Abstract
Objective:
One of four American cancer patients dies of lung cancer. Environmental factors such as tobacco smoking are known to affect lung cancer risk. However, there is a genetic factor to lung cancer risk as well. Here, we perform parametric linkage analysis on family-based genotype data in an effort to find genetic loci linked to the disease.
Methods:
197 individuals from families with a high risk history of lung cancer were recruited and genotyped using an Illumina array. Parametric linkage analyses were performed using an affected-only phenotype model with an autosomal dominant inheritance using a disease allele frequency of 0.01. Three types of analyses were performed: single variant two-point, collapsed haplotype pattern variant two-point, and multipoint analysis.
Results:
Five novel genome-wide significant loci were identified at 18p11.23, 2p22.2, 14q13.1, 16p13, and 20q13.11. The families most informative for linkage were also determined.
Conclusions:
The five novel signals are good candidate regions, containing genes that have been implicated as having somatic changes in lung cancer or other cancers (though not in germ line cells). Targeted sequencing on the significant loci is planned to determine the causal variants at these loci.
Keywords: Family studies, genetic linkage, genome-wide scan, heterogeneity lod score, linkage analysis, lod score, lung cancer, parametric (model-based) analysis
Introduction
Lung cancer is the most lethal cancer in the United States. While mortality for the disease has decreased as we have learned more about the relationship between tobacco smoke and lung cancer, an estimated 158,080 Americans will die of lung cancer in 2016 - approximately 25% of all cancer-related deaths in the country [1].
Environmental exposure to chemical agents found in tobacco smoke [2–5], occupational hazards from mining, asbestos exposure, shipbuilding, and petroleum refining [6] are known to increase the risk of lung cancer. Tobacco smoking is by far the most deleterious; it is directly responsible for approximately 85–90% of lung cancer risk [7–9]. The incidence of lung cancer due to smoking is higher in men (90%) than women (70%) [10].
Though it is evident that the vast majority of lung cancer cases are due to the smoking of tobacco products, this does not account for every case. Approximately 10–15% of nonsmokers develop lung cancer. While a percentage could be due to secondhand smoking, studies have shown that it is responsible for only 16–24% of lung cancer in nonsmokers. Further, the number of lung cancer cases in nonsmokers may actually be increasing, in spite of stricter laws against public tobacco smoking [11].
Lung cancer has been found to have a strong genetic component in addition to its well-publicized environmental components. Familial aggregation of the disease was first identified in 1963 by Tokuhata and Lilienfeld [12,13], who observed that nonsmoking relatives of smoking lung cancer cases had a higher risk of susceptibility than nonsmoking relatives of smoking controls. Further studies in Louisiana [14], Utah [15,16], Texas [17] and Michigan [18] confirmed a higher risk of lung cancer in for individuals with an affected family member after adjusting for smoking histories.
Recently, much work on lung cancer genetics has focused on genome-wide association studies (GWAS). The majority of GWAS are population-based and focus on the identification of common, low penetrance variants with a moderate to small effect on disease risk. Several recent GWAS have provided highly significant and reproducible results for lung cancer. Three studies identified the 15q25 region (which contains the neuronal acetylcholine receptor gene cluster subunits CHRNA3, CHRNA5, and CHRNB4) was associated with increased risk [19–21]. Other GWAS in European populations have found significant associations to 6p21 and 5p15 [19,21–23] while studies in Asian populations have replicated these findings and found new associations at 3q28 [24–26].
Linkage analyses, which use family-based data to find rare, highly penetrant loci, have not been as prevalent as GWAS in the literature. This is likely due to the expensive and time consuming nature of collecting family-based samples instead of population-based samples. The first evidence for genome-wide significant linkage of a lung cancer susceptibility locus was to 6q23–25 [27]. The subjects present in this study had been collected from across the United States by the Genetic Epidemiology of Lung Cancer Consortium (GELCC). The GELCC continues to collect samples from high risk lung cancer families. An update was published 2010 that found further evidence for linkage on 6q and suggestive linkage to chromosomes 1q, 5q, 8q, 9p, 12q, 14q, and 16q [28]. Here, we present linkage analysis on 25 new families that have been recruited by the GELCC from 2008–2014.
Methods
Patient Recruitment and Family Data Description
Participants with a strong familial history of lung cancer were recruited by the GELCC at eight sites across the United States. We defined “strong family history of lung cancer” as having three or more first degree relatives diagnosed with lung cancer. This resulted in the collection of 197 individuals from 25 high risk families. There were 4 two-generation families, 14 three-generation families, 6 four-generation families, and 1 five-generation family. There was an average of approximately 11.04 people per pedigree.
Blood, saliva, and archival tissue were collected for all participants. For the majority of affected participants, cancer status was substantiated through medical records, pathology reports, and death certificates. For the individuals where such documentation did not exist, diagnoses were verified by the reporting of multiple family members. Further information such as birthdays, age at onset, vital statistics, and smoking exposure statistics were also collected.
Genotyping and Quality Control
Genotyping was performed at the Center for Inherited Disease Research (CIDR) at Johns Hopkins University using an Illumina HumanCore-12v1–0 array. 192 of 197 samples were successfully genotyped. 298,830 SNPs were genotyped for each individual. Data cleaning was performed by PLINK [29]; 4,186 SNPs and 0 individuals were removed for having a missingness of 1% or greater. 149 ungenotyped individuals were included in the genotyped pedigrees to create proper familial relationships; these individuals were used to connect pedigrees that would have otherwise been disjointed. Examples would be a child where just one parent was genotyped, or two siblings with neither parent genotyped (likely because the parents were deceased). This also allowed for the calculation of identity-by-descent (IBD) values and to observe any Mendelian inconsistencies in the data. Linkage analysis methods use the genotype information on genotyped family members to calculate the probabilities of specific genotypes/haplotypes of the ungenotyped ancestors in the pedigrees.
IBD values were calculated by PLINK and PRESTPLUS [30] to confirm correct familial relationships; one individual was dropped due to an incorrect relationship (the genotypes for this individual were found to be a duplication of another individual in a different pedigree and thus were most likely due to a pipetting error. This individual was an unaffected child with no offspring in the third generation of a pedigree; thus the loss of genotype information from this person resulted in a small power loss in that family). Sib-pair [31] was used to check all pedigrees for Mendelian errors. SNPs containing Mendelian errors in a single family were removed from the offending family but kept for analyses in the other families. SNPs containing Mendelian errors in two or more families were removed from all families. When there is Mendelian error in only a single family, it is likely due to a single genotype error at that marker in that family. It is not a systemic problem in genotyping the marker but a random, single event error that causes the Mendelian inconsistency. If there is a Mendelian error across multiple families, this is more likely to be a systemic problem in genotyping the marker in any individual. Thus, the genotyping for all individuals is less reliable and thus the SNP is dropped for all families. At this stage, the Mendelian inconsistencies were not caused by familial relationships errors, as we had already checked the IBD values for all individuals and found them to be accurate for the given relationships except for the one person whose genotypes were dropped due to Mendelian inconsistencies (see above). When analyzing SNP array genotype data, it is expected that each family will exhibit a small number of Mendelian inconsistencies due to genotyping errors. True family inconsistencies due to misspecifications of the relationships OR pipetting errors during sample preparation result in very large numbers of Mendelian inconsistencies across many SNPs and changes in the overall IBD sharing values between the relative pairs in question. There were 887 total Mendelian inconsistencies, but only 133 that appeared in multiple families. 48,192 markers that were monomorphic throughout the entire population were also removed. After data cleaning, 246,319 SNPs remained for analysis.
Allele frequencies for the entire data set were then calculated by Sib-pair. Seventeen married-in spouses with genotype information but no offspring were used in the allele frequency calculations but dropped from the linkage analyses. Genetic positions for all SNPs were obtained from the Rutgers Map version 3 [32] using physical positions from GRCh37. Full diagnostics of the samples analyzed, including average age and percent smokers, can be found in Table 1.
Table 1:
Characteristics of Individuals used in Linkage Analyses
| Affected | Unaffected/Unknown | Total | |
|---|---|---|---|
| Genotyped | 35 | 130 | 165 |
| Ungenotyped | 37 | 112 | 149 |
| Average Age | 70 | 63.8 | 66.5 |
| Avg. Age at Onset | 63.7 | N/A | N/A |
| Number Smokers | 55 | 74 | 122 |
| Percentage Smoker | 0.76 | 0.31 | 0.71 |
Diagnostic information on the individuals from the 25 extended families used in the linkage analyses after quality control and removal of married-in spouses. Average age, average age at onset, and smoking statistics were calculated using individuals with available data.
Parametric Linkage Analyses
All linkages analyses were affected-only analyses; affected individuals were coded as affected; unaffected or unknown individuals were coded as having missing phenotypes. This allowed for the high degree of uncertainty between smoking and lung cancer risk as well as jointly allowing for smoking status (80% of affected individuals in the pedigrees smoked). The genetic model assumed a disease allele frequency (DAF) of 1% under an autosomal dominant model.
Historically we have used a low penetrance model of 10% for carriers and a 1% phenocopy rate in linkage analyses of other families because segregation analyses suggest that the lung cancer variant is most likely not highly penetrant in the absence of personal smoking. Given that the linkage analysis methods used do not allow the inclusion of smoking as a covariate in any simple manner, this low penetrance model was used previously to attempt to deal with lack of smoking exposure among many at risk relatives. However, because the families being analyzed in this study consisted of a vast majority of smokers and we coded all unaffected individuals as unknown phenotype, a higher penetrance model made more sense. Thus we performed analyses using our low penetrance model (as done in prior studies) and two higher penetrance models that we believe are more appropriate for this particular data set. As expected, the higher penetrance models produced stronger evidence in favor of linkage in five regions compared to the low penetrance model. However, we found no change in the significant signals between the 40% and 80% penetrance models and the difference between the LOD scores was not statistically significant (though the LOD scores for 80% were slightly higher in magnitude). Given the uncertainty of the correct model to use, we decided to present the results of the more conservative intermediate penetrance model.
Performing an affecteds-only analysis with these penetrance models has the effect that non-smoking unaffected individuals do not contribute information about “not-sharing” genotypes with “affected” individuals in the linkage calculations, thus mitigating the fact that we do not have good age/smoking penetrance distributions to use in our analyses. Furthermore, since most affecteds are smokers and the few non-smokers who are affected are considered to be at very high genetic risk, the moderate phenocopy rate used in the penetrance models allows for the fact that some heavy-smoking affected individuals in these families might not be carrying the same risk variant carried by the other affected members of their family.
Three distinct types of parametric linkage analyses were performed. The first was the standard single variant two-point linkage analysis that observes linkage between a single SNP and the disease trait using an Elston-Stewart algorithm implemented by TwoPointLods [33]. Multipoint linkage analysis was performed by SimWalk2 [34–36]. SNPs were pruned prior to the multipoint linkage analyses in order to remove intermarker linkage disequilibrium that could lead to increased type I error rates. Markers were grouped into 1 cM bins and the SNP with the highest minor allele frequency (thus the highest information content), was chosen to represent the bin. This resulted in approximately 3,000 SNPs for the multipoint analysis. Once linkage analysis was complete, all variants were annotated by ANNOVAR [37,38].
To compensate for some of this loss of information in the multipoint analysis, we used the collapsed haplotype pattern method (CHP) implemented through SEQLinkage [39]. CHP combines SNPs into multiallelic pseudo-markers. These pseudo-markers correspond to annotated genes in RefSeq. The pruning for intermarker LD that is necessary to run programs like SimWalk2 is not needed under this scenario, so more information is retained. This approach has shown to be powerful and maintain proper type I error rates when SNPs with rare minor alleles in the analysis. We restricted CHP analysis to markers with a minor allele frequency of 10% and under (approximately 35,000 SNPs). The regional markers are sometimes further divided into smaller subunits based on observed recombination events within a gene. After the regional pseudo-markers were created, standard two-point linkage analysis was performed on the new markers using MERLIN [40]. This method will henceforth be referred to as CHP two-point linkage.
Results
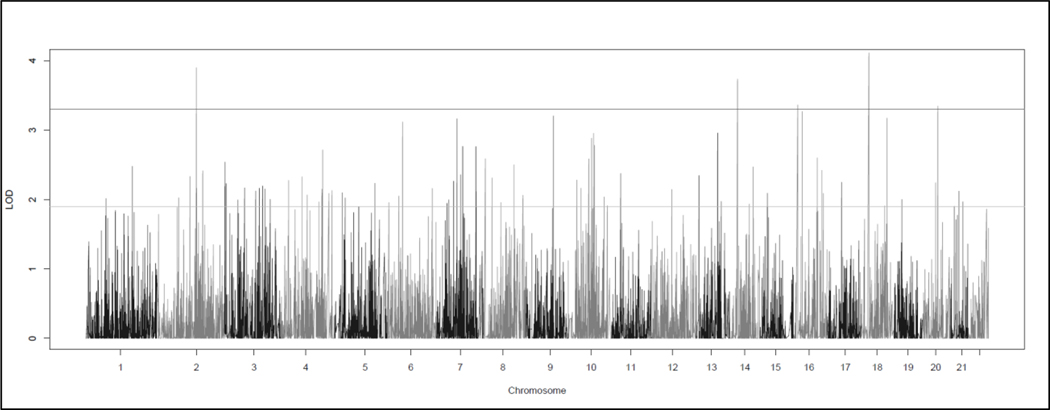
CHP two-point linkage analysis identified five significant linkage signals located on five chromosomes (Figure 1, Table 2). Here, we use the Lander and Kruglyak values of HLOD >= 3.3 and HLOD >=1.9 as the respective thresholds for genome-wide significance and suggestion [41]. A LOD score of 3.3 corresponds to a p-value of 4.9 × 10−5 and a LOD score of 1.9 corresponds to 1.7 × 10-3. The highest HLOD was 4.11 located on 18p11.23 and centered on the PTPRM gene. The other significant signals were located at LRP1B (HLOD = 3.90) at 2p22.2, NPAS3 (HLOD = 3.73) at 14q13.1, RBFOX1 (HLOD = 3.36) at 16p13, and PTPRT (HLOD = 3.34) at 20q13.11. A further 74 suggestive signals were found throughout the genome (Supplemental Table 1).
Figure 1: Genome-wide HLOD Plot of CHP Variant Two-point Linkage Analysis:
The heterogeneity LOD (HLOD) scores calculated across all 25 families for the CHP variant two-point linkage analysis performed by SEQLinkage and MERLIN. The lines at 3.3 and 1.9 represent the thesholds for the respective significant and suggestive LOD scores as recommended by Lander and Kruglylak.
Table 2:
Genome-wide Significant HLOD Scores in CHP Variant Two-point Linkage Analysis
| CHR | POS | GENE | LOD | ALPHA | HLOD |
|---|---|---|---|---|---|
| 18p11.23 | 29.36403 | PTPRM[1] | 4.1099 | 1 | 4.1099 |
| 2p22.2 | 152.1074 | LRP1B[1] | 3.8964 | 1 | 3.8964 |
| 14q13.1 | 32.13378 | NPAS3[1] | 3.7337 | 1 | 3.7337 |
| 16p13 | 18.02736 | RBFOX1[1] | 3.3597 | 1 | 3.3597 |
| 20q13.11 | 62.31841 | PTPRT[1] | 3.3425 | 1 | 3.3425 |
The genome-wide significant (>=3.3) heterogeneity LOD (HLOD) scores from the CHP variant two-point linkage analysis performed by SEQLinkage and MERLIN. CHR stands for chromosome, POS is the start position in cM of the regional marker, GENE is the name of the gene within which the positional marker is located, LOD is the cumulative LOD score across all families, alpha is a measure of the percentage of families linked to that regional marker and is calculated jointly with HLOD, the heterogeneity LOD score. The brackets next to the gene name indicate the gene has been broken into pieces and the number in the bracket represents the particular piece.
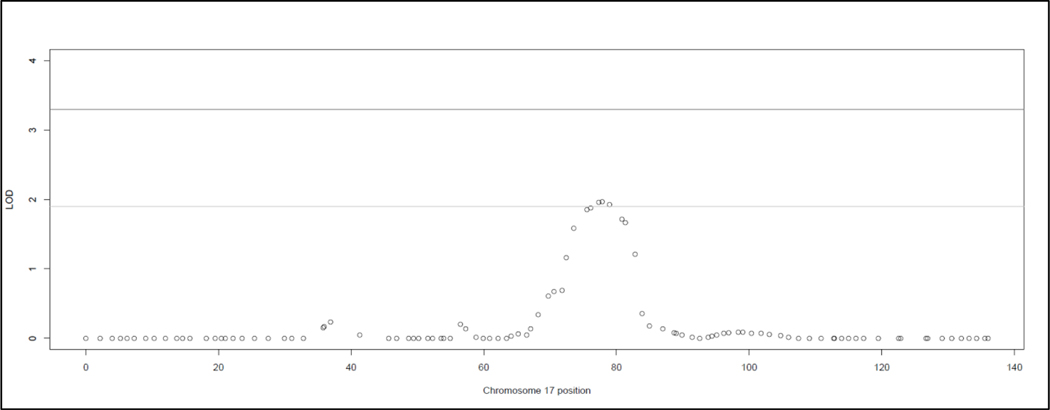
Multipoint analysis yielded no significant linkage signals and three suggestive linkage signals (Figure 2, Table 3). All three suggestive signals were located on 17q21.33. Further the top 9 SNPs were all located in the 17q21.32 – q22 region (Figure 3). The highest HLOD (1.97) was located an intron of CA10; the two other suggestive HLOD scores (1.96 and 1.92) were located in an intron of UTP18 and the intergenic region of CA10 and C17orf112. The highest exonic SNP (HLOD = 1.87) was also located in 17q21.33, in the AMAP1 gene. The 17q21.32-q22 signal was primarily driven by three families – family 138 (HLOD range 0.44 – 0.80), family 147 (HLOD range 0.51 – 0.55), and family 148 (HLOD range 0.34 – 0.45).
Figure 2: Genome-wide HLOD Plot of Multipoint Linkage Analysis:
The heterogeneity LOD (HLOD) scores calculated across all 25 families for the multipoint linkage analysis performed by SimWalk2. SNP pruning was necessary before running SimWalk2, which accounts for the less dense map than the two-point analysis. The lines at 3.3 and 1.9 represent the thesholds for the respective significant and suggestive LOD scores as recommended by Lander and Kruglylak.
Table 3:
Top Nine HLOD Scores in Multipoint Linkage Analysis
| CHR | rsID | POS | LOD | ALPHA | HLOD | FUNCTION | GENE |
|---|---|---|---|---|---|---|---|
| 17q21.33 | rs1263965 | 77.8514 | 1.966 | 1 | 1.966 | intronic | CA10 |
| 17q21.33 | rs6504702 | 77.3418 | 1.957 | 1 | 1.957 | intronic | UTP18 |
| 17q21.33 | rs7218763 | 78.9483 | 1.921 | 1 | 1.921 | intergenic | CA10,C17orf112 |
| 17q21.33 | rs9890721 | 76.1046 | 1.874 | 1 | 1.874 | exonic | AMAP1 |
| 17q21.33 | rs1881140 | 75.5546 | 1.853 | 1 | 1.853 | intergenic | LOC101927230,TMEM92 |
| 17q22 | 12165058 | 80.8282 | 1.715 | 1 | 1.715 | intronic | TOM1L1 |
| 17q22 | rs888207 | 81.3318 | 1.666 | 1 | 1.666 | intergenic | HLF,MMD |
| 17q21.32 | rs4794031 | 73.5562 | 1.584 | 1 | 1.584 | intergenic | FLJ40194,MIR6129 |
| 17q22 | rs9896667 | 82.8156 | 1.2045 | 0.95 | 1.209 | intergenic | PCTP,ANKFN1 |
| 17q21.33 | 11870935 | 72.4112 | 1.0765 | 0.825 | 1.163 | intronic | KPNB1 |
The top nine HLOD scores from the multipoint analysis performed by SimWalk2. All were located between 17q21.32-q22. The top three SNPs are genome-wide suggestive (>= HLOD 1.9) as recommended by Lander and Kruglyak. CHR stands for chromosome, rsID is the SNP name, POS is the start position in cM of the SNP, LOD is the cumulative LOD score across all families, alpha is a measure of the percentage of families linked to the marker and is calculated jointly with HLOD, the heterogeneity LOD score, FUNCTION is the location of the SNP, and GENE is the gene or nearby genes. Annotations for all SNPs were performed by ANNOVAR.
Figure 3: Multipoint HLOD Plot of Chromosome 17:
The heterogeneity LOD (HLOD) scores calculated across all 25 families at chromosome 17 for the multipoint linkage analysis performed by SimWalk2. The lines at 3.3 and 1.9 represent the thesholds for the respective significant and suggestive LOD scores as recommended by Lander and Kruglylak.
Two-point analysis did not reveal any significant or suggestive markers (Supplemental Figure 2). The highest overall HLOD (1.80) was located on 17p12 in an intergenic region between ELAC2 and HS3T3A1.
Since these families had not been previously analyzed, this set of linkage analyses allowed us to determine which families were informative for linkage at all. Five families were not informative for linkage at all (meaning they had no nonzero LOD scores for any of the three types of analyses) (Supplemental Table 2). The other twenty families showed varying degrees of information. From these twenty, there were eight families that had LOD scores above or approximately equal to 0.5 for all three types of analyses. We considered these families highly informative for linkage and will be the most useful for future sequencing studies.
Discussion
CHP two-point analysis located five novel significant linkage signals for familial lung cancer in this genotype data. While these signals had not previously been identified for linkage, all of these signals had been previously implicated in somatic changes in lung cancer in cell lines or in vivo. The protein tyrosine phosphatase gene PTPRM, located on 18p11.23, was the highest linkage peak. Protein tyrosine phosphatases regulate cellular growth and the mitotic cycle and are known oncogenes. PTPRM in particular has been implicated as an oncogene for lung cancer [42]. It has also been found to affect methylation patterns in lung cancer tumor cells compared to non-tumor cells [43] and has been shown to be activated in KRAS mutant lung adenocarcinomas [44].
Another member of the protein tyrosine phosphatase family, PTPRT was also found to be significant for linkage. PTPRT, located on 20q13, has been shown to be mutated in lung cancer cells and may be involved in cellular adhesion and tumor migration [45]. Whole exome sequencing of matched pairs of lung carcinomas and normal tissue found an increase of somatic mutation of this gene [46] and mutational analysis of PTPRT suggested a potential role as a tumor suppressor in colorectal cancer [47].
The low density lipoprotein receptor LRP1B, located at 2p22.2, had the second highest HLOD score and is well documented as being deleted in tumor cells. It is a likely tumor suppressor gene in multiple cancers, including lung cancer [48]. The gene is inactivated in nearly 50% of non-small cell lung cancer cell lines [49]; its normal function when active includes inhibiting cellular migration [50]. All previous reports of LRP1B inactivation are somatic mutations or deletions; this is the first report of an LRP1B mutation in the germ line affecting familial lung cancer risk.
The significant signal at 16p13.3 centered on the RNA binding protein RBFOX1 (HLOD = 3.48). This gene has been found to be deleted in malignant mesothelioma cell lines [51]. Furthermore, RBFOX1 has been linked to disease recurrence in colon cancer in array-CGH [52] and significantly associated with increased survival of chemotherapy treated breast cancer patients in a Finnish GWAS study [53]. Our study is the first to report a familial linkage to the region.
The transcription factor NPAS3 at 14q13.1 has not previously been found to have any links to lung cancer. It has been shown to be critical for lung development [54]. In addition, knockdown of NPAS3 has been shown to induce the growth of malignant astrocytomas in cell lines and overexpressed NPAS3 suppressed transformation in malignant glioma cell lines, leading to speculation that NPAS3 functions as a tumor suppressor [55].
While the multipoint analysis found no significant signals, one suggestive region was found at 17q22.33. This region contains AMAP1, which is overexpressed in breast cancer tumors [56]and has been found to play a role in both metastasis [57] and the epithelial-mesenchymal transition [58]. The membrane trafficking protein TOM1L1 is also located near this region and had been implicated in both breast cancer [59] and colorectal cancer [60].
The single variant two-point analyses found no significant or suggestive variants. This is likely due to the lack of information within these pedigrees for single variant two-point analysis. We had no more than two genotyped affected individuals per family. Therefore, the linkage analysis algorithms use the information from the genotyped affected and unaffected individuals to calculate the probability of a given genotype for additional ancestors in the family (particularly affected family members). This is a standard property of linkage analysis in general. However, imputation of genotypes for the ungenotyped affected individuals is less informative at single SNP than when multiple SNPs are combined into haplotypes. The calculation of genotype probabilities for ungenotyped affecteds is less accurate when using single SNP loci as opposed to multiple SNPs combined into more informative multiallelic haplotypes, as was done in the CHP analysis. This resulted in the higher information content and thus higher power in the CHP two-point analyses.
Another interesting observation is the amount of overlap between the three linkage methods. There was some overlap between the CHP two-point results and the multipoint results, as both analyses localized a signal to the 18q21–23 region, though the magnitude of the signal was much higher in the CHP two-point analysis. The lower magnitude was most likely due to the heavy pruning of the data required to perform the multipoint analysis. Further, large degrees of overlap are unlikely between the CHP two-point and multipoint analyses because the data sets necessitate different types of filtering; multipoint analysis required the binning of SNPs and selection based on the highest MAF, while the CHP two-point analysis required SNPs with a MAF <= 0.1. CHP two-point analysis also used multiallelic pseudo-markers instead of the bilallelic markers used in the multipoint and single variant two-point analyses, resulting in greater information content and consequently higher power for the CHP two-point analysis.
The linkage analyses also allowed us to determine which families were informative for linkage; again critical because no family had more than two genotyped affecteds. Twenty of the twenty-five families were informative for at least one of the linkage analyses. Eight were highly informative. The information content of these families gives them priority for future sequencing studies. We will likely perform targeted sequencing on the five loci of interest identified from this study; the sequencing will focus on these eight families. Similarly, the GELCC is performing whole exome sequencing (WES) on families from throughout its entire data set (not just the new families used here). The information gained from the LOD score metrics from these analyses identified that the top four families (i.e. the 4 most informative of the eight highly informative families identified in this data set) will be included in this WES effort.
We note that we did not see any replication of the previously published linkage signal identified on 6q in these families. Lung cancer (like all cancers) is a highly heterogeneous phenotype and it is not unlikely that the majority of the families here might have different causal loci. In fact, in Bailey-Wilson et al. [27] of the 6q linkage, only a small proportion of the families were strongly linked to this region.
One interesting additional note regarding this data set. We have data for age, age at onset, and smoking status for these families. However, there is currently no reliable way to add covariates to most linkage analysis programs, particularly for multipoint linkage analysis. Development of linkage analysis software stagnated after the explosion of GWAS studies in the early 2000s. Our approach in this study was to control for smoking status by performing affected-only linkage analysis, using the genotypes from unaffected individuals solely to impute genotypes of ungenotyped affecteds and to help compute the probability of identity by descent sharing of alleles by the affected relative pairs using linkage analysis algorithms. This approach was helped by the fact that approximately 80% of the affected individuals were known to smoke. As family-based studies have begun to come back into vogue in recent years, this will hopefully result in the development of additional linkage software that can include covariates.
Despite advances in treatment and prevention, lung cancer still remains the leading cancer killer in the United States. Our linkage analyses identified genome-wide specific signals on 18p11.23, 2p22.2, 14q13.1, 16p13, and 20q13.11. While several of the signals centered on genes with a previous implication to lung cancer (though not in the germ line), we want to further elucidate the causal variant(s) underlying each signal, so targeted sequencing of these regions is planned. The denser map will allow us a greater ability to pinpoint the exact variant(s) that is causing each signal. Once a preliminary causal variant has been identified, laboratory based work will be performed to confirm the finding.
Supplementary Material
Acknowledgments:
This work was funded by the U.S. National Institutes of Health grants U01CA76293, U19CA148127, P30CA22453, HHSN26820100007C, and P30ES006096 and by the Intramural Research Program of the National Human Genome Research Institute. The authors gratefully acknowledge the study participants and their families.
Footnotes
Conflicts of Interest: The authors declare no conflicts of interest.
References
- 1.Society AC: Cancer facts & figures 2016: Atlanta: American Cancer Society, 2016, [Google Scholar]
- 2.Doll R, Peto R: The causes of cancer: Quantitative estimates of avoidable risks of cancer in the united states today. Journal of the National Cancer Institute 1981;66:1191–1308. [PubMed] [Google Scholar]
- 3.Doll R, Peto R, Wheatley K, Gray R, Sutherland I: Mortality in relation to smoking: 40 years’ observations on male british doctors. Bmj 1994;309:901–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carbone D: Smoking and cancer. The American journal of medicine 1992;93:13S–17S. [DOI] [PubMed] [Google Scholar]
- 5.Burch PR: Smoking and lung cancer. Tests of a causal hypothesis. Journal of chronic diseases 1980;33:221–238. [DOI] [PubMed] [Google Scholar]
- 6.Morgan WK, Seaton A: Occupational lung diseases. Philadelphia, W.B. Saunders, 1984. [Google Scholar]
- 7.Peto R, Darby S, Deo H, Silcocks P, Whitley E, Doll R: Smoking, smoking cessation, and lung cancer in the uk since 1950: Combination of national statistics with two case-control studies. Bmj 2000;321:323–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flanders WD, Lally CA, Zhu BP, Henley SJ, Thun MJ: Lung cancer mortality in relation to age, duration of smoking, and daily cigarette consumption: Results from cancer prevention study ii. Cancer research 2003;63:6556–6562. [PubMed] [Google Scholar]
- 9.Mattson ME, Pollack ES, Cullen JW: What are the odds that smoking will kill you? American journal of public health 1987;77:425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shopland DR, Eyre HJ, Pechacek TF: Smoking-attributable cancer mortality in 1991: Is lung cancer now the leading cause of death among smokers in the united states? Journal of the National Cancer Institute 1991;83:1142–1148. [DOI] [PubMed] [Google Scholar]
- 11.Jenks S: Is lung cancer incidence increasing in never-smokers? Journal of the National Cancer Institute 2016;108 [DOI] [PubMed] [Google Scholar]
- 12.Tokuhata GK, Lilienfeld AM: Familial aggregation of lung cancer in humans. Journal of the National Cancer Institute 1963;30:289–312. [PubMed] [Google Scholar]
- 13.Tokuhata GK, Lilienfeld AM: Familial aggregation of lung cancer among hospital patients. Public health reports 1963;78:277–283. [PMC free article] [PubMed] [Google Scholar]
- 14.Ooi WL, Elston RC, Chen VW, Bailey-Wilson JE, Rothschild H: Increased familial risk for lung cancer. Journal of the National Cancer Institute 1986;76:217–222. [PubMed] [Google Scholar]
- 15.Cannon-Albright LA, Thomas A, Goldgar DE, Gholami K, Rowe K, Jacobsen M, McWhorter WP, Skolnick MH: Familiality of cancer in utah. Cancer research 1994;54:2378–2385. [PubMed] [Google Scholar]
- 16.Goldgar DE, Easton DF, Cannon-Albright LA, Skolnick MH: Systematic population-based assessment of cancer risk in first-degree relatives of cancer probands. Journal of the National Cancer Institute 1994;86:1600–1608. [DOI] [PubMed] [Google Scholar]
- 17.Etzel CJ, Amos CI, Spitz MR: Risk for smoking-related cancer among relatives of lung cancer patients. Cancer research 2003;63:8531–8535. [PubMed] [Google Scholar]
- 18.Cote ML, Kardia SL, Wenzlaff AS, Ruckdeschel JC, Schwartz AG: Risk of lung cancer among white and black relatives of individuals with early-onset lung cancer. Jama 2005;293:3036–3042. [DOI] [PubMed] [Google Scholar]
- 19.Hung RJ, McKay JD, Gaborieau V, Boffetta P, Hashibe M, Zaridze D, Mukeria A, Szeszenia-Dabrowska N, Lissowska J, Rudnai P, Fabianova E, Mates D, Bencko V, Foretova L, Janout V, Chen C, Goodman G, Field JK, Liloglou T, Xinarianos G, Cassidy A, McLaughlin J, Liu G, Narod S, Krokan HE, Skorpen F, Elvestad MB, Hveem K, Vatten L, Linseisen J, Clavel-Chapelon F, Vineis P, Bueno-de-Mesquita HB, Lund E, Martinez C, Bingham S, Rasmuson T, Hainaut P, Riboli E, Ahrens W, Benhamou S, Lagiou P, Trichopoulos D, Holcatova I, Merletti F, Kjaerheim K, Agudo A, Macfarlane G, Talamini R, Simonato L, Lowry R, Conway DI, Znaor A, Healy C, Zelenika D, Boland A, Delepine M, Foglio M, Lechner D, Matsuda F, Blanche H, Gut I, Heath S, Lathrop M, Brennan P: A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature 2008;452:633–637. [DOI] [PubMed] [Google Scholar]
- 20.Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP, Manolescu A, Thorleifsson G, Stefansson H, Ingason A, Stacey SN, Bergthorsson JT, Thorlacius S, Gudmundsson J, Jonsson T, Jakobsdottir M, Saemundsdottir J, Olafsdottir O, Gudmundsson LJ, Bjornsdottir G, Kristjansson K, Skuladottir H, Isaksson HJ, Gudbjartsson T, Jones GT, Mueller T, Gottsater A, Flex A, Aben KK, de Vegt F, Mulders PF, Isla D, Vidal MJ, Asin L, Saez B, Murillo L, Blondal T, Kolbeinsson H, Stefansson JG, Hansdottir I, Runarsdottir V, Pola R, Lindblad B, van Rij AM, Dieplinger B, Haltmayer M, Mayordomo JI, Kiemeney LA, Matthiasson SE, Oskarsson H, Tyrfingsson T, Gudbjartsson DF, Gulcher JR, Jonsson S, Thorsteinsdottir U, Kong A, Stefansson K: A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature 2008;452:638–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amos CI, Wu X, Broderick P, Gorlov IP, Gu J, Eisen T, Dong Q, Zhang Q, Gu X, Vijayakrishnan J, Sullivan K, Matakidou A, Wang Y, Mills G, Doheny K, Tsai YY, Chen WV, Shete S, Spitz MR, Houlston RS: Genome-wide association scan of tag snps identifies a susceptibility locus for lung cancer at 15q25.1. Nature genetics 2008;40:616–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Broderick P, Wang Y, Vijayakrishnan J, Matakidou A, Spitz MR, Eisen T, Amos CI, Houlston RS: Deciphering the impact of common genetic variation on lung cancer risk: A genome-wide association study. Cancer research 2009;69:6633–6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Broderick P, Webb E, Wu X, Vijayakrishnan J, Matakidou A, Qureshi M, Dong Q, Gu X, Chen WV, Spitz MR, Eisen T, Amos CI, Houlston RS: Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nature genetics 2008;40:1407–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu Z, Wu C, Shi Y, Guo H, Zhao X, Yin Z, Yang L, Dai J, Hu L, Tan W, Li Z, Deng Q, Wang J, Wu W, Jin G, Jiang Y, Yu D, Zhou G, Chen H, Guan P, Chen Y, Shu Y, Xu L, Liu X, Liu L, Xu P, Han B, Bai C, Zhao Y, Zhang H, Yan Y, Ma H, Chen J, Chu M, Lu F, Zhang Z, Chen F, Wang X, Jin L, Lu J, Zhou B, Lu D, Wu T, Lin D, Shen H: A genome-wide association study identifies two new lung cancer susceptibility loci at 13q12.12 and 22q12.2 in han chinese. Nature genetics 2011;43:792–796. [DOI] [PubMed] [Google Scholar]
- 25.Dong J, Hu Z, Wu C, Guo H, Zhou B, Lv J, Lu D, Chen K, Shi Y, Chu M, Wang C, Zhang R, Dai J, Jiang Y, Cao S, Qin Z, Yu D, Ma H, Jin G, Gong J, Sun C, Zhao X, Yin Z, Yang L, Li Z, Deng Q, Wang J, Wu W, Zheng H, Zhou G, Chen H, Guan P, Peng Z, Chen Y, Shu Y, Xu L, Liu X, Liu L, Xu P, Han B, Bai C, Zhao Y, Zhang H, Yan Y, Amos CI, Chen F, Tan W, Jin L, Wu T, Lin D, Shen H: Association analyses identify multiple new lung cancer susceptibility loci and their interactions with smoking in the chinese population. Nature genetics 2012;44:895–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shiraishi K, Kunitoh H, Daigo Y, Takahashi A, Goto K, Sakamoto H, Ohnami S, Shimada Y, Ashikawa K, Saito A, Watanabe S, Tsuta K, Kamatani N, Yoshida T, Nakamura Y, Yokota J, Kubo M, Kohno T: A genome-wide association study identifies two new susceptibility loci for lung adenocarcinoma in the japanese population. Nature genetics 2012;44:900–903. [DOI] [PubMed] [Google Scholar]
- 27.Bailey-Wilson JE, Amos CI, Pinney SM, Petersen GM, de Andrade M, Wiest JS, Fain P, Schwartz AG, You M, Franklin W, Klein C, Gazdar A, Rothschild H, Mandal D, Coons T, Slusser J, Lee J, Gaba C, Kupert E, Perez A, Zhou X, Zeng D, Liu Q, Zhang Q, Seminara D, Minna J, Anderson MW: A major lung cancer susceptibility locus maps to chromosome 6q23–25. American journal of human genetics 2004;75:460–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amos CI, Pinney SM, Li Y, Kupert E, Lee J, de Andrade MA, Yang P, Schwartz AG, Fain PR, Gazdar A, Minna J, Wiest JS, Zeng D, Rothschild H, Mandal D, You M, Coons T, Gaba C, Bailey-Wilson JE, Anderson MW: A susceptibility locus on chromosome 6q greatly increases lung cancer risk among light and never smokers. Cancer research 2010;70:2359–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC: Plink: A tool set for whole-genome association and population-based linkage analyses. American journal of human genetics 2007;81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McPeek MS, Sun L: Statistical tests for detection of misspecified relationships by use of genome-screen data. American journal of human genetics 2000;66:1076–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duffy D: Sib-pair: A program for simple genetic analysis v1.00.Beta, Queensland Institute of Medical Research, 2008, [Google Scholar]
- 32.Matise TC, Chen F, Chen W, De La Vega FM, Hansen M, He C, Hyland FC, Kennedy GC, Kong X, Murray SS, Ziegle JS, Stewart WC, Buyske S: A second-generation combined linkage physical map of the human genome. Genome research 2007;17:1783–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomas A: Twopointslods: TwoPointLods, http://www-genepi.med.utah.edu/~alun/software/,
- 34.Sobel E, Lange K: Descent graphs in pedigree analysis: Applications to haplotyping, location scores, and marker-sharing statistics. American journal of human genetics 1996;58:1323–1337. [PMC free article] [PubMed] [Google Scholar]
- 35.Sobel E, Papp JC, Lange K: Detection and integration of genotyping errors in statistical genetics. American journal of human genetics 2002;70:496–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sobel E, Sengul H, Weeks DE: Multipoint estimation of identity-by-descent probabilities at arbitrary positions among marker loci on general pedigrees. Human heredity 2001;52:121–131. [DOI] [PubMed] [Google Scholar]
- 37.Wang K, Li M, Hakonarson H: Annovar: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic acids research 2010;38:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang X, Wang K: Wannovar: Annotating genetic variants for personal genomes via the web. Journal of medical genetics 2012;49:433–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang GT, Zhang D, Li B, Dai H, Leal SM: Collapsed haplotype pattern method for linkage analysis of next-generation sequence data. European journal of human genetics : EJHG 2015;23:1739–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abecasis GR, Cherny SS, Cookson WO, Cardon LR: Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nature genetics 2002;30:97–101. [DOI] [PubMed] [Google Scholar]
- 41.Lander E, Kruglyak L: Genetic dissection of complex traits: Guidelines for interpreting and reporting linkage results. Nature genetics 1995;11:241–247. [DOI] [PubMed] [Google Scholar]
- 42.Wang Y, Mei Q, Ai YQ, Li RQ, Chang L, Li YF, Xia YX, Li WH, Chen Y: Identification of lung cancer oncogenes based on the mrna expression and single nucleotide polymorphism profile data. Neoplasma 2015;62:966–973. [DOI] [PubMed] [Google Scholar]
- 43.Mullapudi N, Ye B, Suzuki M, Fazzari M, Han W, Shi MK, Marquardt G, Lin J, Wang T, Keller S, Zhu C, Locker JD, Spivack SD: Genome wide methylome alterations in lung cancer. PloS one 2015;10:e0143826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li J, Sordella R, Powers S: Effectors and potential targets selectively upregulated in human kras-mutant lung adenocarcinomas. Scientific reports 2016;6:27891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu J, Becka S, Zhang P, Zhang X, Brady-Kalnay SM, Wang Z: Tumor-derived extracellular mutations of ptprt /ptprho are defective in cell adhesion. Molecular cancer research : MCR 2008;6:1106–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choi M, Kadara H, Zhang J, Cuentas EP, Canales JR, Gaffney SG, Zhao Z, Behrens C, Fujimoto J, Chow C, Kim K, Kalhor N, Moran C, Rimm D, Swisher S, Gibbons DL, Heymach J, Kaftan E, Townsend JP, Lynch TJ, Schlessinger J, Lee JJ, Lifton RP, Herbst RS, Wistuba, II: Mutation profiles in early-stage lung squamous cell carcinoma with clinical follow-up and correlation with markers of immune function. Annals of oncology : official journal of the European Society for Medical Oncology 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Z, Shen D, Parsons DW, Bardelli A, Sager J, Szabo S, Ptak J, Silliman N, Peters BA, van der Heijden MS, Parmigiani G, Yan H, Wang TL, Riggins G, Powell SM, Willson JK, Markowitz S, Kinzler KW, Vogelstein B, Velculescu VE: Mutational analysis of the tyrosine phosphatome in colorectal cancers. Science 2004;304:1164–1166. [DOI] [PubMed] [Google Scholar]
- 48.Beer AG, Zenzmaier C, Schreinlechner M, Haas J, Dietrich MF, Herz J, Marschang P: Expression of a recombinant full-length lrp1b receptor in human non-small cell lung cancer cells confirms the postulated growth-suppressing function of this large ldl receptor family member. Oncotarget 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu CX, Musco S, Lisitsina NM, Yaklichkin SY, Lisitsyn NA: Genomic organization of a new candidate tumor suppressor gene, lrp1b. Genomics 2000;69:271–274. [DOI] [PubMed] [Google Scholar]
- 50.Li Y, Knisely JM, Lu W, McCormick LM, Wang J, Henkin J, Schwartz AL, Bu G: Low density lipoprotein (ldl) receptor-related protein 1b impairs urokinase receptor regeneration on the cell surface and inhibits cell migration. The Journal of biological chemistry 2002;277:42366–42371. [DOI] [PubMed] [Google Scholar]
- 51.Klorin G, Rozenblum E, Glebov O, Walker RL, Park Y, Meltzer PS, Kirsch IR, Kaye FJ, Roschke AV: Integrated high-resolution array cgh and sky analysis of homozygous deletions and other genomic alterations present in malignant mesothelioma cell lines. Cancer genetics 2013;206:191–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mampaey E, Fieuw A, Van Laethem T, Ferdinande L, Claes K, Ceelen W, Van Nieuwenhove Y, Pattyn P, De Man M, De Ruyck K, Van Roy N, Geboes K, Laurent S: Focus on 16p13.3 locus in colon cancer. PloS one 2015;10:e0131421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fagerholm R, Schmidt MK, Khan S, Rafiq S, Tapper W, Aittomaki K, Greco D, Heikkinen T, Muranen TA, Fasching PA, Janni W, Weinshilboum R, Loehberg CR, Hopper JL, Southey MC, Keeman R, Lindblom A, Margolin S, Mannermaa A, Kataja V, Chenevix-Trench G, kConFab I, Lambrechts D, Wildiers H, Chang-Claude J, Seibold P, Couch FJ, Olson JE, Andrulis IL, Knight JA, Garcia-Closas M, Figueroa J, Hooning MJ, Jager A, Shah M, Perkins BJ, Luben R, Hamann U, Kabisch M, Czene K, Hall P, Easton DF, Pharoah PD, Liu J, Eccles D, Blomqvist C, Nevanlinna H: The snp rs6500843 in 16p13.3 is associated with survival specifically among chemotherapy-treated breast cancer patients. Oncotarget 2015;6:7390–7407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou S, Degan S, Potts EN, Foster WM, Sunday ME: Npas3 is a trachealess homolog critical for lung development and homeostasis. P Natl Acad Sci USA 2009;106:11691–11696. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 55.Moreira F, Kiehl TR, So K, Ajeawung NF, Honculada C, Gould P, Pieper RO, Kamnasaran D: Npas3 demonstrates features of a tumor suppressive role in driving the progression of astrocytomas. The American journal of pathology 2011;179:462–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Onodera Y, Hashimoto S, Hashimoto A, Morishige M, Mazaki Y, Yamada A, Ogawa E, Adachi M, Sakurai T, Manabe T, Wada H, Matsuura N, Sabe H: Expression of amap1, an arfgap, provides novel targets to inhibit breast cancer invasive activities. The EMBO journal 2005;24:963–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sabe H, Hashimoto S, Morishige M, Ogawa E, Hashimoto A, Nam JM, Miura K, Yano H, Onodera Y: The egfr-gep100-arf6-amap1 signaling pathway specific to breast cancer invasion and metastasis. Traffic 2009;10:982–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Matsumoto Y, Sakurai H, Kogashiwa Y, Kimura T, Matsumoto Y, Shionome T, Asano M, Saito K, Kohno N: Inhibition of epithelial-mesenchymal transition by cetuximab via the egfr-gep100-arf6-amap1 pathway in head and neck cancer. Head & neck 2016 [DOI] [PubMed] [Google Scholar]
- 59.Chevalier C, Collin G, Descamps S, Touaitahuata H, Simon V, Reymond N, Fernandez L, Milhiet PE, Georget V, Urbach S, Lasorsa L, Orsetti B, Boissiere-Michot F, Lopez-Crapez E, Theillet C, Roche S, Benistant C: Tom1l1 drives membrane delivery of mt1-mmp to promote erbb2-induced breast cancer cell invasion. Nature communications 2016;7:10765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Emaduddin M, Edelmann MJ, Kessler BM, Feller SM: Odin (anks1a) is a src family kinase target in colorectal cancer cells. Cell communication and signaling : CCS 2008;6:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.