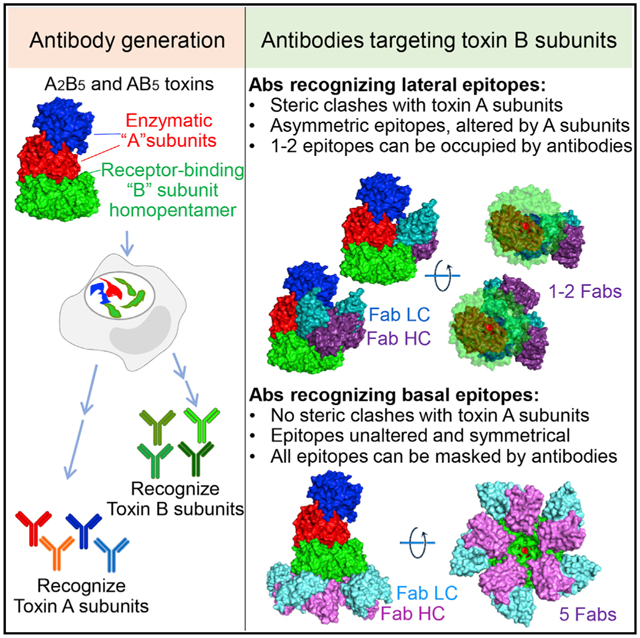
SUMMARY
Many bacterial pathogens secrete A(2)B5 toxins comprising two functionally distinct yet complementary “A” and “B” subunits to benefit the pathogens during infection. The lectin-like pentameric B subunits recognize specific sets of host glycans to deliver the toxin into target host cells. Here, we offer the molecular mechanism by which neutralizing antibodies, which have the potential to bind to all glycan-receptor binding sites and thus completely inhibit toxin binding to host cells, are inhibited from exerting this action. Cryogenic electron microscopy (cryo-EM)-based analyses indicate that the skewed positioning of the toxin A subunit(s) toward one side of the toxin B pentamer inhibited neutralizing antibody binding to the laterally located epitopes, rendering some glycan-receptor binding sites that remained available for the toxin binding and endocytosis process, which is strikingly different from the counterpart antibodies recognizing the far side-located epitopes. These results highlight additional features of the toxin-antibody interactions and offer important insights into anti-toxin strategies.
Graphical Abstract
In brief
Nguyen et al. find that toxin-neutralizing antibodies targeting glycan-receptor binding B subunits can be split into two classes based on their epitope locations. They describe how these two classes exhibit significantly different neutralizing efficacies, a feature that appears to be shared among A(2)B5 toxins, and thus they provide insights into anti-toxin strategies.
INTRODUCTION
The ongoing global spread of antibiotic-resistant bacterial pathogens poses a great public health challenge, and if effective intervention strategies are not implemented on time, this spread will significantly increase the associated morbidity and mortality rates (Feasey et al., 2015; Hernando-Amado et al., 2019; Klemm et al., 2018; Neupane et al., 2021; Parkhill et al., 2001; Centers for Disease Control and Prevention, 2019; Yang et al., 2018a). Bacterial A(2)B5 toxins are asymmetrical multiprotein complex virulence factors secreted by many pathogens causing respiratory, gastrointestinal, and systemic diseases (Beddoe et al., 2010; Lee et al., 2021). Notable examples include cholera toxin from Vibrio cholerae for gastrointestinal disease, pertussis toxin from Bordetella pertussis for whooping cough, Shiga toxin from Shigella dysenteriae, Shiga-like toxin-producing E. coli (STEC) for severe diarrhea and hemolytic uremic syndrome, labile enterotoxin (LT) from E. coli for diarrhea, and subtilase cytotoxin from a subset of STEC (ST) for diarrhea (Beddoe et al., 2010; Fan et al., 2000; Kitov et al., 2000; Zuverink and Barbieri, 2018). These bacteria secrete their AB5 toxins during infection to benefit the pathogens (Beddoe et al., 2010; Lee et al., 2021). The secreted toxins are stable in the local and systemic circulations and recognize specific sets of host cells that are either in the infection site or, in some cases, distant from infected host cells (Lee et al., 2020).
In A(2)B5 toxins, the enzymatic “A” subunits intoxicate host cells by directly altering the function of their target host proteins. The lectin-like pentameric “B” subunits recognize specific sets of host glycans and sialic acids displayed on the surface of target host cells, which therefore determines host cell specificity (Lee et al., 2021). This B subunit recognition of the specific glycan receptors on host cells is also responsible for delivering the A subunits to cellular organelles where host cellular target proteins are located (Lee et al., 2020). Consequently, the specific glycan recognition by the B subunits is associated with toxin tropism (Bourdoulous and Lemichez, 2018; Lee et al., 2020, 2021; Petersen and Miller, 2020) since different cells, tissues, and hosts express structurally different sets of glycans and sialic acids. Furthermore, toxin tropism is often niche specific, corresponding to the primary infection sites of the bacterium producing the toxin, although in some cases, particularly with toxins produced by bacterial pathogens causing systemic infection, toxins can target a broad range of host cells (Lee et al., 2020; Yang et al., 2018b). In the interaction between toxin and host glycan receptor, the homopentameric configuration of the toxin B subunits enables the accommodation of high-avidity multivalent interactions between the toxin and host glycan receptor moieties, contributing significantly to the toxin tropism to specific sets of host cells at the whole-body level (Nguyen et al., 2020; Yang et al., 2018b).
The recent technological advances in toxin/protein biochemistry and glycobiology allowed for determining the interaction between bacterial A(2)B5 toxins and glycan receptor moieties in great detail. Notable examples include Salmonella A2B5 toxins (Lee et al., 2020; Nguyen et al., 2020; Yang et al., 2018b). Typhoid toxin was the first discovered Salmonella A2B5 toxin that is secreted by Salmonella enterica serovar Typhi (S. Typhi) during infection (Deng et al., 2014; Galán, 2016; Song et al., 2013). S. Javiana, a nontyphoidal Salmonella serovar (NTS), secretes Javiana toxin, an intestinal epithelial cell niche-specific typhoid toxin homolog. Javiana toxin receptor-binding subunit JaPltB carries three different amino acid residues located on glycan-receptor binding pockets, resulting in the different glycan-binding profiles that match with glycans expressed on intestinal epithelial cells (Lee et al., 2020).
Typhoid toxin is a pyramid-shaped heptamer consisting of two enzymatic A subunits, CdtB and PltA arranged in tandem, and a homopentamer of receptor-binding B subunits, PltB, on the base of the assembled toxin structure (Song et al., 2013). Pentameric PltB subunits serve as a base of the combined AB toxin complex, where the hydrophobic channel located at the center of the donut-shaped PltB pentamer connects to PltA (mono-ADP-ribosyltransferase [mART]) through the C-terminal α helix structure of PltA (Song et al., 2013). Besides the two Cys residues conserved across members in the mART family, PltA contains an additional Cys residue that links the PltA-PltB5 complex to the nuclease CdtB (Haghjoo and Galán, 2004; Spanò et al., 2008), resulting in its A2B5 stoichiometry (Song et al., 2013). Therefore, there is no direct interaction between PltB pentamer and CdtB located at the base and the vertex of the assembled toxin complex, respectively (Song et al., 2013).
PltB (glycan-receptor binding) and CdtB (nuclease) subunits of the secreted assembled toxin available in the extracellular milieu are responsible for typhoid toxin-mediated cellular and in vivo toxicities (Song et al., 2013; Yang et al., 2018b). In particular, the cellular toxicity of typhoid toxin can be objectively quantified through cell cycle profiling analysis of intoxicated host cells, as previously described (Ahn et al., 2021), also used in this study when evaluating the antibody-mediated neutralizing efficacies. Alternatively, the secreted assembled typhoid toxin mutant containing catalytically inactive PltA available in the extracellular milieu still endocytosed and induced host cell cycle arrests in G2/M and typhoid toxin-mediated in vivo toxicities, indicating that the enzymatic activity of PltA does not directly affect typhoid toxin-mediated endocytosis and intoxication of host cells, although PltA does play an important role at different stages of pathogenesis (unpublished data) (Lee et al., 2020; Song et al., 2013; Yang et al., 2018b).
There is a total of 15 spatially separated glycan-binding pockets on the PltB pentamer of typhoid toxin and, in each PltB monomer, the glycan-receptor binding pocket BS1 is located on the lateral side of PltB, whereas the other two glycan-binding pockets BS2 and BS3 are located on the bottom side of PltB (Lee et al., 2020). The BS1 can accommodate both α2–3 sialosides and α2–6 sialosides with higher affinities, whereas the BS2 and BS3 can accommodate α2–3 sialosides (Lee et al., 2020; Nguyen et al., 2020) (Table S2). In this study, we exploited typhoid toxin in dissecting the neutralizing mechanisms of antibodies targeting glycan-receptor binding subunits. We have obtained the structural basis explaining different toxin neutralization capabilities between two classes of antibodies targeting other regions on glycan-receptor binding subunits. Furthermore, we conducted secondary structure matching (SSM) analyses to determine whether the newly discovered mechanism is also applicable to other toxin-antibody interactions.
RESULTS
Monoclonal antibodies (mAbs) recognizing PltB subunits of typhoid toxin display different toxin neutralization efficacies
To recover a set of neutralizing antibodies targeting PltB subunits of typhoid toxin, we immunized a group of mice with the recombinant typhoid toxoid heptamer and, using splenic B cells, generated a collection of B cell hybridomas producing mAbs against typhoid toxin. In brief, after we obtained recombinant typhoid toxoid preparations, we validated the same A2B5 configuration of the toxoid as wild-type (WT) typhoid toxin via size-exclusion chromatography (SEC). After immunizations when mouse serum reciprocal endpoint titers were >100,000, we generated B cell hybridomas using Hybri-Max polyethylene glycol (PEG)-mediated fusion to Sp2/0-Ag14 myeloma cells, as previously described (Van Slyke et al., 2018). B cell hybridomas were cloned by limiting dilution three times to ensure clonality, resulting in a total of 10 PltB-positive hybridomas, named TyTx1–TyTx10. However, the characterization of hybridoma culture supernatants indicated that TyTx2 was a weak binder to the PltB homopentamer and was excluded from further studies. All of the remaining nine antibodies, that is, TyTx1 and TyTx3–TyTx10, effectively recognized both typhoid toxin and PltB homopentamer evaluated by conducting enzyme-linked immunosorbent assays (ELISAs) (Figures S1A–S1D). To select hybridoma clones producing different antibodies targeting glycan-receptor binding PltB subunits, we sequenced the antigen-recognizing variable regions of nine mAbs by conducting modified reverse transcriptase polymerase chain reaction (RT-PCR) followed by Sanger sequencing, as previously described (Ahn et al., 2021; Meyer et al., 2019). We found from sequencing analysis that seven hybridoma clones producing TyTx4–10 belong to the same family; therefore TyTx1, TyTx3, and TyTx4 (representing 4–10) are three distinct hybridoma clones producing antibodies targeting PltB (Figures S2 and S3). To perform detailed characterizations of antibodies in the context of interactions between toxins and glycan receptor moieties, hybridomas producing TyTx1, TyTx3, and TyTx4 were cultured on a milligram scale. Corresponding mAbs were purified using protein G agarose columns (Figure 1A; Figure S1E), after which we validated that purified TyTx1, TyTx3, and TyTx4 recognize PltB subunits, indicated via western blots using purified typhoid toxin separated by SDS-PAGE (Figure 1B).
Figure 1. mAbs recognizing PltB subunits display different toxin neutralization efficacies.
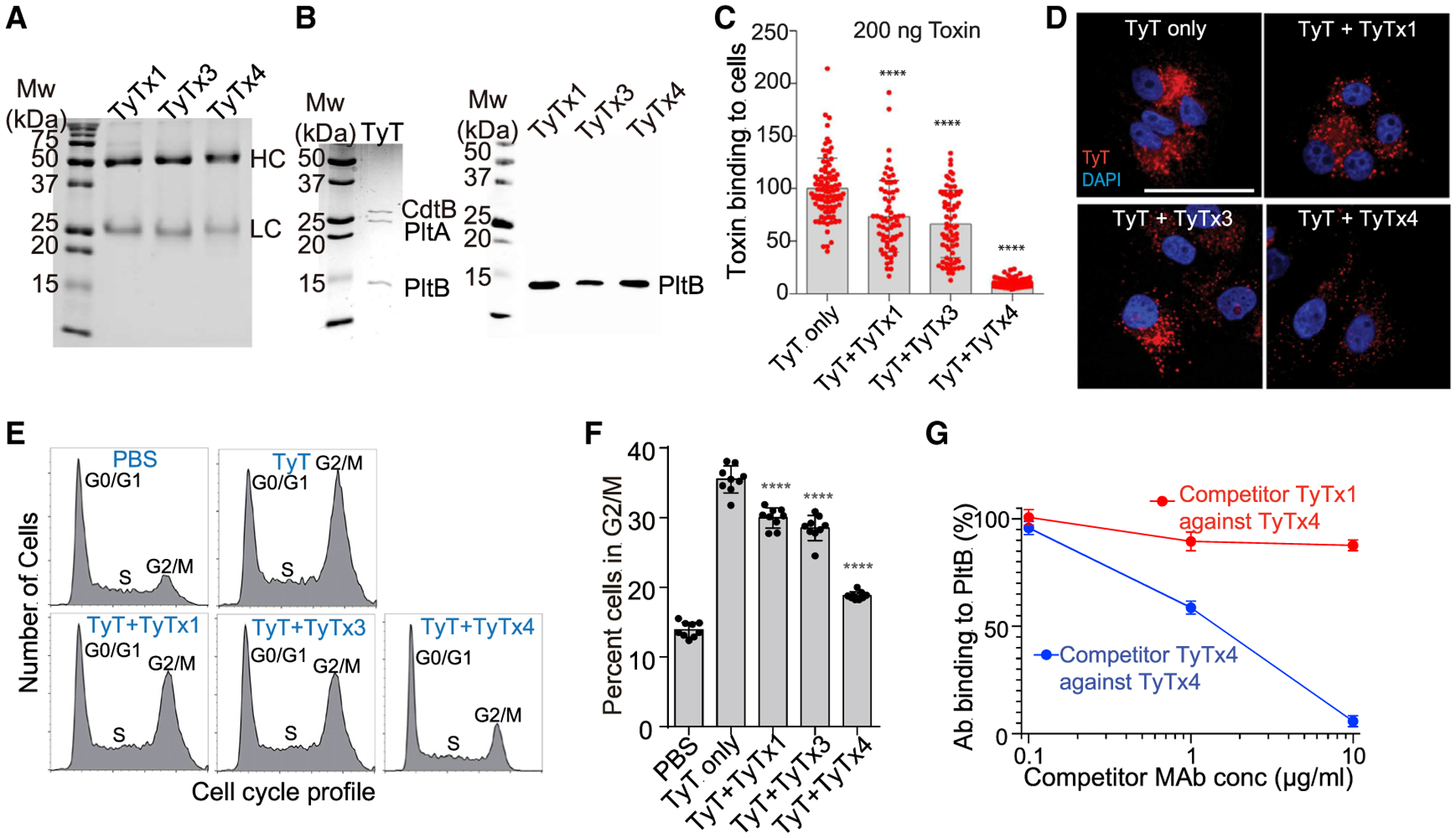
(A) Purified mAbs were separated on SDS-PAGE and stained with Coomassie brilliant blue (CBB). HC, heavy chain. LC, light chain. Superdex 200 chromatograms of these purified mAbs are shown in Figure S1E.
(B) Typhoid toxins (200 ng) were separated on SDS-PAGE and stained with CBB (left panel), when indicated, followed by western blot analysis for each mAb to determine its binding subunits among the three subunits of the typhoid toxin (right panel). Three independent experiments were performed. Representative blot results are shown.
(C) Quantification of mAb-mediated inhibition of typhoid toxin binding to Henle-407 cells. Henle-407 cells grown on coverslips were precooled to 4°C, incubated with 200 ng of typhoid toxin-Alexa Fluor 555 (AF555) in the presence or absence of mAb for 30 min, counterstained with DAPI, and analyzed by fluorescence microscopy. Bars represent the mean ± standard error of the mean (SEM) obtained from three independent experiments. Each dot reflects the typhoid toxin signal intensity per image. n = 65–89. ****p < 0.0001, compared to the TyT-only group. Unpaired two-tailed t tests. Results conducted with 100 ng of typhoid toxin are shown in Figure S1F.
(D) Representative fluorescence microscopy images of Henle-407 cells incubated with 200 ng of typhoid toxin-AF555 with and without mAbs for 4 h. Henle-407 cells were pre-treated with media containing 10 mM NH4Cl for 20 min at 37°C, before chilling and binding of anti-typhoid toxin mAbs and fluorescent-labeled typhoid toxin. TyT, typhoid toxin (red); DAPI (blue), DNA. Scale bar, 10 μm.
(E and F) Jurkat cells were treated with purified typhoid toxin with and without mAbs for 18 h. Cell cycle profiles were analyzed via flow cytometry to assess the typhoid toxin-induced host cell cycle arrest in G2/M. (E) Representative cell cycle profiles of Jurkat cells treated with typhoid toxin with and without mAbs. (F) Percent of cells in the G2/M cell cycle from three independent experiments. Bars represent average ± SEM. ****p < 0.0001. n = 9 per group. Unpaired two-tailed t tests.
(G) Competition ELISA results between TyTx1 and TyTx4. TyTx4 (100 ng) immobilized on protein G ELISA plates was incubated with a mixture of 10 ng of biotinylated PltB pentamer and indicated doses of competitor mAbs. The graph shows the values for the mean ± SEM. Three independent experiments were performed.
See also Figures S1–S3 and Table S2.
PltB subunits recognize specific sets of host glycan moieties expressed on the surface membranes of target cells and subsequently initiate the retrograde toxin trafficking to deliver the toxins into target host cells (Chang et al., 2019; Song et al., 2013). We therefore evaluated whether TyTx1, TyTx3, and TyTx4 could inhibit typhoid toxin binding and toxin trafficking. Quantitative fluorescence microscopy was conducted to measure mAb-mediated inhibition of Alexa Fluor 555-conjugated typhoid toxin binding to host cells. TyTx1 and TyTx3 inhibited toxin binding (200 ng of toxin used) to Henle-407 cells modestly, whereas TyTx4 resulted in a drastic inhibition of the toxin binding to host cells (Figure 1C). Comparable results of mAb-mediated inhibition of the typhoid toxin binding to host cells were observed with a lower dose (100 ng) of typhoid toxin (Figure S1F). Consistently, the toxin amounts delivered inside host cells were slightly decreased in the presence of TyTx1 or TyTx3, whereas TyTx4 markedly decreased toxin delivery into host cells, as compared to typhoid toxin only, which was assessed 4 h after the treatment (Figure 1D). We investigated whether the different outcomes in toxin binding and trafficking inhibition among anti-PltB antibodies correlate to the neutralization outcomes against typhoid toxin-mediated cellular toxicity by analyzing cell cycle profiles of Jurkat cells via flow cytometry. Typhoid toxin induces cell cycle arrests in G2 or M stages in intoxicated host cells due to CdtB-mediated host cell DNA damage (Ahn et al., 2021; Spanò et al., 2008). Consistent with the binding and trafficking inhibition results (Figures 1C and 1D), the effects of TyTx1 and 3 were modest, while the strong neutralizing effect was induced by TyTx4 (Figures 1E and 1F), indicating that TyTx1, TyTx3, and TyTx4 are indeed three different antibodies targeting PltB with different neutralizing efficacies. This result is in line with the different neutralizing in vivo efficacies assessed using a mouse intoxication model challenged with a lethal dose of typhoid toxin (Ahn et al., 2021). Since neutralizing outcomes by TyTx1 and TyTx3 were similar, which is significantly different from TyTx4, we selected TyTx1 and TyTx4 for further detailed atomic-level structure and function analysis. Competition ELISA results further supported the selection of TyTx1 and TyTx4, since the binding of biotinylated PltB homopentamer to TyTx4 immobilized on ELISA plates was completed in the presence of TyTx4 but not TyTx1 (Figure 1G), indicating that TyTx1 and TyTx4 recognize spatially separated epitopes on PltB.
A subunit-mediated asymmetry of the B subunit pentamer interferes with antibody binding to lateral side-located epitopes on PltB
To understand the molecular mechanism by which TyTx1 exhibits a lower neutralizing effect than TyTx4, we used cryogenic electron microscopy (cryo-EM) to analyze multiple complex particles consisting of typhoid toxin and TyTx1, and to determine the amino acid sequence of epitopes on PltB subunits recognized by TyTx1. To eliminate the potential ambiguity in the stoichiometry analysis of complex particles, we prepared TyTx1 Fab fragments for monovalent binding to each PltB subunit, as TyTx mAb would likely result in bivalent interactions with pentameric PltB subunits. TyTx1 Fab fragments were prepared by carrying out papain digestions of TyTx1 immunoglobulin G (IgG), completing protein G-mediated removal of undigested mAb and Fc, and performing SEC of the samples containing Fab using Superdex 75. The prepared TyTx1 Fab fragments were incubated with purified typhoid toxin overnight, resulting in toxin-TyTx1 Fab complex particles. A neutral isotonic buffer (15 mM Tris-HCl [pH 7.5] and 150 mM NaCl) was used to mimic physiologically relevant conditions.
We analyzed 741,088 cryo-EM complex particles of typhoid toxin and TyTx1 Fab (Figure 2A; Figure S4; Table S1). Note that 13% of the 741,088 particles were grouped into a “junk class” since we had an insufficient cryo-EM density required for determining the precise location on PltB homopentamer recognized by TyTx1 (Figure 2A; Figure S4). In the holotoxin structure, the position of PltA leans toward one side of the donut-shaped pentameric PltB subunits (PDB: 4K6L) (Song et al., 2013), resulting in self-control asymmetry of pentameric PltB subunits when PltB subunits are part of the combined holotoxin. Due to this A subunit-induced asymmetry, five subunits of the donut-shaped PltB pentamer are called herein (as well as in PDB: 4K6L crystal structure), in a clockwise manner, chain B (the closest to PltA), chain D, chain E (the second-most distant from PltA), chain C (the most distant from PltA), and chain A. From analyzing 644,747 complex particles (741,088 minus the 13% undefinable junk class), we found that approximately 72.4% of the complexes with sufficient density contained one typhoid toxin molecule and one TyTx1 Fab molecule where a single Fab was found to bind to either the C chain (the most distant from PltA) in ~48.3% of the complexes or the E chain (the second-most distant from PltA) in ~24.1% of the complexes (Figure 2A). The rest (27.6%) of the complexes with sufficient density consisted of typhoid toxin bound to two TyTx1 Fabs via binding of both C and E chains, the most distant and the second-most distant PltB subunits from PltA, respectively (Figure 2A). These results support the hypothesis that the skewed position of the A subunits in the 3D A2B5 toxin structure results in self-control asymmetry of pentameric B subunits and prevents antibodies binding to all five epitopes available on pentameric B subunits.
Figure 2. A subunit-mediated asymmetry of the B subunit pentamer interferes with antibody bindings to lateral side-located epitopes on PltB.
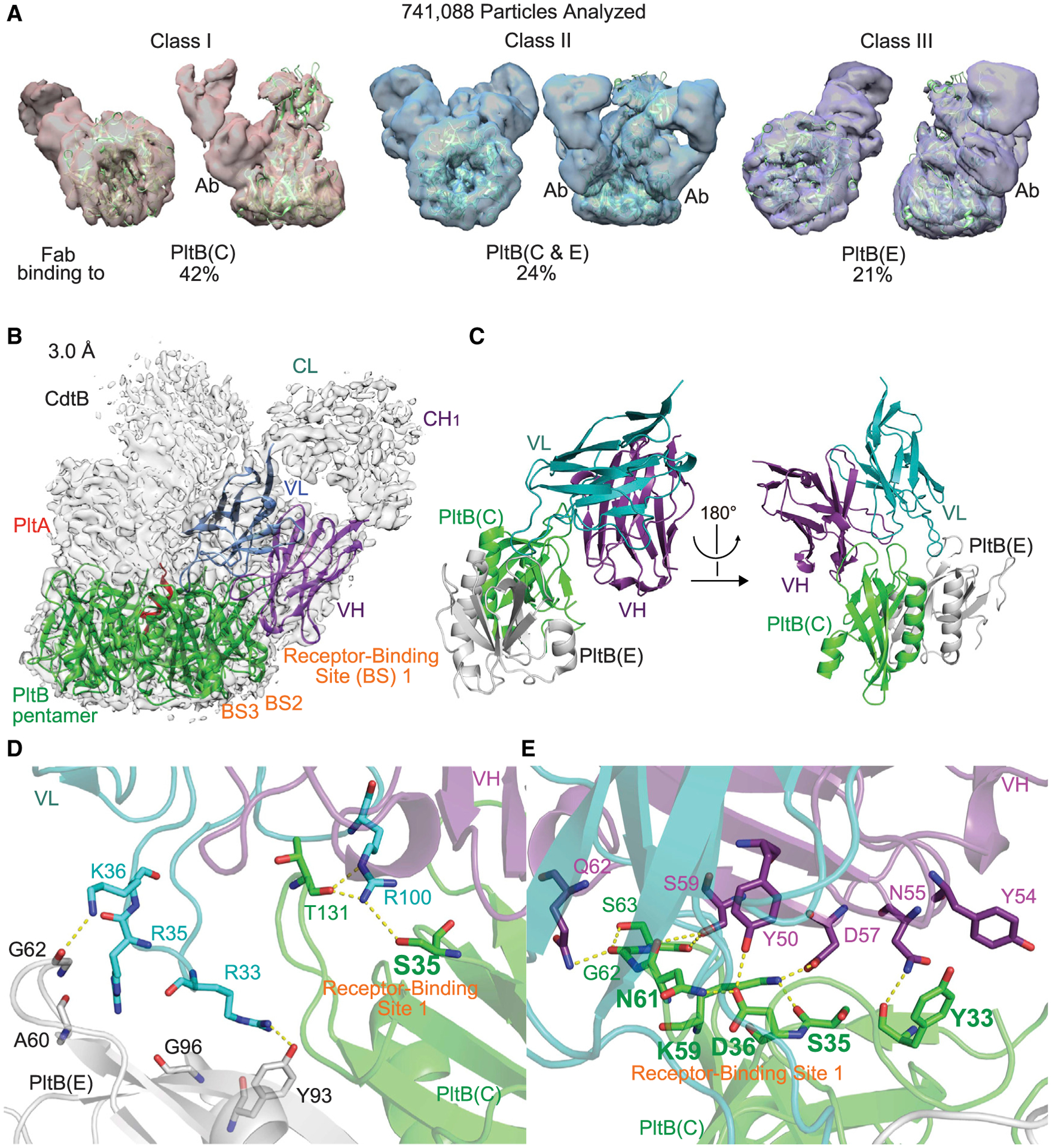
(A) Quantifications of the complexes consisting of typhoid toxin and TyTx1 Fab. The cryo-EM complex structures are overlaid with typhoid toxin PDB: 4K6L (green ribbon diagram). Ab, TyTx1 Fab. The remaining undefinable 13% was classified as “junk class.” See Figure S4 for details.
(B) Sharpened cryo-EM density map (gray) of class I typhoid toxin complexed with TyTx1 Fab (gray) with ribbon diagram of the refined structure of PltB pentamer (green) bound to variable regions of the light chain (VL, blue) and the heavy chain (VH, purple).
(C) Ribbon diagram of the interface between PltB subunits (C chain, green; E chain, gray) and TyTx1 VL and VH.
(D and E) Close-up views of the interactions between PltB subunits and TyTx1 VL (D) and PltB subunits and TyTx1 VH (E). Key residues in the glycan-receptor binding site BS1 of PltB subunits are highlighted in bold.
See also Figures S4 and S5.
To better explain the principle governing this toxin’s A subunit-mediated interference of antibody binding, we refined the class I complex map (TyTx1 Fab bound to the C chain) to 3.0 Å to determine the amino acid sequence of the epitope recognized by TyTx1 (Figures 2B–2E; Figure S4). The color-coded local resolution maps are shown in Figure S4. Typhoid toxin and PltB subunits in the refined cryo-EM structure align well with typhoid toxin and PltB pentamer crystal structures available, that is, PDB: 4K6L and 4RHR, respectively (Deng et al., 2014; Song et al., 2013) (Figure 2B; Figures S4 and S5). The overall and enlarged interfaces between PltB and the variable regions in the heavy and light chains of TyTx1 (VH and VL) are shown in Figure 2C and Figures 2D and 2E, respectively, along with their cryo-EM density maps (Figure S5). There are more interactions between PltB and the heavy chain of TyTx1 than the light chain (Figures 2C–2E; Figures S2 and S3). Intriguingly, many PltB amino acid residues recognized by TyTx1 overlap with PltB amino acid residues used for interacting with its glycan receptors α2–3 and α2–6 sialosides on host cell surfaces.
TyTx1 Fab directly interacts with many key residues of glycan-binding site BS1 on PltB, including Ser35 and Lys59, the most critical amino acid residues on BS1 (Figures 2D and 2E; Figure S5; Table S3). Specifically, among the PltB E chain residues, Ala60* (water-mediated), Gly62* (water-mediated), Tyr93, and Gly96* (water-mediated) form hydrogen bonds with the TyTx1 VL chain Arg35, Lys36, Arg33, and Arg35, respectively. An asterisk indicates an H bond via their main chains. Similarly, Tyr33*, Tyr33, Ser35, Asp36, Lys59, Asn61*, Gly62*, Ser63*, and Thr131* of the PltB C chain interact with Asn55 (VH chain), Tyr54 (VH, π stacks), Arg100 (VL), Tyr50 (VH), Asp57 (VH), Ser59 (VH), Gln62 (VH), Ser59 (VH), and Arg100 (VL) via H bonds or π stacks, respectively (Figures 2D and 2E; Figures S2 and S3). Note that some E chain residues also contribute to TyTx1 binding to the C chain, although these are mostly water-mediated interactions that are in line with the distance to TyTx1 bound to the C chain.
Interference is specific for antibodies recognizing lateral side-located epitopes on PltB subunits
If the A subunit-mediated asymmetry of pentameric PltB subunits is indeed the primary reason for the interference with TyTx1 bindings to its laterally located epitopes on PltB, the counterpart antibodies recognizing epitopes located on the far side of PltB subunits should not be interfered with by the A subunit-mediated asymmetry. Based on the results shown in Figures 1C–1G, we hypothesized that unlike TyTx1 recognizing amino acid residues located on the lateral side of PltB subunits, the epitope recognized by TyTx4 is likely located on the far side of PltB pentamer. We tested this hypothesis by solving the structure of typhoid toxin complexed with TyTx4 Fab via cryo-EM (Figure 3; Table S1). We found that TyTx4 Fab indeed recognizes amino acid residues located on the far side of the PltB pentamer (Figures 3A–3D; Figure S6). Specifically, 83% of the 309,475 complex particles contained one typhoid toxin molecule and five TyTx4 Fab molecules, and the remaining 17% had one typhoid toxin molecule and four TyTx4 Fab molecules (Figure 3A; Figure S6). These results are in sharp contrast to the stoichiometry of the TyTx1 Fab-typhoid toxin complexes and, therefore, support the specificity of self-control asymmetry-mediated interference toward the laterally located epitopes (Figures 2A and 3A; Figures S4 and S6).
Figure 3. Interference is specific for antibodies recognizing lateral side-located epitopes on PltB subunits.
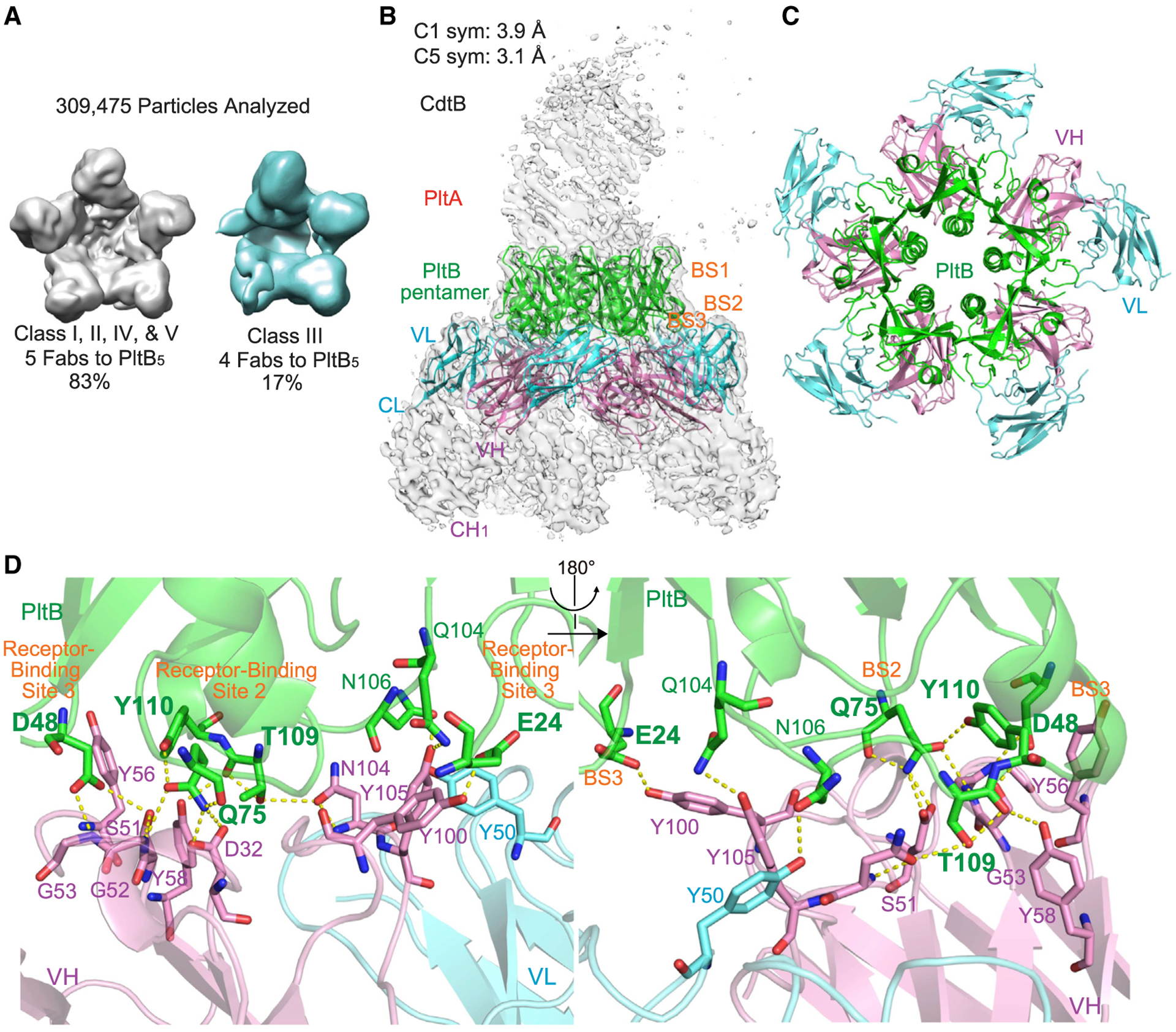
(A) Quantifications of the complexes consisting of typhoid toxin and TyTx4 Fabs. See Figure S6 for details.
(B) Sharpened cryo-EM C1-symmetry density map (gray) of typhoid toxin complexed with TyTx4 Fab with ribbon diagram of the refined structure of PltB pentamer (green) bound to VL (light blue) and VH (pink).
(C) Top view of ribbon diagram of the overall structure of the complex between PltB pentamer and five TyTx4 VLs and VHs.
(D) Close-up views of the interactions between the PltB subunit and TyTx4 VL and VH. Key residues in the glycan-receptor binding sites BS2 and BS3 of PltB subunits are highlighted in bold.
See also Figure S6.
To determine the amino acid sequence recognized by TyTx4, we refined the class I complex structures containing one typhoid toxin molecule and five TyTx4 Fab molecules to 3.1 Å (Figures 3B–3C; Figure S6). The color-coded local resolution map is shown in Figure S6. The epitope sequences for TyTx4 are Glu24, Asp48, Gln75, Gln104, Asn106, Thr109, and Tyr110 on PltB subunits (Figure 3D; Figures S6B–S6D). Significant intermolecular interactions between PltB and TyTx4 Fab are Glu24, Asp48, Asp48, Gln75, Gln75, Gln104, Asn106, Thr109, Thr109*, and Tyr110 of PltB subunits forming either H bond or π stacks by interacting with Tyr100 (VH), Ser51 (VH), Gly53* (VH), Asp32 (VH), Gly52* (VH), Tyr105 (VH), Tyr50 (VL), Asn104 (VH), Tyr58 (VH), and Tyr56 (VH, π stacks), respectively (Figure 3D; Figures S2 and S3). Similar to TyTx1 mAb, most interactions were through the heavy chain of TyTx4 (Figure 3D). Intriguingly, TyTx4 recognition residues also include many amino acid residues critical for glycan-receptor bindings situated on the glycan-binding pockets BS2 and BS3 (highlighted in bold in Figure 3D) that can accommodate α2–3 sialosides (Lee et al., 2020; Nguyen et al., 2020). In particular, Q75, T109, and Y110 in the BS2 and E24 and D48 in the BS3 participate in the direct interaction with α2–3 sialosides (Lee et al., 2020; Nguyen et al., 2020), which are located on the far side of the pentameric PltB subunits (Figure 3B). Therefore, cryo-EM results support the specificity of toxin’s self-control asymmetry-mediated interference toward the laterally located epitopes.
To further support the findings, we prepared TyTx1, TyTx3, and TyTx4 Fabs, incubated them with either typhoid toxin or PltB pentamer, and conducted SECs to estimate the stoichiometry of the complexes formed (Figure 4; Figures S1C and S1D). In line with cryo-EM and other characterization results, SEC analyses indicate that TyTx1 Fab and TyTx3 Fab bound to typhoid toxin in a ratio of 1~2 Fab molecules to 1 typhoid toxin molecule, while TyTx4 Fab bound to the toxin in a ratio of 4~5 TyTx4 Fab molecules to 1 typhoid toxin molecule (Figures 4A–4D). In contrast, when pentameric PltB subunits without the A subunits were used, we found complexes containing Fabs and PltB pentamer in a ratio of about 3~5 TyTx1 Fab and TyTx3 Fab molecules to 1 PltB pentamer and 4~5 TyTx4 Fab molecules to 1 PltB pentamer (Figures 4E–4H; Figures S1C and S1D). Therefore, the SEC results also support the specificity of A subunit-induced, self-control asymmetry-mediated interference toward epitopes laterally located on pentameric B subunits. We further validated the binding of TyTx3 to the lateral side of the PltB subunit by conducting ELISA-based epitope mapping assays using two types of recombinant PltB pentamer preparations, that is, tagless PltB5 and PltB-His6 at the C-terminal end that is located on the lateral side of PltB pentamer in 3D structure (PDB: 4K6L), as previously described (Ahn et al., 2021). TyTx3 recognized tagless PltB (Figure S1) but not PltB-His6, indicating that TyTx3 recognizes epitopes laterally located on PltB.
Figure 4. A subunit-mediated asymmetry is required for toxin interference with antibody binding to the laterally located epitopes on PltB.
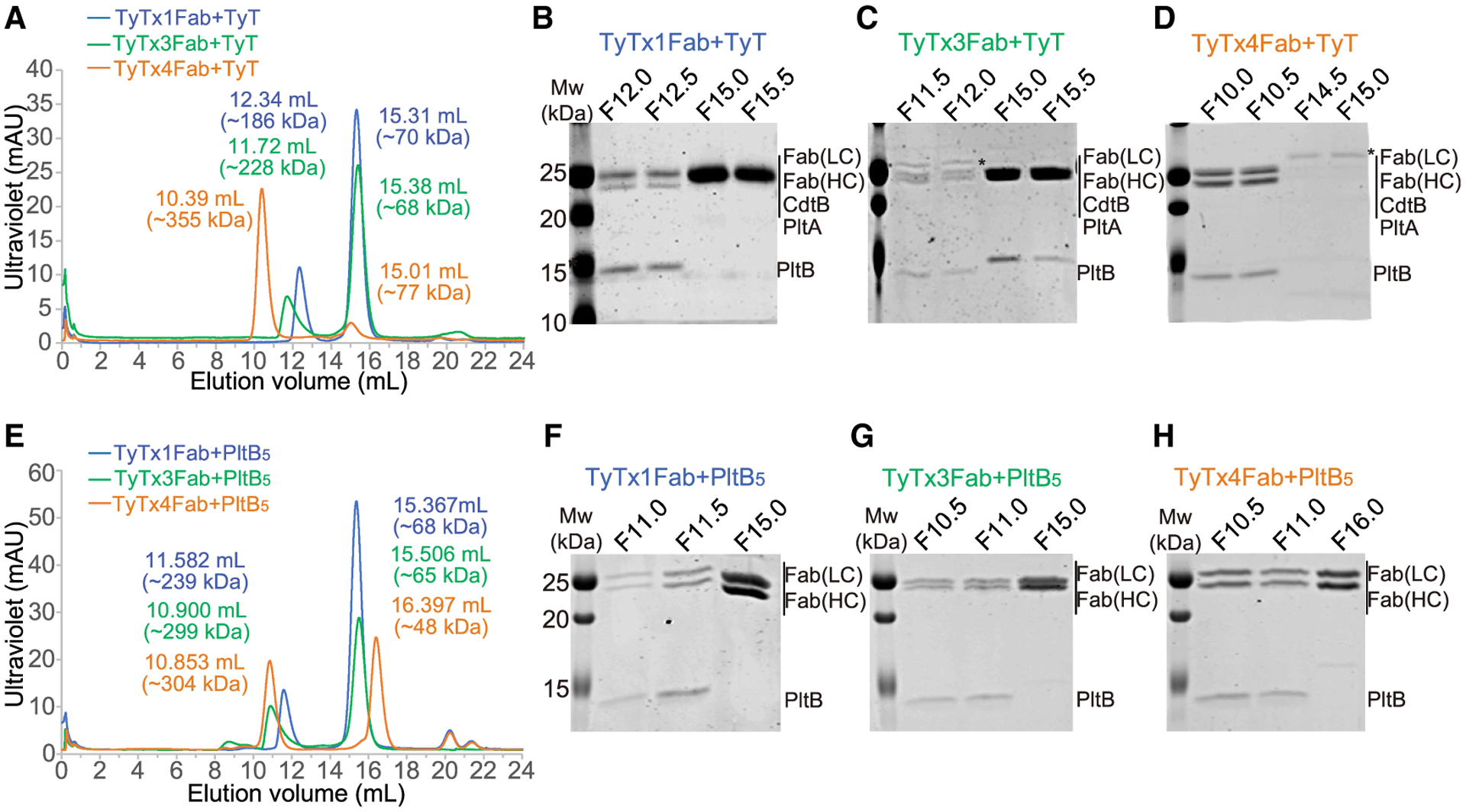
(A–D) Size-exclusion chromatography (SEC) of typhoid toxin complexed with TyTx1 Fab, TyTx3 Fab, or TyTx4 Fab. (A) SD200 chromatograms of typhoid toxin complexed with TyTx1 Fab (blue line), typhoid toxin complexed with TyTx3 Fab (green line), and typhoid toxin complexed with TyTx4 Fab (orange line). (B–D) SDS-PAGE analyses of peak fractions of TyTx1 Fab bound to typhoid toxin (B), TyTx3 Fab bound to typhoid toxin (C), and TyTx4 bound to typhoid toxin (D). Fraction names match with the start elution volume of 0.5 mL of collections (e.g., F12.0 means the 0.5-mL collection corresponding to an elution volume of 12.0–12.5 mL. Mw, molecular weight of standard. Fab (LC), the light (kappa) chain of Fab. Fab (HC), the heavy chain of Fab. Protein bands with asterisks in (C) and (D) indicate Fc contamination.
(E–H) SEC of PltB pentamer complexed with TyTx1 Fab, TyTx3 Fab, or TyTx4 Fab. (E) SD200 chromatograms of tagless PltB pentamer complexed with TyTx1 Fab (blue line), tagless PltB pentamer complexed with TyTx3 Fab (green line), and tagless PltB pentamer complexed with TyTx4 Fab (orange line). (F–H) SDS-PAGE analysis of peak fractions of TyTx1 bound to PltB pentamer (F), TyTx3 bound to PltB pentamer (G), and TyTx4 bound to PltB pentamer (H).
See also Figures S1E and S1F.
mAbs recognizing laterally located epitopes on PltB exhibit markedly reduced toxin-binding avidities
We hypothesized that self-control asymmetry-induced interference toward laterally located epitopes correlates to lower binding avidities for respective antibodies than antibodies that recognize epitopes on the far side. TyTx1 and TyTx3 recognize epitopes on the lateral side of PltB, while TyTx4 binds to the far side of PltB subunits (Figures 1, 2, 3, and 4). To determine the apparent overall affinities or avidities of mAbs to typhoid toxin, we carried out surface plasmon resonance (SPR) analyses, as previously described (Rudolph et al., 2020). Typhoid toxin was immobilized on sensor chips followed by flowing 4-fold serial dilutions of each mAb over the toxin for the apparent affinity/avidity determination of TyTx1, TyTx3, and TyTx4. Consistently with cryo-EM and SEC data indicating ratios of 4~5 TyTx4 Fabs to 1 typhoid toxin (equivalent to 2~3 TyTx IgGs per toxin) and 1~2 TyTx1 Fabs to 1 typhoid toxin (equal to 1 TyTx1 IgG per toxin), SPR results indicate that the apparent affinity of TyTx1 to typhoid toxin was ~71 nM and the apparent avidity of TyTx4 to typhoid toxin was ~7 pM, indicating ~10,000-fold binding affinity/avidity difference between these two antibodies (Figure 5; Table S4). Consistently, the binding affinity of TyTx3 to typhoid toxin was ~145 nM, which is ~20,000-fold lower than the apparent avidity of TyTx4 (Figure 5; Table S4). To better understand the precise contributions of the estimated monovalent versus pentavalent bindings observed between mAbs recognizing the laterally located epitopes and mAbs recognizing the far side-located epitopes to the overall binding affinity/avidity of these mAbs, we aimed to set up additional SPR assays using PltB monomer. Unfortunately, however, we could not obtain the full-length PltB monomer as a soluble protein required to estimate the undisturbed affinity between mAbs and PltB monomer. Therefore, although we were unable to determine the precise contributions of monovalent and pentavalent bindings of TyTx1 and TyTx4 toward their observed binding affinities to typhoid toxin, we demonstrated that the binding affinities of TyTx1 and TyTx3 (recognizing epitopes laterally located on the PltB pentamer and therefore interference) are markedly lower than the overall binding avidity of TyTx4 (recognizing epitopes located on the far side of the PltB pentamer and therefore no interference).
Figure 5. mAbs recognizing laterally located epitopes on PltB exhibit markedly reduced toxin binding avidities.
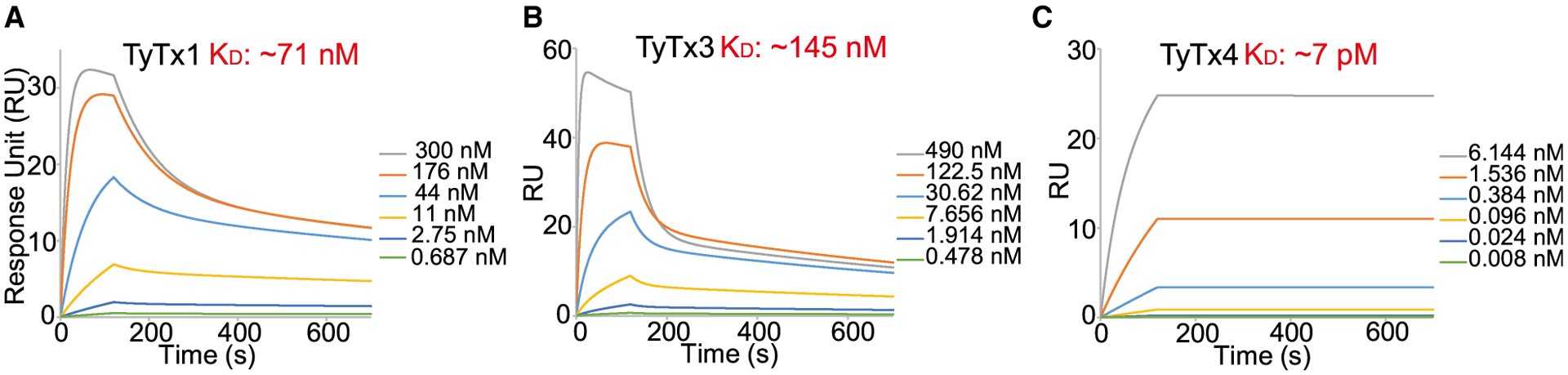
(A–C) Kinetics of TyTx1 (A), TyTx3 (B), and TyTx4 (C) binding to typhoid toxin by SPR assays. Mean sensorgrams from TyTx mAbs binding with typhoid toxin-coated series S CM5 chips (5–10 μg/mL) over a range of high (silver) and low (green) concentrations (nM). Sensorgrams were normalized to fit a bivalent binding model using the Biacore T200 evaluation software (GE Healthcare). Calculated equilibrium constant (KD) values are indicated in the graph.
See also Table S4.
mAbs recognizing laterally located epitopes on PltB exhibit markedly reduced toxin neutralization
We next investigated whether self-control asymmetry of typhoid toxin resulted in a decrease in relevant mAb-mediated toxin neutralization in the context of infection. We carried out in vitro infection and toxin neutralization assays using S. Typhi, as previously described (Ahn et al., 2021). In this system, typhoid toxin was produced continuously by S. Typhi during infection. TyTx4, which is undisturbed by self-control asymmetry, neutralized Salmonella A2B5 toxins much more effectively than did TyTx1 and TyTx3, ones that are disturbed by self-control asymmetry (Figures 6A and 6B). The difference became more apparent when a higher dose of infection was used (Figure 6C). When a multiplicity of infection (MOI) of 30 was used for S. Typhi infection, TyTx4 induced a marked toxin neutralization, but TyTx1 and TyTx3 showed a mild effect in toxin neutralization (Figures 6A and 6B). In comparison to ~50% of host cells in G2/M in the 30 MOI infection, 50 MOI infection of S. Typhi made ~70% of host cells arrested in G2/M (Figure 6C). In this condition, the neutralization effects elicited by TyTx1 and TyTx3 were modest, which was not significantly different from the toxin-only samples without mAbs (Figure 6C). An isogenic S. Typhi strain harboring the CdtBH160Q catalytic mutant subunit indicated that cell cycle arrests in G2/M were specific to the toxicity induced by typhoid toxin (Figures 6A and 6B).
Figure 6. mAbs recognizing laterally located epitopes on PltB exhibit markedly reduced toxin neutralization.
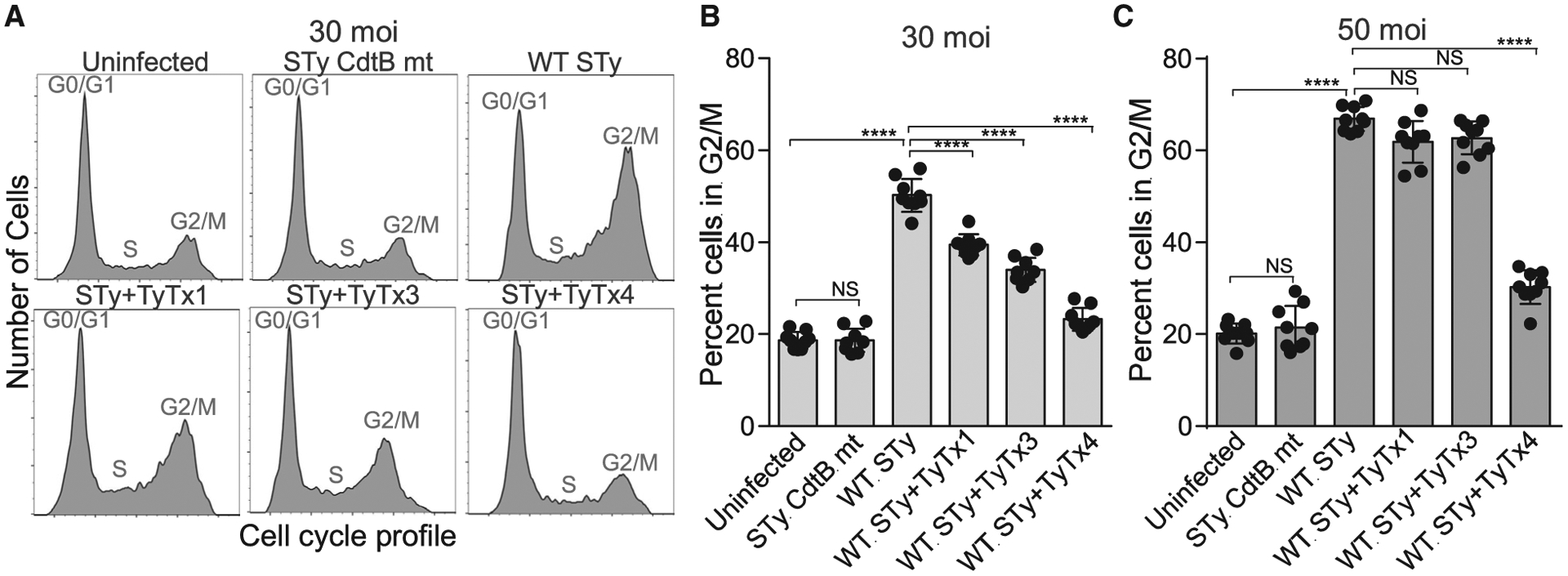
(A–C) Measurements of mAb-mediated toxin neutralization against typhoid toxin continuously produced by S. Typhi during infection. Representative cell cycle histograms (A) and percent cells in the G2/M cell cycle that indicates the typhoid toxin-mediated toxicity (B and C) are shown. Henle-407 cells were left uninfected or infected with S. Typhi (STy) at a multiplicity of infection of 30 (A-B) or 50 (C) for 3 days in the absence or presence of indicated mAbs. A mutant S. Typhi containing CdtBH160Q (STy CdtB mt) was used as an isogenic control for typhoid toxin-mediated G2/M cell cycle arrest. Cell cycle profiles were analyzed via flow cytometry. Three independent experiments were performed. Bars represent average ± SEM. ****p < 0.0001. NS, not significant. Unpaired two-tailed t tests. n = 9 per group.
See also Table S6.
Toxin interference with antibody-binding to laterally located epitopes on pentameric B subunits may be universal among AB5 and A2B5 toxins
Javiana toxin is an epithelial cell niche-specific typhoid toxin homolog exhibiting A2B5 stoichiometry. It contains the homopentameric JaPltB subunits highly homologous to typhoid toxin PltB with a few amino acid sequence variations (Lee et al., 2020). The epitope sequences recognized by TyTx1 and TyTx4 are present in JaPltB. Purified recombinant Javiana toxin was used to evaluate the efficacy of mAbs for Javiana toxin neutralization. Similar to their neutralization efficacies to typhoid toxin, TyTx4 showed the most significant neutralization effect against Javiana toxin, whereas TyTx1 and TyTx3 showed a modest level of Javiana toxin neutralization (Figure 7A). These results indicate that the A subunit-mediated interference with antibody binding to laterally located epitopes on the B pentamer is also applicable to Javiana toxin, another member in the A(2)B5 toxin family.
Figure 7. Toxin interference with antibody binding to laterally located epitopes on pentameric B subunits may be universal among A(2)B5 toxins.
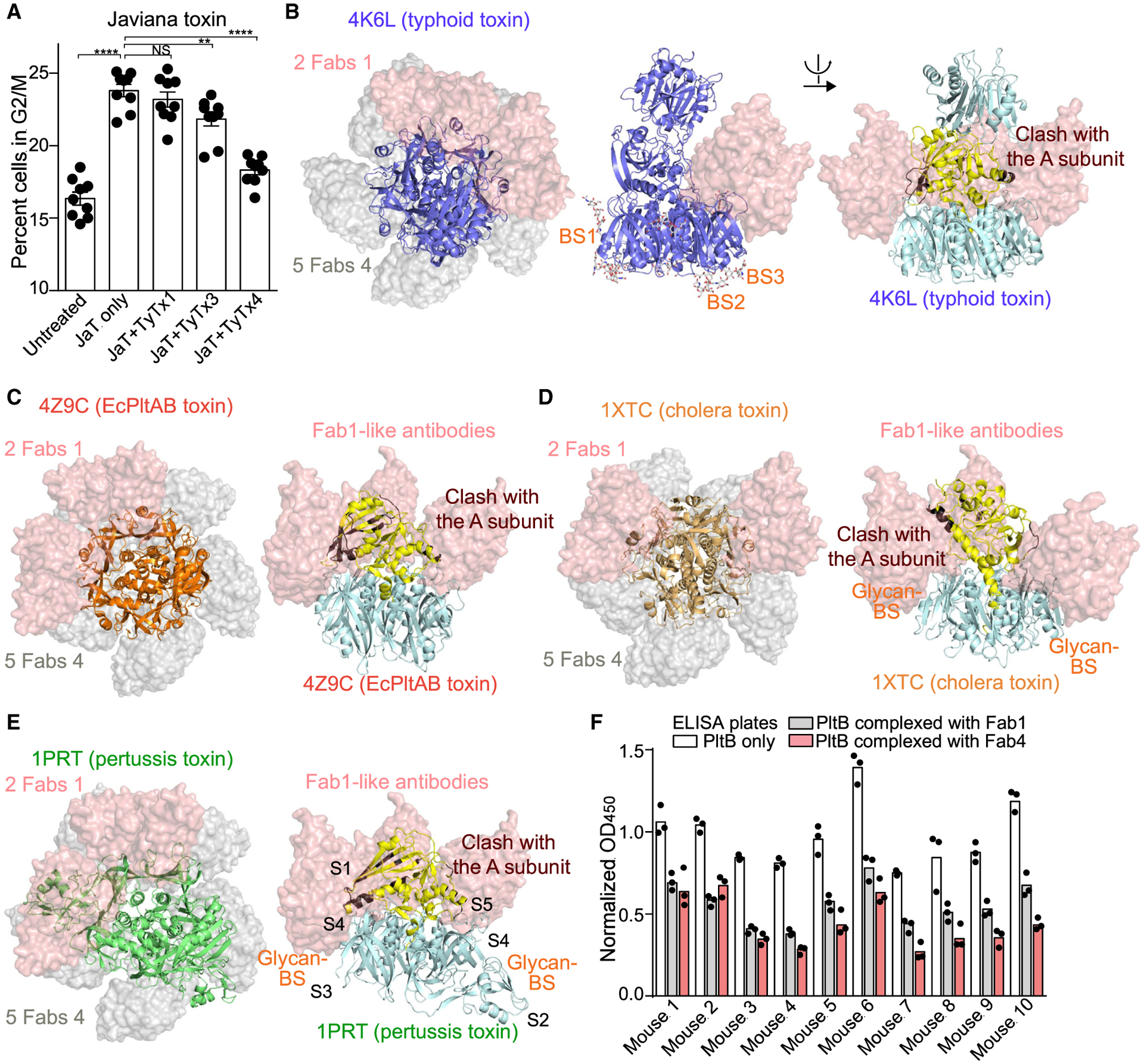
(A) mAb-mediated neutralization of Javiana toxin, an epithelial cell niche-specific typhoid toxin homolog from S. Javiana. Percent of cells in the G2/M cell cycle indicate the Javiana toxin-mediated toxicity. Henle-407 cells were treated for 48 h with PBS (Untreated) or Javiana toxin (0.6 pM) in the absence (JaT only) and the presence of indicated antibodies (300 pM). Cell cycle profiles were analyzed via flow cytometry. Three independent experiments were performed. Bars represent average ± SEM. **p < 0.01, ****p < 0.0001. NS, not significant. Unpaired two-tailed t tests. n = 9 per group.
(B) SSM analyses of typhoid toxin match with the data obtained from the cryo-EM studies, indicating that antibodies recognizing the laterally located epitopes can bind to up to two PltB subunits (indicated as 2 Fabs 1, pink volume), as opposed to antibodies recognizing the bottom-located epitopes can bind to five PltB subunits (5 Fabs 4, gray volume). The left panel shows the top view of typhoid toxin (4K6L), Fab1, and Fab4; the middle panel shows the side view of typhoid toxin complexed with Fab1; and the right panel shows the side view of Fab1 clashed with the toxin A subunit (brown).
(C–E) E. coli PltAB toxin (C), cholera toxin (D), and pertussis toxin (E) with TyTx1 Fabs (pink volume; representing antibodies recognizing the lateral side of the B pentamer) and TyTx4 Fabs (gray volume; representing antibodies recognizing the far side of the B pentamer). Total numbers of Fabs that can bind to the indicated toxins without clash are indicated in the top view images that overlay toxin, Fabs of TyTx1, and Fabs of TyTx4. Yellow indicates A subunits; brown indicates regions on the A subunits that clash with Fab1.
(F) ELISAs determining the relative antibody abundance of antibodies recognizing the laterally located epitopes and antibodies recognizing the far side-located epitopes on PltB. Serum samples from 10 individual mice received two injections with a 2-week interval of typhoid toxoid via the subcutaneous (s.c.) route were analyzed. Bars represent the average of the three independent measurements.
Next, to provide insight into whether the toxin interference mechanism with antibody binding to laterally located epitopes is universal among AB5 and A2B5 toxins, we carried out SSM analyses of four additional AB5 toxins, that is, E. coli PltAB toxin, cholera toxin, Shiga toxin, and pertussis toxin (Figures 7B–7E; Figure S7; Table S5). In 3D structures, such as typhoid toxin, the A subunits of these toxins are asymmetrically positioned toward one side of their donut-shaped pentameric B subunits. Consistently, SSM analyses indicate that antibodies recognizing laterally located epitopes on the B pentamer (such as TyTx1) could not be placed for all five B subunits due to the clashes with the asymmetrically positioned A subunits (Figures 7B–7E; Figure S7; Table S5). In SSM analyses, TyTx1 and TyTx4 Fab structures were computationally placed as representatives to determine whether these two classes of antibodies can fit in other toxin structures without clashes. In contrast, such clashes did not occur with antibodies recognizing the far side of the B pentamer (such as TyTx4) (Figures 7B–7E; Figure S7; Table S5), indicating that the asymmetrically located A subunit-mediated interference with antibody binding to epitopes laterally located on their B subunits may also occur in the case of other AB5 toxins. Lastly, modified ELISAs were conducted to obtain insight into the relative abundance of two types of antibodies targeting glycan-receptor binding subunits after mice were immunized with two-dose typhoid toxoid vaccine shots, as previously described (Yang et al., 2018b). Note that this immunization strategy does not necessarily represent the repertoire of antibodies targeting PltB raised in the context of natural infection. To capture and quantify total anti-PltB antibodies, antibodies recognizing far side-located epitopes on PltB and antibodies recognizing lateral side-located epitopes on PltB, respectively, we probed plasma samples from 10 immunized mice for purified PltB, PltB pre-complexed with TyTx1 Fab, and PltB pre-complexed with TyTx4 Fab and found that all 10 immunized mice had both types of anti-PltB antibodies (Figure 7F).
DISCUSSION
Little is known about the A2B5 toxin neutralizing mechanism by antibodies targeting glycan-receptor binding subunits. We unraveled the structural basis explaining the different toxin neutralizing efficacies among glycan-receptor binding sites targeting antibodies, using wet and dry approaches that integrate structural, biochemical, biological, and computational characterizations at the molecular level. Due to the asymmetrical positioning of the A subunits toward one side of the pentameric B subunits, A2B5 toxins were able to interfere with specific groups of antibody binding, ones that recognize laterally located epitopes on the B pentamer. As a result, this toxin-intrinsic interference made the corresponding glycan-binding sites remain available for toxin binding and endocytosis processes, indicating that A2B5 toxins can tolerate up to a certain threshold of laterally located epitope-binding antibodies. This is supported by the observed difference in the inhibition of toxin binding, trafficking, and in vitro and in vivo neutralization, as well as the apparent binding affinity difference to the toxins among PltB-targeting antibodies (this study and Ahn et al. [2021] for in vivo neutralization studies).
The toxin-intrinsic interference mechanism was specific to epitopes laterally located on the B pentamer of typhoid toxin, consistent with the skewed positioning of the A subunit toward one side of the pentameric PltB subunits. In support of this, antibodies recognizing epitopes located on the far side of the B pentamer indeed bound to all five epitopes available on the B pentamer. Moreover, the observed interference with antibodies recognizing epitopes laterally located on PltB was no longer observed when the A subunits were removed from the assembled toxin complex, indicating that the asymmetrical positioning of the A subunit is essential in this toxin-intrinsic interference against the binding of specific groups of neutralizing antibodies.
Other AB5 and A2B5 toxins likely use this toxin interference mechanism against specific groups of antibodies. Another Salmonella A2B5 Javiana toxin contains the identical epitopes on JaPltB, due to the high sequence similarity between the two. Anti-PltB mAbs also resulted in different toxin-neutralizing efficacies against Javiana toxin, since we observed the modest toxin neutralization by mAbs recognizing laterally located epitopes on JaPltB and the profound neutralizing outcomes by mAbs recognizing far side-located epitopes on JaPltB. Intriguingly, we also demonstrated that cholera toxin, pertussis toxin, E. coli toxin, and Shiga toxin have their A subunits asymmetrically positioned toward one side of the pentameric B subunits. Consistently, SSM analyses were conducted using the crystal structures of these toxins and TyTx1 Fab and TyTx4 Fab structures as model antibodies representing mAbs recognizing laterally located and far side-located epitopes on the B pentamer, respectively. The SSM analysis results indicate that this newly revealed molecular feature may be broadly applicable to the interactions between asymmetrical AB5 and A2B5 toxins and antibodies targeting glycan-receptor binding B subunits.
Based on the less well-resolved density of parts of toxin A subunits (Figures S4 and S6), it is reasonable to think about a possibility that PltA binding to PltB may not be static (e.g., a scenario that PltA rotates around the PltB wheel). However, our recent data indicate that such an effect is negligible even if there is such an effect. This prediction comes from our recent study, where we solved the toxin-TyTx11 mAb complex using cryo-EM (Ahn et al., 2021). We found that TyTx11 stabilizes CdtB and thus the two A subunits of the toxin, resulting in the 3D reconstructed toxin-TyTx11 complex structure without any poor density region and with sufficient density for all three subunits of typhoid toxin. The interface between PltA and PltB pentamer of typhoid toxin complexed with TyTx11 (PDB: 6VX4) is comparable to that of the solved structures of the holotoxin (PDB: 4K6L), strongly supporting the concept that PltA binding to PltB is nearly static or static with no significant flexibility detected in the interface.
Both classes of glycan-receptor binding sites targeting antibodies against typhoid toxin were available at a similar level in all mice vaccinated with two doses of typhoid toxoid, as evaluated by conducting modified ELISAs designed for learning about the relative antibody abundance of two classes of anti-PltB antibodies. In this experiment, inactive typhoid toxoid was used for immunizing mice that received two injections of the toxoid via a subcutaneous route. Thus, this study did provide insight into the relative antibody titers and repertoire about anti-PltB antibodies induced after a two-dose toxoid vaccine but did not provide information about the ratio of these two types of anti-PltB antibodies in other conditions such as natural infection and different vaccination regimens (e.g., different routes and numbers of boosters). However, it is intriguing to speculate that active typhoid toxin secreted by infected S. Typhi during natural infection may result in significantly different antibody titers and repertoire since active typhoid toxin can modulate host immune cell functions (Lee et al., 2020; Nguyen et al., 2020; Yang et al., 2018b). It is also intriguing to learn more in the future about whether the adjustment of toxoid vaccination regimes alters the antibody titers and repertoire toward the prolonged, more efficacious humoral immune responses. The toxin interference phenomenon unraveled in this study is anticipated to be valuable in interpreting data obtained from such future research endeavors with typhoid toxin and other AB5 and A2B5 toxins.
The neutralizing epitopes that we discovered in this study are well conserved among S. Typhi clinical isolates and non-typhoidal Salmonella serovars that harbor an epithelial cell niche-specific typhoid toxin homolog. For instance, all S. Typhi clinical isolates identified thus far, including multidrug-resistant (MDR) and extensively drug-resistant (XDR) S. Typhi strains, encode identical typhoid toxin PltB subunits (Table S6). No sequence variation in the typhoid toxin subunits has been observed among S. Typhi clinical isolates (Table S6), suggesting that the neutralizing epitopes characterized in this study would neutralize typhoid toxin produced by drug-sensitive and resistant S. Typhi during human infection.
In summary, we conducted a series of structural and functional studies designed for learning about the fundamentals of the molecular interactions among bacterial A(2)B5 toxins, glycan receptors, and glycan-receptor binding sites targeting antibodies. The principle governing the toxin-intrinsic interference phenomenon to A(2)B5 toxin neutralization offers insights into the advanced understanding of bacterial AB toxin biology in the context of host-pathogen interactions and the design of the more effective and prolonged anti-toxin strategies that relate to today’s significant public health concerns driven by antibiotic-resistant bacterial pathogens.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further requests for resources and materials should be directed to and will be fulfilled by the Lead Contact, Jeongmin Song (jeongmin.song@cornell.edu).
Materials availability
Recombinant Salmonella A2B5 toxins and antibodies used for this study will be made available to other researchers via the Institutional Material Transfer Agreements for collaborations.
Data and code availability
The published article includes all datasets generated during this study. Coordinates for the atomic structures have been deposited in the RCSB Protein Data Bank: 7K7H and 7K7I (Table S1). Electron microscopy density maps have also been deposited to EMDB: EMDB-22699 and EMDB-22700 (Table S1).
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Microbe strains
WT and CdtB catalytic mutant Salmonella enterica serovar Typhi (S. Typhi) ISP2825 have been described previously (Song et al., 2013). For infection experiments, Salmonella strains were grown at 37°C in 2 mL Luria-Bertani (LB) broth containing 0.3 M NaCl to an OD600nm of ~0.9 after inoculation from an overnight culture in 2 mL LB broth at a dilution of 1:50.
Cell lines
Human intestinal epithelial Henle-407 cells and human peripheral blood T lymphocyte Jurkat cells were cultured in DMEM high glucose (Invitrogen) and RPMI-1640 (Invitrogen) supplemented with 10% FBS (Hyclone cat# SH30396.03, Lot# AD14962284), respectively. Sialic acid contents of the FBS used were validated, which was ~99% Neu5Ac and less than 1% Neu5Gc. Cells were kept at 37°C in a cell culture incubator with 5% CO2. Mycoplasma testing was conducted regularly as part of the cell maintenance practice.
Mouse experiments
Mouse experiments were conducted following the protocol approved by the Wadsworth Center’s and Cornell’s institutional animal care and use committee (IACUC). Protocol #17-428 (Wadsworth) and Protocol 2014-0084 (Cornell) were assigned by the IACUC/ethics committee that approved the animal experiments performed for hybridoma production study and mouse immunization study, respectively. The experiments were followed by IACUC and AAALAC guidelines. For hybridoma production, female BALB/c mice approximately 5–7 weeks of age were purchased from Taconic Biosciences (Albany, NY) and housed under conventional, specific pathogen-free conditions. For immunization study, 5- to 8- week-old male and female CMAH null mice were received two injections with a 2-week interval of typhoid toxoid via the subcutaneous route. The mice were originally purchased from the Jackson Laboratory and bred in a vivarium in the animal facility at Cornell University under conventional, specific pathogen-free conditions.
METHOD DETAILS
Recombinant typhoid toxin, typhoid toxoid, and PltB pentamer
Overexpression and purification of typhoid toxin (containing the CdtB-His6), typhoid toxoid (containing CdtB-His6), and PltB-His6 homopentamer were carried out as previously described (Deng et al., 2014; Song et al., 2013; Yang et al., 2018b). When indicated, purified toxins were fluorescently labeled with either Alexa Fluor-555 dye (Molecular Probes, Thermo Fisher Scientific) according to the vendor’s recommendation.
Cloning, expression, and purification of tagless PltB pentamer
The tagless PltB expression construct, named pJS0051, was generated by removal of the A subunit genes from the pSB5022 plasmid for expressing wild-type typhoid toxin complex with 6xHis tagged CdtB (Song et al., 2013). The following primer pairs were used for the process: F: 5′- GCTTTGGACCCAAGTAATGAGATCCGGCTGCTAACAAAGC-3′ and R: 5′-CTCATTACTTGGGTCCAAAGC-3′. Gibson assembly was used to generate the construct and the sequence was confirmed via Sanger sequencing by the Cornell Biotechnology Resource Center Genomic Core. The following procedures were conducted to purify the tagless PltB pentamer.
Preparation of TyTx1 antibody-conjugated agarose resin
TyTx1 antibody was conjugated onto resin using NHS activated agarose slurry (ThermoFisher Scientific, Pierce #26200) using the provided protocol with minor modification for gravity column chromatography. In brief, 1 mL of settle resin was transferred, washed with PBS, and incubated with PBS containing 14 mg of TyTx1 for 1 hr at room temperature with light rocking for the conjugation to occur. After washing to remove unconjugated proteins, the resin was quenched by repeated washing with 10 mL of 0.5 M Tris-HCl, pH 8.0, and stored at 4°C until use.
Purification of tagless PltB
E. coli BL21(DE3) ΔendA ΔrecA (E. coli Acella strain; Edgebio) carrying pJS0051 was grown in LB broth to approximately OD600 = 0.7 at 37°C, transferred to 28°C, added with 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG), and incubated for 16 hr. The harvested bacteria pellet was resuspended in a buffer containing 15 mM Tris-HCl, pH 8.0 and 150 mM NaCl, 1x EDTA-free protease inhibitor cocktail, 0.2 mg/mL lysozyme, and 80 μg/mL DNase I, subjected to 3 times of the freeze-thaw step in liquid nitrogen and lysed by sonication. The clarified lysate was mixed with the prepared TyTx1 conjugated agarose and incubated at 4°C for 30 minutes with gentle rocking. The resin was recaptured by passing the lysate through a gravity column and washed with 15 mM Tris-HCl, pH 8.0, 150 mM NaCl. The protein was eluted with 5 mL of 100 mM Glycine, pH 3.0, and the eluted protein was quickly neutralized with 1 mL of 1 M Tris-HCl, pH 8.8. The purified protein was examined via 15% SDS-PAGE, SEC, and ELISA.
Generation of hybridomas producing mAbs
Mice were immunized via intraperitoneal injections with 2 μg of ultrapure endotoxin-free recombinant typhoid toxoid without adjuvant at two-week intervals, as previously described (Yang et al., 2018b). Genetically engineered inactive forms of typhoid toxin, carrying all three subunits PltB5, PltA, and CdtB, were used in the study. Specifically, the toxoid carries point mutations in PltAE133A and CdtBH160Q, D195S. The toxoid has the same A2B5 stoichiometry as WT typhoid toxin, as determined via size exclusion chromatography. Serum reciprocal endpoint titers were > 100,000 following three immunizations. B cell hybridomas were generated by performing Hybri-Max PEG fusion procedures using the Sp2/0-Ag14 (ATCC® CRL-1581) myeloma. Similar to the procedure established previously (Van Slyke et al., 2018), we seeded the resulting hybridomas onto wells of 96-well cell culture-treated microtiter plates, and hybridomas were selected in DMEM media supplemented with 10% UltraCruz® Hybridoma Cloning Supplement (HCS) (Santa Cruz Biotechnology, Dallas, TX), 10% fetal calf serum, oxaloacetate, pyruvate, and insulin (OPI), hypoxanthine/aminopterin/thymidine (HAT), and penicillin/streptomycin. During the procedures, HAT was gradually replaced with hypoxanthine-thymidine (HT), and surviving hybridomas secreting antibodies of interest were cloned by limiting dilution and expanded in DMEM media without HT, before being transitioned to either CD Hybridoma AGT (Invitrogen) or DMEM supplemented with 4 mM L-glutamine, 4.5 g/L glucose, 1 mM sodium pyruvate, 1.5 g/L sodium bicarbonate, 10% HCS, 20% Ultra Low IgG Fetal Bovine Serum (Invitrogen), and penicillin/streptomycin. B cell hybridomas were cloned by limiting dilution three times to ensure clonality.
mAb purification and direct enzyme-linked immunosorbent assay (ELISA)
Hybridoma clones were selected upon a positive reaction against typhoid toxoid via direct ELISAs. Murine MAbs were purified from hybridoma supernatants using protein G chromatography and subjected for epitope characterization using ELISAs and western blots. 96-well plates (Costar) were coated with 50 ng of purified typhoid toxoid or tagless PltB pentamer in 100 μL plate-coating buffer (50 mM carbonate-bicarbonate buffer, pH 9.6), and incubated overnight at 4°C. Wells were washed with PBS containing 0.05% Tween 20 and blocked with PBS containing 1% BSA for 1 h at 37°C. Purified mAb contained in 50 μL PBS/0.05% Tween 20/0.5% BSA was added to each well and incubated for 2 h at 37°C. After washing, bound antibodies were detected with horseradish peroxidase (HRP)-conjugated anti-mouse immunoglobulin IgG (Southern Biotech) at a 1:10,000 dilution in PBS/0.05% Tween/0.5% BSA. Wells were then incubated with a HRP substrate, tetramethylbenzidine (Sigma), for 10–30 min and the reaction was stopped by the addition of 100 μL of 1 M H3PO4. The results were assessed by reading a Tecan Infinite 200 Pro microplate reader.
Competition ELISA
Overall procedures were similar to direct ELISA, except for the following. Protein G ELISA plates (Costar) were incubated with 1 μg/mL of capture mAb in 100 μL of 50 mM carbonate-bicarbonate buffer, pH 9.6 for overnight at 4°C. After washing with PBS/0.05% Tween 20 and blocking with 100 μL of PBS/1% BSA for 1 hr at 37 °C, the plates were incubated with 100 μL PBS/0.5% BSA containing 100 ng/mL of biotinylated PltB homopentamer and 0.1–10 μg/mL of competitor mAb for 2 h at 37 °C. Bound biotinylated PltB was detected with streptavidin-HRP-conjugated anti-mouse IgG (Bio-Rad Laboratories) at a 1:10,000 dilution in 100 μL PBS/0.5% BSA.
mAb variable region sequencing
Antibody sequencing was conducted based on a recently published method (Meyer et al., 2019). In brief, total RNA was extracted from the hybridoma cells using the RNeasy mini kit (QIAGEN). The cDNA synthesis and PCR amplification of antibody variable regions were performed. The iScript Select cDNA Synthesis Kit (Bio-Rad) and mouse IgG reverse transcription primers (Table S3) were used for reverse transcription. A touch-down/step-down PCR was occurred with universal forward primer and reverse PCR primer based on the antibody chain (Table S3) to amplify the antibody variable regions. The amplicons were appeared between 550–600 base pairs on 1% agarose gel. DNA was extracted from the gel using a QIAEX II DNA Extraction Kit (QIAGEN). The extracted DNA was Sanger-sequenced using the IgG PCR reverse primers (Table S3). For the TyTx1 heavy chain variable region, total RNA isolated from hybridoma cells was subjected to RT-PCR with AffinityScript (Agilent) reverse transcriptase together with Herculase II polymerase (Agilent) as described in the one-step RT-PCR protocol provided by the vendor. DNA band corresponded to the expected antibody variable region was excised, extracted, further amplified using Herculase II polymerase using the nested primers pair, and Sanger-sequenced.
Typhoid toxin intoxication assays
Jurkat cell intoxication assays
Jurkat cell intoxication assays were performed to evaluate mAb-mediated neutralization in the context of purified typhoid toxin treatment. In brief, Jurkat cells were cultured in RPMI1640 + 10% FBS (HyClone) + 1 mM sodium pyruvate (Invitrogen) + 10 mM HEPES (Invitrogen), and kept at 37°C in a cell culture incubator with 5% CO2. Cells were seeded at 1×105/well into 12-well culture plates and treated with either 0.3 pM 6xHis-tagged typhoid toxin alone or a mixture of typhoid toxin and 30 pM MAbs. After incubation for 18 hr, the cells were collected, washed, and fixed for 2 hr in ~70% ethanol/PBS at −20°C. Fixed cells were washed with PBS and resuspended in 500 μL of PBS containing 50 μg/mL propidium iodide, 0.1 mg/mL DNase-free RNase A, and 0.05% Triton X-100. After incubation for 40 min at 37°C, stained cells were washed with PBS, resuspended in 150 μL PBS, filtered, and read using BD Accuri C6 Plus (BD Biosciences), followed by cell-cycle arrest profile analysis using FlowJo V10 software (Treestar Inc).
Henle-407 cell intoxication assays
Henle-407 cell intoxication assays were performed to evaluate mAb-mediated neutralization in the context of S. Typhi infection. In brief, Henle-407 human intestinal epithelial cells were cultured in DMEM high glucose + 10% FBS (HyClone) and kept at 37°C in a cell culture incubator with 5% CO2. Cells were seeded at 3×104/well into 12-well culture plates and incubated overnight. On the next day, cells were infected with either WT or CdtB catalytic mutant S. Typhi harboring the cdtBH160Q gene in place of the WT cdtB gene at a multiplicity of infection (moi) of 30 or 50 for 1 hr in HBSS (Invitrogen), treated with 100 μg/mL gentamicin to kill all extracellular bacteria for 45 min, and washed with PBS. Infected cells were then incubated in the complete cell culture medium containing 10 μg/mL gentamicin for 72 hr in the presence or absence of 10 nM MAbs. Fresh media containing 10 μg/mL gentamicin ± mAb were provided to the cells every 24 hr during incubation. After incubation for 72 hr, cells were collected, washed, and fixed for 2 hr in 70% ethanol/PBS at −20°C. Other downstream procedures were the same as Jurkat cell assays.
Typhoid toxin binding assay
For typhoid toxin binding assay, Henle-407 cells were seeded onto sterile glass coverslips a day before the experiment. On the following day, cells were cooled to 4 °C for 30 min before the addition of anti-typhoid toxin MAbs (2 μg/coverslip) and incubated for an additional 30 min. The coverslips were then stained with Alexa Fluor-555 conjugated typhoid toxin (200 ng/coverslip) for another 30 min. The coverslips were rinsed with cold PBS three times, fixed with 1% paraformaldehyde (PFA), and the nuclei were counterstained with 4’,6-diamidino-2-phenylindole (DAPI). For fluorescent images in Figure 1, Henle-407 cells were pre-treated with media containing 10 mM NH4Cl for 20 min at 37 °C, before chilling and binding of anti-typhoid toxin MAbs and fluorescent labeled-typhoid toxin. After rinsing the coverslips with PBS, cells were treated with warm media containing 10 mM NH4Cl and transferred to a 37 °C incubator for 4 hr. After the 4 hr-incubation, the cells were fixed with 1% PFA and stained with DAPI for nuclei counterstaining. Fluorescent images were acquired with a Leica DMI6000B/DFC340 FX fluorescence microscope system. 1,600 × 1,200-pixel full-frame pictures of various channels were recorded as 16-bit TIFF files with × 20 (numerical aperture (NA) 0.5) or × 40 (NA 0.75) objectives. The filter wavelengths were as follows: Alexa Fluor-555 – excitation filter 545/30 nm, emission filter 610/75 nm; DAPI – excitation filter 340~380 nm, emission filter 425 nm. The fluorescent signal intensity of images was quantified using the measure function of ImageJ (National Institutes of Health, USA) after subtracting the background. Recorded images were merged using the ImageJ merge channels function and processed further with Adobe Photoshop to adjust the brightness and contrast equally for all recordings.
Cryo-EM, data collection, and structure determination
Preparation of Fab and typhoid toxin complex
Purified MAbs were concentrated to 20 mg/mL in 0.1 mL of a buffer containing 20 mM sodium phosphate, pH 7.0, and 10 mM EDTA, followed by enzyme digestion using immobilized Papain (Thermo Fisher Scientific) overnight at 37°C. Digested MAbs samples were eluted with 10 mM Tris-HCl, pH 7.5, and the Fabs were differentially purified with Protein G resins away from Fc and undigested mAb. Further purification of Fab was carried out using Superdex 75 10/300 Increase column (Cytiva) using a constant flow of 0.5 mL/min of a buffer containing 10 mM Tris-HCl, pH 7.5. Purified Fab was incubated together with purified typhoid toxin (PltB, PltAE133A, CdtBH160Q) overnight at 4°C and submitted to Superdex 200 10/300 increase column (Cytiva) using a constant flow of 0.5 mL/min of a buffer containing 15 mM Tris-HCl, pH 7.5 and 150 mM NaCl. Fractions corresponding to the Fab-toxin complex were collected and used immediately for Cryo-EM grid/sample preparations for TyTx1 and TyTx4.
Cryo-EM grid/sample preparation and data acquisition
For TyTx1 and TyTx4, freshly prepared Fab-toxin complex samples were diluted with a buffer containing 15 mM Tris-HCl, pH 7.5 and 150 mM NaCl to a final concentration of 0.1 and 0.2 mg/mL. Samples (3.5 μL) were applied to glow-discharged copper Quantifoil R1.2/1.3 300 Mesh cryo-EM grid (EMS) mounted, blotted, and plunge frozen in liquid ethane using a Vitribot (ThermoFisher) at 100% humidity at 4°C. Data were acquired at a nominal magnification of 49,000 x for TyTx1 and TyTx4 using a Talos Arctica (Thermofisher) operating at 200 kV equipped with a Gatan energy filter set to a slit width of 20 eV and K3 detector operating in super-resolution counting mode using a defocus range of −0.8 to −1.8 μm. Fifty frame movies with a super-resolution pixel size of 0.83 were collected over a 4 s exposure for both datasets. Both datasets were collected using a similar exposure and dose rates resulting in a total dose of 53 e− Å−2 (1.06 e− Å−2/frame) for the TyTx1/Toxin complex and 51.5 e− Å−2 (1.03 e− Å−2/frame) for the TyTx4/Toxin complex.
Image processing
Motion correction of each image stacks was performed using MotionCor2 function in RELION (Nakane et al., 2018; Zheng et al., 2017).
TyTx1/toxin complex (Figure S4)
Following CTF estimation, 676 images with GCTF (Zhang, 2016) estimated resolution better than 4 Å were selected from a total of 747 movies and subjected to automated Laplacian-of-Gaussian particle picking in RELION 3. A total of 3,159,007 particles were extracted with a binned pixel size of 2.53Å and randomly divided into 3 subsets for faster processing. To generate a reference map, particles from the first 76 micrographs were extracted with a pixel size of 1.66Å and subjected to iterative rounds of heterogeneous and homogeneous refinement in CryoSparc 2 (Punjani et al., 2017; Zheng et al., 2017) using the toxin crystal structure (PDB: 4K6L) lowpass-filtered to 60 Å as an initial reference model. The particle subsets were processed and sorted using several rounds of 3D classification, refinement, per-particle CTF, and beam tilt refinement. Particles from each subset extracted with a pixel size of 0.83Å were subjected to Bayesian polishing and combined to produce a reconstruction from 504,702 particles with a 0.143 FSC cutoff resolution of 3.1 Å. Additional 3D classifications seeking to remove particles containing complexes with two bound antibody molecules or a single antibody bound to an alternative PltB monomer resulted in a lower resolution anisotropic reconstruction, suggesting that these alternate states provide additional particle orientations. 2D classification at this stage revealed 2 classes containing ~81,000 (out of ~500,000) particles that appear to contain complexes with two bound antibody molecules. To improve the reconstruction in the region of interest (PltB subunits and the bound antibody), a mask was created around the density attributed to the PltA subunits and particle subtraction was performed. 3D classification without image alignment of the subtracted particles, using a mask around the PltB subunits and antibody molecule, produced a class of 326,766 particles. Subsequent 3D refinement and CTF refinement of the unsubtracted particle images resulted in a final reconstruction with a 0.143 FSC cutoff resolution of 3.0 Å. Note that this reconstruction surpasses the physical Nyquist limit of 3.32 Å.
To assess the distribution of particles containing one versus two bound antibody fragments without introducing bias from masking, particles from the full dataset extracted with a binned pixel size of 2.53Å were subjected to 2D classification discarding only the classes containing obvious junk particles. A subset of 911,028 particles was selected by excluding particles that originated from micrographs with a GCTF estimated resolution worse than 3.0 Å. This subset was subjected to multiple rounds of 3D classification with a 210 Å circular mask using the toxin crystal structure as a reference model. Only particles in low resolution and low distribution junk classes were discarded after each round of classification. The final round of 3D classification contained 741,088 particles separated into two classes each with a single antibody molecule bound to the PltB(C) subunit or PltB(E) subunit (42% and 21%, respectively), one class containing two antibody molecules bound to the PltB(C) and PltB(E) subunits (24%), and two low-resolution junk classes (7% and 6%). Each of the three main classes failed to further refine to high resolution and produced maps characteristic of orientation bias, again confirming that including particles from alternate states were required to produce isotropic reconstructions.
TyTx4/toxin complex (Figure S6)
Following CTF estimation, a total of 572 movies collected during data acquisition were subjected to Laplacian-of-Gaussian particle picking in RELION 3. Approximately 1000 particles were manually picked, extracted, and 2D classified to serve as initial templates for automated particle picking in RELION 3. 2D classification of the resulting 903,271 particles was used to remove incorrectly picked particles. CryoSparc 2 was used to generate an ab-initio model from the cleaned particle set. This ab-initio model was used as a reference map for subsequent 3D classifications and refinement in RELION 3. Initial 3D classification without symmetry imposed reveals two of five classes contain only 4 Fabs bound constituted for approximately 36% of the initial particles. To exploit the C5 symmetry present when there are 5 Fabs bound, the class with the highest resolution that contains 5 Fabs was used for further refinements. After per-particle CTF refinements and 3D classifications, a Bayesian polishing step followed a final 3D refinement step yield a final reconstruction using 97,594 particles were generated after the masked refinement imposing C5 symmetry. The final 3D reconstruction has a 0.143 FSC cutoff resolution of 3.13 Å according to RELION. We note that this reconstruction also surpasses the physical Nyquist limit of 3.32 Å.
Model building and refinement
Chimera was used to place the crystal structures of typhoid toxin (PDB: 4K6L) and PltB pentamer (PDB: 4RHR) into the sharpened reconstructions as preliminary atomic models. Based on their sequence similarity to the actual sequences of the TyTx1 and TyTx4 antibodies, crystal structures of Fab light and heavy chains were fit into the sharpened reconstructions to use as preliminary atomic models (TyTx1 light chain: PDB: 1MHH; TyTx1 heavy chain: PDB: 4H20; TyTx4 heavy chain: PDB: 4M48, TyTx4 heavy chain: 2OZ4. Coot was used to manually rebuild these preliminary models. The flexibility of the antibody constant regions, CdtB subunit, and the majority of the PltA subunit prevented us from confidently building these portions of the maps, so they were left unbuilt. Models were refined in Phenix using the real-space refinement module. The quality of the final models was validated using MolProbity. Figures were generated using Chimera and Pymol. Cryo-EM density maps and refined complex structures were deposited under EMD-22699 and PDB: 7K7H for the TyTx1 Fab-toxin complex and EMD-22700 and PDB: 7K7I for the TyTx4 Fab-toxin complex (Table S1).
SEC analysis of TyTx1, TyTx3, and TyTx4 Fabs binding to tagless PltB pentamer and typhoid toxin
Before analysis, purified tagless PltB pentamer or typhoid toxin was incubated with a 5-fold molar excess of Fabs and incubated for 16 hours at 4°C. The time was staggered so that the size exclusion chromatography was executed at 16 hour-mark. After 16 hours of incubation, the mixture was loaded onto a Superdex 200 Increase 10/300 GL column (Cytiva scientific, 28990944) equilibrated with PBS. The size exclusion chromatography was carried out at a flow rate of 250 μL/min. Fractions containing peak of interest were analyzed with 15% SDS-PAGE. The elution volume matching the center of the peaks was used to calculate the corresponding molecular weight of the native complex eluted using calibration curves analysis in comparison to known protein standards.
mAb affinity determination: surface plasmon resonance (SPR) assay
TyTx mAb binding kinetics and affinity were determined by performing SPR assays using the Biacore T200 (GE Healthcare), as previously described (Rudolph et al., 2020). Series S CM5 chips were immobilized with WT typhoid toxin to obtain a target response bound of ~100–120 RU. Pilot experiments to optimize experimental parameters were performed using 10-fold serial dilutions of each mAb injected at a flow rate of 50 μL/min with a contact time of 120 s and dissociation time of 600 s, which were conditions deemed appropriate by the Biacore T200 Evaluation software (GE Healthcare) for the affinity determination of each mAb. Following optimization of the concentration range and conditions, 4-fold serial dilutions of each mAb were input for replicate experiments. Sensograms for each replicate were analyzed to fit a bivalent binding model using the Biacore T200 Evaluation software. Data obtained from the Biacore T200 was imported into GraphPad Prism 8.3 for the generation of final sensograms.
SSM analyses of bacterial AB5 toxins
The iterative Cα alignment SSM Algorithm of Eugene Krissinel (https://www.ebi.ac.uk/msd-srv/ssm) which is part of Coot was used to superpose the known toxin structures: S. Typhi typhoid toxin (PDB:4K6L), E. coli PltAB toxin (PDB:4Z9C), V. cholerae cholera toxin (PDB:1XTC), E. coli Shiga toxin type 2 (PDB:1R4P) and B. pertussis pertussis toxin (PDB:1PRT) together with the two Fab-toxin complexes of TyTx1 and TyTx4 (Table S5). The SSM algorithm was also used to superpose glycan-bound toxin structures onto the respective toxins: PDB:6P4M onto the typhoid toxin structure, PDB:5ELB and PDB:2CHB onto the cholera toxin structure, PDB:1BOS onto the Shiga toxin structure, and PDB:1PTO onto the pertussis toxin structure (Table S5). The SSM algorithm was used to generate 4 additional Fabs of the TyTx1-toxin complex for C5 symmetrized overlay purposes. The coordinate of the superposed structures was exported from Coot and figures were generated via Pymol. The region of toxin encroaching the TyTx1 Fab volume was manually identified and highlighted.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data were tested for statistical significance with GraphPad Prism 6 or 8.3 software. The number of replicates for each experiment and the statistical test performed are indicated in the figure legends. Image analysis and quantification and cell cycle profile analysis and quantification were performed using ImageJ and FlowJo V10 software, respectively.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-typhoid toxin MAbs and Fabs | This manuscript | See Method Details |
| Chemicals, peptides, and recombinant proteins | ||
| Typhoid Toxin, Toxoid, & PltB subunits | This manuscript | See Method Details |
| Bacterial and virus strains | ||
| S. Typhi ISP2825 | Galán and Curtiss, 1991 | NCBI Genbank accession number CP080960 |
| S. Typhi ISP2825 carrying cdtBH160Q | Spanò et al., 2008 | N/A |
| E. coli Acella strain (E. coli BL21(DE3) ΔendA ΔrecA) | Originally obtained from EdgeBio. Note that the Acella strain is no longer available from the vendor. | Competent cells were prepared in our laboratory. |
| Critical commercial assays | ||
| Alexa Fluor 555 Protein Labeling Kit | ThermoFisher | A20174 |
| RNeasy mini kit | QIAGEN | 74104 |
| iScript cDNA synthesis kit | Bio-Rad Laboratories | 1708891 |
| Herculase II Fusion DNA polymerase | Agilent | 600677 |
| Phusion DNA polymerase | New England BioLabs | M0530 |
| QIAEX DNA Extraction Kit | QIAGEN | 20021 |
| Deposited data | ||
| Typhoid toxin bound to TyTx1 Fab or TyTx4 Fabs | RCSB Protein Data Bank | PDB: 7K7H and PDB: 7K7I (see Table S1) |
| Complete genome sequence of S. Typhi ISP2825 | GenBank | CP080960 |
| Experimental models: Cell lines | ||
| Henle-407 Cells | Originally obtained from ATCC | Validated using intestinal epithelial markers |
| Jurkat Cells | Originally obtained from ATCC | TIB-152 |
| Sp2/0-Ag14 | Originally obtained from ATCC | CRL-1581 |
| Experimental models: Organisms/strains | ||
| BALB/c | Taconic Biosciences | N/A |
| Oligonucleotides | ||
| See Table S3 for oligonucleotide information | IDT | See Table S3 |
| Software and Algorithms | ||
| GraphPad Prism 6 or 8.3 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| LI-COR Image Studio | LI_COR Image Studio Software | https://www.licor.com/bio/image-studio/resources |
| Excel | Microsoft | Office 365 |
| FlowJo V10 | TreeStar | https://www.flowjo.com/ |
| ImageJ | National Institutes of Health | https://imagej.nih.gov/ij/ |
| Unicorn 6.3 | GE Healthcare Life Sciences | https://www.cytivalifesciences.com/en/us/shop/unicorn-6-3-p-01118 |
| BLASTp | National Institutes of Health | https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins |
| Biacore T200 Evaluation Software | GE Healthcare | https://www.cytivalifesciences.com/en/us/shop/protein-analysis/spr-label-free-analysis/systems/biacore-t200-p-05644 |
| CryoSparc 2 | Punjani et al., 2017 | https://cryosparc.com |
| Relion 3 | Zivanov et al., 2018 | https://www3.mrc-lmb.cam.ac.uk/relion/index.php/Main_Page |
| PyMol | Schrodinger | https://pymol.org/2/ |
| Chimera | UCSF Chimera | https://www.cgl.ucsf.edu/chimera/ |
| PHENIX | Adams et al., 2010 | https://phenix-online.org/ |
| Coot | Emsley et al., 2010 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
Highlights.
Antibodies targeting glycan-receptor binding B subunits can be split into two classes
The two classes are grouped by their epitope locations on the B subunit homopentamer
B homopentamers act similar to heteropentamers due to their skewed A subunit positioning
The two antibody classes exhibit significantly different neutralizing efficacies
ACKNOWLEDGMENTS
We thank the Wadsworth Center’s Media and Tissue Culture Core facility for preparing media and assistance with hybridoma scale-up, and Mariena S. Ramos and Katie Spoth of the Cornell Center for Material Research (CCMR) for help in the use of FEI Vitrobot and Talos Artica TEM/SEM. This work was supported in part by a multi-PI R03 award from NIAID (R03 AI135767) to J.S. and N.J.M.; NIH R01 AI137345, R01 AI139625, and R01 AI141514 to J.S.; NIH R03 AI119647 to N.J.M; NIH R01 GM098621 and R01 GM116942 to J.C.F.; and NSF DMR-1719875 to the CCMR. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109654.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn C, Yang Y-A, Neupane DP, Nguyen T, Richards AF, Sim JH, Mantis NJ, and Song J (2021). Mechanisms of typhoid toxin neutralization by antibodies targeting glycan receptor binding and nuclease subunits. iScience 24, 102454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beddoe T, Paton AW, Le Nours J, Rossjohn J, and Paton JC (2010). Structure, biological functions and applications of the AB5 toxins. Trends Biochem. Sci 35, 411–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdoulous S, and Lemichez E (2018). Decoding glycan recognition by bacterial toxins. Nat. Microbiol 3, 124–126. [DOI] [PubMed] [Google Scholar]
- Chang SJ, Jin SC, Jiao X, and Galán JE (2019). Unique features in the intracellular transport of typhoid toxin revealed by a genome-wide screen. PLoS Pathog. 15, e1007704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Song J, Gao X, Wang J, Yu H, Chen X, Varki N, Naito-Matsui Y, Galán JE, and Varki A (2014). Host adaptation of a bacterial toxin from the human pathogen Salmonella Typhi. Cell 159, 1290–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan E, Merritt EA, Verlinde CL, and Hol WG (2000). AB5 toxins: Structures and inhibitor design. Curr. Opin. Struct. Biol 10, 680–686. [DOI] [PubMed] [Google Scholar]
- Feasey NA, Gaskell K, Wong V, Msefula C, Selemani G, Kumwenda S, Allain TJ, Mallewa J, Kennedy N, Bennett A, et al. (2015). Rapid emergence of multidrug resistant, H58-lineage Salmonella Typhi in Blantyre, Malawi. PLoS Negl. Trop. Dis 9, e0003748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galán JE (2016). Typhoid toxin provides a window into typhoid fever and the biology of Salmonella Typhi. Proc. Natl. Acad. Sci. USA 113, 6338–6344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galán JE, and Curtiss R 3rd. (1991). Distribution of the invA, -B, -C, and -D genes of Salmonella typhimurium among other Salmonella serovars: invA mutants of Salmonella typhi are deficient for entry into mammalian cells. Infect. Immun 59, 2901–2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghjoo E, and Galán JE (2004). Salmonella typhi encodes a functional cytolethal distending toxin that is delivered into host cells by a bacterial-internalization pathway. Proc. Natl. Acad. Sci. USA 101, 4614–4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernando-Amado S, Coque TM, Baquero F, and Martínez JL (2019). Defining and combating antibiotic resistance from One Health and Global Health perspectives. Nat. Microbiol 4, 1432–1442. [DOI] [PubMed] [Google Scholar]
- Kitov PI, Sadowska JM, Mulvey G, Armstrong GD, Ling H, Pannu NS, Read RJ, and Bundle DR (2000). Shiga-like toxins are neutralized by tailored multivalent carbohydrate ligands. Nature 403, 669–672. [DOI] [PubMed] [Google Scholar]
- Klemm EJ, Shakoor S, Page AJ, Qamar FN, Judge K, Saeed DK, Wong VK, Dallman TJ, Nair S, Baker S, et al. (2018). Emergence of an extensively drug-resistant Salmonella enterica serovar Typhi clone harboring a promiscuous plasmid encoding resistance to fluoroquinolones and third-generation cephalosporins. MBio 9, e00105–e00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Yang YA, Milano SK, Nguyen T, Ahn C, Sim JH, Thompson AJ, Hillpot EC, Yoo G, Paulson JC, and Song J (2020). Salmonella typhoid toxin PltB subunit and its non-typhoidal Salmonella ortholog confer differential host adaptation and virulence. Cell Host Microbe 27, 937–949.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Inzerillo S, Lee GY, Bosire EM, Mahato SK, and Song J (2021). Glycan-mediated molecular interactions in bacterial pathogenesis. Trends Microbiol. Published online July 14, 2021. 10.1016/j.tim.2021.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer L, López T, Espinosa R, Arias CF, Vollmers C, and DuBois RM (2019). A simplified workflow for monoclonal antibody sequencing. PLoS ONE 14, e0218717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakane T, Kimanius D, Lindahl E, and Scheres SH (2018). Characterisation of molecular motions in cryo-EM single-particle data by multi-body refinement in RELION. eLife 7, e36861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupane DP, Dulal HP, and Song J (2021). Enteric fever diagnosis: Current challenges and future directions. Pathogens 10, 410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T, Lee S, Yang YA, Ahn C, Sim JH, Kei TG, Barnard KN, Yu H, Millano SK, Chen X, et al. (2020). The role of 9-O-acetylated glycan receptor moieties in the typhoid toxin binding and intoxication. PLoS Pathog. 16, e1008336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhill J, Dougan G, James KD, Thomson NR, Pickard D, Wain J, Churcher C, Mungall KL, Bentley SD, Holden MT, et al. (2001). Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413, 848–852. [DOI] [PubMed] [Google Scholar]
- Petersen E, and Miller SI (2020). Toxin glycan binding: Lectin keys unlocking host and tissue specificity. Cell Host Microbe 27, 851–853. [DOI] [PubMed] [Google Scholar]
- Punjani A, Rubinstein JL, Fleet DJ, and Brubaker MA (2017). cryo-SPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296. [DOI] [PubMed] [Google Scholar]
- Rudolph MJ, Czajka TF, Davis SA, Thi Nguyen CM, Li XP, Tumer NE, Vance DJ, and Mantis NJ (2020). Intracellular neutralization of ricin toxin by single-domain antibodies targeting the active site. J. Mol. Biol 432, 1109–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Gao X, and Galán JE (2013). Structure and function of the Salmonella Typhi chimaeric A2B5 typhoid toxin. Nature 499, 350–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanò S, Ugalde JE, and Galán JE (2008). Delivery of a Salmonella Typhi exotoxin from a host intracellular compartment. Cell Host Microbe 3, 30–38. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (2019). Antibiotic resistance threats in the United States, 2019. https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf.
- Van Slyke G, Angalakurthi SK, Toth RT 4th, Vance DJ, Rong Y, Ehrbar D, Shi Y, Middaugh CR, Volkin DB, Weis DD, and Mantis NJ (2018). Fine-specificity epitope analysis identifies contact points on ricin toxin recognized by protective monoclonal antibodies. Immunohorizons 2, 262–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YA, Chong A, and Song J (2018a). Why is eradicating typhoid fever so challenging: Implications for vaccine and therapeutic design. Vaccines (Basel) 6, E45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YA, Lee S, Zhao J, Thompson AJ, McBride R, Tsogtbaatar B, Paulson JC, Nussinov R, Deng L, and Song J (2018b). In vivo tropism of Salmonella Typhi toxin to cells expressing a multiantennal glycan receptor. Nat. Microbiol 3, 155–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K (2016). Gctf: Real-time CTF determination and correction. J. Struct. Biol 193, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, and Agard DA (2017). MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJ, Lindahl E, and Scheres SH (2018). New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuverink M, and Barbieri JT (2018). Protein toxins that utilize gangliosides as host receptors. Prog. Mol. Biol. Transl. Sci 156, 325–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all datasets generated during this study. Coordinates for the atomic structures have been deposited in the RCSB Protein Data Bank: 7K7H and 7K7I (Table S1). Electron microscopy density maps have also been deposited to EMDB: EMDB-22699 and EMDB-22700 (Table S1).
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


