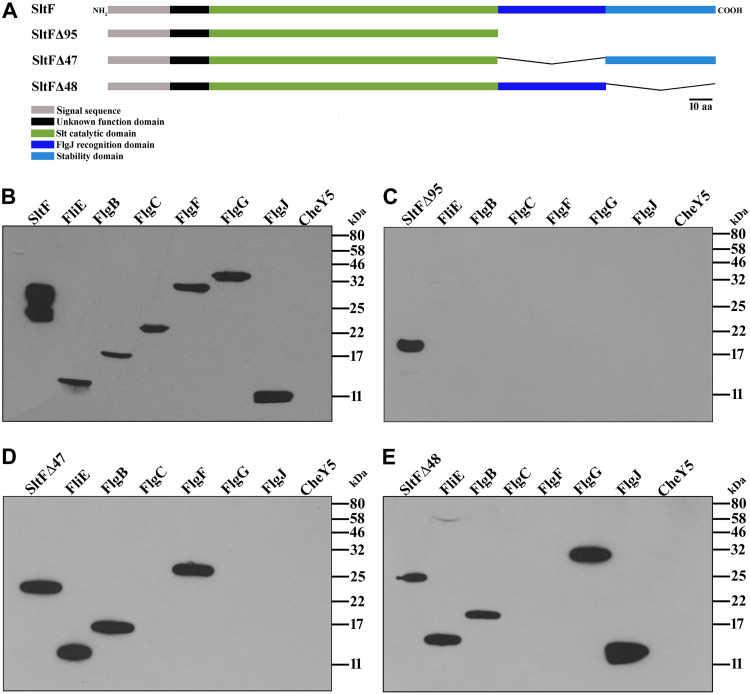
It has been reported that, once SltF reaches the periplasmic space via the Sec pathway, it interacts with FlgJ through its C-terminal domain (41, 48, 49). In addition, it has been shown that SltF interacts with the rod proteins FlgB and FlgF (50). To further analyze these findings and to have a better understanding of the molecular basis of these interactions, we analyzed by means of far-Western blotting, the interactions between the five rod proteins (FliE, FlgB, FlgC, FlgF, and FlgG), and the scaffold protein FlgJ with wild-type SltF, as well as with three mutant versions in which different regions of the C terminus of this protein were deleted (Fig. 2A). In accordance with previous reports, we observed that both the rod proteins and the scaffold protein FlgJ interact with SltF (Fig. 2B) (41, 49). However, the mutant version of SltF that lacks the last 95 residues (SltFΔ95) from its C-terminal domain loses its ability to recognize any of these proteins (Fig. 2C). In an attempt to dissect discrete regions that may interact specifically with these proteins, we tested two additional versions of SltF that dissect in half the C-terminal domain (SltFΔ47 and SltFΔ48) (Fig. 2A). SltFΔ47 recognizes FliE, FlgB, and FlgF, but FlgC, FlgG, and FlgJ are not detected (Fig. 2D). On the other hand, the mutant strain SltFΔ48 interacts with FliE, FlgB, FlgG, and FlgJ but not with FlgC or FlgF (Fig. 2E). Therefore, two proximal rod proteins, i.e., FliE and FlgB, are recognized by either the proximal or distal segments of the C terminus of SltF, whereas FlgG and FlgJ interact with the proximal region, and FlgF interacts with the distal region of the C terminus of SltF. In agreement with previous results (49), we noted that the interaction of SltFΔ48 with FlgJ is stronger than the interaction with wild-type SltF (see Fig. 2A and E), suggesting that the distal segment modulates the binding of FlgJ in a negative manner.
FIG 2.
Far-Western interactions between SltF mutants with the rod proteins. (A) Schematic representation of the mutant constructions of SltF. Mutant SltFΔ95 lacks the 95 residues that conform the C terminus of SltF. The region was divided in two segments, proximal and distal to the N terminus. SltFΔ47 lacks the proximal 47 residues, and SltFΔ48 lacks the distal 48 residues, as indicated. The various domains of the protein are indicated by color. (B to E) Interactions between the rod proteins and FlgJ with SltF were detected by affinity blotting, as described in Materials and Methods. The purified proteins (0.15 nmol) were subjected to electrophoresis in a 17.5% SDS-PAGE and probed with the purified proteins SltF (B), SltFΔ95 (C), SltFΔ47 (D), and SltFΔ48 (E). The identity of the protein loaded in each lane is indicated in the upper part, CheY5 was included as a negative control. Detection was performed using anti-SltF gamma globulins. Molecular mass markers are shown on the right.
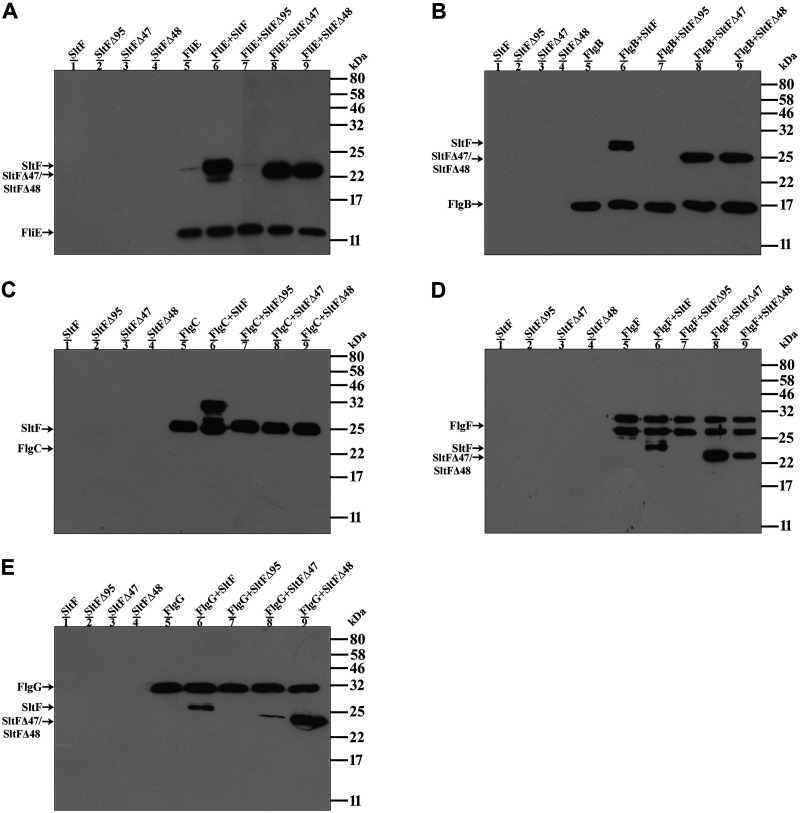
We also carried out coimmunoprecipitation experiments in order to confirm the interactions observed using far-Western analyses. For these experiments, we used specific immunoglobulins for each rod protein to pull down any complex formed between a particular rod protein and SltF. The results of these experiments confirmed that wild-type SltF interacts with the five rod proteins (Fig. 3, lane 6 of each panel), but these interactions did not occur when SltFΔ95 was used in the assay (Fig. 3, lane 7 for each panel). When the mutant versions SltFΔ47 and SltFΔ48 were tested, no interaction between these proteins and FlgC was detected (Fig. 3C, lanes 8 and 9). However, both SltF mutants were able to interact with FliE and FlgB (Fig. 3A and B), as well as with FlgF and FlgG (Fig. 3D and E), but SltFΔ47 showed a strong preference for FlgF and SltFΔ48 for FlgG. These results are in agreement with the results observed using the far-Western assay, but the sensitivity of the coimmunoprecipitation assay seems to be higher. The interaction assays performed suggest that the C terminus of SltF supports the interaction with the structural rod proteins and that the interacting regions for most of the rod proteins are not redundant.
FIG 3.
Interactions between the rod proteins and SltF. Coimmunoprecipitation of the structural rod proteins FliE (A), FlgB (B), FlgC (C), FlgF (D), and FlgG (E) in the presence of wild type SltF and the mutant versions of SltFΔ95, SltFΔ47 and SltFΔ48 was performed as described in Materials and Methods. The assays were performed with anti-FliE (A), anti-FlgB (B), anti-FlgC (C), anti-FlgF (D), and anti-FlgG (E). All purified proteins were used at a concentration of 0.3 μM and loaded onto 17.5% SDS-PAGE gels, and anti-His polyclonal antibodies were used to detect the proteins. Molecular mass markers are shown on the right.
Influence of FlgJ on interactions between SltF and rod proteins.
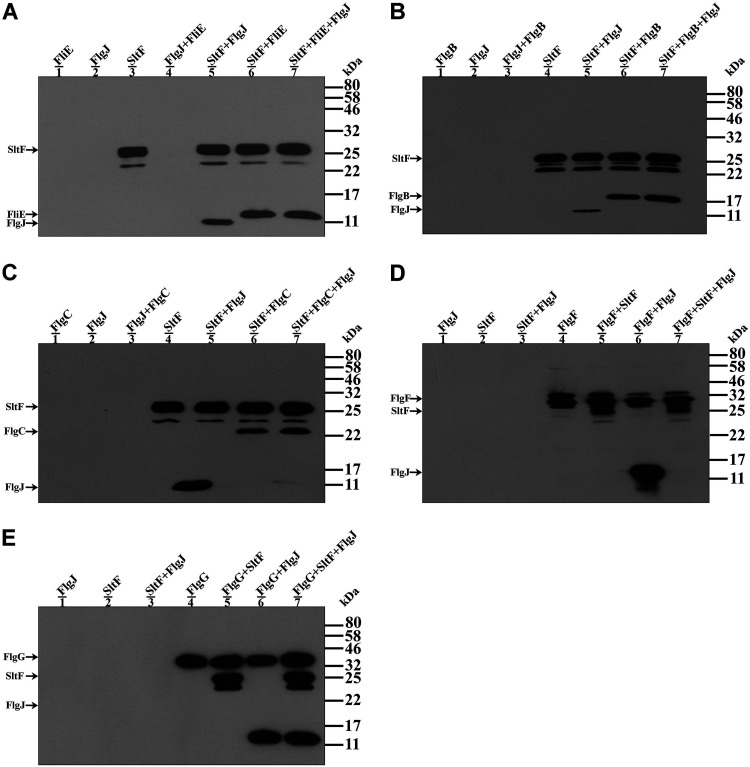
During flagellar biogenesis the rod proteins, the scaffolding protein FlgJ, and the soluble lytic transglycosylase SltF coexist in the periplasmic space, and they probably interact with each other, suggesting that this interaction could recruit SltF, which is exported by the general secretion system, to the growing rod. To analyze whether FlgJ somehow affects the interactions between SltF the rod proteins, we carried out coimmunoprecipitation assays of SltF with the rod proteins FliE, FlgB, and FlgC in the presence of FlgJ. To test the interactions between SltF and the rod proteins FlgF and FlgG in the presence of FlgJ, we used the anti-FlgF or FlgG gamma globulins instead of the anti SltF antibody because in these two cases we detected a high background signal using the latter antibody. The experiments show that the predominant interaction is between SltF and four of the rod proteins (FliE, FlgB, FlgC, and FlgF), as shown in Fig. 4A to D (lanes 7). However, we observed the simultaneous detection of SltF and FlgJ when the rod protein FlgG was immunoprecipitated (Fig. 4E, lane 7). From these results we also detected that the rod proteins FlgF and FlgG are capable of interacting with FlgJ (Fig. 4D and E, lanes 6).
FIG 4.
Interactions between the rod proteins, SltF and FlgJ. (A to C) Coimmunoprecipitations of SltF in the presence of a structural rod protein (FliE, FlgB, and FlgC) and/or FlgJ. The assays were performed using anti-SltF gamma globulins. (D and E) Coimmunoprecipitations of FlgF and FlgG in the presence of SltF and FlgJ. The assays were performed with anti-FlgF and anti-FlgG gamma globulins as described in Materials and Methods. The purified proteins were used at a concentration of 0.3 μM and loaded onto 15 or 17.5% SDS-PAGE gels, and anti-His polyclonal antibodies were used to detect the proteins. Molecular mass markers are shown on the right.
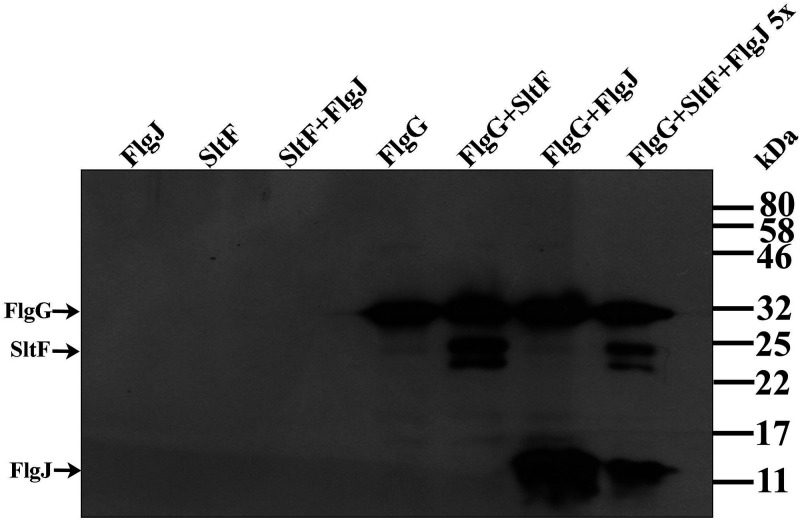
The concomitant precipitation of SltF and FlgJ, together with FlgG, led us to evaluate the interactions between these three proteins by analyzing the effect of including an excess of FlgJ in the assay. When FlgJ is present in a 5-fold excess with respect of SltF and FlgG the three proteins are still coimmunoprecipitated (Fig. 5). This result suggests the existence of a stable ternary complex formed by FlgG/SltF/FlgJ. Alternatively, the affinity of FlgG for FlgJ and SltF could be similar and two different populations of complexes may be present in the reaction. Given that an excess of FlgJ did not affect the apparent equilibrium of the resultant complexes, this explanation seems to be less likely.
FIG 5.
Coimmunoprecipitation of FlgG with SltF in the presence of excess of FlgJ. The proteins were used at a FlgG/SltF/FlgJ molar ratio of 1:1:5. The assays were performed using anti-FlgG gamma globulins as described in Materials and Methods. The purified proteins were used at a concentration of 0.3 or 1.5 μM and loaded onto 17.5% SDS-PAGE gels, and anti-His polyclonal antibodies were used to detect the proteins. Molecular mass markers are shown on the right.
Lytic activity of SltF in the presence of rod proteins and FlgJ.
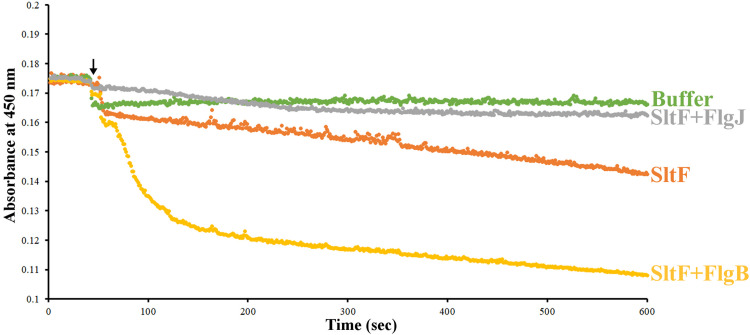
The enzymatic activity of SltF is an important factor during the biogenesis of the flagellum. We therefore tested the effect of the rod proteins and of FlgJ on the enzymatic activity of SltF (Fig. 6). We observed that FlgB stimulates SltF activity by 3.5-fold, whereas the other rod proteins (FliE, FlgC, FlgF, and FlgG) had no effect on SltF activity. The inclusion of FlgJ in these assays showed that this protein by itself inhibits SltF activity; however, this inhibition is relieved by the presence of any rod protein (Table 2). This is in agreement with the coimmunoprecipitation experiments that showed that FlgJ loses its ability to interact with SltF in the presence of the rod proteins FliE, FlgB, FlgC, or FlgF. In the case of the distal rod protein FlgG, which immunoprecipitates SltF and FlgJ simultaneously, the inhibition of SltF activity by FlgJ is also relieved.
FIG 6.
Lytic activity of SltF tested by turbidimetry. The lysis of an insoluble substrate kept in suspension by constant mixing was recorded over time, measuring the absorbance at 450 nm. The arrow indicates the addition of buffer, SltF, SltF plus FlgJ, and SltF plus FlgB. The indicated proteins present in the reaction are in equimolar concentrations. For further details, see Materials and Methods.
TABLE 2.
SltF activity in the absence or presence of rod proteins and scaffolding protein FlgJ
| Sample | ΔOD λ450 nm/s (×10−5) | ±SD (×10−6) | Significancea |
|---|---|---|---|
| Buffer | –1.35 | 1.48 | *** |
| SltF | –3.37 | 2.34 | |
| SltF+FlgJ | –1.03 | 3.69 | *** |
| SltF+FliE | –3.54 | 5.62 | NS |
| SltF+FlgB | –11.9 | 5.12 | *** |
| SltF+FlgC | –3.58 | 3.64 | NS |
| SltF+FlgF | –3.60 | 2.19 | NS |
| SltF+FlgG | –3.47 | 1.00 | NS |
| SltF+FliE+FlgJ | –3.38 | 4.78 | NS |
| SltF+FlgB+FlgJ | –11.2 | 10.3 | *** |
| SltF+FlgC+FlgJ | –3.39 | 1.14 | NS |
| SltF+FlgF+FlgJ | –3.32 | 3.94 | NS |
| SltF+FlgG+FlgJ | –3.53 | 0.762 | NS |
That is, the relationship between the SltF ΔOD and the different samples. ***, P < 0.001; NS, no statistical significance.
DISCUSSION
During the assembly of the flagellar rod, the peptidoglycan layer needs to be remodeled to allow the passage of the growing axial structure. In S. enterica and E. coli, the bidomain protein FlgJ achieves this function through its C-terminal domain, whereas its N-terminal domain acts as a scaffold for the assembly of the rod. In R. sphaeroides, these two domains are split in two different polypeptides, i.e., FlgJ monodomain and the soluble lytic transglycosylase, SltF. It has been demonstrated that SltF is exported by the general secretion system, and the question about how this protein is regulated in a spatiotemporal manner arises. Here, we demonstrate that sltF is expressed along with the early genes of the flagellar hierarchy. The genes of this class include those encoding the flagellar components of the rod that is assembled in the periplasmic space and the hook. Therefore, SltF would be present in the periplasm simultaneously with the rod subunits. It is known that the genes of this class are transcribed by action of the activator proteins FleQ and FleT, along with the RNA polymerase associated with the σ54 factor. In agreement, our results show that the expression of sltF is dependent on FleQ. Even though there is the possibility that sltF could also be expressed from the σ28 promoter located upstream of flgK, therefore, the relevance of expressing sltF from this promoter remains to be determined.
Once SltF is delivered to the periplasmic space via the Sec pathway, we previously proposed that SltF could be recruited to the flagellar rod by its interaction with FlgJ (41, 48). However, a recent report shows that SltF also interacts with the rod proteins FlgB and FlgF (50). In the present study, we extend this observation and provide evidence indicating that SltF can interact with all the structural rod proteins. The multiple interactions in which SltF is involved suggest that different aspects of the functioning of this protein could also be modulated, i.e., recruitment to the flagellar rod, its enzymatic activity, and its possible ride along the growing rod. Importantly, the C terminus of SltF appears to be central for many of these functions. The domain comprises a 95-amino-acid (aa) segment that is essential for the interaction with FlgJ. We determined that the interaction of SltF with the rod proteins is also dependent on this region. It was previously shown that the SltF mutants used here did not restore the swimming phenotype of an sltF mutant strain (41, 49), although the proteins were stable and also retained their enzymatic activity in vitro. Therefore, the ability of SltF to interact with all the different flagellar components is located in its C terminus, and it is required for the proper assembly of the flagellar rod. To learn whether discrete segments of this region are engaged in the specific recognition of its partners, we divided the C-terminal region into two segments, one of 47 residues proximal to the enzymatic domain and a distal segment of 48 aa. Interactions with these mutant versions of SltF revealed that FliE and FlgB are the only proteins that can interact with either of the two C-terminal segments of SltF. This result suggests that these regions could share a repeated motif that could be recognized by these proteins; in agreement with this possibility, the analysis of the last 95 aa from SltF using RADAR software (51) revealed a small imperfect repeat that could explain why FliE and FlgB bind both SltFΔ47 and SltFΔ48 (RGGSRLAEAATDLPDEATL and QGPLELRAAAPDLLAEADL). The proper binding of FlgC requires the integrity of the C-terminal region, suggesting that its interaction site could be around the point in which the 95 residues of the C terminus were divided to obtain SltFΔ47 and SltFΔ48. On the other hand, FlgF preferentially interacts with the distal region of SltF, whereas FlgG mainly interacts with the proximal region; these results suggest that the interaction sites located in the C-terminal domain of SltF are not redundant.
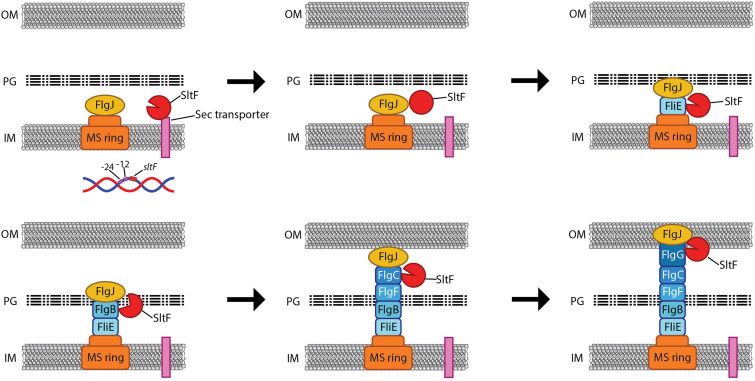
Even in the most thoroughly studied bacterial models, there are many unknown aspects regarding the process of the secretion and assembly of the rod components, including the mechanism of action of FlgJ. Since the rod subunits are secreted through the flagellar secretion system, it is conceivable that once the secreted polypeptides reach the tip of the growing rod, they would be assembled in the proper order probably determined by the previous assembled structure, whereas free rod and hook subunits may be unstable in the periplasm, and they may not be significantly accumulated (52, 53). In this scenario, SltF could be recruited by the proteins of the growing rod. Initially, SltF will interact with FliE, and its enzymatic activity could be moderated; therefore, to efficiently reach the PG, it is possible that SltF could interact with the rod proteins as they are incorporated, and in this manner SltF would ride along the rod during its assembly. It is conceivable that the enzymatic activity of SltF could increase transiently at early stages of flagellar formation, and this situation could be coincident with the presence of FlgB. Subsequently, it is possible that SltF changes its interacting partner for the next rod protein to be assembled, bringing about a reduction in its hydrolytic activity that, according to our assays, seems to be constant after the initial activation by FlgB. During the assembly of the distal rod, both SltF and FlgJ may interact with FlgG, and it is possible that at this point SltF could be released out of the cell, as was reported for FlgJ in S. enterica (28). However, it is also possible that the protein is degraded in the periplasm. In this regard, it has been demonstrated that SltFΔ48 is ∼4-fold more stable than SltF, suggesting that the C-terminal region is required not only to interact with the rod proteins and FlgJ but also to induce a short half-life of the protein (49). A model summarizing the main features of this proposal is shown in Fig. 7.
FIG 7.
Model for the possible role of SltF and FlgJ during rod assembly. The scheme shows the internal membrane (IM), the peptidoglycan layer (PG), and the outer membrane (OM). Arrows indicate the sequence of the events taking place during the assembly of the rod. The rod proteins and FlgJ are secreted to the periplasmic space via the flagellar export apparatus and might remain associated with the growing structure, while SltF is secreted through the Sec pathway. Once in the periplasmic space, SltF can recognize the rod proteins and FlgJ. Interaction with FlgJ inhibits its hydrolytic activity (closed red circle, second panel), and interaction with FlgB promotes its activity (open circle, fourth panel). These interactions ensure that the lytic enzyme is under spatial and temporal control and modulate its enzymatic activity in response to the progress of the growing rod.
In addition, our results confirm that SltF interacts with FlgJ, and reveal that this interaction is dependent on the presence of the 47 residues proximal to the catalytic domain of SltF (41). Importantly, we also observed that the SltF-FlgJ interaction is displaced by the presence of the rod proteins except for FlgG. Therefore, it is possible that the SltF-FlgJ interaction could be very short-lived and displaced by a rod subunit. This arises the relevant question on how FlgJ accomplishes its role as a scaffold for the assembly of the rod proteins. It is also important to consider that the axial proteins are possibly exported in an unfolded state and folded upon assembly (8, 54). There are intermediary structural states that could prove advantageous in the interaction with the various axial components; hence, FlgJ could interact strongly with a prefolded state of the protein but not with a completely folded polypeptide.
Alternatively, FlgJ could be recruited after the completion of the proximal rod, and in this way this protein would serve as a scaffold only for FlgG (28). If this notion is correct, it would agree with the possibility that FlgG is the only protein that could interact simultaneously with SltF and FlgJ.
Finally, it should be stressed that similar mechanisms of transient interactions occur for the assembly of other protein complexes that expand through the bacterial cell envelope and require the activity of lytic enzymes (55). For instance, the type III secretion system (T3SS) of enteropathogenic E. coli requires the periplasmic protein EtgA (56), which acts as a lytic transglycosylase, and its activity is exacerbated by interacting with the axial component EscI of the T3SS (57). A similar situation has been reported for other lytic enzymes involved in the assembly of type IV and type VI secretion systems of other bacteria (37, 58–61), suggesting that this type of control might be extensive to other secretion systems.
MATERIALS AND METHODS
Bacterial strains, plasmids, sequences, and growth conditions.
The bacterial strains and plasmids used here are listed in Table 3. R. sphaeroides was grown photoheterotrophically at 30°C in Sistrom minimal medium (62). Heterotrophic cultures were grown aerobically in flasks and incubated with orbital shaking; photoheterotrophic cultures were grown in completely filled screw-cap tubes under illumination. E. coli was grown in LB medium (63) at 30 or 37°C, with orbital shaking. When necessary, nalidixic acid (20 μg/ml), kanamycin (25 μg/ml), gentamicin (5 μg/ml), and tetracycline (1 μg/ml) were used for R. sphaeroides. For E. coli, spectinomycin (50 μg/ml), tetracycline (25 μg/ml), ampicillin (200 μg/ml), kanamycin (25 μg/ml), gentamicin (30 μg/ml), and chloramphenicol (25 μg/ml) were included in the culture medium when necessary.
TABLE 3.
Strains and plasmids used in this study
| Strains | Relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| TOP10 | F– mcrA Δ(mrr-hsdRMS-mcrBC)ϕ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| M15(pREP4) | thi lac ara gal mtl F′ recA+ uvr+ lon+; pREP4 plasmid; Kanr | Qiagen |
| BL21(DE3)pLysS | F– ompT hsdSB(rB– mB–) gal dcm (DE3)/pLysS Cmr | Novagen |
| S17-1 | recA endA thi hsdR RP4-2-Tc::Mu-Kan::Tn7 Tpr Smr | 66 |
| R. sphaeroides | ||
| WS8N | Wild type; spontaneous Nalr | 64 |
| SltF1 | WS8N derivative ΔsltF(1-336)::aadA Fla– Spcr Nalr | 48 |
| SP13 | WS8N derivative fleQΔ::Kan | 42 |
| Plasmids | ||
| pTZ19R | Cloning vector; Apr ; pUC derivative | Pharmacia |
| pRK415 | pRK404 derivative, for expression in R. sphaeroides, lacZ mob+ Tcr | 65 |
| pBBMCS53 | Transcriptional uidA fusion vector; Gmr | 67 |
| pET19-b | Expression vector 10×His N terminal; Apr | Novagen |
| pQE30 | Expression vector 6×His N terminal; Apr | Qiagen |
| pQE60 | Expression vector 6×His C terminal; Apr | Qiagen |
| pGT001 | 1.4-kb PstI fragment containing sltF wild type cloned into pTZ19R | 40 |
| pRKSltF | sltF wild type cloned into the EcoRI/HindIII sites of pRK415 | 48 |
| pRKrevSltF | sltF wild type cloned into the HindIII/EcoRI sites of pRK415 | This study |
| pBG313 | 3.8 kb BamHI fragment from WS8-N cloned in pTZ19R | 40 |
| Up120sltFprev | pBBMCS53 derivative, sltFp3′-5′–uidA gene fusion | This study |
| Up120sltFpfw | pBBMCS53 derivative, sltFp5′-3′–uidA gene fusion | This study |
| pRS1sltF | sltF cloned into the SacI/HindIII sites of pQE30 | 49 |
| pQE30sltFΔ47 | ΔsltF(510-651) cloned into the SacI/HindIII sites of pQE30 | 41 |
| pQE30sltFΔ48/Δ6 | ΔsltF(651-795) cloned into the SacI/HindIII sites of pQE30 | 49 |
| pQE30sltFΔ95 | ΔsltF(510-795) cloned into the SacI/HindIII sites of pQE30 | 41 |
| pRSJ | flgJ wild type cloned into the NcoI/BglII sites of pQE60 | 48 |
| pRSFliE | fliE wild type cloned into the SacI/HindIII sites of pQE30 | 71 |
| pRSFlgB | flgB wild type cloned into the BamHI/HindIII sites of pQE30 | 71 |
| pRSFlgC | flgC wild type cloned into the NdeI/BamHI sites of pET19-b | 71 |
| pRSFlgF | flgF wild type cloned into the KpnI/HindIII sites of pQE30 | 71 |
| pRSFlgG | flgG wild type cloned into the NdeI/BamHI sites of pET19-b | 71 |
| pRSY5 | cheY5 wild type cloned into the BamHI/PstI sites of pQE30 | 72 |
Str, streptomycin; Ap, ampicillin; Nal, nalidixic acid; Kan, kanamycin; Gm, gentamicin; Spc, spectinomycin; Tc, tetracycline; Cm, chloramphenicol.
Recombinant DNA techniques.
Standard molecular biology techniques were used for the isolation and purification of DNA from R. sphaeroides WS8N (63, 64). Plasmid DNA and PCR fragments were purified by using QIAprep spin and QIAquick PCR kits, respectively (Qiagen GmbH, Germany).
Complementation assays.
The sltF mutant was complemented with plasmids pRKSltF and pRKrevSltF carrying a 1.4-kb PstI DNA fragment obtained from chromosomal DNA. This fragment contains 120 bp upstream of the start of sltF, and it was cloned in two orientations in the multicloning site (MCS) of pTZ19R and subcloned in the restriction sites HindIII and EcoRI of pRK415 (65) (Table 3). Each clone was introduced in the sltF mutant by diparental conjugation using the E. coli strain S17-1 (66).
Conjugation.
Plasmids were introduced into a SltF1 mutant strain by diparental conjugation using E. coli strain S17-1 as described previously (66). Briefly, cultures of E. coli carrying the selected plasmid and R. sphaeroides were grown overnight with orbital shaking at 200 rpm. For E. coli, 50 μl of the overnight culture of was inoculated to 5 ml of LB broth, and growth continued until an optical density at 600 nm (OD600) of 0.5 was reached. For R. sphaeroides, 100 μl of the overnight culture was inoculated into 10 ml of Sistrom medium, and growth was continued until an OD600 of 0.5 was reached. Cultures were centrifuged at 3,000 × g for 5 min, and the pellets were resuspended in 0.5 ml of LB medium. The cells were mixed, placed on nitrocellulose filters on LB medium, and incubated for 16 h. The cells were collected and plated on the suitable selective medium.
Motility assays.
Swimming plates were prepared as described previously (49), spotted with 2 μl of a stationary-phase culture, and incubated aerobically in the dark at 30°C. Swimming capacity was recorded as the ability of bacteria to move away from the inoculation point after 24 h of incubation. Plates were prepared with soft agar (0.25%) in Sistrom minimal medium devoid of succinic acid to which 100 μM sodium propionate was added.
Transcriptional fusion of sltF with uidA.
The transcriptional fusion between the reporter gene uidA and the sltF promoter (sltFp) was carried out using plasmid pBBMCS53 (67). This plasmid contains an MCS upstream of the reporter gene uidA gene that encodes the β-glucuronidase enzyme. The fragment containing sltFp was obtained from plasmid pBG313 (40) by subcloning a 462-bp PstI-ClaI fragment that includes 120 bp upstream of the gene and runs down halfway into the gene. The fragment was cloned in pBBMCS53 in both directions with respect to the reporter gene uidA. These constructions were introduced by conjugation using E. coli S17-1 into the wild-type strain WS8N as well as in the SP13 (ΔfleQ) mutant strain (42).
β-Glucuronidase assays.
Cell-free extracts from exponential phase cultures grown heterotrophically were tested for β-glucuronidase activity according to a previously reported protocol (68, 69). A curve with different concentrations of 4-methyl-umbelliferone (Sigma-Aldrich, St. Louis, MO) was used as the standard. Specific activities are expressed as nanomoles of 4-methyl-umbelliferone formed per minute per milligram of protein. Protein was determined using bovine serum albumin as standard and the method described by Lowry (70).
Overexpression and purification of SltF, FlgJ, SltFΔ47, SltFΔ48, SltFΔ95, and the rod proteins FliE, FlgB, FlgC, FlgF, and FlgG.
Overexpression and purification of His-tagged versions of SltF and FlgJ was carried out as described previously (48). His-tagged versions of SltFΔ47 and SltFΔ95 were overexpressed and purified from E. coli M15(pREP4) as previously reported (41). A His-tagged version of SltFΔ48 was overexpressed and purified similarly to SltFΔ95. His-tagged versions of the rod proteins were overexpressed and purified as described previously (71). A His-tagged version of CheY5 was purified as described previously (72) and included as a negative control. Protein was determined by using bovine serum albumin as a standard (70).
Affinity blotting.
For each protein sample, 0.15-nmol samples were loaded onto 15 and 17.5% SDS-polyacrylamide gels and transferred electrophoretically to nitrocellulose membranes (Bio-Rad, Richmond, CA) and treated as described previously (41). The membranes were probed with SltF, SltFΔ47, SltFΔ48, and SltFΔ95 at a concentration of 3 μg/ml in Tris-buffered saline containing 20 mM Tris-HCl (pH 7.5), 0.01% Tween 20, 500 mM NaCl, and 5% non-fat milk for 1 h at room temperature, followed by incubation with anti-SltF gamma globulins at a 1:10,000 dilution as described previously (48). Detection was carried out using a Thermo SuperSignal kit (Thermo Scientific, Waltham, MA).
Coimmunoprecipitation.
A 25-μl aliquot from a 25-mg/ml stock solution of Sepharose CL-4B was coupled to protein A (Sigma-Aldrich) (48). Once the resin was coupled with the protein A and washed, it was incubated with 5 μg of specific gamma globulins directed against each of the purified rod proteins in 1 ml of 20 mM Tris buffer (pH 7.6) for 14 h at 4°C (71). The mixture was centrifuged at 12,000 × g for 10 min, and the supernatant was carefully discarded. The different versions of SltF at a concentration of 0.3 μM were incubated with Sepharose beads for 1 h at 4°C. The samples were washed four to six times with 1 ml of Tris buffer, and the resulting pellet was resuspended in 30 μl of sample buffer and then boiled for 10 min. Samples were subjected to 17.5% SDS-PAGE, transferred to nitrocellulose membranes, and developed using HisProbe-HRP (Thermo Scientific) at a 1:10,000 dilution.
Competence coimmunoprecipitation.
Coimmunoprecipitation assays of wild-type SltF and FlgJ were carried out in the absence or presence of each of the rod proteins (FliE, FlgB, FlgC, FlgF, and FlgG). For this, a 25-μl aliquot of Sepharose-protein A coupled to 3 μg of anti-SltF gamma globulins were used to detect the interactions with FliE, FlgB, and FlgC. For the interactions between SltF, FlgJ, and FlgF, 6 μg of anti-FlgF gamma globulins was used. For the interactions between SltF, FlgJ, and FlgG, 1.5 μg of anti-FlgG gamma globulins was used. To immunoprecipitate the protein complexes, SltF, FlgJ, and the specific rod protein, suitable for each case, were preincubated at a concentration of 0.15 μM for 1 h at 4°C. This mixture was added to the antibody-bound Sepharose beads, followed by incubation for 1 h at 4°C. The tubes were centrifuged at 12,000 × g for 5 min, and the supernatant was carefully discarded. This washing procedure was repeated four to six times, and the pellet was resuspended in 30 μl of a buffer containing 65 mM Tris-HCl, 10% (vol/vol) glycerol, 2% SDS, and 0.01% bromophenol blue (pH 6.8) and boiled for 10 min. Samples were subjected to SDS-PAGE (15 or 17.5%), transferred to nitrocellulose membranes, and probed against HisProbe-HRP. Horseradish peroxidase (HRP) activity was detected by chemiluminescence.
The interaction between FliG, SltF, and FlgJ was challenged by including a 5-fold molar excess of FlgJ in the assays, and the complexes were immunoprecipitated using 1.5 μg of anti-FlgG gamma globulins.
Muramidase activity determination.
A 0.2-mg/ml suspension of a lyophilized cell extract of Micrococcus lysodeikticus (Sigma-Aldrich) in 50 mM Na2HPO4 (pH 6.7) was used to carry out spectrophotometric analyses of enzymatic activity of SltF, including or not the rod proteins and FlgJ. The measurements were performed at a wavelength of 450 nm with constant agitation at 30°C in a cell containing 2 ml of the suspension and 0.5 mmol of the desired protein. Egg lysozyme (0.5 μg) was used as a positive control. The statistical analysis was carried out using R project (https://www.r-project.org/) and analysis of variance to estimate the statistical significance.
ACKNOWLEDGMENTS
This study was partially supported by DGAPA-PAPIIT grant IG200420.
We thank Aurora Osorio for excellent technical support and the Molecular Biology Unit IFC-UNAM for sequencing facilities.
This study is dedicated to the memory of H. Celis.
Contributor Information
Laura Camarena, Email: rosal@servidor.unam.mx.
Georges Dreyfus, Email: gdreyfus@ifc.unam.mx.
Conrad W. Mullineaux, Queen Mary University of London
REFERENCES
- 1.Szurmant H, Ordal GW. 2004. Diversity in chemotaxis mechanisms among the bacteria and archaea. Microbiol Mol Biol Rev 68:301–319. 10.1128/MMBR.68.2.301-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carroll BL, Liu J. 2020. Structural conservation and adaptation of the bacterial flagella motor. Biomolecules 10:1492. 10.3390/biom10111492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ueno T, Oosawa K, Aizawa S. 1992. M ring, S ring, and proximal rod of the flagellar basal body of Salmonella Typhimurium are composed of subunits of a single protein, FliF. J Mol Biol 227:672–677. 10.1016/0022-2836(92)90216-7. [DOI] [PubMed] [Google Scholar]
- 4.Jones CJ, Macnab RM, Okino H, Aizawa S. 1990. Stoichiometric analysis of the flagellar hook-(basal-body) complex of Salmonella Typhimurium. J Mol Biol 212:377–387. 10.1016/0022-2836(90)90132-6. [DOI] [PubMed] [Google Scholar]
- 5.Makishima S, Komoriya K, Yamaguchi S, Aizawa SI. 2001. Length of the flagellar hook and the capacity of the type III export apparatus. Science 291:2411–2413. 10.1126/science.1058366. [DOI] [PubMed] [Google Scholar]
- 6.Suzuki H, Yonekura K, Namba K. 2004. Structure of the rotor of the bacterial flagellar motor revealed by electron cryomicroscopy and single-particle image analysis. J Mol Biol 337:105–113. 10.1016/j.jmb.2004.01.034. [DOI] [PubMed] [Google Scholar]
- 7.Macnab RM. 2003. How bacteria assemble flagella. Annu Rev Microbiol 57:77–100. 10.1146/annurev.micro.57.030502.090832. [DOI] [PubMed] [Google Scholar]
- 8.Yonekura K, Maki-Yonekura S, Namba K. 2003. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature 424:643–650. 10.1038/nature01830. [DOI] [PubMed] [Google Scholar]
- 9.Yonekura K, Maki S, Morgan DG, DeRosier DJ, Vonderviszt F, Imada K, Namba K. 2000. The bacterial flagellar cap as the rotary promoter of flagellin self-assembly. Science 290:2148–2152. 10.1126/science.290.5499.2148. [DOI] [PubMed] [Google Scholar]
- 10.Maki-Yonekura S, Yonekura K, Namba K. 2003. Domain movements of HAP2 in the cap-filament complex formation and growth process of the bacterial flagellum. Proc Natl Acad Sci USA 100:15528–15533. 10.1073/pnas.2534343100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Irikura VM, Kihara M, Yamaguchi S, Sockett H, Macnab RM. 1993. Salmonella Typhimurium fliG and fliN mutations causing defects in assembly, rotation, and switching of the flagellar motor. J Bacteriol 175:802–810. 10.1128/jb.175.3.802-810.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sockett H, Yamaguchi S, Kihara M, Irikura VM, Macnab RM. 1992. Molecular analysis of the flagellar switch protein FliM of Salmonella Typhimurium. J Bacteriol 174:793–806. 10.1128/jb.174.3.793-806.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sircar R, Borbat PP, Lynch MJ, Bhatnagar J, Beyersdorf MS, Halkides CJ, Freed JH, Crane BR. 2015. Assembly states of FliM and FliG within the flagellar switch complex. J Mol Biol 427:867–886. 10.1016/j.jmb.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson S, Fong YH, Deme JC, Furlong EJ, Kuhlen L, Lea SM. 2020. Symmetry mismatch in the MS ring of the bacterial flagellar rotor explains the structural coordination of secretion and rotation. Nat Microbiol 5:966–975. 10.1038/s41564-020-0703-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lynch MJ, Levenson R, Kim EA, Sircar R, Blair DF, Dahlquist FW, Crane BR. 2017. Co-folding of a FliF-FliG split domain forms the basis of the MS:C ring interface within the bacterial flagellar motor. Structure 25:317–328. 10.1016/j.str.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levenson R, Zhou H, Dahlquist FW. 2012. Structural insights into the interaction between the bacterial flagellar motor proteins FliF and FliG. Biochemistry 51:5052–5060. 10.1021/bi3004582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xue C, Lam KH, Zhang H, Sun K, Lee SH, Chen X, Au SWN. 2018. Crystal structure of the FliF-FliG complex from Helicobacter pylori yields insight into the assembly of the motor MS-C ring in the bacterial flagellum. J Biol Chem 293:2066–2078. 10.1074/jbc.M117.797936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muller V, Jones CJ, Kawagishi I, Aizawa S, Macnab RM. 1992. Characterization of the fliE genes of Escherichia coli and Salmonella Typhimurium and identification of the FliE protein as a component of the flagellar hook-basal body complex. J Bacteriol 174:2298–2304. 10.1128/jb.174.7.2298-2304.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Minamino T, Yamaguchi S, Macnab RM. 2000. Interaction between FliE and FlgB, a proximal rod component of the flagellar basal body of Salmonella. J Bacteriol 182:3029–3036. 10.1128/JB.182.11.3029-3036.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Homma M, Kutsukake K, Hasebe M, Iino T, Macnab RM. 1990. FlgB, FlgC, FlgF, and FlgG: a family of structurally related proteins in the flagellar basal body of Salmonella Typhimurium. J Mol Biol 211:465–477. 10.1016/0022-2836(90)90365-S. [DOI] [PubMed] [Google Scholar]
- 21.Kubori T, Shimamoto N, Yamaguchi S, Namba K, Aizawa S. 1992. Morphological pathway of flagellar assembly in Salmonella Typhimurium. J Mol Biol 226:433–446. 10.1016/0022-2836(92)90958-m. [DOI] [PubMed] [Google Scholar]
- 22.Chevance FF, Takahashi N, Karlinsey JE, Gnerer J, Hirano T, Samudrala R, Aizawa S, Hughes KT. 2007. The mechanism of outer membrane penetration by the eubacterial flagellum and implications for spirochete evolution. Genes Dev 21:2326–2335. 10.1101/gad.1571607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okino H, Isomura M, Yamaguchi S, Magariyama Y, Kudo S, Aizawa SI. 1989. Release of flagellar filament-hook-rod complex by a Salmonella Typhimurium mutant defective in the M ring of the basal body. J Bacteriol 171:2075–2082. 10.1128/jb.171.4.2075-2082.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nambu T, Minamino T, Macnab RM, Kutsukake K. 1999. Peptidoglycan-hydrolyzing activity of the FlgJ protein, essential for flagellar rod formation in Salmonella Typhimurium. J Bacteriol 181:1555–1561. 10.1128/JB.181.5.1555-1561.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirano T, Minamino T, Macnab RM. 2001. The role in flagellar rod assembly of the N-terminal domain of Salmonella FlgJ, a flagellum-specific muramidase. J Mol Biol 312:359–369. 10.1006/jmbi.2001.4963. [DOI] [PubMed] [Google Scholar]
- 26.Herlihey FA, Moynihan PJ, Clarke AJ. 2014. The essential protein for bacterial flagella formation FlgJ functions as a β-N-acetylglucosaminidase. J Biol Chem 289:31029–31042. 10.1074/jbc.M114.603944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DePamphilis ML. 1971. Dissociation and reassembly of Escherichia coli outer membrane and of lipopolysaccharide, and their reassembly onto flagellar basal bodies. J Bacteriol 105:1184–1199. 10.1128/jb.105.3.1184-1199.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cohen EJ, Hughes KT. 2014. Rod-to-hook transition for extracellular flagellum assembly is catalyzed by the L-ring-dependent rod scaffold removal. J Bacteriol 196:2387–2395. 10.1128/JB.01580-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katayama E, Shiraishi T, Oosawa K, Baba N, Aizawa S. 1996. Geometry of the flagellar motor in the cytoplasmic membrane of Salmonella Typhimurium as determined by stereo-photogrammetry of quick-freeze deep-etch replica images. J Mol Biol 255:458–475. 10.1006/jmbi.1996.0038. [DOI] [PubMed] [Google Scholar]
- 30.Makino F, Shen D, Kajimura N, Kawamoto A, Pissaridou P, Oswin H, Pain M, Murillo I, Namba K, Blocker AJ. 2016. The architecture of the cytoplasmic region of type III secretion systems. Sci Rep 6:33341. 10.1038/srep33341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Emerson SU, Tokuyasu K, Simon MI. 1970. Bacterial flagella: polarity of elongation. Science 169:190–192. 10.1126/science.169.3941.190. [DOI] [PubMed] [Google Scholar]
- 32.Iino T. 1969. Polarity of flagellar growth in salmonella. J Gen Microbiol 56:227–239. 10.1099/00221287-56-2-227. [DOI] [PubMed] [Google Scholar]
- 33.Meroueh SO, Bencze KZ, Hesek D, Lee M, Fisher JF, Stemmler TL, Mobashery S. 2006. Three-dimensional structure of the bacterial cell wall peptidoglycan. Proc Natl Acad Sci USA 103:4404–4409. 10.1073/pnas.0510182103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scheurwater EM, Burrows LL. 2011. Maintaining network security: how macromolecular structures cross the peptidoglycan layer. FEMS Microbiol Lett 318:1–9. 10.1111/j.1574-6968.2011.02228.x. [DOI] [PubMed] [Google Scholar]
- 35.Dijkstra AJ, Keck W. 1996. Peptidoglycan as a barrier to transenvelope transport. J Bacteriol 178:5555–5562. 10.1128/jb.178.19.5555-5562.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fisher JF, Meroueh SO, Mobashery S. 2006. Nanomolecular and supramolecular paths toward peptidoglycan structure. Microbe 1:420–427. 10.1128/microbe.1.420.1. [DOI] [Google Scholar]
- 37.Koraimann G. 2003. Lytic transglycosylases in macromolecular transport systems of Gram-negative bacteria. Cell Mol Life Sci 60:2371–2388. 10.1007/s00018-003-3056-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zahrl D, Wagner M, Bischof K, Bayer M, Zavecz B, Beranek A, Ruckenstuhl C, Zarfel GE, Koraimann G. 2005. Peptidoglycan degradation by specialized lytic transglycosylases associated with type III and type IV secretion systems. Microbiology (Reading) 151:3455–3467. 10.1099/mic.0.28141-0. [DOI] [PubMed] [Google Scholar]
- 39.Nambu T, Inagaki Y, Kutsukake K. 2006. Plasticity of the domain structure in FlgJ, a bacterial protein involved in flagellar rod formation. Genes Genet Syst 81:381–389. 10.1266/ggs.81.381. [DOI] [PubMed] [Google Scholar]
- 40.Gonzalez-Pedrajo B, de la Mora J, Ballado T, Camarena L, Dreyfus G. 2002. Characterization of the flgG operon of Rhodobacter sphaeroides WS8 and its role in flagellum biosynthesis. Biochim Biophys Acta 1579:55–63. 10.1016/s0167-4781(02)00504-3. [DOI] [PubMed] [Google Scholar]
- 41.Garcia-Ramos M, de la Mora J, Ballado T, Camarena L, Dreyfus G. 2018. Biochemical and phylogenetic study of SltF, a flagellar lytic transglycosylase from Rhodobacter sphaeroides. J Bacteriol 200. 10.1128/JB.00397-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poggio S, Osorio A, Dreyfus G, Camarena L. 2005. The flagellar hierarchy of Rhodobacter sphaeroides is controlled by the concerted action of two enhancer-binding proteins. Mol Microbiol 58:969–983. 10.1111/j.1365-2958.2005.04900.x. [DOI] [PubMed] [Google Scholar]
- 43.Barrios H, Valderrama B, Morett E. 1999. Compilation and analysis of σ54-dependent promoter sequences. Nucleic Acids Res 27:4305–4313. 10.1093/nar/27.22.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang N, Darbari VC, Glyde R, Zhang X, Buck M. 2016. The bacterial enhancer-dependent RNA polymerase. Biochem J 473:3741–3753. 10.1042/BCJ20160741C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poggio S, Osorio A, Dreyfus G, Camarena L. 2002. The four different σ54 factors of Rhodobacter sphaeroides are not functionally interchangeable. Mol Microbiol 46:75–85. 10.1046/j.1365-2958.2002.03158.x. [DOI] [PubMed] [Google Scholar]
- 46.Poggio S, Osorio A, Dreyfus G, Camarena L. 2006. Transcriptional specificity of RpoN1 and RpoN2 involves differential recognition of the promoter sequences and specific interaction with the cognate activator proteins. J Biol Chem 281:27205–27215. 10.1074/jbc.M601735200. [DOI] [PubMed] [Google Scholar]
- 47.Wilkinson DA, Chacko SJ, Venien-Bryan C, Wadhams GH, Armitage JP. 2011. Regulation of flagellum number by FliA and FlgM and role in biofilm formation by Rhodobacter sphaeroides. J Bacteriol 193:4010–4014. 10.1128/JB.00349-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de la Mora J, Ballado T, Gonzalez-Pedrajo B, Camarena L, Dreyfus G. 2007. The flagellar muramidase from the photosynthetic bacterium Rhodobacter sphaeroides. J Bacteriol 189:7998–8004. 10.1128/JB.01073-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de la Mora J, Osorio-Valeriano M, Gonzalez-Pedrajo B, Ballado T, Camarena L, Dreyfus G. 2012. The C terminus of the flagellar muramidase SltF modulates the interaction with FlgJ in Rhodobacter sphaeroides. J Bacteriol 194:4513–4520. 10.1128/JB.00460-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Herlihey FA, Osorio-Valeriano M, Dreyfus G, Clarke AJ. 2016. Modulation of the lytic activity of the dedicated autolysin for flagellum formation SltF by flagellar rod proteins FlgB and FlgF. J Bacteriol 198:1847–1856. 10.1128/JB.00203-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heger A, Holm L. 2000. Rapid automatic detection and alignment of repeats in protein sequences. Proteins 41:224–237. . [DOI] [PubMed] [Google Scholar]
- 52.Komoriya K, Shibano N, Higano T, Azuma N, Yamaguchi S, Aizawa SI. 1999. Flagellar proteins and type III-exported virulence factors are the predominant proteins secreted into the culture media of Salmonella Typhimurium. Mol Microbiol 34:767–779. 10.1046/j.1365-2958.1999.01639.x. [DOI] [PubMed] [Google Scholar]
- 53.Bonifield HR, Yamaguchi S, Hughes KT. 2000. The flagellar hook protein, FlgE, of Salmonella enterica serovar Typhimurium is posttranscriptionally regulated in response to the stage of flagellar assembly. J Bacteriol 182:4044–4050. 10.1128/JB.182.14.4044-4050.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee PC, Rietsch A. 2015. Fueling type III secretion. Trends Microbiol 23:296–300. 10.1016/j.tim.2015.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vollmer W, Blanot D, de Pedro MA. 2008. Peptidoglycan structure and architecture. FEMS Microbiol Rev 32:149–167. 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- 56.Garcia-Gomez E, Espinosa N, de la Mora J, Dreyfus G, Gonzalez-Pedrajo B. 2011. The muramidase EtgA from enteropathogenic Escherichia coli is required for efficient type III secretion. Microbiology (Reading) 157:1145–1160. 10.1099/mic.0.045617-0. [DOI] [PubMed] [Google Scholar]
- 57.Burkinshaw BJ, Deng W, Lameignere E, Wasney GA, Zhu H, Worrall LJ, Finlay BB, Strynadka NC. 2015. Structural analysis of a specialized type III secretion system peptidoglycan-cleaving enzyme. J Biol Chem 290:10406–10417. 10.1074/jbc.M115.639013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hoppner C, Carle A, Sivanesan D, Hoeppner S, Baron C. 2005. The putative lytic transglycosylase VirB1 from Brucella suis interacts with the type IV secretion system core components VirB8, VirB9 and VirB11. Microbiology (Reading) 151:3469–3482. 10.1099/mic.0.28326-0. [DOI] [PubMed] [Google Scholar]
- 59.Mushegian AR, Fullner KJ, Koonin EV, Nester EW. 1996. A family of lysozyme-like virulence factors in bacterial pathogens of plants and animals. Proc Natl Acad Sci USA 93:7321–7326. 10.1073/pnas.93.14.7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ward DV, Draper O, Zupan JR, Zambryski PC. 2002. Peptide linkage mapping of the Agrobacterium tumefaciens vir-encoded type IV secretion system reveals protein subassemblies. Proc Natl Acad Sci USA 99:11493–11500. 10.1073/pnas.172390299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Santin YG, Cascales E. 2017. Domestication of a housekeeping transglycosylase for assembly of a type VI secretion system. EMBO Rep 18:138–149. 10.15252/embr.201643206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sistrom WR. 1962. The kinetics of the synthesis of photopigments in Rhodopseudomonas sphaeroides. J Gen Microbiol 28:607–616. 10.1099/00221287-28-4-607. [DOI] [PubMed] [Google Scholar]
- 63.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. 1987. Current protocols in molecular biology. John Wiley, New York, NY. [Google Scholar]
- 64.Sockett RE, Foster JCA, Armitage JP. 1990. Molecular biology of the Rhodobacter sphaeroides flagellum. FEMS Symp 53:473–479. [Google Scholar]
- 65.Keen NT, Tamaki S, Kobayashi D, Trollinger D. 1988. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene 70:191–197. 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 66.Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Nat Biotechnol 1:784–745. 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 67.Girard L, Brom S, Davalos A, Lopez O, Soberon M, Romero D. 2000. Differential regulation of fixN-reiterated genes in Rhizobium etli by a novel fixL-fixK cascade. Mol Plant Microbe Interact 13:1283–1292. 10.1094/MPMI.2000.13.12.1283. [DOI] [PubMed] [Google Scholar]
- 68.Jefferson RA, Burgess SM, Hirsh D. 1986. β-Glucuronidase from Escherichia coli as a gene-fusion marker. Proc Natl Acad Sci USA 83:8447–8451. 10.1073/pnas.83.22.8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hernandez-Valle J, Domenzain C, de la Mora J, Poggio S, Dreyfus G, Camarena L. 2017. The master regulators of the Fla1 and Fla2 flagella of Rhodobacter sphaeroides control the expression of their cognate CheY proteins. J Bacteriol 199. 10.1128/JB.00670-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. 1951. Protein measurement with the Folin phenol reagent. J Biol Chem 193:265–275. 10.1016/S0021-9258(19)52451-6. [DOI] [PubMed] [Google Scholar]
- 71.Osorio-Valeriano M, de la Mora J, Camarena L, Dreyfus G. 2016. Biochemical characterization of the flagellar rod components of Rhodobacter sphaeroides: properties and interactions. J Bacteriol 198:544–552. 10.1128/JB.00836-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ferre A, De La Mora J, Ballado T, Camarena L, Dreyfus G. 2004. Biochemical study of multiple CheY response regulators of the chemotactic pathway of Rhodobacter sphaeroides. J Bacteriol 186:5172–5177. 10.1128/JB.186.15.5172-5177.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]