Abstract
The prevalence of obesity has tripled over the past four decades, imposing an enormous burden on people’s health. Polygenic (or common) obesity and rare, severe, early-onset monogenic obesity are often polarized as distinct diseases. However, gene discovery studies for both forms of obesity show that they have shared genetic and biological underpinnings, pointing to a key role for the brain in the control of body weight. Genome-wide association studies (GWAS) with increasing sample sizes and advances in sequencing technology are the main drivers behind a recent flurry of new discoveries. However, it is the post-GWAS, cross-disciplinary collaborations, which combine new omics technologies and analytical approaches, that have started to facilitate translation of genetic loci into meaningful biology and new avenues for treatment.
Subject terms: Disease genetics, Endocrine system and metabolic diseases, Genetic association study, Genetic variation, Genomics
In this Review, Loos and Yeo summarize our current understanding of the genetic underpinnings of monogenic and polygenic obesity. They highlight the commonalities revealed by recent studies and discuss the implications for treatment and prediction of obesity risk.
Introduction
Obesity is associated with premature mortality and is a serious public health threat that accounts for a large proportion of the worldwide non-communicable disease burden, including type 2 diabetes, cardiovascular disease, hypertension and certain cancers1,2. Mechanical issues resulting from substantially increased weight, such as osteoarthritis and sleep apnoea, also affect people’s quality of life3. The impact of obesity on communicable disease, in particular viral infection4, has recently been highlighted by the discovery that individuals with obesity are at increased risk of hospitalization and severe illness from COVID-19 (refs5–7).
On the basis of the latest data from the NCD Risk Factor Collaboration, in 2016 almost 2 billion adults (39% of the world’s adult population) were estimated to be overweight (defined by a body mass index (BMI) of ≥25 kg m−2), 671 million (12% of the world’s adult population) of whom had obesity (BMI ≥30 kg m−2) — a tripling in the prevalence of obesity since 1975 (ref.8) (Fig. 1). Although the rate of increase in obesity seems to be declining in most high-income countries, it continues to rise in many low-income and middle-income countries and prevalence remains high globally8. If current trends continue, it is expected that 1 billion adults (nearly 20% of the world population) will have obesity by 2025. Particularly alarming is the global rise in obesity among children and adolescents; more than 7% had obesity in 2016 compared with less than 1% in 1975 (ref.8).
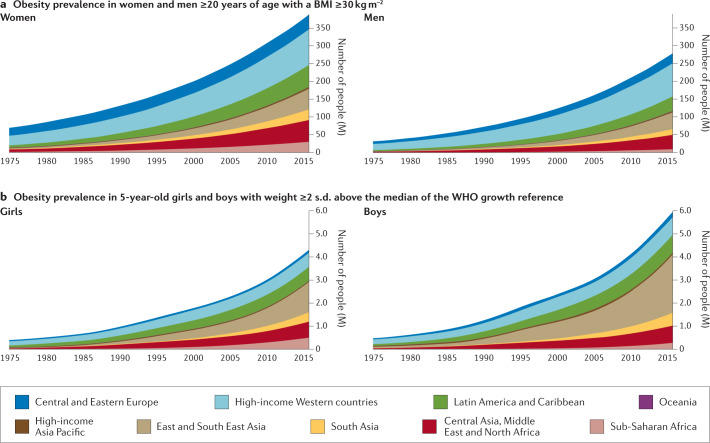
Fig. 1. Prevalence of obesity in males and females according to age and geographical region.
The prevalence of obesity has risen steadily over the past four decades in children, adolescents (not shown) and adults worldwide. a | Prevalence of obesity (body mass index (BMI) ≥30 kg m−2) in women and men ≥20 years of age, from 1975 to 2016. b | Prevalence of obesity (weight ≥2 s.d. above the median of the WHO growth reference) in 5-year-old girls and boys from 1975 to 2016. Geographical regions are represented by different colours. Graphs are reproduced from the NCD Risk Factor Collaboration (NCD RisC) website and are generated from data published in ref.8.
Although changes in the environment have undoubtedly driven the rapid increase in prevalence, obesity results from an interaction between environmental and innate biological factors. Crucially, there is a strong genetic component underlying the large interindividual variation in body weight that determines people’s response to this ‘obesogenic’ environment. Twin, family and adoption studies have estimated the heritability of obesity to be between 40% and 70%9,10. As a consequence, genetic approaches can be leveraged to characterize the underlying physiological and molecular mechanisms that control body weight.
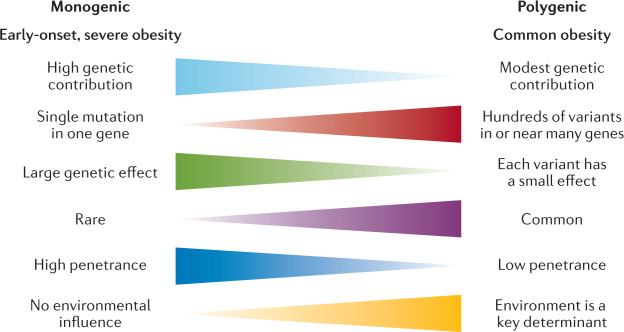
Classically, we have considered obesity in two broad categories (Fig. 2): so-called monogenic obesity, which is inherited in a Mendelian pattern, is typically rare, early-onset and severe and involves either small or large chromosomal deletions or single-gene defects; and polygenic obesity (also known as common obesity), which is the result of hundreds of polymorphisms that each have a small effect. Polygenic obesity follows a pattern of heritability that is similar to other complex traits and diseases. Although often considered to be two distinct forms, gene discovery studies of monogenic and polygenic obesity have converged on what seems to be broadly similar underlying biology. Specifically, the central nervous system (CNS) and neuronal pathways that control the hedonic aspects of food intake have emerged as the major drivers of body weight for both monogenic and polygenic obesity. Furthermore, early evidence shows that the expression of mutations causing monogenic obesity may — at least in part — be influenced by the individual’s polygenic susceptibility to obesity11.
Fig. 2.
Key features of monogenic and polygenic forms of obesity.
In this Review, we summarize more than 20 years of genetic studies that have characterized the molecules and mechanisms that control body weight, specifically focusing on overall obesity and adiposity, rather than fat distribution or central adiposity. Although most of the current insights into the underlying biology have been derived from monogenic forms of obesity, recent years have witnessed several successful variant-to-function translations for polygenic forms of obesity. We also explore how the ubiquity of whole-exome sequencing (WES) and genome sequencing has begun to blur the line that used to demarcate the monogenic causes of obesity from common polygenic obesity. Syndromic forms of obesity, such as Bardet–Biedl, Prader–Willi, among many others12, are not reviewed here. Although obesity is often a dominant feature of these syndromes, the underlying genetic defects are often chromosomal abnormalities and typically encompass multiple genes, making it difficult to decipher the precise mechanisms directly related to body-weight regulation. Finally, as we enter the post-genomic era, we consider the prospects of genotype-informed treatments and the possibility of leveraging genetics to predict and hence prevent obesity.
Gene discovery approaches
The approaches used to identify genes linked to obesity depend on the form of obesity and genotyping technology available at the time. Early gene discovery studies for monogenic forms of obesity had a case-focused design: patients with severe obesity, together with their affected and unaffected family members, were examined for potential gene-disrupting causal mutations via Sanger sequencing. By contrast, genetic variation associated with common forms of obesity have been identified in large-scale population studies, either using a case–control design or continuous traits such as BMI. Gene discovery for both forms of obesity was initially hypothesis driven; that is, restricted to a set of candidate genes that evidence suggests have a role in body-weight regulation. Over the past two decades, however, advances in high-throughput genome-wide genotyping and sequencing technologies, combined with a detailed knowledge of the human genetic architecture, have enabled the interrogation of genetic variants across the whole genome for their role in body-weight regulation using a hypothesis-generating approach.
Gene discovery for monogenic obesity
Many of the candidate genes and pathways linked to body-weight regulation were initially identified in mice, such as the obese (ob)13 and diabetes (db)14 mouse lines, in which severe hyperphagia and obesity spontaneously emerged. Using reverse genetics, the ob gene was shown to encode leptin, a hormone produced from fat, and it was demonstrated that leptin deficiency resulting from a mutation in the ob gene caused the severe obesity seen in the ob/ob mouse15 (Fig. 3). Shortly after the cloning of ob, the db gene was cloned and identified as encoding the leptin receptor (LEPR)16. Reverse genetics was also used to reveal that the complex obesity phenotype of Agouti ‘lethal yellow’ mice is caused by a rearrangement in the promoter sequence of the agouti gene that results in ectopic and constitutive expression of the agouti peptide17,18, which antagonizes the melanocortin 1 and 4 receptors (MC1R and MC4R)19,20. This finding linked the melanocortin pathway to body-weight regulation, thereby unveiling a whole raft of new candidate genes for obesity.
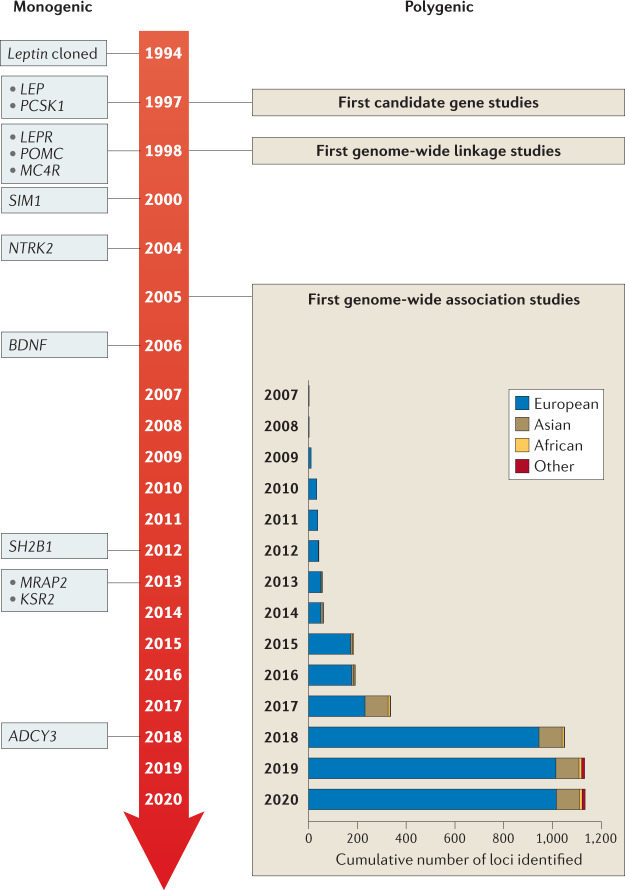
Fig. 3. Timeline of key discoveries in obesity genetics.
Genes identified for monogenic obesity in a given year are shown on the left. Discoveries made for polygenic obesity are shown on the right, including a cumulative count of newly discovered loci per year and by ancestry. Although candidate gene and genome-wide linkage studies became available in the late 1990s, findings were limited, and these study designs are not as frequently used as genome-wide association studies.
Once the genes for leptin and its receptor were identified, they became candidate genes for human obesity, and in 1997 the first humans with congenital leptin deficiency were identified21. This discovery was rapidly followed by the report of humans with mutations in the gene encoding the leptin receptor (LEPR)22, as well as in genes encoding multiple components of the melanocortin pathway, including PCSK1 (ref.23), MC4R24–26 and POMC27–29, all of which were found to result in severe early-onset obesity (Table 1).
Table 1.
Overview of all genes implicated in severe and early-onset obesity
| Gene symbol, name, Gene ID | Location (human, GRCh38/hg38) chr: start…end position | Species in which naturally occurring mutations cause obesity | Tissue expression | Nearby GWAS-identified locus (index SNP) | Role in body-weight regulation |
|---|---|---|---|---|---|
| ADCY3, adenylate cyclase, Gene ID: 109 | 2: 24,819,168…24,920,236 | Humans | Primary cilia of cells | rs6545814 | Disruption of primary cilia in neurons known to influence energy balance67; some evidence of a specific link to the correct function of MC4R67,156 |
| AGRP, agouti-related protein, Gene ID: 181 | 16: 67,482,571…67,483,547 | – | Neurons in the arcuate nucleus of the hypothalamus | – | Endogenous antagonist of MC4R, to which it binds to increase food intake92,94 |
| BDNF, brain-derived neurotrophic factor, Gene ID: 627 | 11: 27,654,893…27,722,030 | Humans | Brain | rs925946 | Probably via its role in regulating neuronal synaptic plasticity111,115 |
| KSR2, kinase suppressor of Ras2, Gene ID: 283455 | 12: 117,453,012…117,968,990 | Humans | Wide expression throughout the body | rs56214831 | Influences both energy intake and expenditure, possibly via interaction with AMPK168 |
| LEP, leptin, Gene ID: 3952 | 7: 128,241,201…128,257,629 | Humans and mice | Fat | rs10487505 | Circulates in proportion to fat mass21,90, and turns on the neuroendocrine starvation response when circulating levels drop below a minimum threshold91 |
| LEPR, leptin receptor, Gene ID: 3953 | 1: 65,420,652…65,641,559 | Humans and mice | The long ‘signalling’ form is expressed widely in the brain | rs11208659 | Cognate receptor for leptin, mediating its downstream neuroendocrine functions16,22 |
| MC4R, melanocortin 4 receptor, Gene ID: 4160 | 18: 60,371,062…60,372,775 | Humans, pigs and blind Mexican cavefish | Central nervous system | rs17782313 | Binds melanocortin peptides and AGRP to mediate appetitive behaviour92 and autonomic output169 |
| MRAP2, melanocortin receptor accessory protein 2, Gene ID: 112609 | 6: 84,032,621…84,146,278 | Humans | Wide expression throughout the body, but highest in the brain | – | An accessory protein that plays a role in trafficking MC4R to the cell surface99 |
| NTRK2, neurotrophic receptor tyrosine kinase 2, Gene ID: 4915 | 9: 84,668,458…85,027,070 | Humans | Brain | rs10868215 | Cognate receptor for BDNF, mediating its downstream effects on synaptic plasticity116 |
| PCSK1, proprotein convertase subtilisin/kexin type 1, Gene ID: 5122 | 5: 96,390,333…96,433,248 | Humans | Endocrine organs, with highest expression in the brain | rs6235 | Encodes one of the prohormone convertases required for processing POMC93 |
| PHIP, pleckstrin homology domain interacting protein, Gene ID: 55023 | 6: 78,934,419…79,078,294 | Humans | Widely expressed | – | Regulates transcription of POMC98 |
| POMC, pro-opiomelanocortin, Gene ID: 5443 | 2: 25,160,860…25,168,580 | Humans and Labrador retriever dogs | Hypothalamus, nucleus tractus solitaris, pituitary, skin, adrenal glands and numerous other tissues | rs6545975 | Complex pro-polypeptide that is processed into melanocortin peptides that signal to MC4R in the brain27,93 |
| SH2B1, SH2B adaptor protein 1 Gene ID: 25970 | 16: 28,846,606…28,874,205 | Humans | Widely expressed | rs7498665 | A signalling molecule downstream of the leptin receptor97 |
| SIM1, SIM bHLH transcription factor 1, Gene ID: 6492 | 6: 100,385,009…100,464,929 | Humans | Hypothalamus, kidney and fat | rs6907240 | A transcription factor crucial for the proper development of the paraventricular nucleus and hence appropriate expression of MC4R, among other genes100 |
AMPK, AMPK-activated kinase; bHLH. basic helix–loop–helix; GWAS, genome-wide association study.
Advances in high-throughput DNA sequencing led to candidate gene screening being replaced by WES, an unbiased approach that allows all coding sequences to be screened for mutations. However, it rapidly became clear that, whereas candidate gene studies yielded few mutations, WES identified too many potential obesity-associated variants such that the noise often masked the true causative mutations. However, with improved algorithms to predict the pathogenicity of mutations, as well as a rapidly expanding toolkit of functional assays, it has become easier to filter the likely pathogenic mutations. Several success stories have been reported in which WES has identified novel pathways and genes linked to obesity, such as the class 3 semaphorins (SEMA3A–G), which have been shown to direct the development of certain hypothalamic neurons, including those expressing pro-opiomelanocortin (POMC)30 (see ‘Other neuronal circuits and molecules linked to severe obesity’).
Most monogenic obesity mutations have been identified in cohorts of patients with severe and early-onset (<10 years old) obesity. Additionally, as monogenic obesity often demonstrates a recessive inheritance pattern31, consanguinity in populations has further increased the chance of identifying mutations, owing to greater chances of homozygosity of deleterious mutations32. For example, studies have reported that mutations in the genes encoding leptin, LEPR and MC4R explain 30% of cases of severe obesity in children from a consanguineous Pakistani population33, and single-gene defects more broadly account for nearly 50%34.
Gene discovery for polygenic obesity
The discovery of genes that influence polygenic obesity, which is common in the general population, started off slowly with candidate gene studies and genome-wide linkage studies. The candidate gene approach was first applied in the mid-1990s and aimed to validate genes identified through human and animal models of extreme obesity for a role in common obesity (Fig. 3). Common variants in such candidate genes were tested for association with obesity risk, BMI or other body composition traits. Over the subsequent 15 years, hundreds of genes were studied as candidates, but variants in only six (ADRB3 (ref.35), BDNF36, CNR1 (ref.37), MC4R38, PCSK1 (ref.39) and PPARG40) showed reproducible association with obesity outcomes. The genome-wide linkage approach made its entrance into the field towards the end of the 1990s (Fig. 3). Genome-wide linkage studies rely on the relatedness of individuals and test whether certain chromosomal regions co-segregate with a disease or trait across generations. Even though more than 80 genome-wide linkage studies identified >300 chromosomal loci with suggestive evidence of linkage with obesity traits, few loci were replicated and none was successfully fine-mapped to pinpoint the causal gene or genes41. Ultimately, candidate gene and genome-wide linkage studies, constrained by small sample sizes, sparse coverage of genetic variation across the genome and lack of replication, only had a marginal impact on the progression of gene discovery for common obesity outcomes.
However, the pace of gene discovery for common diseases accelerated with the advent of genome-wide association studies (GWAS) (Fig. 3). The first GWAS for obesity traits were published in 2007 and identified a cluster of common variants in the first intron of the FTO locus that was convincingly associated with BMI42,43. Many more GWAS followed and, to date, nearly 60 GWAS have identified more than 1,100 independent loci associated with a range of obesity traits44 (Supplementary Tables 1,2).
As sample sizes increase with each consecutive GWAS, the statistical power to identify more loci also increases, in particular for loci that are less common and/or have smaller effects. For example, the first GWAS were relatively small (n = ~5,000) and identified only the FTO locus42,43. The BMI-increasing allele of FTO is common, particularly in populations of European ancestry (minor allele frequency (MAF) 40–45%), and has a relatively large effect on BMI (0.35 kg m−2 per allele; equivalent to 1 kg for a person who is 1.7 m tall). Ten years and numerous GWAS later, the most recent GWAS for BMI included nearly 800,000 individuals, identified more than 750 loci, with MAFs as small as 1.6% and per-allele effects as low as 0.04 kg m−2 per allele (equivalent to 120 g for a person who is 1.7 m tall)45. Combined, these genome-wide significant loci explained 6% of variation in BMI45. Large-scale international collaborations have been formed, such as the Genetic Investigation for Anthropometric Traits (GIANT) consortium, that combine summary statistics of individual GWAS to generate data sets comprising hundreds of thousands of individuals. Furthermore, many GWAS efforts have maximized sample size by focusing on BMI as the primary obesity outcome, an inexpensive and easy-to-obtain measurement that is readily available in most studies. As such, the vast majority of loci have been identified first in GWAS of BMI, but their effects typically transfer to other overall adiposity outcomes.
Even though BMI is widely used, it is considered a crude proxy of overall adiposity because it does not distinguish between lean and fat mass46. Therefore, GWAS have been performed for more refined obesity traits, such as body fat percentage47,48, fat-free mass49, imaging-derived adipose tissue50, circulating leptin levels51 and LEPR levels52. In addition, two GWAS have focused on persistent healthy thinness, assuming that genes that determine resistance to weight gain may also inform obesity prevention and weight loss maintenance53,54. Although GWAS of more refined and alternative obesity outcomes are generally much smaller than those for BMI, the phenotypes are often a more accurate representation of body-weight regulation and, as such, the loci identified tend to more often point to relevant biological pathways that underlie obesity.
Almost all GWAS loci for obesity outcomes were first identified in adults. Most of these loci also associate with obesity and/or BMI in children and adolescents, highlighting the fact that the genetic underpinning of obesity is relatively constant across the course of life55–57. Similarly to gene discovery for other common diseases, the obesity genetics field has suffered from a strong bias in population representation, with the vast majority of GWAS being performed in populations that are exclusively or predominantly of European ancestry. Nevertheless, some loci have first been discovered in populations of Asian58, African59,60, Hispanic or other ancestry61, despite their much smaller sample sizes. Broadly, loci identified in one ancestry demonstrate good transferability (that is, directionally consistent associations) across other ancestries, even though effect sizes and allele frequencies may differ. The modest-to-high genetic correlations across ancestries observed for BMI (r = 0.78) are consistent with good transferability62, but also suggest that ancestry-specific loci remain to be discovered. Besides increasing the sample sizes of GWAS in populations of non-European ancestry, demographic, evolutionary and/or genomic features of specific populations (such as founder, consanguineous or isolated populations) have been leveraged for gene discovery, identifying genetic variants with large effects that are common in the discovery population, such as CREBRF, first identified in Samoan populations, and ADCY3, first identified in the Greenlandic population, but rare or nonexistent in most others63–66. CREBRF has been shown to play a role in cellular energy storage and use, and may be implicated in cellular and organismal adaptation to nutritional stress65. ADCY3 colocalizes with MC4R at the primary cilia of a subset of hypothalamic neurons that have been implicated in body-weight regulation67.
GWAS have typically focused on biallelic, common genetic variation (MAF >5%), but have also been used to screen for the role of copy number variants (CNVs) in obesity. So far, only a few CNVs have been identified that have a convincing association with BMI, such as the 1p31.1 45-kb deletion near NEGR1 (ref.68), which encodes a cell-adhesion molecule expressed in the brain69; the 16p12.3 21-kb deletion upstream of GPRC5B70, which may modulate insulin secretion71; the 10q11.22 CNV in PPYR1 (also known as NPY4R)72, which encodes a potent anti-obesity agent known to inhibit food intake73; and the 1p21.1 multi-allele CNV encompassing AMY1A74, which produces salivary α-amylase, a key enzyme in starch digestion75.
To determine the role of other types of variation in obesity, alternative genome-wide screens have been performed. For example, the impact of low-frequency and rare protein-coding variants has been tested using exome sequencing and exome array data76–79. It was speculated that low-frequency (MAF 1–5%) and rare (MAF <1%) variants would have larger effects than common variants, and thus be easier to detect. Nevertheless, even large-scale studies identified only a few robust associations for rare coding variants. For example, exome-wide screening based on array data from more than 400,000 individuals identified p.Tyr35Ter (rs13447324) in MC4R; p.Arg190Gln (rs139215588) and p.Glu288Gly (rs143430880) in GIPR, which stimulates insulin secretion and mediates fat deposition80; p.Arg95Ter (rs114285050) in GRP151, which modulates habenular function that controls addiction vulnerability81; and p.Arg769Ter (rs533623778) in PKHD1L1, which has been involved in cancer development77,78. A recent study that leveraged WES data for more than 600,000 individuals identified 16 genes for which the burden of rare nonsynonymous variants was associated with BMI, including five brain-expressed G protein-coupled receptors (CALCR, MC4R, GIPR, GPR151 and GPR75)79.
As obesity is a complex, multifactorial condition, some GWAS have integrated demographic factors (such as sex and age82) and environmental factors (such as physical activity83, diet84 or smoking85) into their analyses. Despite sample sizes of more than 200,000 individuals, these genome-wide gene-by-environment (G×E) interaction analyses remain challenging and so far only 12 loci have been identified, the effects of which on obesity are attenuated or exacerbated by non-genetic factors. Nevertheless, the G×E interaction between the FTO locus and a healthy lifestyle has been robustly replicated. Specifically, increased physical activity or a healthy diet can attenuate the effect of the FTO locus on obesity risk by 30–40%86,87.
The increasing availability of large-scale cohorts and biobanks, such as the UK Biobank, the Million Veterans Project, All of Us, Biobank Japan and 23andMe, combined with ongoing work by the GIANT consortium, will boost sample sizes further to easily exceed 4 million participants in meta-analyses, expediting the discovery of many more obesity-associated loci. However, translation of GWAS-identified loci into new biological insights remains a major challenge.
From genes to biology
Despite the difficulties in validating causative mutations and variants, genetic studies into both rare and common obesity over the past two decades have revealed two surprisingly cogent, overarching biological messages: first, the leptin–melanocortin pathway is a key appetitive control circuit31,88 (Fig. 4); and second, genes that are either enriched or exclusively expressed within the brain and CNS have a central role in obesity89.
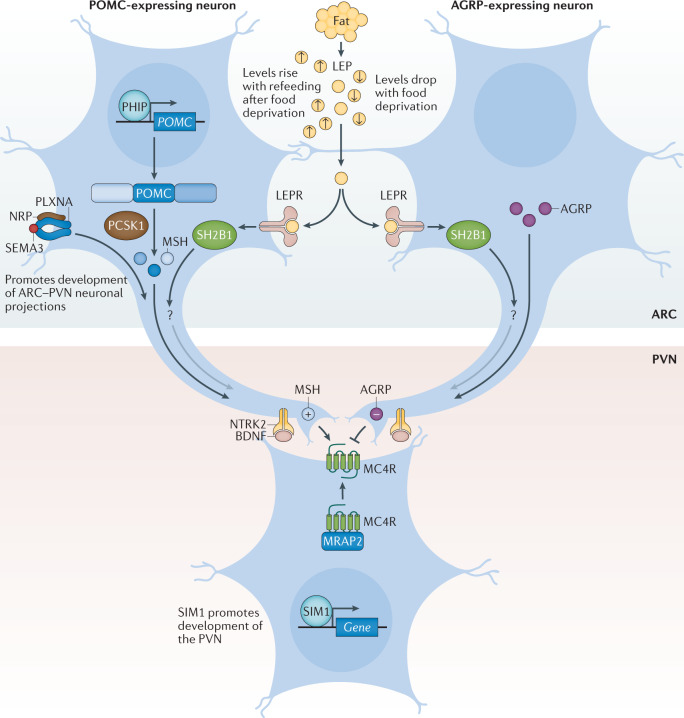
Fig. 4. The leptin–melanocortin pathway.
Pro-opiomelanocortin (POMC)-expressing neurons and agouti-related protein (AGRP)-expressing neurons within the arcuate nucleus of the hypothalamus (ARC) act to sense circulating leptin (LEP) levels, which reflect fat mass. These neurons signal to melanocortin 4 receptor (MC4R)-expressing neurons in the paraventricular nucleus of the hypothalamus (PVN), which controls appetite, thus linking long-term energy stores to feeding behaviour. Binding of class 3 semaphorins (SEMA3) to their receptors NRP and PLXNA influences the projection of POMC neurons to the PVN. Binding of brain-derived neurotrophic factor (BDNF) to its receptor neurotrophic receptor tyrosine kinase 2 (NTRK2) is thought to be an effector of leptin-mediated synaptic plasticity of neurons, including those in the ARC and PVN. The transcription factor SIM1 is crucial for the proper development of the PVN. +, agonist; −, antagonist; LEPR, leptin receptor; MRAP2, melanocortin receptor accessory protein 2; MSH, melanocyte-stimulating hormone; SH2B1, SH2B adaptor protein 1.
The leptin–melanocortin pathway and MC4R
Leptin is a key hormone secreted by adipocytes, which circulates at levels in proportion to fat mass90. Leptin also responds to acute changes in energy state, as its levels decrease with food deprivation and are restored during re-feeding. Administration of leptin to fasted mice abrogates many of the neuroendocrine consequences of starvation, suggesting that the normal biological role of leptin is to initiate the starvation response91. Leptin signals through the LEPR, which exists in several different isoforms. However, obesity-related effects of leptin are predominantly mediated by a long isoform that contains an intracellular domain (LEPRb), which is expressed in various regions of the CNS90.
Within the arcuate nucleus (ARC) of the hypothalamus, LEPRb is found on two populations of neurons at the heart of the melanocortin pathway, one of which expresses POMC and the other agouti-related protein (AGRP)92 (Fig. 4). POMC is post-translationally processed by prohormone convertases to produce several biologically active moieties, including β-lipotrophin and β-endorphin, and, crucially, the melanocortin peptides adrenocorticotrophin (ACTH) and α-, β- and γ-melanocyte-stimulating hormone (MSH)93. The ARC POMC neurons project to MC4R neurons within the paraventricular nucleus (PVN) where melanocortin peptides signal to decrease food intake92. By contrast, AGRP acts as an endogenous antagonist of MC4R to increase food intake92,94. MC3R is another centrally expressed receptor that binds to both melanocortin peptides and AGRP; however, as mice with targeted deletions in the gene are not obese but instead have altered fat to lean mass ratio, MC3R is less likely to be related to food intake and more likely to be involved in nutrient partitioning95,96.
We can state with confidence that the fine balance of melanocortinergic agonism and AGRP antagonism of MC4R, in response to peripheral nutritional cues such as leptin, plays a central part in influencing appetitive drive92. The genetic evidence clearly supports this contention, with mutations in most genes of the melanocortin pathway resulting in hyperphagia and severe obesity in both humans and mice31,88. In fact, the vast majority of single-gene disruptions causing severe early-onset obesity in humans fall within this pathway, including LEPR, POMC, AGRP, MCR4R, PCSK1 (ref.23), SH2B1 (ref.97), PHIP98, MRAP2 (ref.99) and SIM1 (ref.100) (Fig. 4; Table 1). Mutations in MC4R in particular, are the most common single-gene defect leading to hyperphagia and obesity. Pathogenic mutations in MC4R are found in up to 5% of cases of severe childhood obesity101 and up to 0.3% of the general population101,102. Of note, the degree of receptor dysfunction, as measured by in vitro assays, can predict the amount of food eaten at a test meal by an individual harbouring that particular mutation101. Thus MC4R does not act in a binary on/off manner, but as a rheostat; put simply, the melanocortin pathway is a ‘tunable’ system. In addition to regulating food intake, it also regulates food preference, with individuals who carry mutations in MC4R showing a preference for food with higher fat content103.
The importance of the melanocortin pathway in regulating feeding behaviour is highlighted by the identification of naturally occurring mutations in pathway genes in a wide range of different species where the appropriate selection pressure has been present (Table 1). For example, studies have found that 20–25% of Labrador retrievers, which are known to be more food-motivated than other dog breeds, carry a 14-bp deletion in POMC that disrupts the β-MSH and β-endorphin coding sequences and is associated with greater food motivation and increased body weight104. Also, certain breeds of pig have been shown to carry MC4R missense mutations that are associated with fatness, growth and food intake traits105. MC4R mutations even contribute to the adaptation and survival of blind Mexican cavefish to the nutrient-poor conditions of their ecosystem106.
Other neuronal circuits and molecules linked to severe obesity
It is now clear that in addition to engaging classical neuropeptide–receptor systems within the brain, leptin also rapidly modifies synaptic connections between neurons107, and that this structural plasticity is crucial to its downstream functions. One of the ways in which this plasticity is thought to be achieved is via brain-derived neurotrophic factor (BDNF) signalling to its receptor TrkB. BDNF is widely expressed in the CNS where it plays an important part in neuronal development108,109. In the hippocampus, BDNF contributes to synaptic plasticity and long-term potentiation associated with memory and learning110. However, evidence has emerged that implicates BDNF and TrkB in the regulation of mammalian eating behaviour and energy balance111. BDNF is downregulated by nutritional deprivation and upregulated by leptin within the ventromedial nucleus (VMN) of the hypothalamus112, although this regulation is probably indirect, as very few VMN BDNF neurons express the LEPR113 (Fig. 4) and some evidence indicates that it acts at least in part downstream of melanocortin signalling112. Crucially, genetic disruption of BDNF114,115 and TrkB112,116 in both humans and mice results in hyperphagia and severe obesity.
Another group of neuronal proteins important in the development of neuronal circuitry and linked to energy balance are the class 3 semaphorins (SEMA3A–G). A study in humans found that 40 rare loss-of-function variants in SEMA3A–G and their receptors (PLXNA1–4, NRP1 and NRP2) were significantly enriched in 982 individuals with severe obesity compared with 4,449 controls30. Disruption of several of these genes in zebrafish caused increased somatic growth and/or adiposity, and experiments with mouse hypothalamic explants suggest that SEMA3 signalling via NRP2 receptors drives the development of POMC projections from the ARC to the PVN30. However, given that these results are from a single study, more data are required to confirm the exact role of class 3 semaphorins in energy homeostasis.
Insights from genetic loci linked to common obesity
Unlike candidate gene studies, GWAS make no a priori assumptions about the underlying biology that links genetic variants to a disease of interest. While this agnostic approach allows for new biological insights, the vast majority of GWAS-identified variants map to the non-coding parts of genes or to regions between genes. As such, they do not directly disrupt the protein-coding regions, but instead overlap with regulatory elements that influence expression of genes in close proximity or even over long distances.
However, even if the causative genes are unknown, pathway, tissue and functional enrichment analyses based on the genes located in the GWAS loci can provide insights into potential mechanisms. Since the very first GWAS for BMI68,117, such analyses have pointed to the CNS being a key player in body-weight regulation, consistent with insights from human and animal models of extreme obesity. Recent analyses that include the latest BMI-associated loci, combined with updated multi-omics databases and advanced computational tools, have further refined these observations. In addition to the hypothalamus and pituitary gland (which are both known appetite regulation sites), other brain areas have been highlighted, including the hippocampus and the limbic system (which are involved in learning, cognition and emotion) and the insula and the substantia nigra (which are related to addiction and reward)58,89,118,119. The enrichment of immune-related cells (such as lymphocytes and B cells) and adipose tissue was found to be weaker58.
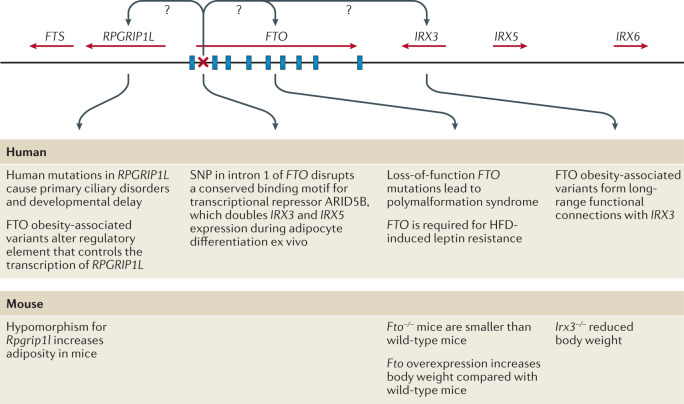
Although enrichment analyses provide preliminary insights into the broad biology represented by genes in the GWAS loci, determining which genes, variants and/or underlying mechanisms are causal has proved an arduous task. For example, the FTO locus, which was identified more than a decade ago and harbours six genes, is the most extensively studied GWAS-identified obesity locus (Fig. 5). Despite its highly significant and widely replicated association with obesity120, the causal variants and/or genes in the FTO locus have not yet been pinpointed with convincing evidence, and the mechanisms by which the locus affects body weight have not been fully elucidated. Early functional follow-up analyses suggested that FTO itself might be responsible, as Fto deficiency in mice results in a lean phenotype, whereas Fto overexpression is associated with increased body weight121,122. Studies in mice have suggested that FTO plays a role in cellular nutrient sensing123,124. Other studies found evidence that FTO influences brain regions that affect appetite, reward processing and incentive motivation by regulating ghrelin levels in humans125 or by controlling dopaminergic signalling in mice126,127. In addition, variants in the FTO locus were shown to alter a regulatory element that controls the transcription of Rpgrip1l in mice, a ciliary gene located immediately upstream of Fto128–130. Mice with reduced Rpgrip1l activity exhibit hyperphagic obesity, possibly mediated through diminished leptin signalling128–130. In recent years, studies in human and animal models have shown that variants in the FTO locus directly interact with the promoter of Irx3, a gene located 0.5 Mb downstream of FTO. Irx3-deficient mice were found to exhibit weight loss and increased metabolic rate with browning of white adipose tissue, without changes in physical activity or appetite131,132. Further in-depth functional characterization showed that rs1421085 in the FTO locus disrupts a conserved binding motif for the transcriptional repressor ARID5B, which leads to a doubling of IRX3 and IRX5 expression during early adipocyte differentiation132. The authors argue that increased expression of these genes results in a developmental shift from energy-dissipating beige adipocytes to energy-storing white adipocytes, a fivefold reduction in mitochondrial thermogenesis and increased lipid storage132. However, given that multiple studies have shown that the FTO locus is robustly associated with food intake, with no evidence to date linking it to changes in energy expenditure, the relevance of this observation to the actual observed human phenotype still needs to be explored133. A recent study reports that the FTO locus affects gene expression in multiple tissues, including adipose tissue and brain, and, more broadly, that the genetic architecture of disease-associated loci may involve extensive pleiotropy and allelic heterogeneity across tissues134.
Fig. 5. Schematic representation of the FTO locus and its neighbouring genes on human chromosome 16q22.
FTO contains nine exons (depicted by blue rectangles) and the body mass index (BMI)-associated SNP identified in genome-wide association studies (depicted by a red ×) maps to intron 1. IRX3 and RPGRIP1L have both been proposed to be the causal genes for obesity within the locus and to act on body weight through distinct mechanisms. HFD, high-fat diet.
Besides the FTO locus, functional follow-up analyses have been performed for only a few obesity-associated GWAS loci. For example, early studies identified a cluster of variants just downstream of TMEM18 (refs68,117). TMEM18 encodes a poorly characterized transmembrane protein that is highly conserved across species and widely expressed across tissues, including in several regions of the brain135,136. Tmem18 deficiency in mice results in a higher body weight owing to increased food intake, whereas Tmem18 overexpression reduces food intake and limits weight gain136. A knockdown experiment in Drosophila melanogaster suggests that TMEM18 affects carbohydrate and lipid levels by disrupting insulin and glucagon signalling137.
Two other GWAS loci for which functional analyses have been performed are located just upstream of CADM1 (ref.82) and in CADM2 (ref.70), genes that encode cell-adhesion proteins of the immunoglobulin superfamily and mediate synaptic assembly in the CNS138. The BMI-increasing alleles at each locus are associated with increased expression of CADM1 and CADM2 in the hypothalamus139,140. Deficiency of either Cadm1 or Cadm2 in mice results in a lower body weight and increased insulin sensitivity, glucose tolerance and energy expenditure without any change in food intake139,140. Conversely, increased neuronal expression of either Cadm1 or Cadm2 is associated with elevated body weight139,140. Furthermore, CADM1 is expressed in POMC neurons and Cadm1 deficiency leads to an increase in the number of excitatory synapses, suggestive of an increased synaptic plasticity140. Cadm2-deficient mice exhibit increased locomotor activity and higher core body temperature139.
Another GWAS locus, just upstream of NEGR1, harbours two deletions associated with increased obesity risk68,117,141. These deletions do not overlap with the coding sequence of NEGR1, but encompass a conserved transcription factor-binding site for NKX6.1, a potent transcriptional repressor68,141. Loss of binding of NKX6.1 leads to higher NEGR1 expression141, which is consistent with the observation that BMI-increasing alleles (that is, deletions) at this locus are associated with higher NEGR1 expression in the brain. Similar to CADM1 and CADM2, NEGR1 is a cell-adhesion molecule of the immunoglobulin superfamily that is expressed in several regions of the brain and has been shown to have a role in brain connectivity69,142, a process believed to be important in obesity143. NEGR1 deficiency in mice was shown to result in lower body weight, mainly due to reduced lean mass, mediated by lower food intake144. However, two other functional studies, one in mice and one in rats, found that knockdown of Negr1 expression resulted in the opposite phenotype — increased body weight and food intake145,146. While NEGR1 deficiency in mice was found to impair core behaviours, so far, findings and proposed mechanisms are not fully aligned69,147–149.
Taken together, functional follow-up analyses for these loci are slowly expanding our understanding of the pathophysiology that drives weight gain. However, many more obesity-associated loci are waiting to be translated into new biological insights. A major hurdle in translating GWAS loci into plausible candidate genes and appropriate paradigms for functional research is the annotation of the associated variants in a locus. Defining the regulatory function of the non-coding variants, identifying their putative effector transcripts and determining their tissues of action remains an ongoing challenge. The advent of high-throughput genome-scale technologies for mapping regulatory elements, combined with comprehensive multi-omics databases, advanced computational tools and the latest genetic engineering and molecular phenotyping approaches, is poised to speed up the translation of GWAS loci into meaningful biology150.
Converging results from monogenic and polygenic forms of obesity
Gene discovery is often dichotomized by allele frequency and disease prevalence; that is, mutations are sought for monogenic forms of obesity and common variants for polygenic obesity (Fig. 2). However, it is increasingly recognized that monogenic and polygenic forms of obesity are not discrete entities. Instead, they lie on a spectrum and share — at least in part — the same biology. As GWAS have continued to discover more obesity-associated loci, an increasing number of these loci harbour genes that were first identified for extreme and early-onset obesity in humans or animal models, including MC4R151,152, BDNF117, SH2B1 (refs68,117), POMC70, LEP51,153, LEPR52,154, NPY155, SIM1 (ref.155), NTRK2 (ref.58), PCSK1 (ref.154) and KSR2 (ref.77). In fact, most of these genes encode components of the leptin–melanocortin and BDNF–TrkB signalling pathways (Table 1). Thus, whereas genetic disruption of components of these pathways results in severe obesity, genetic variants in or near these same genes that have more subtle effects on their expression will influence where an individual might sit in the normal distribution of BMI.
Although most genes have been first identified for extreme forms of obesity, a locus harbouring ADCY3 was first identified in GWAS for common obesity77, and ADCY3 was subsequently confirmed as having a role in extreme obesity63,64. ADCY3 encodes an adenylate cyclase that catalyses the synthesis of cAMP, an important second messenger in signalling pathways. There is some evidence that ADCY3 (adenylate cyclase) colocalizes with MC4R at the primary cilia of PVN neurons67 and that cilia are required specifically on MC4R-expressing neurons for the control of energy homeostasis156. In mice, disruption of Adcy3 or Mc4r in the cilia of these neurons impairs melanocortin signalling, resulting in hyperphagia and obesity67.
As more GWAS loci are reported, we expect that findings across different lines of obesity research will continue to converge, providing accumulating evidence for new biology.
From genes to clinical care
Genetic insights from gene discovery efforts are increasingly being used in the context of precision medicine in ways that directly affect health. Knowing a patient’s genotype may enable a more precise diagnosis of the type of obesity, which in turn allows the prescription of personalized treatment or prevention strategies. Furthermore, knowing an individual’s genetic susceptibility to obesity early in life may help to more accurately predict those most at risk of gaining weight in the future.
Use of genotype information in treatment of obesity
When a disease is caused by a single mutation and the environmental contribution is limited, as is the case for some forms of extreme and early-onset obesity, a genetic test can be instrumental in correctly diagnosing patients. Although no standard genetic testing panel is currently available for extreme and early-onset obesity, some clinics, research centres and pharmaceutical companies sequence well-known candidate genes to identify the functional mutation that may be the cause of a patient’s excess body weight. Such a genetic diagnosis can lessen the feelings of guilt and blame for the patient, and alleviate social stigma and discrimination. Importantly, a genetic diagnosis can inform disease prognosis and, in some cases, it will determine treatment. To date, there are two treatments for obesity that are tailored to patient genotype.
The prototype of genotype-informed treatment for obesity is the administration of recombinant human leptin in patients who are leptin-deficient owing to mutations in the LEP gene157,158. Although congenital leptin deficiency is exceptionally rare (only 63 cases have been reported to date28), leptin replacement therapy has been remarkably beneficial for these patients by substantially reducing food intake, body weight and fat mass, and normalizing endocrine function157,158. It has literally transformed their lives.
The second genotype-informed treatment for obesity is setmelanotide, a selective MC4R agonist that was recently approved by the FDA for rare monogenic obesity conditions including LEPR, PCSK1 and POMC deficiency159. Setmelanotide acts as a substitute for the absent MSH in patients with POMC deficiency owing to mutations in POMC or PCSK1, and in patients with LEPR deficiency owing to mutations in LEPR, which is essential for POMC function160–162. Daily subcutaneous injection of setmelanotide results in substantial weight loss and in reduction of hunger160–162. After a 1-year treatment with setmelanotide in phase III trials, patients with POMC deficiency lost on average 25.6% of their initial weight, with 80% of patients achieving at least a 10% weight loss162. The adverse effects of setmelanotide treatment are minor, and include hyperpigmentation, nausea and/or vomiting, penile erection and injection site reactions. Weight loss in patients with LEPR deficiency was less pronounced; on average, they lost 12.5% of their initial weight, with only 45% of patients achieving at least a 10% weight loss162. The difference in weight loss between the two patient groups may be because POMC deficiency directly affects the production of MC4R ligands (α-MSH and β-MSH), whereas LEPR deficiency affects signalling upstream of POMC162. As such, setmelanotide may be able to completely restore MC4R signalling in POMC deficiency, but only partially in LEPR deficiency. Even though the average weight loss in POMC-deficient patients was twice that in LEPR-deficient patients, the reduction in hunger was substantially larger in LEPR-deficient patients (−43.7%) than in POMC-deficient patients (−27.1%)162. The reasons for the discrepancy between weight loss and reduction in hunger remain to be studied in greater depth. It has been estimated that in the USA, >12,800 individuals carry mutations in the melanocortin pathway for whom setmelanotide may be more effective for weight loss than any other treatment163. Although 12,800 carriers represent only a fraction (0.004%) of the adult population in the USA, and not all of these mutation carriers are overweight or obese, for the patients for whom setmelanotide is effective, it may end a lifelong battle to lose weight163. In patients without genetic defects, neither setmelanotide nor leptin administration have, to date, demonstrated a substantial effect on weight loss164,165.
These two genotype-informed treatments show how insight into the underlying biological mechanisms can guide the development of molecules and medications that restore impaired pathways, at least in monogenic forms of obesity caused by deficiency of one protein. Nevertheless, there remain substantial obstacles in the transition from conventional to precision medicine for monogenic obesity, which would require the adoption of systematic WES for individuals suspected to be carriers of deleterious mutations, and eventually even standardized screening at birth. We are clearly a long way from such a scenario at present.
Use of genotype information in prediction of obesity
As more variants are being discovered for common obesity, there is a growing expectation that genetic information will soon be used to identify individuals at risk of obesity. Knowing a person’s genetic susceptibility would allow for a more accurate prediction of who is at risk of gaining weight and give an opportunity to intervene earlier to prevent obesity more effectively. Genetic susceptibility to complex disease, including obesity, is assessed using a polygenic score (PGS). PGSs to assess obesity susceptibility are based on GWAS for BMI (PGSBMI), the latest of which includes data on more than 2 million variants and explains 8.4% of the variation in BMI166. The average BMI of individuals with a high PGSBMI (top decile) is 2.9 kg m−2 (equivalent to 8 kg in body weight) higher and their odds of severe obesity (BMI ≥40 kg m−2) is 4.2-fold higher than those with a lower PGSBMI (lowest nine deciles)166.
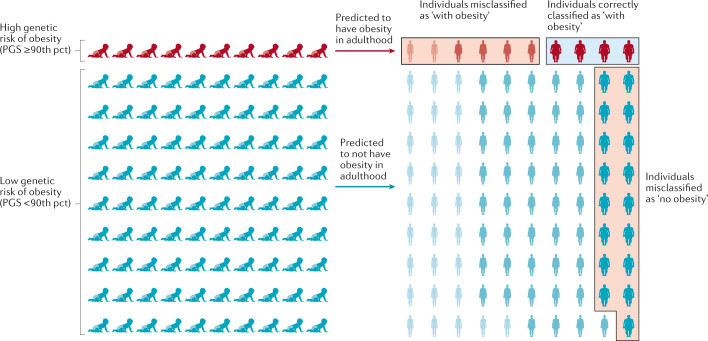
Despite these strong associations with BMI and obesity, the predictive performance of the PGSBMI is weak, which is unsurprising given its limited explained variance. For example, using the same PGSBMI and data from the UK Biobank, we estimate that the area under the receiver operating characteristic curve (AUCROC) is only 0.64 to predict obesity. This means that the probability that an individual with obesity has a higher PGSBMI than an individual without obesity is 0.64. However, for a PGS to have clinical utility, the AUCROC needs to be much higher (>0.80). In addition, we calculated the extent to which a PGSBMI ≥90th percentile correctly classifies individuals with obesity (Fig. 6). We found that such a predictive test (PGSBMI ≥90th percentile) has a positive predictive value of 0.43, meaning that of those who were predicted to develop obesity, only 43% actually developed obesity. Its sensitivity is 0.19, which means that of the individuals who developed obesity, only 19% had been correctly classified by the PGSBMI. Given that the current treatment options for obesity are low risk, or even generally beneficial, the high false-positive rate is less concerning than the low sensitivity, as some at-risk individuals may miss the opportunity for early prevention.
Fig. 6. Predicting obesity using a polygenic score.
The outcome is illustrated for a polygenic score (PGS) that assumes that individuals with a score in the highest decile (≥90th percentile (pct)) will develop obesity, has a positive predictive value of 0.4 and a sensitivity of 0.19. Of ten individuals with a high score classified by the PGS as ‘with obesity’, four will be classified correctly but the other six will be misclassified and will not develop obesity — a positive predictive value of 0.4. Likewise, 17 of the 90 individuals with a score <90th pct who are predicted to not develop obesity, will develop obesity. Thus, only four of the 21 individuals who developed obesity were correctly classified by the PGS — a sensitivity of 0.19. Misclassified individuals are indicated by the red boxes, individuals correctly classified as ‘with obesity’ are indicated by a blue box. Adapted with permission from ref.170, Elsevier.
Thus, the current PGSBMI has a high rate of misclassification and does not reliably predict who is at risk of developing obesity and who is not. The predictive ability of PGSs are expected to improve as GWAS increase in sample size and algorithms to calculate the scores become more refined. Nevertheless, given the importance of socio-demographic, lifestyle and clinical risk factors in the aetiology of obesity, it is unlikely that a PGSBMI will ever be able to accurately predict obesity on its own. Instead, effective prediction models will have to include genetic and non-genetic factors, including a broad spectrum of demographic, environmental, clinical and possibly molecular markers, as well.
Conclusions and future perspectives
What initially began as two apparently distinct approaches, one studying rare Mendelian causes of extreme obesity, and the other exploring complex polygenic influences of population body-weight distribution, have eventually converged on the central role of the brain in regulating body weight. In particular, both approaches have highlighted the roles of the leptin–melanocortin pathway and TrkB–BDNF signalling. Perhaps it seems obvious now, but it was by no means certain that, just because genetic disruption of a pathway resulted in a severe phenotype, polymorphisms within that same pathway would produce a more subtle and nuanced result.
The GWAS approach is hypothesis-free, with the promise to reveal new genes that point to new biology and pathways. However, for the vast majority of the >1,000 GWAS-identified loci, we do not know which genes are causal, what cells, tissues and organs they act in to affect body weight, and we do not understand the underlying mechanisms. The translation from variant to function is a well-known challenge167, but with increasing availability of new omics data, high-throughput technologies and advanced analytical approaches, there is an unprecedented opportunity to speed up the translation of hundreds of GWAS loci.
Sample size remains a major driver for gene discovery. In an ongoing collaboration that combines data from more than 3 million individuals of diverse ancestry from the GIANT consortium, the UK Biobank and 23andMe, the number of BMI-associated GWAS loci is set to double. Also, a recent WES effort of more than 640,000 individuals has demonstrated that rare mutations are discoverable when sample sizes are sufficiently large79. However, alternative study designs, a focus on more refined phenotypes or a focus on population subgroups (that is, more homogeneous groups of individuals with similar outcomes) could further add to gene discovery.
Translation of only a few dozen of the GWAS-identified loci could tremendously improve our insights into the biology of obesity and possibly reveal new therapeutic targets. It would also take us a little closer to the ‘holy grail’ — the ability to move away from a failed ‘one-size-fits-all’ strategy, and towards true precision medicine for obesity, metabolic disease and other diet-related illnesses.
Supplementary information
Acknowledgements
R.J.F.L. is supported by funding from Novo Nordisk Foundation (NNF Laureate Award) and the US National Institutes of Health (R01DK110113; R01DK107786; R01HL142302; R01 DK124097). G.S.H.Y. is supported by the Medical Research Council (MRC Metabolic Diseases Unit (MC_UU_00014/1)). The authors thank M. Guindo Martinez for her help with creating data for Fig. 3 and Supplementary Tables 1 and 2.
Glossary
- Obesogenic environment
An environment that promotes weight gain.
- Monogenic obesity
A severe, early-onset form of obesity, caused by a single-gene mutation, with little or no influence of the environment.
- Polygenic obesity
A common multifactorial form of obesity, resulting from an interaction between the obesogenic environment and hundreds of genetic variants.
- Reverse genetics
An approach used to understand the function of a gene by analysing the consequences of genetically manipulating specific sequences within the gene.
- Candidate gene studies
A hypothesis-driven approach to study the effect of a given gene (chosen based on the current understanding of its biology and pathophysiology) on susceptibility to the phenotype under study.
- Genome-wide linkage studies
A method that relies on the relatedness of study participants to test whether certain chromosomal regions co-segregate with a disease or trait across generations.
- Genome-wide association studies
(GWAS). A hypothesis-generating approach that screens whole genomes for associations between genetic variants and a phenotype of interest at much higher resolution than is possible for genome-wide linkage studies, and is thus better able to narrow down the associated locus.
- Polygenic score
(PGS). A measure used to assess an individual’s genetic susceptibility to disease, calculated by summing the number of disease-increasing alleles, weighted by each variant’s effect size observed in a genome-wide association study.
- Area under the receiver operating characteristic curve
(AUCROC). A metric used to assess the ability of a predictor to discriminate between individuals with and without a disease. The AUC ranges from 0.50 (equal to tossing a coin) to 1.0 (perfect prediction).
Author contributions
The authors contributed equally to all aspects of the article.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information
Nature Reviews Genetics thanks D. Meyre, T. Fall and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
23andMe: https://research.23andme.com/collaborate/
All of Us: https://allofus.nih.gov/
Biobank Japan: http://jenger.riken.jp/en/
Genetic Investigation for ANthropometric Traits (GIANT) consortium: https://portals.broadinstitute.org/collaboration/giant/index.php/Main_Page
Million Veterans Project: https://www.research.va.gov/mvp/
NCD Risk Factor Collaboration (NCD RisC): https://www.ncdrisc.org/data-visualisations.html
UK Biobank: https://www.ukbiobank.ac.uk/
Contributor Information
Ruth J. F. Loos, Email: ruth.loos@sund.ku.dk
Giles S. H. Yeo, Email: gshy2@cam.ac.uk
Supplementary information
The online version contains supplementary material available at 10.1038/s41576-021-00414-z.
References
- 1.GBD 2015 Obesity Collaborators. Health effects of overweight and obesity in 195 countries over 25 years. N. Engl. J. Med. 2017;377:13–27. doi: 10.1056/NEJMoa1614362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Must A, et al. The disease burden associated with overweight and obesity. JAMA. 1999;282:1523–1529. doi: 10.1001/jama.282.16.1523. [DOI] [PubMed] [Google Scholar]
- 3.Fontaine KR, Barofsky I. Obesity and health-related quality of life. Obes. Rev. 2001;2:173–182. doi: 10.1046/j.1467-789x.2001.00032.x. [DOI] [PubMed] [Google Scholar]
- 4.Bhattacharya I, Ghayor C, Perez Dominguez A, Weber FE. From influenza virus to novel corona virus (SARS-CoV-2)—the contribution of obesity. Front. Endocrinol. 2020;11:556962. doi: 10.3389/fendo.2020.556962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petrilli CM, et al. Factors associated with hospital admission and critical illness among 5279 people with coronavirus disease 2019 in New York City: prospective cohort study. BMJ. 2020;369:m1966. doi: 10.1136/bmj.m1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cummings MJ, et al. Epidemiology, clinical course, and outcomes of critically ill adults with COVID-19 in New York City: a prospective cohort study. Lancet. 2020;395:1763–1770. doi: 10.1016/S0140-6736(20)31189-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao X, et al. Obesity increases the severity and mortality of influenza and COVID-19: a systematic review and meta-analysis. Front. Endocrinol. 2020;11:595109. doi: 10.3389/fendo.2020.595109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abarca-Gómez L, et al. Worldwide trends in body-mass index, underweight, overweight, and obesity from 1975 to 2016: a pooled analysis of 2416 population-based measurement studies in 128.9 million children, adolescents, and adults. Lancet. 2017;390:2627–2642. doi: 10.1016/S0140-6736(17)32129-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maes HH, Neale MC, Eaves LJ. Genetic and environmental factors in relative body weight and human obesity. Behav. Genet. 1997;27:325–351. doi: 10.1023/A:1025635913927. [DOI] [PubMed] [Google Scholar]
- 10.Elks CE, et al. Variability in the heritability of body mass index: a systematic review and meta-regression. Front. Endocrinol. 2012;3:29. doi: 10.3389/fendo.2012.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chami N, Preuss M, Walker RW, Moscati A, Loos RJF. The role of polygenic susceptibility to obesity among carriers of pathogenic mutations in MC4R in the UK Biobank population. PLoS Med. 2020;17:e1003196. doi: 10.1371/journal.pmed.1003196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaur Y, de Souza RJ, Gibson WT, Meyre D. A systematic review of genetic syndromes with obesity. Obes. Rev. 2017;18:603–634. doi: 10.1111/obr.12531. [DOI] [PubMed] [Google Scholar]
- 13.Ingalls AM, Dickie MM, Snell GD. Obese, a new mutation in the house mouse. J. Hered. 1950;41:317–318. doi: 10.1093/oxfordjournals.jhered.a106073. [DOI] [PubMed] [Google Scholar]
- 14.Hummel KP, Dickie MM, Coleman DL. Diabetes, a new mutation in the mouse. Science. 1966;153:1127–1128. doi: 10.1126/science.153.3740.1127. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, et al. Positional cloning of the mouse obese gene and its human homologue. Nature. 1994;372:425–432. doi: 10.1038/372425a0. [DOI] [PubMed] [Google Scholar]
- 16.Chen H, et al. Evidence that the diabetes gene encodes the leptin receptor: identification of a mutation in the leptin receptor gene in db/db mice. Cell. 1996;84:491–495. doi: 10.1016/S0092-8674(00)81294-5. [DOI] [PubMed] [Google Scholar]
- 17.Bultman SJ, Michaud EJ, Woychik RP. Molecular characterization of the mouse agouti locus. Cell. 1992;71:1195–1204. doi: 10.1016/S0092-8674(05)80067-4. [DOI] [PubMed] [Google Scholar]
- 18.Miller MW, et al. Cloning of the mouse agouti gene predicts a secreted protein ubiquitously expressed in mice carrying the lethal yellow mutation. Genes Dev. 1993;7:454–467. doi: 10.1101/gad.7.3.454. [DOI] [PubMed] [Google Scholar]
- 19.Lu D, et al. Agouti protein is an antagonist of the melanocyte-stimulating-hormone receptor. Nature. 1994;371:799–802. doi: 10.1038/371799a0. [DOI] [PubMed] [Google Scholar]
- 20.Fan W, Boston BA, Kesterson RA, Hruby VJ, Cone RD. Role of melanocortinergic neurons in feeding and the agouti obesity syndrome. Nature. 1997;385:165–168. doi: 10.1038/385165a0. [DOI] [PubMed] [Google Scholar]
- 21.Montague CT, et al. Congenital leptin deficiency is associated with severe early-onset obesity in humans. Nature. 1997;387:903–908. doi: 10.1038/43185. [DOI] [PubMed] [Google Scholar]
- 22.Clement K, et al. A mutation in the human leptin receptor gene causes obesity and pituitary dysfunction. Nature. 1998;392:398–401. doi: 10.1038/32911. [DOI] [PubMed] [Google Scholar]
- 23.Jackson RS, et al. Obesity and impaired prohormone processing associated with mutations in the human prohormone convertase 1 gene. Nat. Genet. 1997;16:303–306. doi: 10.1038/ng0797-303. [DOI] [PubMed] [Google Scholar]
- 24.Huszar D, et al. Targeted disruption of the melanocortin-4 receptor results in obesity in mice. Cell. 1997;88:131–141. doi: 10.1016/S0092-8674(00)81865-6. [DOI] [PubMed] [Google Scholar]
- 25.Yeo GS, et al. A frameshift mutation in MC4R associated with dominantly inherited human obesity. Nat. Genet. 1998;20:111–112. doi: 10.1038/2404. [DOI] [PubMed] [Google Scholar]
- 26.Vaisse C, Clement K, Guy-Grand B, Froguel P. A frameshift mutation in MC4R is associated with a dominant form of obesity. Nat. Genet. 1998;20:113–114. doi: 10.1038/2407. [DOI] [PubMed] [Google Scholar]
- 27.Krude H, et al. Severe early-onset obesity, adrenal insufficiency and red hair pigmentation caused by POMC mutations in humans. Nat. Genet. 1998;19:155–157. doi: 10.1038/509. [DOI] [PubMed] [Google Scholar]
- 28.Yaswen L, Diehl N, Brennan MB, Hochgeschwender U. Obesity in the mouse model of pro-opiomelanocortin deficiency responds to peripheral melanocortin. Nat. Med. 1999;5:1066–1070. doi: 10.1038/12506. [DOI] [PubMed] [Google Scholar]
- 29.Challis BG, et al. Mice lacking pro-opiomelanocortin are sensitive to high-fat feeding but respond normally to the acute anorectic effects of peptide-YY(3-36) Proc. Natl Acad. Sci. USA. 2004;101:4695–4700. doi: 10.1073/pnas.0306931101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van der Klaauw AA, et al. Human semaphorin 3 variants link melanocortin circuit development and energy balance. Cell. 2019;176:729–742.e18. doi: 10.1016/j.cell.2018.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farooqi S, O’Rahilly S. Genetics of obesity in humans. Endocr. Rev. 2006;27:710–718. doi: 10.1210/er.2006-0040. [DOI] [PubMed] [Google Scholar]
- 32.Saeed S, Arslan M, Froguel P. Genetics of obesity in consanguineous populations: toward precision medicine and the discovery of novel obesity genes. Obesity. 2018;26:474–484. doi: 10.1002/oby.22064. [DOI] [PubMed] [Google Scholar]
- 33.Saeed S, et al. Genetic variants in LEP, LEPR, and MC4R explain 30% of severe obesity in children from a consanguineous population. Obesity. 2015;23:1687–1695. doi: 10.1002/oby.21142. [DOI] [PubMed] [Google Scholar]
- 34.Saeed S, et al. Genetic causes of severe childhood obesity: a remarkably high prevalence in an inbred population of Pakistan. Diabetes. 2020;69:1424–1438. doi: 10.2337/db19-1238. [DOI] [PubMed] [Google Scholar]
- 35.Kurokawa N, et al. The ADRB3 Trp64Arg variant and BMI: a meta-analysis of 44 833 individuals. Int. J. Obes. 2008;32:1240–1249. doi: 10.1038/ijo.2008.90. [DOI] [PubMed] [Google Scholar]
- 36.Shugart YY, et al. Two British women studies replicated the association between the Val66Met polymorphism in the brain-derived neurotrophic factor (BDNF) and BMI. Eur. J. Hum. Genet. 2009;17:1050–1055. doi: 10.1038/ejhg.2008.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benzinou M, et al. Endocannabinoid receptor 1 gene variations increase risk for obesity and modulate body mass index in European populations. Hum. Mol. Genet. 2008;17:1916–1921. doi: 10.1093/hmg/ddn089. [DOI] [PubMed] [Google Scholar]
- 38.Wang D, et al. Association of the MC4R V103I polymorphism with obesity: a Chinese case–control study and meta-analysis in 55,195 individuals. Obesity. 2010;18:573–579. doi: 10.1038/oby.2009.268. [DOI] [PubMed] [Google Scholar]
- 39.Nead KT, et al. Contribution of common non-synonymous variants in PCSK1 to body mass index variation and risk of obesity: a systematic review and meta-analysis with evidence from up to 331 175 individuals. Hum. Mol. Genet. 2015;24:3582–3594. doi: 10.1093/hmg/ddv097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tonjes A, Scholz M, Loeffler M, Stumvoll M. Association of Pro12Ala polymorphism in peroxisome proliferator-activated receptor gamma with pre-diabetic phenotypes: meta-analysis of 57 studies on nondiabetic individuals. Diabetes Care. 2006;29:2489–2497. doi: 10.2337/dc06-0513. [DOI] [PubMed] [Google Scholar]
- 41.Rankinen T, et al. The human obesity gene map: the 2005 update. Obesity. 2006;14:529–644. doi: 10.1038/oby.2006.71. [DOI] [PubMed] [Google Scholar]
- 42.Frayling TM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scuteri A, et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3:e115. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buniello A, et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47:D1005–D1012. doi: 10.1093/nar/gky1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yengo L, et al. Meta-analysis of genome-wide association studies for height and body mass index in approximately 700 000 individuals of European ancestry. Hum. Mol. Genet. 2018;27:3641–3649. doi: 10.1093/hmg/ddy271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Javed A, et al. Diagnostic performance of body mass index to identify obesity as defined by body adiposity in children and adolescents: a systematic review and meta-analysis. Pediatr. Obes. 2015;10:234–244. doi: 10.1111/ijpo.242. [DOI] [PubMed] [Google Scholar]
- 47.Kilpelainen TO, et al. Genetic variation near IRS1 associates with reduced adiposity and an impaired metabolic profile. Nat. Genet. 2011;43:753–760. doi: 10.1038/ng.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu Y, et al. New loci for body fat percentage reveal link between adiposity and cardiometabolic disease risk. Nat. Commun. 2016;7:10495. doi: 10.1038/ncomms10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zillikens MC, et al. Large meta-analysis of genome-wide association studies identifies five loci for lean body mass. Nat. Commun. 2017;8:80. doi: 10.1038/s41467-017-00031-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chu AY, et al. Multiethnic genome-wide meta-analysis of ectopic fat depots identifies loci associated with adipocyte development and differentiation. Nat. Genet. 2017;49:125–130. doi: 10.1038/ng.3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kilpelainen TO, et al. Genome-wide meta-analysis uncovers novel loci influencing circulating leptin levels. Nat. Commun. 2016;7:10494. doi: 10.1038/ncomms10494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun Q, et al. Genome-wide association study identifies polymorphisms in LEPR as determinants of plasma soluble leptin receptor levels. Hum. Mol. Genet. 2010;19:1846–1855. doi: 10.1093/hmg/ddq056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Riveros-McKay F, et al. Genetic architecture of human thinness compared to severe obesity. PLoS Genet. 2019;15:e1007603. doi: 10.1371/journal.pgen.1007603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Orthofer M, et al. Identification of ALK in thinness. Cell. 2020;181:1246–1262.e22. doi: 10.1016/j.cell.2020.04.034. [DOI] [PubMed] [Google Scholar]
- 55.Bradfield JP, et al. A trans-ancestral meta-analysis of genome-wide association studies reveals loci associated with childhood obesity. Hum. Mol. Genet. 2019;28:3327–3338. doi: 10.1093/hmg/ddz161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Felix JF, et al. Genome-wide association analysis identifies three new susceptibility loci for childhood body mass index. Hum. Mol. Genet. 2016;25:389–403. doi: 10.1093/hmg/ddv472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vogelezang S, et al. Novel loci for childhood body mass index and shared heritability with adult cardiometabolic traits. PLoS Genet. 2020;16:e1008718. doi: 10.1371/journal.pgen.1008718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Akiyama M, et al. Genome-wide association study identifies 112 new loci for body mass index in the Japanese population. Nat. Genet. 2017;49:1458–1467. doi: 10.1038/ng.3951. [DOI] [PubMed] [Google Scholar]
- 59.Ng MCY, et al. Discovery and fine-mapping of adiposity loci using high density imputation of genome-wide association studies in individuals of African ancestry: African Ancestry Anthropometry Genetics Consortium. PLoS Genet. 2017;13:e1006719. doi: 10.1371/journal.pgen.1006719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gurdasani D, et al. Uganda genome resource enables insights into population history and genomic discovery in Africa. Cell. 2019;179:984–1002.e36. doi: 10.1016/j.cell.2019.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wojcik GL, et al. Genetic analyses of diverse populations improves discovery for complex traits. Nature. 2019;570:514–518. doi: 10.1038/s41586-019-1310-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Martin AR, et al. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet. 2019;51:584–591. doi: 10.1038/s41588-019-0379-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grarup N, et al. Loss-of-function variants in ADCY3 increase risk of obesity and type 2 diabetes. Nat. Genet. 2018;50:172–174. doi: 10.1038/s41588-017-0022-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Saeed S, et al. Loss-of-function mutations in ADCY3 cause monogenic severe obesity. Nat. Genet. 2018;50:175–179. doi: 10.1038/s41588-017-0023-6. [DOI] [PubMed] [Google Scholar]
- 65.Minster RL, et al. A thrifty variant in CREBRF strongly influences body mass index in Samoans. Nat. Genet. 2016;48:1049–1054. doi: 10.1038/ng.3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Andersen MK, et al. The derived allele of a novel intergenic variant at chromosome 11 associates with lower body mass index and a favorable metabolic phenotype in Greenlanders. PLoS Genet. 2020;16:e1008544. doi: 10.1371/journal.pgen.1008544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Siljee JE, et al. Subcellular localization of MC4R with ADCY3 at neuronal primary cilia underlies a common pathway for genetic predisposition to obesity. Nat. Genet. 2018;50:180–185. doi: 10.1038/s41588-017-0020-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Willer CJ, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat. Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Singh K, et al. Neural cell adhesion molecule Negr1 deficiency in mouse results in structural brain endophenotypes and behavioral deviations related to psychiatric disorders. Sci. Rep. 2019;9:5457. doi: 10.1038/s41598-019-41991-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Speliotes EK, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat. Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Soni A, Amisten S, Rorsman P, Salehi A. GPRC5B a putative glutamate-receptor candidate is negative modulator of insulin secretion. Biochem. Biophys. Res. Commun. 2013;441:643–648. doi: 10.1016/j.bbrc.2013.10.099. [DOI] [PubMed] [Google Scholar]
- 72.Jarick I, et al. Novel common copy number variation for early onset extreme obesity on chromosome 11q11 identified by a genome-wide analysis. Hum. Mol. Genet. 2011;20:840–852. doi: 10.1093/hmg/ddq518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sainsbury A, et al. Synergistic effects of Y2 and Y4 receptors on adiposity and bone mass revealed in double knockout mice. Mol. Cell Biol. 2003;23:5225–5233. doi: 10.1128/MCB.23.15.5225-5233.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Falchi M, et al. Low copy number of the salivary amylase gene predisposes to obesity. Nat. Genet. 2014;46:492–497. doi: 10.1038/ng.2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Meisler MH, Ting CN. The remarkable evolutionary history of the human amylase genes. Crit. Rev. Oral Biol. Med. 1993;4:503–509. doi: 10.1177/10454411930040033501. [DOI] [PubMed] [Google Scholar]
- 76.Hendricks AE, et al. Rare variant analysis of human and rodent obesity genes in individuals with severe childhood obesity. Sci. Rep. 2017;7:4394. doi: 10.1038/s41598-017-03054-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Turcot V, et al. Protein-altering variants associated with body mass index implicate pathways that control energy intake and expenditure in obesity. Nat. Genet. 2018;50:26–41. doi: 10.1038/s41588-017-0011-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Emdin CA, et al. Analysis of predicted loss-of-function variants in UK Biobank identifies variants protective for disease. Nat. Commun. 2018;9:1613. doi: 10.1038/s41467-018-03911-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Akbari P, et al. Sequencing of 640,000 exomes identifies GPR75 variants associated with protection from obesity. Science. 2021;373:eabf8683. doi: 10.1126/science.abf8683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Baggio LL, Drucker DJ. Biology of incretins: GLP-1 and GIP. Gastroenterology. 2007;132:2131–2157. doi: 10.1053/j.gastro.2007.03.054. [DOI] [PubMed] [Google Scholar]
- 81.Antolin-Fontes B, et al. The habenular G-protein-coupled receptor 151 regulates synaptic plasticity and nicotine intake. Proc. Natl Acad. Sci. USA. 2020;117:5502–5509. doi: 10.1073/pnas.1916132117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Winkler TW, et al. The influence of age and sex on genetic associations with adult body size and shape: a large-scale genome-wide interaction study. PLoS Genet. 2015;11:e1005378. doi: 10.1371/journal.pgen.1005378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Graff M, et al. Genome-wide physical activity interactions in adiposity — a meta-analysis of 200,452 adults. PLoS Genet. 2017;13:e1006528. doi: 10.1371/journal.pgen.1006528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Smith CE, et al. Genome-wide interactions with dairy intake for body mass index in adults of European descent. Mol. Nutr. Food Res. 2018;62:1700347. doi: 10.1002/mnfr.201700347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Justice AE, et al. Genome-wide meta-analysis of 241,258 adults accounting for smoking behaviour identifies novel loci for obesity traits. Nat. Commun. 2017;8:14977. doi: 10.1038/ncomms14977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Qi Q, et al. Fried food consumption, genetic risk, and body mass index: gene–diet interaction analysis in three US cohort studies. BMJ. 2014;348:g1610. doi: 10.1136/bmj.g1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kilpelainen TO, et al. Physical activity attenuates the influence of FTO variants on obesity risk: a meta-analysis of 218,166 adults and 19,268 children. PLoS Med. 2011;8:e1001116. doi: 10.1371/journal.pmed.1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yeo GSH. Genetics of obesity: can an old dog teach us new tricks? Diabetologia. 2017;60:778–783. doi: 10.1007/s00125-016-4187-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Locke AE, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Friedman JM, Halaas JL. Leptin and the regulation of body weight in mammals. Nature. 1998;395:763–770. doi: 10.1038/27376. [DOI] [PubMed] [Google Scholar]
- 91.Ahima RS, et al. Role of leptin in the neuroendocrine response to fasting. Nature. 1996;382:250–252. doi: 10.1038/382250a0. [DOI] [PubMed] [Google Scholar]
- 92.Cowley MA, et al. Integration of NPY, AGRP, and melanocortin signals in the hypothalamic paraventricular nucleus: evidence of a cellular basis for the adipostat. Neuron. 1999;24:155–163. doi: 10.1016/S0896-6273(00)80829-6. [DOI] [PubMed] [Google Scholar]
- 93.Bertagna X. Proopiomelanocortin-derived peptides. Endocrinol. Metab. Clin. North Am. 1994;23:467–485. doi: 10.1016/S0889-8529(18)30079-3. [DOI] [PubMed] [Google Scholar]
- 94.Ollmann MM, et al. Antagonism of central melanocortin receptors in vitro and in vivo by agouti-related protein. Science. 1997;278:135–138. doi: 10.1126/science.278.5335.135. [DOI] [PubMed] [Google Scholar]
- 95.Butler AA, et al. A unique metabolic syndrome causes obesity in the melanocortin-3 receptor-deficient mouse. Endocrinology. 2000;141:3518–3521. doi: 10.1210/endo.141.9.7791. [DOI] [PubMed] [Google Scholar]
- 96.Chen AS, et al. Inactivation of the mouse melanocortin-3 receptor results in increased fat mass and reduced lean body mass. Nat. Genet. 2000;26:97–102. doi: 10.1038/79254. [DOI] [PubMed] [Google Scholar]
- 97.Doche ME, et al. Human SH2B1 mutations are associated with maladaptive behaviors and obesity. J. Clin. Invest. 2012;122:4732–4736. doi: 10.1172/JCI62696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Marenne G, et al. Exome sequencing identifies genes and gene sets contributing to severe childhood obesity, linking PHIP variants to repressed POMC transcription. Cell Metab. 2020;31:1107–1119.e12. doi: 10.1016/j.cmet.2020.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Asai M, et al. Loss of function of the melanocortin 2 receptor accessory protein 2 is associated with mammalian obesity. Science. 2013;341:275–278. doi: 10.1126/science.1233000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Michaud JL, et al. Sim1 haploinsufficiency causes hyperphagia, obesity and reduction of the paraventricular nucleus of the hypothalamus. Hum. Mol. Genet. 2001;10:1465–1473. doi: 10.1093/hmg/10.14.1465. [DOI] [PubMed] [Google Scholar]
- 101.Farooqi IS, et al. Clinical spectrum of obesity and mutations in the melanocortin 4 receptor gene. N. Engl. J. Med. 2003;348:1085–1095. doi: 10.1056/NEJMoa022050. [DOI] [PubMed] [Google Scholar]
- 102.Wade KH, et al. Loss-of-function mutations in the melanocortin 4 receptor in a UK birth cohort. Nat. Med. 2021;27:1088–1096. doi: 10.1038/s41591-021-01349-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.van der Klaauw A, et al. Role of melanocortin signalling in the preference for dietary macronutrients in human beings. Lancet. 2015;385:S12. doi: 10.1016/S0140-6736(15)60327-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Raffan E, et al. A deletion in the canine POMC gene is associated with weight and appetite in obesity-prone Labrador retriever dogs. Cell Metab. 2016;23:893–900. doi: 10.1016/j.cmet.2016.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kim KS, Larsen N, Short T, Plastow G, Rothschild MF. A missense variant of the porcine melanocortin-4 receptor (MC4R) gene is associated with fatness, growth, and feed intake traits. Mamm. Genome. 2000;11:131–135. doi: 10.1007/s003350010025. [DOI] [PubMed] [Google Scholar]
- 106.Aspiras AC, Rohner N, Martineau B, Borowsky RL, Tabin CJ. Melanocortin 4 receptor mutations contribute to the adaptation of cavefish to nutrient-poor conditions. Proc. Natl Acad. Sci. USA. 2015;112:9668–9673. doi: 10.1073/pnas.1510802112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pinto S, et al. Rapid rewiring of arcuate nucleus feeding circuits by leptin. Science. 2004;304:110–115. doi: 10.1126/science.1089459. [DOI] [PubMed] [Google Scholar]
- 108.Davies AM. The role of neurotrophins in the developing nervous system. J. Neurobiol. 1994;25:1334–1348. doi: 10.1002/neu.480251103. [DOI] [PubMed] [Google Scholar]
- 109.McAllister AK, Katz LC, Lo DC. Opposing roles for endogenous BDNF and NT-3 in regulating cortical dendritic growth. Neuron. 1997;18:767–778. doi: 10.1016/S0896-6273(00)80316-5. [DOI] [PubMed] [Google Scholar]
- 110.Korte M, et al. Hippocampal long-term potentiation is impaired in mice lacking brain-derived neurotrophic factor. Proc. Natl Acad. Sci. USA. 1995;92:8856–8860. doi: 10.1073/pnas.92.19.8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kernie SG, Liebl DJ, Parada LF. BDNF regulates eating behavior and locomotor activity in mice. EMBO J. 2000;19:1290–1300. doi: 10.1093/emboj/19.6.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Xu B, et al. Brain-derived neurotrophic factor regulates energy balance downstream of melanocortin-4 receptor. Nat. Neurosci. 2003;6:736–742. doi: 10.1038/nn1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Xu B, Xie X. Neurotrophic factor control of satiety and body weight. Nat. Rev. Neurosci. 2016;17:282–292. doi: 10.1038/nrn.2016.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rios M, et al. Conditional deletion of brain-derived neurotrophic factor in the postnatal brain leads to obesity and hyperactivity. Mol. Endocrinol. 2001;15:1748–1757. doi: 10.1210/mend.15.10.0706. [DOI] [PubMed] [Google Scholar]
- 115.Gray J, et al. Hyperphagia, severe obesity, impaired cognitive function, and hyperactivity associated with functional loss of one copy of the brain-derived neurotrophic factor (BDNF) gene. Diabetes. 2006;55:3366–3371. doi: 10.2337/db06-0550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yeo GS, et al. A de novo mutation affecting human TrkB associated with severe obesity and developmental delay. Nat. Neurosci. 2004;7:1187–1189. doi: 10.1038/nn1336. [DOI] [PubMed] [Google Scholar]
- 117.Thorleifsson G, et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat. Genet. 2009;41:18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 118.Finucane HK, et al. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat. Genet. 2018;50:621–629. doi: 10.1038/s41588-018-0081-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ndiaye FK, et al. The expression of genes in top obesity-associated loci is enriched in insula and substantia nigra brain regions involved in addiction and reward. Int. J. Obes. 2020;44:539–543. doi: 10.1038/s41366-019-0428-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Loos RJ, Yeo GS. The bigger picture of FTO: the first GWAS-identified obesity gene. Nat. Rev. Endocrinol. 2014;10:51–61. doi: 10.1038/nrendo.2013.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Church C, et al. Overexpression of Fto leads to increased food intake and results in obesity. Nat. Genet. 2010;42:1086–1092. doi: 10.1038/ng.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Fischer J, et al. Inactivation of the Fto gene protects from obesity. Nature. 2009;458:894–898. doi: 10.1038/nature07848. [DOI] [PubMed] [Google Scholar]
- 123.Cheung MK, Gulati P, O’Rahilly S, Yeo GS. FTO expression is regulated by availability of essential amino acids. Int. J. Obes. 2013;37:744–747. doi: 10.1038/ijo.2012.77. [DOI] [PubMed] [Google Scholar]
- 124.Gulati P, et al. Role for the obesity-related FTO gene in the cellular sensing of amino acids. Proc. Natl Acad. Sci. USA. 2013;110:2557–2562. doi: 10.1073/pnas.1222796110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Karra E, et al. A link between FTO, ghrelin, and impaired brain food-cue responsivity. J. Clin. Invest. 2013;123:3539–3551. doi: 10.1172/JCI44403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hess ME, et al. The fat mass and obesity associated gene (Fto) regulates activity of the dopaminergic midbrain circuitry. Nat. Neurosci. 2013;16:1042–1048. doi: 10.1038/nn.3449. [DOI] [PubMed] [Google Scholar]
- 127.Ruud J, et al. The fat mass and obesity-associated protein (FTO) regulates locomotor responses to novelty via D2R medium spiny neurons. Cell Rep. 2019;27:3182–3198.e9. doi: 10.1016/j.celrep.2019.05.037. [DOI] [PubMed] [Google Scholar]
- 128.Stratigopoulos G, LeDuc CA, Cremona ML, Chung WK, Leibel RL. Cut-like homeobox 1 (CUX1) regulates expression of the fat mass and obesity-associated and retinitis pigmentosa GTPase regulator-interacting protein-1-like (RPGRIP1L) genes and coordinates leptin receptor signaling. J. Biol. Chem. 2011;286:2155–2170. doi: 10.1074/jbc.M110.188482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Stratigopoulos G, et al. Hypomorphism for RPGRIP1L, a ciliary gene vicinal to the FTO locus, causes increased adiposity in mice. Cell Metab. 2014;19:767–779. doi: 10.1016/j.cmet.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Stratigopoulos G, et al. Hypomorphism of Fto and Rpgrip1l causes obesity in mice. J. Clin. Invest. 2016;126:1897–1910. doi: 10.1172/JCI85526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Smemo S, et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature. 2014;507:371–375. doi: 10.1038/nature13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Claussnitzer M, et al. FTO obesity variant circuitry and adipocyte browning in humans. N. Engl. J. Med. 2015;373:895–907. doi: 10.1056/NEJMoa1502214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.O’Rahilly S, Coll AP, Yeo GS. FTO obesity variant and adipocyte browning in humans. N. Engl. J. Med. 2016;374:191. doi: 10.1056/NEJMc1513316. [DOI] [PubMed] [Google Scholar]
- 134.Sobreira DR, et al. Extensive pleiotropism and allelic heterogeneity mediate metabolic effects of IRX3 and IRX5. Science. 2021;372:1085–1091. doi: 10.1126/science.abf1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Almen MS, et al. The obesity gene, TMEM18, is of ancient origin, found in majority of neuronal cells in all major brain regions and associated with obesity in severely obese children. BMC Med. Genet. 2010;11:58. doi: 10.1186/1471-2350-11-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Larder R, et al. Obesity-associated gene TMEM18 has a role in the central control of appetite and body weight regulation. Proc. Natl Acad. Sci. USA. 2017;114:9421–9426. doi: 10.1073/pnas.1707310114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wiemerslage L, et al. The Drosophila ortholog of TMEM18 regulates insulin and glucagon-like signaling. J. Endocrinol. 2016;229:233–243. doi: 10.1530/JOE-16-0040. [DOI] [PubMed] [Google Scholar]
- 138.Biederer T, et al. SynCAM, a synaptic adhesion molecule that drives synapse assembly. Science. 2002;297:1525–1531. doi: 10.1126/science.1072356. [DOI] [PubMed] [Google Scholar]
- 139.Yan X, et al. Cadm2 regulates body weight and energy homeostasis in mice. Mol. Metab. 2018;8:180–188. doi: 10.1016/j.molmet.2017.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Rathjen T, et al. Regulation of body weight and energy homeostasis by neuronal cell adhesion molecule 1. Nat. Neurosci. 2017;20:1096–1103. doi: 10.1038/nn.4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wheeler E, et al. Genome-wide SNP and CNV analysis identifies common and low-frequency variants associated with severe early-onset obesity. Nat. Genet. 2013;45:513–517. doi: 10.1038/ng.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Schafer M, Brauer AU, Savaskan NE, Rathjen FG, Brummendorf T. Neurotractin/kilon promotes neurite outgrowth and is expressed on reactive astrocytes after entorhinal cortex lesion. Mol. Cell. Neurosci. 2005;29:580–590. doi: 10.1016/j.mcn.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 143.Matikainen-Ankney BA, Kravitz AV. Persistent effects of obesity: a neuroplasticity hypothesis. Ann. N. Y. Acad. Sci. 2018;1428:221–239. doi: 10.1111/nyas.13665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lee AW, et al. Functional inactivation of the genome-wide association study obesity gene neuronal growth regulator 1 in mice causes a body mass phenotype. PLoS ONE. 2012;7:e41537. doi: 10.1371/journal.pone.0041537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Joo Y, Kim H, Lee S, Lee S. Neuronal growth regulator 1-deficient mice show increased adiposity and decreased muscle mass. Int. J. Obes. 2019;43:1769–1782. doi: 10.1038/s41366-019-0376-2. [DOI] [PubMed] [Google Scholar]
- 146.Boender AJ, van Gestel MA, Garner KM, Luijendijk MC, Adan RA. The obesity-associated gene Negr1 regulates aspects of energy balance in rat hypothalamic areas. Physiol. Rep. 2014;2:e12083. doi: 10.14814/phy2.12083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Singh K, et al. Neuronal growth and behavioral alterations in mice deficient for the psychiatric disease-associated negr1 gene. Front. Mol. Neurosci. 2018;11:30. doi: 10.3389/fnmol.2018.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Szczurkowska J, et al. NEGR1 and FGFR2 cooperatively regulate cortical development and core behaviours related to autism disorders in mice. Brain. 2018;141:2772–2794. doi: 10.1093/brain/awy190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Noh K, et al. Negr1 controls adult hippocampal neurogenesis and affective behaviors. Mol. Psychiatry. 2019;24:1189–1205. doi: 10.1038/s41380-018-0347-3. [DOI] [PubMed] [Google Scholar]
- 150.Cano-Gamez E, Trynka G. From GWAS to function: using functional genomics to identify the mechanisms underlying complex diseases. Front. Genet. 2020;11:424. doi: 10.3389/fgene.2020.00424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Loos RJ, et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat. Genet. 2008;40:768–775. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Chambers JC, et al. Common genetic variation near MC4R is associated with waist circumference and insulin resistance. Nat. Genet. 2008;40:716–718. doi: 10.1038/ng.156. [DOI] [PubMed] [Google Scholar]
- 153.Yaghootkar H, et al. Genetic studies of leptin concentrations implicate leptin in the regulation of early adiposity. Diabetes. 2020;69:2806–2818. doi: 10.2337/db20-0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kichaev G, et al. Leveraging polygenic functional enrichment to improve GWAS power. Am. J. Hum. Genet. 2019;104:65–75. doi: 10.1016/j.ajhg.2018.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Pulit SL, et al. Meta-analysis of genome-wide association studies for body fat distribution in 694 649 individuals of European ancestry. Hum. Mol. Genet. 2019;28:166–174. doi: 10.1093/hmg/ddy327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wang Y, et al. Melanocortin 4 receptor signals at the neuronal primary cilium to control food intake and body weight. J. Clin. Invest. 2021;131:e142064. doi: 10.1172/JCI142064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Farooqi IS, et al. Effects of recombinant leptin therapy in a child with congenital leptin deficiency. N. Engl. J. Med. 1999;341:879–884. doi: 10.1056/NEJM199909163411204. [DOI] [PubMed] [Google Scholar]
- 158.Farooqi IS, O’Rahilly S. 20 years of leptin: human disorders of leptin action. J. Endocrinol. 2014;223:T63–T70. doi: 10.1530/JOE-14-0480. [DOI] [PubMed] [Google Scholar]
- 159.Yeo GSH, et al. The melanocortin pathway and energy homeostasis: from discovery to obesity therapy. Mol. Metab. 2021;48:101206. doi: 10.1016/j.molmet.2021.101206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kuhnen P, et al. Proopiomelanocortin deficiency treated with a melanocortin-4 receptor agonist. N. Engl. J. Med. 2016;375:240–246. doi: 10.1056/NEJMoa1512693. [DOI] [PubMed] [Google Scholar]
- 161.Clement K, et al. MC4R agonism promotes durable weight loss in patients with leptin receptor deficiency. Nat. Med. 2018;24:551–555. doi: 10.1038/s41591-018-0015-9. [DOI] [PubMed] [Google Scholar]
- 162.Clement K, et al. Efficacy and safety of setmelanotide, an MC4R agonist, in individuals with severe obesity due to LEPR or POMC deficiency: single-arm, open-label, multicentre, phase 3 trials. Lancet Diabetes Endocrinol. 2020;8:960–970. doi: 10.1016/S2213-8587(20)30364-8. [DOI] [PubMed] [Google Scholar]
- 163.Ayers KL, et al. Melanocortin 4 receptor pathway dysfunction in obesity: patient stratification aimed at MC4R agonist treatment. J. Clin. Endocrinol. Metab. 2018;103:2601–2612. doi: 10.1210/jc.2018-00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Heymsfield SB, et al. Recombinant leptin for weight loss in obese and lean adults: a randomized, controlled, dose-escalation trial. JAMA. 1999;282:1568–1575. doi: 10.1001/jama.282.16.1568. [DOI] [PubMed] [Google Scholar]
- 165.Collet TH, et al. Evaluation of a melanocortin-4 receptor (MC4R) agonist (setmelanotide) in MC4R deficiency. Mol. Metab. 2017;6:1321–1329. doi: 10.1016/j.molmet.2017.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Khera AV, et al. Polygenic prediction of weight and obesity trajectories from birth to adulthood. Cell. 2019;177:587–596.e9. doi: 10.1016/j.cell.2019.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Loos RJF. 15 years of genome-wide association studies and no signs of slowing down. Nat. Commun. 2020;11:5900. doi: 10.1038/s41467-020-19653-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Pearce LR, et al. KSR2 mutations are associated with obesity, insulin resistance, and impaired cellular fuel oxidation. Cell. 2013;155:765–777. doi: 10.1016/j.cell.2013.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Greenfield JR, et al. Modulation of blood pressure by central melanocortinergic pathways. N. Engl. J. Med. 2009;360:44–52. doi: 10.1056/NEJMoa0803085. [DOI] [PubMed] [Google Scholar]
- 170.Torkamani A, Topol E. Polygenic risk scores expand to obesity. Cell. 2019;177:518–520. doi: 10.1016/j.cell.2019.03.051. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.