Abstract
The introduction of mutation D119N (or its homolog) in the NKxD nucleotide binding motif of various Ras-like proteins produces constitutively activated or dominant-negative effects, depending on the system and assay. Here we show that Ras(D119N) has an inhibitory effect at a cell-specific concentration in PC12 and NIH 3T3 cells. Biochemical data strongly suggest that the predominant effect of mutation D119N in Ras—a strong decrease in nucleotide affinity—enables this mutant (i) to sequester its guanine nucleotide exchange factor, as well as (ii) to rapidly bind GTP, independent of the regulatory action of the exchange factor. Since mutation D119N does not affect the interaction between Ras and effector molecules, the latter effect causes Ras(D119N) to act as an activated Ras protein at concentrations higher than that of the exchange factor. In comparison, Ras(S17N), which also shows a strongly decreased nucleotide affinity, does not bind to effector molecules. These results point to two important prerequisites of dominant-negative Ras mutants: an increased relative affinity of the mutated Ras for the exchange factor over that for the nucleotide and an inability to interact with the effector or effectors. Remarkably, the introduction of a second, partial-loss-of-function, mutation turns Ras(D119N) into a strong dominant-negative mutant even at high concentrations, as demonstrated by the inhibitory effects of Ras(E37G/D119N) on nerve growth factor-mediated neurite outgrowth in PC12 cells and Ras(T35S/D119N) on fetal calf serum-mediated DNA synthesis in NIH 3T3 cells. Interpretations of these results are discussed.
Growth factor-dependent signal transduction is mediated by Ras and homologous small GTP-binding proteins. These proteins switch between an active GTP-bound form and an inactive GDP-bound form (5). Whereas GTPase-activating proteins (GAPs) inactivate Ras by stimulating the slow intrinsic GTP hydrolysis, the guanine nucleotide exchange factors (GEFs) activate Ras by stimulation of the slow intrinsic GDP dissociation rate, allowing a fast exchange with the cellular pool of nucleotide (3, 23). Mutant Ras(S17N) was shown to be able to block the Ras-dependent signal transduction pathway (12). This mutant, which has a strongly decreased GTP affinity, acts upstream from Ras by sequestering the exchange factor, thereby inhibiting the activation of cellular Ras (11, 31, 43). Although this type of dominant-negative mutant has been used in the elucidation of the role of GTP-binding proteins in a multitude of signal transduction pathways, the mechanism by which the sequestration of GEF occurs is less well understood. It was related to the low nucleotide affinity of Ras(S17N), to a low affinity for GTP in particular, and to a higher affinity for GEF, but other explanations have also been proposed (6, 11, 16, 18, 28, 31, 40, 43). Surprisingly, Rap1A(S17N), while showing a reduced nucleotide affinity similar to Ras(S17N), is unable to inhibit the stimulatory action of the Rap1-specific exchange factor C3G in vitro (45), pointing out that mutants homologous to Ras(S17N) are not per definition dominant-negative mutants. This example underlines the need for a detailed analysis of the mechanism of action of dominant-negative Ras mutants.
We and other groups have demonstrated that Ras(D119N) behaves as an activated Ras mutant (13, 39). This ability is a consequence of the strongly decreased nucleotide affinity, enabling the mutant to bind GTP in a GEF-independent way, as first suggested for Ras(N116I) (49). However, other studies reported dominant-negative effects for Ras(D119N) and homologous mutants (17, 57). Interestingly, Sigal et al. (42) showed that Ras(D119A) has an oncogenic effect in NIH 3T3 fibroblast cells, but causes a dominant temperature-dependent lethality in yeast cells. In addition, mutation D119N in the Caenorhabditis elegans Ras protein let-60 was shown to have a strong dominant-negative effect on let-60-dependent vulval differentiation and larval growth, but was also shown to be able to activate this pathway (15).
In order to resolve the seemingly controversial effects of mutation D119N and to elucidate the prerequisites for a dominant-negative mutant, we have performed detailed in vitro and in vivo analysis. We have microinjected Ras(D119N) at different concentrations in PC12 and NIH 3T3 cells and show that Ras(D119N) is able to inhibit the serum-dependent signal in both cell types when microinjected at a concentration specific for each cell type, presumably enough to sequester all RasGEF molecules. At a higher concentration, however, Ras(D119N) acts as an activated Ras mutant. Biochemical analysis shows that the strongly decreased guanine nucleotide affinity not only begets the activated character of Ras(D119N), but also is the predominant cause of the inhibitory capability of Ras(D119N). Also Ras(S17N) shows a strongly decreased nucleotide affinity, but in contrast to Ras(D119N), Ras(S17N) is no longer capable of interacting with effector molecules, and consequently the dominant-negative effect cannot be masked by an activated character. In apparent agreement, the combination of D119N with partial-loss-of-function mutations, which strongly weakens the interaction with specific downstream effectors, but not with the GEF, turns Ras into a dominant-negative mutant even at high concentrations. On the other hand, alternative interpretations are possible, as pointed out in the Discussion.
MATERIALS AND METHODS
Expression and isolation of proteins.
The wild-type and mutated Ras proteins were expressed from ptacras in E. coli CK600K as described previously (22, 39). The catalytic domain comprising the C-terminal moiety of the mouse guanine nucleotide exchange factor Cdc25Mm, Cdc25Mm285, was expressed as a glutathione S-transferase (GST) fusion protein from pGEX2T-CDC25-12 in the protease-negative E. coli strain AD202 (ompT::Tn5) (1) and purified in the nonfused form as described previously (23). Protein concentrations were determined with Protein Plus solution (Pierce) with bovine serum albumin as a standard.
Synthesis of fluorescently labelled nucleotides carrying an N-methylanthraniloyl group and nucleotide exchange on Ras proteins were performed as described previously (22, 23).
Microinjection of purified proteins.
Microinjection of proteins in PC12 or NIH 3T3 cells was performed as described previously (39). Proteins were diluted to the indicated concentrations with the injection buffer, which was phosphate-buffered saline (PBS) for PC12 cells and a mixture of 10 mM HEPES-NaOH (pH 7.4), 140 mM KCl, 8 mM NaCl, and 1 mM MgCl2 for NIH 3T3 cells.
PC12 cells were grown in Falcon tissue culture flasks in Dulbecco’s modified Eagle’s medium with 1 g of glucose per liter supplemented with 10% horse serum (Gibco) and 5% fetal bovine serum at 37°C in 10% CO2. For microinjection, cells were transferred to Corning polystyrene tissue culture dishes, and the cells were incubated in culture medium buffered with 20 mM Na-HEPES (pH 7.4). For injections, the Zeiss Microinjection Workstation (AIS) and thin borosilicate glass capillaries with filament (Hilgenberg) with a tip diameter smaller than 0.5 μm were used. For each experiment, 100 to 200 cells within a marked region were injected. After microinjection, the cells were further incubated at 37°C, with addition of 100 nM nerve growth factor (NGF) 7S (Boehringer) for the measurement of dominant-negative effects. PC12 cell differentiation was determined by scoring neurite outgrowth 2 days after injection of the protein, measured as the length of the neurites compared to cell body diameter and the number of injected cells possessing neurites. Cells with neurites longer than 1 to 2 times the diameter of the cell body were scored as positive.
NIH 3T3 cells were seeded on glass plates coated with poly-l-lysine (Sigma) and incubated in Dulbecco’s modified Eagle’s medium with 4.5 g of glucose per liter (Gibco) and 7.5% CO2 to a confluence of approximately 50%. In order to achieve cell arrest in the G0 phase, the cells were washed three times with PBS and kept in serum-free medium for 36 h. After microinjection with Ras(D119N), 10 μM bromodeoxyuridine (BrdU) was added to the medium, and the cells were stimulated with 10% fetal calf serum (FCS; Gibco) for measurement of dominant-negative effects or kept in serum-free medium for measurement of the activated effects. After an incubation for 24 h, BrdU incorporation was determined with the BrdU labeling and detection kit I (Boehringer Mannheim Biochemicals) as described by the manufacturer by using a slow-fade TM fluorescence protector (Molecular Probes) and a fluorescence microscope (Axiovert; Zeiss).
Transfection studies.
The pEXV plasmids were constructed from a modified pEXV3 plasmid, a kind gift of S. Hooper and C. J. Marshall. The plasmid was linearized by EcoRI, after which an oligonucleotide with the sequence 5′-AATTgccaccATGGACTACAAGGACGACGATGACAAGGGGAATTCCCGGGC-3′ was inserted (lowercase, Kozak sequence; italic, FLAG tag encoding amino acids Met-Asp-Tyr-Lys-Asp-Asp-Asp-Asp-Lys; underlined, overlapping EcoRI and SmaI sites), thereby disrupting the original EcoRI site. This modified plasmid was named pEXV-KF. Thereafter, EcoRI fragments from ptacras plasmids comprising the mutated Ras genes were transferred into the EcoRI-SmaI-cleaved pEXV-KF. Thus, the pEXV-KF-Ras plasmids expressed N-terminally FLAG-tagged Ras proteins. The pcDNA3-KF-Ras plasmids expressing FLAG-tagged versions of Ras proteins were constructed similarly.
NIH 3T3 cells were seeded on gridded glass coverslips (Eppendorf) and cultivated overnight in Dulbecco’s modified Eagle’s medium (12 mM l-glutamine, 4.5 g of glucose per liter) supplemented with 10% fetal calf serum, penicillin (4 mM), and streptomycin (4 mM) in a humidified atmosphere of 5% CO2 at 37°C. For cotransfection, 2 μg of pEXV-KF-Ras(D119N) (or other Ras mutants) and 2 μg of pEGFP-CI (Clontech) were dissolved in 22 μl of buffer containing 10 mM Tris-HCl–1 mM EDTA (pH 8.0), precipitated by the calcium phosphate method (36), and added to the cells in 0.5 ml of culture medium. The cells were incubated overnight. After undergoing a dimethyl sulfoxide (DMSO) shock (10% DMSO in culture medium, 10 min at 37°C), cells were washed and cultivated for another 24 h. Thereafter, the cells were washed and cultivated for 24 h in serum-free medium followed by 6 h in medium containing 10% serum. For the last 2 h of incubation, BrdU was added. After identification of cells expressing green fluorescence protein (EGFP) by fluorescence microscopy, the cells were fixed with 70% ethanol and stained for incorporated BrdU by using the labeling and detection kit I (Boehringer Mannheim Biochemicals). Only cells expressing EGFP were counted for determination of the effects of the Ras mutants.
Kinetic measurements.
All kinetic measurements were carried out in standard buffer (40 mM Na-HEPES [pH 7.5], 10 mM MgCl2, 5 mM dithioerythritol [DTE]) and at 20°C, unless otherwise indicated.
The intrinsic and Cdc25Mm285-stimulated dissociation rates of Ras-3′mdGDP were measured with an extinction wavelength of 366 nm and an emission wavelength of 450 nm (Perkin-Elmer LS 600 fluorometer). The reaction was started with the addition of the excess unlabeled nucleotide. The data from the dissociation rate determinations were fitted with the program Grafit (Eritacus) to a single exponential function.
Titrations of the Ras-nucleotide complexes with GEF.
Titrations of the Ras-nucleotide complexes with Cdc25Mm285 were carried out in a Fluoromax fluorometer SPEX by pipetting increasing amounts of GEF to 1.2 ml of 86 nM Ras(D119N)-mXpp(NH)p at 20°C in standard buffer as described previously (23). After each addition of GEF, the solution was carefully mixed, and the fluorescent signal was measured until a stable value was obtained (typically 5 to 10 min). At the end of the titration, the end point was determined by adding a 200-fold excess of Xpp(NH)p. The total volume of added GEF and Xpp(NH)p was less than 50 μl.
In order to determine the different dissociation constants characterizing the interactions between Ras, nucleotide, and Cdc25Mm285, the data from the titration were fitted with the multiparameter-fitting program FACSIMILE (AEA Technology, Harwell, Didcot, Oxfordshire, United Kingdom) as described previously (23).
RESULTS
The effects of Ras(D119N) in PC12 and NIH 3T3 cells.
We have reported extensively on the activated character of Ras(D119N) when microinjected at 150 μM into PC12 and NIH 3T3 cells (39). Based on conflicting results in the literature showing either activated or inhibitory action of this mutant, we have carefully analyzed the ability of Ras(D119N) to induce neurite outgrowth in PC12 cells. For this, we microinjected different concentrations of the Ras mutant into unstimulated PC12 cells and measured neurite outgrowth after 24 h of incubation. Whereas no effect was observed with 1 and 5 μM Ras(D119N), microinjection of 10 μM caused neurite outgrowth in 5% of the microinjected cells, and 25 and 50 μM caused neurite outgrowth in more than 60% of the microinjected cells. Hence, at concentrations of 10 μM and higher, Ras(D119N) acts as an activated Ras mutant in PC12 cells. We have argued before that Ras(D119N) can bind GTP independent of the stimulatory action of a GEF and activate c-Raf-1 and/or other effector molecules due to the high intrinsic guanine nucleotide dissociation rate (39). Thus, if Ras(D119N) can act as a dominant-negative mutant, its effect should occur at concentrations below 10 μM.
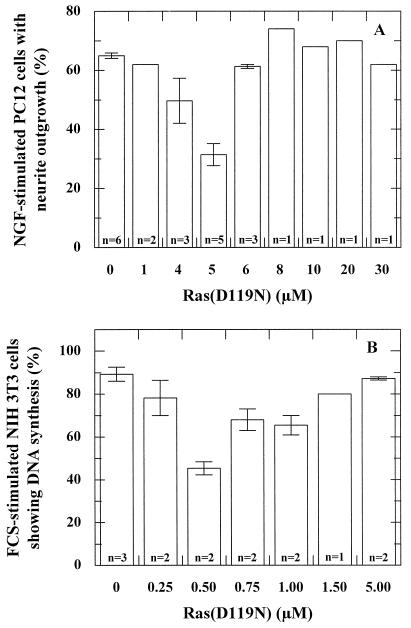
In order to measure a possible dominant-negative effect of Ras(D119N), we microinjected different concentrations of Ras(D119N) into PC12 cells prior to NGF stimulation. Figure 1A shows that after microinjection of 5 μM Ras(D119N), only 30% of the microinjected cells show neurite outgrowth, i.e., approximately 50% of the control level. At lower concentrations, the amount of microinjected Ras(D119N) apparently is not sufficient to sequester all cellular GEF, whereas at higher concentrations, the activated character of Ras(D119N) overrules the inhibitory effect.
FIG. 1.
Ras(D119N) acts at a cell-specific concentration as a dominant-negative Ras mutant in PC12 and NIH 3T3 cells. (A) Histogram showing the effect of microinjection of different concentrations of Ras(D119N) on NGF-stimulated PC12 cells. Depicted is the percentage of microinjected cells showing outgrowth of neurites with at least twice the length of the cell. n, number of experiments (each with 100 to 200 cells). The bars indicate the deviation. (B) Histogram showing the effect of microinjection of different concentrations of Ras(D119N) on FCS-stimulated NIH 3T3 cells. Depicted is the percentage of microinjected cells showing DNA synthesis, as detected with the BrdU assay. n, number of experiments (each with 100 to 200 cells). The bars indicate the deviation.
In order to analyze the effect of Ras(D119N) in another system, we have also tested the influence of microinjection of this mutant on DNA synthesis in NIH 3T3 cells. In resting NIH 3T3 cells, microinjection of 2 μM and higher concentrations of Ras(D119N) induces DNA synthesis as determined by incorporation of BrdU (not shown). Again, any dominant-negative effect of Ras(D119N) should appear after microinjection of smaller concentrations. Indeed, as depicted in Fig. 1B, Ras(D119N) can inhibit the serum-induced DNA synthesis to approximately 50% in NIH 3T3 cells when microinjected at a concentration of 0.5 μM.
It is noteworthy that the bona fide dominant-negative Ras(S17N) mutant has its maximal inhibitory effect when microinjected at concentrations of 50 μM or higher in PC12 cells and 25 μM or higher in NIH 3T3 cells (38). Thus, both in PC12 cells and in NIH 3T3 cells, Ras(D119N) is inhibitory at a concentration at least 10-fold lower than that of Ras(S17N), indicating that Ras(D119N) has a stronger inhibitory potential than Ras(S17N).
Fluorescence analysis of the effects of mutation D119N and comparison with S17N.
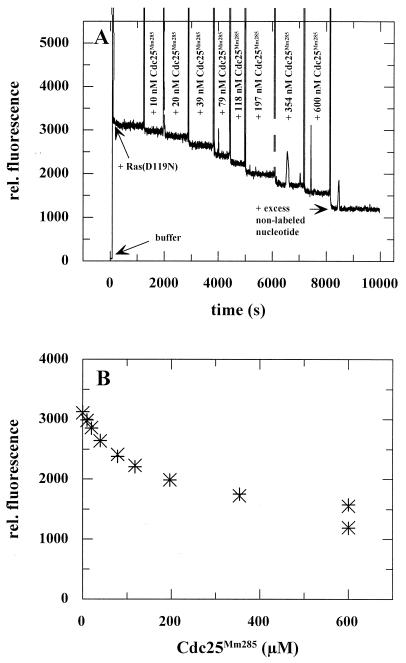
We have shown earlier that mutation D119N strongly affects guanine nucleotide affinity, but that the nucleotide dissociation rate of Ras(D119N) can still be stimulated by Cdc25Mm285 (39). Here, we wanted to analyze whether the mutation affects the interaction between nucleotide-free Ras and GEF. This was done by a fluorescence titration experiment as described previously (23). In this titration, increasing concentrations of the catalytic domain of mouse exchange factor Cdc25Mm, Cdc25Mm285, were added to 86 nM Ras(D119N) in complex with a fluorescently labelled nucleotide (Fig. 2). Addition of the GEF leads to a reduction in the fluorescence signal due to the displacement of the nucleotide from Ras by the GEF. The effectiveness of the competition between Cdc25Mm285 and the nucleotide for binding to Ras depends on the four equilibrium constants that describe the three-component system (Fig. 3). The experimental data were fitted by using a multiparameter fitting program as described previously (23). The best fit of the experimental data was obtained with a Kd2 of 0.4 nM, compared to 4.6 nM for the Ras wild type (23). Thus, the mutation D119N increases the affinity of Cdc25Mm285 for nucleotide-free Ras by a factor of 10. In the experiment depicted in Fig. 2, the fluorescently labelled mXpp(NH)p nucleotide was used, since Ras(D119N) has a higher affinity for xanthosine than guanine nucleotides (39). A comparable value for Kd2 was obtained by a titration carried out with Ras(D119)-mXDP (not shown). Similar to the Ras wild type (23), the data did not allow the determination of Kd3, since under the conditions chosen, no significant amount of the ternary complex Ras(D119N)-mXpp(NH)p-Cdc25Mm285 was formed. For a good fit of the data, a value for Kd3 larger than 2 μM had to be assumed.
FIG. 2.
Equilibrium titration of Ras(D119N) with Cdc25Mm285. Increasing amounts of Cdc25Mm285 were added to 86 nM Ras(D119N)-mXppNHp at 20°C in standard buffer, causing a decrease in relative (rel.) fluorescence related to the liberation of the nucleotide. (A) After each addition of Cdc25Mm285, the fluorescence signal was measured for typically 5 to 10 min in order to ascertain a stable value. At the end of the titration, a 200-fold excess of Xpp(NH)p was added in order to determine the fluorescence of the unbound mXpp(NH)p. (B) Plotting of the fluorescence data as a function of the Cdc25Mm285 concentration and fitting of these data with the multiparameter program Fascimile as described by Lenzen et al. (23). x, experimental data; +, calculated data.
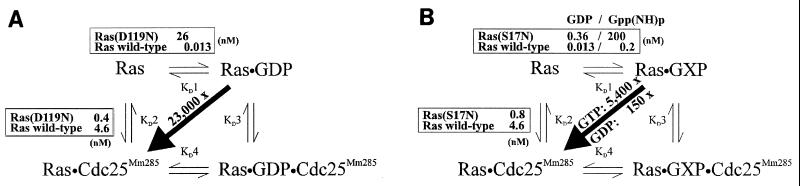
FIG. 3.
Schematic representation of affinity changes brought about by Ras mutations D119N (A) and S17N (B).
Together with the effect on the GDP affinity [Kd1 = 26 nM for Ras(D119N) in comparison 13 pM for the wild type] (39), mutation D119N affects the relative affinity of Ras for GEF relative to that for guanine nucleotide (Kd1/Kd2) by a factor of 23,000 primarily by the change in nucleotide affinity (2,000-fold) and much less by the effect on Kd2 (11.5-fold). It is through this strong effect on relative affinity that Ras(D119N) is able to sequester Cdc25Mm285 and thus to inhibit the GEF-dependent activation of the Ras wild type. This can be demonstrated in vitro: whereas addition of wild-type Ras hardly affects the GEF-stimulated dissociation rate, Ras(D119N) strongly competes with the Ras wild type for binding to Cdc25Mm285 and thus abrogates the Cdc25Mm285-stimulated dissociation of Ras-3′mdGDP (Fig. 4).
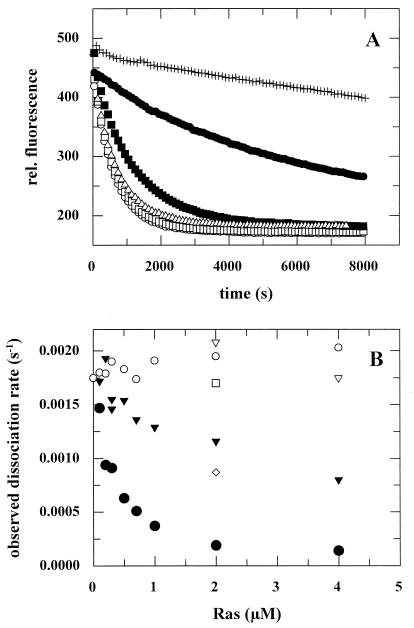
FIG. 4.
Inhibition of the Cdc25Mm285-stimulated dissociation rate of Ras-3′mdGDP by addition of Ras(D119N). (A) The dissociation of 100 nM p21-3′mdGDP in the presence of 20 μM GDP in standard buffer at 20°C (+) was stimulated to 0.0018 s−1 by addition of 163 nM Cdc25Mm285 (○). This stimulation was inhibited by the addition of 2 μM Ras(D119N)-GDP (●), but this inhibition was counteracted by the addition of 20 μM XDP (□), 200 μM GDP (■), or 2 mM GDP (▵). (B) Different representation of measurements performed as described for panel A. Addition of Ras-GDP (○) hardly affected the Cdc25Mm285-stimulated dissociation rate of Ras-3′mdGDP, whereas addition of Ras(D119N)-XDP (⧫) or Ras(D119N)-GDP (●) inhibited this stimulation in a concentration-dependent manner. Replacement of the excess of 20 μM GDP (○, ●, and ⧫) by 20 μM XDP (▿), 200 μM GDP (◊), or 2 mM GDP (□) abrogated the inhibitory action of Ras(D119N).
In agreement with the known nucleotide affinities, Ras(D119N)-XDP has a smaller inhibitory action than Ras(D119N)-GDP (Fig. 4). Since Ras(D119N) has an affinity for XDP of 0.13 nM (39), i.e., of the same order as Kd2 (0.4 nM), only part of the mutant will compete with wild-type Ras for binding to Cdc25Mm285, whereas the rest will stay bound to XDP. Another consequence of the nucleotide affinities is that addition of a 200-fold excess of XDP completely eliminates the inhibitory action of Ras(D119N) (Fig. 4). Similarly, the inhibitory effect of Ras(D119N) can be abrogated by addition of a 20,000-fold excess of GDP (Fig. 4). In agreement with the latter effect, a full exchange of Ras(D119N)-mXDP to Ras(D119N)-GDP can only be achieved by adding an excess of GDP of at least 20,000-fold (not shown). These observations clearly illustrate that the effects of Ras(D119N) depend in a rather delicate way on the concentrations and affinities that characterize the reaction steps of the nucleotide exchange reaction.
For comparative reasons, we have determined the dissociation constants that characterize the commonly used dominant-negative mutant Ras(S17N). As expected and in agreement with earlier observations (11, 16), mutation S17N affects the interaction of Ras with GTP (Kd1[mGppNHp] = 200 nM) much more severely than that with GDP (Kd1[mGDP] = 0.36 nM). Our titration assay revealed a 5.4-fold increase in affinity of nucleotide-free Ras(S17N) towards Cdc25Mm285: Kd2 = 0.85 nM versus 4.6 nM for the Ras wild type. Thus, when looking only at the GDP conformation, mutation S17N causes a 150-fold shift in relative affinity (28-fold in Kd1 and 5.4-fold in Kd2). This is much lower than the effect of mutation D119N. However, taking the effect on the GTP affinity into consideration, one can calculate a shift in relative affinity of 5,400-fold (1,000-fold in Kd1 and 5.4-fold in Kd2). This, together with the observation that the cellular concentration of GTP is 30-fold higher than that of GDP (44), explains why Ras(S17N) is a strong dominant-negative mutant. At the same time, these results also explain why we see a stronger inhibitory effect in vivo with Ras(D119N).
The effects of double mutant Ras(E37G/D119N) in PC12 cells.
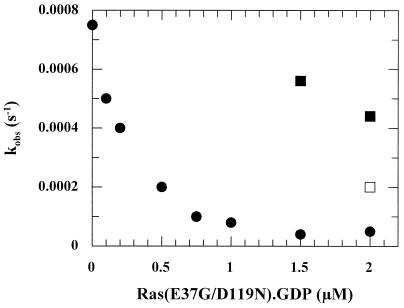
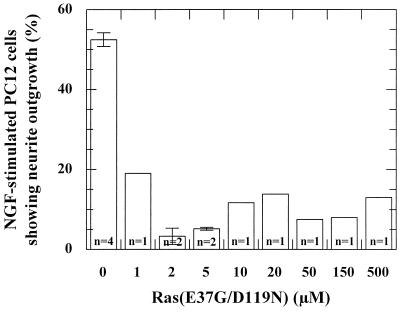
In order to verify our interpretation of the dual effects of Ras(D119N), we introduced a second mutation, E37G, into Ras(D119N). Mutation E37G strongly weakens the interaction with Ras effectors c-Raf-1 and phosphoinositol-3-kinase, but not RalGEFs (10, 20, 26, 29, 30, 35, 50, 51). Since the mitogen-activated protein (MAP) kinase pathway was shown to be essential for PC12 differentiation (7), we expected Ras(E37G/D119N) to behave as a dominant-negative mutant in PC12 differentiation. However, this can only occur under the condition that this mutation does not affect the interaction with the GEF. As can be seen in Table 1, only a twofold decrease in the Cdc25Mm285-dependent stimulation can be measured between the Ras wild type and Ras(E37G). We could show that the double mutant is able to inhibit in vitro the Cdc25Mm285-dependent dissociation of Ras-3′mdGDP and that, similar to Ras(D119N), the inhibitory action of the double mutant can be abrogated almost completely by addition of a 20,000-fold excess of GDP (Fig. 5). Microinjection of the double mutant Ras(E37G/D119N) in NGF-stimulated PC12 cells shows that the double mutant is able to dominantly inhibit neurite outgrowth when microinjected at a concentration above 2 μM, even up to 500 μM (Fig. 6). Thus, the second mutation apparently turns Ras(D119N) into a genuine dominant-negative Ras mutant for PC12 differentiation, indicating that the abrogated interaction with Ras effector(s) is an important aspect of the dominant-negative effect of Ras mutants. It should be noted, however, that other explanations can be found, as discussed below. Even though the double mutant appears to have a slightly stronger inhibitory effect than Ras(D119N), this deviation may very well be caused by experimental differences, e.g., cellular GEF concentration and stability of the mutants.
TABLE 1.
Effect of effector region mutants on GEF activity of the catalytic domain of Cdc25Mma
| Protein type | Rate (10−5 s−1)
|
Fold stimulation | |
|---|---|---|---|
| Intrinsic | Stimulated | ||
| Wild type | 0.7 | 47 | 67 |
| D30K/E31K | 0.4 | 13 | 37 |
| E31K | 0.4 | 29 | 71 |
| T35A | 0.9 | 18 | 20 |
| T35S | 0.6 | 23 | 38 |
| E37G | 2.2 | 64 | 29 |
| D38A | 0.4 | 4 | 10 |
| D38E | 0.4 | 11 | 28 |
| Y40C | 4.4 | 2,558 | 581 |
Ras-3′mdGDP (100 nM) was incubated with Cdc25Mm285 (50 nM) in standard buffer at 20°C, after which the exchange reaction was started by adding GDP (20 μM). The reaction was monitored for approximately 8,000 s and the data were fitted to a single exponential function. Variations between experiments (n = 2) were approximately 10% for the stimulated nucleotide dissociation rate and 20 to 50% for the low intrinsic dissociation rate.
FIG. 5.
Inhibition of the Cdc25Mm285-stimulated dissociation rate of Ras-3′mdGDP by Ras(E37G/D119N). Addition of Ras(E37G/D119N)-GDP to a mixture of 100 nM Ras-3′mdGDP, 100 nM Cdc25Mm285, and 20 μM GDP in standard buffer at 20°C inhibited the Cdc25Mm285-stimulated dissociation rate in a concentration-dependent manner. Replacement of the excess of 20 μM GDP (●) by 200 μM GDP (□) or 2 mM GDP (■) abrogated the inhibitory action of Ras(E37G/D119N). The Cdc25Mm285 preparation used in this experiment was 35% less active than the one used in Fig. 3. kobs, observed dissociation rate constant.
FIG. 6.
Ras(E37G/D119N) acts as a dominant-negative Ras mutant in PC12 cells. Histogram showing the effect of microinjection of different concentrations of Ras(E37G/D119N) in NGF-stimulated PC12 cells. Depicted is the percentage of microinjected cells showing outgrowth of neurites with at least twice the length of the cell. n, number of experiments (each with 100 to 200 cells). The bars indicate the deviation.
The effect of mutations in the Ras effector region on the activity of Cdc25Mm285.
Above, we have shown that mutation E37G hardly affects the interaction of Ras with Cdc25Mm285. In order to know whether other partial-loss-of-function mutations which are commonly used to elucidate different Ras-dependent pathways are involved in the GEF-dependent Ras activation, we have tested the activity of Cdc25Mm285 on Ras proteins mutated in residues 30, 31, 35, 38, and 40 (Table 1). Mutation D38A strongly reduces the Cdc25Mm285-dependent stimulation, which is somewhat less affected by mutations D38E, T35S, and T35A. These results are comparable to the results obtained with the catalytic domain of yeast exchange factor Sdc25p (27). In addition, mutation D30K, but not E31K, appears to reduce the stimulatory action of Cdc25Mm285. Surprisingly, mutation Y40C induces a gain-of-function effect and increases the Cdc25Mm285-stimulatory action by nearly a factor of 10. Nevertheless, whereas microinjection of 150 μM wild-type Ras caused neurite outgrowth in 9% of the injected PC12 cells, Ras(Y40C) is unable to induce neurite outgrowth (0% [data not shown]). This is most likely a consequence of the fact that Ras(Y40C) inefficiently interacts with Raf and RalGEFs and that activation of phosphoinositol-3-kinase alone is not sufficient for the induction of neurite outgrowth.
Our results point out that not all mutations in the effector region can be used in combination with D119N without disturbing the interaction with Cdc25Mm. Moreover, these effects are apparently not identical for all GEFs: mutation E37G hardly affects the interaction of Ras with Cdc25Mm285, but abrogates the interaction with the yeast exchange factor CDC25, as shown by 2H analysis (50).
Effects of Ras(E37G/D119N) and other double mutants in NIH 3T3 cells.
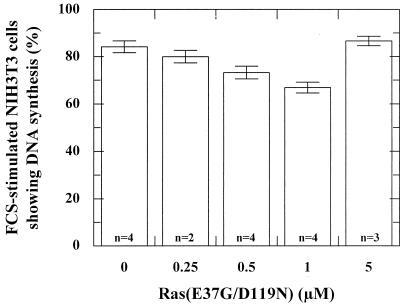
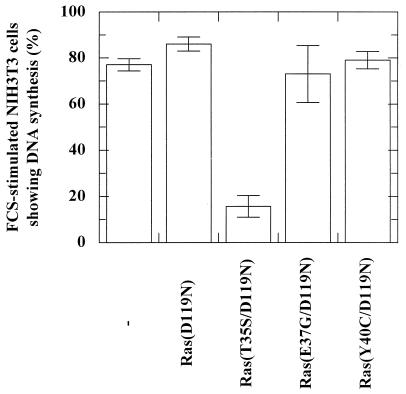
Microinjection of different concentrations of the protein Ras(E37G/D119N) in NIH 3T3 cells showed a weak inhibition of DNA synthesis at 1 μM, whereas no inhibition was observed at higher concentrations (Fig. 7). In order to analyze this further, we have engineered pEXV plasmids expressing Ras(D119N) and three Ras double mutants: Ras(T35S/D119N), Ras(E37G/D119N), and Ras(Y40C/D119N). To ascertain that we counted only transfected cells, we cotransfected the NIH 3T3 cells with pEGFP and selected the cells that showed EGFP fluorescence. As shown in Fig. 8, only the combination of mutations D119N and T35S resulted in a Ras mutant that was able to dominant-negatively abrogate serum-induced DNA synthesis in NIH 3T3 cells. In agreement with the microinjection experiment (Fig. 7), Ras(E37G/D119N) does not inhibit DNA synthesis.
FIG. 7.
Ras(E37G/D119N) and Ras(D119N) have similar effects on the FCS-induced DNA-synthesis in NIH 3T3 cells. Histogram showing the effect of microinjection of different concentrations of Ras(E37G/D119N) in FCS-stimulated NIH 3T3 cells. Depicted is the percentage of microinjected cells showing DNA synthesis as detected with the BrdU assay. n, number of experiments (each with 100 to 200 cells). The bars indicate the deviation.
FIG. 8.
Effector mutation T35S induces a dominant-negative effect in Ras(D119N) as shown by BrdU incorporation in NIH 3T3 cells. Histograms show the effect of transfection of serum-stimulated NIH 3T3 cells by pEXV-KF plasmids expressing Ras(D119N), Ras(T35S/D119N), Ras(E37G/D119N), or Ras(Y40C/D119N). Depicted is the percentage of transfected cells showing DNA synthesis as detected with the BrdU assay. The percentage is the mean of at least two experiments in which 100 to 200 transfected cells were counted. Transfected cells were determined by cotransfection with pEGFP and detection by fluorescence microscopy. The bars indicate the deviation.
The pEXV plasmids were unable to induce an activated phenotype for any of the Ras mutants. This was most likely due to a low expression of the Ras mutants, which allows the dominant-negative but not the activated character of the Ras(D119N) mutants to appear. Therefore, we recloned the Ras mutants in pcDNA3 plasmids, which allow a higher expression. In this context, Ras(E37G/D119N) and Ras(Y40C/D119N) were able to induce DNA synthesis in starving NIH 3T3 cells, whereas Ras(T35S/D119N) could not (data not shown).
DISCUSSION
We show in this study that Ras(D119N) inhibits growth factor stimulation in PC12 and NIH 3T3 cells at a cell-type-specific concentration. In NGF-stimulated PC12 cells, microinjection of 5 μM Ras(D119N) resulted in an approximately 50% inhibition of neurite formation (Fig. 1A). Below this concentration, neurites are probably formed because the amount of Ras(D119N) is not sufficient to sequester all GEF molecules and to prevent activation of endogenous wild-type Ras. Above this concentration, however, neurites are most likely formed, because the amount of mutated protein is in excess over the cellular GEF, and since Ras(D119N) is able to bind GTP in a GEF-independent manner and to bind Ras effectors such as c-Raf-1, GAP, and RalGDS (39, 56), Ras(D119N) acts as an activated Ras mutant. In addition to its effects in PC12 cells, microinjection of 0.5 μM Ras(D119N) inhibits DNA synthesis in FCS-stimulated NIH 3T3 cells by approximately 50% (Fig. 1B). As also shown here, the inhibitory effect of Ras(D119N) is overruled by the activated character of this mutant when higher concentrations of Ras(D119N) are microinjected.
In order to get support for our interpretation of the in vivo effect of Ras(D119N), we have analyzed the biochemical consequences of mutation D119N in more detail. First, we have measured its effect on the interaction of nucleotide-free Ras with the catalytic domain of exchange factor Cdc25Mm. By using a fluorescence titration assay (Fig. 2), we found that mutation D119N increases this affinity by a factor of 10. This was surprising, since no direct interaction of the Ras-specific GEF hSos1 with D119N was observed in the recently solved structure of the complex of nucleotide-free Ras with hSos1 (4) and may indicate a difference between hSos1 and Cdc25Mm or may be a consequence of indirect effects of mutation D119N. In addition to the 11.5-fold increase in the Cdc25Mm285 affinity of nucleotide-free Ras, D119N causes a 2,000-fold decrease in GDP affinity, as shown before (39). Together, this leads to a 23,000-fold increase in relative affinity of the nucleotide-free Ras for GEF over guanine nucleotide (Fig. 3A). The increase in relative affinity enables the mutant to sequester GEF and to prevent GEF stimulation of the dissociation rate of the Ras-GDP wild-type complex. We could show this in an in vitro competition assay (Fig. 4). As expected on the basis of the reaction scheme and from the nucleotide and GEF affinities, this inhibitory effect can be suppressed by the addition of xanthosine nucleotide or high (millimolar) concentrations of guanine nucleotide (Fig. 4). Before extrapolating the latter effect to the in vivo situation, it should be noted that, although the cellular concentration of nucleotide lies in the submillimolar range and thereby is high enough to abrogate the effect of Ras(D119N) in our in vitro studies, in the cellular system, both Ras and the GEF are membrane bound after growth factor stimulation. Consequently, the local concentrations of these interacting proteins are much higher, and the effect of the nucleotide concentration is less dramatic than in solution.
Similar to Ras(D119N), other mutants with reduced nucleotide affinity, e.g., Ras(S17N), Ras(D57Y), and Ras(F28L), inhibit the GEF action of Cdc25Mm285 in vitro (6a, 45). In particular, mutation S17N causes a 150- or 5,400-fold shift in relative affinity, depending on whether it is related to the GDP or the GTP affinity, respectively (Fig. 3B). Together with the observation that the cellular concentration of GTP is 30-fold higher than that of GDP (44), this indicates that the dominant-negative effect of Ras(S17N) is predominantly a consequence of the decrease in GTP affinity. However, a second aspect is equally important for the in vivo inhibitory action of Ras(S17N). Whereas Ras(D119N) can still interact with most, if not all, effector molecules, Ras(S17N), even in its GTP-bound form, can no longer interact with adenylyl cyclase (11), nor with the Ras-binding domains of c-Raf-1 or RalGDS (24). Moreover, it was shown that the GTPase activity of Ras(S17A) is not stimulated by p120GAP (16). Thus, it appears that besides a change in relative affinity, the loss in effector binding is a prerequisite for dominant-negative Ras mutants. Two aspects in the literature underline this argument. First of all, Sigal et al. (42) showed that microinjection of plasmids expressing mutant Ras(D119A) has an oncogenic effect in NIH 3T3 cells, but a dominant-negative effect in yeast cells. Since it was reported that Ha-Ras functions less well than endogenous RAS2 in yeast (8, 19), and assuming that this results from an inefficient interaction with adenylyl cyclase, one would expect that Ras(D119A) will act as a dominant-negative mutant in yeast, because its dominant-negative character can no longer be masked by an activated character at higher concentrations due to inefficient interaction with the effector. As another example, it was reported that mutation of residue Phe28 into Leu or Trp confers weakly transforming properties to Ras, whereas mutation F28D causes cell growth retardation (32, 33). Also here, a difference in effector binding may explain these results: mutation F28D is expected to affect the interaction with effector molecules more severely than mutations F28L and F28W, since a negative charge at position 28 would perturb the Ras structure much more than a hydrophobic residue.
In order to find further support for our interpretation of the concentration-dependent effect of Ras(D119N), we have introduced mutation E37G into Ras(D119N). This effector region mutation abrogates the interaction with c-Raf-1 and phosphoinositol-3-kinase, but not RalGEFs (10, 20, 26, 29, 30, 35, 50, 51). In accordance with the observation that the Raf-MAP kinase and phosphoinositol-3-kinase pathways are essential for PC12 differentiation (7, 21), microinjection of Ras(E37G/D119N) in PC12 cells at concentrations above 2 μM resulted in a dominant-negative effect (Fig. 6). In contrast, Ras(E37G/D119N) could not inhibit FCS-induced DNA synthesis in NIH 3T3 cells and behaved similarly to Ras(D119N) (compare Fig. 1B and 7). This result is consistent with the observation that mutation E37G does not abolish the Ras(G12V)-induced cell growth in NIH 3T3-UNC cells (20) or DNA synthesis in thyrocytes (26). We could, however, show by transfection that the combination of T35S with D119N enables Ras to abrogate DNA synthesis in FCS-stimulated NIH 3T3 cells (Fig. 8). Conflicting reports on the effects of mutation T35S can be found in the literature, but the general line is that this mutation affects binding to most effector molecules, but not c-Raf-1 (20, 30, 35, 41, 50–52). Several reports suggest that the Ral-mediated pathway is essential for the mitogenic response (26, 52), whereas the phosphoinositol-3-kinase-mediated pathway appears to be important for the mitogenic signals of a few, but not all, growth factors (34). Thus, the inhibitory effect of Ras(T35S/D119N) can be explained by assuming an inhibition of the Ral-mediated pathway. On the other hand, Ras(Y40C/D119N), which should also be capable of inhibiting the Ral pathway, was not capable of inhibiting DNA synthesis. Mutation Y40C was reported to weaken the interaction with Raf and RalGEFs, but not phosphoinositol-3-kinase (10, 20, 30, 35, 51, 52). This thus indicates that phosphoinositol-3-kinase may also play a role in the FCS-induced DNA synthesis in NIH 3T3 cells. Our observation that Ras(T35S/D119N) is not capable of inducing DNA synthesis in starving fibroblasts seems to be in conflict with the result that Ras(V12G/T35S) can stimulate cell growth in NIH 3T3(UNC) cells, to an even greater extent than the other two double mutations (20). For the moment, we do not have an explanation for this.
Recently, it has become more and more evident that Ras-dependent pathways are extremely complex. Therefore, it should be stressed that other interpretations of the effects of Ras(D119N) combined with effector mutations cannot be excluded. One aspect is that we have not addressed the question of whether the effector mutations influence the interaction with Sos and other Ras-specific GEFs different from Cdc25Mm. This may affect the phenotypes of these double mutants in different cell types. More importantly, during revision of the manuscript, it was shown that the Ral-mediated pathway inhibits NGF-induced PC12 differentiation (14). This observation would favor an alternative explanation for our results: Ras(E37G/D119N) can inhibit neurite outgrowth by preferentially stimulating the Ral pathway and not so much by sequestering the GEF(s). In addition, in a parallel study, we found that position 37 is one of the tree-determinant positions that determine the specific interaction of Ras-like proteins with effector proteins (2). By using double-hybrid assays, we observed that mutation E37A in Ras induces a weak, but significant, interaction with the Ral effector RLIP, implying that such a mutation may induce erroneous interaction with effector proteins of a different GTP-binding protein. As yet, we cannot distinguish between either of these possibilities. Also in terms of the effect of Ras(T35S/D119N) on DNA synthesis in NIH 3T3 cells, other explanations cannot be ruled out, since the cell cycle is influenced by multiple, partially synergizing pathways (see references 48 and 54 and the references within).
One of the aims of this study was elucidation of the mechanism of dominant-negative mutants. This is of great interest, since Rap1A(S17N) was found not to be able to inhibit the Rap1A-specific exchange factor C3G in vitro (45). Rap1A(S17N) was reported to inhibit cyclic AMP- and NGF-induced stimulation of MAP kinase in PC12 cells (47, 55), but it is less clear whether this is achieved by sequestration of the RapGEF C3G or another Rap-specific GEF (e.g., Epac) (9), or occurs by even another mechanism. Our results suggest that Rap1A(D119N) carrying a suitable partial-loss-of-function mutation may be a good alternative for Rap1A(S17N). However, since alternative interpretations for these results are possible (described above), additional experiments are needed to elucidate the inhibitory effects of the doubly mutated Ras mutants in greater detail before pursuing this approach. As another application of our results, since Ras(D119N) has a 10-fold-stronger inhibitory effect in vivo than Ras(S17N), homologs of Ras(D119N) may be a better bait in double-hybrid screens for new GEFs for GTP-binding proteins.
Recently, the structure of Ras-Sos (4) was determined, showing that Sos interacts with several residues of the switch I region (amino acids 25 to 40) of Ras, as also suggested by mutational analysis of Ras in combination with various RasGEFs (references 25 and 27 and the references within). In accordance with these earlier observations with different GEFs, mutations T35A and D38A strongly reduce the stimulatory action of Cdc25Mm285 (Table 1). Surprisingly, the dissociation rate of mutant Ras(Y40C), a mutant that still binds phosphoinositol-3-kinase and AF6 but that cannot interact with Raf, RalGEFs, adenylyl cyclase, and byr2 (10, 20, 30, 35, 51, 52), is stimulated 10 times more strongly by Cdc25Mm285 than by the Ras wild type. The side chain of Y40 is embedded in both the GDP- and GTP-bound conformations of Ras (37, 53), but contacts Sos in the Ras-Sos complex (4). A diminished interaction of the Ras mutant in the Ras-Cdc25Mm285 complex may allow the exchange reaction to proceed faster. This gain-of-function effect may also be of biological significance, since an increased GEF activity is expected to increase the concentration of Ras-GTP and thus may overcome thresholds of specific cellular pathways. In a parallel work, we could show that the effect of Y40C is associated with Cdc25Mm and does not occur with C3G when introducing the same mutation in Rap (46).
ACKNOWLEDGMENTS
We gratefully thank Christiane Theiß for excellent technical assistance, Margret Schulte Spechtel for help in protein preparations, Brigitte Oeke for help in microinjection experiments, Iris Simon for the fluorescently labeled xanthosine nucleotides, and our colleagues Nina van den Berghe and Christoph Block for fruitful discussion. R.H.C. was supported by the Biotec programs Bio2-CT93-0005 and Bio4-CT96-1110 of the European Community.
REFERENCES
- 1.Akiyamo Y, Ito K. SecY protein, a membrane-embedded secretion factor of E. coli, is cleaved by the OmpT protease in vitro. Biochem Biophys Res Commun. 1990;167:711–715. doi: 10.1016/0006-291x(90)92083-c. [DOI] [PubMed] [Google Scholar]
- 2.Bauer B, Mirey G, Vetter I R, García-Ranea J A, Valencia A, Wittinghofer A, Camonis J H, Cool R H. Effector recognition by the small GTP-binding proteins Ras and Ral. J Biol Chem. 1999;274:17763–17770. doi: 10.1074/jbc.274.25.17763. [DOI] [PubMed] [Google Scholar]
- 3.Boguski M S, McCormick F. Proteins regulating Ras and its relatives. Nature. 1993;366:643–654. doi: 10.1038/366643a0. [DOI] [PubMed] [Google Scholar]
- 4.Boriack-Sjodin P A, Margarit S M, Bar-Sagi D, Kuriyan J. The structural basis of the activation of Ras by Sos. Nature. 1998;394:337–343. doi: 10.1038/28548. [DOI] [PubMed] [Google Scholar]
- 5.Bourne H R, Sanders D A, McCormick F. The GTPase superfamily: a conserved switch for diverse cell functions. Nature. 1990;348:125–132. doi: 10.1038/348125a0. [DOI] [PubMed] [Google Scholar]
- 6.Cai H, Szeberényi J, Cooper G M. Effect of a dominant inhibitory Ha-ras mutation on mitogenic signal transduction in NIH 3T3 cells. Mol Cell Biol. 1990;10:5314–5323. doi: 10.1128/mcb.10.10.5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6a.Cool, R. H. Unpublished data.
- 7.Cowley S, Paterson H, Kemp P, Marshall C J. Activation of MAP kinase kinase is necessary and sufficient for PC12 differentiation and for transformation of NIH 3T3 cells. Cell. 1994;77:841–852. doi: 10.1016/0092-8674(94)90133-3. [DOI] [PubMed] [Google Scholar]
- 8.DeFeo-Jones D, Tatchell K, Robinson L C, Sigal I S, Vass W C, Lowy D R, Scolnick E M. Mammalian and yeast ras gene products: biological function in their heterologous systems. Science. 1985;228:179–184. doi: 10.1126/science.3883495. [DOI] [PubMed] [Google Scholar]
- 9.De Rooij J, Zwartkruis F J T, Verheijen M H G, Cool R H, Nijman S M B, Wittinghofer A, Bos J L. Epac is a Rap1 guanine-nucleotide-exchange factor directly activated by cyclic AMP. Nature. 1998;396:474–477. doi: 10.1038/24884. [DOI] [PubMed] [Google Scholar]
- 10.Esser D, Bauer B, Wolthuis R M F, Wittinghofer A, Cool R H, Bayer P. Structure determination of the Ras-binding domain of the Ral-specific exchange factor Rlf. Biochemistry. 1998;37:13453–13462. doi: 10.1021/bi9811664. [DOI] [PubMed] [Google Scholar]
- 11.Farnsworth C L, Feig L A. Dominant inhibitory mutations in the Mg2+-binding site of RasH prevent its activation by GTP. Mol Cell Biol. 1991;11:4822–4829. doi: 10.1128/mcb.11.10.4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feig L A, Cooper G M. Inhibition of NIH 3T3 cell proliferation by a mutant ras protein with preferential affinity for GDP. Mol Cell Biol. 1988;8:3235–3243. doi: 10.1128/mcb.8.8.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feig L A, Pan B-T, Roberts T M, Cooper G M. Isolation of ras GTP-binding mutants using an in situ colony-binding assay. Proc Natl Acad Sci USA. 1986;83:4607–4611. doi: 10.1073/pnas.83.13.4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goi T, Rusanescu G, Urano T, Feig L A. Ral-specific guanine nucleotide exchange factor activity opposes other Ras effectors in PC12 cells by inhibiting neurite outgrowth. Mol Cell Biol. 1999;19:1731–1741. doi: 10.1128/mcb.19.3.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han M, Sternberg P W. Analysis of dominant-negative mutations of the Caenorhabditis elegans let-60 ras gene. Genes Dev. 1991;5:2188–2198. doi: 10.1101/gad.5.12a.2188. [DOI] [PubMed] [Google Scholar]
- 16.John J, Rensland H, Schlichting I, Vetter I, Borasio G D, Goody R S, Wittinghofer A. Kinetic and structural analysis of the Mg2+-binding site of the guanine nucleotide-binding protein p21H-ras. J Biol Chem. 1993;268:923–929. [PubMed] [Google Scholar]
- 17.Jones S, Litt R J, Richardson C J, Segev N. Requirement of nucleotide exchange factor for Ypt1 GTPase mediated protein transport. J Cell Biol. 1995;130:1051–1061. doi: 10.1083/jcb.130.5.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jung V, Wei W, Ballester R, Camonis J, Mi S, Van Aelst L, Wigler M, Broek D. Two types of RAS mutants that dominantly interfere with activators of RAS. Mol Cell Biol. 1994;14:3707–3718. doi: 10.1128/mcb.14.6.3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kataoka T, Powers S, Cameron S, Fasano O, Goldfarb M, Broach J, Wigler M. Functional homology of mammalian and yeast RAS genes. Cell. 1985;40:19–26. doi: 10.1016/0092-8674(85)90304-6. [DOI] [PubMed] [Google Scholar]
- 20.Khosravi-Far R, White M A, Westwick J K, Solski P A, Chrzanowska-Wodnicka M, Van Aelst L, Wigler M H, Der C J. Oncogenic Ras activation of Raf/mitogen-activated protein kinase-independent pathways is sufficient to cause tumorigenic transformation. Mol Cell Biol. 1996;16:3923–3933. doi: 10.1128/mcb.16.7.3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kimura K, Hattori S, Kabuyama Y, Shizawa Y, Takayanagi J, Nakamura S, Toki S, Matsuda Y, Onodera K, Fukui Y. Neurite outgrowth of PC12 cells is suppressed by wortmannin, a specific inhibitor of phosphatidylinositol 3-kinase. J Biol Chem. 1994;269:18961–18967. [PubMed] [Google Scholar]
- 22.Lenzen C, Cool R H, Wittinghofer A. Analysis of intrinsic and CDC25-stimulated guanine nucleotide exchange of p21ras-nucleotide complexes by fluorescence measurements. Methods Enzymol. 1995;225:95–109. doi: 10.1016/s0076-6879(95)55012-7. [DOI] [PubMed] [Google Scholar]
- 23.Lenzen C, Cool R H, Prinz H, Kuhlmann J, Wittinghofer A. Kinetic analysis by fluorescence of the interaction between Ras and the catalytic domain of mouse guanine nucleotide exchange factor Cdc25Mm. Biochemistry. 1998;37:7420–7430. doi: 10.1021/bi972621j. [DOI] [PubMed] [Google Scholar]
- 24.Lenzen, C. U., and R. H. Cool. Unpublished data.
- 25.Leonardsen L, DeClue J E, Lybaek H, Lowy D R, Willumsen B M. Rasp21 sequences opposite the nucleotide binding pocket are required for GRF-mediated nucleotide release. Oncogene. 1996;13:2177–2187. [PubMed] [Google Scholar]
- 26.Miller M, Prigent S, Kupperman E, Rioux L, Park S-H, Feramisco J R, White M A, Rutkowski J L, Meinkoth J L. RalGDS functions in Ras- and cAMP-mediated growth stimulation. J Biol Chem. 1997;272:5600–5605. doi: 10.1074/jbc.272.9.5600. [DOI] [PubMed] [Google Scholar]
- 27.Mistou M Y, Jacquet E, Poullet P, Rensland H, Gideon P, Schlichting I, Wittinghofer A, Parmeggiani A. Mutations of Ha-ras p21 that define important regions for the molecular mechanism of the SDC25 C-domain, a guanine nucleotide dissociation stimulator. EMBO J. 1992;11:2391–2397. doi: 10.1002/j.1460-2075.1992.tb05303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Munder T, Fürst P. The Saccharomyces cerevisiae CDC25 gene product binds specifically to catalytically inactive Ras proteins in vivo. Mol Cell Biol. 1992;12:2091–2099. doi: 10.1128/mcb.12.5.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murai H, Ikeda M, Kishida S, Ishida O, Okazaki-Kishida M, Matsuura Y, Kikuchi A. Characterization of Ral GDP dissociation stimulator-like (RGL) activities to regulate c-fos promoter and the GDP/GTP exchange of Ral. J Biol Chem. 1997;272:10483–10490. doi: 10.1074/jbc.272.16.10483. [DOI] [PubMed] [Google Scholar]
- 30.Okazaki M, Kishida S, Hinoi T, Hasegawa T, Tamada M, Kataoka T, Kikuchi A. Synergistic activation of c-fos promoter activity by Raf and Ral GDP dissociation stimulator. Oncogene. 1997;14:515–521. doi: 10.1038/sj.onc.1200860. [DOI] [PubMed] [Google Scholar]
- 31.Powers S, O’Neill K, Wigler M. Dominant yeast and mammalian RAS mutants that interfere with the CDC25-dependent activation of wild-type RAS in Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:390–395. doi: 10.1128/mcb.9.2.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reinstein J, Schlichting I, Frech M, Goody R S, Wittinghofer A. p21 with a phenylalanine 28→leucine mutation reacts normally with the GTPase activating protein GAP but nevertheless has transforming properties. J Biol Chem. 1991;266:17700–17706. [PubMed] [Google Scholar]
- 33.Ricketts M H, Durrheim G A, North H M, van der Merwe M J, Levinson A D. Positive and negative modulation of H-ras transforming potential by mutations of phenylalanine-28. Mol Biol Rep. 1996;23:109–117. doi: 10.1007/BF00424436. [DOI] [PubMed] [Google Scholar]
- 34.Roche S, Downward J, Raynal P, Courtneidge S A. A function for phosphatidylinositol 3-kinase β (p85α-p110β) in fibroblasts during mitogenesis: requirement for insulin- and lysophosphatidic acid-mediated signal transduction. Mol Cell Biol. 1998;18:7119–7129. doi: 10.1128/mcb.18.12.7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodriguez-Viciana P, Warne P H, Khwaja A, Marte B M, Pappin D, Das P, Waterfield M D, Ridley A, Downward J. Role of phosphoinositide 3-OH kinase in cell transformation and control of the actin cytoskeleton by Ras. Cell. 1997;89:457–467. doi: 10.1016/s0092-8674(00)80226-3. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. New York, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Schlichting I, Almo S C, Rapp G, Wilson K, Petratos K, Lentfer A, Wittinghofer A, Kabsch W, Pai E M, Petsko G A, Goody R S. Time-resolved X-ray crystallographic study of the conformational change in Ha-Ras p21 protein on GTP hydrolysis. Nature. 1990;345:309–315. doi: 10.1038/345309a0. [DOI] [PubMed] [Google Scholar]
- 38.Schmidt, G. Unpublished results.
- 39.Schmidt G, Lenzen C, Simon I, Deuter R, Cool R H, Goody R S, Wittinghofer A. Biochemical and biological consequences of changing the specificity of p21ras from guanosine to xanthosine nucleotides. Oncogene. 1996;12:87–96. [PubMed] [Google Scholar]
- 40.Schweighoffer F, Cai H, Chevallier-Multon C, Fath I, Cooper G, Tocque B. The Saccharomyces cerevisiae SDC25 C-domain gene product overcomes the dominant inhibitory activity of Ha-Ras Asn-17. Mol Cell Biol. 1993;13:39–43. doi: 10.1128/mcb.13.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shirouzu M, Koide H, Fujita-Yoshigaki J, Oshio H, Toyama Y, Yamasaki K, Fuhrman S, Villafranca E, Kaziro Y, Yokoyama S. Mutations that abolish the ability of Ha-Ras to associate with Raf-1. Oncogene. 1994;9:2153–2157. [PubMed] [Google Scholar]
- 42.Sigal I S, Gibbs J B, D’Alonzo J S, Temeles G L, Wolanski B S, Socher S H, Scolnick E M. Mutant ras-encoded proteins with altered nucleotide binding exert dominant biological effects. Proc Natl Acad Sci USA. 1986;83:952–956. doi: 10.1073/pnas.83.4.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stacey D W, Feig L A, Gibbs J B. Dominant inhibitory Ras mutants selectively inhibit the activity of either cellular or oncogenic Ras. Mol Cell Biol. 1991;11:4053–4064. doi: 10.1128/mcb.11.8.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trahey M, McCormick F. A cytoplasmatic protein stimulates normal N-ras p21 GTPase, but does not affect oncogenic mutants. Science. 1987;238:542–545. doi: 10.1126/science.2821624. [DOI] [PubMed] [Google Scholar]
- 45.Van den Berghe N, Cool R H, Horn G, Wittinghofer A. Biochemical characterization of C3G: an exchange factor that discriminates between Rap1 and Rap2 and is not inhibited by Rap1A(S17N) Oncogene. 1997;15:845–850. doi: 10.1038/sj.onc.1201407. [DOI] [PubMed] [Google Scholar]
- 46.Van den Berghe N, Cool R H, Wittinghofer A. Discriminatory residues in Ras and Rap for guanine nucleotide exchange factor recognition. J Biol Chem. 1999;271:11078–11085. doi: 10.1074/jbc.274.16.11078. [DOI] [PubMed] [Google Scholar]
- 47.Vossler M R, Yao H, York R D, Pan M-G, Rim C S, Stork P J S. cAMP activates MAP kinase and Elk-1 through a B-Raf- and Rap1-dependent pathway. Cell. 1997;89:73–82. doi: 10.1016/s0092-8674(00)80184-1. [DOI] [PubMed] [Google Scholar]
- 48.Walker F, Kato A, Gonez L J, Hibbs M L, Pouliot N, Levitzki A, Burgess A W. Activation of the Ras/mitogen-activated protein kinase pathway by kinase-defective epidermal growth factor receptors results in cell survival but not proliferation. Mol Cell Biol. 1998;18:7192–7204. doi: 10.1128/mcb.18.12.7192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walter M, Clark S G, Levinson A D. The oncogenic activation of human p21ras by a novel mechanism. Science. 1986;233:649–652. doi: 10.1126/science.3487832. [DOI] [PubMed] [Google Scholar]
- 50.White M A, Nicolette C, Minden A, Polverino A, van Aelst L, Karin M, Wigler M H. Multiple Ras functions can contribute to mammalian cell transformation. Cell. 1995;80:533–541. doi: 10.1016/0092-8674(95)90507-3. [DOI] [PubMed] [Google Scholar]
- 51.White M A, Vale T, Camonis J H, Schaefer E, Wigler M H. A role for the Ral guanine nucleotide dissociation stimulator in mediating Ras-induced transformation. J Biol Chem. 1996;271:16439–16442. doi: 10.1074/jbc.271.28.16439. [DOI] [PubMed] [Google Scholar]
- 52.Wolthuis R M F, de Ruiter N, Cool R H, Bos J L. Stimulation of gene induction and cell growth by the Ras effector Rlf. EMBO J. 1997;16:6748–6761. doi: 10.1093/emboj/16.22.6748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamasaki K, Shirouza M, Muto Y, Fujita-Yoshigaki J, Koide H, Ito Y, Kawai G, Hattori S, Yokoyama S, Nishimura S, Miyazawa T. Site-directed mutagenesis, fluorescence, and two-dimensional NMR studies on microenvironments of effector region aromatic residues of human c-Ha-Ras protein. Biochemistry. 1994;33:65–73. doi: 10.1021/bi00167a009. [DOI] [PubMed] [Google Scholar]
- 54.Yang J-J, Kang J-S, Krauss R S. Ras signals to the cell cycle machinery via multiple pathways to induce anchorage-independent growth. Mol Cell Biol. 1998;18:2586–2595. doi: 10.1128/mcb.18.5.2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.York R D, Yao H, Dillon T, Ellig C L, Ecker S P, McCleskey E W, Stork P J S. Rap1 mediates sustained MAP kinase activation induced by nerve growth factor. Nature. 1998;392:622–626. doi: 10.1038/33451. [DOI] [PubMed] [Google Scholar]
- 56.Zhong J-M, Chen-Hwang M-C, Hwang Y-W. Switching nucleotide specificity of Ha-ras p21 by a single amino acid substitution at aspartate 119. J Biol Chem. 1995;270:10002–10007. doi: 10.1074/jbc.270.17.10002. [DOI] [PubMed] [Google Scholar]
- 57.Ziman M, O’Brien J M, Quellette L A, Church W R, Johnson D I. Mutational analysis of CDC42Sc, a Saccharomyces cerevisiae gene that encodes a putative GTP-binding protein involved in the control of cell polarity. Mol Cell Biol. 1991;11:3537–3544. doi: 10.1128/mcb.11.7.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]